Abstract
Humans have two nearly identical copies of survival motor neuron gene: SMN1 and SMN2. Deletion or mutation of SMN1 combined with the inability of SMN2 to compensate for the loss of SMN1 results in spinal muscular atrophy (SMA), a leading genetic cause of infant mortality. SMA affects 1 in ~6000 live births, a frequency much higher than in several genetic diseases. The major known defect of SMN2 is the predominant exon 7 skipping that leads to production of a truncated protein (SMNΔ7), which is unstable. Therefore, SMA has emerged as a model genetic disorder in which almost the entire disease population could be linked to the aberrant splicing of a single exon (i.e. SMN2 exon 7). Diverse treatment strategies aimed at improving the function of SMN2 have been envisioned. These strategies include, but are not limited to, manipulation of transcription, correction of aberrant splicing and stabilization of mRNA, SMN and SMNΔ7. This review summarizes up to date progress and promise of various in vivo studies reported for the treatment of SMA.
Keywords: Spinal muscular atrophy, Survival motor neuron, antisense oligonucleotide, splicing, SMA therapeutics, genetic disease therapy
1. Introduction
Spinal muscular atrophy (SMA) is a genetic disease caused by homozygous deletion, truncation, mutation or gene conversion of survival motor neuron 1 (SMN1) [1–4]. SMN2, a nearly identical copy of SMN1, fails to compensate for the loss of SMN1 owing to a cytosine to thymidine mutation at the 6th position (C6U in the transcript) of exon 7. C6U triggers predominant skipping of SMN2 exon 7 due to disruption of an exonic splicing enhancer and/or creation of an exonic splicing silencer [5–7]. The resultant decrease in full-length transcript reduces functional SMN, since the translated product (SMNΔ7) of the truncated transcript is unstable and rapidly degraded [8–10]. The copy number of SMN2 modulates the severity of SMA: the more SMN2 copies the less severe the disease due to higher levels of the full-length transcript and functional SMN [11–13]. Thus, treatment strategies to halt the disease progression and ameliorate the symptoms have primarily focused on means to increase full-length SMN2 transcript and functional SMN.
The multifunctional SMN has been implicated in snRNP biogenesis [14–17], transcription [18,19], splicing [20], translation [21], signal transduction [22], stress granule formation [23] and intra-cellular trafficking [24]. With respect to neuron-specific functions, SMN facilitates interaction of mRNA binding proteins and participates in mRNA transport across the axonal processes of motor neurons [25–27]. SMN modulates axon outgrowth and cytoskeletal dynamics through β actin localization [28–30]. Preventing SMN transport across axons causes growth cone collapse [31]. SMN also plays an important role in postnatal muscle nerve terminal maturation and reduction in SMN levels is predicted to negatively affect neurotransmission [32]. Defects in snRNP biogenesis correlates with the severity of SMA, although only a subset of snRNPs is preferentially affected [33]. Supporting these arguments, motor neurons of Smn deficient Drosophila show decreased expression of a subset of certain genes containing the U12 type introns [34].
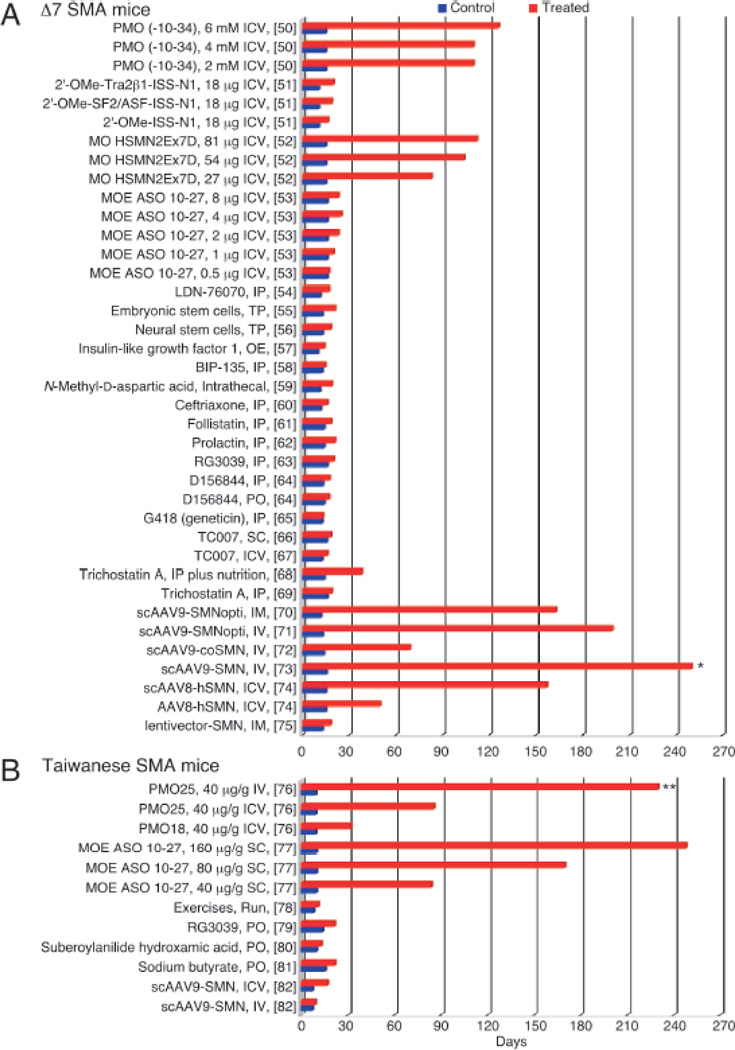
Mice, unlike humans, possess only one Smn gene, and homozygous deletion of Smn is embryonically lethal [35]. Several transgenic mouse models that mimic the SMA pathology have been developed by introducing human SMN2 into the mouse genome in the context of Smn knockout. Two recent excellent reviews describe these models in much detail [36,37]. Preclinical research to identify promising treatments for SMA has relied heavily upon these murine models [22,38–48]. Table 1 lists a few major mouse models utilized in preclinical trials as well as a few other models that may be exploited for these pursuits. Two severe mouse models account for the majority of preclinical studies: the Taiwanese model [38,48] and the Δ7 SMA model [40]. Generally, therapeutic strategies in SMA mice focused on increasing the amount of full-length SMN2 transcript and SMN as a means to extend lifespan and correct tissue and motor function abnormalities. We summarize the major avenues of therapeutic interventions explored for SMA with particular emphasis on small molecules and antisense oligonucleotides (ASOs). This review complements a recent report that describes in detail the progress in the field of ASO-mediated therapy of SMA [49]. Due to lack of space and the staggering number of compounds tested for SMA therapy, we are unable to provide details on dose, duration and frequency of delivery for most of the compounds. For the purposes of comparison, we have put major emphasis on the life expectancy as the primary measure of the therapeutic efficacy in severe SMA mice. Until two years ago there was no report of a therapeutic compound that could extend the life span of a severe SMA mouse beyond 30 days. Recently, independent studies have shown an impressive increase in the life expectancy of severe SMA mice treated with ASOs that specifically target an intronic sequence within SMN2 [refs. in 49]. The noticeable aspect of these studies is the cross validation of ASO efficacy among various mouse models and oligonucleotide chemistries against the same intronic target (described later). However, due to timing of blood brain barrier (BBB) formation and several other features distinct from humans, results in mouse models of SMA should be interpreted with caution. An overwhelming majority of small compounds confer a modest (<1.5-fold) increase in the life expectancy of severe SMA mice (Fig. 1). Consistently, these compounds display poor efficacy in clinical trials. However, possibilities remain that some of these compounds could be further improved to achieve a better therapeutic efficacy.
Table 1.
Mouse models useful for testing drug efficacy for potential SMA therapy
| Model | Genotype | Survival (days) | Outcome Measures | References |
|---|---|---|---|---|
| Taiwanese | Smn−/−; SMN2(2Hung)+/± [Mice carry 1 or 2 copies of transgene] | Type I: ~10 Type II: ~14 Type III: Normal |
Survival; motor function; tail and limb necrosis; motor neuron, NMJ, muscle and heart morphology and/or function | [38,48] |
| Line89 | Smn−/−; SMN2(89Ahmb)+/+ | 5 | Survival; motor function; motor neuron, NMJb and muscle morphology and/or function | [39] |
| Δ7 SMA | Smn−/−; SMN2(89Ahmb)+/+; SMNΔ7+/+ | ~14 | Survival; motor function; motor neuron, NMJ, muscle and cardiac morphology and/or function | [40] |
| 3 copy SMN2 | Smn−/−; SMN2(N11); SMN2(N46) | 14–16 | Survival; motor function; motor neuron, NMJ and muscle morphology and/or function | [41] |
| F7 or exon 7 floxed | SmnF7/Δ7; NSE-Cre [Exon 7 loss in neurons] | 25 | Survival; motor function; motor neuron, NMJ and muscle morphology and/or function | [43] |
| F7 or exon 7 floxed | SmnF7/Δ7; HSA-Cre [Exon 7 loss in skeletal muscle] | 33 | Survival; motor function; motor neuron, NMJ and muscle morphology and/or function | [42] |
| 2B | Smn2B/− [2B is defined by mutation in exon 7 splicing enhancer] | ~30 | Survival; motor function; neuromuscular junction and muscle morphology and/or function | [22] |
| SMNRT | Smn−/−; SMN2(89Ahmb) +/+; SMNΔ7RT +/+ | 34 | Survival; motor function; motor neuron, NMJ and muscle morphology and/or function | [44] |
| Olig2-Cre | SmnF7/−; SMN2(89Ahmb)+/+; Olig2-Cre [Exon 7 loss in motor neuron progenitor cells] | 365a | Motor function; motor neuron, NMJ and muscle morphology and/or function | [45] |
| Smn C>T | SmnC>T [SMN2 mutation inserted in mouse Smn] | Normal | Motor function; motor neuron, NMJ and muscle morphology and/or function | [46] |
| Allele C | SmnC/C [C is defined by a chimeric gene plus SMN2] | Normal | Ear, tail and limb necrosis; cardiac morphology and function | [47] |
70% of these mice survived to 365 days
NMJ, neuromuscular junction
Figure 1. Survival bar charts of treatments in Δ7 SMA mice and Taiwanese SMA mice.
