Abstract
The fibroblast growth factor receptors (FGFRs) regulate important biological processes including cell proliferation and differentiation during development and tissue repair. Over the past decades, numerous pathological conditions and developmental syndromes have emerged as a consequence of deregulation in the FGFRs signaling network. This review aims to provide an overview of FGFR family, their complex signaling pathways in tumorigenesis, and the current development and application of therapeutics targeting the FGFRs signaling for treatment of refractory human cancers.
Keywords: Fibroblast growth factor receptors, Cancer, Signal transduction, Targeted therapy
Introduction
The human fibroblast growth factor receptor (FGFR) family, a subfamily of receptor tyrosine kinases (RTKs), comprises of four family members—FGFR1, FGFR2, FGFR3 and FGFR4. A closely-related receptor which lacks the FGF signaling tyrosine kinase domain, FGFR5, (also known as FGFRL1) was recently discovered on the basis of interaction with FGFR-binding ligands, known as fibroblast growth factors (FGFs) [1]. Collectively, FGFR signaling is associated with the activation of multiple cellular cascades and responses such as cell growth, proliferation, differentiation, and survival [2–4].
FGFR protein structures
The FGFR family members share a high percentage of sequence homology (~55–72 %) [5], and consist of three important domains—extracellular ligand-binding domain, single transmembrane domain, and intracellular tyrosine kinase domain (Fig. 1). The extracellular ligand binding-domain comprises of a hydrophobic signal peptide region and two or three immunoglobulin (Ig)-like domains, designated D1–D3. The acid box is made up of ~30 serine residues and connects D1 and D2. The transmembrane domain facilitates signal transduction from the extracellular region into the cytoplasmic domain. Emerging from the cytoplasmic membrane is a juxtamembrane region, followed by a split tyrosine kinase domain, and finally a COOH tail [2, 5, 6].
Fig. 1.
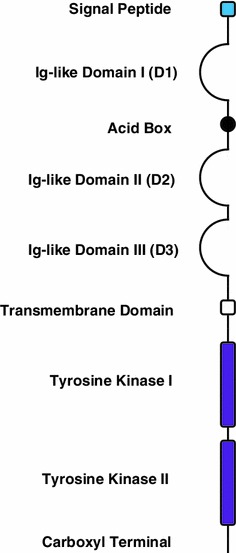
The basic structure of a FGFR. The FGFRs are phylogenetically closely related to the VEGFRs and PDGFRs, consist of three extracellular immunoglobulin (Ig) domains (D1-D3), a single transmembrane helix, an intracellular split tyrosine kinase domain (TK1 and TK2) and an acidic box. D2 and D3 form the ligand-binding pocket and have distinct domains that bind both FGFs and heparan sulfate proteoglycans (HSPGs). Acidic box is required for binding of bivalent cations for optimal interaction between FGFRs and HSPGs
Although FGFR5 is structurally similar to other FGFRs, it lacks the intracellular protein tyrosine kinase domain, which is replaced by a short intracellular tail with a histidine-rich motif [1]. Due to the absence of the tyrosine kinase domain, FGFR5 cannot signal by transautophosphorylation (as other FGFRs), and hence does not function like other FGFRs. Instead, FGFR5 is proposed to act as decoy receptor that binds FGF ligands and sequesters them away from the conventional FGFRs [1]. Additionally, the signal peptide found within the extracellular ligand binding-domain is cleaved off upon insertion into the endoplasmic reticulum, and FGFR5 is occasionally shed from the plasma membrane and is found in a secreted, soluble form [1]. FGFR5 binds to certain FGFs, as do the conventional FGFRs, but it does not bind to FGF1, which is also recognized by all of the other FGFRs [1].
Alternative splicing of FGFRs
Despite the general characteristics shared among the family members of FGFRs, an array of isoforms exist within each family member (Fig. 2). Structural diversity observed across the isoforms of FGFRs, is largely attributed to the alternative splicing of endogenous mRNA sequence. These isoforms include the secreted form of FGFRs that lack the hydrophobic membrane-spanning region and the entire cytoplasmic catalytic domain [7, 8], FGFRs with an extracellular domain composed of either two or three Ig-like domains [7, 9–11], and FGFRs devoid of the acid box [12].
Fig. 2.
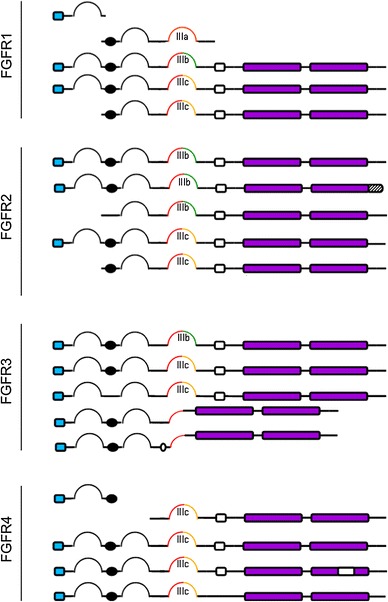
FGFR splice variants. The FGFRs isoforms are generated mainly by alternative splicing of the Ig III domain (D3). The D3 could be encoded by an invariant exon 7 (red) to produce FGFR-IIIa isofom or spliced to either exon 8 (green) or 9 (yellow) to generate the FGFR-IIIb or FGFR-IIIc isoforms, respectively. Epithelial tissues predominantly express the IIIb isoform and mesenchymal tissues express IIIc. FGFR4 is expressed as a single isoform that is paralogous to FGFR-IIIc. Hatched box represents a truncated carboxyl terminal. Clear box indicates a deletion of an exon
One of the most important mechanisms that determine the ligand-binding specificity of FGFRs is by alternate exon usage of the IgIII (D3) domain to produce three possible IgIII domains isoforms, designated IgIIIa, IgIIIb and IgIIIc. IgIIIa is encoded entirely by exon 7 alone while IgIIIb and IgIIIc are derived from alternative splicing of exon 7/8 and exon 7/9, respectively (Fig. 2) [13–16]. The IgIIIb and IgIIIc splice variants are commonly observed in FGFR1, FGFR2 and FGFR3 gene [13, 17]. The FGFR4 gene is unique as only IgIIIc variants are present [18]. FGFR5 isoforms lacking the sequences that encode for either the first Ig domain or the first Ig domain plus the acidic box were also identified, but the properties of these isoforms are not known and no tissue-specific expression has yet been reported [1].
Activation and signaling of FGFR
FGFR signaling is primarily triggered by the binding of the receptors to FGF ligands (Table 1), and the subsequent formation of various complexes to initiate downstream signal transduction including activation of PLCγ, MAPK, AKT, and STAT cascade (Fig. 3) [19].
Table 1.
| FGFR isoform | Ligand specificity |
|---|---|
| FGFR1, IIIb | FGF-1, 2, 3, 10, and 22 |
| FGFR1, IIIc | FGF-1, 2, 4, 5, 6, 19, 20, and 21 |
| FGFR2, IIIb | FGF-1, 3, 4, 6, 7, 10, and 22 |
| FGFR2, IIIc | FGF-1, 2, 4, 5, 6, 8, 9, 17, 18, 19, 21, and 23 |
| FGFR3, IIIb | FGF-1 and 9 |
| FGFR3, IIIc | FGF-1, 2, 4, 8, 9, 17, 18 19, 21, and 23 |
| FGFR4 | FGF-1, 2, 4, 6, 8, 9, 16, 17, 18, and 19 |
Fig. 3.
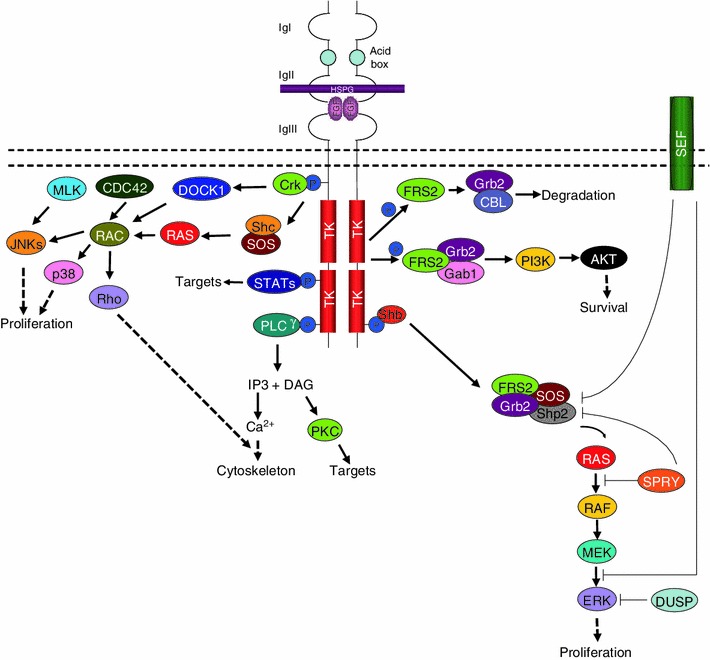
FGFR signaling pathway. FGFs induce FGFR-mediated signaling pathway by interacting with specific FGFRs and HSPGs. The macromolecular interactions mediate FGFRs dimerization or oligomerization and activate multiple signal transduction pathways, including those involving FRS2, RAS, p38 MAPKs, ERKs, JNKs, Src, PLCγ, Crk, PKC and PI3K. These pathways are negatively regulated in part by the activities of DUSPs, SPRY, SEF and CBL
The phosphotyrosine residues in the carboxy-terminal regions of FGFR confer selective and strong binding to Src homolog 2 (SH2) domain-containing molecules, such as phospholipase Cγ (PLCγ) [20, 21]. This interaction results in the hydrolysis of phosphatidylinositol 4,5-biphosphate (PIP2) to generate two effectors, inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG) [22]. Accumulation of IP3 further stimulates calcium release from internal stores, whereas DAG mediates the activation of protein kinase C (PKC) and other downstream targets such as the Ras/MEK/MAPK pathway (Fig. 3) [22].
One of the many adaptor proteins which facilitates signal transduction from FGFRs is the v-crk sarcoma virus CT10 oncogene homolog (avian) (Crk). Upon exposure to growth factors, the juxtamembrane tyrosine residue 463 of FGFR1 is phosphorylated, followed by the transient phosphorylation of Crk to mediate the co-complexing of FGFR1 and Crk [23]. Crk has been reported to associate with various signaling molecules such as the guanine nucleotide exchange factor, SOS [24] and C3G [25], as well as the dedicator of cytokinesis 1 (DOCK1) [26]. SOS then activates JNK via Ras [27] and Rac [28, 29]. In addition to Rac, cdc42 has also been proposed as an intermediate to the JNK and p38 activation cascades [27]. Direct interaction of DOCK180 to Rac1 have been reported, and DOCK180 activates JNK in a manner dependent on Cdc42Hs, and SEK [30]. On the other hand, C3G activates JNK1 through a pathway involving the MLK family of protein [31].
Activation of FGFRs also lead to phosphorylation of the docking protein FGFR substrate 2 (FRS2) followed by the recruitment of Shp2 tyrosine phosphatase, whereby the subsequent phosphorylation of Shp2 facilitates its association with growth factor receptor-bound 2 (Grb2) and SOS [32–35]. This complex triggers the induction of the Ras/MEK/MAPK signaling pathway [32]. In addition, the tyrosine phosphorylation of FRS2α also mediates the recruitment of Grb2 and Gab1, resulting in the recruitment and activation of PI3-kinase [36]. Alternatively, the Grb2/FRS2α complex interacts with Cbl via the SH3 domains resulting in the ubiquitination of FGFRs and FRS2α for the attenuation of growth factor signaling [37].
Accessory proteins such as SH2 domain-containing adaptor protein B (Shb) and SH2 domain-containing collagen (Shc) also interact with FGFRs to facilitate signal transduction [22, 38–43]. Shb2 binds to tyrosine 766 in FGFR1 to facilitate the phosphorylation of FRS2 for the subsequent activation of the Ras/MEK/MAPK pathway [22]. Similar to Shp2, Shc is an intermediate molecule which mediates the assembly of FGFR and Grb2-SOS complexes, also to activate the Ras/Raf/MEK/MAPK pathway [3, 41, 44]. Shc co-localizes exclusively with FGFR2 at the plasma membrane and intracellular membranous compartment [45]. The localization of Shc and the stabilization of the binding of its SH2 domain to FGFR2 is attributed to the presence of the phosphotyrosine-binding (PTB) domain [45, 46].
FGFR also binds to signal transducers and activators of transcription (STAT) and ribosomal protein S6 kinase 2 (RSK2). The interaction between FGFR and STAT was first observed in polymorphism studies, in which the point mutation—K650E of FGFR3, and other FGFR isoforms, led to the constitutive activation of both FGFR and STAT [47–49]. STAT3 was further documented to bind to phosphorylated Tyr677 of FGFR1 [50]. In addition, tyrosine activation of STAT3 requires the overexpression of FGFR1 or FGFR2 (Fig. 3) [50].
Negative regulation of FGFR signaling
Given the fact that FGFRs activate an array of signaling pathways, it is crucial to have a regulatory system in place to circumvent unnecessary signal transmission. FGF signaling is negatively modulated by regulators such as Cbl proto-oncogene E3 ubiquitin protein ligase (CBL), sprouty homolog (SPRY), similar expression to fgf genes (SEF) and MAPK phosphatases (MKP) (Fig. 3).
Following activation, FGFRs are often ubiquitinated by CBL to facilitate clathrin-mediated endocytosis and receptor degradation [37, 51]. Intriguingly, different FGFR isoforms have discrete propensities for ubiquitination. FGFR1 is more susceptible to ubiquitination whereas FGFR4 has only a few lysine ubiquitination sites [52, 53]. The difference in ubiquitination is thought to account for the different sorting of FGFRs to the lysosome [52].
A second pathway that negatively regulates FGFR signaling involves the inhibition of RAS/RAF/ERK downstream pathway through SPRY [54–59]. Upon FGF ligand stimulation, SPRY isoforms form homo- and hetero-oligomers via their C-terminal domains and attenuate ERK activation through interaction with FRS2–Grb2-SOS complex or direct inhibition of RAS/RAF signaling pathway (Fig. 3) [56, 60–64]. Among all the SPRY isoforms, SPRY2 is considered more inhibitory than SPRY1 and SPRY 4 [61], but the SPRY1/SPRY4 hetero-oligomer exhibits the most potent inhibitory effect on ERK [60]. However, the binding of SPRYs to Grb2 does not always result in ERK inhibition [65, 66]. This implies that SPRY can inhibit signal transduction in a Grb2-independent pathway, and the molecular machinery that is involved in this context is worth investigating. In this context, it is likely that the functional role of SPRY in FGFR signaling is highly dependent on the cell type and stimuli, and might play a different role in different cell types.
In addition to CBL and SPRY, SEF has also been shown to negatively modulates FGF-mediated ERK and AKT activation [67–70]. In addition to receptor inhibition, SEF could also hinder the nuclear translocation of ERK in certain cell lines (Fig. 3) [71, 72].
The role of dual phosphatases in the negative feedback of growth factor signaling has also been reported by many researches. For instance, the MAP kinase phosphatase -1 (MKP-1) has been discovered on the basis of its ability to dephosphorylate MAP kinase in vivo [73]. Similarly, MKP-3 blocks both the phosphorylation and enzymatic activation of ERK2 by mitogens [74]. Consistent with this, the targeted inactivation of Dusp6, which encodes MKP-3, led to the increase of phosphorylated ERK and pERK targets [75]. However, not every member of this class of dual phosphatase inhibits ERK. Although MKP-3 blocks ERK1 activation by oncogenic p21(ras), but ERK1 activated by p21(ras) (G12V) is insensitive to the M3/6 dual phosphatase [76].
FGFRs and human cancer
Numerous human pathological conditions are tightly associated with the deregulation of FGFR signaling. Aberrant FGFR signaling is largely attributed to several underlying mechanisms involving gene amplification, single nucleotide polymorphism (SNP), chromosomal translocation, ligand availability and impaired termination program in FGF-mediated signaling, which is reviewed in the following section. In addition, a further layer of complexity is added by the fact that FGFRs are subjected to alternative splicing, giving rise to multiple isoforms which may promote or repress tumorigenesis, under different circumstances.
Gene amplification
Enhanced FGFR expression is commonly observed in various types of human malignancies. Such elevations in FGFRs expression could be due to gene amplification or deregulation at the transcriptional level (Table 2) [77–80]. In line with this, the chromosomal regions 8p11-12 and 10q26 are consistently amplified in human breast cancers, and some of the genes within this region that are amplified are FGFR1 and FGFR2 [77–83]. Amplifications of 8p11 and 8p12 are associated with early relapse, poor prognosis and survival, especially in ER-positive breast cancer patients where FGFR1 amplification drives resistance to hormonal therapy [84–86]. Although high copy numbers and expression levels of FGFR1 has been shown to promote tumorigenesis [79, 87–90], several studies also show that amplification of the focal region 8p11-12 does not always result in the overexpression of FGFR1 [91–93]. In this case, other genes located at the same locus may also be amplified and contribute to the oncogenesis of human mammary carcinomas [92], independent of FGFR1. Indeed, a few candidate genes identified from the recurrent amplicon 8p11-12 were overexpressed and promotes the survival of the breast tumors [93].
