The dynamics of topoisomerase IIα binding to DNA and histones are important for successful mitosis and are regulated by a novel chromatin tether (ChT) domain in topoisomerase IIα.
Abstract
DNA topoisomerase IIα (Topo IIα) is the target of an important class of anticancer drugs, but tumor cells can become resistant by reducing the association of the enzyme with chromosomes. Here we describe a critical mechanism of chromatin recruitment and exchange that relies on a novel chromatin tether (ChT) domain and mediates interaction with histone H3 and DNA. We show that the ChT domain controls the residence time of Topo IIα on chromatin in mitosis and is necessary for the formation of mitotic chromosomes. Our data suggest that the dynamics of Topo IIα on chromosomes are important for successful mitosis and implicate histone tail posttranslational modifications in regulating Topo IIα.
Introduction
In preparation for chromosome segregation, fibers of interphase chromatin are remodeled to form rod-shaped chromonemas of mitotic chromosomes (Swedlow and Hirano, 2003; Eltsov et al., 2008; Nishino et al., 2012). This dramatic transformation of interphase chromatin to a set of physically tractable condensed chromosomes is complete within minutes, yet must achieve not just linear compaction, but also individualization of each chromosome (Giménez-Abián et al., 1995) and resolution of the two sisters within the pair (Sumner, 1991). The extreme fidelity with which cells carry out this process of mitotic chromosome formation is essential for preventing chromosome segregation errors.
Chromosome morphological changes in mitosis have been suggested to depend on the reorganization of chromatin on a proteinaceous axial core, first revealed in electron micrographs of dehistonized condensed chromosomes (Paulson and Laemmli, 1977; Mullinger and Johnson, 1979). Although the “axial core” is a cytologically defined structure, it likely corresponds to the chromosome scaffold, a highly stable structure that remains intact after treatment of chromosomes with micrococcal nuclease and 2 M NaCl. This biochemical fraction contains DNA topoisomerase IIα (Topo IIα) and 13S condensin (Adolph et al., 1977; Earnshaw et al., 1985; Gasser and Laemmli, 1987), enzymes that function in mitotic chromosome formation. In mitosis, Topo IIα is largely restricted to the axial core (Tavormina et al., 2002; Maeshima and Laemmli, 2003), and the residence time of Topo IIα on chromosomes is very short (∼15 s) in live cells (Tavormina et al., 2002). However, little is known about the mechanism that localizes Topo IIα to chromosomes, and it is not known if the highly dynamic property of the enzyme is biologically important.
Previous studies raised the possibility that there are distinct factors conferring Topo IIα localization upon the axial core. In either Drosophila melanogaster or chicken cells depleted of condensin, Topo IIα is targeted to mitotic chromosomes but core enrichment is abolished (Coelho et al., 2003; Hudson et al., 2003). This function of condensin involves its ability to generate positively supercoiled DNA, the preferred topological substrate of Topo IIα (Kimura and Hirano, 1997; McClendon et al., 2008). It is not known if the chromosome core region is enriched with DNA in a positively supercoiled topological state, but this can be inferred from the fact that condensin localization is mostly restricted to the core region of chromosomes (Maeshima and Laemmli, 2003; Ono et al., 2003). These data are therefore consistent with a multi-mechanism process in which, independent of condensin, Topo IIα can bind to chromatin but, influenced by condensin activity, Topo IIα becomes enriched at the axial core.
The enzyme activity of Topo IIα is to perform a strand passage reaction that allows transit of one double helix of DNA through another, allowing the removal of entanglements, supercoils, and catenations. It achieves this by making a transient double-strand break in one helix, passing a second helix through the break, then re-ligating the first (Wang, 2002). Strikingly, however, the domain of the enzyme sufficient for this reaction in vitro is not sufficient for localizing Topo IIα to chromosomes in cells (Linka et al., 2007). In fact, human cells contain two genetically distinct isoforms of Topo II (α and β) that have indistinguishable catalytic cycles, but only Topo IIα is localized to mitotic chromosomes, a property conferred by its divergent C-terminal region (CTR; Linka et al., 2007). As suggested by their respective localization patterns, Topo IIα is essential for chromosome condensation and segregation, whereas Topo IIβ is dispensable (Grue et al., 1998; Sakaguchi and Kikuchi, 2004).
Here we describe a novel element in the CTR that dictates the dynamics of Topo IIα on chromosomes and is required for mitotic chromosome formation. We refer to this component as the chromatin tether (ChT) domain because it facilitates stable binding to chromatin. In vitro, the ChT domain is required for both interaction with DNA and histone H3 N-terminal tails, the latter being enhanced by methylation of Arg 26 and Lys 27 and inhibited by phosphorylation of Ser 28. To probe the biological importance of the ChT domain, we established a knockdown/rescue system for functional analysis of Topo IIα mutants in human cells and used this system to determine whether the ChT domain is essential for normal mitotic chromosome formation.
Results
Human Topo IIα is enriched within the axial core of Muntiacus muntjak chromosomes
To determine the mechanism of Topo IIα localization to mitotic chromosomes in intact mammalian cells, we used a previously characterized human fusion protein (GFP-hTopo IIα; Tavormina et al., 2002) and expressed it in M. muntjak cells. These cells are well suited to this study because M. muntjak has the largest mammalian chromosomes and the lowest diploid chromosomal number in mammals (2n = 6, females), facilitating detailed visualization of individual arms and centromeres. Further, we found that human Topo IIα cannot be immunoprecipitated with the M. muntjak orthologue, which indicates that these interspecies isoforms cannot dimerize (Fig. 1 A). This was important because type II topoisomerases function as stable homodimeric enzymes (Tennyson and Lindsley, 1997), raising the concern that a transfected mutant Topo IIα would dimerize with the endogenous wild-type (WT) protein and localize passively, masking effects of mutations. We thus analyzed mutant transfected forms of GFP-hTopo IIα that apparently localized independently of the WT endogenous M. muntjak protein.
Figure 1.
Topo IIα localizes to the axial core of M. muntjak mitotic chromosomes. (A) GFP–human Topo IIα transfected into M. muntjak cells does not coimmunoprecipitate with endogenous M. muntjak Topo IIα when precipitated using an anti-GFP antibody (right). GFP–human Topo IIα ΔCTR (i.e., amino acids 1–1,321) transfected into HeLa cells and immunoprecipitated in the same way coprecipitates the endogenous, untagged protein (left). (The anti–Topo IIα antibody used for Western blotting recognizes only WT human or M. muntjak Topo IIα and not Topo IIα ΔCTR.) (B) Human Topo IIα localization in M. muntjak cells transfected with a construct encoding GFP–Topo IIα, fixed and stained with DAPI, demonstrating densely punctate localization of GFP–Topo IIα to the axial core of mitotic chromosomes (bottom) and nuclear localization during interphase (top). Bars, 10 µm. (See also Video 1.) (C, left) Enlarged portion of region marked by the broken-line box in B to show apparent coiling of the axial core. (C, right) A representative (more than three experimental repeats) plot across mitotic chromosome arms showing that the GFP signal (Topo IIα) occupies a narrower region than the DNA (DAPI) signal (corresponds to the solid boxed region in B). Bars, 1 µm.
Transfected GFP-hTopo IIα was primarily nuclear in interphase M. muntjak cells (Fig. 1 B, top). In mitotic cells, GFP-hTopo IIα decorated the length of chromosomes with a distinct enrichment at centromeres and at the axial core (Fig. 1 B, bottom; and Fig. 1 C). In some regions, apparent coiling of the core was observed (Fig. 1 C and Video 1). The enhanced localization at the core was evident as a structure narrower than the full width of the mitotic chromosome (Fig. 1 C, right). This matches the pattern seen in other mammalian cells when observing GFP–Topo IIα directly or using anti–Topo IIα antibodies against the endogenous protein (Earnshaw et al., 1985; Giménez-Abián et al., 1995; Tavormina et al., 2002). The heterologous system using M. muntjak cells is thus ideal for determining the mechanism of Topo IIα recruitment to mitotic chromosomes.
Topo IIα associates with mitotic chromosomes independently of nuclear localization during S phase
Based on results in the Xenopus laevis egg extract system (Cuvier and Hirano, 2003), the mechanism of Topo IIα recruitment to mitotic chromosomes appears to depend on a prior association with chromatin in S phase, during DNA replication. For example, the protein may acquire a specific posttranslational modification while on interphase chromatin that licenses the later binding to mitotic chromosomes. Indeed, previous studies have shown that mouse Topo IIα must be sumoylated for mitotic kinetochore localization (Dawlaty et al., 2008).
In intact mammalian cells, the nuclear envelope breaks down at M phase. We reasoned that an NLS mutant of Topo IIα would be cytoplasmic in S phase, but would have the opportunity to associate with chromosomes during M phase. Furthermore, the presence of endogenous M. muntjak Topo IIα in interphase would allow normal assembly of the chromosome axial core in mitosis.
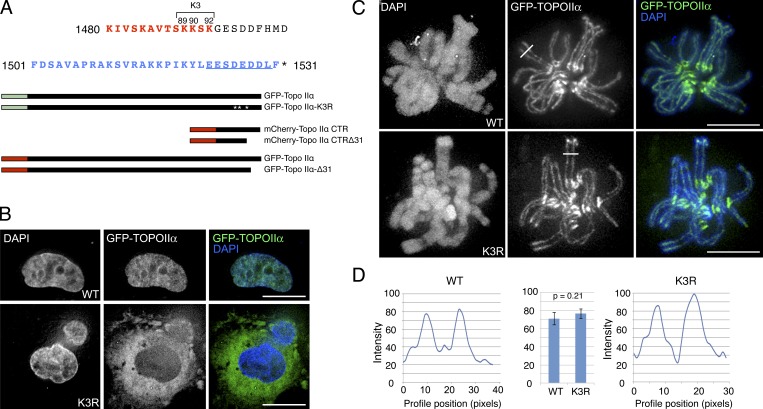
Near the C terminus of Topo IIα, a cluster of basic residues (aa 1,490–1,492) has been reported to be part of a functional bipartite NLS (Mirski et al., 1997; Wessel et al., 1997; Fig. 2 A, red text). Because NLS sequences can rely on stretches of positively charged residues (Conti et al., 1998; Cokol et al., 2000; Nitiss, 2009), we generated a triple mutant at lysines 1,489, 1,490, and 1,492, substituting with the conservative change to arginine that presumably minimizes changes to the overall structure of the protein. This mutant (Topo IIα K3R) was indeed defective for nuclear import, remaining predominantly cytosolic in interphase (Fig. 2 B).
Figure 2.
