Abstract
The accuracy of an ultrasound (US) computer-aided diagnosis (CAD) system was evaluated for the classification of BI-RADS category 3, probably benign masses. The US database used in this study contained 69 breast masses (21 malignant and 48 benign masses) that at blinded retrospective interpretation were assigned to BI-RADS category 3 by at least one of five radiologists. For computer-aided analysis, multiple morphology (shape, orientation, margin, lesions boundary, and posterior acoustic features) and texture (echo patterns) features based on BI-RADS lexicon were implemented, and the binary logistic regression model was used for classification. The receiver operating characteristic curve analysis was used for statistical analysis. The area under the curve (Az) of morphology, texture, and combined features were 0.90, 0.75, and 0.95, respectively. The combined features achieved the best performance and were significantly better than using texture features only (0.95 vs. 0.75, p value = 0.0163). The cut-off point at the sensitivity of 86 % (18/21), 95 % (20/21), and 100 % (21/21) achieved the specificity of 90 % (43/48), 73 % (35/48), and 33 % (16/48), respectively. In conclusion, the proposed CAD system has the potential to be used in upgrading malignant masses misclassified as BI-RADS category 3 on US by the radiologists.
Keywords: Breast cancer, BI-RADS, Ultrasound, Computer-assisted diagnosis
Introduction
As an adjunct to mammography, ultrasound (US) is widely used in breast imaging. Stavros et al. [1] proposed several US characteristics to describe a mass and achieved a sensitivity of 98.4 % (123 of 125) and a specificity of 67.8 % (424 of 625) for the classification of 750 solid breast masses. In 2003, Breast Imaging Reporting and Data System (BI-RADS) lexicon was developed to provide the descriptors for tumor characterization on breast US [2]. The BI-RADS lexicon defined for US findings in masses includes shape, orientation, margin, lesion boundary, echo pattern, and posterior acoustic features. According to the BI-RADS, the management strategy of breast masses classified as BI-RADS category 3 is follow up rather than core needle or surgical biopsy since the likelihood of malignancy is very low (less than 2 %) [3–5]. The results of recent studies support this strategy [6–9]. Circumscribed margin and parallel orientation of the mass has been emphasized as benign US findings like fibroadenomas. However, to avoid misclassifying carcinomas into the category 3, the radiologists should thoroughly evaluate lesions before placing them in this category [10, 11].
Various computer-aided diagnosis (CAD) systems were proposed to quantify the characteristics of benign and malignant breast tumors [12–19]. In several studies, the quantitative features used in the CAD systems were combined to classify tumors according to BI-RADS assessment categories [14]. The performance similar to expert radiologists was achieved with the CAD system. Consequently, the CAD system was suggested as a second reader for tumor assessment and used to decrease false positive rates at US [16]. Previous US CAD studies, however, used image database including wide spectrum of breast masses belonged to typically benign lesion (category 2 by BI-RADS) to lesions with suspicious (category 4) or typical malignant (category 5) findings. With increasing use of breast US in screening women with dense breasts, breast lesions classified as category 3 are found more often [6, 8]. Therefore, CAD system can play a role in classifying benign and malignant among category 3 lesions.
In this study, the benign and malignant breast masses assessed as BI-RADS category 3 at US by radiologists were evaluated using the proposed CAD system. To provide probability of malignancy in breast masses, various quantitative features connected to the BI-RADS lexicon were implemented in the proposed CAD system. This system is likely going to be used while a radiologist detects a lesion with uncertainty about whether the lesion is benign or malignant. In this situation, the proposed CAD can provide a quantitative suggestion.
