Abstract
Abasic sites in genomic DNA can be a significant source of mutagenesis in biological systems, including human cancers. Such mutagenesis requires translesion DNA synthesis (TLS) bypass of the abasic site by specialized DNA polymerases. The abasic site bypass specificity of TLS proteins had been studied by multiple means in vivo and in vitro, although the generality of the conclusions reached have been uncertain. Here, we introduce a set of yeast reporter strains for investigating the in vivo specificity of abasic site bypass at numerous random positions within chromosomal DNA. When shifted to 37°C, these strains underwent telomere uncapping and resection that exposed reporter genes within a long 3′ ssDNA overhang. Human APOBEC3G cytosine deaminase was expressed to create uracils in ssDNA, which were excised by uracil-DNA N-glycosylase. During repair synthesis, error-prone TLS bypassed the resulting abasic sites. Because of APOBEC3G’s strict motif specificity and the restriction of abasic site formation to only one DNA strand, this system provides complete information about the location of abasic sites that led to mutations. We recapitulated previous findings on the roles of REV1 and REV3. Further, we found that sequence context can strongly influence the relative frequency of A or C insertion. We also found that deletion of Pol32, a non-essential common subunit of Pols δ and ζ, resulted in residual low-frequency C insertion dependent on Rev1 catalysis. We summarize our results in a detailed model of the interplay between TLS components leading to error-prone bypass of abasic sites. Our results underscore the utility of this system for studying TLS bypass of many types of lesions within genomic DNA.
Keywords: abasic site, translesion DNA synthesis, APOBEC, single-strand DNA, deoxycytidyltransferase
1. Introduction
DNA is under constant threat of damage by both endogenous and environmental agents [1]. Damage to the nitrogenous bases that result in miscoding can be particularly deleterious if left unrepaired before a subsequent round of replication, as this can result in mutation fixation [1]. A principal means of correcting various adduct lesions is by base excision repair (BER), which is initiated when a DNA N-glycosylase specifically recognizes its cognate adducted base substrate and excises it to generate an abasic site [2]. Abasic sites also are generated when glycosylases erroneously excise normal bases [3] or by spontaneous breakage of the N-glycosidic bond, especially in adducted nucleotides where this bond is destabilized [4]. Usually, short- or long-patch BER replaces the damaged nucleotide (along with some neighboring residues, in the latter case), using the intact complementary strand as a template [2].
Abasic sites are thought to be one of the most common lesions within DNA, under steady state, unstressed conditions [5]. While one would expect that the majority of such lesions should be repaired correctly by BER, abasic sites formed within single-strand DNA (ssDNA) likely would not be subject to such error-free repair. This is because the complementary strand to template repair synthesis is, by definition, absent. Moreover, it would be counterproductive to attempt BER on an abasic site within ssDNA of a replication fork, as doing so would risk strand breakage and fork collapse [6]. Similarly, BER would be unavailable for repairing abasic sites formed by strand resection from double-strand breaks or within subtelomeric ssDNA following telomere uncapping. In the latter scenario, we previously observed clusters of multiple base substitutions originating from abasic sites on the 3′ ssDNA overhang strand [7]. More recently, Neuberger and colleagues expressed hyperactive cytosine deaminases in yeast and found similar mutation showers, which were associated with breakpoints and thought to result from damage to ssDNA at resected double strand breaks [8]. Such formations as observed by both groups are reminiscent of similar clusters of multiple point mutations identified among various human cancers, also likely to have originated from many abasic sites within long stretches of ssDNA [9–11].
When confronted with abasic sites in a DNA configuration that is not amenable to BER, the cell resorts to using mechanisms of DNA damage tolerance that result in bypass of the lesion without repairing it (reviewed recently in [6, 12]). One such mechanism is translesion DNA synthesis (TLS) by specialized DNA polymerases, which can be error-prone, i.e. mutagenic (reviewed recently in [12–14]). This is because TLS polymerases can contain relatively large active sites that accommodate non-canonical bases (e.g., [15, 16]) and lack the proofreading activity of the higher fidelity replicative polymerases (reviewed in [17, 18]). As a result, TLS polymerases are considerably more error-prone, even when copying undamaged DNA templates (e.g., [19, 20]). Thus, cells have evolved these low fidelity polymerases as a means to bypass genomic lesions that would otherwise block replication, but at the cost of possible mutation fixation [12–14].
Conserved proteins that take part in TLS have been identified in many organisms, including budding yeast [21–24]. Yeast has a repertoire of three specialized TLS polymerases whose functional specificity in bypassing abasic sites has been investigated by the following approaches: in vitro TLS assays (e.g., [25–27]); transfecting cells with plasmids carrying a single, engineered abasic site [28–30]; expressing mutator DNA glycosylases that generate excess abasic sites in BER-deficient strains [31, 32]; and analyzing unselected abasic site-associated TLS events within a frameshift reversion reporter, in excision repair defective backgrounds [33]. Pol η, which catalyzes error-free bypass of UV-induced cyclobutane pyrimidine dimers, is encoded by the RAD30 gene [34]. Pol ζ is proficient at extending from various mismatched termini during TLS in vitro (e.g., [27, 35, 36]) and includes a catalytic subunit encoded by REV3 along with an accessory subunit encoded by REV7 [37]. There is also evidence that Pol ζ, acting alone, can bypass some lesions in vitro [37, 38]. REV1 encodes a protein with deoxycytidyltransferase activity [26] and provides a structural function crucial for TLS bypass of certain lesions (e.g., [39–41]). Additionally, the replicative polymerase δ has been implicated in the insertion of A opposite abasic sites [27]. A non-essential accessory subunit of both Pol δ and Pol ζ, encoded by POL32 [42–44], is necessary for efficient TLS bypass of various lesions, including abasic sites [29, 30].
Previously, we used a subtelomeric ssDNA mutagenesis reporter system to confirm that the mutagenic bypass of abasic sites (resulting from Ung1-catalyzed excision of uracils [45] that were formed by cytosine deamination) within chromosomal DNA required the TLS activity of Pol ζ [7]. In contrast, when uracils were left intact in ung1Δ cells, mutagenesis was completely independent of TLS, as deletion of REV3 did not affect the frequency of gene inactivation at all in this background [7]. Thus, the TLS dependence in cells with Ung1 argues that mutagenesis due to residual amounts of unexcised uracils was at most, a minor factor. Here, we apply the subtelomeric ssDNA mutagenesis reporter system to investigate the roles of TLS proteins in the error-prone bypass of randomly generated abasic sites within chromosomal DNA. We found that similar to human cancers [9–11], A and C tend to be incorporated opposite abasic sites at similar frequencies. Based on our results, we infer the respective roles and relative contributions of the various TLS proteins toward the error-prone bypass of abasic sites within genomic DNA.
2. Materials and Methods
2.1. Yeast Strains
Yeast strains used in this study are listed in Table 1. All are isogenic to CG379 [46] with the following common markers: MATα his7-2 leu2-3,112 trp1-289. WT and rev3 subtelomeric reporter strains bearing tetracycline regulatable APOBEC3G plasmids were described previously [7]. Other TLS gene deletion strains were constructed by one step gene replacement [47]. The rev1-AA allele (with D647A and E648A amino acid substitutions [27]) was constructed by the delitto perfetto approach [48]. Strain constructions were verified by diagnostic replica plating, PCR, and sequencing (for rev1-AA). Strains were maintained on TRP dropout media to select for plasmid retention.
Table 1.
