Abstract
Cancer cells can survive through the upregulation of cell cycle and the escape from apoptosis induced by numerous cellular stresses. In the normal cells, these biological cascades depend on scheduled proteolytic degradation of regulatory proteins via the ubiquitin–proteasome pathway. Therefore, interruption of regulated proteolytic pathways leads to abnormal cell‐proliferation. Ubiquitin ligases called SCF complex (consisting of Skp‐1, cullin, and F‐box protein) or CRL (cullin‐RING ubiquitin ligase) are predominant in a family of E3 ubiquitin ligases that control a final step in ubiquitination of diverse substrates. To a great extent, the ubiquitin ligase activity of the SCF complex requires the conjugation of NEDD8 to cullins, i.e. scaffold proteins. This review is anticipated to review the downregulation system of NEDD8 conjugation by several factors including a chemical compound such as MLN4924 and protein molecules (e.g. COP9 signalosome, inactive mutant of Ubc12, and NUB1/NUB1L). Since the downregulation of NEDD8 conjugation affects cell‐cycle progression by inhibiting the ligase activity of SCF complexes, such knowledge in the NEDD8‐conjugation pathway will contribute to the more magnificent therapies that selectively suppress tumorigenesis.
Keywords: Ubiquitination, SCF complex, NEDD8, MLN4924, Ubc12, NUB1
Highlights
We have provided recent insights in the NEDD8‐associated researches.
We have revealed NEDD8‐associated molecules regulating the SCF ubiquitin E3 ligases.
Deneddylation‐related proteins are candidates in inhibiting the SCF ligase.
MLN4924 is a compound involved in initial inhibition of the neddylation cascade.
1. Introduction
Programmed degradation of regulatory proteins associated with diverse biological events (e.g. cell‐cycle regulation, cell‐proliferation, intracellular signaling, DNA repair and apoptosis) is crucial for cellular homeostasis (Ciechanover, 1998; Ciechanover and Schwartz, 1998; Hershko, 2005; Hershko and Ciechanover, 1998). The ubiquitin–proteasome pathway is a major clearance system associated with proteolysis inside the cell. The dysregulation is involved in the pathogenesis of cancer and other diseases via inappropriate loss of regulatory proteins or permanent activation of specific signal cascade. Therefore, it is possible that these diseases can be treated by modulating the ubiquitin–proteasome pathway. Indeed, a proteasome inhibitor Bortezomib has broadly been used for the treatment of patients with multiple myeloma, which has gained good response (Jagannath et al., 2005; Richardson et al., 2005; San Miguel et al., 2008).
A ubiquitin‐like protein NEDD8 has high similarity with ubiquitin in the amino acid sequence. It activates the ubiquitin E3 ligase activity of SCF complex by covalently binding to cullins (Kamitani et al., 1997; Morimoto et al., 2000; Osaka et al., 1998; Podust et al., 2000; Read et al., 2000; Wada et al., 1999a). Thus, the NEDD8‐mediated activation of SCF complex is necessary for the aforementioned biological functions regulated by the ubiquitin–proteasome pathway. Here, we review the NEDD8‐conjugation/‐deconjugation system and novel strategies to possible target molecules in anticancer therapy.
2. Ubiquitination pathway
The proteasomal degradation pathway begins by conjugating a chain of polyubiquitin to a target molecule (Ciechanover, 1998; Ciechanover and Schwartz, 1998; Hershko and Ciechanover, 1998). The first step in the production of this chain is to connect a single ubiquitin molecule to E1 (ubiquitin‐activating enzyme) through a thioester bond in an ATP‐dependent manner. Next, E2 (ubiquitin‐conjugating enzyme) receives the activated ubiquitin from E1 and transfers the ubiquitin molecule to a lysine residue in a target protein with the assistance of an E3 ubiquitin ligase. Repeated cycles via the E1–E2–E3 cascade generate a polyubiquitin chain, namely a death signal, which is subsequently recognized by the regulatory subunit of the 26S proteasome machinery (Figure 1).
Figure 1.
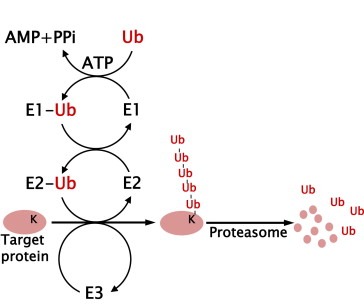
Schematic summary of ubiquitin–proteasome pathway. Lysine residue (K) of the substrate is conjugated with ubiquitin (Ub).