Nuclear localization of Topo IIα during interphase is not necessary for localization to chromosomes in mitosis. (A, top) Sequence of the extreme CTR of Topo IIα. Red residues indicate putative constituents of the bipartite NLS. Blue residues indicate the ChT domain described herein. (A, bottom) Schematic showing GFP- and mCherry-tagged WT Topo IIα and mutants and truncation constructs used herein. GFP–Topo IIα K3R comprised of K1489, K1490, and K1492 mutated to Arg. (B) M. muntjak cells transfected with GFP–Topo IIα or GFP–Topo IIα K3R imaged in live interphase cells. The WT protein is predominantly nuclear; the K3R mutant is predominantly cytoplasmic. Bars, 10 µm. (C) GFP–Topo IIα K3R localizes similarly to WT GFP–Topo IIα in live mitotic M. muntjak cells. Lines indicate locations of line profile plots in Fig 2 D. See also Video 2. Bars, 10 µm. (D) Quantification of WT Topo IIα and Topo IIα K3R abundance at mitotic chromosome cores in live M. muntjak cells. Both localize to the chromosome core with equal intensity. Shown are the mean of maximum intensities across n = 73 (K3R) and n = 53 (WT) core regions after background subtraction. P-value indicates two-tailed unpaired samples t test. Error bars indicate SD.
Despite the lack of access to replicating chromatin in S phase, GFP-hTopo IIα K3R relocalized from the cytosol to chromosomes upon nuclear envelope breakdown (Video 2). We quantified chromosomal localization in live cells and found that Topo IIα K3R localized to axial cores during mitosis with abundance similar to that of the WT protein (Fig. 2, C and D). This argues against the hypothesis that the presence of Topo IIα in the nucleus during interphase is critical for its chromosomal localization in mitosis (Wang, 2002; Cuvier and Hirano, 2003). Similarly ruled out are mechanisms wherein passage through the nuclear pore during interphase is a necessary step for chromosome localization in mitosis, e.g., via posttranslational modification by the nuclear pore SUMO ligase RanBP2 (Dawlaty et al., 2008).
The CTR of Topo IIα mediates chromatin association
Because the data did not support a mechanistic link to DNA replication or transit through nuclear pores, we sought to identify residues of Topo IIα that are required for mitotic chromosome localization.
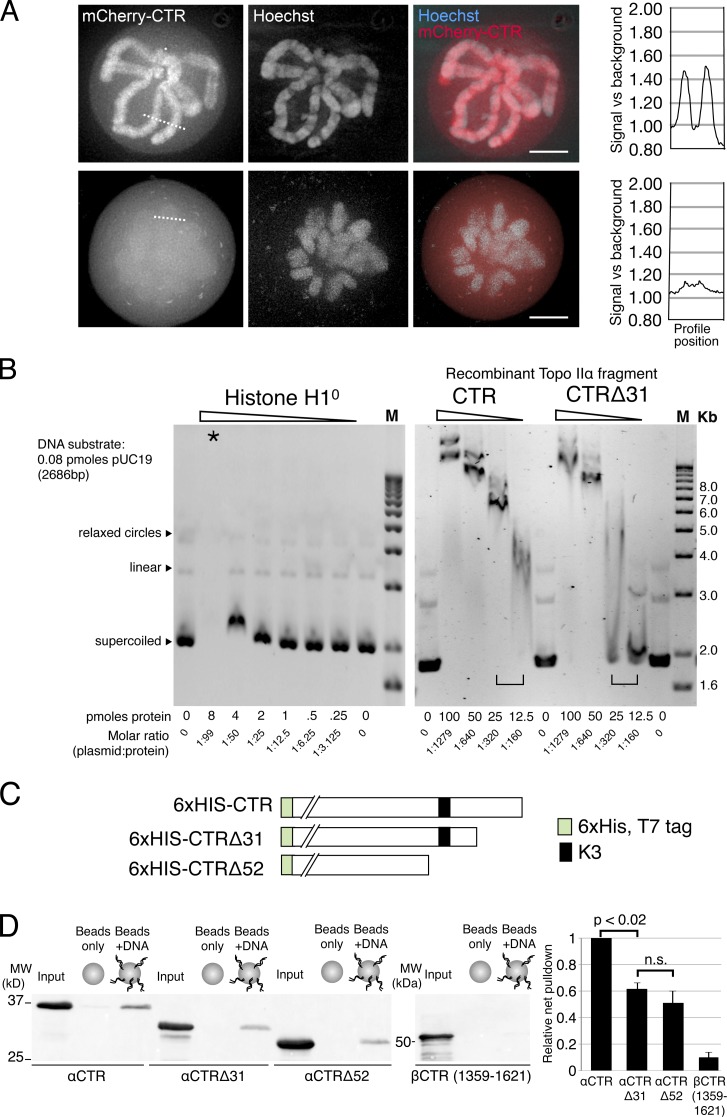
Linka et al. (2007) have shown that the C-terminal 211 residues of hTopo IIα (1,321–1,531) are sufficient to direct localization of a fluorescent protein to mitotic human chromosomes. However, it is not known if this fragment can bind to the endogenous full-length human Topo IIα and be carried to chromosomes passively. We fused this region, referred to as the CTR of hTopo IIα, to mCherry and expressed it in M. muntjak cells. We found that mCherry-CTR localized efficiently to mitotic M. muntjak chromosomes (Fig. 3 A). Although the CTR was enriched at the centromere regions, like full-length Topo IIα, the CTR occupied a broader localization on the chromosome arms, rather than being enriched at the axial core. This indicates that the CTR mediates a more broad interaction with chromosomes, similar to the observed localization of full-length Topo IIα seen in the absence of condensin (Coelho et al., 2003). Together, the data suggest that the CTR could provide a condensin-independent mechanism of chromatin association. Strikingly, a truncated CTR protein lacking only the extreme C-terminal 31 residues of the Topo IIα CTR (CTRΔ31) showed only minimal enrichment on mitotic chromosomes, which indicates that these are key residues that facilitate chromatin association (Fig. 3 A).
Figure 3.
The C-terminal 31 residues of Topo IIα are important for CTR chromosomal association and association with DNA in vitro. (A) M. muntjak cells transfected with mCherry–Topo IIα CTR (Topo IIα residues 1,321–1,531) or mCherry–Topo IIα CTRΔ31 (Topo IIα residues 1,321–1,500) imaged in live metaphase cells. mCherry–Topo IIα CTR localizes to mitotic chromosomes, whereas mCherry–Topo IIα CTRΔ31 is localized diffusely through the nucleoplasm. (A, right) A representative quantification (>3 experimental repeats) of mCherry signal across mitotic chromosomes (the broken lines in the images). Bars, 5 µm. (B) The CTR of Topo IIα binds to plasmid DNA in vitro. EMSA analysis of supercoiled, linear, and relaxed pUC19 DNA mixed with recombinant histone H1° or CTR fragments of Topo IIα (defined in A) and resolved on agarose gels. M = 1 kb ladder. The star indicates the lane where the H1°–DNA complex is assumed to be net positively charged and has reversed its migration direction. Brackets highlight the mobility shifts in the 12.5–25 pmol range for CTR and CTRΔ31. (C) Schematic showing recombinant Topo IIα fragments used in DNA–beads binding assays (D). (D, left) Anti-His tag immunoblot of Topo IIα fragment pull-downs using DNA-coated beads and purified HIS-tagged Topo IIα CTR fragments. (D, right) Quantification of pull-down efficiency when uncoated bead background is subtracted (percentage of input). Topo IIα CTRΔ31 and Topo IIα CTRΔ52 bind DNA with reduced efficiency. The CTR of Topo IIβ (βCTR, residues 1,359–1,621) has negligible DNA-binding activity. n = 3. Error bars indicate SD. n.s., not a statistically significant difference.
Topo IIα CTR binds to DNA in vitro with an affinity similar to H1 isoforms
We next sought to determine the physical mechanism of association between the Topo IIα CTR and chromatin, and first asked if recombinant Topo IIα CTR fragments bind directly to DNA: either plasmid DNA or 60-bp oligonucleotides (Fig. 3, B–D).
To investigate binding of supercoiled and relaxed plasmid DNA, we used the electromobility shift assay (EMSA) in agarose gels (Fig. 3 B). Purified recombinant Topo IIα CTR fragments or purified histone H1° was incubated with pUC19 DNA containing a mixture of supercoiled, open circular, and linear forms. An electromobility shift was seen in all three forms of DNA, decreasing in magnitude as the molar ratio of Topo IIα CTR protein to DNA decreased (Fig. 3 B, right). Topo IIα CTRΔ31 lacking the last 31 residues of the CTR showed a decrease in the magnitude of the mobility shift, which was particularly evident between 12.5 and 25 pmol of protein. Importantly, at 25 pmol of protein, the DNA shifted by Topo IIα CTR largely remained as a tight band, which indicates a maintained interaction with DNA throughout the electrophoretic migration. In contrast, smearing through the lane was observed in the case of the truncated mutant. This demonstrates that the CTRΔ31 protein dissociated from the plasmid during electrophoresis (Tseng et al., 1999; Kaer et al., 2008; Park et al., 2008). Compared with histone H1°, the Topo IIα CTR required ∼6–12-fold more protein to produce a shift. However, H1 subtypes vary widely in their affinity for nonchromatinized DNA, with H1° having up to 19-fold higher affinity than H1a (Orrego et al., 2007). Thus, the DNA affinity of the Topo IIα CTR is in a similar range to histone H1 isoforms.
To gain a relative quantification of the interaction between DNA and the Topo IIα CTR versus truncated versions, we used a pull-down strategy in which recombinant Topo IIα CTR fragments were incubated with 60-bp oligonucleotide-coated beads. The Topo IIα CTR, bound to the DNA beads, was consistently recovered using this assay (Fig. 3, C and D). However, Topo IIα CTR mutants truncated by 31 or 52 residues (CTRΔ31 and CTRΔ52) both showed a reduction in DNA binding efficiency (Fig. 3, C and D). Because these two mutants were precipitated with the DNA beads with similar efficiencies, this suggests that the K3 lysine residues within the bipartite NLS are not important for the interaction with DNA, as those residues are lacking in the CTRΔ52 mutant. Furthermore, a recombinant fragment of the CTR of Topo IIβ (Topo IIβ1359-1621) with the highest homology to the Topo IIα CTR and which does not show strong chromosomal localization in live cells (Linka et al., 2007) was recovered with only 10% of the efficiency of the Topo IIα CTR. This indicates that DNA binding may be a property of the α-CTR, not shared by the β-CTR, which promotes association with chromatin.
Together with the in vivo experiments in M. muntjak cells, these data suggest that the extreme 31 residues of the CTR contain residues that confer chromatin association via stable binding to DNA.