(A) Survival bar charts comparing control and treatment in Δ7 SMA mice. *, still alive at > 250 days. (B) Survival bar charts comparing control and treatment in Taiwanese SMA mice. **, still alive at > 230 days. References for the studies are denoted in the brackets. Abbreviations: AAV, adeno-associated virus; scAAV, self-complementary adeno-associated virus; coSMN, codono-ptimized SMN; scAAV9-SMNopti, an optimized SMN-encoding scAAV9; IM, intramuscular injection; ICV, intracerebroventricular injection; IV, intravenous injection; IP, intraperitoneal injection; SC, subcutaneous injection; OE, overexpression; PO, per os; TP, transplantation; ASO, antisense oligonucleotide; MOE, an ASO with phophorothioate backbone and 2′-O-methoxyethyl modification; MO, morpholino; 2′-OMe, an ASO with phophorothioate backbone and 2′-O-methyl modification; ISS-N1, intronic splicing silencer N1; PMO, phosphorodiamidate morpholino oligonucleotides; Tra2β1, transformer-2 protein homolog β; SF2/ASF, splicing factor2/alternative splicing factor.
2. Treatment with small compounds
Small compounds offer several advantages, including an easy transport across biological barriers. Considering SMA is a neurodegenerative disease, compounds that are transported across BBB would be best suited for an effective therapy. A summary of the relative efficacy of small compounds and other treatments is given in Figure 1 [based on refs. 50–82]. Available reports underscore the diversity of processes that may impact SMN2 transcription, SMN2 exon 7 splicing and/or SMN levels within a cell. Given below are the major classes of compounds that have been tested for their efficacy for SMA therapy.
2.1. Histone deacetylase (HDAC) inhibitors
HDAC inhibitors prevent deacetylation of histones and increase gene expression through chromatin remodeling [83,84]. Various chemical classes of HDAC inhibitors have been shown to enhance the expression of SMN2 in SMA patient cells and in mouse models of SMA (Fig. 1) [68,69,80,81,85–88]. Sodium butyrate modestly increased survival in Taiwanese type II mice and reduced tail necrosis [81]. Valproic acid (VPA), a FDA-approved compound with multiple functions including HDAC inhibition, increased motor neuron density in the lumbar spinal cord and ameliorated necrosis of the tail and ears of Taiwanese type III mice [89]. In a follow-up study, type III SMA mice treated with VPA exhibited decreased spinal cord motor neuron degeneration, decreased muscle atrophy and improved neuromuscular junction innervation [90]. However, the effects of VPA in severe SMA mice were less pronounced. Nevertheless, VPA has been extensively examined as a treatment for types I, II and III SMA patients in several clinical trials (Clinicaltrials.gov ID numbers NCT00661453, NCT00227266, NCT00374075, NCT00481013, and NCT01671384). The beneficial effects of VPA in SMA patients, however, have been nominal [91–95]. Another HDAC inhibitor, phenylbutyrate, has been trialed in SMA patients with modest results [96], although more extensive clinical trials have been terminated due to poor treatment compliance or slow enrollment (Clinicaltrials.gov ID numbers NCT00439569 and NCT00439218).
Trichostatin A (TSA), a second-generation HDAC inhibitor, improved motor function and modestly increased the survival of Δ7 SMA mice (Fig. 1) [69]. Addition of a nutritional supplement with TSA treatment augmented the beneficial effects, including a ~2.5-fold increase in lifespan of Δ7 SMA mice (Fig. 1) [68]. Suberoylanilide hydroxamic acid (SAHA), another second-generation HDAC inhibitor, rescued the embryonic lethality and modestly increased survival of Taiwanese type I SMA mice (Fig. 1) [80]. However, higher doses of SAHA resulted in toxicity even in heterozygous mice [80]. A recent study with SAHA showed weight gain and improved motor function in Taiwanese type I SMA mice, although the effect on lifespan was not reported [97]. Treatment of Taiwanese type I SMA mice with JNJ-26481585, a novel second-generation HDAC inhibitor, did not provide any lifespan benefit [98]. Thus far, only modest benefits have been observed in SMA patients treated with HDAC inhibitors. HDAC inhibitors continue to be evaluated as potential SMA therapeutics.
2.2. Translation read-through compounds
Skipping of SMN2 exon 7 produces a truncated protein (SMNΔ7) with a novel extension of four amino acids (EMLA) coded by exon 8. EMLA serves as a degradation signal within SMNΔ7 [10]. Several aminoglycosides are known to interact with ribosomes and allow readthrough at stop codons [99]. Read-through of the first stop codon in the exon 7-deleted transcript of SMN2 is predicted to add another five amino acids to the C-terminus of SMNΔ7. Incidentally, SMA patient cells treated with aminoglycosides showed increased SMN bodies (gems), suggesting that addition of extra amino acids due to read-through has an stabilizing effect on SMNΔ7 [100,101]. A follow up study confirmed stabilization of SMNΔ7 upon addition of the five extra amino acids at the C-terminus of SMNΔ7 [102]. TC007, a novel aminoglycoside (an analog of neomycin not expected to cross the BBB), modestly extended the mean lifespan of Δ7 SMA mice when administered through intracerebroventricular (ICV) route (Fig. 1) [67]. However, chronic subcutaneous (SC) administration of this compound did not produce any life expectancy benefit (Fig. 1) [66]. Geneticin (G418), another aminoglycoside, improved motor function but did not provide survival benefit in Δ7 SMA mice (Fig.1) [65]. Further, chronic administration of G418 turned out to be toxic even for the wild type mice [65]. These findings underscore the limitations of current generation of aminoglycosides as potential drug candidate for SMA therapy.
2.3. Quinazolines
Quinazolines are a family of compounds that inhibit RNA decapping enzyme DcpS (involved in RNA turnover), and this inhibition can consequently increase SMN2 expression [103]. A high-throughput screening identified a few quinazolines as lead compounds that increased full-length SMN2 transcript and SMN [104]. These compounds had poor BBB penetration and required high doses to achieve sufficient upregulation of SMN. Based on a lead quinazoline, derivatives were developed to overcome the obstacles associated with the low BBB penetration [105]. One of these derivatives, D156844, crossed the BBB without any adverse effect in Δ7 SMA neonates. However, D156844 produced only a modest lifespan extension in Δ7 SMA mice (Fig. 1) [64]. Further optimization of this compound led to the identification of D157495, also called RG3039. In one study, daily intraperitoneal (IP) administration of RG3039 modestly extended the lifespan of Δ7 SMA mice and improved the morphology and function of the neuromuscular junction (NMJ) [63]. In another study, daily oral administration of RG3039 modestly extended the lifespan of Taiwanese type I mice (Fig. 1) [79]. Additionally, RG3039 markedly increased the lifespan in less severe Smn2B/− mouse model (median lifespan increased from 18 to 112 days) [79]. Currently, RG3039 is undergoing through phase I clinical trial in which safety and efficacy of various doses will be evaluated.
2.4. Hydroxyurea
Hydroxyurea has been shown to increase SMN2 expression in SMA patient fibroblasts, purportedly via an increase in nitric oxide [106]. Due to the positive in vitro effects and the BBB permeability of hydroxyurea, this compound was tested in SMA patients in several clinical trials (Clinicaltrials.gov ID numbers NCT00485511, NCT00568698, and NCT00568802). While one small study registered encouraging results in SMA patients treated with hydroxyurea [107], a larger, placebo-controlled study did not reveal any significant effects in SMA treated patients [108]. These findings limit the scope of hydroxyurea as an effective drug for SMA therapy.
2.5. Compounds from cell-based drug screenings
A luciferase reporter system coupled to SMN2 minigene carrying CMV promoter was employed in a high-throughput screening to identify indoprofen, a non-steroidal anti-inflammatory drug (NSAID) that promoted SMN2 exon 7 inclusion in C33a cervical carcinoma cells [109]. Other NSAID compounds failed to stimulate SMN2 exon 7 inclusion, suggesting that the effect of indoprofen is mediated through a cyclooxygenase-independent pathway. Subsequent evaluation of indoprofen in SMA patient cells failed to show any stimulatory effect on SMN2 exon 7 splicing from endogenous SMN2. Interestingly, through an unknown mechanism indoprofen produced a noticeable upregulation of SMN (protein) in SMA patient cells [109]. In vivo efficacy of indoprofen was tested employing a highly severe SMA model in which all SMA pups die by embryonic day 11 (E11). A single IP administration of indoprofen (5 mg/kg) was able to increase the mean litter size of embryos at E14 [109]. However, efficacy of indoprofen in less severe mouse models remains to be evaluated. Another reporter assay that used SMN2 minigene coupled with SMN2 promoter identified a set of compounds including LDN-75654 and LDN-76070. These compounds were found to increase SMN levels in SMA patient cells [54]. IP administration of LDN-76070 in Δ7 SMA mice modestly extended lifespan compared to untreated mice (Fig. 1) [54]. However, in vivo efficacy of other LDN compounds could not be evaluated due to their extremely poor water solubility.
2.6. Antibiotics
Several antibiotics have been evaluated as potential therapeutic compounds for the treatment of SMA [60,110,111]. Screening of tetracycline-like compounds using a cell-free splicing assay identified PTK-SMA1 [110]. PTK-SMA1 increased SMN levels SMA patient cells as well as in a type III SMA mouse model [110]. However, therapeutic efficacy of tetracycline-like compounds on phenotype and life expectancy of a mouse model of SMA is yet to be evaluated. Ceftriaxone is a third-generation cephalosporin antibiotic that has been tested for its therapeutic efficacy in a mouse model of amyotrophic lateral sclerosis (ALS), a devastating neurodegenerative disease [111]. Ceftriaxone prevented motor neuron loss and increased survival of ALS mice possibly through increased glutamate clearance and reduced excitotoxicity [111]. Treatment of Δ7 SMA mice with high doses of ceftriaxone (200 mg/kg) showed some benefits on muscle and NMJ morphology, however, the survival benefit was very modest (Fig. 1) [60].
2.7. Signal transducing molecules
Splicing factor Tra2β1 stimulates SMN2 exon 7 inclusion by directly binding to exon 7 [112]. Dephosphorylation of Tra2β1 by protein phosphatase 1 (PP1) was implicated in the promotion of SMN2 exon 7 skipping [113]. Recently, cantharidin analogs that modulate PP1 activity were shown to increase SMN2 exon 7 inclusion and SMN levels in SMA patient cells [114]. Like several phosphatase inhibitors, cantharidins are known to affect transcription [115]. However, it remains to be seen if cantharidin analogs that promote SMN2 exon 7 inclusion also stimulate SMN2 transcription. Sodium vanadate is a tyrosine phosphatase inhibitor that markedly increases SMN2 exon 7 inclusion in a cell-based splicing assay [116]. However, sodium vanadate was considered inappropriate for SMA therapy due to high toxicity. To overcome this toxicity, Taiwanese type III SMA mice were simultaneously treated with sodium vanadate and the detoxification agent L-ascorbic acid [117]. Indeed, the combination therapy prevented the morbidity and mortality induced by sodium vanadate. Also, the combination therapy delayed the onset of tail necrosis, increased lumbar spinal cord motor neurons and improved muscle histology. However, effects in a more severe mouse model of SMA remain to be determined.