Table 2.
Gene amplifications of FGFRs in human cancers
| FGFRs | Cancer types | References |
|---|---|---|
| FGFR1 | Breast, ovarian, bladder, lung and rhabdomyosarcoma | [77–83, 86, 201–206] |
| FGFR2 | Gastric, breast, lung, endometrial and esophageal cancer | [78, 80, 82, 95–102, 207–211] |
| FGFR3 | Bladder and salivary adenoid cystic cancer | [212, 213] |
| FGFR4 | Gastric, breast and ovarian cancer | [80, 83, 94, 214] |
Amplification of the FGFR2 gene is identified in gastric, lung, and endometrial primary tumors and biologically-transformed cell lines [19, 78, 80, 82, 83, 94–102]. Furthermore, this event is occasionally accompanied by other forms of genetic alteration such as base deletion. For example, scirrhous-type gastric cancer cells which harbor amplification in the region 10q26, were also found to contain deletions in the C terminal of the FGFR2 exons [103]. It is postulated that the presence of highly amplified copies of the truncated receptor could promote tumor proliferation and oncogenesis [104–106].
It is also common to observe secondary chromosomal locus amplification in parallel to elevated FGFR expression. For instance, 30–40 % of breast tumors with amplification of the FGFR1-containing chromosomal region 8p12, were concurrently presented with CCND1 (Cyclin D1) gene amplification at the 11q13 locus [77]. Since the number of genetic alterations observed in tumor is associated with poor prognosis [107], there could be a synergistic effect from the concomitant amplification of both oncogenes to drive tumorigenesis.
Point mutations
Various human diseases are driven, in part, by point mutations. The sequencing of cancer genomes has uncovered over a thousand somatic mutations in the coding exons of 518 human kinase genes [108]. Most of the non-synonymous mutations involve FGF signaling [108]. Oncogenic point mutations of FGFRs could essentially include any parts of the receptor, as summarized in Table 3. Some of these mutations have been shown to exert a gain-of-function effect, contributing to the developmental abnormalities, uncontrolled growth, and metastasis in a various cancerous cells, the role of other mutations in tumorigenesis remained unknown [108].
Table 3.
Point mutations of FGFRs in human cancers
| FGFR | Cancers | Mutations | References |
|---|---|---|---|
| FGFR1a | Lung | G70Re, T141Re, P252T/S, P576H, V664L | [215, 216] |
| Prostate | R78H | [217] | |
| Breast | S125L | [108, 218, 219] | |
| Skin | P252T/S | [108, 217, 220] | |
| Stomach | A268S | [217] | |
| Colon | A268S, A429S | [217, 221] | |
| Brain | N546Kf, R576W, K656Ef | [222] | |
| FGFR2b | Skin | S24F, V77M, E160A, H213Y, E219K, G227E, V248D, R251Q, G271E, G305R, T370R, W474X, E475K, D530N, E574K, E636K, M640I, I642V, A648T, S688F, G701S, P708S, R759X/Q, L770V | [223] |
| Bladder | M71T | [217] | |
| Lymphoma | M71T | [217] | |
| Cervix | A97T | [115] | |
| Endometrial | D101Y, S252Wf, P253Rf, K310R, A314D, A315T, S372C, Y375C, C382R, A389T, M391R, I547, N549, K659/M/E | [115–117, 201, 224] | |
| Breast | R203C | [108, 218, 225] | |
| Lung | N211I, D283, W290C, I380, H544Qe, R612Tg | [108, 115, 201, 216, 220, 226] | |
| Brain | Q212, G462E | [227, 228] | |
| Ovary | S252Wf, G272V, Y375C | [229] | |
| Stomach | S267Pf | [230] | |
| Colon | Q361Rh, P582L | [228] | |
| FGFR3c | Lung | T79S | [108] |
| Multiple myeloma | G197Se, Y241C, R248Cf, P250Re, Y373Cf, G380Rf, G382Df, F384Le, S433C, A441Te, A452Se, K650E/Q/M/N/Tf, A717Te, I726Fe | [137, 201, 231–239] | |
| Colon | C228R, E322 | [108, 230] | |
| Bladder | R248C, S249C, G370C, S371C, Y373C, I376C, G380R, G382D, F384Le, A391E, D646Y, K650E/Q/M/N/T | [120, 201, 240–250] | |
| Head and neck | R248Cf, S249Cf, D617G, V630, E686, G697Cf | [251–253] | |
| Cervix | S249Cf | [119, 254, 255] | |
| Prostate | S249Cf, F384Le, A391Ef | [256] | |
| Brain | E466 | [257] | |
| Testis | K650E/Q/M/N/Tf | [258] | |
| FGFR4d | Rhabdomyosarcoma | C56S, R72L, T122A, A175T, R234H, G388Re, N535/D, V550Ef/L/Mi, A554, G576D | [259] |
| Lung | R183Se, S232Ie, G388Re, R616Ge, E681, P712Tg, A729Ge, S772 | [108, 127, 201, 216, 217, 220, 260–262] | |
| Breast | Y367Cf, G388Re, V550Ef/L/Ma | [108, 124, 217, 218, 263] | |
| Stomach | G388Re | [264] | |
| Skin | G388Re, P716R | [217, 223, 265] | |
| Brain | G388Re | [217] | |
| Colon | G388Re | [124] | |
| Liver | G388Re | [266] | |
| Soft tissue sarcoma | G388Re | [267] | |
| Prostate | G388Re | [217, 263, 268–271] | |
| Head and neck | G388Re | [217, 272–274] |
aMutations relative to the FGFR1 IIIc (GenBank accession number NM_023110)
bMutations relative to the FGFR2 IIIc (GenBank accession number NP_000132)
cMutations relative to the FGFR3 IIIc (GenBank accession number NP_000133)
dMutations relative to the FGFR4 (GenBank accession number X57205)
eSingle nucleotide polymorphism (SNP)
fGain-of-function mutations have been demonstrated experimentally
gR612T is referred to as R496T in the literature due to a numbering relative to FGFR2 isoform 7 precursor which lacks two exons compared with transcript variant 1
hGlu361 is only present in FGFR2 IIIb
iFGFR4 V550M, P712T and S772N are referred to as V510M, P672T and S732N (respectively) in the literature and in COSMIC due to a numbering relative to FGFR4 transcript variant 2, which lacks 40 amino acids (including the transmembrane domain) compared with X57205
Various hotspots for point mutations have been characterized in FGFR2. Substitution of S252W and P253R alter the ligand binding specificities of FGFR2b and FGFR2c, but retain the ligand-dependent activation properties [109], whereas K659N causes receptor hyperactivation [110]. In terms of disease pathogenesis, point mutations of S252W and P253R in FGFR2 are associated with Apert syndrome [111], and some of these led to a crouzonoid phenotype [111–114]. Similar point mutations were also observed in various cancer types, such as uterine carcinoma and endometrial carcinoma [115–117], suggesting that point mutations which causes skeletal disorders are also causally linked to tumorigenesis.
In addition, activating mutant forms of FGFR3 attributed to point mutations are frequently detected in bladder cancer [118–120]. The substitution of cysteine residues at the extracellular domain and juxtamembrane region enhances intermolecular disulfide bonding and ligand-independent receptor dimerization [121–123]. These events eventually led to the continuous activation of FGFR3 and its downstream signaling pathways.
FGFR4 G388R is one of the most common nucleotide polymorphisms (SNPs), with at least one copy identified in nearly 50 % of the population [124]. G388R sustains the activation of FGFR4 [125], and human cancers characterized by this SNP were reported to be highly aggressive and metastatic in nature [124, 126–129]. Additionally, the FGFR4 R388 allele is also associated with invasion and metastasis by stabilizing the endosomal MMP14, thus promoting collagen degradation [126, 130]. The elevated expression of MMP1 further stimulates the autophosphorylation of FGFR4 R388, and collectively these regulators act synergistically to promote tumor invasion and metastasis [126, 130].
Chromosomal translocation
Fusion proteins arise from intragenic chromosomal rearrangements that commonly involve reciprocal translocations. As a result of chromosomal fusions, the mutated proteins gain new functions which dominate the functions of wild-type proteins. Human FGFR fusion proteins generally consist of two main segments—the anterior being a dimerized domain from a partnering gene and tyrosine kinase domain at the posterior [131]. Unlike wild type receptors, mutant FGFRs are expressed intracellularly and retained in the cytosol, thus they escape the typical receptor degradation processes, further prolonging the activation signal [3].
FGFR-related fusion proteins which are caused by intragenic chromosomal translocation have been detected in hematological cancers (Table 4). These fusion proteins are mainly found in patients diagnosed with stem cell leukemia lymphoma (SCLL) (also known as 8p11 myeloproliferative) syndrome. FGFR fusion proteins are able to transform normal cell lines into SCLL or chronic myelogenous leukemia-like disease (CMLL) [132–136]. Interestingly, newly diagnosed multiple myeloma (MM) patients that harbor the t(4;14) translocation often exhibit overexpression of FGFR3 in the absence of activating mutations, while late stage MM patients carrying the same translocation were observed to have activated FGFR3 and multiple myeloma SET domain (MMSET) mutations [137]. It is believed that MMSET might contribute to cellular adhesion, clonogenic growth, and tumorigenicity [138]. The mechanism which drives the change in FGFR3 activation status during the early and late onset of MM has yet to be elucidated. One plausible explanation is that other forms of mutations that accompany FGFR3 translocation may trigger and maintain the abnormal signaling of FGFR3 [137].
Table 4.
Fusion proteins of FGFRs found in human cancers
| FGFRs | Fusion partners | Cancers |
|---|---|---|
| FGFR1 | ZNF198/RAMP/FIM/ZMYM2a | SCLL [275–281] |
| FOP/FGFR1OP1a | SCLL [282–284], lung [285] | |
| CEP110/CEP1a | SCLL [284, 286–289] | |
| BCR | SCLL [136, 290–293] | |
| LRRFIP1 | SCLL [294] | |
| FGFR1OP2a | SCLL [295], AML [296] | |
| TRIM24/TIF1 | SCLL [297] | |
| MYO18A | SCLL [298] | |
| CPSF6 | SCLL [299] | |
| HERV-K | SCLL [300] | |
| PLAG1 | H&N [301] | |
| CUX1 | L/EMS/L [302] | |
| TACC1 | Glioblastoma [303] | |
| FOXO1 | Rhabdomyosarcoma [304] | |
| FGFR3 | TEL/ETV6a | T-cell Lymphoma [305] |
| TACC3a | Glioblastoma [303], bladder [306] |
aFusion proteins which have been demonstrated to be functionally oncogenic in cell lines and mouse models
Addiction to FGF ligand via autocrine and paracrine signalling
Soluble mitogenic growth factors can be synthesized by one cell type and signals the proliferation of another cell type, and the intersignaling between two different cells are termed paracrine signaling [139]. Alternatively, cancer cells may also produce growth factors to which they are responsive, creating a positive feedback signaling loop often termed autocrine stimulation [139].
One example of autocrine FGF-signaling is the elevated expression of FGFR1 and its high-affinity ligand, FGF2 in human melanoma xenografts. The silencing of either FGFR1 or FGF2 arrests tumor growth, suggesting the presence of the FGFR1–FGF2 autocrine loop in the subset of human subcutaneous carcinomas [140]. Similarly, in multiple basal-like breast cancer cells, tumor growth was suppressed following RNAi-mediated silencing of endogenous FGF2 [141]. In human non-small-cell lung carcinomas (NSCLC), FGF2, FGF9, and their respective receptors, were also reported to mediate autocrine signaling which drives tumor resistance to specific kinase targeted therapy [142].
Oncogenic transformation that is facilitated by paracrine signaling is reflected in the formation of multifocal prostate adenocarcinomas whereby FGF10, which is highly expressed by mesenchymal cells, histologically transforms and stimulates the expression of the epithelial androgen receptor on the adjacent wild type epithelium [143]. In addition, paracrine signaling of FGF10 promoted androgen independent survival of a subset of prostate adenocarcinoma, and also synergizes with epithelial autonomous AKT signaling, leading to high-grade carcinoma [143]. FGF–FGFR paracrine signaling is also identified in a subset of breast cancer stem-like cells. The uncontrolled growth of this subpopulation was proposed to be driven by estrogen hormone stimulation, which in turn, regulates the paracrine signaling of FGF9–FGFR3 [144].
Impaired negative feedback mechanisms in FGFR signaling
The deregulation of negative regulators of FGFR signaling has been associated with the pathogenesis of various malignancies. For instance, lack of SEF expression has been observed in primary tumors of the breast, ovary, and thyroid [145], while high-grade prostate carcinomas have much lower SEF and SPRY expression compared to healthy individuals [146, 147]. Consistent with this, the levels of SEF is downregulated, whereas FGF2, FGF8, and FGFR4 levels are upregulated in aggressive prostate cancer specimens [148]. Taken together, these data imply that the loss of SEF might contribute to the hyperactivation of the FGF/FGFR signaling axis, thus leading to oncogenesis.
Localization of FGFRs to the surface of the plasma membrane also facilitates FGF ligand binding and intracellular tyrosine kinase transphosphorylation, an event which is required to trigger signal transduction [149]. Under normal circumstances, ligand-receptor complexes are usually endocytosed and transported to lysosomes for degradation. Thus, disruption in the endocytic pathway is expected to alter FGFR signaling [149, 150]. Indeed, in patients suffering from achondroplasia (ACH) and related chondrodysplasia, the substitution of K650E and G380R in FGFR3 protects the surface growth-receptor from being sorted to the lysosomes [151]. The accumulation of these FGFR3 variants, in turn, boosts the signaling capacity of the receptor as their retained half-life is twice that of the wild-type receptor [151].
Similarly, the FGFR4 Arg388 SNP variant which is found in most prostate cancer, has been reported not only to protect FGFR from lysosomal degradation, but also to sustain receptor phosphorylation [125]. Likewise, the FGFR2 IIIb C3 variant which harbors nucleotide deletion at the cytoplasmic C-terminal sequences known to code for the endocytic motif, exhibited aberrant receptor trafficking and stability, and thus, enhanced receptor signaling capacity [152].
Alternative splicing of FGFRs
Alternative splicing of the IgIII (D3) domain generates the IIIb and IIIc isoforms in FGFRs1–3 [142], and each of these isoforms display different affinity to bind to their FGF family members. For instance, FGFR1 IIIb, FGFR1 IIIc, and FGFR2 IIIc bind FGF2 and FGF9 with high affinity, whereas FGFR2 IIIb preferably binds FGF7 and FGF10 [142].
These IIIb and IIIc isoforms of FGFRs have different roles in cancer. This is reflected in various cancers, including breast, endometrial, cervical, lung, esophageal, gastric, pancreatic, and colorectal cancer [153], which displayed overexpression of FGFR2 IIIb. The role of FGFR2 IIIb, together with its major ligands, such as FGF7 and FGF10, in this context, promotes tumor angiogenesis and migration in pancreatic cancers [154, 155]. However, the tumor promoting roles of FGFR2 IIIb is thought to be exclusive to different cancer types. In line with this, diminished expression of FGFR2 IIIb in gastric cancer cells results in hyperproliferation and invasion [153, 156]. Furthermore, cell lines derived from bladder cancers of lower stage and grade expressed FGFR3 IIIb as their major transcript, while cells derived from high grade tumors exhibit a switch to favour FGFR3 IIIc expression, suggesting that FGFR3 IIIb may have tumor-restrictive properties in bladder cancer [157]. These lines of evidence imply that FGFR2 IIIb and FGFR IIIb play dual roles in tumorigenesis.
Likewise, FGFR1 mRNA is exclusively spliced to generate FGFR1 IIIc in small cell lung carcinoma (NSCLC) cells lines [142]. Additionally, FGFR1 IIIc and FGFR2 IIIc are frequently co-expressed with their complementary ligands—FGF2 and FGF9 in primary NSCLC tumors [142]. The role of IIIc and its respective ligands is thought to compromise a growth factor autocrine loop in a subset of NSCLC cells [142]. FGFR2 IIIc is also associated with the progression of prostate cancers [158], and confers growth advantage to cervical cancer cell lines [159].
Apart from the D3 domain variants, deregulated splicing mechanism affecting other domains are also associated with tumorigenesis. The increased expression of FGFR1β isoform, which lacks the IgI domain, confers increased sensitivity to FGF1, leading to poor prognosis in breast tumors and malignant of astrocytomas [160]. Similarly, a pituitary tumor-derived, N-terminally truncated isoform of FGF receptor-4 (ptd-FGFR4) which lacks the signal peptide and the first two extracellular Ig-like domains, possesses high transforming properties in vitro and in vivo [161]. The targeted expression of ptd-FGFR4 results in pituitary tumors in transgenic mice [161].
