Abstract
Killer cell immunoglobulin-like receptor (KIR) genes are expressed by natural killer cells and encoded by a family of genes exhibiting considerable haplotypic and allelic variation. HLA-C molecules, the dominant ligands for KIR, are present in all individuals and are discriminated by two KIR epitopes, C1 and C2. We studied the frequencies of KIR genes and HLA-C1 and C2 groups in a large cohort (n = 492) from Kampala, Uganda, East Africa and compared our findings with published data from other populations in sub-Saharan Africa (SSA) and several European populations. We find considerably more KIR diversity and weaker linkage disequilibrium in SSA compared to the European populations and describe several novel KIR genotypes. C1 and C2 frequencies were similar to other SSA populations with a higher frequency of the C2 epitope (54.9 %) compared to Europe (average 39.7 %). Analysis of this large cohort from Uganda in the context of other African populations reveals variations in KIR and HLA-C1 and C2 that are consistent with migrations within Africa and potential selection pressures on these genes. Our results will help understand how KIR/HLA-C interactions contribute to resistance to pathogens and reproductive success.
Electronic supplementary material
The online version of this article (doi:10.1007/s00251-013-0724-7) contains supplementary material, which is available to authorized users.
Keywords: KIR, HLA-C, Uganda, Africa, Population study, NK cell
Introduction
Killer cell immunoglobulin-like receptor genes (KIR) are expressed by natural killer (NK) cells and function in infection, cancer, and reproduction. KIR bind some human leukocyte antigen (HLA) class I molecules and other unknown ligands, and a balance between activating and inhibitory signals mediated by KIR regulates NK cell functions. Both KIR and HLA genetic regions are highly variable and can be used as tools in anthropological studies (Parham and Moffett 2013) revealing the history of populations, migration, admixture, or pathogen pressure (Hedrick and Thomson 1983; Piazza et al. 1980). African populations have higher levels of genetic diversity (Tishkoff and Verrelli 2003), which has been documented by many studies using mitochondrial DNA and/or Y chromosome markers and neutral markers as well as HLA polymorphisms (Pilkington et al. 2008; Prugnolle et al. 2005; Tishkoff et al. 2009). KIR variability across the African continent is still not well documented despite the burden of infectious disease including HIV/AIDS, malaria, and tuberculosis. In addition, maternal and reproductive health disorders are a major problem in SSA (WHO databases, www.who.int), and KIR and HLA have been associated with these diseases (Chazara et al. 2011; Hiby et al. 2010; Kulkarni et al. 2008; Parham 2005).
The KIR gene family is located on chromosome 19 and contains 12 genes and 2 pseudogenes (Trowsdale et al. 2001; Wende et al. 1999) The KIR region exhibits great diversity through both the presence and absence of genes as well as allelic polymorphism of individual genes. KIR nomenclature is based on the number of extracellular immunoglobulin-like domains, two domains (2D) or three domains (3D), followed by the length of their cytoplasmic tail, short or long. HLA-C is the major ligand for KIR and a dimorphism at position 80 of the α1 domain divides all HLA-C allotypes into two groups, C1 and C2 (Colonna et al. 1993; Mandelboim et al. 1996). In general, KIR2DL2 and KIR2DL3 interact with C1 allotypes, while KIR2DL1 and KIR2DS1 bind specifically to C2 allotypes.
Soon after KIR genes were discovered, it was appreciated that they could be subdivided into two main haplotypes, “A” and “B,” distinguished by gene content (Uhrberg et al. 1997). KIR A haplotypes have a fixed gene content of seven genes encoding mostly inhibitory receptors, whereas KIR B haplotypes have a variable gene content that includes more genes encoding activating receptors. KIR2DL4, 3DL2, and 3DL3 are framework genes common to both haplotypes (Wilson et al. 2000). A recombination breakpoint between KIR2DL1 and KIR2DL4 allows a classification of KIR haplotypes in Eurasians into different combinations of centromeric A or B (cA, cB) and telomeric A or B (tA, tB) regions. These regions are characterized by the presence of certain KIR genes: cA (KIR2DL1, 2DL3), cB (KIR2DL2, 2DS2), tA (KIR3DL1, 2DS4), and tB (KIR3DS1, 2DS1). HLA and KIR genes are the most polymorphic and variable gene systems in humans with no “wild-type” genes (Parham et al. 2011). Though KIR and HLA class I genes are located on different chromosomes (and therefore segregate independently), they have been found to be coevolving, accordingly to the receptor–ligand interactions between them (Guinan et al. 2010; Single et al. 2007).
Our aim was to investigate the variability of the KIR region in Africa, as the findings will have relevance to population evolutionary history, disease, and vaccine studies for populations in Africa and those with African ancestry elsewhere in the world.
Materials and methods
Study subjects
Blood samples were obtained from 492 unrelated healthy female donors at Mulago Hospital, Kampala between 2009 and 2011. The majority of individuals (61.5 %) reported one or both parent speaking the Ganda language, classified with the Bantu languages (Table S 1). The subjects were healthy with no known chronic disease, apart from a HIV prevalence of approximately 5 %. Healthy controls from the UK were used for comparison (Hiby et al. 2010). KIR and HLA-C gene frequencies and genotypes frequencies for other African populations were obtained from the Allele Frequency Net database and previous reports (Gonzalez-Galarza et al. 2011; Hollenbach et al. 2012; Omosun et al. 2012; Single et al. 2007; Yindom et al. 2010).