2.8. Neuroprotective compounds
The neuroprotection conferred by small molecules can occur via several mechanisms and could potentially spare or protect motor neurons and thus ameliorate the disease. Glycogen synthase kinase-3 (GSK-3) has been linked to human pathologies, and inhibition of this signaling molecule is purported to activate cyclic AMP response element binding protein (CREB) and increase transcription of neurotrophic factors. Indeed, GSK-3 inhibition extended lifespan of a mouse model of ALS [118]. When BIP-135 (a GSK-3 inhibitor) was administered to Δ7 SMA mice, median lifespan increased modestly compared to untreated mice (Fig. 1) [58]. Further studies would be needed to appropriately evaluate the effect of various treatment regimes. As a potential treatment for SMA, activation of the N-methyl-D-aspartic acid (NMDA) receptor has also been examined. Intrathecal NMDA administration in two SMA mouse models accelerated maturation of motor units, conferred neuroprotection to motor neurons and significantly extended the lifespan (Fig. 1) [59]. NMDA also led to CREB activation with a resultant increase in SMN levels [59]. However, the clinical utility of intrathecal NMDA administration is yet to be evaluated in SMA patients.
Olesoxime is a mitochondrial pore modulator that provides protective effects in motor neurons [119]. While the neuroprotective effect of olesoxime has not been tested in SMA mouse models, it is currently undergoing a phase 2 clinical trial for the treatment of SMA (Clinicaltrials.gov ID number NCT01302600). SMA type II and III patients are enrolled in this ongoing study; each patient will undergo two years of treatment to examine the beneficial effects of this compound.
Riluzole is the only FDA-approved compound for ALS treatment. Although the mechanism of riluzole action remains vague, it appears to exert its neuroprotective effect by modulating glutamate transmission [120]. Therapeutic efficacy of riluzole was evaluated in a SMA mouse model that was generated by neuron-specific knockout of Smn exon 7 [121]. Riluzole improved the architecture of motor neuron synaptic terminals and modestly increased the lifespan of SMA mice [121]. A phase I clinical trial in which infants with SMA type I were treated with riluzole produced intriguing results. Three of the seven patients lived well past the usual age of death with limited need for respiratory support, although conclusions were not definitive due to very small sample size [122]. A phase 2/3 clinical trial of riluzole with type II and type III patients has been conducted and findings of this study are still awaited (Clinicaltrials.gov ID number NCT00774423).
3. Polypeptide and protein-based therapies
Transcription of SMN2 is modulated by transcription factor STAT5, a phosphorylation substrate of Janus Kinase (JAK) [123]. Prolactin, a polypeptide hormone that activates JAK2/STAT5 signaling pathway has been shown to improve motor function and modestly increase the lifespan of Δ7 SMA mice (Fig. 1) [62]. Prolactin also increased SMN levels in the brain possibly through activation of transcription of SMN2 [62]. However, it remains to be seen if other members of the JAK/STAT signaling pathway would have a better therapeutic efficacy.
Myostatin, a member of the transforming growth factor beta (TGFβ) family, is expressed almost exclusively in muscle and functions as a negative regulator of skeletal muscle growth [124]. Follistatin, a known myostatin inhibitor, improved motor function and modestly increased lifespan of Δ7 SMA mice (Fig. 1) [61]. In another study, Δ7 SMA mice treated with activin IIB (ActIIB-Fc), a soluble receptor of myostatin, increased the mass of several muscles, but did not rescue motor function or extend survival [125]. Furthermore, knocking out of myostatin gene in Δ7 SMA model did not reduce the severity of disease [126]. These findings rule out the TGFβ signaling pathway modulating compounds as the potential therapeutic candidates of SMA.
Insulin-like growth factor 1 (IGF-1), a protein hormone, serves as an important regulator of skeletal muscle development and function [127]. Incidentally, IGF-1 serum levels are significantly reduced in both Taiwanese type I SMA [77] and Δ7 SMA mice [128]. To investigate the role of IGF-1 in SMA pathology, a novel mouse model that overexpressed a rat muscle-specific isoform of IGF-1 in skeletal muscle of Δ7 SMA model was developed [57]. IGF-1 overexpression modestly affected the phenotype: median lifespan was extended ~1.4 fold and several muscles increased in weight (Fig. 1) [57]. In a separate study, treatment of Δ7 SMA mice with IPLEX (recombinant human IGF-1 complexed with recombinant IGF binding protein 3) prevented muscle wasting and neuromuscular junction abnormalities [128]. However, this approach did not improve motor function and lifespan of Δ7 SMA mice [128].
4. Gene therapy and trans-splicing
Gene therapy promises a permanent solution for SMA through viral delivery and insertion of the entire SMN1 gene or cDNA sequence into the genome of SMA patients. This process is generally irreversible and there are apparent risks if the exogenously delivered gene is inserted at a wrong location and/or overexpressed. The first gene therapy, conducted in Δ7 SMA mice, introduced the entire human SMN1 gene and conferred some protection to brainstem motor neurons and increased lifespan from 13 to 18 days (Fig. 1) [75]. Despite the modest outcomes, findings provided the first direct evidence that the postnatal increase in SMN levels confers definite therapeutic benefits. In another study, ICV administration of the SMN1 gene substantially (~10-fold) increased the lifespan of Δ7 SMA mice (Fig. 1) [74]. This finding suggested that increasing SMN levels in the brain is critical for prolonging the life of SMA patients. However, numerous studies that delivered SMN1 intravenously (IV) also demonstrated impressive results [71,72,129]. In particular, a codon optimized SMN1 sequence injected into the temporal vein increased median survival ~15-fold (Fig.1) [71]. Intramuscular delivery of the same sequence produced comparable results (Fig.1) [70]. Another study conducted in Δ7 SMA mice demonstrated similar benefits from IV and ICV administrations of scAAV9-SMN [130]. On the other hand, experiments conducted in Taiwanese type I SMA mice yielded variable results and less promising survival benefits with ICV administration of SMN1 sequence (Fig. 1) [82]. These findings underscore the caution that must be exercised when extrapolating the results of pre-clinical studies conducted in a specific mouse model to design clinical trials in humans. Overall, the results of gene therapy appear promising and may offer one of the best therapeutic alternatives to a specific group of SMA patients who are too weak to receive frequent invasive treatments.
Trans-splicing is a gene-therapy-like approach with the potential to treat SMA patients. This approach involves exogenous delivery of a partial gene or mRNA that triggers production of a chimeric transcript that is equivalent to the full-length mRNA generated from the endogenous gene [131,132]. Considering that the trans-splicing process uses endogenous pre-mRNA as a substrate, it caps the level of mRNAs and protein produced. Therefore, trans-splicing has inbuilt potential to fully avoid complications of protein overexpression, which is an anticipated risk associated with the gene therapy. Of note, it must be brought forth that there is no evidence to suggest that gene therapy-based approaches would cause SMN overexpression that result into a recognizable harmful effect in pre-clinical studies. Experiments conducted in SMA mouse models have shown encouraging results of a trans-splicing-based approach [133]. However, the improvements in lifespan were less impressive [133]. In general, trans-splicing is a very inefficient process that remains an option only when other therapies fail to meet the high expectations.
5. Stem cell based therapy
Stem cell therapy is based on the assumption that intrathecal transplantation of healthy donor stem cells in the spinal cord of SMA patients would compensate for the eventual death of the endogenous motor neurons. Two major studies evaluated the effect of stem cells in mouse models of SMA [55, 56]. In one study, spinal cord neural stem cells injected into cerebrospinal fluid increased the mean survival of Δ7 SMA mice ~39% (Fig. 1) [56]. In a separate study, intrathecal transplantation of embryonic stem cell-derived neural stem cells augmented the number and size of motor neurons, improved muscle innervation and increased mean survival of Δ7 SMA mice by 64% (Fig. 1) [55]. These modest benefits in pre-clinical studies somewhat undermine the promise of stem cell therapy for the treatment of SMA.
6. ASO-based therapy
ASO-based strategies employ specific SMN2 sequences as targets for SMA therapy [49]. Earlier studies focused on bifunctional ASOs that blocked the C6U mutation site within SMN2 exon 7 and provided a tailed sequence for the recruitment of the positive splicing factors at the weak 3′ splice site (3′ ss) of exon 7 [134,135]. These studies were driven by a well-founded belief that a weak 3′ ss was the sole cause of SMN2 exon 7 skipping and forced recruitment of a positive regulator at the 3′ ss was the best mechanistic solution for the restoration of SMN2 exon 7 inclusion. Subsequent studies shifted attention towards the 5′ ss as it became obvious that a series of inhibitory elements are positioned in the vicinity of the 5′ ss [136–139]. A major breakthrough came with the discovery of intronic splicing silencer N1 (ISS-N1), a fifteennucleotide sequence spanning from the 10th to the 24th positions of SMN2 intron 7 (Fig. 2) [137]. Since an ASO annealing to an intronic sequence does not interfere with the transport and translational machinery, ISS-N1 serves as an ideal target for an ASO-mediated splicing correction in SMA. Indeed, sequestration of ISS-N1 by an ASO fully restored SMN2 exon 7 inclusion and substantially increased levels of SMN in SMA patient cells [137].
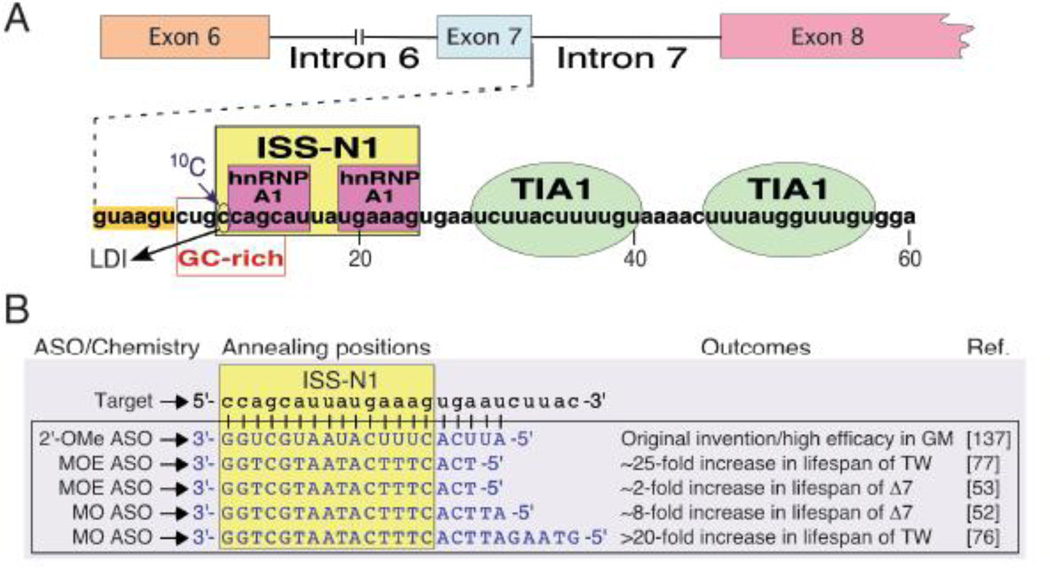
Figure 2. ASO-mediated SMN2 splicing correction as a therapy for SMA.