List of yeast strains used in this study.
| strain name(s) | genotype | plasmid |
|---|---|---|
| yKC066, yKC067 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 lys2::CAN1-URA3-ADE2 (in sub-telomere 5L)a | pCM252-A3G |
| yKC068, yKC069 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 lys2::CAN1-URA3-ADE2 (in sub-telomere 5L)a | pCM252 |
| yKC213, yKC214 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rad30::HygR lys2::CAN1- URA3-ADE2 (in sub-telomere 5L) | pCM252-A3G |
| yKC217, yKC218 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rad30::HygR lys2::CAN1- URA3-ADE2 (in sub-telomere 5L) | pCM252 |
| yKC131, yKC135 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rev3::NatR lys2::CAN1-URA3- ADE2 (in sub-telomere 5L)a | pCM252-A3G |
| yKC133, yKC137 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rev3::NatR lys2::CAN1-URA3- ADE2 (in sub-telomere 5L)a | pCM252 |
| yKC205, yKC206 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rev1::HygR lys2::CAN1-URA3- ADE2 (in sub-telomere 5L) | pCM252-A3G |
| yKC209, yKC210 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rev1::HygR lys2::CAN1-URA3- ADE2 (in sub-telomere 5L) | pCM252 |
| yKC385 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rev1-AA lys2::CAN1-URA3- ADE2 (in sub-telomere 5L) | pCM252-A3G |
| yKC387, yKC388 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rev1-AA lys2::CAN1-URA3- ADE2 (in sub-telomere 5L) | pCM252 |
| yKC299, yKC230 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rad30::HygR rev3::NatR lys2::CAN1-URA3-ADE2 (in sub-telomere 5L) | pCM252-A3G |
| yKC233, yKC234 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rad30::HygR rev3::NatR lys2::CAN1-URA3-ADE2 (in sub-telomere 5L) | pCM252 |
| yKC245, yKC246 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rad30::KanMX4 rev1::HygR lys2::CAN1-URA3-ADE2 (in sub-telomere 5L) | pCM252-A3G |
| yKC249, yKC250 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 rad30::KanMX4 rev1::HygR lys2::CAN1-URA3-ADE2 (in sub-telomere 5L) | pCM252 |
| yKC261, yKC262 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 pol32::HygR lys2::CAN1- URA3-ADE2 (in sub-telomere 5L) | pCM252-A3G |
| yKC265 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 pol32::HygR lys2::CAN1- URA3-ADE2 (in sub-telomere 5L) | pCM252 |
| yKC399 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 pol32::HygR rev1-AA lys2::CAN1-URA3-ADE2 (in sub-telomere 5L) | pCM252-A3G |
| yKC400, yKC402 | MATα his7-2 leu2-3,112 trp1-289 cdc13-1 pol32::HygR rev1-AA lys2::CAN1-URA3-ADE2 (in sub-telomere 5L) | pCM252 |
The WT and rev3 strains were described previously in [7]. All other yeast strains originate from this study.
2.2. Determination of CAN1 Inactivation Frequency
For each combination of genotype and plasmid, multiple independent colonies each were inoculated into 5 mL of YPDA liquid (1% yeast extract, 2% peptone, 2% dextrose, supplemented with 0.01% adenine sulfate, filter-sterilized) and grown at 23°C for two days. Then, 500 μL of each culture was added to 4.5 mL of fresh YPDA with 10 μg/mL doxycycline hyclate [49] (Sigma-Aldrich, St. Louis, MO) and shifted to 37°C for six hours. Cells were counted and G2 arrest was monitored by examining cell morphology. Cells were washed once in water. 500 cells were plated, in triplicate, onto synthetic complete to determine plating efficiency. An appropriate dilution for each strain was plated, in triplicate, onto ARG dropout plates with 60 mg/mL canavanine sulfate and 20 mg/mL adenine sulfate to select for mutants. Plates were counted after incubation at 23°C for five days. Canavanine selection plates were replica plated onto glycerol plates to eliminate cultures started by petite colonies from further consideration.
2.3. Collection and Sequencing of CAN1 Inactivation Mutants
Cultures derived from independent colonies were started and temperature shifted as described in 2.2. From each culture, an appropriate dilution was plated onto a single canavanine selection plate and incubated for five days at 23°C. A single colony, located closest to a fixed point on each plate, was streaked onto YPDA. A single colony from each streak was patched onto YPDA, grown for two days at 23°C, and replica plated onto canavanine media to verify loss of CAN1 function, and onto glycerol media to verify normal aerobic respiration capacity. Genomic DNA was prepared from each mutant isolate using a QIAcube robot (QIAGEN, Valencia, CA), according to the manufacturer’s DNeasy protocol. Using standard methods, CAN1 was PCR amplified and sequenced (primers are listed in Supplementary Table 1), and mutations were identified, as described previously [7].
2.4. Statistical Analyses
Prism 6 software (GraphPad Software, LaJolla, CA) was used to carry out statistical tests on the data. The Kolmogorov-Smirnov test was used for pairwise comparisons (between TLS genotypes) of CAN1 inactivation frequency and plating efficiency. The two-tailed 2-by-2 Fisher’s exact test or the Chi-square test was used to compare relative proportions of different mutation types between TLS genotypes, or between APOBEC3G vs. empty vector in the same TLS genotype.
3. Results
3.1. A model system to assess the roles of TLS proteins in the mutagenic bypass of abasic sites within chromosomal DNA
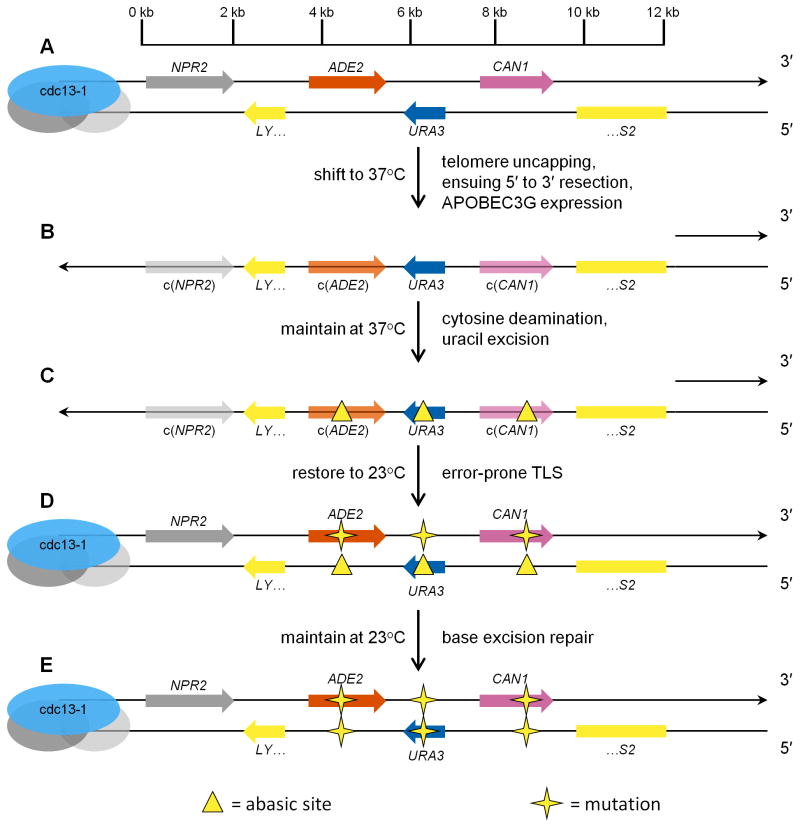
The experimental design is summarized in Figure 1. When reporter cells are shifted from 23° to 37°C, telomere uncapping and ensuing enzymatic resection produce long 3′ ssDNA overhangs [50]. The sequences that either code for, or are complementary to, three reporter genes are located within the left subtelomeric region of Chromosome V. We expressed human APOBEC3G [51, 52] to generate abasic sites in the subtelomeric ssDNA overhang. APOBEC3G deaminates the 3′ C within CC motifs [53] of the ssDNA reporter region at random, but does not react with duplex DNA [54]. Ung1 then excises the resulting uracils to generate abasic sites [45], which are bypassed by error-prone TLS during re-synthesis of the resected strand, when cells are returned to 23°C. This series of events results in mutations that are identified by plating on media that selects cells with inactivated CAN1 [55]. A unique strength of this experimental design is that it enables the characterization of mutagenic bypass at endogenously generated abasic sites, at a well-defined sequence motif, in numerous random locations within chromosomal DNA. This approach should yield results of broader generality than other methods, e.g. surveying TLS opposite plasmid-borne single, engineered abasic sites that are introduced into the cell by transfection [28–30].
Figure 1.
Genetic construct and experimental design to assess mutagenesis arising from random abasic sites within subtelomeric ssDNA. (A) Each reporter strain bears the cdc13-1 mutation and three reporter genes (ADE2, URA3, and CAN1), near a de novo telomere on the left arm of Chromosome V. (B) Temperature shift results in dissociation of the protective proteinaceous cap of the telomere and enzymatic 5′ to 3′ resection, generating a long 3′ overhang of ssDNA. Simultaneously, APOBEC3G cytosine deaminase expression is driven by adding doxycycline. c(ADE2) and c(CAN1) denote the complement of the two genes. (C) APOBEC3G deaminates the 3′ cytosines of CC motifs at random within the ssDNA overhang. Ung1 generates abasic sites by excising the resulting uracils. (D) When restored to permissive temperature, error-prone TLS usually inserts A or C opposite the abasic sites. The restored duplex, containing abasic site(s), is a substrate for base excision repair. (E) After base excision repair, mutations are fixed on both strands.