Ethics statement
Informed written consent was obtained, and the study was conducted under ethical approval from the Higher Degrees Research and Ethics Committee of Makerere University College of Health Sciences and the Uganda National Council for Science and Technology.
DNA isolation and genotyping
Genomic DNA was isolated from 5 mL of blood using the QIAmp DNA Maxi Blood Kit (Qiagen). Polymerase chain reaction-sequence-specific primer (PCR-SSP) was performed to amplify the genomic DNA for presence or absence of 12 KIR genes (2DL1, 2DL2, 2DL3, 2DL5, 3DL1, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, and 3DS1) and the pseudogene 2DP1. KIR2DS4 alleles were also typed as being either full length or having the 22-bp deletion that prevents cell surface expression. Two pairs of primers were used for each gene, selected to give relatively short amplicons of 100–800 bp, as previously described in full detail (Hiby et al. 2004). HLA-C alleles were genotyped as either group C1 or C2 allotypes. Genotyping was performed by PCR-SSP using two pairs of primers for both HLA-C allotypes, as previously detailed (Hiby et al. 2008). Extra KIR primers were designed using sequence information from the IPD-KIR database (release 2.4.0) to detect rare alleles of KIR2DS5 and KIR2DL3 (KIR2DS5, 2DS5rev2: TCC AGA GGG TCA CTG GGA and KIR2DL3, 2DL3rev3: AGA CTC TTG GTC CAT TAC CG) (Robinson et al. 2011). Selected samples (based on their unusual KIR gene content) were typed for copy number by multiplexed quantitative PCR for all 16 KIR genes (KIR2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 2DP1, 3DP1, 3DL1, 3DL2, 3DL3, and 3DS1) (Jiang et al. 2012). KIR haplotypes were defined by matrix subtraction of gene copy numbers using previously characterized common and contracted KIR haplotypes using the KIR Haplotype Identifier software (www.bioinformatics.cimr.cam.ac.uk/haplotypes).
Data analysis and statistical methods
KIR gene carrier frequencies (F) and HLA-C1 and C2 group frequencies were calculated by direct counting. KIR gene frequencies were estimated by the formula , where F is the KIR gene carrier frequency in the population. Differences between the Ugandan and the UK populations were compared by χ 2. A centered unscaled principal components analysis was conducted as implemented in the adegenet R package (Jombart and Ahmed 2011). A map was generated using GMT using a modified script available online (www.pypop.org/popdata, Owen Solberg) (Wessel and Smith 1991). Pairwise linkage disequilibrium (Wn*) was calculated for the 12 KIR loci (Single et al. 2008). Statistical significance was obtained using Fisher's exact test, as implemented in PyPop (Lancaster et al. 2007). A graphical view of the linkage disequilibrium (LD) from our study data and that of Single et al. (2008) was achieved using Gold (Abecasis and Cookson 2000).
Results and discussion
KIR and HLA-C group carrier frequencies
We first compared the carrier frequencies of the 12 KIR genes in the Ugandan population with frequencies in the UK cohort (Table 1). The Ugandan population showed marked differences in individual KIR gene frequencies; in particular, KIR3DS1 and KIR2DS1, both associated with the KIR telomeric B region, were significantly less frequent. Additionally, comparatively higher frequencies of KIR2DL2, KIR2DL5, and KIR2DS5 and lower frequencies of KIR2DL3 and KIR2DS3 were seen (KIR2DS3 and KIR2DS5 are alleles of the same gene). Increased KIR2DL2 and decreased KIR2DL3 frequency suggests that the centromeric B region is abundant in the Ugandan population. KIR2DS5 can be found on either the centromeric or telomeric B regions in Africans, similarly to KIR2DL5 in both European and African populations, making precise interpretation of centromeric B frequencies difficult (Hou et al. 2010). KIR gene frequencies in the Uganda population are similar to those reported for the South African Xhosa and San population (Middleton et al. 2007) indicating that African populations have lower frequencies of the telomeric B region and higher frequencies of the centromeric B region compared with European populations.
Table 1.