(A) Diagrammatic representation of splicing cis-elements in SMN2 intron 7. Numbering begins from the first position of intron 7. The negative cis-element ISS-N1 (yellow box) contains two hnRNP A1 binding sites (magenta boxes) and has emerged as a leading ASO target for splicing correction in SMA. Green ovals represent TIA-1 binding sites, an enhancer of exon 7 splicing. (B) Representative ISS-N1-targeting ASOs tested in SMA mouse models. Intron 7 sequence is in lower caps and ASO sequences are in upper caps. The chemistry for each ASO is listed to the left of the sequence. The outcomes for studies in which the ASOs were tested are listed to the right of the sequence; references are denoted in brackets. Abbreviations: ASO, antisense oligonucleotide; ISS-N1, intronic splicing silencer N1; hnRNP A1, heterogenous ribonucleoprotein A1; TIA1, T-cell restricted intracellular antigen 1; LDI, long-distance interaction; 2′-OMe, an ASO with phosphorothioate backbone and 2′-O-methyl modification; MOE, an ASO with phosphorothioate backbone and 2′-O-methoxyethyl modification; MO, morpholino; GM, SMA patient fibroblast cells (cell line GM03813); TW, Taiwanese SMA mouse model; Δ7, Δ7 SMA mouse model.
A vast number of independent reports have put ISS-N1 as one of the best-studied antisense targets for splicing correction in a human disease [49]. Therefore, lessons learned from these reports have wide implications for SMA therapy as well as for therapy of many other genetic diseases. Our understanding of the mechanism by which an ISS-N1-targeting ASO promotes SMN2 exon 7 inclusion continues to evolve. ISS-N1 harbors two hnRNP A1/A2 binding sites that do not include a cytosine residue (10C) at the 10th intronic position (the first nucleotide of ISS-N1) (Fig. 2). It was initially hypothesized that an ASO-mediated sequestration of hnRNP A1 motifs of ISS-N1 is necessary and sufficient to confer full stimulatory effect on SMN2 exon 7 splicing [139]. However, a subsequent study suggested that the sequestration of 10C is also critical for the robust stimulatory effect on SMN2 exon 7 splicing [140]. Incidentally, 10C falls within an 8-nucleotide GC-rich sequence that partially overlaps with the first hnRNP A1 motif of ISS-N1 (Fig. 2A) [141]. Sequestration of this GC-rich sequence alone by an ASO fully restored SMN2 exon 7 inclusion [141,142]. A recent study puts 10C and GC-rich sequence in a structural context that involves a unique long-distance interaction [143]. This study also brings a new perspective by demonstrating an ASO-mediated remodeling of RNA structure in the vicinity of the 5′ ss [143]. Intronic sequences downstream of ISS-N1 contain binding sites for T-cell restricted intracellular antigen 1 (TIA1), which is a positive regulator of SMN2 exon 7 splicing [144]. A single amino acid substitution within TIA1 has been shown to negatively affect SMN2 exon 7 splicing in patients of Welander distal myopathy [145]. Therefore, TIA1 emerges as the first and the only splicing factor whose mutation could be directly linked to SMN2 exon 7 splicing in the context of a human disease. Based on a recent study, an ASO targeting ISS-N1 or GC-rich sequence brings a structural change that promotes TIA1 recruitment and consequently favors inclusion of SMN2 exon 7 [143].
In order to validate the therapeutic efficacy of ISS-N1-targeting ASOs in vivo, oligonucleotides with three different modifications have been tested (Fig. 2) [49]. These modifications are: 2′-O-methyl with phosphorothioate backbone (2′-OMe), 2′-O-methoxyethyl with phosphorothioate backbone (MOE) and morpholino. A systematic study reported by Krainer and colleagues employed an ISS-N1-targeting 18-mer MOE ASO and demonstrated ~25-fold increase in the life expectancy of the severe Taiwanese model of SMA (Fig. 1) [77]. Surprisingly, subcutaneous injections produced the most beneficial effects, whereas ICV delivery resulted in only a moderate response. These findings supported the peripheral requirement of SMN for the avoidance of SMA. However, results of other studies conducted recently with ISS-N1-targeting morpholino ASOs were somewhat different. In particular, the Burghes group used a 20-mer morpholino ASO and showed ~8-fold increase in the life expectancy of Δ7 SMA mouse upon ICV administration (Fig. 1) [52]. The Muntoni group observed similar lifespan benefits in Taiwanese model mice upon ICV administration of a 25-mer morpholino ASO (PMO25) (Fig. 1) [76]. These findings are consistent with the early requirement of SMN in motor neurons. Interestingly, a single facial injection of PMO25 was found to be highly efficacious as it led to ~22-fold increase in the life expectancy of Taiwanese mouse model (Fig. 1) [76]. While these results support the peripheral requirement of SMN, they are confounded by the fact that BBB in mice remains permeable for a few days after birth. Therefore, it is likely that the beneficial effect at high doses of peripheral administration is due to leakage of ASOs into the brain. Overall, the results of these studies showed an unprecedented therapeutic benefit of ASOs targeting ISS-N1.
The observation that the efficacy of an ISS-N1-targeting ASO is on par with the gene replacement therapy underscores that ASO-mediated splicing correction is among the most promising options of SMA therapy (Fig. 1). Moving forward with clinical trials, concerns related to ASO delivery and toxicity would have to be addressed. ISIS Pharmaceuticals has recently concluded phase 1 and initiated the phase 2 clinical trial of ISIS-SMNRx, an ISS-N1 targeting MOE ASO (Clinicaltrials.gov ID NCT01839656). Encouraging outcomes of ISIS-SMNRx in a phase 1 clinical trial demands an expansion of ASO-based strategies to include additional targets and different oligonucleotide chemistries. Of note, a slight change in ASO sequence could confer entirely different pharmacokinetic and pharmacodynamic properties. Also, a change in the ASO backbone would generate an entirely new class of compound. Given the variable severity and age diversity among SMA patients, a variety of chemotherapy options would be needed for the treatment of SMA. In principle, multiple ASO-based drugs with different pharmacokinetic and pharmacodynamic properties could be developed against a single target, provided issues related to the proprietary rights on antisense target, oligonucleotide chemistry and delivery protocols are appropriately reconciled. Two recent clinical trials of a morpholino ASO have shown promising results for the treatment of Duchenne Muscular Dystrophy [146,147]. Incidentally, ISS-N1 targeting morpholino ASOs have demonstrated substantially better ICV efficacy than MOE ASOs (Fig.1). Therefore, employment of morpholino chemistry for the development of the next ASO-based drug for SMA treatment remains an attractive proposition.
7. Other therapies
There are very-limited studies that utilize treatments that do not ostensibly affect SMN. Of these studies, the effects of physical exercise on the survival and phenotype of SMA mouse models has been most widely examined. Taiwanese type II SMA mice trained to run on an exercise wheel showed increased full-length SMN2 transcript in the spinal cord, decreased neuronal loss and an extended lifespan (Fig. 1) [78]. In another study, forced running enhanced the development of several muscles compared to sedentary Δ7 SMA mice [148]. Interestingly, administration MK-801, a NMDA receptor antagonist, abolished the lifespan extension conferred by exercise [148]. These results suggest the involvement of glutamate neurotransmission as an important mediator of exercise benefits. A recent study in Δ7 SMA mice showed tangible benefits of exercise on cardiac function such as improved conduction velocity, reduced fibrosis, improved bradycardia and decreased arrhythmias [149]. These findings support that exercise could be very useful as an adjunct therapy of SMA [150].
Proper supportive therapy is crucial to increase the survival and quality of life of SMA patients. Infants diagnosed with type I SMA will eventually require some sort of ventilation support to facilitate their breathing. Studies in type I SMA patients have shown respiratory interventions (invasive or noninvasive) increase survival compared to those patients who do not receive respiratory support [151,152]. Of course, patients with the more severe manifestations of the disease will likely require more invasive interventions. Respiratory support, as well as other supportive therapy such as nutritional supplementation or total parenteral nutrition, can certainly improve the quality of life for SMA patients.
8. Conclusions
SMA is a progressive neurodegenerative disease of infants and children. Although SMA has no known cure, the patient population possesses SMN2, which can be potentially targeted for therapy. The last ten years have witnessed a dramatic progress in the development of therapeutic strategies for SMA. Much of the success in the field of SMA therapy could be attributed to a better understanding of SMN function, SMN2 exon 7 splicing regulation, SMA pathogenesis and the developments in the related fields. Small compounds continue to attract attention due to expected ease of their delivery across the BBB. However, all of small compounds tested thus far showed a very small lifespan extension benefit. SMN is an essential protein, which is involved in regulation of a number of vital processes within a cell. The low efficacy of small compounds could be due to modulation of a limited number of downstream events regulated by SMN. It is also likely that some of these compounds have antagonistic effects on various critical pathways regulated by SMN. Therefore, there is a need for additional drug screening assays, which identify new classes of small compounds that affect upstream events such as transcription and splicing of SMN2. Exceptional pre-clinical results of ISS-N1-targeting ASOs and promising outcome of phase 1 clinical trail of ISIS-SMNRx provide one of the best hopes for SMA therapy. Recent discovery of ISS-N2, a novel antisense target [143], further expands the potential for the development of additional antisense-based drugs. Currently, invasive intrathecal administration remains the most effective mode of delivery of ASOs to SMA patients. Therefore, there is a need to work on modifications and non-invasive delivery protocols that allow an easy transport of ASOs cross the BBB. A success in this direction would dramatically elevate the status of ASO-based therapy of SMA and possibly several other diseases requiring transport of a small nucleic acid molecule across BBB.
