Abstract
Molecular evolution is driven by mutations, which may affect the fitness of an organism and are then subject to natural selection or genetic drift. Analysis of primary protein sequences and tertiary structures has yielded valuable insights into the evolution of protein function, but little is known about evolution of functional mechanisms, protein dynamics and conformational plasticity essential for activity. We characterized the atomic-level motions across divergent members of the dihydrofolate reductase (DHFR) family. Despite structural similarity, E. coli and human DHFRs use different dynamic mechanisms to perform the same function, and human DHFR cannot complement DHFR-deficient E. coli cells. Identification of the primary sequence determinants of flexibility in DHFRs from several species allowed us to propose a likely scenario for the evolution of functionally important DHFR dynamics, following a pattern of divergent evolution that is tuned by the cellular environment.
INTRODUCTION
Diversification of gene families and their resulting protein products through mutation, random genetic drift, and natural selection has resulted in the wide spectrum of enzymes, signal transducers, cellular scaffolds, and other molecular machines that are found in the diverse species represented in all kingdoms of life. The effects of such diversification on three-dimensional protein structures are addressed in many studies that provide fundamental insights into evolutionary pressures that drive diversification of protein folds1–3. However, motions and flexibility are also essential for the function of proteins and macromolecular machines and, just as protein structures are subject to natural selection, evolutionary pressures might also be expected to tune protein dynamics to adapt proteins to new environments and facilitate the emergence of novel functionalities. Indeed, comparisons between thermophilic and mesophilic enzymes reveal that their dynamics and activity are adapted to the thermal environment of the organism4,5. In principle, the adaptation of enzymes to different environments or to specialized functions may involve a radical reconfiguration of the dynamic landscape. Understanding how new dynamic modes arise would provide fundamental insight into the evolution of novel functionality, and is addressed here in the context of the enzyme dihydrofolate reductase (DHFR).
DHFR catalyzes the NADPH-dependent reduction of dihydrofolate (DHF) to tetrahydrofolate (THF), an essential precursor for thymidylate synthesis in cells6. The evolution of DHFR is of great interest, both in the context of understanding how the enzyme has adapted to different cellular environments, as well as in predicting its evolution in drug-resistant pathogens7. E. coli DHFR (ecDHFR, ecE) has long served as a paradigm for understanding enzyme mechanisms8–12. Although human DHFR (hDHFR, hE) is structurally similar to ecDHFR (Fig. 1a), their primary sequences are highly divergent, which is reflected in subtle changes in the catalytic cycle9,10,13 with different kinetics and different rate-limiting step under physiological concentrations of ligands (Fig. 1b). We hypothesized that ecDHFR and hDHFR may have evolved different dynamic mechanisms within the constraints of the same fold and the same key catalytic residues. To address this hypothesis we used an integrated approach including structural biology, mutagenesis, bioinformatic analyses and cell biology, which allowed us to uncover evolutionary aspects of the motions present in the dihydrofolate reductase (DHFR) enzyme family.
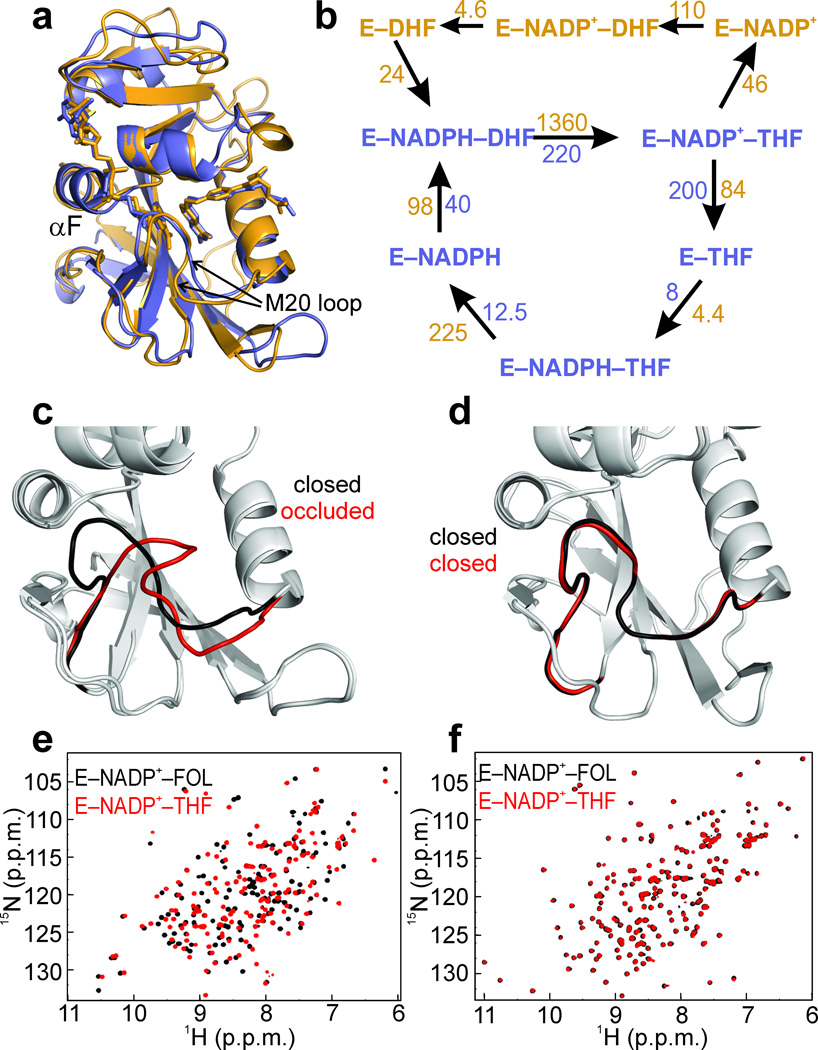
Figure 1. Human and E. coli DHFRs are structurally conserved, but have different active site loop movements.
(a) Superposition of hDHFR (orange) and ecDHFR (purple), bound to NADP+ and FOL. Ligands are shown as sticks. (b) Catalytic cycles of ecDHFR and hDHFR. Both enzymes share a similar catalytic cycle, involving five observable intermediates (purple). In addition, the human enzyme also traverses a second catalytic cycle (orange), with E–NADP+–THF being the branch point. Approximately 65% of the flux proceeds through the same catalytic cycle as ecDHFR (purple), while 35% proceeds through the upper cycle (orange)10. Units are in s−1 for first order rates and M−1s−1 for bimolecular rates. (c) Crystal structures of ecDHFR bound to NADP+ and FOL (1RX218, Met20 loop shown in black) or NADP+ and ddTHF (1RX418, Met20 loop shown in red). The ecDHFR Met20 loop shifts from the closed (black) to occluded (red) conformations depending on the ligand bound. (d) Crystal structures of hDHFR bound to NADP+ and FOL (Met20 loop shown in black) or NADP+ and THF (Met20 loop shown in red). (e) 15N HSQC spectra of ecDHFR bound to NADP+ and FOL (black) or NADP+ and THF (red), showing chemical shift changes between the closed Michaelis model complex and the occluded product ternary complex. (f) 15N HSQC of hDHFR bound to NADP+ and FOL (black) or NADP+ and THF (red). The active site loops of hDHFR remain in the closed position across the hydride transfer step.
RESULTS
Active site loop motions in human DHFR
Given the well-established role that dynamics plays in ecDHFR function14–16, we hypothesized that altered dynamics in hDHFR might account for its unique catalytic properties. ecDHFR undergoes conformational changes, involving rearrangement of its active site loops17–21, as it proceeds through five observable intermediates in the catalytic cycle (Fig. 1b). To investigate and characterize key intermediates in the catalytic cycle of hDHFR, we determined crystal structures (Supplementary Figs. 1,2 and Table 1) of hDHFR in complex with NADP+ and folic acid (hE–NADP+–FOL, 1.4 Å resolution) and in complex with NADP+ and 5,10-dideazatetrahydrofolate (hE–NADP+–ddTHF, 1.7 Å resolution), which model the Michaelis complex and product ternary complex, respectively. In contrast to ecDHFR, in which the Met20 loop moves from the closed conformation in the E–NADPH and E–NADP+–FOL complexes to the occluded conformation in the three product complexes (Fig. 1c)18, thereby facilitating ligand flux14,21–23, hDHFR remains in the closed conformation in both ligand-bound states, without any apparent structural change in the active site loops (Fig. 1d). Thus, in hDHFR, the Met20 loop appears to be locked in place and unable to undergo this conformation change. Consistent with our findings, the active site loops adopt the closed conformation in all available crystal structures of vertebrate DHFRs, including complexes of hDHFR with small molecule inhibitors and a substrate (folate)24. Importantly, the closed to occluded conformational transition in ecDHFR can also be visualized directly in solution by comparing the 15N HSQC spectra of the ecE–NADP+–FOL and ecE–NADP+–THF complexes, which differ due to the conformational change in the Met20 loop (Fig. 1e)14,18,20. In marked contrast to ecDHFR, the 15N HSQC spectra of the hE–NADP+–FOL and hE–NADP+–THF complexes are almost identical (Fig. 1f), showing that in solution, as well as in the crystal structures, no backbone conformational changes are observed for the human enzyme.