KIR carrier and estimated gene frequencies in the Ugandan and UK populations
| 2DS2 | 2DL2 | 2DL3 | 2DP1 | 2DL1 | 3DL1 | 3DS1 | 2DL5 | 2DS5 | 2DS3 | 2DS1 | 2DS4 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uganda | Carrier frequency | 0.54 | 0.61 | 0.85 | 0.98 | 0.99 | 0.98 | 0.12 | 0.65 | 0.50 | 0.24 | 0.24 | 0.96 |
| Estimated gene frequency | 0.32 | 0.37 | 0.62 | 0.87 | 0.88 | 0.86 | 0.06 | 0.41 | 0.29 | 0.13 | 0.13 | 0.79 | |
| UK | Carrier frequency | 0.52 | 0.51 | 0.91 | 0.96 | 0.96 | 0.96 | 0.44 | 0.54 | 0.36 | 0.28 | 0.42 | 0.96 |
| Estimated gene frequency | 0.30 | 0.30 | 0.70 | 0.79 | 0.80 | 0.79 | 0.25 | 0.32 | 0.20 | 0.15 | 0.24 | 0.79 | |
| Chi-square p value | 0.00437 | 0.01177 | <1E-07 | 0.00055 | 1.2E-05 | <1E-07 | |||||||
We compared these results with data from a range of African populations where frequencies for KIR2DL1, KIR2DL2, KIR2DL3, KIR3DL1, and KIR3DS1 have been reported (Table 2). Of note is that the sample size was under 100 in 19 out of 22 studies. All African populations share similar characteristics compared to UK populations with lower KIR2DS1 and KIR3DS1 carrier frequencies; the highest value for KIR2DS1 was 0.28 (Congo Mbuti) and 0.23 for KIR3DS1 (Ethiopian) compared with, respectively, 0.42 and 0.44 for the UK population. The variability of these frequencies between the African populations may be visualized using principal component analysis (Fig. 1). The Ugandan population localizes to an intermediate position, between Central and West African populations and the East African populations. Therefore, in terms of KIR gene frequencies, the Ugandan population can be regarded as intermediate to these populations. The history of human migration in Africa is consistent with this observation because Uganda is an East African country known to have been populated from the Bantu migrations from West and Central Africa (Campbell and Tishkoff 2008). Furthermore, it is in both the northern and southern hemispheres, being situated on the equatorial line.
Table 2.
KIR carrier frequencies in the Ugandan and 22 African populations
| Population | 2DL1 | 2DL2 | 2DL3 | 2DL4 | 2DL5 | 2DP1 | 2DS1 | 2DS2 | 2DS3 | 2DS4 | 2DS5 | 3DL1 | 3DL2 | 3DL3 | 3DP1 | 3DS1 | Location | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele Frequency Net | Uganda Kampala (n = 492) | 0.99 | 0.61 | 0.85 | 0.65 | 0.98 | 0.24 | 0.54 | 0.24 | 0.96 | 0.50 | 0.98 | 0.12 | 0°18′ N 32°34′ E | ||||
| Equatorial Guinea Bioko Island Bubi (n = 95) | 0.96 | 0.57 | 0.80 | 0.99 | 0.52 | 0.97 | 0.20 | 0.48 | 0.26 | 0.97 | 0.34 | 0.99 | 1.00 | 1.00 | 0.99 | 0.05 | 1°55′ N 10°6′ E | |
| Gabon (n = 54) | 1.00 | 0.65 | 0.80 | 1.00 | 0.70 | 1.00 | 0.19 | 0.57 | 0.35 | 1.00 | 0.33 | 1.00 | 1.00 | 1.00 | 1.00 | 0.07 | 1°38′ S 13°35′ E | |
| Ghana (n = 41) | 0.93 | 0.54 | 0.85 | 1.00 | 0.56 | 1.00 | 0.05 | 0.56 | 0.34 | 0.98 | 0.22 | 1.00 | 1.00 | 1.00 | 1.00 | 0.05 | 5°33′ N 0°15′ W | |
| Ivory Coast Abidjan (n = 25) | 0.80 | 0.64 | 0.80 | 0.56 | 0.16 | 0.60 | 0.40 | 1.00 | 0.28 | 1.00 | 1.00 | 0.16 | 5°20′ N 4°1′ W | |||||
| Senegal (n = 118) | 1.00 | 0.55 | 0.90 | 1.00 | 0.52 | 1.00 | 0.13 | 0.42 | 0.24 | 1.00 | 0.30 | 0.99 | 1.00 | 1.00 | 1.00 | 0.04 | 14°0′ N 16°0′ W | |
| South Africa San (n = 91) | 0.96 | 0.74 | 0.60 | 1.00 | 0.68 | 0.92 | 0.14 | 0.73 | 0.25 | 1.00 | 0.64 | 0.99 | 1.00 | 1.00 | 1.00 | 0.02 | 34°21′ S 18°28′ E | |