Despite admirable attempts by a number of investigators [153–155], one of the major challenges for the therapeutic development in SMA has been the availability of the robust biomarkers. The first therapy with substantial lifespan enhancements in SMA would likely offer additional biomarkers. Knowledge of such biomarkers would be extremely helpful for future drug screenings. Therapeutic progress in SMA thus far supports that compounds specifically aimed at correction of SMN2 exon 7 splicing offer the most promising outcome. The challenge remains how to identify/design a small compound that fully corrects SMN2 exon 7 splicing as well as is able to cross BBB. The answer could lie within intronic sequences that fold into secondary and high-order RNA structures. Indeed, small compounds can affect splicing through specific interactions with a unique RNA structure [156]. The complex process of pre-mRNA splicing of a specific exon comes at the expense of hundreds of structural rearrangements. As we move forward with uncovering of structural rearrangements during pre-mRNA splicing of SMN2 exon 7, we hope to see new avenues open up for discovery of unique compounds for SMA therapy. Thanks to the partnership between academia and industry, SMA has an impressive track record of clinical trials of a variety of compounds covering different pathways. Many of these compounds are being tested for the first time to cure a genetic disease. Therefore, a potential success will likely translate into therapeutic development for a number of genetic diseases requiring splicing modulation.
Table 2.
Pathways affected by compounds used for potential SMA therapy
| Treatment | Process | Models used | References |
|---|---|---|---|
| Histone deacetylase inhibitors | |||
| Trichostatin A | Transcription | Δ7 SMA mice | [68,69] |
| Suberoylanilide hydroxamic acid | Transcription | Taiwanese mice | [80] |
| Sodium butyrate | Transcription | Taiwanese mice | [81] |
| Valproic acid | Transcription | Taiwanese mice | [90] |
| Translation read-through compounds | |||
| G418 (geneticin) | Translation | Δ7 SMA mice | [65] |
| TC007 | Translation | Δ7 SMA mice | [66,67] |
| Quinazolines | |||
| RG3039 | Transcription | Δ7 SMA mice Taiwanese mice 2B |
[63] [79] [79] |
| D156844 | Transcription | Δ7 SMA mice | [64] |
| Other small molecules that increase SMN2 expression | |||
| LDN-76070 | Transcription | Δ7 SMA mice | [54] |
| Indoprofen | Splicing and/or translation | Line 89 | [109] |
| Antibiotics | |||
| Ceftriaxone | Cell signaling | Δ7 SMA mice | [60] |
| PTK-SMA1 (tetracycline-like) | Splicing | Taiwanese mice | [110] |
| Signal transducing molecules | |||
| Sodium vanadate | Splicing | Taiwanese mice | [117] |
| Neuroprotective compounds | |||
| BIP-135 | Cell signaling | Δ7 SMA mice | [58] |
| N-Methyl-D-aspartic acid | Cell signaling | Δ7 SMA mice | [59] |
| Riluzole | Cell signaling | F7 or exon 7 floxed | [121] |
| Polypeptides and proteins | |||
| Insulin-like growth factor 1 | Cell signaling | Δ7 SMA mice | [57] |
| Follistatin | Cell signaling | Δ7 SMA mice | [61] |
| Prolactin | Transcription | Δ7 SMA mice | [62] |
| Gene therapy and trans-splicing | |||
| SMN1 gene therapy | SMN-associated functions | Δ7 SMA mice Taiwanese mice |
[70–75] [82] |
| Trans-splicing | Splicing | Δ7 SMA mice | [133] |
| Stem cell based therapy of SMA | |||
| Stem cell transplantation | SMN-associated functions | Δ7 SMA mice | [55,56] |
| SMN-independent treatment strategies | |||
| Forced running | Multiple | Δ7 SMA mice Taiwanese mice |
[148] [78] |
| ASO-based therapy | |||
| Antisense oligonucleotides | Splicing | Δ7 SMA mice Taiwanese mice |
[50–53] [76,77] |
Highlights.
We compare various pre-clinical studies for therapy of spinal muscular atrophy.
Antisense oligonucleotide-based therapy has shown the best promise.
Clinical trials with oligonucleotides should include additional targets and chemistries.
RNA structure-based assays should be developed for future drug screenings.
Acknowledgements
Funding: This work was supported by grants from the National Institutes of Health (R01 NS055925, R21 NS072259 and R21 NS080294) and Salsbury Endowment (Iowa State University, Ames, IA, USA) to RNS.
Abbreviations
- ALS
amyotrophic lateral sclerosis
- ASO
antisense oligonucleotide
- BBB
blood brain barrier
- CREB
cyclic AMP response element binding protein
- FDA
Food and Drug Administration
- GSK-3
glycogen synthase kinase 3
- HDAC
histone deacetylase
- hnRNP A1
heterogenous ribonucleoprotein A1
- ICV
intracerebroventricular
- IGF-1
insulin-like growth factor 1
- IP
intraperitoneal
- ISS-N1
intronic splicing silencer N1
- ISS-N2
intronic splicing silencer N2
- IV
intravascular
- JAK
Janus Kinase
- NMDA
N-methyl D-aspartic acid
- NSAID
non-steroidal anti-inflammatory drug
- SAHA
suberoylanilide hydroxamic acid
- SC
subcutaneous
- scAAV
self-complimentary adeno-associated virus
- SMA
spinal muscular atrophy
- SMN
survival motor neuron
- snRNP
small nuclear ribonucleoprotein
- STAT5
signal transducer and activator of transcription 5
- TIA1
T-cell restricted intracellular antigen 1
- TSA
trichostatin A
- VPA
valproic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures and competing interests: ISS-N1 target (US patent # 7,838,657) was discovered in the Singh lab at UMASS Medical School (Worcester, MA, USA). Inventors, including RNS and UMASS Medical School, are currently benefiting from licensing of ISS-N1 target (US patent # 7,838,657) to ISIS Pharmaceuticals. A GC-rich target for a small ASO (Patent# US 20110269820) was discovered in the Singh lab at Iowa State University (Ames, IA, USA). Therefore, inventors including RNS and Iowa State University could potentially benefit from any future commercial exploitation of the above-mentioned target.
References
- 1.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Prior TW. Spinal muscular atrophy diagnostics. J. Child Neurol. 2007;22:952–956. doi: 10.1177/0883073807305668. [DOI] [PubMed] [Google Scholar]
- 5.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 6.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 7.Singh NN, Singh RN. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 2011;8:600–606. doi: 10.4161/rna.8.4.16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, Brahe C, Saugier-Veber P, Bonnefont JP, Melki J. Refined characterization of the expression and stability of the SMN gene products. Am. J. Pathol. 2007;171:1269–1280. doi: 10.2353/ajpath.2007.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol. Cell. Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirth B, Brichta L, Schrank B, Lochmüller H, Blick S, Baasner A, Heller R. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 13.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 14.Yong J, Pellizzoni L, Dreyfuss G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 2002;21:1188–1196. doi: 10.1093/emboj/21.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell. Biol. 2002;22:6533–6541. doi: 10.1128/MCB.22.18.6533-6541.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 17.Battle DJ, Kasim M, Yong J, Lotti F, Lau C-K, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- 18.Strasswimmer J, Lorson CL, Breiding DE, Chen JJ, Le T, Burghes AH, Androphy EJ. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum. Mol. Genet. 1999;8:1219–1226. doi: 10.1093/hmg/8.7.1219. [DOI] [PubMed] [Google Scholar]
- 19.Campbell L, Hunter KM, Mohaghegh P, Tinsley JM, Brasch MA, Davies KE. Direct interaction of Smn with dp103, a putative RNA helicase: a role for Smn in transcription regulation? Hum. Mol. Genet. 2000;9:1093–1100. doi: 10.1093/hmg/9.7.1093. [DOI] [PubMed] [Google Scholar]
- 20.Rossoll W, Kröning A-K, Ohndorf U-M, Steegborn C, Jablonka S, Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum. Mol. Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez G, Dury AY, Murray LM, Biondi O, Tadesse H, El Fatimy R, Kothary R, Charbonnier F, Khandjian EW, Côté J. A novel function for the survival motoneuron protein as a translational regulator. Hum. Mol. Genet. 2013;22:668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- 22.Bowerman M, Shafey D, Kothary R. Smn depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J. Mol. Neurosci. 2007;32:120–131. doi: 10.1007/s12031-007-0024-5. [DOI] [PubMed] [Google Scholar]
- 23.Zou T, Yang X, Pan D, Huang J, Sahin M, Zhou J. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell. Mol. Neurobiol. 2011;31:541–550. doi: 10.1007/s10571-011-9647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peter CJ, Evans M, Thayanithy V, Taniguchi-Ishigaki N, Bach I, Kolpak A, Bassell GJ, Rossoll W, Lorson CL, Bao Z-Z, Androphy EJ. The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum. Mol. Genet. 2011;20:1701–1711. doi: 10.1093/hmg/ddr046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagliardini S, Giavazzi A, Setola V, Lizier C, Di Luca M, DeBiasi S, Battaglia G. Subcellular localization and axonal transport of the survival motor neuron (SMN) protein in the developing rat spinal cord. Hum. Mol. Genet. 2000;9:47–56. doi: 10.1093/hmg/9.1.47. [DOI] [PubMed] [Google Scholar]