Topo IIα CTR binds histone H3 in vitro
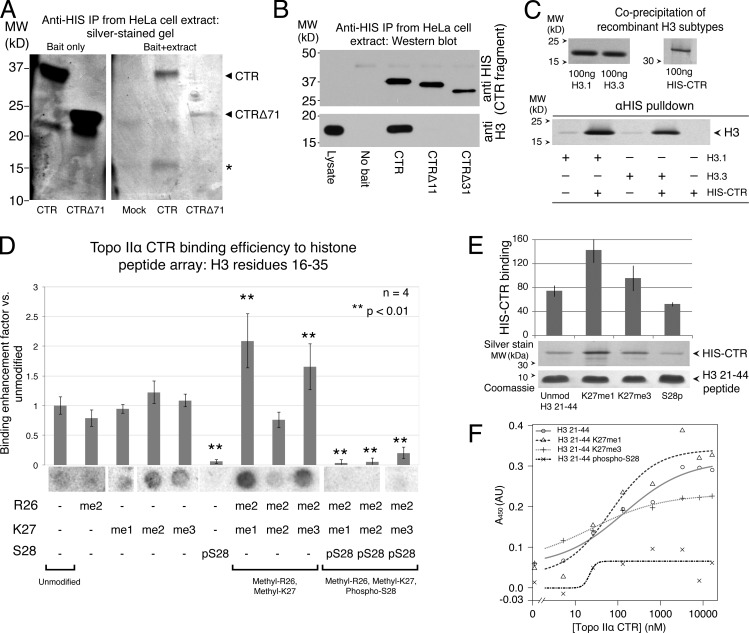
We next considered whether the Topo IIα CTR interacts with chromatin proteins in addition to binding DNA. To address this, we used anti-HIS magnetic beads to which HIS-tagged Topo IIα CTR fragments were bound. We incubated these “bait” beads with stringently nuclease-treated HeLa cell lysates in a pull-down assay. Silver-staining of the resulting SDS-PAGE gels revealed a low–molecular weight protein at ∼15 kD that was only visible when the Topo IIα CTR was used as bait (Fig. 4 A), but not in a bead-only control (mock) or in samples using a truncated CTR fragment (Topo IIα CTRΔ71).
Figure 4.
The Topo IIα CTR binds to the N-terminal tail of histone H3 in vitro. (A) Silver-stained SDS-PAGE gel reveals an ∼15-kD protein (asterisk) coprecipitated from HeLa lysates with HIS-tagged Topo IIα CTR (bait). The low–molecular weight protein was not precipitated in samples using HIS–Topo IIα CTRΔ71. (B) Immunoblot detecting immunoprecipitated recombinant HIS–Topo IIα CTR fragments (top) and coprecipitated histone H3 from HeLa cell lysates (bottom). Topo IIα CTR, but not Topo IIα Δ11 or Δ31, copurifies histone H3. Note that the data for the left three control lanes are presented again in Fig. S4 A. (C) Silver-stained protein gel detecting recombinant histone H3 subtypes (H3.1 and H3.3) coprecipitated with recombinant HIS–Topo IIα CTR using anti-HIS beads. (D) Topo IIα CTR binding to a peptide array containing histone H3 residues 16–35 with combinatorial posttranslational modifications (as indicated at the bottom; R, Arg; K, Lys; S, Ser; me1, mono-methylated; me2, di-methylated; me3, tri-methylated; p, phosphorylated). Purified recombinant HIS–Topo IIα CTR was detected strongly at spots (beneath the histograms) containing specific isoforms methylated at R26 and K27, but was not detected when S28 was also phosphorylated. Histograms show signals measured as integrated intensity of spots, subtracting local background. Error bars indicate ±SEM; n = 4. Asterisks indicate a significant difference (P < 0.01) compared with the mean of single-modified peptides. Fig. S1 shows an expanded array. (E) Detection of recombinant Topo IIα CTR coprecipitated with biotinylated histone H3 N-terminal tail (aa 21–44) peptides, either unmodified (Unmod), mono-methylated at Lys 27 (K27me1), tri-methylated at Lys 27 (K27me3), or phosphorylated at Ser 28 (S28p). CTR and peptides were mixed then precipitated with Streptavidin beads. n = 3. Error bars indicate SD. (F) ELISA assay showing binding of HIS–Topo IIα CTR to immobilized H3 (21–44) peptides, fit to a 4 parameter logistic (4PL) curve. Note that H3 (21–44) S28p binding fits the 4PL curve poorly, which suggests a low specificity of binding. A representative example from three experimental repeats is shown. The x axis is a log scale, except to the left of hash marks to allow inclusion of 0 nM HIS–Topo IIα data points on the same plot.
Given the apparent molecular weight of this coprecipitated protein, we performed Western blots on the pull-down samples and immunoblotted with antibodies recognizing histone proteins, revealing that histone H3 is specifically present in samples in which Topo IIα CTR was used as bait, and not in samples incubated with beads only (Fig. 4 B). Strikingly, H3 was not recovered in samples using truncated fragments of the CTR in which as few as 11 residues had been removed from the C terminus (CTRΔ31 and CTRΔ11). The inclusion of nucleases in the lysis buffer indicates that the interaction with H3 is unlikely to be mediated by DNA. More importantly, recombinant CTRΔ31 did interact with DNA in vitro (with ∼60% of the efficiency of the full-length CTR; Fig. 3 D) but was not able to precipitate any detectable H3 from lysates. Thus, interactions with DNA and with H3 are likely to be independent of each other. To test this directly, we mixed recombinant histone H3 subtypes with the recombinant HIS-tagged CTR, revealing that both H3.1 and H3.3 coprecipitate with the CTR protein (Fig. 4 C).
There is a wealth of evidence to suggest that histone-interacting proteins primarily recognize histone N-terminal tails (Ruthenburg et al., 2007). Further, because H3 tails are extensively posttranslationally modified, we were interested in the possibility that the Topo IIα CTR may bind in an H3 tail modification–specific manner. To test this, we incubated a histone H3 tail peptide array with purified recombinant Topo IIα CTR. The array contained H3 tail peptides with combinations of posttranslational modifications, spotted onto a glass surface. After incubation with Topo IIα CTR and extensive washing, we detected bound 6×His-tagged CTR with anti-HIS antibody. Four replicates of the array gave similar results, and no signal was observed when the array was incubated with antibody alone. This analysis revealed a profile of interaction between the CTR and H3 tail isoforms (Fig. 4 D). Binding was enhanced by specific methylation modifications at Arg 26 and Lys 27, and was inhibited by phosphorylation at Ser 28.
To confirm the peptide array data using a more quantifiable approach, we used biotin-labeled H3 tail peptides at concentrations determined accurately by mass spectrometry. After mixing the tail peptides with recombinant CTR protein, we precipitated the peptides with Streptavidin beads and estimated the amount of coprecipitated CTR on protein gels. We observed a similar trend to that seen in the peptide arrays, where methylated Lys 27 isoforms preferentially coprecipitated the CTR, whereas Ser 28 phosphorylation had an inhibitory effect (Fig. 4 E). ELISA analysis of binding to the same peptides indicated that HIS–Topo IIα CTR binds robustly to unmodified and both K27me1 and K27me3 isoforms of H3 (peptides 21–44). In particular, the affinity of K27me1-modified peptides was slightly higher than that of unmodified peptide. Strikingly, HIS–Topo IIα CTR did not bind strongly to H3 (21–44) phospho-S28 peptides, in agreement with both peptide array and beads pull-down analysis.
We conclude that in vitro, the Topo IIα CTR binds directly to histone H3 and that modified isoforms of the H3 tail either enhance or inhibit binding. Because the Topo IIα CTR can bind to DNA and histone H3 at least in part through independent mechanisms, we refer to the last 31 residues of Topo IIα as the ChT domain.
Topo IIα and histone H3 phospho–Ser 28 are localized differentially on mitotic chromosomes
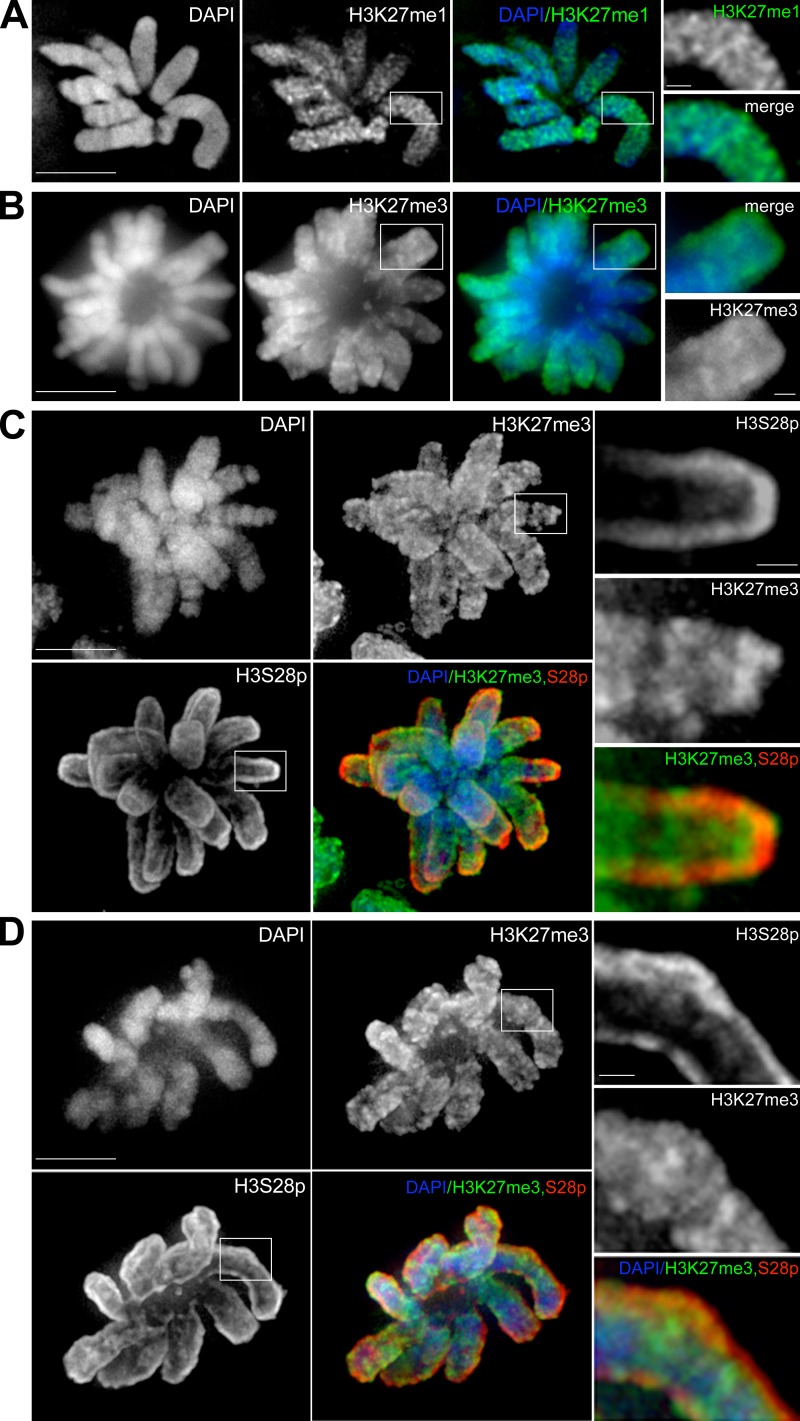
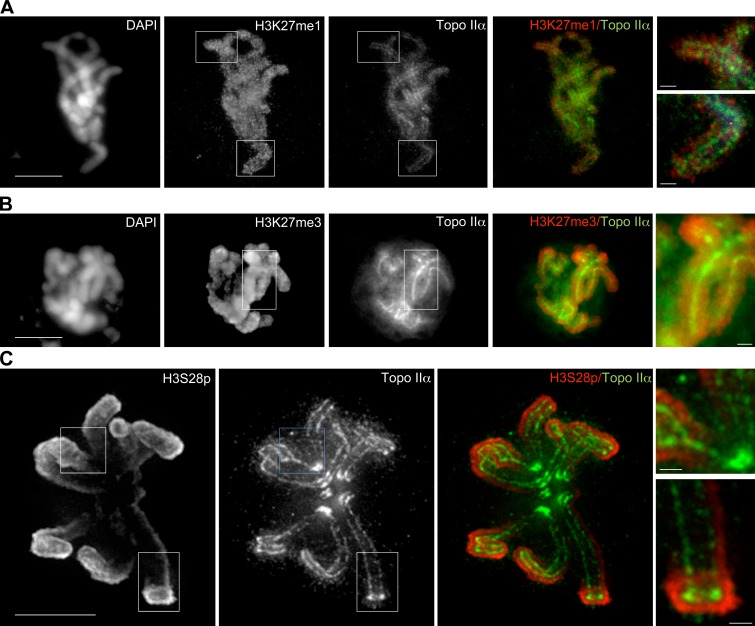
If Topo IIα binds preferentially to certain histone H3 isoforms in vivo, then one possibility is that the localization patterns of such isoforms and of Topo IIα are related. Of particular interest is that phosphorylation of Ser 28 is associated with mitotic chromosomes (Goto et al., 2002), and it was therefore counterintuitive that this modification inhibits the interaction of H3 and Topo IIα CTR in vitro (Fig. 4). To investigate this we immunostained the large mitotic chromosomes of M. muntjak using antibodies that have specificity for K27me1, K27me3, or S28p. Both K27 methylation-specific antibodies stained the entire width of mitotic chromosomes, with K27me1 localizing with a more punctate pattern than K27me3 (Fig. 5, A and B). In contrast, S28p antibodies stained the periphery of chromosomes intensely, with weak staining within the perimeter of each chromosome. To ensure that this pattern was not caused by poor penetration of antibody into chromosomes, we costained with antibodies against K27me3 and S28p. This revealed a clear contrast in localization patterns, where S28p was peripheral and K27me3 was present throughout the width of chromosome arms (Fig. 5, C and D).