In general, the alternative splicing of FGFRs generates multiple isoforms, which are selectively expressed in different types of cancer. These isoforms have tumor promoting and suppressive roles in different cancer types. Further efforts to characterize the novel FGFRs isoforms, and to determine if these isoforms antagonize, or work in synergy to promote or repress cancer cell growth is necessary to improve the design of cancer therapeutics.
Overall, the deregulation of FGFR can occur at several tiers ranging from gene to protein translation and trafficking, and that aberrant FGFR signaling is causally linked to tumorigenesis.
Current targeted therapies for FGFRs/FGFs signaling pathway
Given the important role of FGFR signaling in tumorigenesis, various approaches have been developed to target the upstream and downstream axis of this signaling pathway. A number of novel therapeutic molecules have been introduced and are currently undergoing preclinical and clinical trials in various FGFRs-related tumors (Table 5). These small molecules can be generally classified as receptor tyrosine kinase inhibitors (RTKIs) which are mainly ATP-competitive molecules; or antagonistic antibodies which target FGF ligands or receptors [162].
Table 5.
| Drugs | Target (s) IC50 [nM]a | Clinical trials | Manufacturer | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FGFR1 | FGFR2 | FGFR3 | FGFR4 | VEGFR1 | VEGFR2 | VEGFR3 | PDGFRα | PDGFRβ | Others | |||
| BIBF1120 (Vargatef) | 69 | 37 | 108 | – | 34 | 13 | 13 | 59 | 60 |
Src: 156 Lck: 16 Lyn: 195 |
I–III | Boehringer-Ingelheim |
| TKI258 (Dovotinib) | 8 | – | 9 | – | 10 | 13 | 8 | 21 | 27 |
CSF1: 36 c-KIT: 2 |
I–III | Novartis |
| BMS 582664 (Brivanib) | 148 | 125 | 68 | – | 380 | 25 | 10 | – | >6000 | I–II | Bristol-Myers Squibb | |
| E7080 (Lenvantinib) | 46 | – | – | – | 22 | 4 | 5 | 51 | 39 | c-KIT: 100 | I–II | Eisai |
| TSU-68 (Orantinib) | 1,200 | – | – | – | – | 2,100 | – | – | 8 | – | I–II | Taiho Pharm. |
| AB1010 (Masantanib) | – | – | 5,500 | – | – | – | – | 300 | 50 |
c-KIT: 150 Lyn: 510 |
Preclinical | AB Science |
| AZD4547 | 0.2 | 2.5 | 1.8 | 164.8 | – | – | – | – | – | – | I–II | AstraZeneca |
| BGJ398 | 0.9 | 1.4 | 1.0 | 60 | – | – | – | – | – | – | I | Novartis |
aIC50 as determined by in vitro kinase assay
Ab antibody, CSF1 colony-stimulating factor 1, FGF fibroblast growth factor, FGFR fibroblast growth factor receptor, PDGFR platelet-derived growth factor receptor, VEGFR vascular endothelial growth factor receptor
Most of the RTKIs assessed to date are non-selective FGFRs inhibitors. Despite targeting the FGFR family, they also inhibit other RTKs such as vascular endothelial growth factor receptors (VEGFRs) and platelet-derived growth factor receptors (PDGFRs) [163, 164]. These include compounds like the specific FGFR inhibitor, PD173074; and second-generation FGFR inhibitors that target FGFRs and other RTKs such as dovitinib (TKI258), AZD4547, Ki23057, E7080, brivanib alaninate, intedanib, ponatinib, MK-2461, and E-3810 [162]. Some of these compounds, such as TKI258 and AZD4547 have demonstrated promising potentials as inhibitors of FGFRs, are being tested in advanced clinical trials.
Dovitinib shows high potency against most FGFRs besides targeting c-KIT, CSF-1, VEGFRs and PDGFRs (Table 5) [19, 165]. This molecule is currently being tested in phase III clinical trial for renal cell carcinoma and phase II clinical trials for advanced breast and endometrial cancers, relapsed MM, and urothelial cancer [162]. Given that dovitinib may exert its anticancer effects either by directly targeting FGFRs or regulators of angiogenesis, endometrial cancer patients with or without FGFR2 mutations are separately enrolled in a clinical to prove its mechanisms of functions [166].
Unlike TKI258 which target multiple kinases, AZD4547 specifically targets FGFR family proteins. Pre-clinical studies have recently demonstrated the selectivity and potency of AZD4547 in FGFR-driven breast tumors cancer models with minimal to none adverse drug reactions at effective doses [167]. In addition, pre-clinical studies on xenograft models transplanted with transformed cells derived from FGFR1-amplified non-small cell lung cancer (NSCLC) patients demonstrated that AZD4547 stops tumor growth and promotes regression [168]. AZD4547 is currently under Phase II clinical trial for breast cancer and phase I for solid tumors [162].
In addition to RTKIs, the development of therapeutic monoclonal antibodies targeting the FGFRs signaling have been demonstrated to exhibit specific antitumor activity in cancer cell lines and animal models. For instance, KM1334 neutralizing antibodies targeting the FGF8 isoform b significantly hinder FGF ligand-mediated signaling in mammary tumorigenesis [169] and FGF8b-expressing clinical prostate cancers [170] in addition to, inflammatory responses and bone damages in rat model of rheumatoid arthritis [169–171]. GP369, an isoform-specific blocking antibody that binds to FGFR2 IIIb have been shown to inhibit the proliferation of several human cancer cell lines and tumor xenografts harboring FGFR2 gene amplification and FGFR2 activating point mutation (S252W and N550K) [99]. R3Mab, an antibody specifically acts on FGFR3 (IIIb and IIIc isoforms) but has no effect on FGFR1, FGFR2, or FGFR4 has been shown to exhibits significant inhibitory effects on FGF1-induced cancer cell proliferation and inhibitory effects on both bladder cancer and MM in mice [172]. Several other antibodies such as IMC-A1 [173], PRO-001 [174], R3Mab [172, 175] and 1A6 [176] also show potentials in treating cancers driven by aberrant FGFR signaling.
Another emerging treatment option is the FGF ligand traps. FGF ligand trap is a fusion of Ig Fc with a soluble FGFR construct that captures FGF1, 2, 3, 7, 10 and inhibits ligand-dependent FGFR signaling [177]. One example of this class of molecule which has been developed as cancer therapeutics is FP-1039, which comprises of the extracellular ligand-binding domain of FGFR1c fused to the crystallizable fragment regions of human immunoglobulin G [178, 179]. The specificity of this ligand is reflected by the observation that the growth rate of head and neck squamous cell carcinoma cells expressing abundant FGF2 is significantly inhibited upon treatment with FP-1039, whereas HNSCC cells that express little FGF2 is not affected [179]. FP-1039 is currently in a Phase II trial for endometrial cancers with the S252W FGFR2 mutation which confers increased affinity and altered specificity of FGF binding [178].
Other strategies targeting the FGFR signaling such as peptide mimetics, RNA aptamers, siRNAs and miRNAs have also been investigated intensively merits further improvements [180].
Challenges in the targeting FGFRs in human cancers
Given the broad expression of FGFRs and their key role in development and physiology, toxicity issues are to be expected from FGFR inhibition. The FGFR pathway is involved in normal phosphate and vitamin D homeostasis. The preclinical development of FGFR inhibitors has been complicated by hyperphosphatemia-mediated tissue calcification, due to the blockade of FGF23 release from bone and of FGF23 signal in kidney [181]. In preclinical models, dynamic modulation of circulating FGF23 levels was reported following FGFR inhibition. FGF23 levels was suppressed during the periods of drug exposure attributed to direct inhibition of FGF23 release from the bone and elevated upon drug withdrawal driven by increased plasma phosphate and vitamin D levels acting on bone to stimulate FGF23 production [182]. FGF23 may also bind FGFR4, FGFR1 IIIc and FGFR3 IIIc [183, 184], but the relative contribution of individual FGFR subtypes to hyperphosphatemia remains unclear [185–187].
Similarly, administration of an anti-FGFR1 IIIc antibody has been shown to result in profound weight loss in preclinical in vivo models, potentially due to FGFR1 targeting in the hypothalamus [173], and this has prevented further clinical development. It remains to be ascertained whether this would be a class effect for all FGFR1 IIIc antibodies.
Finally, the great challenge for future development of specific FGFR inhibitors in the clinic is to carefully determine a therapeutic dose which will balance efficacy against gene-addicted tumors with a manageable tolerability profile. Continued clinical research may identify which FGFR isoforms have the greatest efficacy potential, and whether inhibition of particular isoforms can avoid side effects associated with broad specificity small-molecule FGFR inhibitors.
Conclusions
In conclusion, the role of FGFRs signaling in promoting tumorigenesis is well established. As such, novel molecules which inhibit FGF or FGFR interactions have been introduced and many of them are currently undergoing clinical trials for treating various types of human malignancies associated with hyperactivation of FGFR signaling. Given that the standard chemotherapies in cancer patients are often associated with adverse toxicity, such targeted-therapies could present a more viable option in addressing this issue. In addition, the inhibition of molecules downstream of FGFRs network could also serve as a secondary approach as many of these molecules regulate various cellular processes and functions as well. With the emergence of drug resistance and disease recurrence issues, the current trend of clinical approach is moving towards multi-targeted drugs and combination therapies. This requires in-depth understanding of the signaling pathways in order to formulate an appropriate strategy which focuses on cancer cells. The clinical application of these therapeutic strategies warrants further studies to ensure maximum clinical benefit in cancer patients.
Conflict of interest
The authors declare that they have no competing interests.
Contributor Information
Kai Hung Tiong, Email: kaihung_tiong@imu.edu.my.
Li Yen Mah, Email: MahLiYen@imu.edu.my.
Chee-Onn Leong, Phone: +603-2731-7528, FAX: +603-8656-7229, Email: cheeonn_leong@imu.edu.my.
References
- 1.Trueb B. Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell Mol Life Sci. 2011;68(6):951–964. doi: 10.1007/s00018-010-0576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287(2):390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437(2):199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 4.Haugsten EM, Wiedlocha A, Olsnes S, Wesche J. Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res. 2010;8(11):1439–1452. doi: 10.1158/1541-7786.MCR-10-0168. [DOI] [PubMed] [Google Scholar]
- 5.Johnson DE, Williams LT. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 6.Citores L, Khnykin D, Sorensen V, Wesche J, Klingenberg O, Wiedlocha A, Olsnes S. Modulation of intracellular transport of acidic fibroblast growth factor by mutations in the cytoplasmic receptor domain. J Cell Sci. 2001;114(Pt 9):1677–1689. doi: 10.1242/jcs.114.9.1677. [DOI] [PubMed] [Google Scholar]
- 7.Johnson DE, Lee PL, Lu J, Williams LT. Diverse forms of a receptor for acidic and basic fibroblast growth factors. Mol Cell Biol. 1990;10(9):4728–4736. doi: 10.1128/mcb.10.9.4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katoh M, Hattori Y, Sasaki H, Tanaka M, Sugano K, Yazaki Y, Sugimura T, Terada M. K-sam gene encodes secreted as well as transmembrane receptor tyrosine kinase. Proc Natl Acad Sci USA. 1992;89(7):2960–2964. doi: 10.1073/pnas.89.7.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid HH, Wilks AF, Bernard O. Two forms of the basic fibroblast growth factor receptor-like mRNA are expressed in the developing mouse brain. Proc Natl Acad Sci USA. 1990;87(4):1596–1600. doi: 10.1073/pnas.87.4.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dionne CA, Crumley G, Bellot F, Kaplow JM, Searfoss G, Ruta M, Burgess WH, Jaye M, Schlessinger J. Cloning and expression of two distinct high-affinity receptors cross-reacting with acidic and basic fibroblast growth factors. EMBO J. 1990;9(9):2685–2692. doi: 10.1002/j.1460-2075.1990.tb07454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crumley G, Bellot F, Kaplow JM, Schlessinger J, Jaye M, Dionne CA. High-affinity binding and activation of a truncated FGF receptor by both aFGF and bFGF. Oncogene. 1991;6(12):2255–2262. [PubMed] [Google Scholar]
- 12.Shimizu A, Tada K, Shukunami C, Hiraki Y, Kurokawa T, Magane N, Kurokawa-Seo M. A novel alternatively spliced fibroblast growth factor receptor 3 isoform lacking the acid box domain is expressed during chondrogenic differentiation of ATDC5 cells. J Biol Chem. 2001;276(14):11031–11040. doi: 10.1074/jbc.M003535200. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DE, Lu J, Chen H, Werner S, Williams LT. The human fibroblast growth factor receptor genes: a common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol Cell Biol. 1991;11(9):4627–4634. doi: 10.1128/mcb.11.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dell KR, Williams LT. A novel form of fibroblast growth factor receptor 2. Alternative splicing of the third immunoglobulin-like domain confers ligand binding specificity. J Biol Chem. 1992;267(29):21225–21229. [PubMed] [Google Scholar]
- 15.Werner S, Duan DS, de Vries C, Peters KG, Johnson DE, Williams LT. Differential splicing in the extracellular region of fibroblast growth factor receptor 1 generates receptor variants with different ligand-binding specificities. Mol Cell Biol. 1992;12(1):82–88. doi: 10.1128/mcb.12.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avivi A, Yayon A, Givol D. A novel form of FGF receptor-3 using an alternative exon in the immunoglobulin domain III. FEBS Lett. 1993;330(3):249–252. doi: 10.1016/0014-5793(93)80882-u. [DOI] [PubMed] [Google Scholar]
- 17.Chellaiah AT, McEwen DG, Werner S, Xu J, Ornitz DM. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J Biol Chem. 1994;269(15):11620–11627. [PubMed] [Google Scholar]
- 18.Vainikka S, Partanen J, Bellosta P, Coulier F, Birnbaum D, Basilico C, Jaye M, Alitalo K. Fibroblast growth factor receptor-4 shows novel features in genomic structure, ligand binding and signal transduction. EMBO J. 1992;11(12):4273–4280. doi: 10.1002/j.1460-2075.1992.tb05526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A, Ashworth A, van de Vijver MJ, Reis-Filho JS. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29(14):2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammadi M, Honegger AM, Rotin D, Fischer R, Bellot F, Li W, Dionne CA, Jaye M, Rubinstein M, Schlessinger J. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol Cell Biol. 1991;11(10):5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361(6410):315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 22.Cross MJ, Lu L, Magnusson P, Nyqvist D, Holmqvist K, Welsh M, Claesson-Welsh L. The Shb adaptor protein binds to tyrosine 766 in the FGFR-1 and regulates the Ras/MEK/MAPK pathway via FRS2 phosphorylation in endothelial cells. Mol Biol Cell. 2002;13(8):2881–2893. doi: 10.1091/mbc.E02-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson H, Klint P, Landgren E, Claesson-Welsh L. Fibroblast growth factor receptor-1-mediated endothelial cell proliferation is dependent on the Src homology (SH) 2/SH3 domain-containing adaptor protein Crk. J Biol Chem. 1999;274(36):25726–25734. doi: 10.1074/jbc.274.36.25726. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda M, Hashimoto Y, Muroya K, Hasegawa H, Kurata T, Tanaka S, Nakamura S, Hattori S. CRK protein binds to two guanine nucleotide-releasing proteins for the Ras family and modulates nerve growth factor-induced activation of Ras in PC12 cells. Mol Cell Biol. 1994;14(8):5495–5500. doi: 10.1128/mcb.14.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K, et al. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc Natl Acad Sci USA. 1994;91(8):3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasegawa H, Kiyokawa E, Tanaka S, Nagashima K, Gotoh N, Shibuya M, Kurata T, Matsuda M. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol Cell Biol. 1996;16(4):1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81(7):1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 28.Mochizuki N, Ohba Y, Kobayashi S, Otsuka N, Graybiel AM, Tanaka S, Matsuda M. Crk activation of JNK via C3G and R-Ras. J Biol Chem. 2000;275(17):12667–12671. doi: 10.1074/jbc.275.17.12667. [DOI] [PubMed] [Google Scholar]