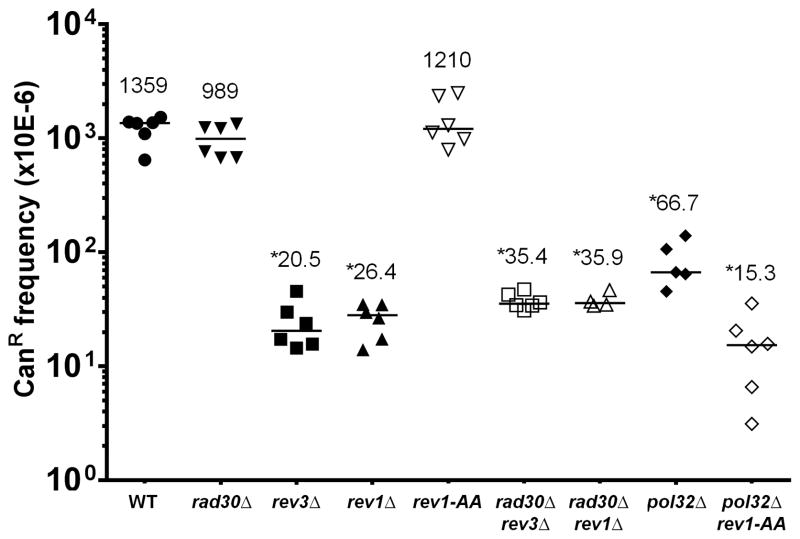
To characterize the roles of proteins involved in TLS, we first determined the frequency of CAN1 inactivation among cells expressing APOBEC3G during temperature shift, as a means to estimate the frequency of mutagenic TLS at abasic sites (see Figure 2). Data on plating efficiencies and CAN1 inactivation frequencies of empty vector controls are in Supplementary Figures 1 and 2, respectively. Consistent with in vivo studies based on transfection of plasmids bearing a single, designed abasic site [29, 30], we found that REV3 and REV1 both were necessary for the vast majority of mutagenic TLS, while deletion of RAD30 did not affect the frequency of TLS significantly compared to WT. In contrast to cells deleted for REV1, we found that cells lacking only Rev1 catalytic deoxycytidyltransferase activity [27] (i.e. rev1-AA cells) exhibited a gene inactivation frequency that was indistinguishable from WT, supporting prior observations that only the structural function of Rev1 is required for WT levels of TLS at a model abasic site [30]. Deletion of POL32 also resulted in a significant decrease in frequency of TLS when compared to WT, consistent with previous reports [29, 30]. Finally, deletion of POL32 in the rev1-AA background resulted in a further 4.4-fold decrease in frequency of TLS when compared to the pol32 single mutant (P = 0.0043 by Kolmogorov-Smirnov test), suggesting that the majority of low frequency, residual TLS in pol32 cells involved Rev1 catalytic activity.
Figure 2.
CAN1 inactivation frequencies in cdc13-1 reporter strains expressing APOBEC3G cytosine deaminase during temperature shift-induced generation of subtelomeric ssDNA. Each data point represents an independent replicate derived from a single colony, numbers indicate median values, and asterisks denote significant difference (P < 0.05) compared to WT by the Kolmogorov-Smirnov test. Canavanine selection plates were counted after five days of growth at 23°C following mutagenic exposure. See Materials and Methods for details of experimental procedures.
3.2. Sequence context can influence the choice of nucleotide inserted opposite abasic sites in WT cells
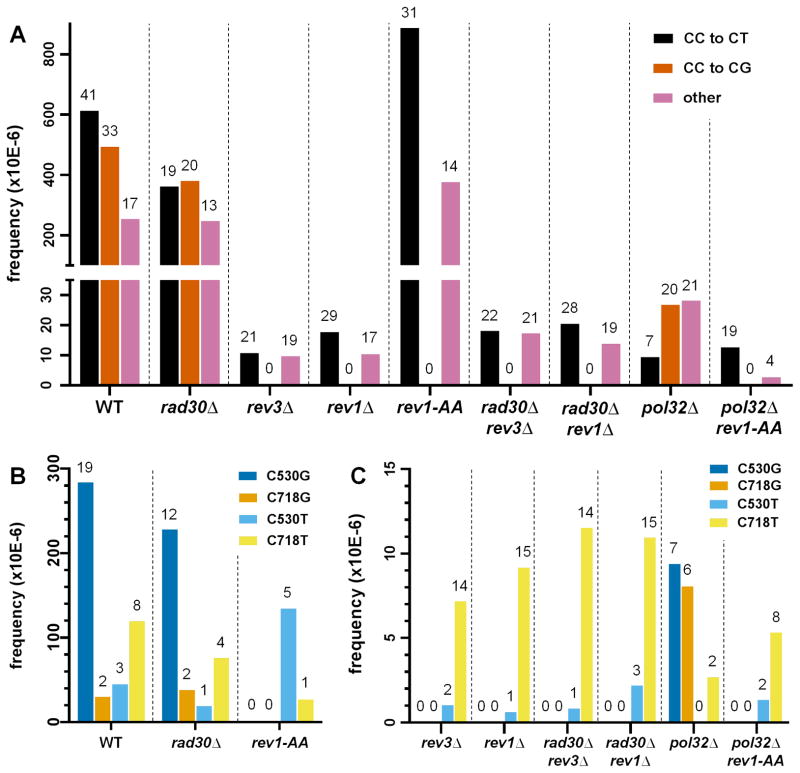
To glean further insight into the molecular mechanisms underlying mutagenic abasic site bypass, we sequenced the CAN1 ORF from independent mutant isolates. Starting with cells WT for TLS, we sequenced 84 isolates, which yielded 91 mutations in total within CAN1. Detailed mutation data for all genotypes, either expressing APOBEC3G or bearing empty vector, are in Supplementary Tables 2 and 3, respectively. 74 of the mutations occurred at the 3′ C of CC motifs, i.e. at the cognate motif of APOBEC3G, and thus were likely to have originated from abasic sites. 41 were CC to CT transitions (i.e., insertion of A opposite the abasic site), while 33 were CC to CG transversions (insertion of C). To obtain the frequency of a given class of mutations, we multiplied the overall CAN1 inactivation frequency (1.36×10−3 for WT) by the ratio of the number of mutations of that class to the total number of mutations (see Figure 3A).
Figure 3.
Frequencies of various types of mutations. Numbers above each bar indicate the number of independent mutant isolates within each category. (A) Mutations are grouped as CC to CT, CC to CG, and all others. Note that significant proportions of mutations occurred at the APOBEC3G cognate motif, namely CC. (B) Two mutational hotspots, C530 and C718 are shown for WT, rad30, and rev1-AA, three genetic backgrounds that are proficient for TLS bypass opposite abasic sites. Abasic sites at C530 favored TLS insertion of C. In contrast at C718, insertion of A was favored. (C) Frequencies of substitutions at C530 and C718 for TLS deficient backgrounds. The prevalence of C530G and C718G substitutions in pol32 was unique among TLS deficient genotypes.