| South Africa Xhosa (n = 50) | 0.96 | 0.72 | 0.64 | 1.00 | 0.82 | 0.98 | 0.10 | 0.64 | 0.38 | 1.00 | 0.62 | 1.00 | 1.00 | 1.00 | 1.00 | 0.04 | 34°0′ S 19°0′ E | |
| Single et al. (2007) | Central African Republic Bagandu Biaka (n = 69) | 0.97 | 0.58 | 0.80 | 0.01 | 0.57 | 1.00 | 0.03 | 5°0′ N 15°0′ E | |||||||||
| Congo Kinshasa Ituri Forest Mbuti (n = 38) | 0.94 | 0.66 | 0.62 | 0.28 | 0.66 | 0.97 | 0.11 | 3°0′ N 26°0′ E | ||||||||||
| Ethiopian (n = 31) | 0.90 | 0.81 | 0.77 | 0.26 | 0.77 | 0.97 | 0.23 | 9°1′ N 38°44′ E | ||||||||||
| Nigeria Benin Yoruba (n = 75) | 1.00 | 0.38 | 0.92 | 0.15 | 0.35 | 0.99 | 0.12 | 10°0′ N 2°0′ E | ||||||||||
| Nigeria Enugu Ibo (n = 48) | 0.96 | 0.61 | 0.76 | 0.08 | 0.58 | 1.00 | 0.06 | 7°0′ N 5°0′ E | ||||||||||
| Nigeria Zaria Hausa (n = 37) | 1.00 | 0.37 | 0.97 | 0.14 | 0.32 | 1.00 | 0.03 | 11°4′ N 7°42′ E | ||||||||||
| Hollenbach et al. (2012) | Biaka Pygmies (n = 29) | 1.00 | 0.62 | 0.90 | 1.00 | 0.76 | 1.00 | 0.03 | 0.59 | 0.17 | 1.00 | 0.66 | 1.00 | 1.00 | 1.00 | 1.00 | 0.07 | 30–31°66–67′ E |
| Mandenka (n = 23) | 1.00 | 0.70 | 0.91 | 1.00 | 0.57 | 1.00 | 0.22 | 0.52 | 0.39 | 0.96 | 0.30 | 1.00 | 1.00 | 1.00 | 1.00 | 0.17 | 12°N 12°W | |
| Yoruba (n = 23) | 1.00 | 0.35 | 0.87 | 1.00 | 0.30 | 1.00 | 0.17 | 0.30 | 0.17 | 0.96 | 0.17 | 0.96 | 1.00 | 1.00 | 1.00 | 0.13 | 6–10°N 2–8°E | |
| Yindom et al. (2012) | Gambia Mandingo (n = 199) | 1.00 | 0.77 | 0.90 | 1.00 | 0.60 | 1.00 | 0.21 | 0.60 | 0.43 | 1.00 | 0.28 | 1.00 | 1.00 | 1.00 | 0.11 | 13°28′ N 16°36′ W | |
| Gambia Wollof (n = 88) | 1.00 | 0.69 | 0.91 | 1.00 | 0.53 | 0.99 | 0.24 | 0.57 | 0.42 | 1.00 | 0.25 | 1.00 | 1.00 | 1.00 | 0.06 | 13°28′ N 16°36′ W | ||
| Gambia Fula (n = 137) | 1.00 | 0.74 | 0.88 | 1.00 | 0.59 | 1.00 | 0.21 | 0.58 | 0.45 | 1.00 | 0.33 | 1.00 | 1.00 | 1.00 | 0.07 | 13°28′ N 16°36′ W | ||
| Gambia Jola (n = 89) | 1.00 | 0.79 | 0.83 | 1.00 | 0.56 | 0.99 | 0.25 | 0.64 | 0.49 | 1.00 | 0.26 | 1.00 | 1.00 | 1.00 | 0.08 | 13°28′ N 16°36′ W | ||
| Gambia Serere (n = 29) | 1.00 | 0.59 | 0.90 | 1.00 | 0.55 | 0.97 | 0.24 | 0.52 | 0.31 | 1.00 | 0.35 | 1.00 | 1.00 | 1.00 | 0.14 | 13°28′ N 16°36′ W | ||
| Omosun et al. (2012) | Western Kenya (predominantly Luo) (n = 93) | 0.94 | 0.57 | 0.84 | 0.99 | 0.61 | 0.20 | 0.50 | 0.25 | 0.91 | 0.40 | 0.96 | 0.11 | 1°16′ S 36°48′ E |
Fig. 1.
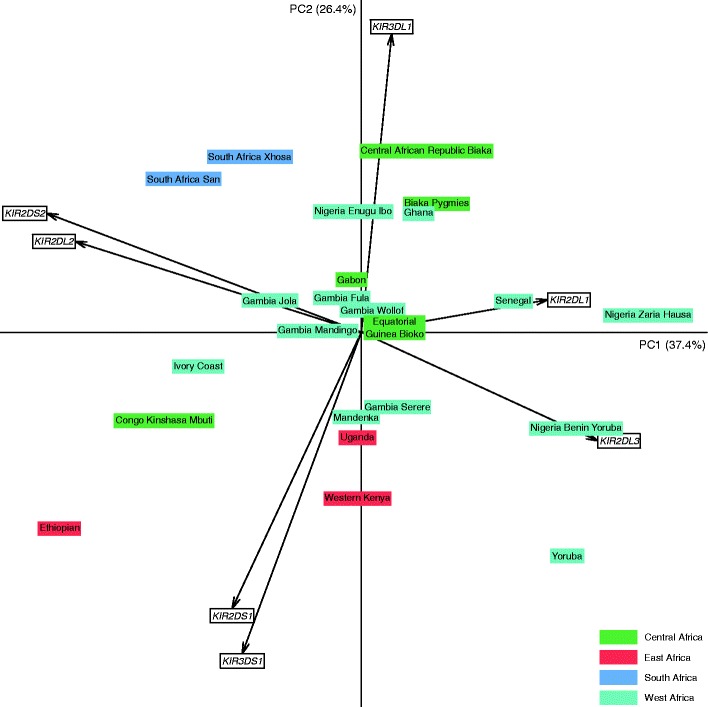
Principal component analysis based on seven KIR gene carrier frequencies for the 23 African populations described in Table 2. Arrows represent the relative contribution of each KIR gene frequency to the variability along the first two axes (PC1 and PC2)
Previous reports have revealed extensive allelic diversity at individual KIR loci in African populations. To ensure all variants were detected, we used additional new primers that were added when positive results were obtained with only one pair of primers out of the two classically used or when the individual KIR gene content was unusual. Allele “dropout” was most commonly suspected for KIR2DL3, KIR2DS1, and KIR2DS5. This could be seen where the allele occurred in the hemizygous or homozygous state and was detected in 2.4 % of the individuals. For KIR2DS5, this was usually observed on a contracted haplotype. Without using additional primers, published results for other African populations may not have detected these unusual alleles.