- 29.Dolfi F, Garcia-Guzman M, Ojaniemi M, Nakamura H, Matsuda M, Vuori K. The adaptor protein Crk connects multiple cellular stimuli to the JNK signaling pathway. Proc Natl Acad Sci USA. 1998;95(26):15394–15399. doi: 10.1073/pnas.95.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiyokawa E, Hashimoto Y, Kobayashi S, Sugimura H, Kurata T, Matsuda M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12(21):3331–3336. doi: 10.1101/gad.12.21.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka S, Hanafusa H. Guanine-nucleotide exchange protein C3G activates JNK1 by a ras-independent mechanism. JNK1 activation inhibited by kinase negative forms of MLK3 and DLK mixed lineage kinases. J Biol Chem. 1998;273(3):1281–1284. doi: 10.1074/jbc.273.3.1281. [DOI] [PubMed] [Google Scholar]
- 32.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89(5):693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 33.Gotoh N, Laks S, Nakashima M, Lax I, Schlessinger J. FRS2 family docking proteins with overlapping roles in activation of MAP kinase have distinct spatial-temporal patterns of expression of their transcripts. FEBS Lett. 2004;564(1–2):14–18. doi: 10.1016/S0014-5793(04)00287-X. [DOI] [PubMed] [Google Scholar]
- 34.Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18(7):3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harada A, Katoh H, Negishi M. Direct interaction of Rnd1 with FRS2 beta regulates Rnd1-induced down-regulation of RhoA activity and is involved in fibroblast growth factor-induced neurite outgrowth in PC12 cells. J Biol Chem. 2005;280(18):18418–18424. doi: 10.1074/jbc.M411356200. [DOI] [PubMed] [Google Scholar]
- 36.Ong SH, Hadari YR, Gotoh N, Guy GR, Schlessinger J, Lax I. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc Natl Acad Sci USA. 2001;98(11):6074–6079. doi: 10.1073/pnas.111114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong A, Lamothe B, Lee A, Schlessinger J, Lax I. FRS2 alpha attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proc Natl Acad Sci USA. 2002;99(10):6684–6689. doi: 10.1073/pnas.052138899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. 1996;16(3):977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raffioni S, Thomas D, Foehr ED, Thompson LM, Bradshaw RA. Comparison of the intracellular signaling responses by three chimeric fibroblast growth factor receptors in PC12 cells. Proc Natl Acad Sci USA. 1999;96(13):7178–7183. doi: 10.1073/pnas.96.13.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JK, Gao G, Goldfarb M. Fibroblast growth factor receptors have different signaling and mitogenic potentials. Mol Cell Biol. 1994;14(1):181–188. doi: 10.1128/mcb.14.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klint P, Kanda S, Claesson-Welsh L. Shc and a novel 89-kDa component couple to the Grb2-Sos complex in fibroblast growth factor-2-stimulated cells. J Biol Chem. 1995;270(40):23337–23344. doi: 10.1074/jbc.270.40.23337. [DOI] [PubMed] [Google Scholar]
- 42.Spivak-Kroizman T, Mohammadi M, Hu P, Jaye M, Schlessinger J, Lax I. Point mutation in the fibroblast growth factor receptor eliminates phosphatidylinositol hydrolysis without affecting neuronal differentiation of PC12 cells. J Biol Chem. 1994;269(20):14419–14423. [PubMed] [Google Scholar]
- 43.Foehr ED, Raffioni S, Fuji R, Bradshaw RA. FGF signal transduction in PC12 cells: comparison of the responses induced by endogenous and chimeric receptors. Immunol Cell Biol. 1998;76(5):406–413. doi: 10.1046/j.1440-1711.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- 44.Zakrzewska M, Haugsten EM, Nadratowska-Wesolowska B, Oppelt A, Hausott B, Jin Y, Otlewski J, Wesche J, Wiedlocha A. ERK-mediated phosphorylation of fibroblast growth factor receptor 1 on Ser777 inhibits signaling. Sci Signal. 2013;6(262):11. doi: 10.1126/scisignal.2003087. [DOI] [PubMed] [Google Scholar]
- 45.Schuller AC, Ahmed Z, Levitt JA, Suen KM, Suhling K, Ladbury JE. Indirect recruitment of the signalling adaptor Shc to the fibroblast growth factor receptor 2 (FGFR2) Biochem J. 2008;416(2):189–199. doi: 10.1042/BJ20080887. [DOI] [PubMed] [Google Scholar]
- 46.Skjerpen CS, Nilsen T, Wesche J, Olsnes S. Binding of FGF-1 variants to protein kinase CK2 correlates with mitogenicity. EMBO J. 2002;21(15):4058–4069. doi: 10.1093/emboj/cdf402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su WC, Kitagawa M, Xue N, Xie B, Garofalo S, Cho J, Deng C, Horton WA, Fu XY. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature. 1997;386(6622):288–292. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]
- 48.Li C, Chen L, Iwata T, Kitagawa M, Fu XY, Deng CX. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet. 1999;8(1):35–44. doi: 10.1093/hmg/8.1.35. [DOI] [PubMed] [Google Scholar]
- 49.Hart KC, Robertson SC, Kanemitsu MY, Meyer AN, Tynan JA, Donoghue DJ. Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene. 2000;19(29):3309–3320. doi: 10.1038/sj.onc.1203650. [DOI] [PubMed] [Google Scholar]
- 50.Dudka AA, Sweet SM, Heath JK. Signal transducers and activators of transcription-3 binding to the fibroblast growth factor receptor is activated by receptor amplification. Cancer Res. 2010;70(8):3391–3401. doi: 10.1158/0008-5472.CAN-09-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jean S, Mikryukov A, Tremblay MG, Baril J, Guillou F, Bellenfant S, Moss T. Extended-synaptotagmin-2 mediates FGF receptor endocytosis and ERK activation in vivo. Dev Cell. 2010;19(3):426–439. doi: 10.1016/j.devcel.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 52.Haugsten EM, Sorensen V, Brech A, Olsnes S, Wesche J. Different intracellular trafficking of FGF1 endocytosed by the four homologous FGF receptors. J Cell Sci. 2005;118(Pt 17):3869–3881. doi: 10.1242/jcs.02509. [DOI] [PubMed] [Google Scholar]
- 53.Haugsten EM, Malecki J, Bjorklund SM, Olsnes S, Wesche J. Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol Biol Cell. 2008;19(8):3390–3403. doi: 10.1091/mbc.E07-12-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gross I, Bassit B, Benezra M, Licht JD. Mammalian sprouty proteins inhibit cell growth and differentiation by preventing ras activation. J Biol Chem. 2001;276(49):46460–46468. doi: 10.1074/jbc.M108234200. [DOI] [PubMed] [Google Scholar]
- 55.Impagnatiello MA, Weitzer S, Gannon G, Compagni A, Cotten M, Christofori G. Mammalian sprouty-1 and -2 are membrane-anchored phosphoprotein inhibitors of growth factor signaling in endothelial cells. J Cell Biol. 2001;152(5):1087–1098. doi: 10.1083/jcb.152.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yusoff P, Lao DH, Ong SH, Wong ES, Lim J, Lo TL, Leong HF, Fong CW, Guy GR. Sprouty2 inhibits the Ras/MAP kinase pathway by inhibiting the activation of Raf. J Biol Chem. 2002;277(5):3195–3201. doi: 10.1074/jbc.M108368200. [DOI] [PubMed] [Google Scholar]
- 57.Reich A, Sapir A, Shilo B. Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development. 1999;126(18):4139–4147. doi: 10.1242/dev.126.18.4139. [DOI] [PubMed] [Google Scholar]
- 58.Casci T, Vinos J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96(5):655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- 59.Li X, Brunton VG, Burgar HR, Wheldon LM, Heath JK. FRS2-dependent SRC activation is required for fibroblast growth factor receptor-induced phosphorylation of Sprouty and suppression of ERK activity. J Cell Sci. 2004;117(Pt 25):6007–6017. doi: 10.1242/jcs.01519. [DOI] [PubMed] [Google Scholar]
- 60.Ozaki K, Miyazaki S, Tanimura S, Kohno M. Efficient suppression of FGF-2-induced ERK activation by the cooperative interaction among mammalian Sprouty isoforms. J Cell Sci. 2005;118(Pt 24):5861–5871. doi: 10.1242/jcs.02711. [DOI] [PubMed] [Google Scholar]
- 61.Lao DH, Chandramouli S, Yusoff P, Fong CW, Saw TY, Tai LP, Yu CY, Leong HF, Guy GR. A Src homology 3-binding sequence on the C terminus of Sprouty2 is necessary for inhibition of the Ras/ERK pathway downstream of fibroblast growth factor receptor stimulation. J Biol Chem. 2006;281(40):29993–30000. doi: 10.1074/jbc.M604044200. [DOI] [PubMed] [Google Scholar]
- 62.Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16(1):45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 63.Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4(11):850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 64.Lee SH, Schloss DJ, Jarvis L, Krasnow MA, Swain JL. Inhibition of angiogenesis by a mouse sprouty protein. J Biol Chem. 2001;276(6):4128–4133. doi: 10.1074/jbc.M006922200. [DOI] [PubMed] [Google Scholar]
- 65.Martinez N, Garcia-Dominguez CA, Domingo B, Oliva JL, Zarich N, Sanchez A, Gutierrez-Eisman S, Llopis J, Rojas JM. Sprouty2 binds Grb2 at two different proline-rich regions, and the mechanism of ERK inhibition is independent of this interaction. Cell Signal. 2007;19(11):2277–2285. doi: 10.1016/j.cellsig.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 66.Mason JM, Morrison DJ, Bassit B, Dimri M, Band H, Licht JD, Gross I. Tyrosine phosphorylation of Sprouty proteins regulates their ability to inhibit growth factor signaling: a dual feedback loop. Mol Biol Cell. 2004;15(5):2176–2188. doi: 10.1091/mbc.E03-07-0503. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol. 2002;4(2):165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- 68.Furthauer M, Lin W, Ang SL, Thisse B, Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol. 2002;4(2):170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- 69.Ren Y, Li Z, Rong Z, Cheng L, Li Y, Wang Z, Chang Z. Tyrosine 330 in hSef is critical for the localization and the inhibitory effect on FGF signaling. Biochem Biophys Res Commun. 2007;354(3):741–746. doi: 10.1016/j.bbrc.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 70.Kovalenko D, Yang X, Nadeau RJ, Harkins LK, Friesel R. Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J Biol Chem. 2003;278(16):14087–14091. doi: 10.1074/jbc.C200606200. [DOI] [PubMed] [Google Scholar]
- 71.Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell. 2004;7(1):33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 72.Ziv I, Fuchs Y, Preger E, Shabtay A, Harduf H, Zilpa T, Dym N, Ron D. The human sef-a isoform utilizes different mechanisms to regulate receptor tyrosine kinase signaling pathways and subsequent cell fate. J Biol Chem. 2006;281(51):39225–39235. doi: 10.1074/jbc.M607327200. [DOI] [PubMed] [Google Scholar]
- 73.Sun H, Charles CH, Lau LF, Tonks NK. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75(3):487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 74.Muda M, Boschert U, Dickinson R, Martinou JC, Martinou I, Camps M, Schlegel W, Arkinstall S. MKP-3, a novel cytosolic protein-tyrosine phosphatase that exemplifies a new class of mitogen-activated protein kinase phosphatase. J Biol Chem. 1996;271(8):4319–4326. doi: 10.1074/jbc.271.8.4319. [DOI] [PubMed] [Google Scholar]
- 75.Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134(1):167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muda M, Theodosiou A, Rodrigues N, Boschert U, Camps M, Gillieron C, Davies K, Ashworth A, Arkinstall S. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J Biol Chem. 1996;271(44):27205–27208. doi: 10.1074/jbc.271.44.27205. [DOI] [PubMed] [Google Scholar]
- 77.Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, Rodriguez C, Theillet C. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57(19):4360–4367. [PubMed] [Google Scholar]
- 78.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, Ethier SP. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81(8):1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacquemier J, Adelaide J, Parc P, Penault-Llorca F, Planche J, deLapeyriere O, Birnbaum D. Expression of the FGFR1 gene in human breast-carcinoma cells. Int J Cancer. 1994;59(3):373–378. doi: 10.1002/ijc.2910590314. [DOI] [PubMed] [Google Scholar]
- 80.Penault-Llorca F, Bertucci F, Adelaide J, Parc P, Coulier F, Jacquemier J, Birnbaum D, deLapeyriere O. Expression of FGF and FGF receptor genes in human breast cancer. Int J Cancer. 1995;61(2):170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- 81.Theillet C, Adelaide J, Louason G, Bonnet-Dorion F, Jacquemier J, Adnane J, Longy M, Katsaros D, Sismondi P, Gaudray P, et al. FGFRI and PLAT genes and DNA amplification at 8p12 in breast and ovarian cancers. Genes Chromosom Cancer. 1993;7(4):219–226. doi: 10.1002/gcc.2870070407. [DOI] [PubMed] [Google Scholar]
- 82.Adnane J, Gaudray P, Dionne CA, Crumley G, Jaye M, Schlessinger J, Jeanteur P, Birnbaum D, Theillet C. BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers. Oncogene. 1991;6(4):659–663. [PubMed] [Google Scholar]
- 83.Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, Brennan C, Polyak K. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66(8):4065–4078. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- 84.Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9(2):R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Letessier A, Sircoulomb F, Ginestier C, Cervera N, Monville F, Gelsi-Boyer V, Esterni B, Geneix J, Finetti P, Zemmour C, Viens P, Charafe-Jauffret E, Jacquemier J, Birnbaum D, Chaffanet M. Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer. 2006;6:245. doi: 10.1186/1471-2407-6-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, Gillett C, Grigoriadis A, Tutt A, Reis-Filho JS, Ashworth A. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70(5):2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, Lasorsa L, Letessier A, Ginestier C, Monville F, Esteyries S, Adelaide J, Esterni B, Henry C, Ethier SP, Bibeau F, Mozziconacci MJ, Charafe-Jauffret E, Jacquemier J, Bertucci F, Birnbaum D, Theillet C, Chaffanet M. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3(12):655–667. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- 88.Ugolini F, Adelaide J, Charafe-Jauffret E, Nguyen C, Jacquemier J, Jordan B, Birnbaum D, Pebusque MJ. Differential expression assay of chromosome arm 8p genes identifies frizzled-related (FRP1/FRZB) and fibroblast growth factor receptor 1 (FGFR1) as candidate breast cancer genes. Oncogene. 1999;18(10):1903–1910. doi: 10.1038/sj.onc.1202739. [DOI] [PubMed] [Google Scholar]
- 89.Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, Delaloge S, Hortobagyi GN, Symmans WF, Lazar V, Pusztai L. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15(2):441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 90.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10(6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 91.Ray ME, Yang ZQ, Albertson D, Kleer CG, Washburn JG, Macoska JA, Ethier SP. Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer Res. 2004;64(1):40–47. doi: 10.1158/0008-5472.can-03-1022. [DOI] [PubMed] [Google Scholar]