Table 1.
Data collection and refinement statistics for crystal structures of hDHFR complexes.
| hE–NADPH | hE–NADP+–FOL | hE–NADP+–ddTHF | |
|---|---|---|---|
| Data collection | |||
| Space group | C2221 | P6322 | P6322 |
| Cell dimensions | |||
| a, b, c (Å) | 39.0, 65.5, 152.3 | 68.2, 68.2, 160.2 | 67.8, 67.8, 160.4 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50-1.20 (1.23-1.20)* | 50-1.40 (1.42-1.40) | 50-1.70 (1.73-1.70) |
| Rsym | 0.11 (0.54) | 0.08 (0.79) | 0.13 (0.85) |
| I / σI | 14.0 (2.0) | 24.4 (2.1) | 25.4 (2.2) |
| Completeness (%) | 96.8 (76.9) | 99.6 (92.3) | 99.9 (99.3) |
| Redundancy | 6.5 (2.9) | 19.3 (8.7) | 18.3 (8.4) |
| Refinement | |||
| Resolution (Å) | 50-1.20 | 50-1.40 | 50-1.70 |
| No. reflections | 59,264 | 44,348 | 24,628 |
| Rwork / Rfree | 16.3 / 18.5 | 14.5 / 18.4 | 19.2 / 24.0 |
| No. atoms | |||
| Protein | 1,639 | 1,635 | 1,547 |
| Ligand/ion | 48 | 86 | 93 |
| Water | 196 | 206 | 143 |
| B-factors | |||
| Protein | 24.0 | 20.2 | 34.4 |
| Ligand/ion | 20.8 | 19.4 | 43.0 |
| Water | 38.8 | 32.7 | 40.4 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.009 | 0.009 | 0.007 |
| Bond angles (°) | 1.48 | 1.41 | 1.25 |
Values in parentheses are for highest-resolution shell.
Active site packing and preorganization in hDHFR
The hDHFR active site cleft in the model Michaelis complex, E–NADP+–FOL, is more tightly packed than that of ecDHFR bound to the same ligands (Fig. 2a, b) and likely plays an important role in optimal positioning of the donor and acceptor atoms for catalysis, thereby contributing to its increased rate of hydride transfer8,10. However, the tight packing of the hDHFR active site, coupled with the apparent lack of active site loop motions to facilitate ligand flux, raises a critical question: how do the ligands get into and out of the hDHFR active site?
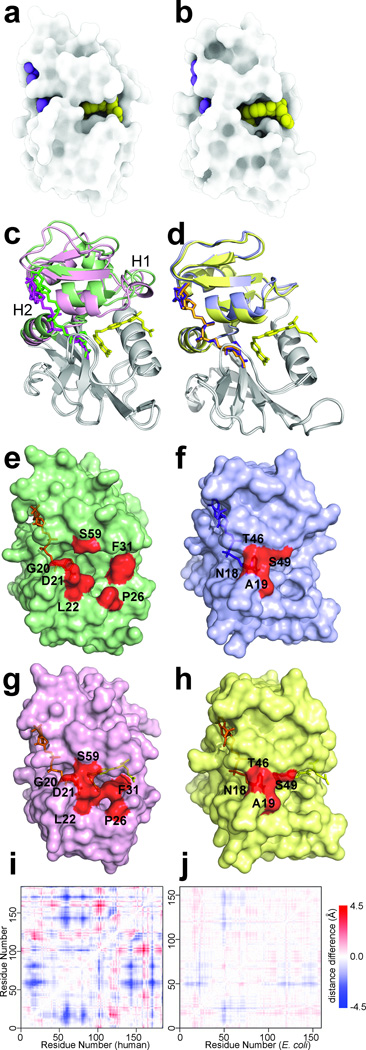
Figure 2. Active site packing and hinge motions in hDHFR.
a,b Surface rendition of hDHFR–NADP+–FOL (a) and ecDHFR:NADP+–FOL (b) generated using only ambient occlusion, a 3D light attenuation calculation where deep pockets render dark and exposed surfaces render light35. c,d Superposition of crystal structures, aligned on the loop subdomain (gray), of hE–NADPH and hE–NADP+–FOL (c) and ecE–NADPH (PDB code: 1RX118) and ecE–NADP+–FOL (PDB code: 1RX218) (d). The adenosine-binding subdomain is colored green for hE–NADPH and pink for hE–NADP+–FOL. Ligands are shown as sticks, with NADPH in green, NADP+ in magenta and FOL in yellow. The adenosine-binding subdomain is colored purple for ecE–NADPH and yellow for ecE–NADP+–FOL, with NADPH in purple, NADP+ in orange and folate in yellow. e,f,g,h Surface representations of hE–NADPH (e), ecE–NADPH (f), hE–NADP+–FOL (g) and ecE–NADP+–FOL (h). Residues highlighting the opening and closing of the active site cleft are colored in red. i,j Difference distance matrix for hE–NADPH and hE–NADP+–FOL (i) and ecE–NADPH and ecE–NADP+–FOL (j), showing the magnitude and character of the conformational changes associated with the hinge motions.
Differences in dynamics between ecDHFR and hDHFR
In ecDHFR, millisecond timescale fluctuations in the active site contribute to efficient ligand flux and catalysis; mutations that perturb the dynamic equilibrium between the closed and occluded conformations of the Met20 loop notably alter substrate and cofactor flux14,23,25,26. Although the crystal structures and 15N HSQC spectra of hE–NADP+–FOL and hE–NADP+–THF (or hE–NADP+–ddTHF) suggest that the active site loops of hDHFR are predominantly closed, we used Carr-Purcell-Meiboom-Gill (CPMG)–based 15N R2 relaxation dispersion NMR experiments to assess whether transient loop fluctuations might facilitate ligand flux. In stark contrast to ecDHFR, fluctuations on the millisecond time scale are not observed in the hE–NADP+–FOL, hE–NADP+–THF, hE–FOL, E–THF, or hE–NADPH complexes, suggesting that the human enzyme utilizes a different mechanism or different motions to mediate ligand flux. Indeed, 15N R1ρ relaxation dispersion experiments revealed pervasive motions on a faster, microsecond time scale in the hE–NADP+–FOL complex (Supplementary Fig. 3a–c). These rapid conformational fluctuations (at rates ranging from ~15,000–30,000 s−1) occur in many regions of the enzyme, including regions that line one edge of the active site, and may play a role in ligand binding and release. Remarkably, despite the structural similarity between human and E. coli DHFRs, both the nature and time scale of the ground-state conformational fluctuations have diverged considerably, bolstering the hypothesis that the dynamic mechanisms of ecDHFR and hDHFR are fundamentally different.
Exaggerated hinge movements in hDHFR
To gain further insights into how hDHFR exchanges substrate in the absence of flexible active site loops, we determined the crystal structure of the hE–NADPH binary complex at 1.2 Å resolution. Notably, the active site cleft in the hE–NADPH structure is more open than that in the hE–NADP+–FOL complex (Fig. 2c,e,g). Opening of the active site is accomplished not by the motion of flexible loops, as in ecDHFR, but by a rigid body, twisting-hinge motion that rotates the adenosine-binding subdomain (defined as Thr40-Gly129) ~10° away from the loop subdomain (Gly2-Thr39 and His130-Asp186). These subdomains are connected by two hinges: hinge 1 (Thr39-Leu49) and hinge 2 (Gly129-Leu131). In the hE–NADPH complex, the hinges stabilize the active site cleft in an open (hinge-open) conformation that likely facilitates substrate entry (Fig. 2e). Upon binding of substrate (folate), the adenosine-binding subdomain rotates inward to tightly close the active site (Fig. 2g, Supplementary Fig. 2a–c and Supplementary Table 1). Changes in chemical shifts between the NMR spectra of the hE–NADPH and hE–NADP+–FOL complexes (Supplementary Fig. 3d,e) suggest that this hinge movement also occurs in solution. In both hinge-open and hinge-closed conformations, an extensive hydrogen bonding network stabilizes the hinge 1 structure and anchors it to the adenosine-binding subdomain and to the C-terminal end of the αB helix (Supplementary Note, Supplementary Fig. 2d,e and Supplementary Table 2). Hinge 1 moves as a rigid body to accommodate the subdomain rotation and hinge 2 allows the sliding motion of helix αF that opens and closes the active site cleft27.