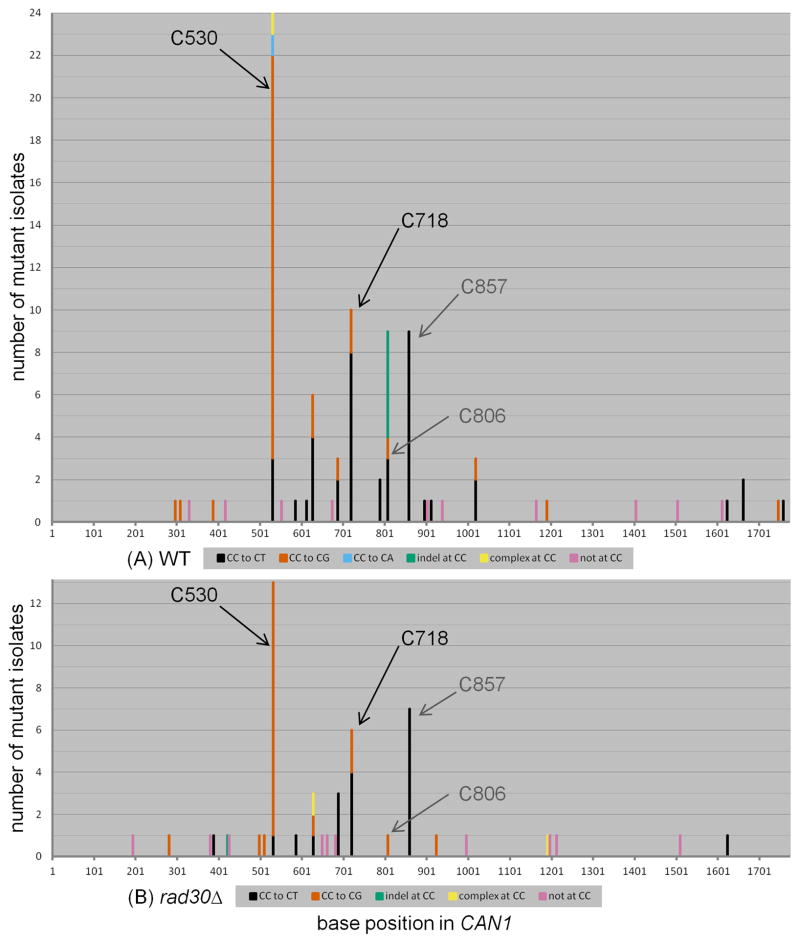
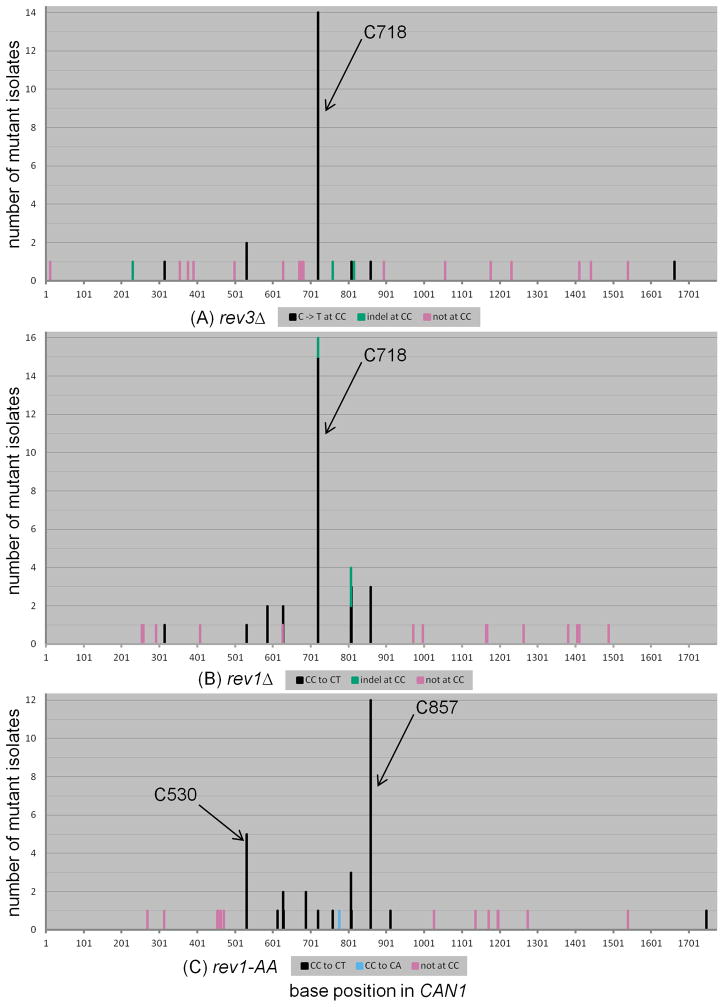
While mutations were found at 19 CC sites spread across the CAN1 reporter, we noticed that some positions contained considerably more mutations than others (Figure 4A). Specifically, 32 out of 74 mutations occurred in just two positions, C530 and C718, where both CC to CT and CC to CG substitutions were observed (see also Supplementary Table 4). The C to G allele at position 530 results in a tryptophan to serine substitution, while the C to T allele results in an amber stop codon. Both the C to G and C to T alleles at position 718 yield glycine to arginine substitutions. (Since the coding strand for CAN1 is complementary to the ssDNA overhang left by resection from the uncapped telomere, the actual coding changes are complementary to the mutations occurring at CC motifs in the ssDNA overhang.) All four alleles (C530G, C530T, C718G, and C718T) were clearly canavanine resistant. Interestingly, these positions differed for the relative proportion of CC to CT vs. CC to CG (P = 0.0006 by Fisher’s exact test; see also Figure 4A). There was a clear preference for insertion of C across an abasic site at position 530 versus a preference for insertion of A across an abasic site at position 718, suggesting a genuine bias in favor of one type of TLS transaction over the other. We propose that the local sequence context at these two positions can influence the choice of nucleotide inserted opposite the abasic site. However, it is not clear which differences in sequence could be influencing this choice, as there are many differences in the respective flanking sequences (see Discussion).
Figure 4.
The mutation spectra of (A) WT and (B) rad30 strains expressing APOBEC3G during temperature shift. Position 530 exhibited a strong preference for inserting C opposite the abasic site, resulting in frequent CC to CG transversions. Position 718 exhibited a preference for inserting A opposite the abasic site, resulting in CC to CT transitions. C857 exhibited CC to CT transitions exclusively, possibly because the C857G transversion, resulting in a tryptophan to serine amino acid change, did not inactivate CAN1. The respective X- and Y-axes in Figures 4, 5, and 6 are of the same scale.
We also noticed that the highest number of CC to CT transitions was observed in position 857, which creates an amber stop codon. However the C857G allele, which would result in a tryptophan to serine substitution, was never isolated as an inactivating mutation in any of numerous studies with published CAN1 mutation spectra [53, 56–60]. Similarly, we have never observed a C857G allele resulting in gene inactivation among existing datasets in our group ([10, 61, 62] and unpublished observations). As it is quite possible that the C857G allele does not inactivate CAN1, we do not consider position 857 to be a site where any preference for insertion of A (instead of C) can be inferred. Nonetheless, the sharp contrast in nucleotide insertion preferences between positions 530 and 718 argues that not all abasic sites are processed in the same manner by the TLS machinery.
3.3. Rad30 (Pol η) does not play a significant role in mutagenic TLS opposite abasic sites
Consistent with previous studies [29, 30], we found that deletion of RAD30 did not alter the frequency of mutagenic TLS opposite abasic sites significantly, when compared to WT (see Figure 2). If Rad30 does not play any significant role in TLS opposite abasic sites, then the mutation spectrum of rad30 cells also should be essentially the same as that of WT. We sequenced 48 independent isolates from rad30 cultures treated with APOBEC3G, to obtain 52 mutations. Similar to WT, the large majority of mutations (39 of 52) occurred at CC motifs and there were similar numbers of CC to CT transitions (19) and CC to CG transversions (20) (see Figure 3A). Also similar to WT, the rad30 mutation spectrum revealed a CC to CG preference at position 530 and a CC to CT preference at position 718 (P = 0.0006 when comparing these two sites by Fisher’s exact test; see Figure 3B and Supplementary Table 4). Application of the Chi-square test to compare relative proportions of mutation types confirmed that there was no significant difference between WT and rad30 (see Table 2).
Table 2.
Comparison of proportions of mutation types between WT and each TLS mutant.
| genotype | CC to CG transversions | CC to CT transitions | all other mutations | Chi-square P-value vs. WT |
|---|---|---|---|---|
| WT | 33 | 41 | 17 | n/a |
| rad30 | 19 | 20 | 13 | 0.6164 |
| rev3 | 0 | 21 | 19 | <0.0001 |
| rev1 | 0 | 29 | 17 | <0.0001 |
| rev1-AA | 0 | 31 | 14 | <0.0001 |
| rad30 rev3 | 0 | 22 | 22 | <0.0001 |
| rad30 rev1 | 0 | 28 | 19 | <0.0001 |
| pol32 | 20 | 7 | 21 | 0.0004 |
| pol32 rev1-AA | 0 | 19 | 4 | 0.0013 |
The only apparent possible difference between the WT and rad30 spectra was at position 806 (compare Figure 4A to 4B). In WT, this site exhibited three CC to CT transitions, one CC to CG transversion, and five single base deletions of C, i.e. nine mutations in total. By contrast in rad30, we observed a lone CC to CG transversion. Yet, comparing the relative ratios of mutations at C806, between WT and rad30 by two-tailed Fisher’s exact test, we obtained a P-value that was not quite statistically significant (P = 0.0936). Altogether, we conclude that Rad30 plays, at most, a negligible role in the bypass of abasic sites in vivo.
3.4. Rev3 and Rev1 are required for efficient TLS, and for the insertion of C opposite abasic sites
Deletion of either REV3 (encoding the catalytic subunit of Pol ζ) or REV1 resulted in 66- and 51-fold decreases in TLS frequency when compared to WT, respectively (see Figure 2). Dividing the CAN1 inactivation frequency of each mutant by that of WT, we infer that rev3 retained 1.5% of the TLS capacity of WT, while rev1 retained 1.9% of WT capacity. This is in good agreement with the results of Lawrence and colleagues, who obtained values of 4.9% and 3.4% for rev3 and rev1, respectively, using a plasmid-based in vivo assay [28]. Prakash and colleagues, using a similar approach as Lawrence, derived values of 8% and 41% of WT TLS capacity for rev3 and rev1, respectively [30]. Thus, the rev3 data for all three studies were similar, while the higher TLS capacity observed by Prakash and colleagues for rev1 was not observed in Lawrence’s system or ours.