- 92.Garcia MJ, Pole JC, Chin SF, Teschendorff A, Naderi A, Ozdag H, Vias M, Kranjac T, Subkhankulova T, Paish C, Ellis I, Brenton JD, Edwards PA, Caldas C. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24(33):5235–5245. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- 93.Bernard-Pierrot I, Gruel N, Stransky N, Vincent-Salomon A, Reyal F, Raynal V, Vallot C, Pierron G, Radvanyi F, Delattre O. Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res. 2008;68(17):7165–7175. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
- 94.Jaakkola S, Salmikangas P, Nylund S, Partanen J, Armstrong E, Pyrhonen S, Lehtovirta P, Nevanlinna H. Amplification of fgfr4 gene in human breast and gynecological cancers. Int J Cancer. 1993;54(3):378–382. doi: 10.1002/ijc.2910540305. [DOI] [PubMed] [Google Scholar]
- 95.Luqmani YA, Graham M, Coombes RC. Expression of basic fibroblast growth factor, FGFR1 and FGFR2 in normal and malignant human breast, and comparison with other normal tissues. Br J Cancer. 1992;66(2):273–280. doi: 10.1038/bjc.1992.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heiskanen M, Kononen J, Barlund M, Torhorst J, Sauter G, Kallioniemi A, Kallioniemi O. CGH, cDNA and tissue microarray analyses implicate FGFR2 amplification in a small subset of breast tumors. Anal Cell Pathol. 2001;22(4):229–234. doi: 10.1155/2001/981218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dahlberg PS, Ferrin LJ, Dahal G. FGFR2 amplification in gastric cancer. J Surg Res. 2004;121(2):331. [Google Scholar]
- 98.Kunii K, Davis L, Gorenstein J, Hatch H, Yashiro M, Di Bacco A, Elbi C, Lutterbach B. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008;68(7):2340–2348. doi: 10.1158/0008-5472.CAN-07-5229. [DOI] [PubMed] [Google Scholar]
- 99.Bai A, Meetze K, Vo NY, Kollipara S, Mazsa EK, Winston WM, Weiler S, Poling LL, Chen T, Ismail NS, Jiang J, Lerner L, Gyuris J, Weng Z. GP369, an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer Res. 2010;70(19):7630–7639. doi: 10.1158/0008-5472.CAN-10-1489. [DOI] [PubMed] [Google Scholar]
- 100.Mor O, Ranzani GN, Ravia Y, Rotman G, Gutman M, Manor A, Amadori D, Houldsworth J, Hollstein M, Schwab M, Shiloh Y. DNA amplification in human gastric carcinomas. Cancer Genet Cytogenet. 1993;65(2):111–114. doi: 10.1016/0165-4608(93)90217-a. [DOI] [PubMed] [Google Scholar]
- 101.Yoshida T, Sakamoto H, Terada M. Amplified genes in cancer in upper digestive tract. Semin Cancer Biol. 1993;4(1):33–40. [PubMed] [Google Scholar]
- 102.Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, Nakanishi I. Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest. 1998;78(9):1143–1153. [PubMed] [Google Scholar]
- 103.Ueda T, Sasaki H, Kuwahara Y, Nezu M, Shibuya T, Sakamoto H, Ishii H, Yanagihara K, Mafune K, Makuuchi M, Terada M. Deletion of the carboxyl-terminal exons of K-sam/FGFR2 by short homology-mediated recombination, generating preferential expression of specific messenger RNAs. Cancer Res. 1999;59(24):6080–6086. [PubMed] [Google Scholar]
- 104.Lorenzi MV, Castagnino P, Chen Q, Chedid M, Miki T. Ligand-independent activation of fibroblast growth factor receptor-2 by carboxyl terminal alterations. Oncogene. 1997;15(7):817–826. doi: 10.1038/sj.onc.1201242. [DOI] [PubMed] [Google Scholar]
- 105.Ishiwata T, Matsuda Y, Yamamoto T, Uchida E, Korc M, Naito Z. Enhanced expression of fibroblast growth factor receptor 2 IIIc promotes human pancreatic cancer cell proliferation. Am J Pathol. 2012;180(5):1928–1941. doi: 10.1016/j.ajpath.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matsuda Y, Ishiwata T, Yamahatsu K, Kawahara K, Hagio M, Peng WX, Yamamoto T, Nakazawa N, Seya T, Ohaki Y, Naito Z. Overexpressed fibroblast growth factor receptor 2 in the invasive front of colorectal cancer: a potential therapeutic target in colorectal cancer. Cancer Lett. 2011;309(2):209–219. doi: 10.1016/j.canlet.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 107.Cuny M, Kramar A, Courjal F, Johannsdottir V, Iacopetta B, Fontaine H, Grenier J, Culine S, Theillet C. Relating genotype and phenotype in breast cancer: an analysis of the prognostic significance of amplification at eight different genes or loci and of p53 mutations. Cancer Res. 2000;60(4):1077–1083. [PubMed] [Google Scholar]
- 108.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu K, Herr AB, Waksman G, Ornitz DM. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc Natl Acad Sci USA. 2000;97(26):14536–14541. doi: 10.1073/pnas.97.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, Miller WT, Mohammadi M. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27(5):717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wilkie AO, Slaney SF, Oldridge M, Poole MD, Ashworth GJ, Hockley AD, Hayward RD, David DJ, Pulleyn LJ, Rutland P, et al. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat Genet. 1995;9(2):165–172. doi: 10.1038/ng0295-165. [DOI] [PubMed] [Google Scholar]
- 112.Park WJ, Theda C, Maestri NE, Meyers GA, Fryburg JS, Dufresne C, Cohen MM, Jr, Jabs EW. Analysis of phenotypic features and FGFR2 mutations in Apert syndrome. Am J Hum Genet. 1995;57(2):321–328. [PMC free article] [PubMed] [Google Scholar]
- 113.Meyers GA, Day D, Goldberg R, Daentl DL, Przylepa KA, Abrams LJ, Graham JM, Jr, Feingold M, Moeschler JB, Rawnsley E, Scott AF, Jabs EW. FGFR2 exon IIIa and IIIc mutations in Crouzon, Jackson-Weiss, and Pfeiffer syndromes: evidence for missense changes, insertions, and a deletion due to alternative RNA splicing. Am J Hum Genet. 1996;58(3):491–498. [PMC free article] [PubMed] [Google Scholar]
- 114.Kan SH, Elanko N, Johnson D, Cornejo-Roldan L, Cook J, Reich EW, Tomkins S, Verloes A, Twigg SR, Rannan-Eliya S, McDonald-McGinn DM, Zackai EH, Wall SA, Muenke M, Wilkie AO. Genomic screening of fibroblast growth-factor receptor 2 reveals a wide spectrum of mutations in patients with syndromic craniosynostosis. Am J Hum Genet. 2002;70(2):472–486. doi: 10.1086/338758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, Nicoletti R, Winckler W, Grewal R, Hanna M, Wyhs N, Ziaugra L, Richter DJ, Trovik J, Engelsen IB, Stefansson IM, Fennell T, Cibulskis K, Zody MC, Akslen LA, Gabriel S, Wong KK, Sellers WR, Meyerson M, Greulich H. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci USA. 2008;105(25):8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pollock PM, Gartside MG, Dejeza LC, Powell MA, Mallon MA, Davies H, Mohammadi M, Futreal PA, Stratton MR, Trent JM, Goodfellow PJ. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26(50):7158–7162. doi: 10.1038/sj.onc.1210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu T, Willmore-Payne C, Wallander ML, Layfield LJ. Utilization of unlabeled probes for the detection of fibroblast growth factor receptor 2 exons 7 and 12 mutations in endometrial carcinoma. Appl Immunohistochem Mol Morphol. 2011;19(4):341–346. doi: 10.1097/PAI.0b013e318201dae8. [DOI] [PubMed] [Google Scholar]
- 118.Pandith AA, Shah ZA, Siddiqi MA. Oncogenic role of fibroblast growth factor receptor 3 in tumorigenesis of urinary bladder cancer. Urol Oncol. 2010 doi: 10.1016/j.urolonc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 119.Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23(1):18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 120.Hernandez S, Lopez-Knowles E, Lloreta J, Kogevinas M, Amoros A, Tardon A, Carrato A, Serra C, Malats N, Real FX. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006;24(22):3664–3671. doi: 10.1200/JCO.2005.05.1771. [DOI] [PubMed] [Google Scholar]
- 121.d’Avis PY, Robertson SC, Meyer AN, Bardwell WM, Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by mutations responsible for the lethal skeletal dysplasia thanatophoric dysplasia type I. Cell Growth Differ. 1998;9(1):71–78. [PubMed] [Google Scholar]
- 122.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13(2):233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 123.Adar R, Monsonego-Ornan E, David P, Yayon A. Differential activation of cysteine-substitution mutants of fibroblast growth factor receptor 3 is determined by cysteine localization. J Bone Miner Res. 2002;17(5):860–868. doi: 10.1359/jbmr.2002.17.5.860. [DOI] [PubMed] [Google Scholar]
- 124.Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, Knyazeva T, Muller S, Gartner S, Sures I, Wang H, Imyanitov E, Haring HU, Knayzev P, Iacobelli S, Hofler H, Ullrich A. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62(3):840–847. [PubMed] [Google Scholar]
- 125.Wang J, Yu W, Cai Y, Ren C, Ittmann MM. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia. 2008;10(8):847–856. doi: 10.1593/neo.08450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sugiyama N, Varjosalo M, Meller P, Lohi J, Chan KM, Zhou Z, Alitalo K, Taipale J, Keski-Oja J, Lehti K. FGF receptor-4 (FGFR4) polymorphism acts as an activity switch of a membrane type 1 matrix metalloproteinase-FGFR4 complex. Proc Natl Acad Sci USA. 2010;107(36):15786–15791. doi: 10.1073/pnas.0914459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U, Dragani TA. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23(29):7307–7311. doi: 10.1200/JCO.2005.17.350. [DOI] [PubMed] [Google Scholar]
- 128.Spinola M, Leoni VP, Tanuma J, Pettinicchio A, Frattini M, Signoroni S, Agresti R, Giovanazzi R, Pilotti S, Bertario L, Ravagnani F, Dragani TA. FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol Rep. 2005;14(2):415–419. [PubMed] [Google Scholar]
- 129.Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, Ulm K, Kiechle M, Hoefler H, Ullrich A, Harbeck N. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol. 2006;24(23):3747–3755. doi: 10.1200/JCO.2005.04.8587. [DOI] [PubMed] [Google Scholar]
- 130.Sugiyama N, Varjosalo M, Meller P, Lohi J, Hyytiainen M, Kilpinen S, Kallioniemi O, Ingvarsen S, Engelholm LH, Taipale J, Alitalo K, Keski-Oja J, Lehti K. Fibroblast growth factor receptor 4 regulates tumor invasion by coupling fibroblast growth factor signaling to extracellular matrix degradation. Cancer Res. 2010;70(20):7851–7861. doi: 10.1158/0008-5472.CAN-10-1223. [DOI] [PubMed] [Google Scholar]
- 131.Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordestgaard BG, Axelsson CK, Arias JI, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Zamora P, Brauch H, Justenhoven C, Hamann U, Ko YD, Bruening T, Haas S, Dork T, Schurmann P, Hillemanns P, Bogdanova N, Bremer M, Karstens JH, Fagerholm R, Aaltonen K, Aittomaki K, von Smitten K, Blomqvist C, Mannermaa A, Uusitupa M, Eskelinen M, Tengstrom M, Kosma VM, Kataja V, Chenevix-Trench G, Spurdle AB, Beesley J, Chen X, Devilee P, van Asperen CJ, Jacobi CE, Tollenaar RA, Huijts PE, Klijn JG, Chang-Claude J, Kropp S, Slanger T, Flesch-Janys D, Mutschelknauss E, Salazar R, Wang-Gohrke S, Couch F, Goode EL, Olson JE, Vachon C, Fredericksen ZS, Giles GG, Baglietto L, Severi G, Hopper JL, English DR, Southey MC, Haiman CA, Henderson BE, Kolonel LN, Le Marchand L, Stram DO, Hunter DJ, Hankinson SE, Cox DG, Tamimi R, Kraft P, Sherman ME, Chanock SJ, Lissowska J, Brinton LA, Peplonska B, Hooning MJ, Meijers-Heijboer H, Collee JM, van den Ouweland A, Uitterlinden AG, Liu J, Lin LY, Yuqing L, Humphreys K, Czene K, Cox A, Balasubramanian SP, Cross SS, Reed MW, Blows F, Driver K, Dunning A, Tyrer J, Ponder BA, Sangrajrang S, Brennan P, McKay J, Odefrey F, Gabrieau V, Sigurdson A, Doody M, Struewing JP, Alexander B, Easton DF, Pharoah PD. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4(4):e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Roumiantsev S, Krause DS, Neumann CA, Dimitri CA, Asiedu F, Cross NC, Van Etten RA. Distinct stem cell myeloproliferative/T lymphoma syndromes induced by ZNF198-FGFR1 and BCR-FGFR1 fusion genes from 8p11 translocations. Cancer Cell. 2004;5(3):287–298. doi: 10.1016/s1535-6108(04)00053-4. [DOI] [PubMed] [Google Scholar]
- 133.Guasch G, Delaval B, Arnoulet C, Xie MJ, Xerri L, Sainty D, Birnbaum D, Pebusque MJ. FOP-FGFR1 tyrosine kinase, the product of a t(6;8) translocation, induces a fatal myeloproliferative disease in mice. Blood. 2004;103(1):309–312. doi: 10.1182/blood-2003-05-1690. [DOI] [PubMed] [Google Scholar]
- 134.Chen J, Deangelo DJ, Kutok JL, Williams IR, Lee BH, Wadleigh M, Duclos N, Cohen S, Adelsperger J, Okabe R, Coburn A, Galinsky I, Huntly B, Cohen PS, Meyer T, Fabbro D, Roesel J, Banerji L, Griffin JD, Xiao S, Fletcher JA, Stone RM, Gilliland DG. PKC412 inhibits the zinc finger 198-fibroblast growth factor receptor 1 fusion tyrosine kinase and is active in treatment of stem cell myeloproliferative disorder. Proc Natl Acad Sci USA. 2004;101(40):14479–14484. doi: 10.1073/pnas.0404438101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ren M, Li X, Cowell JK. Genetic fingerprinting of the development and progression of T-cell lymphoma in a murine model of atypical myeloproliferative disorder initiated by the ZNF198-fibroblast growth factor receptor-1 chimeric tyrosine kinase. Blood. 2009;114(8):1576–1584. doi: 10.1182/blood-2009-03-212704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Demiroglu A, Steer EJ, Heath C, Taylor K, Bentley M, Allen SL, Koduru P, Brody JP, Hawson G, Rodwell R, Doody ML, Carnicero F, Reiter A, Goldman JM, Melo JV, Cross NC. The t(8;22) in chronic myeloid leukemia fuses BCR to FGFR1: transforming activity and specific inhibition of FGFR1 fusion proteins. Blood. 2001;98(13):3778–3783. doi: 10.1182/blood.v98.13.3778. [DOI] [PubMed] [Google Scholar]
- 137.Onwuazor ON, Wen XY, Wang DY, Zhuang L, Masih-Khan E, Claudio J, Barlogie B, Shaughnessy JD, Jr, Stewart AK. Mutation, SNP, and isoform analysis of fibroblast growth factor receptor 3 (FGFR3) in 150 newly diagnosed multiple myeloma patients. Blood. 2003;102(2):772–773. doi: 10.1182/blood-2003-04-1204. [DOI] [PubMed] [Google Scholar]
- 138.Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q, Arceci RJ, Matsui W, Park BH. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111(2):856–864. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 140.Wang Y, Becker D. Antisense targeting of basic fibroblast growth factor and fibroblast growth factor receptor-1 in human melanomas blocks intratumoral angiogenesis and tumor growth. Nat Med. 1997;3(8):887–893. doi: 10.1038/nm0897-887. [DOI] [PubMed] [Google Scholar]
- 141.Sharpe R, Pearson A, Herrera-Abreu MT, Johnson D, Mackay A, Welti JC, Natrajan R, Reynolds AR, Reis-Filho JS, Ashworth A, Turner NC. FGFR signaling promotes the growth of triple-negative and basal-like breast cancer cell lines both in vitro and in vivo. Clin Cancer Res. 2011;17(16):5275–5286. doi: 10.1158/1078-0432.CCR-10-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, McDermott LA, Coldren CD, Nemenoff RA, Merrick DT, Helfrich BA, Bunn PA, Jr, Heasley LE. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75(1):196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Memarzadeh S, Xin L, Mulholland DJ, Mansukhani A, Wu H, Teitell MA, Witte ON. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12(6):572–585. doi: 10.1016/j.ccr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA. 2010;107(50):21737–21742. doi: 10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zisman-Rozen S, Fink D, Ben-Izhak O, Fuchs Y, Brodski A, Kraus MH, Bejar J, Ron D. Downregulation of Sef, an inhibitor of receptor tyrosine kinase signaling, is common to a variety of human carcinomas. Oncogene. 2007;26(41):6093–6098. doi: 10.1038/sj.onc.1210424. [DOI] [PubMed] [Google Scholar]