Interestingly, subtle hinge movements have also been reported for ecDHFR18. However, unlike the human enzyme, the subdomain rotation in ecDHFR displays a much smaller range of motion of the active site cleft, (Fig. 2d,f,h). At the widest point, the hDHFR active site cleft opens by ~3 Å (comparing hE–NADPH and hE–NADP+–FOL; Fig. 2e,g), while ecDHFR opens ~0.5 Å (comparing 1RX118 and 1RX218; Fig. 2f,h). We further quantified the difference between binary and ternary structures for hDHFR and ecDHFR by calculating the distance difference matrix (Fig. 2i,j), which shows clearly that the extent of motion in the human enzyme is greater at this step of the catalytic cycle than observed for the E. coli enzyme. The length of the hinges is seemingly a key factor in determining the magnitude of the hinge bending motions. The ecDHFR hinge 1 and 2 regions are tightly packed and may not be long enough to provide the structural framework for the larger rigid-body motion observed in hDHFR (Fig. 3). In particular, hinge 1 in ecDHFR is “embedded” in a tight groove where any large movement would be highly constrained by clashes with both the adenosine-binding and loop subdomains. The observation that a subtle subdomain rotation is possible in the E. coli enzyme18 provides a framework for understanding how alternative dynamic mechanisms may arise and change during the course of evolution. Just as new functionalities are most frequently derived from existing protein folds, new dynamic mechanisms likely derive from existing modes of protein motions. The subtle hinge motion in ecDHFR may represent an ancestral motion that has been accentuated in hDHFR through the course of evolution, increasing the amplitude of domain movement to permit ligand flux and thus eliminating the need for the closed-occluded transition of the Met20 loop that is utilized by ecDHFR.
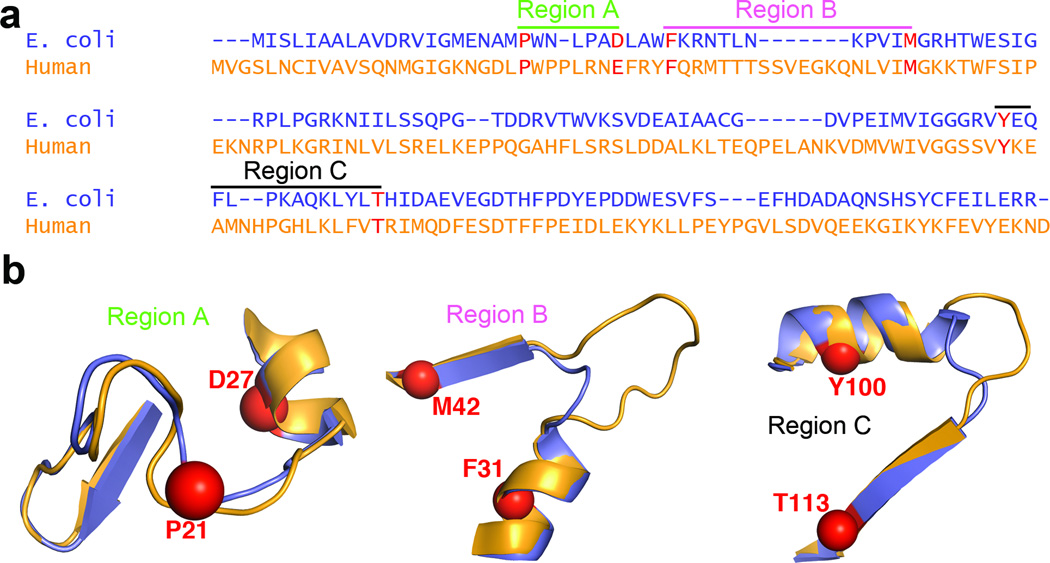
Figure 3. Primary sequence features related to flexibility and conformational change in E. coli and human DHFR.
(a) Sequence alignment of ecDHFR and hDHFR showing three regions of the sequence related to dynamic mechanism. The anchor residues for sequence alignment are shown in red. (b) Structure of regions highlighted in a, with anchor residues shown as spheres. ecDHFR is shown in purple, and hDHFR in orange. Regions A, B and C correspond to the “Met20” loop, hinge 1 and hinge 2, respectively. The following anchor residues were chosen for sequence alignments (E. coli numbering): P21 and D27 for Region A, F31 and M42 for Region B, and Y100 and T113 for Region C.
Sequence determinants and evolution of dynamic mechanisms
To further explore the evolution of dynamics in the DHFR family, we conducted a comprehensive comparative analysis of all available DHFR sequences (~1800 sequences), including eukaryotic DHFRs from all available fully sequenced genomes (172 sequences) (Supplementary Table 3). In particular, we focused on three regions that we hypothesized would best account for the mechanistic differences between ecDHFR and hDHFR: Region A, at the end of the Met20 loop, and Regions B and C, which contain the hinges implicated in subdomain rotation (Fig. 3). The analysis reveals a clear pattern in sequence and length of Regions A, B, and C that we link to flexibility, allowing us to propose that these regions of the DHFR sequence determine, at least in part, the protein motions that mediate the flux of substrate, product, and cofactor at various stages of the catalytic cycle.
Region A loop length influences conformational flexibility
Region A contains 7 residues in ecDHFR and 8 residues in hDHFR. This region became proline-rich in recent evolutionary times28. Incorporation of the PWPP motif from Region A of the human enzyme into ecDHFR, through the mutation N23PP (which changes 21PWNL24 to 21PWPPL24) alters the motions in the Met20 loop and active site14. To determine whether this polyproline motif in Region A affects flexibility in ecDHFR, or whether the loop length itself contributes to differences in conformational sampling, we characterized an ecDHFR mutant with an alanine insertion (21PWNAL24). 15N HSQC spectra of the pre- and post-hydride transfer complexes (modeled by E–NADP+–FOL, and E–NADP+–THF) show that insertion of a single residue (Ala) is sufficient to prevent the transition to the occluded conformation (Fig. 4a–c) and severely dampens millisecond timescale motions in the active site (Supplementary Fig. 4). Thus, it is primarily the length of Region A and not its proline content that influences the conformational flexibility of the Met20 loop.
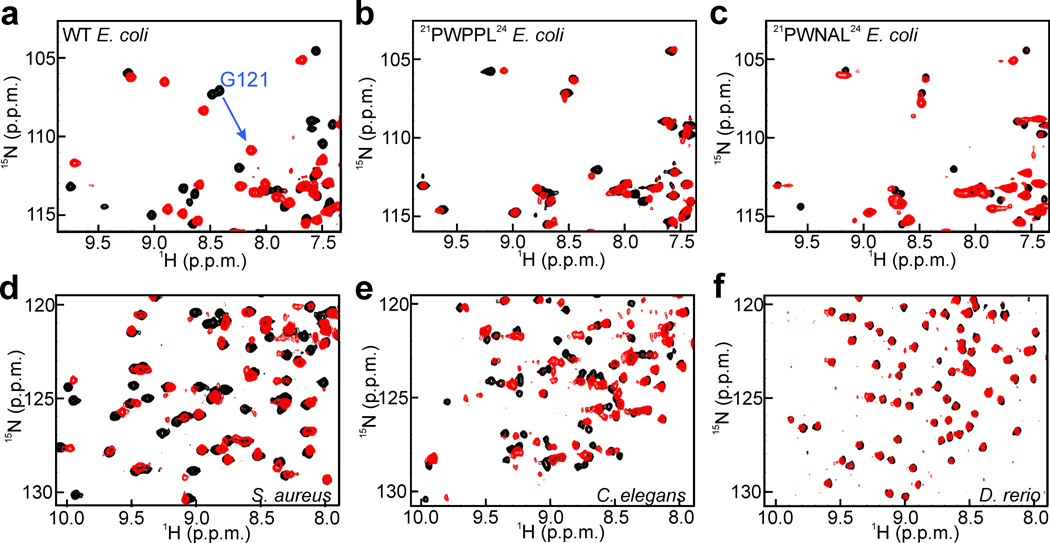
Figure 4. Conformational changes between reactant and product complexes.
Cropped regions of 15N-HSQC spectra of DHFRs: (a) wild-type (WT) E. coli; (b) PWPPL E. coli; (c) PWNAL E. coli; (d) S. aureus; (e) C. elegans; (f) D. rerio. Each panel shows a superposition of E– NADP+–FOL (model Michaelis complex, black) and E–NADP+– THF (product ternary complex, red). A blue arrow marks the change in the position of the G121 cross peak for wild type E. coli DHFR (a). There is no change in the chemical shift of G121 upon formation of the product complexes of the 21PWPPL24 and 21PWNAL24 mutants (b and c). Full 15N HSQC spectra for DHFR from different species are shown in Supplementary Fig. 6.
The majority of prokaryotic DHFRs have 7 residues in Region A and we hypothesized that they would have conformational flexibility similar to that of ecDHFR. Region A in eukaryotic DHFRs is more heterogeneous, with sequences containing 7 residues (E. coli-like), 8 residues (human-like), and >8 residues (Fig. 5). We chose representative DHFRs from several species (Supplementary Fig. 5a–d), in which Region A contains 7, 8 or 10 residues, and investigated their flexibility using NMR spectroscopy to obtain insights into ligand-dependent conformational sampling. We compared 15N HSQC spectra of the DHFRs from these species bound either to NADP+ and FOL (model Michaelis complex) or to NADP+ and THF (product ternary complex) to identify conformational changes across the hydride transfer step. For four DHFRs which contain 7 residues in Region A, i.e. Bacillus anthracis (baDHFR), Staphylococcus aureus (saDHFR), Streptococcus pneumoniae (spDHFR) and Vibrio cholerae (vcDHFR) (34% to 51% identity with ecDHFR), the 15N HSQC spectra reveal conformational changes between the model Michaelis complex and the product ternary complex (Fig. 4 and Supplementary Fig. 6a–d).