We sequenced 40 rev3- and 46 rev1-derived isolates, each with one mutation in CAN1 and found that the mutation spectra for both rev3 and rev1 were completely devoid of CC to CG transversions (see Figure 3A). In contrast, CC to CT transitions comprised over half of total mutations for both genotypes. The differences in proportion of mutation types were highly significant when compared to WT (P < 0.0001 by Chi-square test for either rev3 or rev1 vs. WT, see Table 2). Only two out of 21 independent isolates each from rev3 and rev1 bearing empty vector exhibited CC to CT transitions (no CC to CG transversions were observed; see Supplementary Table 3), even in genetic backgrounds grossly deficient for TLS. Fisher’s exact test P-values comparing the proportion of CC to CT mutations in rev3 cells expressing APOBEC3G vs. empty vector is 0.0009. The corresponding P-value for rev1 cells is 0.0001. Thus, the comparison between APOBEC3G-expressing strains to empty vector controls clearly showed the mutagenic specificity of this cytosine deaminase, even in genetic backgrounds that were grossly TLS deficient.
The lack of C insertion in our system is in good agreement with results from Lawrence’s group [29], but less so with Prakash’s group [30], who observed 31% and 10% insertion of C opposite their model abasic site in the rev3 and rev1 backgrounds, respectively. In addition, our mutation spectra exhibited a strong CC to CT hotspot at position 718 (see Figures 5A, 5B, and 3C). Also note that position 718 tended to favor insertion of A (CC to CT transitions) in WT (see Figures 3B, 4A, and Supplementary Table 4). Our results suggest that position 718 might be relatively easy to bypass, even in genotypes with severe TLS deficiency, perhaps by the action of replicative polymerase δ alone. While we cannot rule out the possibility that residual CC to CT transitions observed in rev3 or rev1 could be due to small numbers of unexcised uracils, the overall agreement with the results of Lawrence [29] and Prakash [30] is consistent with low-frequency TLS.
Figure 5.
The mutation spectra of (A) rev3, (B) rev1, and (C) rev1-AA strains expressing APOBEC3G during temperature shift. All three genotypes were devoid of CC to CG transversions. Deletion of either REV3 (A) or REV1 (B) resulted in a prominent CC to CT hotspot at position 718. In contrast, rev1-AA cells (C) exhibited frequent CC to CT transitions at position 857. In addition, CC to CT transitions were observed at position 530, whereas in WT cells this was a strong CC to CG hotspot. These data suggest that insertion of C opposite abasic sites was entirely attributable to the deoxycytidyltransferase activity of Rev1.
Finally, we found that rad30 rev3 and rad30 rev1 double mutants behaved similarly to rev3 and rev1 single mutants, respectively (see Figures 2, 3A, 3C, and Supplementary Table 4). These results suggest that Rad30 truly does not play any significant role during mutagenic bypass of abasic sites, not even in a backup capacity when either Pol ζ or Rev1 are unavailable.
3.5. The structural function of Rev1 enables efficient TLS, but catalytic function is necessary for insertion of C opposite abasic sites
Rev1 is thought to serve a dual role in enabling TLS. It possesses a deoxycytidyltransferase activity and a structural role (independent of its enzymatic function) that facilitates the activity of other TLS proteins (reviewed in [12–14]). Since no CC to CG mutations were found among the 46 independent rev1-derived isolates with inactivated CAN1, we tested whether Rev1 catalytic activity was responsible for insertion of C opposite abasic sites. As mentioned, rev1-AA cells exhibited WT levels of overall TLS frequency (see Figure 2). This indicates that Rev1’s structural function is in itself, sufficient to support efficient TLS, in some form.
Although the overall TLS frequencies were similar between rev1-AA and WT, the compositions of the mutation spectra were radically different. We sequenced 42 independent rev1-AA isolates to obtain 45 mutations in CAN1. 31 mutations were CC to CT while none were CC to CG (see Figure 3A). With the loss of Rev1 catalytic activity, the prominent CC to CG signature at position 530 was abolished, leaving only the residual CC to CT transitions also observed at this site in WT (compare Figure 5C to Figure 4A and see Supplementary Table 4). Another consequence was that C857T became the most prominent hotspot in the rev1-AA spectrum (see Figure 5C). Altogether, these data suggest that the deoxycytidyltransferase activity of Rev1 is necessary for the vast majority of (if not all) instances where C is inserted opposite abasic sites, consistent with previous reports based on assays of TLS opposite a single, engineered abasic site in plasmid substrates [30] and analysis of unselected TLS events in a genetic reversion system within excision repair deficient backgrounds [33].
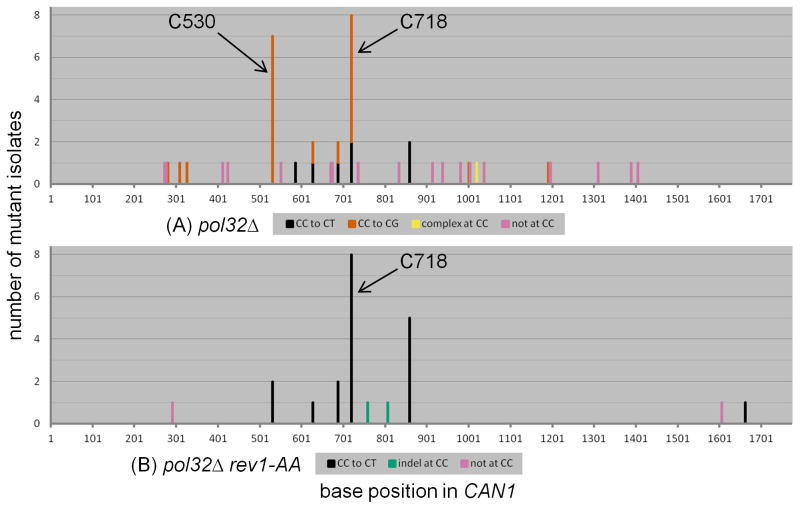
3.6. In contrast to rev3 and rev1, pol32 cells retain a residual capacity to insert C opposite abasic sites
Pol32 is an accessory subunit of both Pol δ (a replicative polymerase) and Pol ζ (a TLS polymerase) [42–44]. As mentioned previously, deletion of POL32 resulted in a TLS defect that was comparable to that seen in rev3 and rev1 (see Figure 2). But given the dual role of Pol32, we surmised that there might be differences in its mutation spectrum when compared to either rev3 or rev1. We sequenced 48 independent pol32 isolates, each containing a single mutation in CAN1 (see Figure 6A). In contrast to the absence of CC to CG transversions in rev3 and rev1, we observed 20 such substitutions in pol32 (see Figure 3A). 13 of these CC to CG transversions occurred at positions 530 or 718 (see Figure 3C and Supplementary Table 4). In addition, the relative proportions of CC to CT, CC to CG, and all other mutations were significantly different compared to WT (P = 0.0004 by Chi-square test, see Table 2). Thus, pol32 exhibited a mutation signature that was unique among the various genetic backgrounds examined.
Figure 6.
The mutation spectra of (A) pol32 and (B) pol32 rev1-AA subtelomeric ssDNA reporter cells expressing APOBEC3G during temperature shift. (A) In contrast to all other genotypes examined, pol32 cells exhibited CC to CG hotspots at both positions 530 and 718. Also, CC to CT transitions were relatively infrequent. (B) CC to CG transversions were abolished in pol32 rev1-AA cells. There were frequent CC to CT transitions at position 718.
The relative prominence of CC to CG transversions in pol32 led us to test whether Rev1deoxycytidyltransferase activity was necessary for these substitutions. As already mentioned, loss of Rev1 catalytic activity in the pol32 background resulted in a further significant decrease of TLS activity compared to the pol32 single mutant (see Figure 2). We sequenced 23 independent isolates from the pol32 rev1-AA background, each bearing a single mutation in CAN1. As expected, CC to CG transversions were abolished (see Figures 3A and 6B). CC to CT transitions were by far the predominant type of mutation, seen in 19 isolates. Eight of these events were observed at position 718 (see Figure 3C), a site that also favored insertion of A opposite the abasic site in WT. Taken together, we conclude that the residual TLS in pol32 cells (evident when compared to rev3 or rev1) was attributable mostly to a capacity for inserting C opposite abasic sites, in a Rev1 catalysis-dependent manner.