2.1. Diversity of SCF complexes and their target proteins
Most SCF complexes or CRLs (cullin‐RING ubiquitin ligases) consist of Skp‐1, cullin, F‐box protein and RING finger protein Rbx/Roc. They are predominate among family members of E3 ubiquitin ligase that promote ubiquitination of substrate proteins regulating various biological processes, including cell‐cycle progression, signal transduction, and differentiation. The substrate specificity of SCF ligase depends on the combination pattern of its components, particularly the F‐box protein. Numerous regulatory proteins targeted by SCF complexes have been reported (Table 1). Therefore, dysregulation of SCF complexes impairs many biological events, resulting in cell‐cycle arrest, apoptosis, tumorigenesis, etc.
Table 1.
SCF E3 ubiquitin ligases and their substrates
| Scaffold | Adapter | Receptor | Ring box | Substrates |
|---|---|---|---|---|
| Cul‐1 | Skp1 | Skp2 | Rbx1 | p21, p27, p73, p130, Cydin A/D |
| Cul‐1 | Skp1 | β‐TrCP | Rbx1 | lkβα, β‐catenin, BimEL, Weel, p53 |
| Cul‐1 | Skp1 | Fbxw7 | Rbx1 | Cydin E, o‐Myc, o‐Jun, Notch |
| Cul‐2 | Elongin BC | VHL | Rbx1 | HIF1α |
| Cul‐3 | BTB‐domain proteins | Rbx1 | Cydin E, Mei‐1, Dsh, NRF2 | |
| Cul‐4 | DDBI | DCAF | Rbx1 | CDT1, p21, Histone H2A/H3/H4, XPC |
| Cul‐5 | Elongin BC | SOCS | Rbx1 | TEL‐JAK2, JAK‐STAT family proteins |
| Cul‐7 | Skp1 | Fbxw8 | Rbx1 | IRS1, Cydin D |
2.2. Conjugation pathway of a ubiquitin‐like protein NEDD8
Several ubiquitin‐like proteins (UBLs), including NEDD8, SUMO, ISG15, FAT10, Atg8 and Atg12, have been demonstrated to conjugate to target proteins in a manner analogous to ubiquitination (Bawa‐Khalfe and Yeh, 2010; Haas et al., 1987; Ichimura et al., 2000; Kamitani et al., 1997; Liu et al., 1999; Loeb and Haas, 1992; Mizushima et al., 1998; Yeh et al., 2000). NEDD8 (neural precursor cell‐expressed developmentally downregulated protein 8) was originally reported as a novel gene highly enriched in fetal mouse brain (Kumar et al., 1992). NEDD8 encodes a small protein of 81 amino acids, which is 60% identical and 80% homologous to ubiquitin, and equivalently conjugates to substrates (Kamitani et al., 1997). The crystal structure of NEDD8 is quite analogous to that of ubiquitin with the exception of two surface regions (Rao‐Naik et al., 1998; Whitby et al., 1998).
The NEDD8‐conjugation cascade, called neddylation, is mediated by E1 NEDD8‐activating enzyme (NAE), E2 NEDD8‐conjugating enzyme (Ubc12), and E3 NEDD8 ligase‐like protein, which successively activate and transfer NEDD8 to a target molecule. First, the C‐terminal glycine of NEDD8 is adenylated by the NAE, which is a heterodimer consisting of APP‐BP1 and Uba3, in an ATP‐dependent manner and covalently conjugated to the NAE via a thiolester linkage (Walden et al., 2003). Second, the activated NEDD8 is consecutively transferred to the E2 NEDD8‐conjugation enzyme (Gong and Yeh, 1999; Huang et al., 2005). As regards the transfer of NEDD8 from the E2 NEDD8‐conjugating enzyme to the specific substrates (i.e. cullins) via an isopeptide bond, some RING (really interesting novel gene) finger domain‐containing proteins (such as Rbx1/Roc1, MDM2, FBXO11, and c‐Cbl) have been shown to function in an E3 ligase‐like fashion for cullin neddylation (Abida et al., 2007; Kamura et al., 1999a; Morimoto et al., 2003; Oved et al., 2006; Xirodimas et al., 2004). Furthermore, DCN1 (defective in cullin neddylation 1), also called SCCRO (squamous cell carcinoma‐related oncogene), has been identified as a scaffold‐like protein of E3 ligase for cullin neddylation (Kurz et al., 2005, Kurz et al., 2008; Yang et al., 2007). The yeast DCN1 homolog has been shown to act together with Hrt1 (Rbx1/Roc1 in mammals) for cdc53 neddylation (Scott et al., 2010). In addition, the von Hippel‐Lindau gene product (pVHL) has been shown to promote Cul‐2 neddylation, suggesting the NEDD8 E3 ligase‐like activity of pVHL (Wada et al., 1999b). On the contrary, the covalently conjugated NEDD8 on the substrate is deconjugated by the deneddylation activity of several proteins such as COP9 signalosome, NEDP1/DEN1, and USP21 (Chan et al., 2008; Gong et al., 2000; Lyapina et al., 2001; Mendoza et al., 2003; Rabut and Peter, 2008; Schwechheimer et al., 2001). The neddylation is also inhibited by CAND1 (cullin‐associated and neddylation‐dissociate 1) binding to cullins (Goldenberg et al., 2004; Liu et al., 2002) or is negatively downregulated by NUB1 (NEDD8 ultimate buster 1) linked to the 26S proteasome (Kamitani et al., 2001; Kito et al., 2001) (Figure 2).