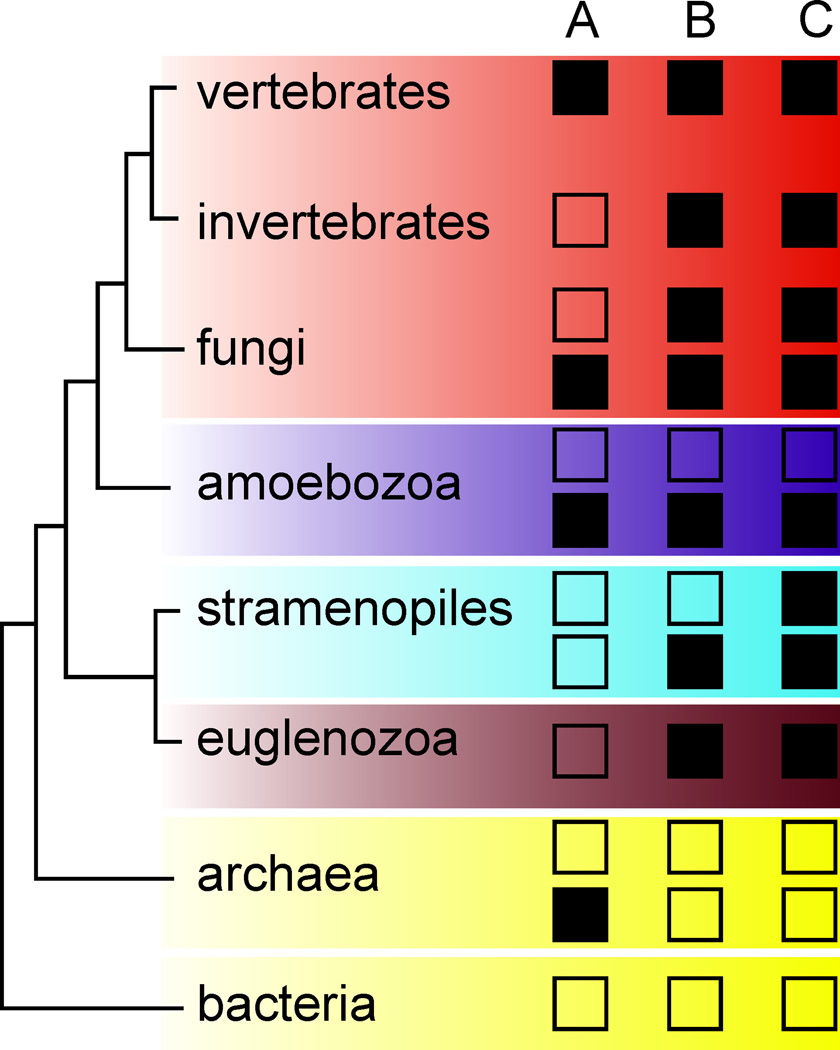
Figure 5. Overview of patterns in length of Met20 loop and hinges.
A, B and C refer to Regions A (Met20 loop), B (hinge 1), and C (hinge 2) of the DHFR primary sequence as described in the text and Fig. 3. For A, open squares indicate 7 residues and filled squares indicate 8 or 10 residues in Region A. Enzymes with 7 residues in Region A undergo conformational changes across the hydride transfer step. An increase in the length of Region A (to 8 or 10 residues) is associated with limited flexibility in the active site loops and the absence of conformational change upon formation of product. For B, open squares indicate a short hinge (<15 residues in Region B) and filled squares indicate a long hinge (≥15 residues in Region B). For C, open squares indicate a short hinge (12 residues in Region C) and filled squares indicate a long hinge (≥14 residues in Region C). Long hinges facilitate the exaggerated hinge-twisting motion observed in hDHFR. While in some groups (e.g. fungi) more than one combination of features can be found, their distributions within the group do not follow any well-established phylogenetic divisions.
HSQC spectra were also acquired for the NADP+–FOL and NADP+–THF complexes of several vertebrate DHFRs that vary in length and sequence of Region A. For Sus scrofa (pig) and Bos taurus (cow) DHFR, Region A is identical to human and the NMR experiments confirm that no conformational changes occur across the hydride transfer step (Supplementary Fig. 5e–i). Rattus norvegicus (rat) DHFR (rDHFR) also contains 8 residues, but Pro26 of hDHFR is replaced by Leu. Danio rerio (zebrafish) DHFR (zDHFR) contains a two-residue insertion in Region A, making it 10 residues in length, with His in place of Pro25 of hDHFR (Supplementary Fig. 5). Again, the almost-identical 15N HSQC spectra of pre- and post-hydride transfer complexes of rDHFR and zDHFR confirm that the enzymes do not undergo conformational changes across the chemical step (Fig. 4f and Supplementary Fig. 6e–i). We therefore conclude that proline at position 25 or 26 (hDHFR numbering) is not essential for stabilization of the closed conformation and that the length of loop A is the primary determinant of conformational change in the active site loops between pre- and post-hydride transfer complexes.
The Caenorhabditis elegans DHFR (ceDHFR) sequence is an example of a eukaryotic DHFR in which Region A is 7 residues (as in bacterial enzymes), but the hinges have similar lengths to hDHFR. Several chemical shift differences were observed between pre- and post-hydride transfer complexes of ceDHFR in the 15N HSQC spectra, indicating similar conformational flexibility to bacterial DHFRs (Fig. 4e). ceDHFR is prototypical of a large subset of eukaryotic DHFRs, in which the Region A length resembles ecDHFR, while Regions B and C are similar to hDHFR.
Properties of the hinges
Hinge 1 is located in Region B. Bacterial DHFR sequences predominantly contain short Region B sequences, as in ecDHFR (12 residues), while in eukaryotic DHFRs, with very few exceptions found in unicellular species (e.g. some amoebozoans and stramenopiles), Region B is ≥19 residues (Fig. 5 and Supplementary Fig. 5). In hDHFR, Asn48 hydrogen bonds to the backbone of Thr38 and Thr40 in the hinge and to Met111 in the adenosine-binding domain, maintaining the hinge structure during subdomain rotation (Supplementary Fig. 2d and Supplementary Table 2). Interestingly, this Asn is invariant in all eukaryotic 19-residue Region B sequences (Supplementary Table 4) and is likely a key mediator of the hinge motion.
Hinge 2, within Region C, is formed by His127-Leu131 for hDHFR and Pro105-Ala107 for ecDHFR. Region C contains 14 residues in E. coli DHFR and 16 residues in hDHFR (Fig. 3). In hDHFR, helix αF slides 2.5 Å towards the active site in the hinge-open conformation, which is stabilized by hydrogen bonds between NADP and Ser119 at the N-terminus of helix αF that are not formed in the hinge-closed conformation. In ecDHFR, the shorter hinge forms hydrogen bonds that involve exclusively backbone atoms, which limit flexibility and restrict αF movement.
Although the detailed molecular mechanism is not yet fully understood, the long hinges in hDHFR appear to provide flexibility that allows the adenosine-binding and loop subdomains to rotate away from each other in the hE–NADPH complex, thereby opening the active site cleft and facilitating substrate access. Subdomain rotation is limited in DHFRs with shorter hinges, such as ecDHFR, where ligand flux is facilitated by flexible active-site loops. Mutations that perturb the conformational fluctuations of the active site loops in ecDHFR result in altered ligand flux, suggesting that either flexible loops or long hinges are required for optimal function14. Consistent with this finding, the sequence patterns in Regions A, B and C indicate that, while some DHFRs contain both flexible loops (7 residues in Region A) and long hinges, a few archaeal enzymes contain both a rigidified active site (8 residues in Region A) and short hinges (Fig. 5); these enzymes are predicted to exchange ligands inefficiently.
Enzymes with intermediate dynamic mechanisms
Although bacterial and human DHFRs have similar three-dimensional structures, their intrinsic dynamic properties are quite different and are associated with important differences in their catalytic mechanisms. We can only speculate as to the mechanism of an ancestral DHFR, but it is natural to expect that enzymes with intermediate mechanisms existed at some point in the evolution of the DHFR family, either as the ancestral enzyme or as intermediates between the ancestral enzyme and the other extant DHFR. DHFRs with 7 residues in region A (E. coli-like) and long, human DHFR-like hinges are found in various invertebrate animals, fungi, and members of the flagellate unicellular eukaryotes euglenozoa (Fig. 5), and potentially represent enzymes with such intermediate dynamic and mechanistic characteristics. For example C. elegans DHFR displays dynamic characteristics that are a mix of hDHFR and ecDHFR. Like E. coli DHFR, the C. elegans protein samples different ground state conformations across the hydride transfer step (Fig. 4), but 15N relaxation dispersion experiments show that millisecond timescale motions are absent, as for hDHFR. Unfortunately, ceDHFR structures are not available; however, the NADPH complex of Candida albicans DHFR (caDHFR, PDB code: 1AI929), with similar sequence features to ceDHFR with respect to regions A, B and C, adopts a conformation equivalent to the hinge-open state of hDHFR (Supplementary Fig. 2f). We conclude that this class of enzymes can, in fact, support the hinge-open conformation. The combined data from ceDHFR and caDHFR then provide insights into the mosaic of dynamic features present in these enzymes. Further studies will undoubtedly reveal the finer features of dynamic tuning in the DHFR family.