Figure 5.
Immunolocalization of histone H3 isoforms in mitotic M. muntjak chromosomes. M. muntjak cells stained with antibodies against H3K27me1 (A) or H3K27me3 (B), or costained with H3K27me3 and H3S28p (C and D). H3K27me1 and H3K27me3 are distributed throughout chromosome arms, whereas H3S28p is more peripheral. Flattened z stacks are shown. DAPI, DNA stain. Bars, 10 µm. Images on the right show enlarged segments of the boxed regions (bars, 1.25 µm).
We next asked how H3 isoforms localize relative to Topo IIα. Both K27 methylated isoforms localized with a pattern partially overlapping with the axial core localization of Topo IIα (Fig. 6, A and B), but S28p was clearly excluded from the centrally localized axial core (Fig. 6 C and Video 3). These immunolocalization studies of H3 isoforms are thus consistent with the biochemical analysis of binding between H3 isoforms and the Topo IIα CTR in vitro.
Figure 6.
Immunolocalization of histone H3 isoforms and Topo IIα in mitotic M. muntjak chromosomes. M. muntjak cells stained with anti-GFP antibody to recognize GFP–Topo IIα and costained with antibodies against H3K27me1 (A), H3K27me3 (B), or H3S28p (C). H3K27me1 and H3K27me3 are distributed throughout chromosome arms and partially colocalize with Topo IIα. H3S28p localizes peripherally to Topo IIα. Flattened z stacks are shown. Bar, 10 µm. The full z-series of C is shown in Video 3. DAPI, DNA stain. Images on the right show enlarged segments of the boxed regions (bars, 1.25 µm).
A novel knockdown/rescue system for analysis of Topo IIα mutants
The data described so far indicate that the CTR, and in particular the ChT domain, contains residues important for the association of Topo IIα with mitotic chromosomes, at least in part through direct binding to DNA and histone H3. Because these interactions suggest a previously unknown mechanism of Topo IIα association with chromatin, we sought to test the functional relevance of the ChT domain in human cells. This was important for a further reason: In Saccharomyces cerevisiae, mutants lacking the CTR are viable (Jensen et al., 1996). A complication in addressing this question is that functional analysis of Topo IIα mutants in human cultured cells has previously been technically challenging. Transient transfection typically results in overexpression of Topo IIα, leading to cell death. Carpenter and Porter (2004) previously generated a doxycycline-repressible Topo IIα human cell line and described in detail the consequences of loss of Topo IIα on chromosome condensation and segregation. However, this system did not permit controlled expression of Topo IIα rescue constructs. Here, we developed a system in which Topo IIα (and mutants) are inducibly expressed close to endogenous levels and the endogenous Topo IIα can be efficiently depleted.
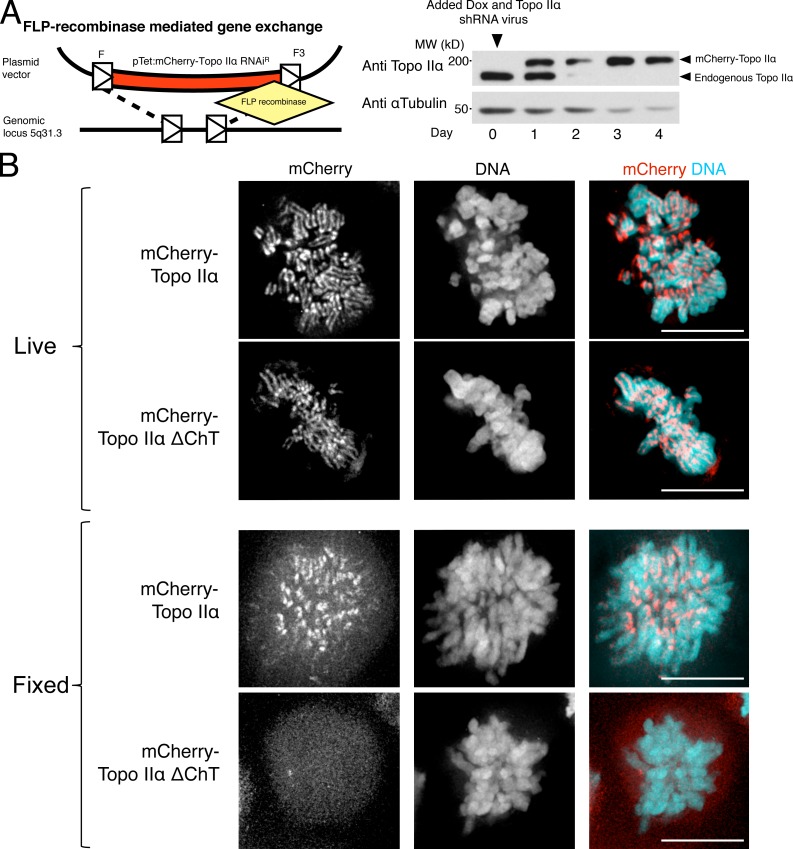
The system employs a HeLa cell line (EM2-11ht) in which a gene of interest is integrated stably at the well-characterized 5q31.3 “silent-but-activatable” genomic locus using the FLP recombinase (Fig. 7 A, left; Weidenfeld et al., 2009). This avoids detectable expression in the absence of doxycycline (Fig. 7 A, right). Constructs were inserted encoding WT mCherry–Topo IIα, mCherry–Topo IIα ΔChT, or mCherry–Topo IIα Y804F (a catalytic mutant of the active site tyrosine, serving as a negative control), and clonal lines were isolated. Because the locus of integration was the same for all cell lines generated, the induction characteristics were similar, permitting a direct comparison of independently derived transgenic cell lines. This enabled us to determine a concentration of doxycycline at which the level of transgenic mCherry–Topo IIα was similar to that of the endogenous protein (Fig. 7 A, right). Lentivirus-mediated shRNA resulted in depletion of endogenous Topo IIα to levels undetectable by immunoblotting (Fig. 7 A, right). Transgenic mCherry–Topo IIα and mutants of interest were rendered insensitive to the shRNA construct by introducing silent mutations before integration.
Figure 7.
An inducible system for analysis of Topo IIα mutants. (A, left) Schematic describing the strategy to derive HeLa EM2-11ht cells expressing RNAi-resistant mCherry–Topo IIα at close to endogenous levels. Single copy insertion of Topo IIα alleles at locus 5q31.3 was achieved using asymmetric FLP (F/F3) recombinase sites (see Materials and methods and Weidenfeld et al., 2009). (A, right) Immunoblot detecting endogenous Topo IIα and exogenously expressed mCherry–Topo IIα. HeLa EM2-11 cells were infected with lentiviral particles encoding knockdown shRNA constructs at day 0 against Topo IIα and Topo IIβ simultaneously with the addition of 250 ng/ml doxycycline to induce expression of the exogenous allele. Endogenous Topo IIα is effectively depleted, whereas the exogenous protein is expressed to approximately endogenous levels within 24 h. (B) Localization of mCherry–Topo IIα and mCherry–Topo IIα ΔChT after 48 h of induction with doxycycline in live HeLa EM2-11 cells (top) or after fixation with methanol (bottom). Topo IIα ΔChT is labile upon fixation. Bars, 10 µm.
ΔChT domain mutants have altered dynamics on mitotic chromosomes
We first compared the localization pattern of mCherry–Topo IIα and mCherry–Topo IIα ΔChT. Although ΔChT clearly localized to interphase nuclei (Fig. S2) and to mitotic chromosomes in live cells (Fig. 7 B, top), it was entirely lost from chromosomes after fixation in formaldehyde or methanol (Fig. 7 B, bottom), whereas the full-length mCherry–Topo IIα was fully retained at centromeres and, to a lesser extent, at the axial core. Thus, topoisomerases lacking the ChT domain must associate with chromatin in a manner that becomes labile upon fixation. This result was the same whether or not the endogenous Topo IIα protein was depleted using shRNA (unpublished data). This indicates that the ChT domain is necessary for a stable association with chromosomes even in the presence of endogenous Topo IIα, where the mutant is presumably able to form a heterodimer.