- 26.Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossoll W, Jablonka S, Andreassi C, Kröning A-K, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Xing L, Rossoll W, Wichterle H, Singer RH, Bassell GJ. Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J. Neurosci. 2006;26:8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Todd AG, Morse R, Shaw DJ, McGinley S, Stebbings H, Young PJ. SMN, Gemin2 and Gemin3 associate with beta-actin mRNA in the cytoplasm of neuronal cells in vitro. J. Mol. Biol. 2010;401:681–689. doi: 10.1016/j.jmb.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 31.Ting C-H, Wen H-L, Liu H-C, Hsieh-Li H-M, Li H, Lin-Chao S. The spinal muscular atrophy disease protein SMN is linked to the Golgi network. PLoS One. 2012;7:e51826. doi: 10.1371/journal.pone.0051826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres-Benito L, Neher MF, Cano R, Ruiz R, Tabares L. SMN requirement for synaptic vesicle, active zone and microtubule postnatal organization in motor nerve terminals. PLoS One. 2011;6:e26164. doi: 10.1371/journal.pone.0026164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabanella F, Butchbach MER, Saieva L, Carissimi C, Burghes AHM, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lotti F, Imlach WL, Saieva L, Beck ES, Hao LT, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, Pellizzoni L. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrank B, Götz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sleigh JN, Gillingwater TH, Talbot K. The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy. Dis. Model. Mech. 2011;4:457–467. doi: 10.1242/dmm.007245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bebee TW, Dominguez CE, Chandler DS. Mouse models of SMA: tools for disease characterization and therapeutic development. Hum. Genet. 2012;131:1277–1293. doi: 10.1007/s00439-012-1171-5. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 39.Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossoll W, Rossol W, Prior TW, Morris GE, Burghes AH. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 40.Le TT, Pham LT, Butchbach MER, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AHM. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 41.Michaud M, Arnoux T, Bielli S, Durand E, Rotrou Y, Jablonka S, Robert F, Giraudon-Paoli M, Riessland M, Mattei M-G, Andriambeloson E, Wirth B, Sendtner M, Gallego J, Pruss RM, Bordet T. Neuromuscular defects and breathing disorders in a new mouse model of spinal muscular atrophy. Neurobiol. Dis. 2010;38:125–135. doi: 10.1016/j.nbd.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Cifuentes-Diaz C, Frugier T, Tiziano FD, Lacène E, Roblot N, Joshi V, Moreau MH, Melki J. Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J. Cell Biol. 2001;152:1107–1114. doi: 10.1083/jcb.152.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cifuentes-Diaz C, Nicole S, Velasco ME, Borra-Cebrian C, Panozzo C, Frugier T, Millet G, Roblot N, Joshi V, Melki J. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Hum. Mol. Genet. 2002;11:1439–1447. doi: 10.1093/hmg/11.12.1439. [DOI] [PubMed] [Google Scholar]
- 44.Cobb MS, Rose FF, Rindt H, Glascock JJ, Shababi M, Miller MR, Osman EY, Yen P-F, Garcia ML, Martin BR, Wetz MJ, Mazzasette C, Feng Z, Ko C-P, Lorson CL. Development and characterization of an SMN2-based intermediate mouse model of Spinal Muscular Atrophy. Hum. Mol. Genet. 2013;22:1843–1855. doi: 10.1093/hmg/ddt037. [DOI] [PubMed] [Google Scholar]
- 45.Park G-H, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J. Neurosci. 2010;30:12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gladman JT, Bebee TW, Edwards C, Wang X, Sahenk Z, Rich MM, Chandler DS. A humanized Smn gene containing the SMN2 nucleotide alteration in exon 7 mimics SMN2 splicing and the SMA disease phenotype. Hum. Mol. Genet. 2010;19:4239–4252. doi: 10.1093/hmg/ddq343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osborne M, Gomez D, Feng Z, McEwen C, Beltran J, Cirillo K, El-Khodor B, Lin M-Y, Li Y, Knowlton WM, McKemy DD, Bogdanik L, Butts-Dehm K, Martens K, Davis C, Doty R, Wardwell K, Ghavami A, Kobayashi D, Ko C-P, et al. Characterization of behavioral and neuromuscular junction phenotypes in a novel allelic series of SMA mouse models. Hum. Mol. Genet. 2012;21:4431–4447. doi: 10.1093/hmg/dds285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gogliotti RG, Hammond SM, Lutz C, Didonato CJ. Molecular and phenotypic reassessment of an infrequently used mouse model for spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2010;391:517–522. doi: 10.1016/j.bbrc.2009.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivanesan S, Howell MD, DiDonato CJ, Singh RN. Antisense oligonucleotide mediated therapy of spinal muscular atrophy. Translat. Neurosci. 2013;4:1–7. doi: 10.2478/s13380-013-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitrpant C, Porensky P, Zhou H, Price L, Muntoni F, Fletcher S, Wilton SD, Burghes AHM. Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy. PLOS One. 2013;8:e62114. doi: 10.1371/journal.pone.0062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osman EY, Yen P-F, Lorson CL. Bifunctional RNAs targeting the intronic splicing silencer N1 increase SMN levels and reduce disease severity in an animal model of spinal muscular atrophy. Mol. Ther. 2012;20:119–126. doi: 10.1038/mt.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porensky PN, Mitrpant C, McGovern VL, Bevan AK, Foust KD, Kaspar BK, Wilton SD, Burghes AHM. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, Hua Y, Rigo F, Matson J, Hung G, Kaye EM, Shihabuddin LS, Krainer AR, Bennett CF, Cheng SH. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cherry JJ, Osman EY, Evans MC, Choi S, Xing X, Cuny GD, Glicksman MA, Lorson CL, Androphy EJ. Enhancement of SMN protein levels in a mouse model of spinal muscular atrophy using novel drug-like compounds. EMBO Mol. Med. 2013;5:1103–1118. doi: 10.1002/emmm.201202305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Ronchi D, Simone C, Falcone M, Papadimitriou D, Locatelli F, Mezzina N, Gianni F, Bresolin N, Comi GP. Embryonic stem cell-derived neural stem cells improve spinal muscular atrophy phenotype in mice. Brain. 2010;133:465–481. doi: 10.1093/brain/awp318. [DOI] [PubMed] [Google Scholar]
- 56.Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Ronchi D, Saladino F, Bordoni A, Fortunato F, Del Bo R, Papadimitriou D, Locatelli F, Menozzi G, Strazzer S, Bresolin N, Comi GP. Neural stem cell transplantation can ameliorate the phenotype of a mouse model of spinal muscular atrophy. J. Clin. Invest. 2008;118:3316–3330. doi: 10.1172/JCI35432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bosch-Marcé M, Wee CD, Martinez TL, Lipkes CE, Choe DW, Kong L, Van Meerbeke JP, Musarò A, Sumner CJ. Increased IGF-1 in muscle modulates the phenotype of severe SMA mice. Hum. Mol. Genet. 2011;20:1844–1853. doi: 10.1093/hmg/ddr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen PC, Gaisina IN, El-Khodor BF, Ramboz S, Makhortova NR, Rubin LL, Kozikowski AP. Identification of a Maleimide-Based Glycogen Synthase Kinase-3 (GSK-3) Inhibitor, BIP-135, that Prolongs the Median Survival Time of Δ7 SMA KO Mouse Model of Spinal Muscular Atrophy. ACS Chem. Neurosci. 2012;3:5–11. doi: 10.1021/cn200085z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biondi O, Branchu J, Sanchez G, Lancelin C, Deforges S, Lopes P, Pariset C, Lécolle S, Côté J, Chanoine C, Charbonnier F. In vivo NMDA receptor activation accelerates motor unit maturation, protects spinal motor neurons, and enhances SMN2 gene expression in severe spinal muscular atrophy mice. J. Neurosci. 2010;30:11288–11299. doi: 10.1523/JNEUROSCI.1764-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nizzardo M, Nardini M, Ronchi D, Salani S, Donadoni C, Fortunato F, Colciago G, Falcone M, Simone C, Riboldi G, Govoni A, Bresolin N, Comi GP, Corti S. Beta-lactam antibiotic offers neuroprotection in a spinal muscular atrophy model by multiple mechanisms. Exp. Neurol. 2011;229:214–225. doi: 10.1016/j.expneurol.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 61.Rose FF, Jr, Mattis VB, Rindt H, Lorson CL. Delivery of recombinant follistatin lessens disease severity in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2009;18:997–1005. doi: 10.1093/hmg/ddn426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farooq F, Molina FA, Hadwen J, MacKenzie D, Witherspoon L, Osmond M, Holcik M, MacKenzie A. Prolactin increases SMN expression and survival in a mouse model of severe spinal muscular atrophy via the STAT5 pathway. J. Clin. Invest. 2011;121:3042–3050. doi: 10.1172/JCI46276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Meerbeke JP, Gibbs RM, Plasterer HL, Miao W, Feng Z, Lin M-Y, Rucki AA, Wee CD, Xia B, Sharma S, Jacques V, Li DK, Pellizzoni L, Rusche JR, Ko C-P, Sumner CJ. The DcpS inhibitor RG3039 improves motor function in SMA mice. Hum. Mol. Genet. 2013 doi: 10.1093/hmg/ddt257. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butchbach MER, Singh J, Thorsteinsdóttir M, Saieva L, Slominski E, Thurmond J, Andrésson T, Zhang J, Edwards JD, Simard LR, Pellizzoni L, Jarecki J, Burghes AHM, Gurney ME. Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum. Mol. Genet. 2010;19:454–467. doi: 10.1093/hmg/ddp510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heier CR, DiDonato CJ. Translational readthrough by the aminoglycoside geneticin (G418) modulates SMN stability in vitro and improves motor function in SMA mice in vivo. Hum. Mol. Genet. 2009;18:1310–1322. doi: 10.1093/hmg/ddp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mattis VB, Fosso MY, Chang C-W, Lorson CL. Subcutaneous administration of TC007 reduces disease severity in an animal model of SMA. BMC Neurosci. 2009;10:142. doi: 10.1186/1471-2202-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mattis VB, Ebert AD, Fosso MY, Chang C-W, Lorson CL. Delivery of a read-through inducing compound, TC007, lessens the severity of a spinal muscular atrophy animal model. Hum. Mol. Genet. 2009;18:3906–3913. doi: 10.1093/hmg/ddp333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Narver HL, Kong L, Burnett BG, Choe DW, Bosch-Marcé M, Taye AA, Eckhaus MA, Sumner CJ. Sustained improvement of spinal muscular atrophy mice treated with trichostatin A plus nutrition. Ann. Neurol. 2008;64:465–470. doi: 10.1002/ana.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Avila AM, Burnett BG, Taye AA, Gabanella F, Knight MA, Hartenstein P, Cizman Z, Di Prospero NA, Pellizzoni L, Fischbeck KH, Sumner CJ. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. Clin. J. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benkhelifa-Ziyyat S, Besse A, Roda M, Duque S, Astord S, Carcenac R, Marais T, Barkats M. Intramuscular scAAV9-SMN Injection Mediates Widespread Gene Delivery to the Spinal Cord and Decreases Disease Severity in SMA Mice. Mol. Ther. 2013;21:282–290. doi: 10.1038/mt.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dominguez E, Marais T, Chatauret N, Benkhelifa-Ziyyat S, Duque S, Ravassard P, Carcenac R, Astord S, Pereira de Moura A, Voit T, Barkats M. Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum. Mol. Genet. 2011;20:681–693. doi: 10.1093/hmg/ddq514. [DOI] [PubMed] [Google Scholar]