Materials and Methods
Patients and Data Acquisition
The approval for this retrospective study was obtained from our institution review board, and informed consent was waived. During a 2-year period, US images of 100 consecutive solid tumors were obtained before needle biopsy or surgery. These cases were initially assessed as BI-RADS category 3 in 32, category 4 in 56, and category 5 in 12 cases. The acquired breast US images were collected using a Voluson 530 scanner (Kretz Technik, Zipf, Austria) or an ATL HDI 3000 scanner (Philips, Bothell, WA, USA) and a linear transducer with a frequency of 5–10 MHz. For each lesion, blinded retrospective interpretation was performed by five radiologists to provide BI-RADS category. There were 69 masses (21 malignant and 48 benign masses) that were assigned to BI-RADS category 3 by at least one of five radiologists included in this study. The age of the patients ranged 20–84 years (mean age, 43). All 21 malignant lesions were invasive ductal carcinoma and were histologically grade 2 (n = 6) or grade 3 (n = 15). For benign lesions, 34 cases of fibroadenoma, 13 cases of fibrocystic changes, and one case of papilloma were included. The size of malignant masses had a mean value of 2.7 cm (range, 1.2–4.7 cm), and the size of benign masses had a mean value of 2.6 cm (range 1.4–4.3 cm). Lesions were palpable in 15 cases and nonpalpable in 54 cases.
Feature Extraction
In the proposed CAD system, a tumor has to be segmented first for feature extraction. To segment a tumor from background tissues, level-set segmentation method was used [20]. Using gradient as the criterion in differential equation, level-set function evolves the user-defined seed into a complex shape to get the approach for tumor contour. After tumor segmentation, the contour of a tumor (Fig. 1a) was delineated to specify tumor area as show in Fig. 1b. With the specification of tumor area, two-dimensional (2D) spatial information was employed for image interpretation. According to the suggested six descriptive categories in BI-RADS lexicon including shape, orientation, margin, lesion boundary, echo pattern, and posterior acoustic features, the corresponding measurements were quantified in the CAD systems for classifying breast masses. They were listed in Table 1. In each descriptive category, there were one or more quantitative features.
Fig. 1.
The illustration of the quantitative features for describing BI-RADS US lexicon. a The original US image of a BI-RADS category 3 mass confirmed as benign fibroadenoma at core needle biopsy. b The segmentation result of (a). c Tumor perimeter (Tumor_p) was compared to the length of the major axis of the best-fit ellipse (Ellipse_a) to quantify the shape features. d The angle between the major axis of the best-fit ellipse and the horizontal line (Ellipse_theta) was used to quantify the orientation feature. e The undulations on tumor boundary (MU, MNS) were used to quantify the margin features. f The gray-level intensity difference between the inner and outer bands around the tumor boundary (LB) was used to quantify the boundary feature. g The gray-level intensity difference between the region under the tumor and the tumor (PS) was used to quantify the posterior acoustic feature. h The spatial correlations between pixels inside a tumor (16 GLCM texture features) were used to quantify the echo pattern features
Table 1.
Quantitative BI-RADS features
| Category | Feature | Description |
|---|---|---|
| Shape | Tumor_a, Tumor_p | Tumor area (Tumor_a) and tumor perimeter (Tumor_p) [14] |
| Ellipse_a, Ellipse_b, Ellipse_a/b | The length of the major axis (a) and minor axis (b) of the best-fit ellipse [14] | |
| Ep/Tp | The ratio of the ellipse perimeter (Ep) and the tumor perimeter (Tp) [14] | |
| Ellipse_compactness | The overlap between the tumor area and the ellipse area [14] | |
| NRL entropy, NRL variance | The statistics of the distances between boundary points and tumor center | |
| Compactness | Tumor roundness [22] | |
| Orientation | Ellipse_theta | The angle between the major axis of the best-fit ellipse and the horizontal line [14] |
| Margin | Undulation, Sharp, MU | The number of undulations on tumor boundary [13] |
| NS | The number of spicules on tumor boundary | |
| MNS | NS × Compactness | |
| MaxSpicule | Length of the longest spicule of NS | |
| Lesion boundary | LB | Intensity difference around tumor boundary [13] |
| Echo pattern | EPc | Intensity difference between the 25 % brighter pixels and whole tumor pixels [13] |
| EP_diff | Intensity difference between the tumor and the surrounding tissues | |
| Energy avg., Energy std., Entropy avg., Entropy std., Correlation avg., Correlation std., Inverse Difference Moment avg., Inverse Difference Moment std., Inertia avg., Inertia std., Cluster Shade avg., Cluster Shade std., Cluster Prominence avg., Cluster Prominence std., Haralick Correlation avg., Haralick Correlation std. | 16 GLCM texture features [23], the statistics of the correlations between neighbor pixels | |
| Posterior acoustic features | PS | The average intensity difference between the tumor and the region under the tumor [13] |
| PS_diff | The average intensity difference between the surrounding tissues and the region under the tumor |
The quantitative features developed in previous CAD systems [15] were classified into two categories: morphology features and texture features. In this study, the performance of morphology features, texture features, and the combination of both feature sets were evaluated. Figure 1 shows the subgroups of morphology features included shape (Fig. 1c), orientation (Fig. 1d), margin (Fig. 1e), lesion boundary (Fig. 1f), and posterior acoustic features (Fig. 1g). The texture features took the echogenicity of echo pattern (Fig. 1h) into consideration. The measurements indicated in the figures were described below.