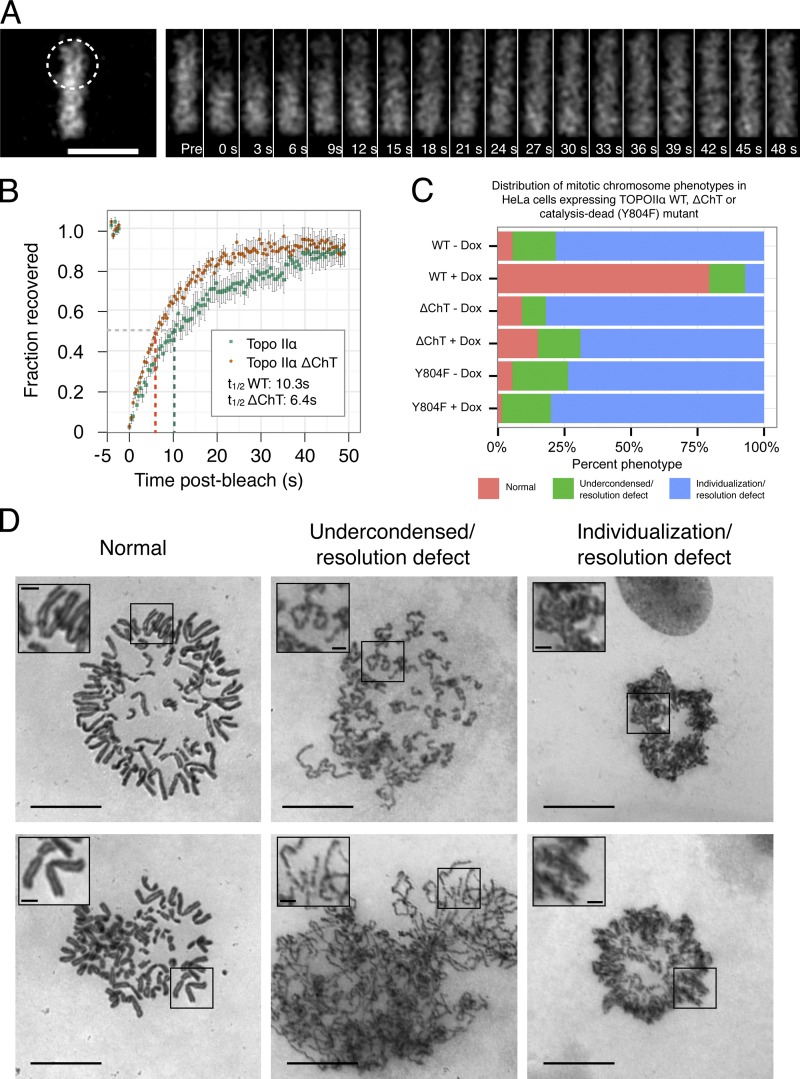
To study this phenomenon using a quantifiable approach in live cells, we used FRAP analysis. Previous work studying the dynamics of EGFP–Topo IIα used cells that overexpress the protein by an estimated several-fold (Tavormina et al., 2002), which may affect the apparent recovery dynamics due to, e.g., saturation of chromosomal binding sites. In addition to expression at near-endogenous levels, our strategy used the mCherry fluorescent protein, which is more likely to remain monomeric at high concentrations of Topo IIα seen on mitotic chromosomes than the weakly dimeric EGFP (Zacharias et al., 2002).
We found that the mCherry-tagged WT protein is highly mobile on chromatin in metaphase cells, with a recovery t1/2 of 10.3 s (Fig. 8, A and B). This is slightly faster than the published data that yielded a t1/2 of 15.5 s (Tavormina et al., 2002). Importantly, however, we observed that mCherry–Topo IIα ΔChT had an increased mobility such that the recovery t1/2 was reduced to 6.4 s. This is consistent with a higher Koff than the WT protein and the loss of chromatin interactions when compared with the WT protein. Again, these experiments were performed without depletion of the endogenous Topo IIα, which indicates that the difference in mobility is evident even when there presumably exists a population of WT/mutant heterodimers. For this reason, our estimate of the reduction in residence time of Topo IIα lacking the ChT domain is likely to be conservative. Moreover, the data suggest that in WT cells, two intact ChT domains are required for the stable association of the Topo IIα homodimer with chromatin.
Figure 8.
The ChT domain controls the dynamics of Topo IIα on mitotic chromosomes and is necessary for mitotic chromosome individualization and condensation. (A) Images of a metaphase plate from a representative FRAP experiment using HeLa-EM2-11 mCherry–Topo IIα cells 48 h after induction with doxycycline. A laser-scanning confocal microscope was used to bleach a circular region (broken line) encompassing ∼50% of the metaphase plate, and recovery was monitored over time (seconds). Pre, prebleach image. Bar, 10 µm. (B) Mean FRAP of mCherry–Topo IIα (n = 11 cells) and mCherry–Topo IIα ΔChT (n = 21) in HeLa-EM2-11 cells, as described in A. After collection of prebleach data (top left of plot), images were collected every 0.43 s. Recovery was quantified in the bleached area over 50 s and normalized as described previously for analysis of chromatin proteins (Phair et al., 2004a). Error bars indicate standard error. (C) Distribution of phenotypes seen in cells depleted of endogenous Topo II and induced to express mCherry–Topo IIα, mCherry–Topo IIα ΔChT, or mCherry–Topo IIαY804F. Induction of mCherry–Topo IIα rescues chromosome condensation, individualization, and resolution defects, whereas Topo IIα ΔChT and Topo IIαY804F fail to rescue. Cells were assayed after a double thymidine synchrony and release protocol in conjunction with shRNA-mediated depletion of the endogenous Topo II (see Materials and methods). WT − Dox, n = 129; WT + Dox, n = 141; ΔChT − Dox, n = 100; ΔChT + Dox, n = 100; Y804F − Dox, n = 148; Y804F + Dox, n = 146. (D) Classification used in C. Spreads of chromosomes after depletion of endogenous Topo II show a range of mitotic phenotypes: “Normal” metaphase plates containing individualized chromosomes with visibly resolved sister chromatids (left, red in C), chromosomes that failed to complete linear condensation and sister chromatid resolution (middle, green in C), and failed chromosome individualization and sister chromatid resolution (right, blue in C). Bars, 10 µm. Insets show enlarged views of the boxed regions (bars, 1.25 µm).
In sum, the interaction of mCherry–Topo IIα ΔChT is more labile upon fixation of cells, and in live mitotic cells it is more mobile, which is consistent with weaker affinity interactions with chromatin. The ChT domain dictates the dynamics of Topo IIα on mitotic chromosomes.
ΔChT domain mutants cannot facilitate chromosome individualization, resolution, or condensation
We next investigated the biological consequences of the altered dynamics of mCherry–Topo IIα ΔChT in terms of mitotic chromosome formation. Because previous studies have shown that Topo IIβ can weakly compensate for the lack of Topo IIα (Linka et al., 2007), we codepleted both isoforms, using shRNA lentiviral transduction, to achieve a minimum base level of type II topoisomerase activity (Fig. S3). We simultaneously induced expression of mCherry–Topo IIα, mCherry–Topo IIα ΔChT, or mCherry–Topo IIα Y804F (as a negative control) in the isogenic cell lines and monitored changes in metaphase chromosome morphology when cells reached mitosis after release from a double thymidine synchrony. Giemsa staining of Carnoy’s fixed cells allowed fine-scale examination of changes in chromosome morphology (Giménez-Abián et al., 1995; Giménez-Abián and Clarke, 2009; Fig. 8, C and D). In addition, we filmed cells, treated in the same manner, by digital time-lapse microscopy to observe single live cells as they entered and progressed through mitosis (Fig. 9).
Figure 9.
The ChT domain of Topo IIα is necessary for chromosome condensation and segregation in mitosis. (A and B) Still images taken from digital time-lapse imaging of live cells after depletion of endogenous Topo IIα and induced with doxycycline to express mCherry–Topo IIα or mCherry–Topo IIα ΔChT. Cells were assayed after a double thymidine synchrony and release protocol in conjunction with shRNA-mediated depletion of the endogenous Topo IIα and DNA observed via imaging of Hoechst. Red boxed regions are enlarged in B to highlight the differences in metaphase chromatin morphology. Frames were taken at 5-min intervals. Time is indicated (hours:minutes) in each movie frame. Bars, 10 µm. (C) Distribution of phenotypes seen in live single cells described in A and B. Induction of mCherry–Topo IIα rescues chromosome condensation and segregation defects, whereas Topo IIα ΔChT fails to rescue. WT − Dox, n = 96; WT + Dox, n = 100; ΔChT − Dox, n = 61; ΔChT + Dox, n = 82.
In cells depleted of type II topoisomerases, we saw large numbers of mitotic cells with chromosomes that failed to become individualized (Fig. 8 C and Fig. 8 D, right). This is identical to the morphology observed when cells attempt to condense their chromosomes in the presence of high doses of Topo IIα catalytic inhibitors (Giménez-Abián and Clarke, 2009). Less frequently, we observed undercondensed chromosomes in which the sister chromatids had failed to become resolved into discrete pairs (Fig. 8 C and Fig. 8 D, middle). This is identical to the results of treatment with lower doses of Topo II catalytic inhibitors that restrict but do not eliminate Topo II activity (Giménez-Abián and Clarke, 2009). Control cells treated with a nontargeted shRNA virus showed normal mitotic chromosome morphologies with well-individualized and condensed chromosomes that had fully resolved sister chromatids (Fig. 8 D, left). The time-lapse analysis of single cells was consistent with these data and in addition revealed obvious defects in chromosome segregation and progression through mitosis (Fig. 9 and Videos 4–9).
Strikingly, the induction of mCherry-tagged Topo IIα rescued the condensation-, individualization-, and resolution-defective phenotypes in >75% of the fixed cells analyzed (Fig. 8 C). Similarly, based on the analysis of single live cells, the mitotic defects were rescued in ∼60% of the cells (Fig. 9 C). Thus, the mCherry-tagged protein can functionally complement the loss of endogenous Topo IIα, and the shRNA vectors did not have significant off-target effects that affected chromosome morphology. However, neither the induction of mCherry–Topo IIα ΔChT or of mCherry–Topo IIα Y804F was able to rescue the chromosome morphology defects, which indicates that they are each deficient in the formation of mitotic chromosomes (Fig. 8 C and Fig. 9 C). This is the first indication that the precise dynamics of Topo IIα on mitotic chromosomes may be required for the chromosome structural changes needed for successful mitosis in mammalian cells.
Discussion
Our data demonstrate that a novel ChT domain at the extreme C terminus of Topo IIα increases the residency time on mitotic chromosomes in a manner that correlates with histone H3 and DNA binding in vitro and which is essential for chromosome condensation. This suggests that the precise dynamics of Topo IIα are crucial for assembly of mitotic chromosomes. Insight into the interaction between Topo IIα and chromatin is significant for ongoing research efforts to maximize the efficacy of Topo II–targeted cancer drugs because tumor cells with reduced strand passage activity become drug resistant (Nitiss, 2009), and one such route to loss of activity is through mutations that reduce chromatin binding.