- 146.Darby S, Murphy T, Thomas H, Robson CN, Leung HY, Mathers ME, Gnanapragasam VJ. Similar expression to FGF (Sef) inhibits fibroblast growth factor-induced tumourigenic behaviour in prostate cancer cells and is downregulated in aggressive clinical disease. Br J Cancer. 2009;101(11):1891–1899. doi: 10.1038/sj.bjc.6605379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fritzsche S, Kenzelmann M, Hoffmann MJ, Muller M, Engers R, Grone HJ, Schulz WA. Concomitant down-regulation of SPRY1 and SPRY2 in prostate carcinoma. Endocr Relat Cancer. 2006;13(3):839–849. doi: 10.1677/erc.1.01190. [DOI] [PubMed] [Google Scholar]
- 148.Murphy T, Darby S, Mathers ME, Gnanapragasam VJ. Evidence for distinct alterations in the FGF axis in prostate cancer progression to an aggressive clinical phenotype. J Pathol. 2010;220(4):452–460. doi: 10.1002/path.2657. [DOI] [PubMed] [Google Scholar]
- 149.Abella JV, Park M. Breakdown of endocytosis in the oncogenic activation of receptor tyrosine kinases. Am J Physiol Endocrinol Metab. 2009;296(5):E973–E984. doi: 10.1152/ajpendo.90857.2008. [DOI] [PubMed] [Google Scholar]
- 150.Haglund K, Rusten TE, Stenmark H. Aberrant receptor signaling and trafficking as mechanisms in oncogenesis. Crit Rev Oncog. 2007;13(1):39–74. doi: 10.1615/critrevoncog.v13.i1.20. [DOI] [PubMed] [Google Scholar]
- 151.Cho JY, Guo C, Torello M, Lunstrum GP, Iwata T, Deng C, Horton WA. Defective lysosomal targeting of activated fibroblast growth factor receptor 3 in achondroplasia. Proc Natl Acad Sci USA. 2004;101(2):609–614. doi: 10.1073/pnas.2237184100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cha JY, Maddileti S, Mitin N, Harden TK, Der CJ. Aberrant receptor internalization and enhanced FRS2-dependent signaling contribute to the transforming activity of the fibroblast growth factor receptor 2 IIIb C3 isoform. J Biol Chem. 2009;284(10):6227–6240. doi: 10.1074/jbc.M803998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Matsuda Y, Yamamoto T, Kudo M, Kawahara K, Kawamoto M, Nakajima Y, Koizumi K, Nakazawa N, Ishiwata T, Naito Z. Expression and roles of lumican in lung adenocarcinoma and squamous cell carcinoma. Int J Oncol. 2008;33(6):1177–1185. [PubMed] [Google Scholar]
- 154.Cho K, Ishiwata T, Uchida E, Nakazawa N, Korc M, Naito Z, Tajiri T. Enhanced expression of keratinocyte growth factor and its receptor correlates with venous invasion in pancreatic cancer. Am J Pathol. 2007;170(6):1964–1974. doi: 10.2353/ajpath.2007.060935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nomura S, Yoshitomi H, Takano S, Shida T, Kobayashi S, Ohtsuka M, Kimura F, Shimizu H, Yoshidome H, Kato A, Miyazaki M. FGF10/FGFR2 signal induces cell migration and invasion in pancreatic cancer. Br J Cancer. 2008;99(2):305–313. doi: 10.1038/sj.bjc.6604473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Matsunobu T, Ishiwata T, Yoshino M, Watanabe M, Kudo M, Matsumoto K, Tokunaga A, Tajiri T, Naito Z. Expression of keratinocyte growth factor receptor correlates with expansive growth and early stage of gastric cancer. Int J Oncol. 2006;28(2):307–314. [PubMed] [Google Scholar]
- 157.Tomlinson DC, L’Hote CG, Kennedy W, Pitt E, Knowles MA. Alternative splicing of fibroblast growth factor receptor 3 produces a secreted isoform that inhibits fibroblast growth factor-induced proliferation and is repressed in urothelial carcinoma cell lines. Cancer Res. 2005;65(22):10441–10449. doi: 10.1158/0008-5472.CAN-05-1718. [DOI] [PubMed] [Google Scholar]
- 158.Carstens RP, Eaton JV, Krigman HR, Walther PJ, Garcia-Blanco MA. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene. 1997;15(25):3059–3065. doi: 10.1038/sj.onc.1201498. [DOI] [PubMed] [Google Scholar]
- 159.Kawase T, Matsuo K, Suzuki T, Hiraki A, Watanabe M, Iwata H, Tanaka H, Tajima K. FGFR2 intronic polymorphisms interact with reproductive risk factors of breast cancer: results of a case control study in Japan. Int J Cancer. 2009;125(8):1946–1952. [Google Scholar]
- 160.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64(21):7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 161.Ezzat S, Zheng L, Zhu XF, Wu GE, Asa SL. Targeted expression of a human pituitary tumor-derived isoform of FGF receptor-4 recapitulates pituitary tumorigenesis. J Clin Investig. 2002;109(1):69–78. doi: 10.1172/JCI14036. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 162.Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2013 doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 163.Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 2013;3(3):264–279. doi: 10.1158/2159-8290.CD-12-0362. [DOI] [PubMed] [Google Scholar]
- 164.Shiang CY, Qi Y, Wang B, Lazar V, Wang J, Fraser Symmans W, Hortobagyi GN, Andre F, Pusztai L. Amplification of fibroblast growth factor receptor-1 in breast cancer and the effects of brivanib alaninate. Breast Cancer Res Treat. 2010;123(3):747–755. doi: 10.1007/s10549-009-0677-6. [DOI] [PubMed] [Google Scholar]
- 165.Daniele G, Corral J, Molife LR, de Bono JS. FGF receptor inhibitors: role in cancer therapy. Curr Oncol Rep. 2012;14(2):111–119. doi: 10.1007/s11912-012-0225-0. [DOI] [PubMed] [Google Scholar]
- 166.Konecny GE, Kolarova T, O’Brien NA, Winterhoff B, Yang G, Qi J, Qi Z, Venkatesan N, Ayala R, Luo T, Finn RS, Kristof J, Galderisi C, Porta DG, Anderson L, Shi MM, Yovine A, Slamon DJ. Activity of the fibroblast growth factor receptor inhibitors dovitinib (TKI258) and NVP-BGJ398 in human endometrial cancer cells. Mol Cancer Ther. 2013;12(5):632–642. doi: 10.1158/1535-7163.MCT-12-0999. [DOI] [PubMed] [Google Scholar]
- 167.Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, Rooney C, Coleman T, Baker D, Mellor MJ, Brooks AN, Klinowska T. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72(8):2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 168.Zhang J, Zhang L, Su X, Li M, Xie L, Malchers F, Fan S, Yin X, Xu Y, Liu K, Dong Z, Zhu G, Qian Z, Tang L, Zhan P, Ji Q, Kilgour E, Smith PD, Brooks AN, Thomas RK, Gavine PR. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18(24):6658–6667. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 169.Shimada N, Ishii T, Imada T, Takaba K, Sasaki Y, Maruyama-Takahashi K, Maekawa-Tokuda Y, Kusaka H, Akinaga S, Tanaka A, Shitara K. A neutralizing anti-fibroblast growth factor 8 monoclonal antibody shows potent antitumor activity against androgen-dependent mouse mammary tumors in vivo. Clin Cancer Res. 2005;11(10):3897–3904. doi: 10.1158/1078-0432.CCR-04-2358. [DOI] [PubMed] [Google Scholar]
- 170.Maruyama-Takahashi K, Shimada N, Imada T, Maekawa-Tokuda Y, Ishii T, Ouchi J, Kusaka H, Miyaji H, Akinaga S, Tanaka A, Shitara K. A neutralizing anti-fibroblast growth factor (FGF) 8 monoclonal antibody shows anti-tumor activity against FGF8b-expressing LNCaP xenografts in androgen-dependent and -independent conditions. Prostate. 2008;68(6):640–650. doi: 10.1002/pros.20728. [DOI] [PubMed] [Google Scholar]
- 171.I. Miki TT, M. Uchii, T. Suda, A. Tanaka (2008) An anti-FGF8 antibody reduced joint inflammation and bone destruction in Rat adjuvant-induced arthritis and mouse type II collagen-induced arthritis. In: American College of Rheumatology 2008 annual scientific meeting
- 172.Qing J, Du X, Chen Y, Chan P, Li H, Wu P, Marsters S, Stawicki S, Tien J, Totpal K, Ross S, Stinson S, Dornan D, French D, Wang QR, Stephan JP, Wu Y, Wiesmann C, Ashkenazi A. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009;119(5):1216–1229. doi: 10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sun HD, Malabunga M, Tonra JR, DiRenzo R, Carrick FE, Zheng H, Berthoud HR, McGuinness OP, Shen J, Bohlen P, Leibel RL, Kussie P. Monoclonal antibody antagonists of hypothalamic FGFR1 cause potent but reversible hypophagia and weight loss in rodents and monkeys. Am J Physiol Endocrinol Metab. 2007;292(3):E964–E976. doi: 10.1152/ajpendo.00089.2006. [DOI] [PubMed] [Google Scholar]
- 174.Trudel S, Stewart AK, Rom E, Wei E, Li ZH, Kotzer S, Chumakov I, Singer Y, Chang H, Liang SB, Yayon A. The inhibitory anti-FGFR3 antibody, PRO-001, is cytotoxic to t(4;14) multiple myeloma cells. Blood. 2006;107(10):4039–4046. doi: 10.1182/blood-2005-10-4179. [DOI] [PubMed] [Google Scholar]
- 175.Hadari Y, Schlessinger J. FGFR3-targeted mAb therapy for bladder cancer and multiple myeloma. J Clin Invest. 2009;119(5):1077–1079. doi: 10.1172/JCI38948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, Ross J, Carano R, D’Souza A, Qing J, Mohtashemi I, Ashkenazi A, French DM. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27(1):85–97. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- 177.Allen E, Walters IB, Hanahan D. Brivanib, a dual FGF/VEGF inhibitor, is active both first and second line against mouse pancreatic neuroendocrine tumors developing adaptive/evasive resistance to VEGF inhibition. Clin Cancer Res. 2011;17(16):5299–5310. doi: 10.1158/1078-0432.CCR-10-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Harding TC, Long L, Palencia S, Zhang H, Sadra A, Hestir K, Patil N, Levin A, Hsu AW, Charych D, Brennan T, Zanghi J, Halenbeck R, Marshall SA, Qin M, Doberstein SK, Hollenbaugh D, Kavanaugh WM, Williams LT, Baker KP. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Sci Transl Med. 2013;5(178):178ra39. doi: 10.1126/scitranslmed.3005414. [DOI] [PubMed] [Google Scholar]
- 179.Marshall ME, Hinz TK, Kono SA, Singleton KR, Bichon B, Ware KE, Marek L, Frederick BA, Raben D, Heasley LE. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2011;17(15):5016–5025. doi: 10.1158/1078-0432.CCR-11-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Katoh Y, Katoh M. FGFR2-related pathogenesis and FGFR2-targeted therapeutics (Review) Int J Mol Med. 2009;23(3):307–311. doi: 10.3892/ijmm_00000132. [DOI] [PubMed] [Google Scholar]
- 181.Brown AP, Courtney CL, King LM, Groom SC, Graziano MJ. Cartilage dysplasia and tissue mineralization in the rat following administration of a FGF receptor tyrosine kinase inhibitor. Toxicol Pathol. 2005;33(4):449–455. doi: 10.1080/01926230590961845. [DOI] [PubMed] [Google Scholar]
- 182.Wohrle S, Bonny O, Beluch N, Gaulis S, Stamm C, Scheibler M, Muller M, Kinzel B, Thuery A, Brueggen J, Hynes NE, Sellers WR, Hofmann F, Graus-Porta D. FGF receptors control vitamin D and phosphate homeostasis by mediating renal FGF-23 signaling and regulating FGF-23 expression in bone. J Bone Miner Res. 2011;26(10):2486–2497. doi: 10.1002/jbmr.478. [DOI] [PubMed] [Google Scholar]
- 183.Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, Fujita T, Fukumoto S, Yamashita T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444(7120):770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 184.Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, Kuro-o M. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem. 2006;281(10):6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu S, Vierthaler L, Tang W, Zhou J, Quarles LD. FGFR3 and FGFR4 do not mediate renal effects of FGF23. J Am Soc Nephrol JASN. 2008;19(12):2342–2350. doi: 10.1681/ASN.2007121301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gattineni J, Twombley K, Goetz R, Mohammadi M, Baum M. Regulation of serum 1,25(OH)2 vitamin D3 levels by fibroblast growth factor 23 is mediated by FGF receptors 3 and 4. Am J Physiol Renal physiol. 2011;301(2):F371–F377. doi: 10.1152/ajprenal.00740.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gattineni J, Bates C, Twombley K, Dwarakanath V, Robinson ML, Goetz R, Mohammadi M, Baum M. FGF23 decreases renal NaPi-2a and NaPi-2c expression and induces hypophosphatemia in vivo predominantly via FGF receptor 1. Am J Physiol Renal physiol. 2009;297(2):F282–F291. doi: 10.1152/ajprenal.90742.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ibrahimi OA, Zhang F, Eliseenkova AV, Itoh N, Linhardt RJ, Mohammadi M. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Hum Mol Genet. 2004;13(19):2313–2324. doi: 10.1093/hmg/ddh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.MacArthur CA, Lawshe A, Xu J, Santos-Ocampo S, Heikinheimo M, Chellaiah AT, Ornitz DM. FGF-8 isoforms activate receptor splice forms that are expressed in mesenchymal regions of mouse development. Development. 1995;121(11):3603–3613. doi: 10.1242/dev.121.11.3603. [DOI] [PubMed] [Google Scholar]
- 190.Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20(2):185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Miralles F, Czernichow P, Ozaki K, Itoh N, Scharfmann R. Signaling through fibroblast growth factor receptor 2b plays a key role in the development of the exocrine pancreas. Proc Natl Acad Sci USA. 1999;96(11):6267–6272. doi: 10.1073/pnas.96.11.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Igarashi M, Finch PW, Aaronson SA. Characterization of recombinant human fibroblast growth factor (FGF)-10 reveals functional similarities with keratinocyte growth factor (FGF-7) J Biol Chem. 1998;273(21):13230–13235. doi: 10.1074/jbc.273.21.13230. [DOI] [PubMed] [Google Scholar]
- 193.Hecht D, Zimmerman N, Bedford M, Avivi A, Yayon A. Identification of fibroblast growth factor 9 (FGF9) as a high affinity, heparin dependent ligand for FGF receptors 3 and 2 but not for FGF receptors 1 and 4. Growth Factors. 1995;12(3):223–233. doi: 10.3109/08977199509036882. [DOI] [PubMed] [Google Scholar]
- 194.Santos-Ocampo S, Colvin JS, Chellaiah A, Ornitz DM. Expression and biological activity of mouse fibroblast growth factor-9. J Biol Chem. 1996;271(3):1726–1731. doi: 10.1074/jbc.271.3.1726. [DOI] [PubMed] [Google Scholar]
- 195.Fortin D, Rom E, Sun H, Yayon A, Bansal R. Distinct fibroblast growth factor (FGF)/FGF receptor signaling pairs initiate diverse cellular responses in the oligodendrocyte lineage. J Neurosci. 2005;25(32):7470–7479. doi: 10.1523/JNEUROSCI.2120-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xu J, Lawshe A, MacArthur CA, Ornitz DM. Genomic structure, mapping, activity and expression of fibroblast growth factor 17. Mech Dev. 1999;83(1–2):165–178. doi: 10.1016/s0925-4773(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 197.Nezu M, Tomonaga T, Sakai C, Ishii A, Itoga S, Nishimura M, Matsuo Y, Tagawa M, Nomura F. Expression of the fetal-oncogenic fibroblast growth factor-8/17/18 subfamily in human hematopoietic tumors. Biochem Biophys Res Commun. 2005;335(3):843–849. doi: 10.1016/j.bbrc.2005.07.153. [DOI] [PubMed] [Google Scholar]
- 198.Ohmachi S, Mikami T, Konishi M, Miyake A, Itoh N. Preferential neurotrophic activity of fibroblast growth factor-20 for dopaminergic neurons through fibroblast growth factor receptor-1c. J Neurosci Res. 2003;72(4):436–443. doi: 10.1002/jnr.10592. [DOI] [PubMed] [Google Scholar]
- 199.Ren G, Yin J, Wang W, Li L, Li D. Fibroblast growth factor (FGF)-21 signals through both FGF receptor-1 and 2. Sci China Life Sci. 2010;53(8):1000–1008. doi: 10.1007/s11427-010-4035-z. [DOI] [PubMed] [Google Scholar]