Morphology Features
To describe the regularity of tumor boundary, normalized radial length (NRL) [21] was defined according to the distance between the center of tumor area and each boundary pixel. NRL entropy and NRL variance are two of the measurements to exhibit the uniformity degree of the lengths. Typical benign lesions, which have ellipse-like boundaries, have less variance of NRL than malignant lesions. The length of tumor boundary was also compared with the perimeter of best-fit ellipse (Fig. 1c) [14] to obtain the contour difference. Typical malignant lesions with irregular shape have longer perimeter than its best-fit ellipse.
In addition to the boundary pixels, compactness measurement of the perimeter and area of the segmented tumor was used to estimate whether the shape is oval or irregular [22]. The overlap between the tumor area and the corresponding best-fit ellipse was also suggested for compactness. Malignant lesions have typically taller than wide appearance. Consequently, calculating the angle between the major axis of the best-fit ellipse and the horizontal achieved the measurement and was used as the orientation features shown in Fig. 1d [14].
To describe tumor margin, a maximum circle inside a tumor was built according to the distance between the center and the boundary pixels. Several parts of tumor that exceeded the circle were partitioned into the lobulate regions, which were used to determine the variability as shown in Fig. 1e [14]. Spicules on tumor boundary can also be counted to express the smoothness level. With a similarity threshold, peaks that had relatively longer distance than their neighbors to the tumor center were regarded as spicules.
The lesion boundary was measured using two bands as shown in Fig. 1f. Whether there was a sharp demarcation between the lesion and surrounding tissue can be estimated by the intensity difference between the inner band and outer band of tumor contour [14].
For measuring the posterior acoustic feature, a posterior area was defined behind the mass as shown in Fig. 1g. The width of the posterior area was defined by two-thirds of the mass width. At both sides of posterior area, a gap which had one-sixth of the mass width was preserved for avoiding the edge shadowing. The height of posterior area was equal to the mass height but did not exceed 100 pixels. The average intensity difference between the tumor and the region under the tumor (PS) and the average intensity difference between the surrounding tissues and the region under the tumor (PS_diff) were used as posterior acoustic features [13].
Texture Features
For tissue characterization, texture analysis of breast tumors on B-mode images was proposed, and gray-level co-occurrence matrix (GLCM) [23] was used in this study. That is, the correlations between pixels and their neighbors inside the tumor region were calculated via the statistics of their gray-level values. The average and standard deviation (SD) of eight GLCM metrics including energy, entropy, correlation, inverse difference moment, inertia, cluster shade, cluster prominence, and Haralick’s correlation were calculated to be the texture features which were used to interpret echo pattern. The distance for the occurrence of two pixels was one, and four offset directions (0°, 45°, 90°, and 135°) were used. For rotation invariance, the four directions were combined to one matrix. In Fig. 1h, the correlation between a pixel and its neighbors inside a tumor interpreted the echogenicity for texture features.