- 72.Valori CF, Ning K, Wyles M, Mead RJ, Grierson AJ, Shaw PJ, Azzouz M. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci. Transl. Med. 2010;2:35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- 73.Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, Le TT, Morales PR, Rich MM, Burghes AHM, Kaspar BK. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Passini MA, Bu J, Roskelley EM, Richards AM, Sardi SP, O’Riordan CR, Klinger KW, Shihabuddin LS, Cheng SH. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Azzouz M, Le T, Ralph GS, Walmsley L, Monani UR, Lee DCP, Wilkes F, Mitrophanous KA, Kingsman SM, Burghes AHM, Mazarakis ND. Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2004;114:1726–1731. doi: 10.1172/JCI22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou H, Janghra N, Mitrpant C, Dickinson RL, Anthony K, Price L, Eperon IC, Wilton SD, Morgan J, Muntoni F. A Novel Morpholino Oligomer Targeting ISS-N1 Improves Rescue of Severe Spinal Muscular Atrophy Transgenic Mice. Hum. Gene Ther. 2013;24:331–342. doi: 10.1089/hum.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grondard C, Biondi O, Armand A-S, Lécolle S, Della Gaspera B, Pariset C, Li H, Gallien C-L, Vidal P-P, Chanoine C, Charbonnier F. Regular exercise prolongs survival in a type 2 spinal muscular atrophy model mouse. J. Neurosci. 2005;25:7615–7622. doi: 10.1523/JNEUROSCI.1245-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gogliotti RG, Cardona H, Singh J, Bail S, Emery C, Kuntz N, Jorgensen M, Durens M, Xia B, Barlow C, Heier C, Plasterer HL, Jacques V, Kiledjian M, Jarecki J, Rusche J, Didonato CJ. The DcpS inhibitor RG3039 improves survival, function and motor unit pathologies in two SMA mouse models. Hum. Mol. Genet. 2013 doi: 10.1093/hmg/ddt258. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riessland M, Ackermann B, Förster A, Jakubik M, Hauke J, Garbes L, Fritzsche I, Mende Y, Blumcke I, Hahnen E, Wirth B. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum. Mol. Genet. 2010;19:1492–1506. doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 81.Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Glascock JJ, Osman EY, Wetz MJ, Krogman MM, Shababi M, Lorson CL. Decreasing disease severity in symptomatic, Smn(−/−);SMN2(+/+), spinal muscular atrophy mice following scAAV9-SMN delivery. Hum. Gene Ther. 2012;23:330–335. doi: 10.1089/hum.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 84.Chuang D-M, Leng Y, Marinova Z, Kim H-J, Chiu C-T. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kernochan LE, Russo ML, Woodling NS, Huynh TN, Avila AM, Fischbeck KH, Sumner CJ. The role of histone acetylation in SMN gene expression. Hum. Mol. Genet. 2005;14:1171–1182. doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- 86.Brichta L, Hofmann Y, Hahnen E, Siebzehnrubl FA, Raschke H, Blumcke I, Eyupoglu IY, Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 87.Sumner CJ, Huynh TN, Markowitz JA, Perhac JS, Hill B, Coovert DD, Schussler K, Chen X, Jarecki J, Burghes AHM, Taylor JP, Fischbeck KH. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 88.Andreassi C, Angelozzi C, Tiziano FD, Vitali T, De Vincenzi E, Boninsegna A, Villanova M, Bertini E, Pini A, Neri G, Brahe C. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 89.Tsai L-K, Tsai M-S, Lin T-B, Hwu W-L, Li H. Establishing a standardized therapeutic testing protocol for spinal muscular atrophy. Neurobiol. Dis. 2006;24:286–295. doi: 10.1016/j.nbd.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 90.Tsai L-K, Tsai M-S, Ting C-H, Li H. Multiple therapeutic effects of valproic acid in spinal muscular atrophy model mice. J. Mol. Med. 2008;86:1243–1254. doi: 10.1007/s00109-008-0388-1. [DOI] [PubMed] [Google Scholar]
- 91.Brichta L, Holker I, Haug K, Klockgether T, Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann. Neurol. 2006;59:970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 92.Swoboda KJ, Scott CB, Reyna SP, Prior TW, LaSalle B, Sorenson SL, Wood J, Acsadi G, Crawford TO, Kissel JT, Krosschell KJ, D’Anjou G, Bromberg MB, Schroth MK, Chan GM, Elsheikh B, Simard LR. Phase II open label study of valproic acid in spinal muscular atrophy. PLoS One. 2009;4:e5268. doi: 10.1371/journal.pone.0005268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swoboda KJ, Scott CB, Crawford TO, Simard LR, Reyna SP, Krosschell KJ, Acsadi G, Elsheik B, Schroth MK, D’Anjou G, LaSalle B, Prior TW, Sorenson SL, Maczulski JA, Bromberg MB, Chan GM, Kissel JT. SMA CARNI-VAL trial part I: double-blind, randomized, placebo-controlled trial of L-carnitine and valproic acid in spinal muscular atrophy. PLoS One. 2010;5:e12140. doi: 10.1371/journal.pone.0012140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kissel JT, Scott CB, Reyna SP, Crawford TO, Simard LR, Krosschell KJ, Acsadi G, Elsheik B, Schroth MK, D’Anjou G, LaSalle B, Prior TW, Sorenson S, Maczulski JA, Bromberg MB, Chan GM, Swoboda KJ. SMA CARNIVAL TRIAL PART II: a prospective, single-armed trial of L-carnitine and valproic acid in ambulatory children with spinal muscular atrophy. PLoS One. 2011;6:e21296. doi: 10.1371/journal.pone.0021296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darbar IA, Plaggert PG, Resende MBD, Zanoteli E, Reed UC. Evaluation of muscle strength and motor abilities in children with type II and III spinal muscle atrophy treated with valproic acid. BMC Neurol. 2011;11:36. doi: 10.1186/1471-2377-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brahe C, Vitali T, Tiziano FD, Angelozzi C, Pinto AM, Borgo F, Moscato U, Bertini E, Mercuri E, Neri G. Phenylbutyrate increases SMN gene expression in spinal muscular atrophy patients. Eur. J. Hum. Genet. 2005;13:256–259. doi: 10.1038/sj.ejhg.5201320. [DOI] [PubMed] [Google Scholar]
- 97.Somers E, Riessland M, Schreml J, Wirth B, Gillingwater TH, Parson SH. Increasing SMN levels using the histone deacetylase inhibitor SAHA ameliorates defects in skeletal muscle microvasculature in a mouse model of severe spinal muscular atrophy. Neurosci. Lett. 2013;544:100–104. doi: 10.1016/j.neulet.2013.03.052. [DOI] [PubMed] [Google Scholar]
- 98.Schreml J, Riessland M, Paterno M, Garbes L, Roßbach K, Ackermann B, Krämer J, Somers E, Parson SH, Heller R, Berkessel A, Sterner-Kock A, Wirth B. Severe SMA mice show organ impairment that cannot be rescued by therapy with the HDACi JNJ-26481585. Eur. J. Hum. Genet. 2012;21:643–652. doi: 10.1038/ejhg.2012.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mankin AS, Liebman SW. Baby, don’t stop! Nat. Genet. 1999;23:8–10. doi: 10.1038/12600. [DOI] [PubMed] [Google Scholar]
- 100.Wolstencroft EC, Mattis V, Bajer AA, Young PJ, Lorson CL. A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum. Mol. Genet. 2005;14:1199–1210. doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- 101.Mattis VB, Rai R, Wang J, Chang C-WT, Coady T, Lorson CL. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum. Genet. 2006;120:589–601. doi: 10.1007/s00439-006-0245-7. [DOI] [PubMed] [Google Scholar]
- 102.Mattis VB, Bowerman M, Kothary R, Lorson CL. A SMNDelta7 read-through product confers functionality to the SMNDelta7 protein. Neurosci. Lett. 2008;442:54–58. doi: 10.1016/j.neulet.2008.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singh J, Salcius M, Liu S-W, Staker BL, Mishra R, Thurmond J, Michaud G, Mattoon DR, Printen J, Christensen J, Bjornsson JM, Pollok BA, Kiledjian M, Stewart L, Jarecki J, Gurney ME. DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem. Biol. 2008;3:711–722. doi: 10.1021/cb800120t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jarecki J, Chen X, Bernardino A, Coovert DD, Whitney M, Burghes A, Stack J, Pollok BA. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum. Mol. Genet. 2005;14:2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- 105.Thurmond J, Butchbach MER, Palomo M, Pease B, Rao M, Bedell L, Keyvan M, Pai G, Mishra R, Haraldsson M, Andresson T, Bragason G, Thosteinsdottir M, Bjornsson JM, Coovert DD, Burghes AHM, Gurney ME, Singh J. Synthesis and biological evaluation of novel 2,4-diaminoquinazoline derivatives as SMN2 promoter activators for the potential treatment of spinal muscular atrophy. J. Med. Chem. 2008;51:449–469. doi: 10.1021/jm061475p. [DOI] [PubMed] [Google Scholar]
- 106.Xu C, Chen X, Grzeschik SM, Ganta M, Wang CH. Hydroxyurea enhances SMN2 gene expression through nitric oxide release. Neurogenetics. 2011;12:19–24. doi: 10.1007/s10048-010-0268-z. [DOI] [PubMed] [Google Scholar]
- 107.Liang W-C, Yuo C-Y, Chang J-G, Chen Y-C, Chang Y-F, Wang H-Y, Ju Y-H, Chiou S-S, Jong Y-J. The effect of hydroxyurea in spinal muscular atrophy cells and patients. J. Neurol. Sci. 2008;268:87–94. doi: 10.1016/j.jns.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 108.Chen T-H, Chang J-G, Yang Y-H, Mai H-H, Liang W-C, Wu Y-C, Wang H-Y, Huang Y-B, Wu S-M, Chen Y-C, Yang S-N, Jong Y-J. Randomized, double-blind, placebo-controlled trial of hydroxyurea in spinal muscular atrophy. Neurology. 2010;75:2190–2197. doi: 10.1212/WNL.0b013e3182020332. [DOI] [PubMed] [Google Scholar]