The ChT domain is required for a full-affinity interaction with both DNA and histone H3 in vitro and with mitotic chromosomes in the context of full-length Topo IIα. It possesses an N-terminal stretch of nonpolar residues interspersed with Arg and Lys, and a C-terminal stretch of acidic amino acids (Fig. 2 A). Because the ChT domain is implicated in an interaction with both DNA and histone H3, a simple predication is that the acidic stretch contacts the basic H3 N-terminal tail and that the DNA associated with or adjacent to the corresponding nucleosome interacts with the Lys and Arg residues of the ChT domain. In agreement with this possibility, we observed that the acidic residues are required for the in vitro interaction with H3. However, because a crystal structure including the entire CTR of human Topo IIα does not exist, we are limited to speculating on its likely tertiary structure. Moreover, our data indicate that the interaction surfaces between chromatin and the CTR are still more complex than the residues within the ChT domain that we have shown to be crucial. First, the CTR lacking the ChT domain retains 60% of the binding capacity for DNA in vitro, indicating that other CTR residues contact DNA. Second, we observed that the Topo IIα K3R mutant has slightly reduced affinity for histone H3 in vitro (Fig. S4). A structural approach will be needed to determine all of the contact residues between DNA, histone H3, and the Topo IIα CTR.
The K3R mutant studies revealed that Topo IIα does not need to be in the nucleus during interphase in order for it to localize to mitotic chromosomes in mitosis (Fig. 2). However, in support of the biological importance of the K3 residues, Topo IIα K3R had altered dynamics on mitotic chromosomes and was defective for chromosome formation in mitosis in the absence of the endogenous Topo IIα (Fig. S4). We cannot distinguish whether these defects arise as a consequence of the lack of Topo IIα K3R in the nucleus during DNA replication, which is consistent with previous studies (Cuvier and Hirano, 2003), or are due to reduced affinity for histone H3.
We also provided evidence that the precise dynamics of Topo IIα, dictated by the ChT domain, are important for chromosome individualization and condensation, and for sister chromatid resolution in mitosis. This indicates that the dissociation frequency of the enzyme from chromatin may dictate the ability of the enzyme to perform its mitotic functions. The ΔChT mutant had about a 40% decrease in recovery t1/2 versus WT Topo IIα. However, this is likely to be a conservative estimate of the defect observed because the FRAP studies were performed in the presence of the endogenous WT protein, which potentially allowed the formation of Topo IIα ΔChT/Topo IIα heterodimers with an intermediate affinity for chromatin. Recent work led to the estimate that Topo IIα performs around 10 enzyme cycles before releasing from chromatin (Yogo et al., 2012). The increased dissociation rate of the Topo IIα ΔChT mutant, as measured by FRAP, presumably decreases the processivity of the enzyme, most likely via the loss of interactions with histone H3 and DNA. It will be of interest to determine how binding of the ChT domain to chromatin is coordinated in the context of the enzyme cycle: whether the ChT domains of the homodimeric enzyme associate with both G-segment and T-segment chromatin or specifically with just one of the segments, and if the interactions are lost during any of the large conformational changes through which the enzyme transits during strand passage. Intriguingly, the acidic stretch of residues essential for histone H3 binding contains a nested serine (Ser 1524), which, upon phosphorylation stimulated by Topo IIα catalytic inhibitors, interacts with the cell cycle checkpoint protein MDC1 (Luo et al., 2009). Presumably, there must be a competition in vivo between histone H3 and MDC1 for ChT domain occupancy. If H3 binding is intimately linked with strand passage, then interruption of the reaction cycle may lead to release of H3, allowing phosphorylation of Ser 1524 and checkpoint activation via MDC1 binding. Why does Topo IIα interact with histone H3? Estimates of a mean internucleosomal linker DNA length of ∼54 bp (Schones et al., 2008), of which Topo IIα contacts 26 bp in coordinating a double-strand break, suggest that highly nucleosome-occupied areas of chromatin may present relatively limited opportunities for Topo IIα activity. Indeed, it has been reported that Topo IIα activity is inhibited on nucleosome-occupied DNA in vitro (Galande and Muniyappa, 1997). An attractive hypothesis is that the ChT domain serves to mitigate the inhibitory effect of nucleosomes, perhaps by positioning the enzyme adjacent to a histone octamer, or more simply, increasing the residence time of the enzyme at nucleosome-rich regions such that its rate of catalytically productive engagement is raised nearby.
Immunostaining of M. muntjak chromosomes revealed strikingly different localization patterns of H3 isoforms in mitotic chromosomes. The methylated K27 isoforms partially overlapped with Topo IIα within the axial core (Figs. 5 and 6). Similarly, superresolution microscopy studies have shown that the K27me1 isoform is partly confined to the interior of Drosophila mitotic chromosomes (Strukov et al., 2011). Most interestingly, we observed that H3 phosphor-Ser 28 is largely excluded from the core region of M. muntjak chromosomes. The Aurora B kinase that modifies H3 Ser 28 has multiple substrates. Nevertheless, it is intriguing that inhibition of Aurora B alters the localization of Topo IIα on mitotic chromosomes (Fig. S5). Together, these studies raise the possibility that the status of histone tail modifications controls the association of Topo IIα with chromatin. It will be of interest to determine if other chromosomal proteins possess ChT domains and whether they bind to particular histone isoforms.
Materials and methods
Cloning
The pT104-1 EGFP–Topo IIα expression vector contains a SacII–HindIII fragment encoding human Topo IIα cloned in-frame into pEGFP-C3 under a cytomegalovirus promoter (Takara Bio Inc.; Mo et al., 1998). Site-directed mutagenesis to generate the K3R, Y804F, and truncation mutants used the method of Liu and Naismith (2008). mCherry-tagged CTR constructs were constructed using pmCherry-C1 (Takara Bio Inc.), and PCR-amplified products encoding C-terminal fragments from pT104-1 (or mutants) were cloned in at BglII–SacII; the resultant constructs are under a cytomegalovirus promoter. S2F-IMCg-F-EGFP/luc-F3 vectors were a gift from K. Schoenig (Zentralinstitut für Seelische Gesundheit [ZI], Mannheim, Germany; Weidenfeld et al., 2009). To generate FLP-in Topo IIα vectors, EGFP was removed from the self-inactivating viral vector S2F-IMCg-F-EGFP/luc-F3 using SacII–NotI sites and replaced with an mCherry PCR product containing an additional 3′ MluI site to render S2F-IMCg-F-mCherry/luc-F3. Topo IIα was amplified by PCR to contain MluI sites at both ends and cloned into S2F-IMCg-mCherry/luc-F3 to render S2F-IMCg-F-mCherry–Topo IIα/luc-F3; expression is thus under an rtTA (Tet-On) promoter. All growth of S2F-IMCg vectors was performed at 30°C to reduce the frequency of plasmid rearrangement. Constructs used for expression of Topo IIα/β fragments in Escherichia coli were generated using PCR-amplified fragments from pT104-1 or Topo IIB 1186–1621 (Linka et al., 2007) cloned into pET28a (EMD Millipore) at SacI–HindIII under a T7/lac promoter. Constructs encoding Topo IIα with silent mutations conferring resistance to shRNA constructs V3LHS_327878 and V3LHS_327876 were constructed as follows. Fragments of pT104-1 were amplified using primers containing a 5′ synthetic shRNA-resistant sequence and a 3′ adjoining Topo IIα sequence. Vector IMCg-F-mCherry–Topo IIα/luc-F3 was amplified using similar primers, designed to amplify around the plasmid. The resulting PCR products thus contained homology in the ∼20 bp shRNA-resistance regions at the ends of each product and could be combined using sequence and ligation-independent (SLIC) cloning (Li and Elledge, 2007). shRNA-resistant regions were then subcloned out of the resulting vector and cloned back into IMCg-F-mCherry–Topo IIα/luc-F3 by traditional (restriction enzyme–mediated) methods to avoid mutations generated during vector PCR. This process generated IMCg-F-mCherry–Topo IIα/luc-F3 shRNAR (thus all Topo IIα constructs also express luciferase from a coregulated promoter).
FLP-in cell line generation
HeLa EM2-11ht cells were a gift from K. Schoenig. Recombination-mediated cassette exchange was achieved using cotransfection of S2f-IMCg–Topo IIα/luc-F3 and pPGKFLPobpA (Flp recombinase encoding plasmid; Raymond and Soriano, 2007), and Addgene plasmid 13793 and pTRE2-pur (Takara Bio Inc.) in a 6-well dish. At t = 24 h, cells were trypsinized and plated in 4 µg/ml puromycin. At t = 48 h, existing media was replaced with media containing 50–100 µM ganciclovir. Cells were cultured with ganciclovir for 10–14 d until colonies were visible, with media changes every 48 h.
Topo IIα shRNA knockdowns
Knockdowns used GIPZ clones Topo IIα V3LHS_327878, Topo IIβ V2LHS_94084, or accompanying GIPZ nonsilencing control sequence vector (catalog no. RHS4346; Thermo Fisher Scientific). Viruses were produced by cotransfection of HEK293T cells with GIPZ vector and pMDG and ΔNRF packaging constructs using linear polyethylenimine (PEI) at a 3:1 DNA/PEI ratio. Viral transductions were performed using standard protocols in the presence of 8 µg/ml Polybrene.
DNA transfections
Transfections were performed using GenJet Plus reagent (SignaGen Laboratories) according to the manufacturer’s instructions.
Immunostaining
Mitotic cells were obtained by shake-off, then allowed to adhere onto poly-l-lysine–coated coverslips before fixation with 4% paraformaldehyde for 10 min, post-fix extraction with 0.5% Triton X-100 for 5 min, and further fixation in −20°C methanol for 10 min. Blocking was achieved in wash buffer (PBS with 0.01% Triton X-100) with 3% BSA (Sigma-Aldrich). Antibodies used were: α-H3S28phos (catalogue no. H9908; Sigma-Aldrich), α-H3K27me1 (catalogue no. A-4037; Epigentek), α-H3K27me3 (catalogue no. 07-449; EMD Millipore), α-GFP (catalogue no. 1814460; Roche), goat α-rat Cy3 (catalogue no. 112-165-143; Jackson ImmunoResearch Labs), goat α-rabbit IgG Alexa Fluor 488 (catalogue no. A11008; Molecular Probes; Invitrogen), goat α-mouse IgG1 Alexa Fluor 488 (catalogue no. A21121; Molecular Probes, Invitrogen), and goat α-rabbit IgG (H+L) Alexa Fluor 568 (catalogue no. A11011; Molecular Probes, Invitrogen).
Fluorescence and bright field microscopy
Fluorescence imaging was performed using a DeltaVision PersonalDV microscope system (Applied Precision), a 100× Uplan S Apochromat (NA 1.4) objective lens (Olympus), and a CoolSNAP HQ2 camera (Photometrics). Fixed material was imaged at ambient temperature after mounting in Vectashield (Vector Laboratories). Live cell time-lapse analysis was conducted using the DeltaVision weather station chamber at 37°C in CO2-independent medium, and images were collected at 5-min intervals. Images were acquired using DeltaVision SoftWoRx software (Applied Precision) and processed using ImageJ (Abràmoff et al., 2004). If necessary, camera hot pixels removed using Remove Outliers; care was taken to avoid altering chromosome signal. Line profiles in Figs. 1–3 were calculated using the Plot Profile tool in ImageJ with a line width of 20 pixels and plotted relative to background (nonchromosomal) areas. Where indicated, z stacks were deconvolved using DeltaVision SoftWoRx software (Applied Precision). Fluorochromes used for immunostaining were as follows: Cy3 (goat α-rat conjugate, catalogue no. 112-165-143; Jackson ImmunoResearch Laboratories, Inc.), Alexa Fluor 488 (goat α-rabbit IgG conjugated, catalogue no. A11008; Molecular Probes; Invitrogen), Alexa Fluor 488 (goat α-mouse IgG1 conjugate, catalogue. no. A21121; Molecular Probes; Invitrogen), Alexa Fluor 568 (goat α-rabbit IgG [H+L] conjugate; catalogue no. A11011; Molecular Probes; Invitrogen). Bright-field microscopy: Giemsa-stained chromosomes (mounted in Entellan; EM Science) were imaged using an Axioplan2 microscope (Carl Zeiss), a 100× α-Plan Fluor (NA 1.45) objective lens, and an AxioCam HRm camera (Carl Zeiss) at ambient temperature.