Classification
To evaluate the likelihood of malignancy, the proposed quantitative features were all used in a classifier for classifying BI-RADS category 3 masses. Besides, the performances of texture features and morphology features were also evaluated, respectively. GLCM features were combined to be the texture feature set. The rest of the features extracted from the segmented tumor contour were combined to be the morphology feature set. The classifier used in the experiment was binary logistic regression [24]. Based on the stepwise procedure, backward elimination evaluated the capabilities of features in distinguishing between benign and malignant lesions for feature selection. While the least error rate was obtained in the classifier, the corresponding feature set was selected to be relevant. The total 69 specimens were all used for the feature selection process. Therefore, the performance of the classifier was evaluated by using leave-one-out cross-validation method. A total of 69 iterations of case training and testing were performed in the method. In each iteration, one individual case was separated from the total cases and was used to test the result trained by the rest 68 cases. The final performance of the classifier was the average of 69 iterations. The leave-one-out cross-validation was used to make the validation more rigorous and to show the true generalization ability of the CAD to new cases. The number of cases was not a limitation to the number of quantitative features.
Statistical Analysis
According to the biopsy-proven pathology, the cases were labeled as true positive, true negative, false positive, and false negative. Then, five performance indices, accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), were calculated to evaluate the classification result. In addition to the individual morphology and texture feature set, the performance of combining both feature set was evaluated in the comparison. The difference between two feature sets for the five performance indices was analyzed using chi-square test. With respect to the trade-offs between sensitivity and specificity, receiver operating characteristic (ROC) curve was drawn. The area under the ROC curve (Az) was also calculated and compared using the z-test in ROCKIT software (C. Metz, University of Chicago, Chicago, IL, USA).
Furthermore, individual quantitative feature implemented in the proposed CAD system was evaluated. Student’s t test [25] was used to evaluate features with normal distribution. Features with non-normal distribution were evaluated using the Mann–Whitney U test [25]. As the evaluation result, features obtained a p value less than 0.05 were significant statistically in distinguishing between benign and malignant lesions. To quantize the correlation between the numeric distribution of each feature and the biopsy-proven malignancy, Pearson correlation [25] was also used in the analysis. Finally, the classification accuracy of the individual quantitative feature in the binary logistic regression model was obtained, and the most significant features were ranked to provide their dominances in classifying BI-RADS category 3 masses. Pearson correlation and test methods were performed using SPSS software (version 16 for Windows; SPSS, Chicago, IL, USA).
Results
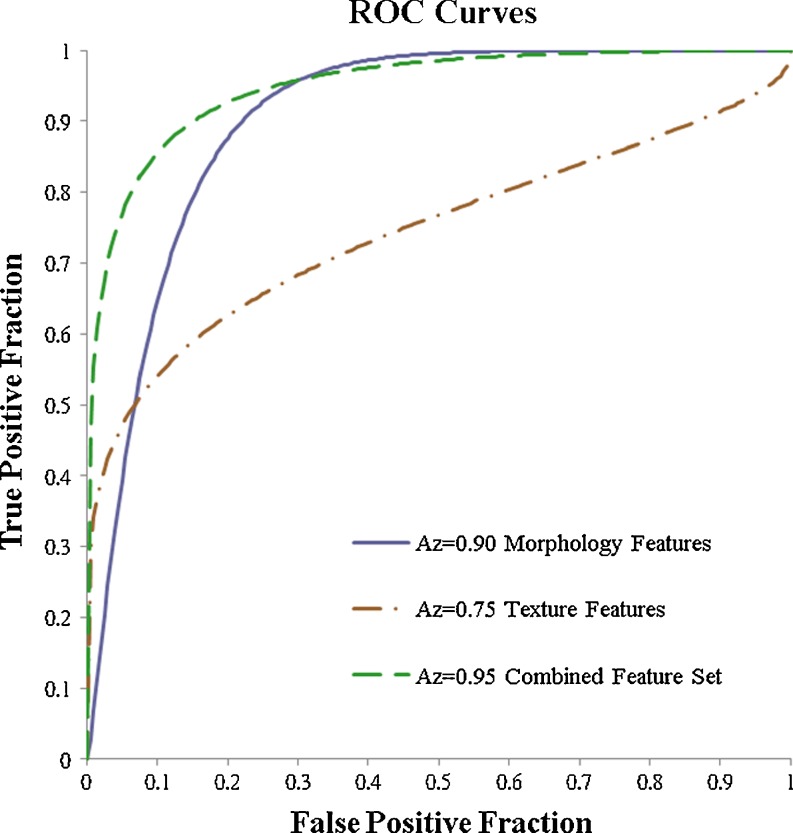
In Table 2, the performance of the morphology features, the texture features, and the combined feature set are listed. The performance achieved by the morphology features was better than the performance of the texture features. However, the difference was not statistically different (Az 0.90 vs. 0.75; p value, 0.0703). The performance achieved by the combined features set was statistically significantly better than the texture features (Az 0.95 vs. 0.75; p value, 0.0163; Fig. 2). The cut-off point at the sensitivity of 86 % (18/21), 95 % (20/21), and 100 % (21/21) achieved the specificity of 90 % (43/48), 73 % (35/48), and 33 % (16/48), respectively.