4. Discussion
4.1. An in vivo model system to investigate TLS bypass of lesions in DNA
In this work, we describe the use of a subtelomeric ssDNA reporter system to define the roles of proteins involved in error-prone TLS bypass of abasic sites generated at random CC motifs in chromosomal DNA. Abasic sites are well-studied lesions in a variety of systems. Using the ssDNA reporter, we have recapitulated previous findings on the roles of TLS proteins in yeast, thus validating our overall approach. Additionally, we obtained evidence that sequence context can strongly influence the relative likelihood of A vs. C insertion opposite abasic sites, a possibility that had hardly been investigated previously. We also found that deletion of POL32 results in a mutation signature entirely distinct from that of any other genetic defect in TLS.
Yeast has a relatively small number of proteins that take part in TLS. Thus, it was straightforward to construct a set of relevant mutants to investigate the relative contribution of each protein toward overall TLS activity to bypass abasic sites. In principle, the same approach can be applied to determine the TLS bypass specificity of a large variety of lesions. In this study, we used only one reporter gene (CAN1) to assess TLS bypass because the low frequency of TLS in the various mutant backgrounds required the mutagenization of, and selection from, large numbers of TLS mutant cells to obtain meaningful data. Under conditions of efficient WT TLS, we had easily identified many instances of inactivating simultaneous mutations in multiple reporter genes (i.e. mutation clusters or showers) within the same mutant isolate [7]. In effect, TLS events were sampled at even larger numbers of different sequence contexts without having to select from an impractically large number of mutagenized cells. Indeed, we have used this system to show that TLS is necessary for bypass of sulfite-induced lesions at cytosines in ssDNA [7], although the relative contributions of the various TLS proteins toward this type of lesion bypass have yet to be investigated.
The subtelomeric ssDNA reporter also has two key advantages over conventional mutagenesis reporter systems. First, the single-strand state of the reporter DNA enables detection of mutagenesis arising from agents that are not reactive enough to damage dsDNA appreciably, but might be able to form adducts in ssDNA. Such agents usually exhibit only weak activity in conventional mutagenesis reporters [63]. Secondly, the known strandedness of the reporter DNA in this system enables unambiguous assignment of the actual base change. In contrast, conventional mutagenesis reporters typically are unable to distinguish whether an agent induces a given mutation or its complement, and are reliant upon other experimental approaches to provide such information.
4.2. The importance of Rev1 catalytic activity
Our results provide clear evidence that Rev1 catalytic activity is required for the insertion of C opposite abasic sites randomly generated in chromosomal DNA, which is consistent with previous findings in yeast [30, 33] and vertebrates [64, 65]. When Rev1 catalytic activity is removed, the overall frequency of TLS does not decrease significantly compared to WT, i.e. the rev1-AA cells are not defective for TLS opposite abasic sites, per se. However, the rev1-AA mutation spectrum is completely devoid of CC to CG transversions, indicating that the TLS bypass mechanism which is left intact (requiring Rev1 structural function and Pol ζ) does not insert C opposite abasic sites at a detectable frequency. Further, the loss of Rev1 catalytic activity does not have any apparent effect on the ability to insert A opposite abasic sites, indicating that polymerase activities besides that of Rev1 are entirely responsible for A insertion events.
As noted, abasic sites can be formed by enzymatic excision of damaged bases [2]. One of the most commonly occurring damaged bases is 8-oxo-7,8-dihydroguanine (8-oxoG), which results from the reaction of G with reactive oxygen species and can mispair with A [2]. 8-oxoG is excised by conserved DNA N-glycosylases, e.g. Ogg1 in yeast, to generate an abasic site [66]. Also, alkylating agents are particularly reactive toward N7 of guanine, such that adducts in this position destabilize the N-glycosidic bond, resulting in spontaneous depurination, i.e. abasic site formation [67]. Since abasic sites resulting from damage to guanine are likely to be especially common, it is plausible that evolution selected for an enzyme specifically with deoxycytidyltransferase activity (Rev1) to counteract the possible mutagenic effects. Such a hypothesis also would be consistent with the notion that abasic sites are cognate lesions for Rev1 [68].
4.3. The nucleotide inserted opposite an abasic site is indicative of the set of TLS activities at work
By analyzing the outcomes of TLS events in various TLS deficient backgrounds at numerous randomly generated abasic sites, we have gained important insight into the underlying molecular mechanisms. Sequence context can have a striking effect on the choice of nucleotide inserted opposite an abasic site. Specifically, abasic sites at position 530 of CAN1 tend to be bypassed by C insertion (resulting in CC to CG transversions), while abasic sites at position 718 tend to be bypassed by A insertion (resulting in CC to CT transitions) (see Figure 3A). The local sequence context of position 530 is 5′-AGTGATTGCCCAAGAAAACCA-3′ while the context of position 718 is 5′-ATTAGAAACCCGATAATGGCT-3′ (positions 530 and 718 are underlined, respectively). As there are numerous differences between these two sequence contexts, further work is necessary to determine which sequence features influence the choice of nucleotide insertion. Although Lawrence’s group had observed previously that varying the position of an engineered abasic site by one base can result in statistically significant differences in the insertion frequencies of A vs. C [28, 29], the effects that we have observed are much more pronounced. Differences in sequence context also might be a factor underlying the Lawrence group’s observation of frequent (76%) insertion of C opposite their model abasic sites [29] while Prakash’s group observed frequent (66%) insertion of A opposite their model abasic site [30].
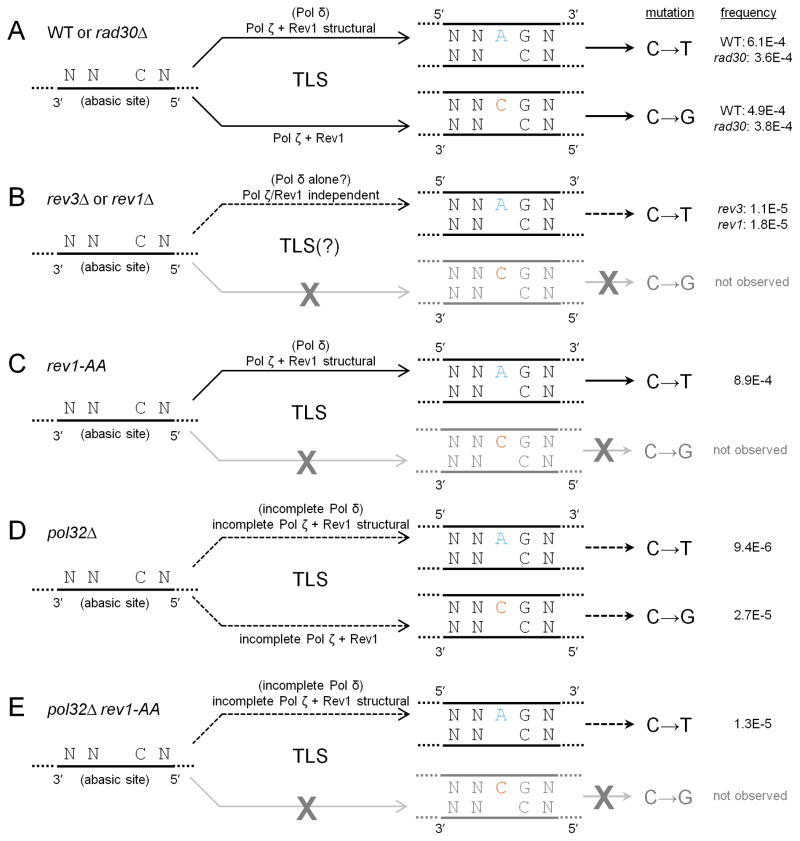
Our results are summarized in Figure 7. In WT (and rad30) cells (Figure 7A), where TLS bypass of abasic sites is fully proficient, TLS involving Rev1 structural function and Pol ζ enables insertion of A, while TLS involving Pol ζ and both Rev1 structural and catalytic functions enables insertion of C. When either REV3 or REV1 is deleted (Figure 7B), only a residual amount of low-frequency TLS (possibly by Pol δ acting alone) that always inserts A opposite abasic sites is observed. Ablation of Rev1 catalytic function (Figure 7C) leaves the Rev1 structural function- and Pol ζ-dependent pathway intact, which explains the observation of only A insertions (and a complete lack of C insertions) in rev1-AA cells. Deletion of POL32 (Figure 7D) leaves both Pols δ and ζ as structurally incomplete complexes, resulting in a severe TLS defect. But in contrast to rev3 and rev1, a readily detectable proportion of C insertions occur. Finally, the C insertions in pol32 cells are completely dependent on Rev1 catalytic function (Figure 7E), since only A insertion events are found in the pol32 rev1-AA double mutant.