- 109.Lunn MR, Root DE, Martino AM, Flaherty SP, Kelley BP, Coovert DD, Burghes AH, Man NT, Morris GE, Zhou J, Androphy EJ, Sumner CJ, Stockwell BR. Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism. Chem. Biol. 2004;11:1489–1493. doi: 10.1016/j.chembiol.2004.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hastings ML, Berniac J, Liu YH, Abato P, Jodelka FM, Barthel L, Kumar S, Dudley C, Nelson M, Larson K, Edmonds J, Bowser T, Draper M, Higgins P, Krainer AR. Tetracyclines that promote SMN2 exon 7 splicing as therapeutics for spinal muscular atrophy. Sci. Transl. Med. 2009;1:5ra12. doi: 10.1126/scitranslmed.3000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rothstein JD, Patel S, Regan MR, Haenggeli C, Huang YH, Bergles DE, Jin L, Dykes Hoberg M, Vidensky S, Chung DS, Toan SV, Bruijn LI, Su Z-Z, Gupta P, Fisher PB. Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature. 2005;433:73–77. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- 112.Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl. Acad. Sci. U.S.A. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach MER, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AHM, Bollen M, Stamm S. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum. Mol. Genet. 2008;17:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Z, Kelemen O, van Santen MA, Yelton SM, Wendlandt AE, Sviripa VM, Bollen M, Beullens M, Urlaub H, Lührmann R, Watt DS, Stamm S. Synthesis and characterization of pseudocantharidins, novel phosphatase modulators that promote the inclusion of exon 7 into the SMN (survival of motoneuron) pre-mRNA. J. Biol. Chem. 2011;286:10126–10136. doi: 10.1074/jbc.M110.183970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dorn DC, Kou CA, Png KJ, Moore MAS. The effect of cantharidins on leukemic stem cells. Int. J. Cancer. 2009;124:2186–2199. doi: 10.1002/ijc.24157. [DOI] [PubMed] [Google Scholar]
- 116.Zhang ML, Lorson CL, Androphy EJ, Zhou J. An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA: potential therapy of SMA. Gene Ther. 2001;8:1532–1538. doi: 10.1038/sj.gt.3301550. [DOI] [PubMed] [Google Scholar]
- 117.Liu H-C, Ting C-H, Wen H-L, Tsai L-K, Hsieh-Li H-M, Li H, Lin-Chao S. Sodium vanadate combined with L-ascorbic acid delays disease progression, enhances motor performance, and ameliorates muscle atrophy and weakness in mice with spinal muscular atrophy. BMC Med. 2013;11:38. doi: 10.1186/1741-7015-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Koh S-H, Kim Y, Kim HY, Hwang S, Lee CH, Kim SH. Inhibition of glycogen synthase kinase-3 suppresses the onset of symptoms and disease progression of G93A-SOD1 mouse model of ALS. Exp. Neurol. 2007;205:336–346. doi: 10.1016/j.expneurol.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 119.Bordet T, Buisson B, Michaud M, Drouot C, Galéa P, Delaage P, Akentieva NP, Evers AS, Covey DF, Ostuni MA, Lacapère J-J, Massaad C, Schumacher M, Steidl E-M, Maux D, Delaage M, Henderson CE, Pruss RM. Identification and characterization of cholest-4-en-3-one, oxime (TRO19622), a novel drug candidate for amyotrophic lateral sclerosis. J. Pharmacol. Exp. Ther. 2007;322:709–720. doi: 10.1124/jpet.107.123000. [DOI] [PubMed] [Google Scholar]
- 120.Dimitriadi M, Kye MJ, Kalloo G, Yersak JM, Sahin M, Hart AC. The neuroprotective drug riluzole acts via small conductance Ca2+-activated K+ channels to ameliorate defects in spinal muscular atrophy models. J. Neurosci. 2013;33:6557–6562. doi: 10.1523/JNEUROSCI.1536-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haddad H, Cifuentes-Diaz C, Miroglio A, Roblot N, Joshi V, Melki J. Riluzole attenuates spinal muscular atrophy disease progression in a mouse model. Muscle Nerve. 2003;28:432–437. doi: 10.1002/mus.10455. [DOI] [PubMed] [Google Scholar]
- 122.Russman BS, Iannaccone ST, Samaha FJ. A phase 1 trial of riluzole in spinal muscular atrophy. Arch. Neurol. 2003;60:1601–1603. doi: 10.1001/archneur.60.11.1601. [DOI] [PubMed] [Google Scholar]
- 123.Ting C-H, Lin C-W, Wen S-L, Hsieh-Li H-M, Li H. Stat5 constitutive activation rescues defects in spinal muscular atrophy. Hum. Mol. Genet. 2007;16:499–514. doi: 10.1093/hmg/ddl482. [DOI] [PubMed] [Google Scholar]
- 124.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 125.Sumner CJ, Wee CD, Warsing LC, Choe DW, Ng AS, Lutz C, Wagner KR. Inhibition of myostatin does not ameliorate disease features of severe spinal muscular atrophy mice. Hum. Mol. Genet. 2009;18:3145–3152. doi: 10.1093/hmg/ddp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rindt H, Buckley DM, Vale SM, Krogman M, Rose FF, Jr, Garcia ML, Lorson CL. Transgenic inactivation of murine myostatin does not decrease the severity of disease in a model of Spinal Muscular Atrophy. Neuromuscul. Disord. 2012;22:277–285. doi: 10.1016/j.nmd.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 127.Duan C, Ren H, Gao S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. Gen. Comp. Endocrinol. 2010;167:344–351. doi: 10.1016/j.ygcen.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 128.Murdocca M, Malgieri A, Luchetti A, Saieva L, Dobrowolny G, de Leonibus E, Filareto A, Quitadamo MC, Novelli G, Musarò A, Sangiuolo F. IPLEX administration improves motor neuron survival and ameliorates motor functions in a severe mouse model of spinal muscular atrophy. Mol. Med. 2012;18:1076–1085. doi: 10.2119/molmed.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Glascock JJ, Shababi M, Wetz MJ, Krogman MM, Lorson CL. Direct central nervous system delivery provides enhanced protection following vector mediated gene replacement in a severe model of spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2012;417:376–381. doi: 10.1016/j.bbrc.2011.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hammond SM, Wood MJA. Genetic therapies for RNA mis-splicing diseases. Trends Genet. 2011;27:196–205. doi: 10.1016/j.tig.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 132.Havens MA, Duelli DM, Hastings ML. Targeting RNA splicing for disease therapy. Wiley Interdiscip Rev RNA. 2013;4:247–266. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shababi M, Glascock J, Lorson CL. Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy. Hum. Gene Ther. 2011;22:135–144. doi: 10.1089/hum.2010.114. [DOI] [PubMed] [Google Scholar]
- 134.Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- 135.Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Singh NN, Hollinger K, Bhattacharya D, Singh RN. An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing. RNA. 2010;16:1167–1181. doi: 10.1261/rna.2154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Singh NN, Seo J, Rahn SJ, Singh RN. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS One. 2012;7:e49595. doi: 10.1371/journal.pone.0049595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt609. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol. Cell. Biol. 2011;31:935–954. doi: 10.1128/MCB.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Klar J, Sobol M, Melberg A, Mäbert K, Ameur A, Johansson ACV, Feuk L, Entesarian M, Orlén H, Casar-Borota O, Dahl N. Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum. Mutat. 2013;34:572–577. doi: 10.1002/humu.22282. [DOI] [PubMed] [Google Scholar]
- 146.Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C, Guglieri M, Ashton E, Abbs S, Nihoyannopoulos P, Garralda ME, Rutherford M, McCulley C, Popplewell L, Graham IR, Dickson G, Wood MJ, Wells DJ, Wilton SD, Kole R, Straub V, Bushby K, Sewry C, Morgan JE, Muntoni F. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ, Dickson G, Wood MJ, Wilton SD, Straub V, Kole R, Shrewsbury SB, Sewry C, Morgan JE, Bushby K, Muntoni F. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Biondi O, Grondard C, Lécolle S, Deforges S, Pariset C, Lopes P, Cifuentes-Diaz C, Li H, della Gaspera B, Chanoine C, Charbonnier F. Exercise-induced activation of NMDA receptor promotes motor unit development and survival in a type 2 spinal muscular atrophy model mouse. J. Neurosci. 2008;28:953–962. doi: 10.1523/JNEUROSCI.3237-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Biondi O, Lopes P, Desseille C, Branchu J, Chali F, Salah AB, Pariset C, Chanoine C, Charbonnier F. Physical exercise reduces cardiac defects in type 2 spinal muscular atrophy-like mice. J. Physiol. (Lond.) 2012;590:5907–5925. doi: 10.1113/jphysiol.2012.238196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lewelt A, Newcomb TM, Swoboda KJ. New therapeutic approaches to spinal muscular atrophy. Curr. Neurol. Neurosci. Rep. 2012;12:42–53. doi: 10.1007/s11910-011-0240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bach JR, Baird JS, Plosky D, Navado J, Weaver B. Spinal muscular atrophy type 1: management and outcomes. Pediatr. Pulmonol. 2002;34:16–22. doi: 10.1002/ppul.10110. [DOI] [PubMed] [Google Scholar]
- 152.Gregoretti C, Ottonello G, Chiarini Testa MB, Mastella C, Ravà L, Bignamini E, Veljkovic A, Cutrera R. Survival of patients with spinal muscular atrophy type 1. Pediatrics. 2013;131:e1509–e1514. doi: 10.1542/peds.2012-2278. [DOI] [PubMed] [Google Scholar]
- 153.Finkel RS, Crawford TO, Swoboda KJ, Kaufmann P, Juhasz P, Li X, Guo Y, Li RH, Trachtenberg F, Forrest SJ, Kobayashi DT, Chen KS, Joyce CL, Plasterer T. Candidate proteins, metabolites and transcripts in the Biomarkers for Spinal Muscular Atrophy (BforSMA) clinical study. PLoS One. 2012;7:e35462. doi: 10.1371/journal.pone.0035462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Crawford TO, Paushkin SV, Kobayashi DT, Forrest SJ, Joyce CL, Finkel RS, Kaufmann P, Swoboda KJ, Tiziano D, Lomastro R, Li RH, Trachtenberg FL, Plasterer T, Chen KS. Evaluation of SMN protein, transcript, and copy number in the biomarkers for spinal muscular atrophy (BforSMA) clinical study. PLoS One. 2012;7:e33572. doi: 10.1371/journal.pone.0033572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kobayashi DT, Shi J, Stephen L, Ballard KL, Dewey R, Mapes J, Chung B, McCarthy K, Swoboda KJ, Crawford TO, Li R, Plasterer T, Joyce C, Chung WK, Kaufmann P, Darras BT, Finkel RS, Sproule DM, Martens WB, McDermott MP, et al. SMA-MAP: A Plasma Protein Panel for Spinal Muscular Atrophy. PLoS One. 2013;8:e60113. doi: 10.1371/journal.pone.0060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shen M, Bellaousov S, Hiller M, de La Grange P, Creamer TP, Malina O, Sperling R, Mathews DH, Stoilov P, Stamm S. Pyrvinium pamoate changes alternative splicing of the serotonin receptor 2C by influencing its RNA structure. Nucleic Acids Res. 2013;41:3819–3832. doi: 10.1093/nar/gkt063. [DOI] [PMC free article] [PubMed] [Google Scholar]