Table 2.
The comparisons of performance indices and p values (chi-square test or z test) among the morphology features, the texture features, and the combined feature set
| Morphology | Texture | Combined | Morphology vs. texture (p value) | Combined vs. morphology (p value) | Combined vs. texture (p value) | |
|---|---|---|---|---|---|---|
| Accuracy | 84 % (58/69) | 77 % (53/69) | 88 % (61/69) | 0.2833 | 0.4586 | 0.0724 |
| Sensitivity | 86 % (18/21) | 62 % (13/21) | 86 % (18/21) | 0.0793 | 1.0000 | 0.0793 |
| Specificity | 83 % (40/48) | 83 % (40/48) | 90 % (43/48) | 1.0000 | 0.3709 | 0.3709 |
| PPV | 69 % (18/26) | 62 % (13/21) | 78 % (18/23) | 0.5982 | 0.4749 | 0.2349 |
| NPV | 93 % (40/43) | 83 % (40/48) | 93 % (43/46) | 0.1569 | 0.9318 | 0.1261 |
| Az | 0.90 | 0.75 | 0.95 | 0.0703 | 0.2898 | 0.0163* |
Combined feature set includes the morphology features and the texture features
*p < 0.05 (indicates a statistically significant difference)
Fig. 2.
The receive operating characteristic (ROC) curves of the morphology features, the texture feature, and the combined feature set
The best eight features were illustrated in Fig. 3 according to their accuracy, Pearson correlation, and t test. The first four features, Ellipse_a/b, Ellipse_theta, Ep/Tp, and Ellipse_b, achieved the accuracy near 80 % (55/69). For Pearson correlation, the result is not the same as the classification accuracy. That is, a feature that has a higher classification accuracy may not also have a higher Pearson correlation than other features. This was the reason why we provided three different performance metrics to generate a more objective evaluation. Nevertheless, the first four features all have higher Pearson correlation value than the last four features. For the t test, only Tumor_P and EPc have the p values higher than 0.001. The other six features all have the p value less than 0.001 which represents better discriminability.
Fig. 3.
The performances of the top eight features evaluated by using accuracy, Pearson correlation, and t test. The first four features (Ellipse_a/b, Ellipse_theta, Ep/Tp, and Ellipse_b) achieved better accuracy and Pearson correlation value than the last four. Ellipse_a/b, Ellipse_theta, Ep/Tp, and Ellipse_b were extracted from the best-fit ellipse. Tumor_p was the tumor perimeter. EPc was the contrast value of tumor. NRL_entropy was the regularity measurement of tumor boundary. Undulation described tumor margin
Figure 4 shows one case of malignant lesions which were correctly classified by the proposed CAD system. This malignant lesion with round shape had quantitative features of Ellipse_a/b = 1.1 (mean = 1.3) and Ellipse_theta = 0.4 (mean = 0.3). In Fig. 5, one ellipse-like benign lesion with Ellipse_a/b = 2.0 (mean = 1.7) and Ellipse_theta = 0.1 (mean = 0.1) was evaluated to have low likelihood of malignancy. For all lesions, the mean values of Ellipse_a/b and Ellipse_theta were 1.6 and 0.2, respectively.