Analysis of the KIR2DL1 and KIR2DS1 ligand, HLA-C alleles encoding the C2 allotype, reveals that in general C2 are more frequent than C1 alleles in Africa compared with elsewhere in the world (Fig. 2). The Ugandan population, with a C2 frequency of 54.9 % and a C1 frequency of 45.1 % is similar to the neighboring populations such as Kenyan Luo (C2 = 59 % and C1 = 41 %) or Rwandan (C2 = 55 % and C1 = 45 %) (Cao et al. 2004; Tang et al. 2000). The distribution of the C2 group frequencies shows limited variation across the continent (Fig. 2). Therefore, in contrast to its activating receptor, KIR2DS1, African populations have high frequencies of HLA-C2, with comparatively little variation, in accordance with previous reports (Single et al. 2007). There must be strong selective pressures within Africa, either from pathogens or reproductive success that are maintaining this high level of C2.
Fig. 2.
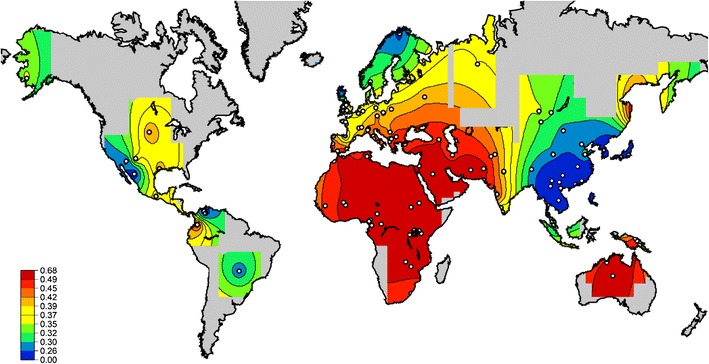
Worldwide HLA-C2 allele frequencies for 208 populations from the Allele Frequency Net database and the Ugandan population. Only non-migrant populations are included
KIR genotypes
Genotyping of the entire cohort for the presence/absence of the 12 variable KIR genes allowed the definition of individual KIR genotypes. Fifty-two KIR gene content profiles were observed in the Ugandan population (Fig. 3). Nineteen genotypes had a frequency above 1 %, accounting for 90.0 % of individuals. Half of the individuals were classified as Ganda (both parents speaking the Ganda language, 50.9 %), but the other groups were broadly similar in KIR genotype frequencies (Table S 1). As in other African populations, the most frequent KIR genotype in the Ugandan population is homozygosity for the KIR A haplotype (28.1 %). The reported frequencies for this KIR AA genotype in Africa are variable, from 12 % in a South Africa Xhosa population to 42 % in Senegal (Williams et al. 2004; Yindom et al. 2010).
Fig. 3.

KIR genotype profiles and frequencies observed in the Ugandan population. Reference numbers are according to the Allele Frequency Net database. New genotypes identified in this study are described as “novel.” Genes are presented in the order observed on sequenced KIR haplotypes except for KIR2DS3S5 and KIR2DL5. Inhibitory genes are in red, activating genes are in blue, and the pseudo gene KIR2DP1 is in gray
The remaining 33 genotypes are rare (<1 %), comprising 9.9 % of the total population. These represent unusual genotypes, including contracted or extended KIR haplotypes containing an unusual combination of KIR genes. In order to investigate these haplotypes, which arise from ancestral unequal recombination events, 15 individuals were fully typed for KIR gene copy number. Firstly, different regions of KIR3DL1 (exons 4 and 9) and of KIR3DL2 (exons 4 and 9) were separately typed for copy number to detect the KIR3DL1/3DL2 hybrid gene (Shilling et al. 2002). Nine out of 15 individuals had one or two copies of this telomeric region (tA01-hybd1; individuals 1–6 and 13–15, Fig. 4). Two additional genotypes (numbers 438 and 252/72/83, Fig. 3) have KIR3DL1 present without KIR2DS4, and these individuals are also likely to have the same contracted telomeric region (tA01-hybd1). This contracted haplotype was observed in this Ugandan cohort at an underestimated frequency of 1.7 %, which is lower than the 6.5 % from other African populations (Norman et al. 2009; Norman et al. 2007), because the common telomeric A region (tA01) will hide this deletion, as seen with individual 6. Secondly, nine different individuals (individuals 7–15) have one or two copies of other contracted haplotypes defined by a deletion of the central framework genes, KIR3DP1, KIR2DL4, and KIR3DS1, defined as del6 (Pyo et al. 2010) and previously reported in individuals of African ancestry (Gómez-Lozano et al. 2003; Norman et al. 2002). Using these individuals and the similar KIR genotypes with KIR2DS1 present without KIR3DS1, the frequency of this deletion in Uganda is greater than 7.25 %.
Fig. 4.