Cytological analysis of chromosome structure
shRNA-transduced cells were synchronized using the double-thymidine arrest protocol. In brief, beginning 52 h after shRNA transduction/doxycycline addition, cells were incubated for 16 h in 2 mM thymidine. Thymidine was then washed out and the cells cultured for 10 h. The culture medium was then resupplemented with 2 mM thymidine for 16 h. Thymidine was washed out a second time and cells were harvested 10 h later in M phase. Chromosome preparations were prepared by osmotic swelling in 50% medium/50% tap water for 7 min, followed by three washed in Carnoy’s fixative (75% methanol, 25% glacial acetic acid; Giménez-Abián and Clarke, 2009). After dropping the material onto cleaned glass slides, chromosomes were stained 5% Giemsa (EMD) in phosphate buffer, pH 6.8 (Harleco; EMD).
Protein expression and purification
Fragments of human Topo IIα or Topo IIβ were amplified from pT104-1 or from YFP–Topo IIβ expression plasmids as used in Linka et al. (2007), cloned into pET28a, and transformed into BL21 (DE3). Expression was induced using the autoinduction method (Studier, 2005) in rich media, cells were lysed using lysozyme and sonication, and the supernatant was purified on Ni-NTA resin (QIAGEN) under nondenaturing conditions. Proteins were quantified using a BCA assay (Thermo Fisher Scientific).
Electromobility shift assays
Purified recombinant Topo IIα fragments were incubated with purified pUC19 DNA in bandshift buffer (Campoy et al., 1995), prepared without E. coli competitor DNA, for 30 min at RT. Samples were loaded onto 1% agarose gels in 0.5× Tris/Borate/EDTA (TBE) and run at 1.2 V/cm at 4°C for 20 h, then post-stained with ethidium bromide, based on the protocol of Yang and Champoux (2009).
DNA binding assays by pull-down
Assays were performed using a modified version of Wu (2006), as follows. Complimentary 60-bp 5′ biotin-conjugated primers (Integrated DNA Technologies) were annealed by mixing in an equimolar ratio, heating to 100°C, and cooling to RT. The resulting double-stranded DNA (dsDNA) was incubated with Streptavidin-coated Magnabeads (Genscript) for 30 min at RT, rotating end over end. The resulting oligonucleotide-coated beads were then blocked with 10% milk (filtered to remove particles) and 1% NP-40 in PBS for 50 min at RT. Beads were washed once in PBS/1% NP-40/1% milk, incubated with purified recombinant Topo IIα or Topo IIβ fragments in the same solution for 3 h at RT, washed twice in PBS/1% NP-40, once in PBS, and then boiled in SDS sample buffer and resolved on a 15% SDS-PAGE gel. The protein was transferred to polyvinylidene fluoride (PVDF) and subjected to immunodetection using anti-His primary antibody H1029 (Sigma-Aldrich) and anti–mouse secondary antibody conjugated to IR680 dye and scanned using a LI-COR Odyssey blot scanner (LI-COR Biosciences).
Western blotting and protein extraction
HeLa cells were harvested and lysed in TBS/2% SDS, 1× Complete protease inhibitor (Roche), and sonicated to shear DNA. Protein was quantified using a BCA protein assay kit (Thermo Fisher Scientific) and loaded onto SDS-PAGE gels, run as standard. Protein was transferred to PVDF and probed using anti–Topo IIα antibody sc-13058 (Santa Cruz Biotechnology, Inc.) or anti–α-Tubulin antibody ab15246 (Abcam).
Modified histone arrays
Histone peptide arrays (Active Motif) were blocked using 5% nonfat dried milk and 1% porcine gelatin for 2 h. Arrays were then incubated with 200 nM HIS–Topo IIα CTR for 1 h. Bound Topo IIα was detected using anti-HIS monoclonal antibody at 1:5,000 (H1029; Sigma-Aldrich) and HRP-conjugated anti–mouse secondary antibody at 1:3,000 (Invitrogen) for chemiluminescent detection. All solutions were prepared in TBS containing 0.02% Tween 20, and washes in this buffer were performed between steps.
Topo IIα fragment pull-downs from HeLa cell lysates
HeLa cells were lysed using NP-40 buffer (20 mM Tris, pH 8, 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, 1× Complete protease inhibitors [Roche], 420 µg/ml NaF, 1.84 mg/ml Na3VO4, 10 mM N-ethyl-maleimide, 5 U/ml cynase [Ribosolutions, Inc.], and 4 mM MgCl2). Lysates were incubated with anti-HIS Magnabeads at 4°C overnight, then washed in PBS/0.1% NP-40. Bound proteins were eluted in SDS-PAGE loading buffer and analyzed by SDS-PAGE and silver staining (Bio-Rad Laboratories).
In vitro pull-down assays
To study interactions between Topo IIα CTR and biotinylated H3 peptides (residues 21–44), 25 µl of Streptavidin Dynabeads MyOne T1 were washed in binding buffer (50 mM NaH2PO4, pH 7.5, 50 mM NaCl, 0.05% NP-40, and 10 mg/ml BSA) three times before resuspending in a complex of 3 µg peptide and 10 µg purified HIS–Topo IIα CTR that had been incubated overnight at 4°C in a volume of 50 µl. Peptides/Topo IIα CTR complexes were conjugated to beads in a 2 h, 30 min incubation at RT. Beads were washed three times in 150 µl of binding buffer without BSA. For analysis, beads were boiled in 15 µl SDS-PAGE loading buffer and separated on 4–12% SDS-PAGE gels.
To study interactions between HIS–Topo IIα CTR and full-length H3, 2.5 µg of mouse anti-HIS antibody (Takara Bio Inc.) per sample was incubated with 15 µl Protein A Dynabeads in 25 µl of 50 mM NaH2PO4, pH 8, 0.05% NP-40, 50 mM NaCl, and 10 mg/ml BSA per sample overnight at 4°C. Beads were washed in 150 µl of the same buffer and resuspended in 50 µl. Simultaneous to antibody–bead conjugation, HIS–Topo IIα CTR (0.75 µg) and/or H3.1 or H3.3 (1.5 µg) as appropriate were incubated in a volume of 25 µl of 50 mM NaH2PO4, pH 7.5, 50 mM NaCl, 0.05% NP-40, and 10 mg/ml BSA overnight at 4°C. HIS–Topo IIα CTR/H3 complexes were incubated with antibody-conjugated beads for 2 h and 30 min at RT. Beads were washed three times in 150 µl of 50 mM NaH2PO4, pH 7.5, 50 mM NaCl, and 0.05% NP-40, then resuspended in 15 µl SDS-PAGE loading buffer and separated on 4–12% SDS-PAGE gels.
ELISA assays
96-well MaxiSorp plates (Thermo Fisher Scientific) were coated with 5 µg/ml NeutrAvidin in 200 mM carbonate/bicarbonate buffer, pH 9.4, for 48 h at 4°C. Wells were blocked with 300 µl PBS containing 5 mg/ml BSA for 45 min. Unmodified, methylated, or phosphorylated histone H3 (21-44) C-biotinylated peptides (AnaSpec) were resuspended to 100 ng/ml in PBS containing 10 mg/ml BSA and 0.05% Tween 20, and incubated (100 µl per well) for 1 h at RT. Wells were washed using 300 µl of wash buffer (PBS containing 0.05% Tween 20) three times before adding 100 µl HIS–Topo IIα CTR (1321-1530) at various concentrations in 50 mM NaH2PO4, pH 7.5, 50 mM NaCl, 10 mg/ml BSA, and 0.05% NP-40 and incubating for 1 h at RT. After washing three times with 300 µl wash buffer, bound HIS–Topo IIα CTR was detected using 1:2,500 anti-HIS antibody (Takara Bio Inc.) in PBS containing 10 mg/ml BSA and 0.05% NP-40 for 1 h. After washing again, wells were incubated for 30 min with 1:2,500 goat anti–mouse ECL (HRP-conjugated) secondary antibody (GE Healthcare). After an additional wash step, HRP signal was developed using TMB substrate (R&D Systems) according to the manufacturer’s protocol. Plates were read at 450 nm on a SpectraMax M2 plate reader (Molecular Devices). After subtraction of negative control values to account for nonspecific binding, absorbance values were fitted to a four parameter logistic curve using the drc package in the R Statistical Computing environment.
FRAP
FRAP using HeLa EM2-11ht cells was performed using a point-scanning confocal microscope (FluoView 1000 IX2; Olympus), 100×/1.3 NA UPlanFL objective lens, and FluoView acquisition software. Cells were maintained in DMEM containing Hepes buffer on a Delta-T heated dish system (Bioptechs, Inc.) at 37°C. After data collection, images were analyzed using ImageJ, and intensity values were double-normalized as per Phair et al. (2004b) and scaled to between 0 and 1.
Online supplemental material
Fig. S1 shows Topo IIα CTR binding to an expanded H3 tail peptide array. Fig. S2 shows localization of Topo IIα ΔChT in interphase. Fig. S3 shows a Western blot of HeLa cells expressing exogenous Topo IIα mutants and depleted of endogenous Topo IIα. Fig. S4 shows analysis of Topo IIα K3R mutant. Fig. S5 shows localization of Topo IIα after Aurora B inhibition in mitosis. Video 1 shows a 3D reconstruction of Topo IIα localization in M. muntjak cells. Video 2 shows Topo IIα K3R localization in a live cell entering mitosis. Video 3 shows z sections through a fixed M. muntjak cell costained to localize Topo IIα and H3S28p. Videos 4–9 show live HeLa cells depleted of Topo IIα and induced to express either Topo IIα or Topo IIα ΔChT. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201303045/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201303045.dv.
Supplementary Material
Acknowledgments
We thank Ryoko Kuriyama, Melissa Gardner, Meg Titus, Dennis Livingston, Gant Luxton, and Rebecca Heald for insightful comments on the manuscript; Hung-Ji Tsai, Jeremy Wilbur, Ina Weidenfeld, Kai Schonig, and Andrew Grenfell for input on experimental procedures; and Mark Sanders, Guillermo Marques, and the University of Minnesota Biomedical Image Processing Laboratory for assistance with the FRAP studies.