Fig. 4.
One malignant lesion (invasive ductal carcinoma) classified correctly by the proposed CAD system. a The original US image. b The segmentation result of (a). c The corresponding best-fit ellipse compared to the segmentation result of (b)
Fig. 5.
One benign lesion (fibrocystic changes) classified correctly by the proposed CAD system. a The original US image. b The segmentation result of (a). c The corresponding best-fit ellipse compared to the segmentation result of (b)
Discussion
The proposed CAD system based on the morphology and texture features was used for estimating the likelihood of malignancy of masses assessed as BI-RADS category 3, probably benign by at least one of five radiologists. With the performance (Az) of 0.95, the US CAD system achieved the sensitivity of 95 % and the specificity of 73 %. That is, 20 of 21 BI-RADS category 3 malignant masses were recognized as carcinomas, while 35 of 48 BI-RADS category 3 benign masses were correctly classified as benign by the US CAD system. Since the CAD could not be perfect (100 % sensitivity and 100 % specificity), the cut-off point with sensitivity of 95 % (20/21, one malignant case misclassified) can be chosen for the classification in a clinical setting. Recognizing the carcinomas among the BI-RADS category 3 lesions is important since circumscribed malignant lesions are often high-grade tumors [26–28]. The proposed CAD system can correctly upgrade most of malignant masses misclassified as BI-RADS category 3 on US by radiologists while downgrade majority of benign lesions classified as BI-RADS category 3. To the best of our knowledge, this is the first study to evaluate the BI-RADS category 3, probably benign lesions with a US CAD system.
In this study, the quantitative features based on the standardized BI-RADS lexicon were implemented and investigated to classify BI-RADS category 3 masses into benign or malignant. The result suggested that the features extracted from the best-fit ellipse would be useful to estimate the likelihood of malignancy in BI-RADS category 3 masses. According to the described characteristics, the BI-RADS category 3 masses showing round shape tend to be malignant compared to masses showing ellipse-like shape. The orientation and the length of tumor boundary features were also helpful for the classification [29]. Consequently, the developed CAD may provide complementary diagnostic assessment for radiologists to reduce the interobserver and intraobserver variability [11, 30].
With respect to the performances achieved in the previous CAD systems [12–19], the Az values presented in the literature were in the range of 0.92–0.97. The suggested quantitative features were significant in distinguishing between benign and malignant lesions. However, the BI-RADS category classification of the collected specimens was only shown in several studies [13, 14, 18]. For the others, the descriptions of specimens just focused on the types of malignant and benign lesions. All of the studies suggested their CAD systems to classify the tumors into malignant and benign without considering whether the CAD systems performed well in the BI-RADS 3 masses. As a second viewer, the proposed CAD system which upgraded the malignant masses misclassified as BI-RADS category 3 on US by radiologists is more meaningful. Especially, we found that the features extracted from the best-fit ellipse such as Ellipse_a/b, Ellipse_theta, Ep/Tp, and Ellipse_b were useful. In previous CAD systems without BI-RADS classification, round and oval shapes were considered to be benign findings. For BI-RADS 3 masses used in this study, round shape was more likely to be a malignant finding. The quantitative features used in our experiment included morphology and texture features, which were already suggested in the previous CAD systems. As the results shown in the literature, the best diagnostic performance can be achieved by combining both morphology and texture features.
The limitations of this study included the number of specimens and the types of malignant tumors. The number of masses assessed as BI-RADS category 3 for evaluating the proposed CAD system in this study was only 69, and all malignant cases belonged to the invasive ductal carcinomas. To apply the proposed CAD system on clinical examination, more specimens and various types of cancers should be included in further experiments for evaluation. The criterion for classifying the truth in our study was very liberal since only one of five radiologists had to call it a category 3. The results could have changed if some other stricter criteria were used instead of using this criterion. However, previous studies consistently showed that CAD is more useful to improve performance in the less experienced than in the experienced radiologists [16]. For the improvement of the CAD system, more robust quantitative features can be developed. In the experiment, the performance of texture features was not as good as that of morphology features. The possible reason may be the variance of gray-level intensities among US images. Combining more intensity-invariant features with the existed quantitative features would enhance the CAD system further.