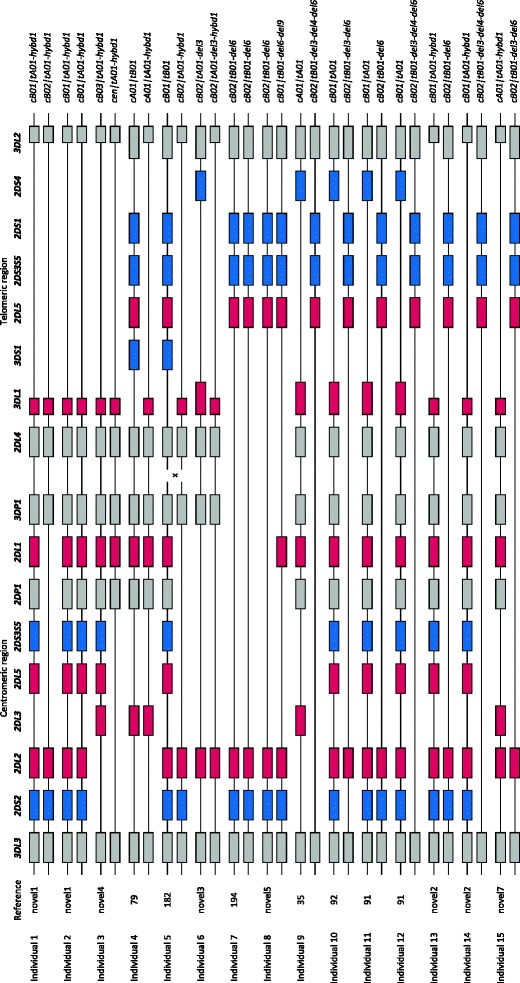
Inferred KIR haplotypes for 15 individuals representing 13 KIR genotypes. Genes are presented in the order observed on published sequenced KIR haplotypes. Inhibitory genes are in red, activating genes are in blue, and pseudo genes are in gray. Haplotype names are according to Pyo et al. (2013)
More KIR profiles were found in the Ugandan cohort (52 genotypes) when compared to a similar sized cohort of women from the UK (40 genotypes) (Fig. 5). The two populations only shared 25 genotypes present in 72.6 % of Ugandans and 88.3 % of UK. Similarly to the six other African populations (from the Allele Frequency Net database: Gabon, Ghana, Senegal, South Africa San, and South Africa Xhosa), the Ugandan population displays a higher diversity at the genotype level, and more than a quarter of these genotypes are not observed in Europeans. Forty-eight genotypes are unique to the African populations (including Uganda) and therefore might be African genotypes that have not spread out of Africa to Europe.
Fig. 5.
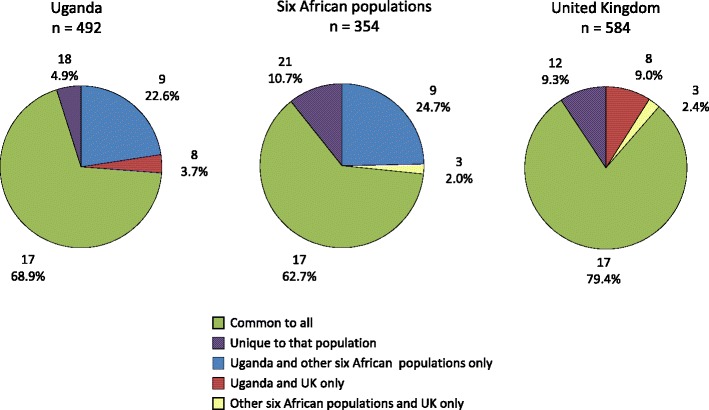
Genotypes observed for the Ugandan population (n = 492), six other African populations from the Allele Frequency Net database (Gabon, Ghana, Senegal, South Africa San, and South Africa Xhosa, n = 354), and the UK population (n = 584), showing proportions of common and unique genotypes for each group
Linkage disequilibrium
The LD between KIR genes was calculated for the 12 KIR loci in order to observe the pairwise association of genes in this population (Table 3). The strongest positive and negative LD values were observed between KIR2DL1 and KIR2DP1 (0.88) and KIR2DL2 and KIR2DL3 (−0.35), consistent with a previous report for six African populations (Single et al. 2008). A graphical analysis of Ugandan compared to these African populations and seven European populations is shown (Fig. 6). The positive LD between KIR2DS1, KIR2DS5, and KIR3DS1 observed in Europeans is absent in Africans; in contrast, a positive association is seen between KIR2DS5 and KIR2DS2-KIR2DL2, consistent with the location of KIR2DS5 gene in the centromeric B region where KIR2DS3 is generally located in Europeans. Overall, the LD values observed for the KIR genes are less extreme in Africa.
Table 3.