Figure 7.
Summary of the relative contributions of alternative TLS mechanisms toward mutagenesis at abasic sites. (A) In WT and rad30, TLS dependent on Pol ζ and the structural function of Rev1 is necessary for the majority of A insertion events, while C insertion events require the presence of Pol ζ and fully functional Rev1. The relative frequencies of the two outcomes are roughly equal. (B) In rev3 and rev1, C insertion events are not observed. The observed A insertion events are independent of Pol ζ and Rev1, and might represent low-frequency TLS bypass by a replicative polymerase (presumably Pol δ) acting alone. (C) In rev1-AA, only A insertion events, which require Pol ζ and Rev1 structural function, are observed. The complete lack of C insertion events in rev1-AA implies that Rev1’s deoxycytidyltransferase function is responsible for such events in WT. (D) In pol32, both Pols δ and ζ are structurally incomplete. This presumably results in low-frequency, but observable TLS events that insert either A or C. (E) In pol32 rev1-AA, only low-frequency A insertion events are observed, involving incomplete Pol ζ and Rev1 structural function.
Taken together, we conclude that in cells fully proficient for TLS, certain sequence contexts (e.g. around C530) tend either to promote the catalytic activity of Rev1 to insert C, or inhibit the activity of Pol δ to insert A, or some combination of both effects. Conversely, other sequence contexts, such as near C718, either promote the activity of Pol δ, inhibit the catalytic activity of Rev1, or some combination of both. We speculate that contexts favoring C insertion tend to stabilize Rev1 (by some combination of hydrogen bonding, salt bridges, and van der Waals interactions) in a configuration favorable for catalysis, while the opposite is true for contexts favoring A insertion. As with any mutagenesis reporter system, selection influences which mutations are recovered most frequently and necessarily leads to the assessment of only a subset of all possible mutations within a reporter gene (reviewed in [69]). Thus, it is important to extend this work to a genomewide scale, sampling TLS events over a larger number of sequence contexts that are not under selection pressure as CAN1 inactivation was. This could identify the sequence determinants that influence which contexts favor one type of TLS event over the other.
4.4. Abasic site bypass within long regions of ssDNA in cancer genomes
Recent work has shown that mutations at cytosines in 5′-TC-3′ motifs were significantly enriched in sequenced tumors from multiple types of cancer [9–11]. Further, clusters of multiple point mutations (C to G/T substitutions) that all occurred at TC motifs were identified [10, 11]. These observations suggest that at some point during tumorigenesis, long stretches of ssDNA were exposed to deamination by at least one enzyme among the TC-specific subclass of APOBEC deaminases (reviewed in [70]). Presumably, UNG glycosylases then excised the resulting uracils to generate abasic sites, which were bypassed in an error-prone manner by TLS.
Humans possess a larger complement of TLS polymerases than yeast [12–14]. It might be possible to carry out a detailed analysis of the role of each human TLS protein in the bypass of random abasic sites in vivo by heterologous expression of each human TLS polymerase (or combinations of multiple human TLS polymerases), in a yeast reporter deleted for the yeast TLS polymerases while expressing APOBEC3G. If this heterologous expression were successful, mutation spectrum analysis to infer the roles of individual human TLS proteins should be possible. Such work could shed important new light on molecular mechanisms of carcinogenesis.
5. Conclusions
Genomic DNA is susceptible to mutation arising from damage caused by endogenous and environmental agents [1]. Since genomewide sequencing of human cancers already has revealed the pervasive, distinctive mutagenic signature of endogenous ssDNA damaging agents (the APOBEC cytosine deaminases) [9–11], we propose that other agents also would have the opportunity to react with long stretches of ssDNA at some point(s) during carcinogenesis. Having validated our experimental approach by analyzing abasic sites, the ssDNA reporter strains now can be used to identify and characterize ssDNA-specific mutagens, i.e. agents that are not reactive enough to damage dsDNA significantly, but potentially can react with ssDNA in vivo to form lesions that lead to mutation fixation. Detailed understanding of the underlying mutagenic mechanisms could be obtained by determining the TLS bypass specificity of such lesions. In turn, this knowledge would help decipher the complex superimposition of myriad DNA damage, repair, and damage tolerance processes that acted to create any given cancer genome [71].
Supplementary Material
Plating efficiencies of reporter strains. For each sample, 500 cells were plated onto synthetic complete in triplicate, incubated for five days at 23°C, and then counted. (A) Plating efficiencies of APOBEC3G expressing strains that underwent 6 hour shift to 37°C. Median plating efficiency for pol32 and pol32 rev1-AA were 17.4 and 20.9%, respectively. Both were significantly less than WT (P < 0.05 by Kolmogorov-Smirnov test, denoted by asterisk). Median plating efficiency for all other strains ranged between 45 to 60%. (B) Plating efficiencies of empty vector strains that underwent 6 hour shift to 37°C. Again, median plating efficiency for pol32 and pol32 rev1-AA were significantly lower than for the WT empty vector strain (denoted by asterisk). Daggers indicate significant difference compared to the same genetic background expressing APOBEC3G during temperature shift (compare to panel A). APOBEC3G expression induced slight, but statistically significant, toxicity in WT, rad30, and rev3.
CAN1 inactivation frequencies of reporter strains bearing empty vector (pCM252) that were temperature shifted to generate subtelomeric ssDNA. Each data point represents an independent replicate derived from a single colony, numbers indicate median values, and asterisks denote significant difference (P < 0.05) compared to WT by Kolmogorov-Smirnov test. Canavanine selection plates were counted after five days of growth at 23°C, following the temperature shift-induced generation of subtelomeric ssDNA. See Materials and Methods for details of experimental procedures.
Highlights.
Sequence context influences A and C insertion frequencies across abasic sites.
Residual TLS by Rev1 can occur in the absence of the Pol32 subunit of Pols δ and ζ.
Confirmation of previous findings on roles of TLS proteins validates ssDNA reporter.
The ssDNA reporter can be used to study TLS bypass of other DNA lesions.
Acknowledgments
We thank Drs. T.A. Kunkel, D.R. Menendez, T.T. Nguyen, and S.A. Roberts for critical reading of the manuscript. This work was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences (NIH, DHHS) project ES065073 to MAR.
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kin Chan, Email: chank2@niehs.nih.gov.
Michael A. Resnick, Email: resnick@niehs.nih.gov.