DHFRs may be tuned to cellular ligand concentrations
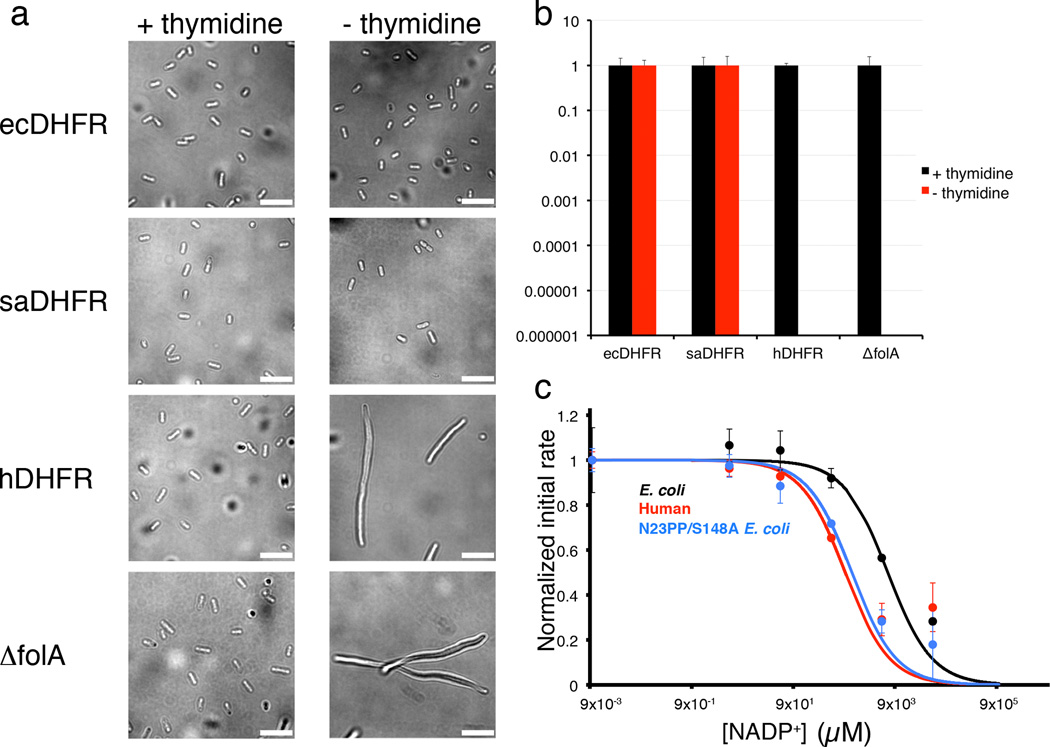
Why have DHFRs evolved different dynamics? We hypothesized that the divergent dynamics of ecDHFR and hDHFR may represent important functional adaptations to the disparate intracellular conditions in bacterial versus mammalian cells10. To test this hypothesis, we assessed the ability of hDHFR to complement a folA null allele in the wild-type E. coli strain MG1655 (Online methods). We replaced the folA open reading frame with that of hDHFR, and found that expression of soluble hDHFR failed to support cell proliferation. Growth of the resulting ΔfolA::hDHFR strain was strictly dependent upon the addition of exogenous thymidine to the culture medium, similar to a complete DHFR knockout (ΔfolA) (Fig. 6a,b). Further, culture of the ΔfolA::hDHFR strain in the absence of supplemental thymidine resulted in the arrest of cytokinesis and rapid filamentation (Fig. 6a), reminiscent of “thymineless death” in thymidylate synthase (thyA) mutants30. One possible explanation is that the highly divergent hDHFR (28% sequence identity with ecDHFR) is unable to make essential interactions with other E. coli proteins. However, S. aureus DHFR (only 35% identity with ecDHFR) fully complements the ecDHFR knockout (ΔfolA::saDHFR) (Fig. 6a,b), suggesting that differences intrinsic to hDHFR limit its efficiency in a bacterial cell, even though its hydride transfer rate is faster than that of ecDHFR10.
Figure 6. Human DHFR cannot complement DHFR-knockout E. coli cells, and is more sensitive to product inhibition than E. coli DHFR.
(a) DIC micrographs of MG1655 ΔfolA and DHFR knock-in strains temporarily grown with or without thymidine, after initial growth in media supplemented with thymidine. The morphology of MG1655 ΔfolA cells expressing ecDHFR or saDHFR are similar with or without thymidine (short rods). In contrast, MG1655 ΔfolA cells expressing hDHFR filament extensively when grown in the absence of thymidine, similar to the DFHR knockout cell line, MG1655 ΔfolA. The scale bar corresponds to 10 µm. Images were obtained using the open-source microscopy software, µManager36. (b) Relative plating efficiency of MG1655 ΔfolA and DHFR knock-in strains on LB medium with or without 100 µg/mL thymidine. Plating efficiency for each strain on LB with thymidine is normalized to 1. While both ecDHFR and saDHFR restore the ability to grow in the absence of thymidine, hDHFR fails to complement and resembles the folA null mutant, both of which are not viable in the absence of thymidine. The mean plating efficiency (n=3) is reported here, with error bars indicating the standard deviation. c. Initial kinetic rates for ecDHFR (black), hDHFR (red) and E. coli N23PP S148A mutant (blue) enzyme activity plotted as a function of increasing NADP+ concentrations. The IC50 for human DHFR is 948 µM, for ecDHFR, 6518 µM, and for the mutant N23PP S148A ecDHFR 1274 µM, closer to that of hDHFR. The experiment was carried out in duplicate, and values for the mean initial rates are plotted in the figure, with error bars indicating the range of values measured.
Vertebrate DHFRs function at much lower cellular concentrations of NADP+ (~20 µM vs ~2 mM) and THF (~0.3 µM vs ~13 µM) than the E. coli enzyme10,31–33 and the ratio of NADPH to NADP+ differs greatly between mammalian (~100:1) and E. coli (~1:1) cells. Consequently, the E. coli enzyme may have evolved a dynamic mechanism that allows it to avoid end product inhibition due to the high concentrations of NADP+ in bacterial cells. In contrast, hDHFR was not subjected to the same evolutionary pressure, as the large excess of NADPH over NADP+ in mammalian cells favors efficient exchange of NADPH for NADP+. To assess whether the human and E. coli enzymes are differentially susceptible to end product inhibition by NADP+, we monitored the effect of increasing concentrations of NADP+ on the initial catalytic rate for both ecDHFR and hDHFR. As predicted, hDHFR is ~10 times more sensitive to inhibition by NADP+ (IC50 ~620 µM for hDHFR versus ~5 mM for ecDHFR) (Fig. 6c). Thus, at concentrations of NADP+ and NADPH similar to those found in E. coli, ecDHFR retains near maximal activity, while hDHFR is strongly inhibited. Additionally, the N23PP S148A mutant in which the M20 loop is stabilized in the closed conformation shows increased NADP+ inhibition compared to ecDHFR, almost to the same extent as hDHFR (Fig. 6c). This confirms that the increased product inhibition of hDHFR and N23PP S148A ecDHFR can be largely attributed to the altered dynamics of the M20 loop, which remains in the closed position.
DISCUSSION
With new insights in hand, we are poised to begin addressing why hDHFR cannot complement DHFR-deficient E. coli cells. We propose that differences in the dynamic mechanisms of the two enzymes are responsible, at least in part, for the inability of hDHFR to function efficiently in the environment of an E. coli cell. In ecDHFR, formation of product is accompanied by a shift in the ground state conformation from closed to occluded, with concomitant expulsion of the nicotinamide ring from the active site18. This process is dynamic, with conformational fluctuations between the occluded and closed states and exchange of nicotinamide out of and into the active site occurring at a rate of 1300 s−1 (refs. 16,25). The transition to the occluded ground state conformation in the E–NADP+–THF product complex of ecDHFR is intimately linked to NADP dissociation, and mutations that perturb the flexibility of the Met20 loop, or alter the closed to occluded equilibrium perturb the NADP flux14,26,34. In particular, mutations such as N23PP that “lock” ecDHFR in the closed conformation and alter millisecond time scale fluctuations of the Met20 loop greatly decrease the rate of NADP+ dissociation14. Since vertebrate DHFRs function at much lower cellular concentrations of NADP+ and THF10,31–33, they have evolved to bind cofactor more tightly and release it more slowly, primarily by eliminating coordinated conformational fluctuations on the millisecond time scale and stabilizing the active site loops in the closed conformation. The permanently closed conformation comes at the expense of slower on or off rates for substrates and products, and suggests that the exaggerated domain twisting might compensate for reduced ligand accessibility to the active site. While hDHFR has higher activity10, it is unable to rescue an E. coli cell when expressed under the endogenous promoter because the concentrations of NADP+ and THF in E. coli are inhibitory. Thus, our data strongly suggest that protein dynamics in DHFR play a major role in tuning the catalytic mechanism and ligand flux, and have evolved to allow optimal enzyme function in a given cellular environment.