Linkage disequilibrium observed in the KIR region in the Ugandan population
| 2DS2 | 2DL2 | 2DL3 | 2DL5 | 2DS3 | 2DS5 | 2DP1 | 2DL1 | 3DL1 | 3DS1 | 2DS1 | 2DS4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wn* | ||||||||||||
| 2DS2 | 0.867 | −0.347 | 0.569 | 0.397 | 0.355 | −0.095 | −0.076 | −0.104 | 0.033 | 0.224 | −0.114 | |
| 2DL2 | −0.334 | 0.669 | 0.349 | 0.486 | −0.110 | −0.097 | −0.087 | 0.011 | −0.213 | −0.109 | ||
| 2DL3 | −0.278 | −0.223 | −0.320 | 0.330 | 0.290 | 0.063 | 0.116 | −0.082 | 0.140 | |||
| 2DL5 | 0.412 | 0.733 | −0.036 | −0.016 | −0.105 | 0.254 | 0.405 | −0.154 | ||||
| 2DS3 | −0.130 | 0.077 | 0.068 | 0.081 | 0.044 | −0.090 | 0.119 | |||||
| 2DS5 | −0.075 | −0.051 | −0.143 | 0.288 | 0.534 | −0.210 | ||||||
| 2DP1 | 0.880 | 0.303 | 0.050 | −0.139 | 0.196 | |||||||
| 2DL1 | 0.226 | 0.044 | −0.135 | 0.144 | ||||||||
| 3DL1 | −0.260 | −0.259 | 0.682 | |||||||||
| 3DS1 | 0.495 | −0.203 | ||||||||||
| 2DS1 | −0.262 | |||||||||||
| 2DS4 | ||||||||||||
| p value | ||||||||||||
| 2DS2 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | NS | NS | 0.005 | NS | <0.0001 | NS | |
| 2DL2 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.003 | <0.0001 | NS | NS | <0.0001 | 0.006 | ||
| 2DL3 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | NS | 0.001 | NS | 0.002 | |||
| 2DL5 | <0.0001 | <0.0001 | NS | NS | 0.004 | <0.0001 | <0.0001 | <0.0001 | ||||
| 2DS3 | 0.001 | NS | NS | NS | NS | NS | <0.0001 | |||||
| 2DS5 | NS | NS | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||||||
| 2DP1 | <0.0001 | <0.0001 | NS | <0.0001 | 0.001 | |||||||
| 2DL1 | 0.005 | NS | <0.0001 | 0.001 | ||||||||
| 3DL1 | <0.0001 | <0.0001 | <0.0001 | |||||||||
| 3DS1 | <0.0001 | <0.0001 | ||||||||||
| 2DS1 | <0.0001 | |||||||||||
| 2DS4 | ||||||||||||
Fig. 6.
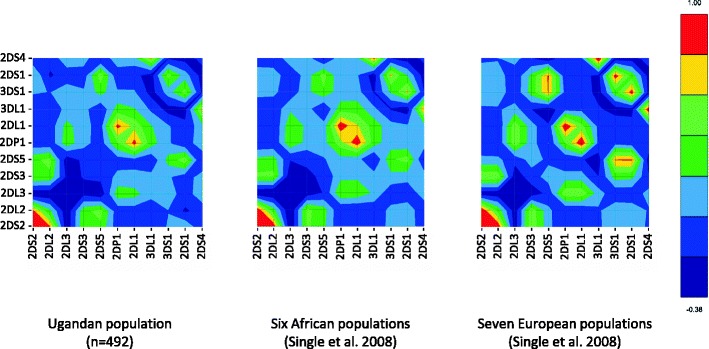
Comparison of the linkage disequilibrium observed in the KIR region between the Ugandan population and six African and seven European populations. The LD values calculated for 11 KIR gene loci for Ugandan population are compared with the values obtained using the same method by Single et al. (2008). Red represents strong positive LD, blue represents strong negative LD
Conclusions
This Ugandan cohort is the largest African population typed for KIR genes, and when considered with other published data, the remarkable extent of KIR diversity in Africa is clear. The KIR and HLA-C frequencies observed in this population are consistent with other studies of African populations. A number of novel KIR gene profiles are described, including new genotypes and rare patterns. This East African population will be an important reference group for future disease and anthropological studies in African or African-derived populations. Our study has not considered the extensive allelic variation seen in Africans, for example, for KIR3DL1 or KIR2DL1, where the great KIR diversity will become even more apparent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 14 kb)
Acknowledgments
We thank all the clinical and laboratory staff who acquired the DNA samples: Margaret Sewagaba, Florence Mugema, Dorothy Mugabi, Anastanzia Karungi, and Prossy Namukwaya. We are grateful to all study participants for voluntary consent. We also thank Paul Norman for his helpful comments on the draft manuscript and Jill Hollenbach for her help with the HLA-C frequency map. This work was funded by the Wellcome Trust (090108/Z/09/Z, 085992/Z/08/Z, and a vacation scholarship in 2011), the British Heart Foundation (PG/09/077/27964), the Center for Trophoblast Research at the University of Cambridge, and a Wellcome Trust Uganda PhD Fellowship in Infection and Immunity held by Annettee Nakimuli, funded by a Wellcome Trust Strategic Award, grant number 084344.
Conflict of interest
The authors have declared that no competing interests exist.
Footnotes
Annettee Nakimuli and Olympe Chazara contributed equally to this work.