Conclusions
Using morphology and texture features to estimate the likelihood of malignancy of BI-RADS category 3 masses in the proposed CAD obtained the performance (Az) of 0.95 with 95 % sensitivity and 73 % specificity. The proposed CAD system has the potential to be used in upgrading malignant masses misclassified as BI-RADS category 3 on US by the radiologists. However, an observer study needs to be done to see if it actually does impact decision making clinically.
Acknowledgments
The authors thank the National Science Council (NSC 101-2221-E-002-068-MY3), Ministry of Economic Affairs (100-EC-17-A-19-S1-164), and Ministry of Education (AE-00-00-06) of the Republic of China for the financial support. This study was also supported by a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (A01185).
References
- 1.Stavros AT, Thickman D, Rapp CL, Dennis MA, Parker SH, Sisney GA. Solid breast nodules: use of sonography to distinguish between benign and malignant lesions. Radiology. 1995;196:123–134. doi: 10.1148/radiology.196.1.7784555. [DOI] [PubMed] [Google Scholar]
- 2.American College of Radiology, editor. Breast Imaging Reporting and Data System. 4. Reston: American College of Radiology; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg WA. Supplemental screening sonography in dense breasts. Radiol Clin North Am. 2004;42:845–851. doi: 10.1016/j.rcl.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Sickles EA. Nonpalpable, circumscribed, noncalcified solid breast masses—likelihood of malignancy based on lesion size and age of patient. Radiology. 1994;192:439–442. doi: 10.1148/radiology.192.2.8029411. [DOI] [PubMed] [Google Scholar]
- 5.Leung JW, Sickles EA. The probably benign assessment. Radiol Clin North Am. 2007;45:773–789. doi: 10.1016/j.rcl.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Berg WA, et al. Combined screening with ultrasound and mammography vs mammography alone in women at elevated risk of breast cancer. Jama-J Am Med Assoc. 2008;299:2151–2163. doi: 10.1001/jama.299.18.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber R, Jaromi S, Rudas M, Pfarl G, Riedl CC, Flöry D, Graf O, Sickles EA, Helbich TH. Histologic work-up of non-palpable breast lesions classified as probably benign at initial mammography and/or ultrasound (BI-RADS category 3) Eur J Radiol. 2013;82:398–403. doi: 10.1016/j.ejrad.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Kim SJ, Chang JM, Cho N, Chung SY, Han W, Moon WK. Outcome of breast lesions detected at screening ultrasonography. Eur J Radiol. 2012;81:3229–3233. doi: 10.1016/j.ejrad.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Graf O, Helbich TH, Hopf G, Graf C, Sickles EA. Probably benign breast masses at US: is follow-up an acceptable alternative to biopsy? Radiology. 2007;244:87–93. doi: 10.1148/radiol.2441060258. [DOI] [PubMed] [Google Scholar]
- 10.Lazarus E, Mainiero MB, Schepps B, Koelliker SL, Livingston LS. BI-RADS lexicon for US and mammography: interobserver variability and positive predictive value. Radiology. 2006;239:385–391. doi: 10.1148/radiol.2392042127. [DOI] [PubMed] [Google Scholar]
- 11.Lee HJ, et al. Observer variability of Breast Imaging Reporting and Data System (BI-RADS) for breast ultrasound. Eur J Radiol. 2008;65:293–298. doi: 10.1016/j.ejrad.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Jesneck JL, Lo JY, Baker JA. Breast mass lesions: computer-aided diagnosis models with mammographic and sonographic descriptors. Radiology. 2007;244:390–398. doi: 10.1148/radiol.2442060712. [DOI] [PubMed] [Google Scholar]