ONLINE METHODS
Cloning, expression and purification of DHFR
Wild-type human DHFR (hDHFR) was cloned into a pET21a vector and transformed into E. coli BL21 (DE3) (DNAY) cells for expression. Cells were grown in M9 minimal media containing 2 mM folic acid, 0.5 g/L 15N ammonium sulfate, 0.5 g/L 15N ammonium chloride, 3g/L 13C glucose or deuterium oxide for uniform labeling. 13C15N or 2H13C15N uniformly labeled samples were used for triple resonance experiments. Cells were harvested after ~24 hours by centrifugation at 3000 g at 4 °C for 15 minutes and frozen at −20 °C. Frozen cell pellets can be stored for several weeks at −20 °C. hDHFR was purified and the desired complex was prepared by refolding as previously described37.
Wild type reference sequences (as listed in the NCBI database) were synthesized by the company Mr. Gene for DHFR from the following species: Bacillus anthracis, Staphylococcus aureus, Streptococcus pneumoniae, Vibrio cholerae, Sus scrofa, Bos taurus, Danio rerio, Caenorhabditis elegans, and Rattus norvegicus. Constructs were codon-optimized for expression in E. coli, and contained an N-terminal His6 tag. Constructs were cloned into the pET15b vector and expressed in the host E. coli BL21 (DE3) (DNAY). Cells were grown at 37°C in M9 minimal medium containing 15N ammonium sulfate and 15N ammonium chloride, induced with 1mM IPTG at an OD600 of ~0.9 at 15 °C, and harvested after ~24 hours by centrifugation at 3000x g and 4°C for 15 minutes. Cell pellets were frozen at −20°C and can be stored up to several weeks.
Cell pellets from 2 L of cell culture were resuspended in 80 mL lysis buffer containing 50 mM Tris pH 8.0, 200 mM NaCl, 1mM folic acid, 10 mM imidazole and one EDTA-free protease inhibitor cocktail tablet (Roche) and lysed by two passes through an EmulsiFlex C-3 cell disrupter (Avestin). Affinity purification was carried out using NiNTA resin (Qiagen), and protein was eluted with 50 mM Tris pH 8.0, 200 mM NaCl, 1mM folic acid, 250 mM imidazole. Fractions containing DHFR were further purified by reversed-phase HPLC, and refolded previously described37. DHFR from each species was divided into two aliquots; one was refolded with NADP+ and FOL, while the other was refolded with NADP+ and THF.
21PWNAL24 ecDHFR was expressed and purified by anion exchange and gel filtration as previously described37.
Preparation of NMR samples
NADP+, NADPH and folic acid were purchased from Sigma. (6S)-THF was purchased from Schircks Laboratories. Folic acid is light sensitive, and THF and NADPH are both light and oxygen sensitive, and need to be treated accordingly. Buffer was extensively degassed using freeze-pump-thaw cycles on a vacuum line, after which 1 mM DTT was added. Protein was exchanged into the degassed NMR buffer (50 mM potassium phosphate, pH 6.5 or pH 8.0, 50 mM KCl, 1mM EDTA, 1 mM DTT, 0.02% NaN3) in an inert environment in a glove box using a NAP-5 column, and fresh ligands were added in 10-fold excess. All spectra were recorded at pH 6.5 except for the hE–NADPH complex, for which data were acquired at pH 8.0. The samples were placed in amber NMR tubes with a vacuum line adapter, subjected to gentle vacuum, overlaid with argon, and flame-sealed to prevent oxidation of ligands.
NMR experiments
The hE–NADP+–FOL and hE–NADP+–THF complexes are stable, and could be concentrated to ~1 mM for NMR experiments. Due to the low yield after refolding and instability of the complexes, hE–NADPH and hE–FOL or hE–THF were concentrated to ~300 µM and 500 µM respectively for NMR experiments. Standard 3D HNCA, HNCOCA, HNCACB and HNCOCACB triple resonance experiments were used to assign the spectra of all complexes formed with either uniformly 2H13C15N labeled protein or 13C15N uniformly labeled protein. All NMR spectra were processed using NMRPipe and analyzed with NMRView.
15N R2 relaxation dispersion for the 21PWNAL24 mutant of ecDHFR bound to NADP+ and FOL was measured at 500 MHz and 750 MHz using constant-time CPMG experiments, as described previously38. Dispersion measurements for hDHFR were made at 800 MHz and 599 MHz, and at 800 MHz for C. elegans DHFR. All data were collected at 300K with a total relaxation period (Tcpmg) of 40 ms. Data were fitted as previously described16,39, using an in-house fitting program, GLOVE40.
Off-resonance 15N R1ρ relaxation dispersion experiments for the E-NADP+-FOL complex of hDHFR were carried out at 280 K using 900 MHz and 750 MHz spectrometers. A TROSY version of the pulse sequence described in ‘Scheme 2’ of Massi et al. was used41. R1ρ relaxation rate constants for each residue were determined by acquiring a series of 2-dimensional spectra with different relaxation delays, from 6 to 160 ms. Intensities of cross peaks were fitted using a 2-parameter exponential decay function to give relaxation rate constants. Spectra were processed and analyzed using NMRPipe, NMRView and Curvefit. Ten different effective fields were obtained using a spin lock field of 1000 Hz and different resonance offsets. On average, for each residue a maximum effective field (ωe2) of between 4 and 6 was obtained. R1 and R2 values were measured at 280 K using established inversion recovery and CPMG experiments42. Maximum relaxation delays of 2000 ms and 240 ms were used for R1 and R2 respectively. Heteronuclear NOE measurements were made in triplicate, using established methods43,44. R2 values were calculated and used instead of using R1ρ values directly according to the following relationship:
R1ρ = R1 cos2 θ + R2 sin2 θ
where θ is the tilt angle between the static magnetic field and the effective field in the rotating frame. Dispersion curve fitting was carried out as described by Massi et al.44, using the fitting program, GLOVE40.
Crystallography
Purified hDHFR was concentrated to ~12 mg/mL, exchanged into buffer containing potassium phosphate (50 mM, pH 7.5), and DTT (1 mM) and ~3-fold excess of the desired ligand (NADP+, NADPH, FOL and/or ddTHF) was added. Initial crystallization trials were performed using the automated Rigaku Crystalmation robotic system at the Joint Center for Structural Genomics (JCSG, www.jcsg.org). Crystals were optimized in sitting drops by setting up a fine screen around the conditions of the original hit, using 0.5 µL protein + 0.5 µL precipitant per drop, and varying pH and precipitant concentrations. For the E–NADP+–FOL complex, no optimization was necessary, and data were collected on a crystal obtained directly from the robotic screen. Crystals were cryoprotected in the reservoir solution supplemented with ~30% glycerol, then flash cooled and stored in liquid nitrogen until data collection. All diffraction data were collected at the Advanced Photon Source (APS) General Medical Sciences and National Cancer Institutes Collaborative Access Team (GM/CA-CAT) beamlines 23ID-B or 23ID-D.
hE–NADP+–FOL was crystallized in 2 M ammonium sulfate and 100 mM phosphate-citrate pH 4.2 at 4 °C. The 1.4 Å resolution dataset was indexed and integrated in spacegroup P6322. The hE–NADPH complex was crystallized in 24% PEG 6000 and 100 mM Tris pH 9.5 at 4 °C, and data were collected to 1.2 Å resolution and indexed and integrated in spacegroup C2221. The hE–NADP+–ddTHF complex was crystallized in 2.2 M ammonium sulfate and 100 mM sodium citrate pH 4.0 at 4 °C. A 1.7 Å resolution dataset was indexed and integrated in spacegroup P6322. Initial data processing was done using HKL2000 (HKL Research) and merged with Xprep (Bruker). Data collection statistics for all structures are summarized in Table 1.
The structures were solved by molecular replacement using Phaser with 1DLS45 (hE–NADPH–MTX, L22Y variant) as a search model for hE–NADP+–FOL, and with the hE–NADP+–FOL structure as a search model for the other two structures. Ligands and waters were removed from all the search models prior to molecular replacement. One copy of the DHFR complex was found in the asymmetric unit for all structures. Rigid body, restrained refinement and simulated annealing were carried out in Phenix46. After manual adjustment of the model in Coot47, including ligand placement, and the addition of waters, two TLS groups were defined for hE–NADP+–FOL and hE–NADP+–ddTHF restrained TLS refinement of the structure was completed using Phenix version 1.7. Anisotropic ADP refinement was carried out for the hE–NADPH structure at 1.2 Å resolution. The structures were validated using the Quality Control Check v2.7 developed by the JCSG, which included Molprobity48 (publically available at http://smb.slac.stanford.edu/jcsg/QC/). Final refinement statistics are shown in Table 1. The wavelength of data collection was 0.980 for hE–NADPH, and 1.033 for hE–NADP+–FOL and hE–NADP+–ddTHF. Ramachandran statistics are 98.5% favored, 0.0% disallowed for hE–NADPH and hE–NADP+–FOL, and 98.9% favored, 0.0% disallowed for hE–NADP+–ddTHF.