References
- Abecasis GR, Cookson WO. GOLD—graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Moormann AM, Lyke KE, et al. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens. 2004;63:293–325. doi: 10.1111/j.0001-2815.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- Chazara O, Xiong S, Moffett A. Maternal KIR and fetal HLA-C: a fine balance. J Leukoc Biol. 2011;90:703–716. doi: 10.1189/jlb.0511227. [DOI] [PubMed] [Google Scholar]
- Colonna M, Borsellino G, Falco M, et al. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Lozano N, De Pablo R, Puente S, Vilches C. Recognition of HLA-G by the NK cell receptor KIR2DL4 is not essential for human reproduction. Eur J Immunol. 2003;33:639–644. doi: 10.1002/eji.200323741. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:913–919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinan KJ, Cunningham RT, Meenagh A, et al. Signatures of natural selection and coevolution between killer cell immunoglobulin-like receptors (KIR) and HLA class I genes. Genes Immun. 2010;11:467–478. doi: 10.1038/gene.2010.9. [DOI] [PubMed] [Google Scholar]
- Hedrick PW, Thomson G. Evidence for balancing selection at HLA. Genetics. 1983;104:449–456. doi: 10.1093/genetics/104.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiby S, Apps R, Sharkey A, et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2 -J Clin Invest. 2010;120:4102. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiby SE, Regan L, Lo W, et al. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Human Reprod. 2008;23:972–976. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- Hiby SE, Walker JJ, O'shaughnessy KM, et al. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J of Exp Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach JA, Nocedal I, Ladner MB, et al. Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations. Immunogenetics. 2012;64:719–737. doi: 10.1007/s00251-012-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Chen M, Jiang B, et al. Thirty allele-level haplotypes centered around KIR2DL5 define the diversity in an African–American population. Immunogenetics. 2010;62:491–498. doi: 10.1007/s00251-010-0458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Johnson C, Jayaraman J, et al. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012;22:1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics (Oxford, England) 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Single RM, Solberg OD, et al. PyPop update—a software pipeline for large-scale multilocus population genomics. Tissue Antigens. 2007;69(Suppl 1):192–197. doi: 10.1111/j.1399-0039.2006.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelboim O, Reyburn HT, Valés-Gómez M, et al. Protection from lysis by natural killer cells of groups 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D, Meenagh A, Moscoso J. Killer immunoglobulin receptor gene and allele frequencies in Caucasoid. Oriental and Black Populations from Differ Continents. 2007 doi: 10.1111/j.1399-0039.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- Norman PJ, Abi-Rached L, Gendzekhadze K, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman PJ, Abi-Rached L, Gendzekhadze K, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- Norman PJ, Carrington CV, Byng M, et al. Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun. 2002;3:86–95. doi: 10.1038/sj.gene.6363836. [DOI] [PubMed] [Google Scholar]
- Omosun YO, Blackstock AJ, Gatei W, et al. Differential association of gene content polymorphisms of killer cell immunoglobulin-like receptors with placental malaria in HIV− and HIV+ mothers. PLoS One. 2012 doi: 10.1371/journal.pone.0038617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P. Immunogenetics of killer cell immunoglobulin-like receptors. Mol Immunol. 2005;42:459–462. doi: 10.1016/j.molimm.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction, and human evolution. Nat Rev Immunol. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Variable NK cell receptors exemplified by human KIR3DL1/S1. J of Immunol (Baltimore, Md: 1950) 2011;187:11–19. doi: 10.4049/jimmunol.0902332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Menozzi P, Cavalli-Sforza LL. The HLA-A, B gene frequencies in the world: migration or selection? Hum Immunol. 1980;1:297–304. doi: 10.1016/0198-8859(80)90105-6. [DOI] [PubMed] [Google Scholar]
- Pilkington MM, Wilder JA, Mendez FL, et al. Contrasting signatures of population growth for mitochondrial DNA and Y chromosomes among human populations in Africa. Mol Biol Evol. 2008;25:517–525. doi: 10.1093/molbev/msm279. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Manica A, Charpentier M, et al. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 2005;15:1022–1027. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- Pyo C-W, Guethlein LA, Vu Q, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo C-W, Wang R, Vu Q, et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC genomics. 2013;14:89. doi: 10.1186/1471-2164-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Mistry K, McWilliam H, et al. The IMGT/HLA database. Nucleic Acids Res. 2011;39:1171–1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilling HG, Guethlein LA, Cheng NW, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- Single RM, Martin MP, Gao X, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- Single RM, Martin MP, Meyer D, et al. Methods for assessing gene content diversity of KIR with examples from a global set of populations. Immunogenetics. 2008;60:711–725. doi: 10.1007/s00251-008-0331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Naik E, Costello C, et al. Characteristics of HLA class I and class II polymorphisms in Rwandan women. Exp Clin Immunogenet. 2000;17:185–198. doi: 10.1159/000019138. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African–Americans. Sci (New York, NY) 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Barten R, Haude A, et al. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065X.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/S1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mammalian Genome. 1999;10:154. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- Wessel P, Smith WHF. Free software helps map and display data. Eos, Trans Am Geophys Union. 1991;72:441–446. doi: 10.1029/90EO00319. [DOI] [Google Scholar]
- Williams F, DuToit ED, Middleton D. KIR allele frequencies in a Xhosa population from South Africa. Hum Immunol. 2004;65:1084–1085. doi: 10.1016/j.humimm.2004.08.120. [DOI] [Google Scholar]
- Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yindom LM, Forbes R, Aka P et al (2012) Killer-cell immunoglobulin-like receptors and malaria caused by Plasmodium falciparum in The Gambia. Tissue Antigens 79:104–113. doi:10.1111/j.1399-0039.2011.01818.x [DOI] [PMC free article] [PubMed]
- Yindom L-M, Leligdowicz A, Martin MP, et al. Influence of HLA class I and HLA-KIR compound genotypes on HIV-2 infection and markers of disease progression in a Manjako community in West Africa. J Virol. 2010;84:8202–8208. doi: 10.1128/JVI.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 14 kb)