Dmitry A. Gordenin, Email: gordenin@niehs.nih.gov.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. ASM Press; Washington, DC: 2006. [Google Scholar]
- 3.Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc Natl Acad Sci U S A. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loeb LA, Preston BD. Mutagenesis by Apurinic/Apyrimidinic Sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 5.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Sale JE. Competition, collaboration and coordination -- determining how cells bypass DNA damage. J Cell Sci. 2012;125:1633–1643. doi: 10.1242/jcs.094748. [DOI] [PubMed] [Google Scholar]
- 7.Chan K, Sterling JF, Roberts SA, Bhagwat AS, Resnick MA, Gordenin DA. Base Damage within Single-Strand DNA Underlies In Vivo Hypermutability Induced by a Ubiquitous Environmental Agent. PLoS Genet. 2012;8:e1003149. doi: 10.1371/journal.pgen.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2 doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jönsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerød A, Tutt A, Martens JWM, Aparicio SAJR, Borg Å, Salomon AV, Thomas G, Børresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak Leszek J, Kryukov Gregory V, Malc E, Mieczkowski Piotr A, Resnick Michael A, Gordenin Dmitry A. Clustered Mutations in Yeast and in Human Cancers Can Arise from Damaged Long Single-Strand DNA Regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts SA, Lawrence MS, Klimczak Leszek J, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov Gregory V, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin Dmitry A. An APOBEC Cytidine Deaminase Mutagenesis Pattern is Ubiquitous in Human Cancers. Nat Genet. doi: 10.1038/ng.2702. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boiteux S, Jinks-Robertson S. DNA Repair Mechanisms and the Bypass of DNA Damage in Saccharomyces cerevisiae. Genetics. 2013;193:1025–1064. doi: 10.1534/genetics.112.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sale JE. Translesion DNA Synthesis and Mutagenesis in Eukaryotes. Cold Spring Harb Perspect Biol. 2012;5 doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma S, Helchowski CM, Canman CE. The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability. Mutat Res. 2013;743–744:97–110. doi: 10.1016/j.mrfmmm.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 Employs a Novel Mechanism of DNA Synthesis Using a Protein Template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 16.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-synthymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 17.Goodman MF. Error-Prone Repair DNA Polymerases In Prokaryotes And Eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 18.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Washington MT, Johnson RE, Prakash S, Prakash L. Fidelity and Processivity of Saccharomyces cerevisiae DNA Polymerase η. J Biol Chem. 1999;274:36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- 20.Zhong X, Garg P, Stith CM, McElhinny SAN, Kissling GE, Burgers PMJ, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 Gene, a Homologue of Escherichia coli dinB and umuC, Is DNA Damage Inducible and Functions in a Novel Error-Free Postreplication Repair Mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larimer FW, Perry JR, Hardigree AA. The REV1 gene of Saccharomyces cerevisiae: isolation, sequence, and functional analysis. J Bacteriol. 1989;171:230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerik KJ, Li X, Pautz A, Burgers PMJ. Characterization of the Two Small Subunits of Saccharomyces cerevisiae DNA Polymerase δ. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 25.Haracska L, Washington MT, Prakash S, Prakash L. Inefficient Bypass of an Abasic Site by DNA Polymerase η. J Biol Chem. 2001;276:6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 26.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 27.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PMJ, Prakash S, Prakash L. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbs PEM, Lawrence CW. Novel Mutagenic Properties of Abasic Sites in Saccharomyces cerevisiae. J Mol Biol. 1995;251:229–236. doi: 10.1006/jmbi.1995.0430. [DOI] [PubMed] [Google Scholar]
- 29.Gibbs PEM, McDonald J, Woodgate R, Lawrence CW. The Relative Roles in Vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 Protein and Pol32 in the Bypass and Mutation Induction of an Abasic Site, T-T (6–4) Photoadduct and T-T cis-syn Cyclobutane Dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagès V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci U S A. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auerbach P, Bennett RAO, Bailey EA, Krokan HE, Demple B. Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc Natl Acad Sci U S A. 2005;102:17711–17716. doi: 10.1073/pnas.0504643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auerbach PA, Demple B. Roles of Rev1, Pol ζ, Pol32 and Pol η in the bypass of chromosomal abasic sites in Saccharomyces cerevisiae. Mutagenesis. 2010;25:63–69. doi: 10.1093/mutage/gep045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim N, Mudrak SV, Jinks-Robertson S. The dCMP transferase activity of yeast Rev1 is biologically relevant during the bypass of endogenously generated AP sites. DNA Repair. 2011;10:1262–1271. doi: 10.1016/j.dnarep.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson RE, Prakash S, Prakash L. Efficient Bypass of a Thymine-Thymine Dimer by Yeast DNA Polymerase, Polη. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 35.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 36.Guo D, Wu X, Rajpal DK, Taylor JS, Wang Z. Translesion synthesis by yeast DNA polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 2001;29:2875–2883. doi: 10.1093/nar/29.13.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson JR, Lawrence CW, Hinkle DC. Thymine-Thymine Dimer Bypass by Yeast DNA Polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 38.Stone JE, Kumar D, Binz SK, Inase A, Iwai S, Chabes A, Burgers PM, Kunkel TA. Lesion bypass by S. cerevisiae Pol ζ alone. DNA Repair. 2011;10:826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson JR, Gibbs PEM, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, Wang J, Zhang Y, Wang Z. The catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis. Nucleic Acids Res. 2010;38:5036–5046. doi: 10.1093/nar/gkq225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiltrout ME, Walker GC. The DNA Polymerase Activity of Saccharomyces cerevisiae Rev1 is Biologically Significant. Genetics. 2011;187:21–35. doi: 10.1534/genetics.110.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci U S A. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA Polymerase δ and ζ Switch by Sharing Accessory Subunits of DNA Polymerase δ. J Biol Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crosby B, Prakash L, Davis H, Hinkle DC. Purification and characterization of a uracil-DNA glycosylase from the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1981;9:5797–5810. doi: 10.1093/nar/9.21.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′--->5′ exonuclease activity. Proc Natl Acad Sci U S A. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 48.Storici F, Resnick MA, Judith LC, Paul M. Methods in Enzymology. Academic Press; 2006. The Delitto Perfetto Approach to In Vivo Site-Directed Mutagenesis and Chromosome Rearrangements with Synthetic Oligonucleotides in Yeast; pp. 329–345. [DOI] [PubMed] [Google Scholar]
- 49.Bellí G, Garí E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Booth C, Griffith E, Brady G, Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen KM, Martemyanova N, Lu Y, Shindo K, Matsuo H, Harris RS. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Letters. 2007;581:4761–4766. doi: 10.1016/j.febslet.2007.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. An Extended Structure of the APOBEC3G Catalytic Domain Suggests a Unique Holoenzyme Model. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schumacher AJ, Nissley DV, Harris RS. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc Natl Acad Sci U S A. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 55.Grenson M, Mousset M, Wiame JM, Bechet J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae: I. Evidence for a specific arginine-transporting system. Biochim Biophy Acta. 1966;127:325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- 56.Lang GI, Murray AW. Estimating the Per-Base-Pair Mutation Rate in the Yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic Processing of Ribonucleotides in DNA by Yeast Topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2-Msh3 Acts in Repair of Base-Base Mispairs. Mol Cell Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Brien TJ, Witcher P, Brooks B, Patierno SR. DNA polymerase ζ is essential for hexavalent chromium-induced mutagenesis. Mutat Res. 2009;663:77–83. doi: 10.1016/j.mrfmmm.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohnishi G, Endo K, Doi A, Fujita A, Daigaku Y, Nunoshiba T, Yamamoto K. Spontaneous mutagenesis in haploid and diploid Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2004;325:928–933. doi: 10.1016/j.bbrc.2004.10.120. [DOI] [PubMed] [Google Scholar]
- 61.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of Damaged Single-Strand DNA Formed at Double-Strand Breaks and Uncapped Telomeres in Yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair. 2010;9:914–921. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singer B, Kusmierek JT. Chemical Mutagenesis. Annu Rev Biochem. 1982;51:655–691. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- 64.Ross AL, Sale JE. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 65.Masuda K, Ouchida R, Li Y, Gao X, Mori H, Wang JY. A Critical Role for REV1 in Regulating the Induction of C:G Transitions and A:T Mutations during Ig Gene Hypermutation. J Immunol. 2009;183:1846–1850. doi: 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- 66.van der Kemp PA, Thomas D, Barbey R, de Oliveira R, Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc Natl Acad Sci U S A. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singer B, Grünberger D. Molecular Biology of Mutagens and Carcinogens. Plenum; New York: 1983. [Google Scholar]
- 68.Pryor JM, Washington MT. Pre-steady state kinetic studies show that an abasic site is a cognate lesion for the yeast Rev1 protein. DNA Repair. 2011;10:1138–1144. doi: 10.1016/j.dnarep.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rogozin IB, Pavlov YI. Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutat Res. 2003;544:65–85. doi: 10.1016/s1383-5742(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 70.Prochnow C, Bransteitter R, Chen X. APOBEC deaminases-mutases with defensive roles for immunity. Sci China C Life Sci. 2009;52:893–902. doi: 10.1007/s11427-009-0133-1. [DOI] [PubMed] [Google Scholar]
- 71.Stratton MR. Exploring the Genomes of Cancer Cells: Progress and Promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plating efficiencies of reporter strains. For each sample, 500 cells were plated onto synthetic complete in triplicate, incubated for five days at 23°C, and then counted. (A) Plating efficiencies of APOBEC3G expressing strains that underwent 6 hour shift to 37°C. Median plating efficiency for pol32 and pol32 rev1-AA were 17.4 and 20.9%, respectively. Both were significantly less than WT (P < 0.05 by Kolmogorov-Smirnov test, denoted by asterisk). Median plating efficiency for all other strains ranged between 45 to 60%. (B) Plating efficiencies of empty vector strains that underwent 6 hour shift to 37°C. Again, median plating efficiency for pol32 and pol32 rev1-AA were significantly lower than for the WT empty vector strain (denoted by asterisk). Daggers indicate significant difference compared to the same genetic background expressing APOBEC3G during temperature shift (compare to panel A). APOBEC3G expression induced slight, but statistically significant, toxicity in WT, rad30, and rev3.
CAN1 inactivation frequencies of reporter strains bearing empty vector (pCM252) that were temperature shifted to generate subtelomeric ssDNA. Each data point represents an independent replicate derived from a single colony, numbers indicate median values, and asterisks denote significant difference (P < 0.05) compared to WT by Kolmogorov-Smirnov test. Canavanine selection plates were counted after five days of growth at 23°C, following the temperature shift-induced generation of subtelomeric ssDNA. See Materials and Methods for details of experimental procedures.