Model Building
Strong, clear electron density was observed for all ligands in each structure (Supplementary Fig. 1), with the exception of NADP+ in the hE–NADP+–ddTHF structure (Supplementary Fig. 1f). The hE–NADP+–FOL and hE–NADP+–ddTHF complexes crystallized under similar conditions and in the same space group (P6322), and diffracted to similar resolutions. However, whereas the electron density for NADP+ is clear and well-defined in the hE–NADP+–FOL structure (Supplementary Fig. 1c), it is weaker and discontinuous in the hE–NADP+–ddTHF structure, indicating disorder or low occupancy of the ligand. For the comparable ecDHFR structure (PDB code 1RX418), the electron density for the cofactor is well defined except for the nicotinamide ring, which resides outside the active site and for which no electron density is observed. Electron density that would place NADP+ outside the active site (as in ecE–NADP+–ddTHF) was not observed in the hE–NADP+–ddTHF electron density map. In both the hE–NADP+–ddTHF and hE–NADP+–FOL structures, 3-fold excess NADP+ was added to the sample. Electron density is observed in the NADP binding site in hE–NADP+–ddTHF; however, in the 2Fo-Fc map contoured at σ = 1, the electron density is broken in the region of the adenosyl ribose, despite clear density for the phosphates on either side (Supplementary Fig. 1f). In order to assess whether the observed electron density represents NADP+ or molecules of free buffer components, we modeled in NADP+ and, in parallel, carried out refinement of the structure in which we modeled in 2 phosphates, 2 glycerol molecules and waters to fit the electron density as best as possible in the absence of NADP+. The density is satisfied better with NADP+ modeled in to the active site, with the occupancy refined to ~0.8 (Supplementary Fig. 1g,h). Given the strong density for the phosphate groups, we conclude that NADP+ is bound, but is disordered in several regions. The B-values of the NADP+ ligand vary greatly, ranging from 24 Å2 for the more ordered phosphate to 61 Å2 in the adenosyl ribose. The lower B-values of the NADP+ are comparable to those in the surrounding regions of the protein, consistent with the notion that the ligand is present, but not well ordered. These structures suggest that at low pH (4–4.5), NADP+ is well ordered in the ternary Michaelis model complex, hE–NADP+–FOL, but is disordered in the product ternary complex, hE–NADP+–ddTHF, for which the next step is co-factor or product release. hDHFR was co-crystallized with 5,10-dideazatetrahydrofolate. While the configuration of the C6 stereocenter was unknown in the starting material (potentially 6R, 6S, or a racemic mixture), the electron density in the hDHFR folate binding site indicates that the 6S enantiomer is the predominant form bound in the crystal structure (Supplementary Fig. 1i,j). As our hE–NADPH crystal structure is in a different space group from the other structures and at a different pH, we supplemented the crystal structure analysis with solution NMR data (Supplementary Fig. 3d,e and Supplementary Note) in order to elucidate the conformational changes in the human enzyme as it progresses through the catalytic cycle. The two subdomains in hDHFR were identified and defined using Dyndom49.
Bioinformatic analysis
~1800 DHFR sequences deposited in the UniProt database50 were aligned using MUSCLE51. In all of our analyses, DHFR-thymidylate synthase (DHFR-TS) enzymes were excluded, as these enzymes represent a distinct evolutionary lineage and the dynamics of these bifunctional enzymes have not been extensively characterized. In addition, a separate analysis was carried out in which eukaryotic DHFR sequences from 172 completely sequenced genomes were analyzed by aligning their DHFR domains (as defined by the Pfam HMM model of the DHFR family, PF00186) using MAFFT52 followed by phylogenetic inference with the minimal evolution approach implemented in FastME53. The details of the analysis are as follows. Protein predictions for 172 completely sequenced eukaryotic genomes were obtained from a variety of sources, for details see Supplementary Table 3. The domain repertoire for each genome was determined by hmmscan (with default options, except for an E-value cutoff of 2.0 and 'nobias') from the HMMER 3.0 package (http://hmmer.org/) using hidden Markov models from the Pfam database. In a second step, the hmmscan results were filtered by the domain-specific 'gathering' (GA) cutoff scores provided by Pfam. From this set of analyzed proteins, we selected those that contained DHFR domains for phylogenetic analysis. The extent of the DHFR domain for the purpose of multiple alignments was defined by the Pfam 25.0 model of the DHFR domain. Multiple sequence alignments were then produced by MAFFT 6.240 (localpair, maxiterate 1000)52. Multiple sequence alignment columns with a gap in more than 50% of sequences were deleted and not used in further analysis. Phylogenetic trees were then calculated using FastME 1.153. All sequence, alignment, and phylogeny files are available upon request.
Complementation assays
A ΔfolA MG1655 strain was generated using recombineering, essentially as previously described54. The folA region was replaced with a Kanamycin resistance cassette. In other strains, the folA gene was replaced with genes expressing wild type E. coli DHFR (folA, control), human DHFR, or S. aureus DHFR. Wild type MG1655 E. coli cells and ΔfolA, ΔfolA::ecDHFR, ΔfolA::hDHFR or ΔfolA::saDHFR strains were grown in media supplemented with 100 µg/mL thymidine, gently pelleted, resuspended in LB media without thymidine, and plated either on plates containing LB+100 µg/mL thymidine or LB only. Plates were incubated at 37 °C overnight, and colonies were counted the following morning to determine plating efficiency. Each experiment was performed in triplicate. Note that for ΔfolA::hDHFR and ΔfolA strains, zero colonies grew in the absence of thymidine; therefore, errors could not be determined for these samples.
For filamentation assays, strains were grown in LB media supplemented with 100 µg/mL thymidine (Sigma) until an OD600 of ~0.6 was reached. Cells were gently pelleted (~2000 rpm, 5 min), and washed twice in LB media to remove traces of thymidine. Cells were diluted to an OD600 ~0.1, and grown at 37 °C, 250 rpm for 4 hours. Cell were fixed directly in the culture medium by the addition of paraformaldehyde to a final concentration of 4% and incubated at room temperature for 30 minutes. 10 µL of each cell suspension was spotted on poly-lysine coated coverslip and mounted in 80% glycerol. Samples were imaged on an inverted fluorescence microscope (Olympus model IX71) using a PlanApo N 60X (NA 1.42; Olympus) objective, equipped with a Hamamatsu Photonics ORCA-ER camera (model C4742-80-12AG). The open source microscopy software µManager36 was used to control image acquisition.
To assess the solubility of human DHFR, we used western blotting. Endogenous DHFR is expressed at very low levels, and hDHFR expression is nearly undetectable in human cell lysates unless the cells are transfected with a hDHFR expressing plasmid (Manufacturer's product information, Abnova, Catalog#: H00001719-M01). Consequently, prior to Western blotting using an α-hDHFR antibody (kindly supplied by X.Zhang), soluble fractions of MG1655 ΔfolA::hDHFR and MG1655 ΔfolA lysates were enriched in His-tagged proteins by binding to NiNTA resin (Qiagen). After elution, the Ni-enriched soluble fractions were subjected to Western blotting using standard methods.
Activity Assays
All kinetic measurements were made at pH 7.0 in buffer containing 50 mM potassium phosphate, 100 mM NaCl and 2mM DTT at room temperature. The enzyme at 400 nM was pre-equilibrated with 500 µM NADPH in a 100 µL reaction volume for 3 min in a 96-well plate. The reaction was initiated by adding DHF to a final concentration of 200 µM. Initial rates were calculated from the change in absorbance at 340 nm, which was monitored immediately after addition of DHFR for 120 s with a cycle time of 10 s. Reaction rates were measured in the presence of 0, 5, 50, 500, 5000 and 50,000 µM NADP+ to assess product inhibition. All experiments were done in duplicate, and were interleaved to minimize the effect of potential degradation of the unstable ligands, which were incubated on ice for the duration of the experiment. Control experiments were carried out in which each protein or ligand was omitted from the reaction, and resulted in rates of ~0. Data were fit using Matlab.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge M. Yamout for assistance with design and preparation of DHFR expression constructs, G. Johnson for assistance with figure preparation, X. Zhang (The Scripps Research Institute) for providing α-hDHFR antibody, D. Boger (The Scripps Research Institute) for kindly providing ddTHF, X. Dai for assistance with crystallography data collection, M. Mettlen for assistance with microscopy, and J. James and E. Jonsson for assistance with analysis of kinetic data. This work was supported by the National Institutes of Health (NIH) grant GM75995 and the Skaggs Institute of Chemical Biology (P.E.W). G.B. is the Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2136-12). D.C.E is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2140-12). D.C.E. was supported by a predoctoral fellowship from the Achievement Rewards for College Scientists Foundation, grant GM080209 from the NIH Molecular Evolution Training Program. The Joint Center for Structural Genomics is supported by NIH National Institute of General Medical Sciences (NIGMS) (U54 GM094586). C.M.Z. was supported by National Institutes of Health (NIH) grant GM087218. The GM/CA CAT 23-ID-D has been funded in whole or in part with federal funds from National Cancer Institute (Y1-CO-1020) and NIGMS (Y1-GM-1104). Use of the Advanced Photon Source (APS) was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract DE-AC02-06CH11357.