- 200.Wu AL, Coulter S, Liddle C, Wong A, Eastham-Anderson J, French DM, Peterson AS, Sonoda J. FGF19 regulates cell proliferation, glucose and bile acid metabolism via FGFR4-dependent and independent pathways. PLoS ONE. 2011;6(3):e17868. doi: 10.1371/journal.pone.0017868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, Mohammadi M, Rosenblatt KP, Kliewer SA, Kuro-o M. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem. 2007;282(37):26687–26695. doi: 10.1074/jbc.M704165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gorringe KL, Jacobs S, Thompson ER, Sridhar A, Qiu W, Choong DY, Campbell IG. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res. 2007;13(16):4731–4739. doi: 10.1158/1078-0432.CCR-07-0502. [DOI] [PubMed] [Google Scholar]
- 203.Simon R, Richter J, Wagner U, Fijan A, Bruderer J, Schmid U, Ackermann D, Maurer R, Alund G, Knonagel H, Rist M, Wilber K, Anabitarte M, Hering F, Hardmeier T, Schonenberger A, Flury R, Jager P, Fehr JL, Schraml P, Moch H, Mihatsch MJ, Gasser T, Sauter G. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res. 2001;61(11):4514–4519. [PubMed] [Google Scholar]
- 204.Missiaglia E, Selfe J, Hamdi M, Williamson D, Schaaf G, Fang C, Koster J, Summersgill B, Messahel B, Versteeg R, Pritchard-Jones K, Kool M, Shipley J. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: an approach to identify candidate genes involved in tumor development. Genes Chromosom Cancer. 2009;48(6):455–467. doi: 10.1002/gcc.20655. [DOI] [PubMed] [Google Scholar]
- 205.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, Chande A, Tanaka KE, Stransky N, Greulich H, Gray NS, Meyerson M. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6(6):e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Turner NC, Seckl MJ. A therapeutic target for smoking-associated lung cancer. Sci Transl Med. 2010;2(62):62ps56. doi: 10.1126/scitranslmed.3001942. [DOI] [PubMed] [Google Scholar]
- 207.Takeda M, Arao T, Yokote H, Komatsu T, Yanagihara K, Sasaki H, Yamada Y, Tamura T, Fukuoka K, Kimura H, Saijo N, Nishio K. AZD2171 shows potent antitumor activity against gastric cancer over-expressing fibroblast growth factor receptor 2/keratinocyte growth factor receptor. Clin Cancer Res. 2007;13(10):3051–3057. doi: 10.1158/1078-0432.CCR-06-2743. [DOI] [PubMed] [Google Scholar]
- 208.Tannheimer SL, Rehemtulla A, Ethier SP. Characterization of fibroblast growth factor receptor 2 overexpression in the human breast cancer cell line SUM-52PE. Breast Cancer Res. 2000;2(4):311–320. doi: 10.1186/bcr73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Matsumoto K, Arao T, Hamaguchi T, Shimada Y, Kato K, Oda I, Taniguchi H, Koizumi F, Yanagihara K, Sasaki H, Nishio K, Yamada Y. FGFR2 gene amplification and clinicopathological features in gastric cancer. Br J Cancer. 2012;106(4):727–732. doi: 10.1038/bjc.2011.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Azuma K, Tsurutani J, Sakai K, Kaneda H, Fujisaka Y, Takeda M, Watatani M, Arao T, Satoh T, Okamoto I, Kurata T, Nishio K, Nakagawa K. Switching addictions between HER2 and FGFR2 in HER2-positive breast tumor cells: FGFR2 as a potential target for salvage after lapatinib failure. Biochem Biophys Res Commun. 2011;407(1):219–224. doi: 10.1016/j.bbrc.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 211.Kato H, Arao T, Matsumoto K, Fujita Y, Kimura H, Hayashi H, Nishiki K, Iwama M, Shiraishi O, Yasuda A, Shinkai M, Imano M, Imamoto H, Yasuda T, Okuno K, Shiozaki H, Nishio K. Gene amplification of EGFR, HER2, FGFR2 and MET in esophageal squamous cell carcinoma. Int J Oncol. 2013;42(4):1151–1158. doi: 10.3892/ijo.2013.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nord H, Segersten U, Sandgren J, Wester K, Busch C, Menzel U, Komorowski J, Dumanski JP, Malmstrom PU, Diaz de Stahl T. Focal amplifications are associated with high grade and recurrences in stage Ta bladder carcinoma. Int J Cancer. 2010;126(6):1390–1402. doi: 10.1002/ijc.24954. [DOI] [PubMed] [Google Scholar]
- 213.Vekony H, Ylstra B, Wilting SM, Meijer GA, van de Wiel MA, Leemans CR, van der Waal I, Bloemena E. DNA copy number gains at loci of growth factors and their receptors in salivary gland adenoid cystic carcinoma. Clin Cancer Res. 2007;13(11):3133–3139. doi: 10.1158/1078-0432.CCR-06-2555. [DOI] [PubMed] [Google Scholar]
- 214.Shin EY, Lee BH, Yang JH, Shin KS, Lee GK, Yun HY, Song YJ, Park SC, Kim EG. Up-regulation and co-expression of fibroblast growth factor receptors in human gastric cancer. J Cancer Res Clin Oncol. 2000;126(9):519–528. doi: 10.1007/s004320000128. [DOI] [PubMed] [Google Scholar]
- 215.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A, Moch H, Wagener P, Fischer F, Heynck S, Koker M, Schottle J, Leenders F, Gabler F, Dabow I, Querings S, Heukamp LC, Balke-Want H, Ansen S, Rauh D, Baessmann I, Altmuller J, Wainer Z, Conron M, Wright G, Russell P, Solomon B, Brambilla E, Brambilla C, Lorimier P, Sollberg S, Brustugun OT, Engel-Riedel W, Ludwig C, Petersen I, Sanger J, Clement J, Groen H, Timens W, Sietsma H, Thunnissen E, Smit E, Heideman D, Cappuzzo F, Ligorio C, Damiani S, Hallek M, Beroukhim R, Pao W, Klebl B, Baumann M, Buettner R, Ernestus K, Stoelben E, Wolf J, Nurnberg P, Perner S, Thomas RK. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ruhe JE, Streit S, Hart S, Wong CH, Specht K, Knyazev P, Knyazeva T, Tay LS, Loo HL, Foo P, Wong W, Pok S, Lim SJ, Ong H, Luo M, Ho HK, Peng K, Lee TC, Bezler M, Mann C, Gaertner S, Hoefler H, Iacobelli S, Peter S, Tay A, Brenner S, Venkatesh B, Ullrich A. Genetic alterations in the tyrosine kinase transcriptome of human cancer cell lines. Cancer Res. 2007;67(23):11368–11376. doi: 10.1158/0008-5472.CAN-07-2703. [DOI] [PubMed] [Google Scholar]
- 218.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, O’Meara S, Parker A, Tarpey P, Avis T, Barthorpe A, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh B, Yuen ST, Nicholson AG, Lakhani S, Easton DF, Weber BL, Stratton MR, Futreal PA, Wooster R. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37(6):590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 219.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, Hodi FS, Dranoff G, Sellers WR, Thomas RK, Meyerson M, Golub TR, Dummer R, Herlyn M, Getz G, Garraway LA. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68(3):664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, Parker A, O’Meara S, Avis T, Barthorpe S, Brackenbury L, Buck G, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh BT, Yuen ST, Lakhani SR, Easton DF, Weber BL, Goldstraw P, Nicholson AG, Wooster R, Stratton MR, Futreal PA. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65(17):7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 221.Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300(5621):949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 222.Rand V, Huang J, Stockwell T, Ferriera S, Buzko O, Levy S, Busam D, Li K, Edwards JB, Eberhart C, Murphy KM, Tsiamouri A, Beeson K, Simpson AJ, Venter JC, Riggins GJ, Strausberg RL. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci USA. 2005;102(40):14344–14349. doi: 10.1073/pnas.0507200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gartside MG, Chen H, Ibrahimi OA, Byron SA, Curtis AV, Wellens CL, Bengston A, Yudt LM, Eliseenkova AV, Ma J, Curtin JA, Hyder P, Harper UL, Riedesel E, Mann GJ, Trent JM, Bastian BC, Meltzer PS, Mohammadi M, Pollock PM. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Mol Cancer Res. 2009;7(1):41–54. doi: 10.1158/1541-7786.MCR-08-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Byron SA, Gartside MG, Wellens CL, Mallon MA, Keenan JB, Powell MA, Goodfellow PJ, Pollock PM. Inhibition of activated fibroblast growth factor receptor 2 in endometrial cancer cells induces cell death despite PTEN abrogation. Cancer Res. 2008;68(17):6902–6907. doi: 10.1158/0008-5472.CAN-08-0770. [DOI] [PubMed] [Google Scholar]
- 225.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 226.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The catalogue of somatic mutations in cancer (COSMIC) Curr Protoc Hum Genet. 2008;10:11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hansen RM, Goriely A, Wall SA, Roberts IS, Wilkie AO. Fibroblast growth factor receptor 2, gain-of-function mutations, and tumourigenesis: investigating a potential link. J Pathol. 2005;207(1):27–31. doi: 10.1002/path.1816. [DOI] [PubMed] [Google Scholar]
- 229.Byron SA, Gartside MG, Wellens CL, Goodfellow PJ, Birrer MJ, Campbell IG, Pollock PM. FGFR2 mutations are rare across histologic subtypes of ovarian cancer. Gynecol Oncol. 2010;117(1):125–129. doi: 10.1016/j.ygyno.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 230.Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001;61(9):3541–3543. [PubMed] [Google Scholar]
- 231.Claudio JO, Zhan F, Zhuang L, Khaja R, Zhu YX, Sivananthan K, Trudel S, Masih-Khan E, Fonseca R, Bergsagel PL, Scherer SW, Shaughnessy J, Stewart AK. Expression and mutation status of candidate kinases in multiple myeloma. Leukemia. 2007;21(5):1124–1127. doi: 10.1038/sj.leu.2404612. [DOI] [PubMed] [Google Scholar]
- 232.Soverini S, Terragna C, Testoni N, Ruggeri D, Tosi P, Zamagni E, Cellini C, Cavo M, Baccarani M, Tura S, Martinelli G. Novel mutation and RNA splice variant of fibroblast growth factor receptor 3 in multiple myeloma patients at diagnosis. Haematologica. 2002;87(10):1036–1040. [PubMed] [Google Scholar]
- 233.Intini D, Baldini L, Fabris S, Lombardi L, Ciceri G, Maiolo AT, Neri A. Analysis of FGFR3 gene mutations in multiple myeloma patients with t(4;14) Br J Haematol. 2001;114(2):362–364. doi: 10.1046/j.1365-2141.2001.02957.x. [DOI] [PubMed] [Google Scholar]
- 234.Richelda R, Ronchetti D, Baldini L, Cro L, Viggiano L, Marzella R, Rocchi M, Otsuki T, Lombardi L, Maiolo AT, Neri A. A novel chromosomal translocation t(4; 14)(p16.3; q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood. 1997;90(10):4062–4070. [PubMed] [Google Scholar]
- 235.Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, Bergsagel PL. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16(3):260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ronchetti D, Greco A, Compasso S, Colombo G, Dell’Era P, Otsuki T, Lombardi L, Neri A. Deregulated FGFR3 mutants in multiple myeloma cell lines with t(4;14): comparative analysis of Y373C, K650E and the novel G384D mutations. Oncogene. 2001;20(27):3553–3562. doi: 10.1038/sj.onc.1204465. [DOI] [PubMed] [Google Scholar]
- 237.Chesi M, Brents LA, Ely SA, Bais C, Robbiani DF, Mesri EA, Kuehl WM, Bergsagel PL. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97(3):729–736. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- 238.Reddy PL, Grewal RP. The G1138A mutation rate in the fibroblast growth factor receptor 3 (FGFR3) gene is increased in cells carrying the t (4; 14) translocation. Genet Mol Res. 2009;8(2):435–439. doi: 10.4238/vol8-2gmr564. [DOI] [PubMed] [Google Scholar]
- 239.Fracchiolla NS, Luminari S, Baldini L, Lombardi L, Maiolo AT, Neri A. FGFR3 gene mutations associated with human skeletal disorders occur rarely in multiple myeloma. Blood. 1998;92(8):2987–2989. [PubMed] [Google Scholar]
- 240.Tomlinson DC, Baldo O, Harnden P, Knowles MA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol. 2007;213(1):91–98. doi: 10.1002/path.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Serizawa RR, Ralfkiaer U, Steven K, Lam GW, Schmiedel S, Schuz J, Hansen AB, Horn T, Guldberg P. Integrated genetic and epigenetic analysis of bladder cancer reveals an additive diagnostic value of FGFR3 mutations and hypermethylation events. Int J Cancer. 2011;129(1):78–87. doi: 10.1002/ijc.25651. [DOI] [PubMed] [Google Scholar]
- 242.Kompier LC, Lurkin I, van der Aa MN, van Rhijn BW, van der Kwast TH, Zwarthoff EC. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One. 2010;5(11):e13821. doi: 10.1371/journal.pone.0013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zieger K, Dyrskjot L, Wiuf C, Jensen JL, Andersen CL, Jensen KM, Orntoft TF. Role of activating fibroblast growth factor receptor 3 mutations in the development of bladder tumors. Clin Cancer Res. 2005;11(21):7709–7719. doi: 10.1158/1078-0432.CCR-05-1130. [DOI] [PubMed] [Google Scholar]
- 244.van Oers JM, Lurkin I, van Exsel AJ, Nijsen Y, van Rhijn BW, van der Aa MN, Zwarthoff EC. A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clin Cancer Res. 2005;11(21):7743–7748. doi: 10.1158/1078-0432.CCR-05-1045. [DOI] [PubMed] [Google Scholar]
- 245.Hernandez S, Lopez-Knowles E, Lloreta J, Kogevinas M, Jaramillo R, Amoros A, Tardon A, Garcia-Closas R, Serra C, Carrato A, Malats N, Real FX. FGFR3 and Tp53 mutations in T1G3 transitional bladder carcinomas: independent distribution and lack of association with prognosis. Clin Cancer Res. 2005;11(15):5444–5450. doi: 10.1158/1078-0432.CCR-05-0122. [DOI] [PubMed] [Google Scholar]
- 246.Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24(33):5218–5225. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 247.van Rhijn BW, van der Kwast TH, Vis AN, Kirkels WJ, Boeve ER, Jobsis AC, Zwarthoff EC. FGFR3 and P53 characterize alternative genetic pathways in the pathogenesis of urothelial cell carcinoma. Cancer Res. 2004;64(6):1911–1914. doi: 10.1158/0008-5472.can-03-2421. [DOI] [PubMed] [Google Scholar]
- 248.Bakkar AA, Wallerand H, Radvanyi F, Lahaye JB, Pissard S, Lecerf L, Kouyoumdjian JC, Abbou CC, Pairon JC, Jaurand MC, Thiery JP, Chopin DK, de Medina SG. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer Res. 2003;63(23):8108–8112. [PubMed] [Google Scholar]
- 249.Rieger-Christ KM, Mourtzinos A, Lee PJ, Zagha RM, Cain J, Silverman M, Libertino JA, Summerhayes IC. Identification of fibroblast growth factor receptor 3 mutations in urine sediment DNA samples complements cytology in bladder tumor detection. Cancer. 2003;98(4):737–744. doi: 10.1002/cncr.11536. [DOI] [PubMed] [Google Scholar]
- 250.van Rhijn BW, Vis AN, van der Kwast TH, Kirkels WJ, Radvanyi F, Ooms EC, Chopin DK, Boeve ER, Jobsis AC, Zwarthoff EC. Molecular grading of urothelial cell carcinoma with fibroblast growth factor receptor 3 and MIB-1 is superior to pathologic grade for the prediction of clinical outcome. J Clin Oncol. 2003;21(10):1912–1921. doi: 10.1200/JCO.2003.05.073. [DOI] [PubMed] [Google Scholar]
- 251.Chou A, Dekker N, Jordan RC. Identification of novel fibroblast growth factor receptor 3 gene mutations in actinic cheilitis and squamous cell carcinoma of the lip. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107(4):535–541. doi: 10.1016/j.tripleo.2008.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shotelersuk V, Ittiwut C, Shotelersuk K, Triratanachat S, Poovorawan Y, Mutirangura A. Fibroblast growth factor receptor 3 S249C mutation in virus associated squamous cell carcinomas. Oncol Rep. 2001;8(6):1301–1304. doi: 10.3892/or.8.6.1301. [DOI] [PubMed] [Google Scholar]
- 253.Zhang Y, Hiraishi Y, Wang H, Sumi KS, Hayashido Y, Toratani S, Kan M, Sato JD, Okamoto T. Constitutive activating mutation of the FGFR3b in oral squamous cell carcinomas. Int J Cancer. 2005;117(1):166–168. doi: 10.1002/ijc.21145. [DOI] [PubMed] [Google Scholar]
- 254.Sibley K, Stern P, Knowles MA. Frequency of fibroblast growth factor receptor 3 mutations in sporadic tumours. Oncogene. 2001;20(32):4416–4418. doi: 10.1038/sj.onc.1204543. [DOI] [PubMed] [Google Scholar]
- 255.Wu R, Connolly D, Ngelangel C, Bosch FX, Munoz N, Cho KR. Somatic mutations of fibroblast growth factor receptor 3 (FGFR3) are uncommon in carcinomas of the uterine cervix. Oncogene. 2000;19(48):5543–5546. doi: 10.1038/sj.onc.1203934. [DOI] [PubMed] [Google Scholar]
- 256.Hernandez S, de Muga S, Agell L, Juanpere N, Esgueva R, Lorente JA, Mojal S, Serrano S, Lloreta J. FGFR3 mutations in prostate cancer: association with low-grade tumors. Mod Pathol. 2009;22(6):848–856. doi: 10.1038/modpathol.2009.46. [DOI] [PubMed] [Google Scholar]