- 13.Shen WC, Chang RF, Moon WK, Chou YH, Huang CS. Breast ultrasound computer-aided diagnosis using BI-RADS features. Acad Radiol. 2007;14:928–939. doi: 10.1016/j.acra.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Shen WC, Chang RF, Moon WK. Computer aided classification system for breast ultrasound based on breast imaging reporting and data system (BI-RADS) Ultrasound Med Biol. 2007;33:1688–1698. doi: 10.1016/j.ultrasmedbio.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Kim KG, Cho SW, Min SJ, Kim JH, Min BG, Bae KT. Computerized scheme for assessing ultrasonographic features of breast masses. Acad Radiol. 2005;12:58–66. doi: 10.1016/j.acra.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Jiang YL, Nishikawa RM, Schmidt RA, Metz CE, Giger ML, Doi K. Improving breast cancer diagnosis with computer-aided diagnosis. Acad Radiol. 1999;6:22–33. doi: 10.1016/S1076-6332(99)80058-0. [DOI] [PubMed] [Google Scholar]
- 17.Drukker K, Giger ML, Metz CE. Robustness of computerized lesion detection and classification scheme across different breast US platforms. Radiology. 2005;237:834–840. doi: 10.1148/radiol.2373041418. [DOI] [PubMed] [Google Scholar]
- 18.Moon WK, Lo CM, Huang CS, Chen JH, Chang RF. Computer-aided diagnosis based on speckle patterns in ultrasound images. Ultrasound Med Biol. 2012;38:1251–1261. doi: 10.1016/j.ultrasmedbio.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 19.Moon WK, Lo CM, Chang JM, Huang CS, Chen JH, Chang RF. Computer-aided classification of breast masses using speckle features of automated breast ultrasound images. Med Phys. 2012;39:6465–6473. doi: 10.1118/1.4754801. [DOI] [PubMed] [Google Scholar]
- 20.Moon WK, Chang SC, Huang CS, Chang RF. Breast tumor classification using fuzzy clustering for breast elastography. Ultrasound Med Biol. 2011;37:700–708. doi: 10.1016/j.ultrasmedbio.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol. 2008;15:1513–1525. doi: 10.1016/j.acra.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangayyan RM, Mudigonda NR, Desautels JEL. Boundary modelling and shape analysis methods for classification of mammographic masses. Med Biol Eng Comput. 2000;38:487–496. doi: 10.1007/BF02345742. [DOI] [PubMed] [Google Scholar]
- 23.Haralick RM, Shanmuga K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern Smc. 1973;3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 24.Hosmer DW. Applied Logistic Regression. 2. New York: Wiley; 2000. [Google Scholar]
- 25.Field AP. Discovering Statistics Using SPSS. 3. Los Angeles: SAGE; 2009. [Google Scholar]
- 26.Schrading S, Kuhl CK. Mammographic, US, and MR imaging phenotypes of familial breast cancer. Radiology. 2008;246:58–70. doi: 10.1148/radiol.2461062173. [DOI] [PubMed] [Google Scholar]
- 27.Bae MS, et al. Characteristics of breast cancers detected by ultrasound screening in women with negative mammograms. Cancer Sci. 2011;102:1862–1867. doi: 10.1111/j.1349-7006.2011.02034.x. [DOI] [PubMed] [Google Scholar]
- 28.Rubin E. Six-month follow-up: an alternative view. Radiology. 1999;213:15–18. doi: 10.1148/radiology.213.1.r99oc4715. [DOI] [PubMed] [Google Scholar]
- 29.Fornage BD, Sneige N, Faroux MJ, Andry E. Sonographic appearance and ultrasound-guided fine-needle aspiration biopsy of breast carcinomas smaller than 1 cm3. J Ultrasound Med. 1990;9:559–568. doi: 10.7863/jum.1990.9.10.559. [DOI] [PubMed] [Google Scholar]
- 30.Sahiner B, et al. Malignant and benign breast masses on 3D US volumetric images: effect of computer-aided diagnosis on radiologist accuracy. Radiology. 2007;242:716–724. doi: 10.1148/radiol.2423051464. [DOI] [PMC free article] [PubMed] [Google Scholar]