Footnotes
Accession codes. Coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession codes 4M6J (hDHFR-NADPH), 4M6K (hDHFR-NADP+-FOL) and 4M6L (hDHFR-NADP+-ddTHF)
AUTHOR CONTRIBUTIONS
G.B. and P.E.W. designed the research. G.B., M.J and G.K. collected the data. G.B., D.C.E., C.M.Z, L.T., I.A.W. and P.E.W. analyzed the data. All authors contributed to writing the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. A minimal sequence code for switching protein structure and function. Proc. Natl. Acad. Sci. USA. 2009;106:21149–21154. doi: 10.1073/pnas.0906408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson J, Baker D. Incorporation of evolutionary information into Rosetta comparative modeling. Proteins. 2011;79:2380–2388. doi: 10.1002/prot.23046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohen A, Klinman JP. Protein flexibility correlates with degree of hydrogen tunneling in thermophilic and mesophilic alcohol dehydrogenases. J. Am Chem. Soc. 2000;122:10738–10739. [Google Scholar]
- 5.Wolf-Watz M, et al. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat. Struct. Mol. Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 6.Schnell JR, Dyson HJ, Wright PE. Structure, dynamics and catalytic function of dihydrofolate reductase. Ann. Rev. Biophys. Biomol. Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 7.Brown KM, et al. Compensatory mutations restore fitness during the evolution of dihydrofolate reductase. Mol. Biol. Evol. 2010;27:2682–2690. doi: 10.1093/molbev/msq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fierke CA, Johnson KA, Benkovic SJ. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry. 1987;26:4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 9.Appleman JR, et al. Unusual transient- and steady-state kinetic behavior is predicted by the kinetic scheme operational for recombinant human dihydrofolate reductase. J. Biol. Chem. 1990;265:2740–2748. [PubMed] [Google Scholar]
- 10.Appleman JR, et al. Atypical transient state kinetics of recombinant human dihydrofolate reductase produced by hysteretic behavior. Comparison with dihydrofolate reductases from other sources. J. Biol. Chem. 1989;264:2625–2633. [PubMed] [Google Scholar]
- 11.Beard WA, Appleman JR, Delcamp TJ, Freisheim JH, Blakley RL. Hydride transfer by dihydrofolate reductase. Causes and consequences of the wide range of rates exhibited by bacterial and vertebrate enzymes. J. Biol. Chem. 1989;264:9391–9399. [PubMed] [Google Scholar]
- 12.Matthews DA, et al. Dihydrofolate reductase: X-ray structure of the binary complex with methotrexate. Science. 1977;197:452–455. doi: 10.1126/science.17920. [DOI] [PubMed] [Google Scholar]
- 13.Beard WA, et al. Role of the conserved active site residue tryptophan-24 of human dihydrofolate reductase as revealed by mutagenesis. Biochemistry. 1991;30:1432–1440. doi: 10.1021/bi00219a038. [DOI] [PubMed] [Google Scholar]
- 14.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammes GG, Benkovic SJ, Hammes-Schiffer S. Flexibility, diversity, and cooperativity: pillars of enzyme catalysis. Biochemistry. 2011;50:10422–10430. doi: 10.1021/bi201486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 17.Bystroff C, Kraut J. Crystal structure of unliganded Escherichia coli dihydrofolate reductase. Ligand-induced conformational changes and cooperativity in binding. Biochemistry. 1991;30:2227–2239. doi: 10.1021/bi00222a028. [DOI] [PubMed] [Google Scholar]
- 18.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 19.Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Backbone dynamics in dihydrofolate reductase complexes: Role of loop flexibility in the catalytic mechanism. Biochemistry. 2001;40:9846–9859. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 20.Osborne MJ, Venkitakrishnan RP, Dyson HJ, Wright PE. Diagnostic chemical shift markers for loop conformation and cofactor binding in dihydrofolate reductase complexes. Protein Sci. 2003;12:2230–2238. doi: 10.1110/ps.03219603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkitakrishnan RP, et al. Conformational changes in the active site loops of dihydrofolate reductase during the catalytic cycle. Biochemistry. 2004;43:16046–16055. doi: 10.1021/bi048119y. [DOI] [PubMed] [Google Scholar]
- 22.Miller GP, Benkovic SJ. Stretching exercises--flexibility in dihydrofolate reductase catalysis. Chem. Biol. 1998;5:R105–R113. doi: 10.1016/s1074-5521(98)90616-0. [DOI] [PubMed] [Google Scholar]
- 23.Miller GP, Wahnon DC, Benkovic SJ. Interloop contacts modulate ligand cycling during catalysis by Escherichia coli dihydrofolate reductase. Biochemistry. 2001;40:867–875. doi: 10.1021/bi001608n. [DOI] [PubMed] [Google Scholar]
- 24.Davies JF, II, et al. Crystal structures of recombinant human dihydrofolate reductase complexed with folate and 5-deazafolate. Biochemistry. 1990;29:9467–9479. doi: 10.1021/bi00492a021. [DOI] [PubMed] [Google Scholar]
- 25.Boehr DD, McElheny D, Dyson HJ, Wright PE. Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc. Natl. Acad. Sci. USA. 2010;107:1373–1378. doi: 10.1073/pnas.0914163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller GP, Benkovic SJ. Strength of an interloop hydrogen bond determines the kinetic pathway in catalysis by Escherichia coli dihydrofolate reductase. Biochemistry. 1998;37:6336–6342. doi: 10.1021/bi973065w. [DOI] [PubMed] [Google Scholar]
- 27.Cody V, Pace J, Rosowsky A. Structural analysis of a holoenzyme complex of mouse dihydrofolate reductase with NADPH and a ternary complex with the potent and selective inhibitor 2,4-diamino-6-(2'-hydroxydibenz[b,f]azepin-5-yl)methylpteridine. Acta Crystallogr. D. Biol. Crystallogr. 2008;64:977–984. doi: 10.1107/S0907444908022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CT, et al. Functional significance of evolving protein sequence in dihydrofolate reductase from bacteria to humans. Proc. Natl. Acad. Sci. USA. 2013;110:10159–10164. doi: 10.1073/pnas.1307130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitlow M, et al. X-ray crystallographic studies of Candida albicans dihydrofolate reductase. J. Biol. Chem. 1997;272:30289–30298. doi: 10.1074/jbc.272.48.30289. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 31.Fierke CA, Kuchta RD, Johnson KA, Benkovic SJ. Implications for enzymic catalysis from free-energy reaction coordinate profiles. Cold Spring Harb. Symp. Quant. Biol. 1987;52:631–638. doi: 10.1101/sqb.1987.052.01.072. [DOI] [PubMed] [Google Scholar]
- 32.Allegra CJ, Fine RL, Drake JC, Chabner BA. The effect of methotrexate on intracellular folate pools in human MCF-7 breast cancer cells. Evidence for direct inhibition of purine synthesis. J. Biol. Chem. 1986;261:6478–6485. [PubMed] [Google Scholar]
- 33.Greenbaum AL, Gumaa KA, Mclean P. The distribution of hepatic metabolites and the control of the pathways of carbohydrate metabolism in animals of different dietary and hormonal status. Arch. Biochem. Biophys. 1971;143:617–663. doi: 10.1016/0003-9861(71)90247-5. [DOI] [PubMed] [Google Scholar]
- 34.Cameron CE, Benkovic SJ. Evidence for a functional role of the dynamics of glycine-121 of Escherichia coli dihydrofolate reductase obtained from kinetic analysis of a site-directed mutant. Biochemistry. 1997;36:15792–15800. doi: 10.1021/bi9716231. [DOI] [PubMed] [Google Scholar]
- 35.Johnson GT, Autin L, Goodsell DS, Sanner MF, Olson AJ. ePMV embeds molecular modeling into professional animation software environments. Structure. 2011;19:293–303. doi: 10.1016/j.str.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. 2010;Chapter 14(Unit14) doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-Only References
- 37.Bhabha G, Tuttle L, Martinez-Yamout MA, Wright PE. Identification of endogenous ligands bound to bacterially expressed human and E. coli dihydrofolate reductase by 2D NMR. FEBS Letters. 2011;585:3528–3532. doi: 10.1016/j.febslet.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loria JP, Rance M, Palmer AG., III A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am Chem. Soc. 1999;121:2331–2332. [Google Scholar]
- 39.McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc. Natl. Acad. Sci. USA. 2005;102:5032–5037. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugase K, Konuma T, Lansing JC, Wright PE. Fast and accurate fitting of relaxation dispersion data using the flexible software package GLOVE. J. Biomol. NMR. 2013;56:275–283. doi: 10.1007/s10858-013-9747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Massi F, Johnson E, Wang C, Rance M, Palmer AG., III NMR R1ρ rotating-frame relaxation with weak radio frequency fields. J. Am Chem. Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- 42.Farrow NA, et al. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 43.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 44.Massi F, Grey MJ, Palmer AG., III Microsecond timescale backbone conformational dynamics in ubiquitin studied with NMR R1ρ relaxation experiments. Protein Sci. 2005;14:735–742. doi: 10.1110/ps.041139505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis WS, et al. Methotrexate-resistant variants of human dihydrofolate reductase with substitutions of leucine 22. Kinetics, crystallography, and potential as selectable markers. J. Biol. Chem. 1995;270:5057–5064. doi: 10.1074/jbc.270.10.5057. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 50.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J. Comput. Biol. 2002;9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- 54.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.