- 257.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Goriely A, Hansen RM, Taylor IB, Olesen IA, Jacobsen GK, McGowan SJ, Pfeifer SP, McVean GA, Rajpert-De Meyts E, Wilkie AO. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat Genet. 2009;41(11):1247–1252. doi: 10.1038/ng.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Taylor JG, VI, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, Yu Y, Chen QR, Shah K, Youngblood V, Fang J, Kim SY, Yeung C, Helman LJ, Mendoza A, Ngo V, Staudt LM, Wei JS, Khanna C, Catchpoole D, Qualman SJ, Hewitt SM, Merlino G, Chanock SJ, Khan J. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119(11):3395–3407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Falvella FS, Frullanti E, Galvan A, Spinola M, Noci S, De Cecco L, Nosotti M, Santambrogio L, Incarbone M, Alloisio M, Calabro E, Pastorino U, Skaug V, Haugen A, Taioli E, Dragani TA. FGFR4 Gly388Arg polymorphism may affect the clinical stage of patients with lung cancer by modulating the transcriptional profile of normal lung. Int J Cancer. 2009;124(12):2880–2885. doi: 10.1002/ijc.24302. [DOI] [PubMed] [Google Scholar]
- 261.Sasaki H, Okuda K, Kawano O, Yukiue H, Yano M, Fujii Y. Fibroblast growth factor receptor 4 mutation and polymorphism in Japanese lung cancer. Oncol Rep. 2008;20(5):1125–1130. [PubMed] [Google Scholar]
- 262.Marks JL, McLellan MD, Zakowski MF, Lash AE, Kasai Y, Broderick S, Sarkaria IS, Pham D, Singh B, Miner TL, Fewell GA, Fulton LL, Mardis ER, Wilson RK, Kris MG, Rusch VW, Varmus H, Pao W. Mutational analysis of EGFR and related signaling pathway genes in lung adenocarcinomas identifies a novel somatic kinase domain mutation in FGFR4. PLoS One. 2007;2(5):e426. doi: 10.1371/journal.pone.0000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Xu W, Li Y, Wang X, Chen B, Wang Y, Liu S, Xu J, Zhao W, Wu J. FGFR4 transmembrane domain polymorphism and cancer risk: a meta-analysis including 8555 subjects. Eur J Cancer. 2010;46(18):3332–3338. doi: 10.1016/j.ejca.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 264.Ye Y, Shi Y, Zhou Y, Du C, Wang C, Zhan H, Zheng B, Cao X, Sun MH, Fu H. The fibroblast growth factor receptor-4 Arg388 allele is associated with gastric cancer progression. Ann Surg Oncol. 2010;17(12):3354–3361. doi: 10.1245/s10434-010-1323-6. [DOI] [PubMed] [Google Scholar]
- 265.Streit S, Mestel DS, Schmidt M, Ullrich A, Berking C. FGFR4 Arg388 allele correlates with tumour thickness and FGFR4 protein expression with survival of melanoma patients. Br J Cancer. 2006;94(12):1879–1886. doi: 10.1038/sj.bjc.6603181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ho HK, Pok S, Streit S, Ruhe JE, Hart S, Lim KS, Loo HL, Aung MO, Lim SG, Ullrich A. Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J Hepatol. 2009;50(1):118–127. doi: 10.1016/j.jhep.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 267.Morimoto Y, Ozaki T, Ouchida M, Umehara N, Ohata N, Yoshida A, Shimizu K, Inoue H. Single nucleotide polymorphism in fibroblast growth factor receptor 4 at codon 388 is associated with prognosis in high-grade soft tissue sarcoma. Cancer. 2003;98(10):2245–2250. doi: 10.1002/cncr.11778. [DOI] [PubMed] [Google Scholar]
- 268.Wang J, Stockton DW, Ittmann M. The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clin Cancer Res. 2004;10(18 Pt 1):6169–6178. doi: 10.1158/1078-0432.CCR-04-0408. [DOI] [PubMed] [Google Scholar]
- 269.Xu B, Tong N, Chen SQ, Hua LX, Wang ZJ, Zhang ZD, Chen M. FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC Cancer. 2011;11:84. doi: 10.1186/1471-2407-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ma Z, Tsuchiya N, Yuasa T, Inoue T, Kumazawa T, Narita S, Horikawa Y, Tsuruta H, Obara T, Saito M, Satoh S, Ogawa O, Habuchi T. Polymorphisms of fibroblast growth factor receptor 4 have association with the development of prostate cancer and benign prostatic hyperplasia and the progression of prostate cancer in a Japanese population. Int J Cancer. 2008;123(11):2574–2579. doi: 10.1002/ijc.23578. [DOI] [PubMed] [Google Scholar]
- 271.FitzGerald LM, Karlins E, Karyadi DM, Kwon EM, Koopmeiners JS, Stanford JL, Ostrander EA. Association of FGFR4 genetic polymorphisms with prostate cancer risk and prognosis. Prostate Cancer Prostatic Dis. 2009;12(2):192–197. doi: 10.1038/pcan.2008.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Streit S, Bange J, Fichtner A, Ihrler S, Issing W, Ullrich A. Involvement of the FGFR4 Arg388 allele in head and neck squamous cell carcinoma. Int J Cancer. 2004;111(2):213–217. doi: 10.1002/ijc.20204. [DOI] [PubMed] [Google Scholar]
- 273.da Costa Andrade VC, Parise O, Jr, Hors CP, de Melo Martins PC, Silva AP, Garicochea B. The fibroblast growth factor receptor 4 (FGFR4) Arg388 allele correlates with survival in head and neck squamous cell carcinoma. Exp Mol Pathol. 2007;82(1):53–57. doi: 10.1016/j.yexmp.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 274.Tanuma J, Izumo T, Hirano M, Oyazato Y, Hori F, Umemura E, Shisa H, Hiai H, Kitano M. FGFR4 polymorphism, TP53 mutation, and their combinations are prognostic factors for oral squamous cell carcinoma. Oncol Rep. 2010;23(3):739–744. [PubMed] [Google Scholar]
- 275.Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, Stone R, Weissman SM, Hudson TJ, Fletcher JA. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 1998;18(1):84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 276.Popovici C, Adelaide J, Ollendorff V, Chaffanet M, Guasch G, Jacrot M, Leroux D, Birnbaum D, Pebusque MJ. Fibroblast growth factor receptor 1 is fused to FIM in stem-cell myeloproliferative disorder with t(8;13) Proc Natl Acad Sci USA. 1998;95(10):5712–5717. doi: 10.1073/pnas.95.10.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Reiter A, Sohal J, Kulkarni S, Chase A, Macdonald DH, Aguiar RC, Goncalves C, Hernandez JM, Jennings BA, Goldman JM, Cross NC. Consistent fusion of ZNF198 to the fibroblast growth factor receptor-1 in the t(8;13)(p11;q12) myeloproliferative syndrome. Blood. 1998;92(5):1735–1742. [PubMed] [Google Scholar]
- 278.Smedley D, Hamoudi R, Clark J, Warren W, Abdul-Rauf M, Somers G, Venter D, Fagan K, Cooper C, Shipley J. The t(8;13)(p11;q11-12) rearrangement associated with an atypical myeloproliferative disorder fuses the fibroblast growth factor receptor 1 gene to a novel gene RAMP. Hum Mol Genet. 1998;7(4):637–642. doi: 10.1093/hmg/7.4.637. [DOI] [PubMed] [Google Scholar]
- 279.Wong WS, Cheng KC, Lau KM, Chan NP, Shing MM, Cheng SH, Chik KW, Li CK, Ng MH. Clonal evolution of 8p11 stem cell syndrome in a 14-year-old Chinese boy: a review of literature of t(8;13) associated myeloproliferative diseases. Leuk Res. 2007;31(2):235–238. doi: 10.1016/j.leukres.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 280.Matsumoto K, Morita K, Takada S, Sakura T, Shiozaki H, Murakami H, Miyawaki S. A chronic myelogenous leukemia-like myeloproliferative disorder accompanied by T-cell lymphoblastic lymphoma with chromosome translocation t(8;13)(p11;q12): a Japanese case. Int J Hematol. 1999;70(4):278–282. [PubMed] [Google Scholar]
- 281.Etienne A, Gelsi-Boyer V, Carbuccia N, Adelaide J, Barba G, La Starza R, Murati A, Eclache V, Birg F, Birnbaum D, Mozziconacci MJ, Mecucci C, Chaffanet M. Combined translocation with ZNF198-FGFR1 gene fusion and deletion of potential tumor suppressors in a myeloproliferative disorder. Cancer Genet Cytogenet. 2007;173(2):154–158. doi: 10.1016/j.cancergencyto.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 282.Popovici C, Zhang B, Gregoire MJ, Jonveaux P, Lafage-Pochitaloff M, Birnbaum D, Pebusque MJ. The t(6;8)(q27;p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor 1. Blood. 1999;93(4):1381–1389. [PubMed] [Google Scholar]
- 283.Vizmanos JL, Hernandez R, Vidal MJ, Larrayoz MJ, Odero MD, Marin J, Ardanaz MT, Calasanz MJ, Cross NC. Clinical variability of patients with the t(6;8)(q27;p12) and FGFR1OP-FGFR1 fusion: two further cases. Hematol J. 2004;5(6):534–537. doi: 10.1038/sj.thj.6200561. [DOI] [PubMed] [Google Scholar]
- 284.Sohal J, Chase A, Mould S, Corcoran M, Oscier D, Iqbal S, Parker S, Welborn J, Harris RI, Martinelli G, Montefusco V, Sinclair P, Wilkins BS, van den Berg H, Vanstraelen D, Goldman JM, Cross NC. Identification of four new translocations involving FGFR1 in myeloid disorders. Genes Chromosom Cancer. 2001;32(2):155–163. doi: 10.1002/gcc.1177. [DOI] [PubMed] [Google Scholar]
- 285.Mano Y, Takahashi K, Ishikawa N, Takano A, Yasui W, Inai K, Nishimura H, Tsuchiya E, Nakamura Y, Daigo Y. Fibroblast growth factor receptor 1 oncogene partner as a novel prognostic biomarker and therapeutic target for lung cancer. Cancer Sci. 2007;98(12):1902–1913. doi: 10.1111/j.1349-7006.2007.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Park TS, Song J, Kim JS, Yang WI, Song S, Kim SJ, Suh B, Choi JR. 8p11 myeloproliferative syndrome preceded by t(8;9)(p11;q33), CEP110/FGFR1 fusion transcript: morphologic, molecular, and cytogenetic characterization of myeloid neoplasms associated with eosinophilia and FGFR1 abnormality. Cancer Genet Cytogenet. 2008;181(2):93–99. doi: 10.1016/j.cancergencyto.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 287.Guasch G, Mack GJ, Popovici C, Dastugue N, Birnbaum D, Rattner JB, Pebusque MJ. FGFR1 is fused to the centrosome-associated protein CEP110 in the 8p12 stem cell myeloproliferative disorder with t(8;9)(p12;q33) Blood. 2000;95(5):1788–1796. [PubMed] [Google Scholar]
- 288.Yamamoto K, Kawano H, Nishikawa S, Yakushijin K, Okamura A, Matsui T. A biphenotypic transformation of 8p11 myeloproliferative syndrome with CEP1/FGFR1 fusion gene. Eur J Haematol. 2006;77(4):349–354. doi: 10.1111/j.1600-0609.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 289.Mozziconacci MJ, Carbuccia N, Prebet T, Charbonnier A, Murati A, Vey N, Chaffanet M, Birnbaum D. Common features of myeloproliferative disorders with t(8;9)(p12;q33) and CEP110-FGFR1 fusion: report of a new case and review of the literature. Leuk Res. 2008;32(8):1304–1308. doi: 10.1016/j.leukres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 290.Kim SY, Oh B, She CJ, Kim HK, Jeon YK, Shin MG, Yoon SS, Lee DS. 8p11 Myeloproliferative syndrome with BCR-FGFR1 rearrangement presenting with T-lymphoblastic lymphoma and bone marrow stromal cell proliferation: a case report and review of the literature. Leuk Res. 2011;35(5):e30–e34. doi: 10.1016/j.leukres.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 291.Agerstam H, Lilljebjorn H, Lassen C, Swedin A, Richter J, Vandenberghe P, Johansson B, Fioretos T. Fusion gene-mediated truncation of RUNX1 as a potential mechanism underlying disease progression in the 8p11 myeloproliferative syndrome. Genes Chromosom Cancer. 2007;46(7):635–643. doi: 10.1002/gcc.20442. [DOI] [PubMed] [Google Scholar]
- 292.Murati A, Arnoulet C, Lafage-Pochitaloff M, Adelaide J, Derre M, Slama B, Delaval B, Popovici C, Vey N, Xerri L, Mozziconacci MJ, Boulat O, Sainty D, Birnbaum D, Chaffanet M. Dual lympho-myeloproliferative disorder in a patient with t(8;22) with BCR-FGFR1 gene fusion. Int J Oncol. 2005;26(6):1485–1492. [PubMed] [Google Scholar]
- 293.Fioretos T, Panagopoulos I, Lassen C, Swedin A, Billstrom R, Isaksson M, Strombeck B, Olofsson T, Mitelman F, Johansson B. Fusion of the BCR and the fibroblast growth factor receptor-1 (FGFR1) genes as a result of t(8;22)(p11;q11) in a myeloproliferative disorder: the first fusion gene involving BCR but not ABL. Genes Chromosom Cancer. 2001;32(4):302–310. doi: 10.1002/gcc.1195. [DOI] [PubMed] [Google Scholar]
- 294.Soler G, Nusbaum S, Varet B, Macintyre EA, Vekemans M, Romana SP, Radford-Weiss I. LRRFIP1, a new FGFR1 partner gene associated with 8p11 myeloproliferative syndrome. Leukemia. 2009;23(7):1359–1361. doi: 10.1038/leu.2009.79. [DOI] [PubMed] [Google Scholar]
- 295.Grand EK, Grand FH, Chase AJ, Ross FM, Corcoran MM, Oscier DG, Cross NC. Identification of a novel gene, FGFR1OP2, fused to FGFR1 in 8p11 myeloproliferative syndrome. Genes Chromosom Cancer. 2004;40(1):78–83. doi: 10.1002/gcc.20023. [DOI] [PubMed] [Google Scholar]
- 296.Gu TL, Goss VL, Reeves C, Popova L, Nardone J, Macneill J, Walters DK, Wang Y, Rush J, Comb MJ, Druker BJ, Polakiewicz RD. Phosphotyrosine profiling identifies the KG-1 cell line as a model for the study of FGFR1 fusions in acute myeloid leukemia. Blood. 2006;108(13):4202–4204. doi: 10.1182/blood-2006-06-026666. [DOI] [PubMed] [Google Scholar]
- 297.Belloni E, Trubia M, Gasparini P, Micucci C, Tapinassi C, Confalonieri S, Nuciforo P, Martino B, Lo-Coco F, Pelicci PG, Di Fiore PP. 8p11 myeloproliferative syndrome with a novel t(7;8) translocation leading to fusion of the FGFR1 and TIF1 genes. Genes Chromosom Cancer. 2005;42(3):320–325. doi: 10.1002/gcc.20144. [DOI] [PubMed] [Google Scholar]
- 298.Walz C, Chase A, Schoch C, Weisser A, Schlegel F, Hochhaus A, Fuchs R, Schmitt-Graff A, Hehlmann R, Cross NC, Reiter A. The t(8;17)(p11;q23) in the 8p11 myeloproliferative syndrome fuses MYO18A to FGFR1. Leukemia. 2005;19(6):1005–1009. doi: 10.1038/sj.leu.2403712. [DOI] [PubMed] [Google Scholar]
- 299.Hidalgo-Curtis C, Chase A, Drachenberg M, Roberts MW, Finkelstein JZ, Mould S, Oscier D, Cross NC, Grand FH. The t(1;9)(p34;q34) and t(8;12)(p11;q15) fuse pre-mRNA processing proteins SFPQ (PSF) and CPSF6 to ABL and FGFR1. Genes Chromosom Cancer. 2008;47(5):379–385. doi: 10.1002/gcc.20541. [DOI] [PubMed] [Google Scholar]
- 300.Guasch G, Popovici C, Mugneret F, Chaffanet M, Pontarotti P, Birnbaum D, Pebusque MJ. Endogenous retroviral sequence is fused to FGFR1 kinase in the 8p12 stem-cell myeloproliferative disorder with t(8;19)(p12;q13.3) Blood. 2003;101(1):286–288. doi: 10.1182/blood-2002-02-0577. [DOI] [PubMed] [Google Scholar]
- 301.Persson F, Winnes M, Andren Y, Wedell B, Dahlenfors R, Asp J, Mark J, Enlund F, Stenman G. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene. 2008;27(21):3072–3080. doi: 10.1038/sj.onc.1210961. [DOI] [PubMed] [Google Scholar]
- 302.Wasag B, Lierman E, Meeus P, Cools J, Vandenberghe P. The kinase inhibitor TKI258 is active against the novel CUX1-FGFR1 fusion detected in a patient with T-lymphoblastic leukemia/lymphoma and t(7;8)(q22;p11) Haematologica. 2011;96(6):922–926. doi: 10.3324/haematol.2010.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, Li X, Gumin J, Zheng H, Hu L, Yli-Harja O, Haapasalo H, Visakorpi T, Liu X, Liu CG, Sawaya R, Fuller GN, Chen K, Lang FL, Nykter M, Zhang W. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013 doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Liu J, Guzman MA, Pezanowski D, Patel D, Hauptman J, Keisling M, Hou SJ, Papenhausen PR, Pascasio JM, Punnett HH, Halligan GE, de Chadarevian JP. FOXO1-FGFR1 fusion and amplification in a solid variant of alveolar rhabdomyosarcoma. Mod Pathol. 2011;24(10):1327–1335. doi: 10.1038/modpathol.2011.98. [DOI] [PubMed] [Google Scholar]
- 305.Yagasaki F, Wakao D, Yokoyama Y, Uchida Y, Murohashi I, Kayano H, Taniwaki M, Matsuda A, Bessho M. Fusion of ETV6 to fibroblast growth factor receptor 3 in peripheral T-cell lymphoma with a t(4;12)(p16;p13) chromosomal translocation. Cancer Res. 2001;61(23):8371–8374. [PubMed] [Google Scholar]
- 306.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22(4):795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]