This work was supported in part by National Institutes of Health grant CA099033, National Science Foundation grant MCB-0842157 (DJC), and the Minnesota Medical Foundation (all to D.J. Clarke).
Footnotes
Abbreviations used in this paper:
- ChT
- chromatin tether
- CTR
- C-terminal region
- Topo II
- topoisomerase II
- WT
- wild type
References
- Abràmoff M.D., Magalhães P.J., Ram S.J. 2004. Image processing with ImageJ. Biophotonics International. 11:36–42 [Google Scholar]
- Adolph K.W., Cheng S.M., Paulson J.R., Laemmli U.K. 1977. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc. Natl. Acad. Sci. USA. 74:4937–4941 10.1073/pnas.74.11.4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoy F.J., Meehan R.R., McKay S., Nixon J., Bird A. 1995. Binding of histone H1 to DNA is indifferent to methylation at CpG sequences. J. Biol. Chem. 270:26473–26481 10.1074/jbc.270.44.26473 [DOI] [PubMed] [Google Scholar]
- Carpenter A.J., Porter A.C.G. 2004. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol. Biol. Cell. 15:5700–5711 10.1091/mbc.E04-08-0732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho P.A., Queiroz-Machado J., Sunkel C.E. 2003. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 116:4763–4776 10.1242/jcs.00799 [DOI] [PubMed] [Google Scholar]
- Cokol M., Nair R., Rost B. 2000. Finding nuclear localization signals. EMBO Rep. 1:411–415 10.1093/embo-reports/kvd092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 94:193–204 10.1016/S0092-8674(00)81419-1 [DOI] [PubMed] [Google Scholar]
- Cuvier O., Hirano T. 2003. A role of topoisomerase II in linking DNA replication to chromosome condensation. J. Cell Biol. 160:645–655 10.1083/jcb.200209023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M.M., Malureanu L., Jeganathan K.B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J.M. 2008. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 133:103–115 10.1016/j.cell.2008.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Halligan B., Cooke C.A., Heck M.M., Liu L.F. 1985. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 100:1706–1715 10.1083/jcb.100.5.1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltsov M., Maclellan K.M., Maeshima K., Frangakis A.S., Dubochet J. 2008. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. USA. 105:19732–19737 10.1073/pnas.0810057105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galande S., Muniyappa K. 1997. Effects of nucleosomes and anti-tumor drugs on the catalytic activity of type II DNA topoisomerase from rat testis. Biochem. Pharmacol. 53:1229–1238 10.1016/S0006-2952(96)00897-0 [DOI] [PubMed] [Google Scholar]
- Gasser S.M., Laemmli U.K. 1987. Improved methods for the isolation of individual and clustered mitotic chromosomes. Exp. Cell Res. 173:85–98 10.1016/0014-4827(87)90334-X [DOI] [PubMed] [Google Scholar]
- Giménez-Abián J.F., Clarke D.J. 2009. Cytological analysis of chromosome structural defects that result from topoisomerase II dysfunction. Methods Mol. Biol. 582:189–207 10.1007/978-1-60761-340-4_15 [DOI] [PubMed] [Google Scholar]
- Giménez-Abián J.F., Clarke D.J., Mullinger A.M., Downes C.S., Johnson R.T. 1995. A postprophase topoisomerase II-dependent chromatid core separation step in the formation of metaphase chromosomes. J. Cell Biol. 131:7–17 10.1083/jcb.131.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Yasui Y., Nigg E.A., Inagaki M. 2002. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 7:11–17 10.1046/j.1356-9597.2001.00498.x [DOI] [PubMed] [Google Scholar]
- Grue P., Grässer A., Sehested M., Jensen P.B., Uhse A., Straub T., Ness W., Boege F. 1998. Essential mitotic functions of DNA topoisomerase IIalpha are not adopted by topoisomerase IIbeta in human H69 cells. J. Biol. Chem. 273:33660–33666 10.1074/jbc.273.50.33660 [DOI] [PubMed] [Google Scholar]
- Hudson D.F., Vagnarelli P., Gassmann R., Earnshaw W.C. 2003. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 5:323–336 10.1016/S1534-5807(03)00199-0 [DOI] [PubMed] [Google Scholar]
- Jensen S., Andersen A.H., Kjeldsen E., Biersack H., Olsen E.H., Andersen T.B., Westergaard O., Jakobsen B.K. 1996. Analysis of functional domain organization in DNA topoisomerase II from humans and Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3866–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaer K., Mätlik K., Metsis M., Speek M. 2008. Combination of native and denaturing PAGE for the detection of protein binding regions in long fragments of genomic DNA. BMC Genomics. 9:272 10.1186/1471-2164-9-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Hirano T. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 90:625–634 10.1016/S0092-8674(00)80524-3 [DOI] [PubMed] [Google Scholar]
- Linka R.M., Porter A.C.G., Volkov A., Mielke C., Boege F., Christensen M.O. 2007. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 35:3810–3822 10.1093/nar/gkm102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.Z., Elledge S.J. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 4:251–256 10.1038/nmeth1010 [DOI] [PubMed] [Google Scholar]
- Liu H., Naismith J.H. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8:91 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Yuan J., Chen J., Lou Z. 2009. Topoisomerase IIalpha controls the decatenation checkpoint. Nat. Cell Biol. 11:204–210 10.1038/ncb1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K., Laemmli U.K. 2003. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell. 4:467–480 10.1016/S1534-5807(03)00092-3 [DOI] [PubMed] [Google Scholar]
- McClendon A.K., Gentry A.C., Dickey J.S., Brinch M., Bendsen S., Andersen A.H., Osheroff N. 2008. Bimodal recognition of DNA geometry by human topoisomerase II alpha: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain. Biochemistry. 47:13169–13178 10.1021/bi800453h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirski S.E., Gerlach J.H., Cummings H.J., Zirngibl R., Greer P.A., Cole S.P. 1997. Bipartite nuclear localization signals in the C terminus of human topoisomerase II alpha. Exp. Cell Res. 237:452–455 10.1006/excr.1997.3805 [DOI] [PubMed] [Google Scholar]
- Mo Y.Y., Ameiss K.A., Beck W.T. 1998. Overexpression of human DNA topoisomerase II alpha by fusion to enhanced green fluorescent protein. Biotechniques. 25:1052–1057 [DOI] [PubMed] [Google Scholar]
- Mullinger A.M., Johnson R.T. 1979. The organization of supercoiled DNA from human chromosomes. J. Cell Sci. 38:369–389 [DOI] [PubMed] [Google Scholar]
- Nishino Y., Eltsov M., Joti Y., Ito K., Takata H., Takahashi Y., Hihara S., Frangakis A.S., Imamoto N., Ishikawa T., Maeshima K. 2012. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 31:1644–1653 10.1038/emboj.2012.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J.L. 2009. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 9:338–350 10.1038/nrc2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 115:109–121 10.1016/S0092-8674(03)00724-4 [DOI] [PubMed] [Google Scholar]
- Orrego M., Ponte I., Roque A., Buschati N., Mora X., Suau P. 2007. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 5:22 10.1186/1741-7007-5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-W., Parrott A.M., Fritz D.T., Park Y., Mathews M.B., Lee C.-G. 2008. Regulation of the catalytic function of topoisomerase II alpha through association with RNA. Nucleic Acids Res. 36:6080–6090 10.1093/nar/gkn614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J.R., Laemmli U.K. 1977. The structure of histone-depleted metaphase chromosomes. Cell. 12:817–828 10.1016/0092-8674(77)90280-X [DOI] [PubMed] [Google Scholar]
- Phair R.D., Scaffidi P., Elbi C., Vecerová J., Dey A., Ozato K., Brown D.T., Hager G., Bustin M., Misteli T. 2004a. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402 10.1128/MCB.24.14.6393-6402.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R.D., Gorski S.A., Misteli T. 2004b. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375:393–414 10.1016/S0076-6879(03)75025-3 [DOI] [PubMed] [Google Scholar]
- Raymond C.S., Soriano P. 2007. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS ONE. 2:e162 10.1371/journal.pone.0000162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg A.J., Li H., Patel D.J., Allis C.D. 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8:983–994 10.1038/nrm2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A., Kikuchi A. 2004. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J. Cell Sci. 117:1047–1054 10.1242/jcs.00977 [DOI] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.-Y., Barski A., Wang Z., Wei G., Zhao K. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell. 132:887–898 10.1016/j.cell.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strukov Y.G., Sural T.H., Kuroda M.I., Sedat J.W. 2011. Evidence of activity-specific, radial organization of mitotic chromosomes in Drosophila. PLoS Biol. 9:e1000574 10.1371/journal.pbio.1000574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Sumner A.T. 1991. Scanning electron microscopy of mammalian chromosomes from prophase to telophase. Chromosoma. 100:410–418 10.1007/BF00337519 [DOI] [PubMed] [Google Scholar]
- Swedlow J.R., Hirano T. 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell. 11:557–569 10.1016/S1097-2765(03)00103-5 [DOI] [PubMed] [Google Scholar]
- Tavormina P.A., Côme M.-G., Hudson J.R., Mo Y.-Y., Beck W.T., Gorbsky G.J. 2002. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J. Cell Biol. 158:23–29 10.1083/jcb.200202053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson R.B., Lindsley J.E. 1997. Type II DNA topoisomerase from Saccharomyces cerevisiae is a stable dimer. Biochemistry. 36:6107–6114 10.1021/bi970152f [DOI] [PubMed] [Google Scholar]
- Tseng H., Biegel J.A., Brown R.S. 1999. Basonuclin is associated with the ribosomal RNA genes on human keratinocyte mitotic chromosomes. J. Cell Sci. 112:3039–3047 [DOI] [PubMed] [Google Scholar]
- Wang J.C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430–440 10.1038/nrm831 [DOI] [PubMed] [Google Scholar]
- Weidenfeld I., Gossen M., Löw R., Kentner D., Berger S., Görlich D., Bartsch D., Bujard H., Schönig K. 2009. Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res. 37:e50 10.1093/nar/gkp108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel I., Jensen P.B., Falck J., Mirski S.E., Cole S.P., Sehested M. 1997. Loss of amino acids 1490Lys-Ser-Lys1492 in the COOH-terminal region of topoisomerase IIalpha in human small cell lung cancer cells selected for resistance to etoposide results in an extranuclear enzyme localization. Cancer Res. 57:4451–4454 [PubMed] [Google Scholar]
- Wu K.K. 2006. Analysis of protein-DNA binding by streptavidin-agarose pulldown. Methods Mol. Biol. 338:281–290 [DOI] [PubMed] [Google Scholar]
- Yang Z., Champoux J.J. 2009. Assays for the preferential binding of human topoisomerase I to supercoiled DNA. Methods Mol. Biol. 582:49–57 10.1007/978-1-60761-340-4_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo K., Ogawa T., Hayashi M., Harada Y., Nishizaka T., Kinosita K., Jr 2012. Direct observation of strand passage by DNA-topoisomerase and its limited processivity. PLoS ONE. 7:e34920 10.1371/journal.pone.0034920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.