Figure 2.
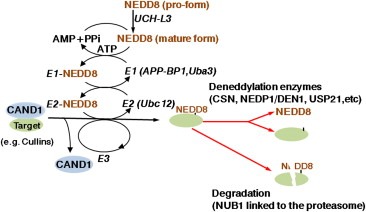
Schematic summary of NEDD8‐conjugation and ‐deconjugation pathway.
2.3. Substrates of NEDD8 conjugation: cullins and other molecules
Cullins are scaffold proteins assembling SCF complexes with other components. They are known as the substrates conjugated with NEDD8 (Table 1). To date, seven cullin family members (Cul‐1, ‐2, ‐3, ‐4A, ‐4B, ‐5, and ‐7) have been identified (Hori et al., 1999; Kipreos et al., 1996; Osaka et al., 1998; Wada et al., 1999a). Cul‐1‐based SCF complexes (CRL1), such as SCFSkp 2, SCFβ‐TrCP and SCFFbw7, are the most studied ones in terms of their cancer‐related actions. SCFSkp 2 is involved in the degradation of several cell‐cycle regulators including cyclin D, p27Kip1, p21Cip1, p73, and p130 (Guardavaccaro and Pagano, 2004; Laney and Hochstrasser, 1999; Pagano, 1997). SCFβ‐TrCP promotes degradation of β‐catenin and the NF‐κB inhibitor IκBα (Skaar et al., 2009). Moreover, SCFFbw7 has been reported to promote degradation of cyclin E, c‐myc oncoprotein and Notch (Guardavaccaro and Pagano, 2004). A recent study identified more than 350 possible substrates of CRL1 by employing global protein stability profiling method (Yen and Elledge, 2008). Cul‐2 interacts with pVHL through elongins B and C to properly generate a CRL2 complex (also known as VBC) (Kamura et al., 1999b; Lisztwan et al., 1999; Stebbins et al., 1999). This complex induces degradation of hypoxia‐inducible factor 1α (HIF1α), of which proline residues are hydroxylated by prolyl hydroxylase in an oxygen‐dependent manner and then targeted to pVHL for ubiquitination and subsequent proteasomal degradation (Kaelin and Ratcliffe, 2008; Willam et al., 2004). Bialleic deletion of the VHL gene mainly results in the stability of HIF1α and thereby ultimately contributes to tumorigenesis of sporadic clear‐cell type renal cell carcinoma (RCC). The Cul‐3‐based SCF complexes (CRL3), which are produced with Rbx1 and BTB‐domain protein, promote degradation of cyclin E, Mei‐1 (a component of mitotic spindle), Dsh (a regulator of Wnt‐β‐catenin pathway) and NRF2 (a transcriptional factor associated with an anti‐oxidant response) (Angers et al., 2006; Furukawa et al., 2003; Furukawa and Xiong, 2005; Li and Kong, 2009; Pintard et al., 2003; Shibata et al., 2008; Singer et al., 1999). The CRL4 complexes, consisting of Cul‐4 (A or B), a damaged DNA binding protein (DDB1), and a DDB1 and Cul‐4‐associated factor (DCAF), control DNA replication and nucleotide excision repair through ubiquitination of CDT1, p21, Histone H2A/H3/H4, XPC, TSC2, etc (Hu et al., 2004, Hu et al., 2008; Jackson and Xiong, 2009; Kapetanaki et al., 2006; Sugasawa et al., 2005; Zhong et al., 2003). Interestingly, the knockdown effect of Cul‐4A and effect of Cul‐4A‐specific inhibitor have attracted research attention as a strategy for treating Cul‐4‐amplified breast cancer (Chen et al., 1998; Melchor et al., 2009) and UV‐induced skin cancer (Liu et al., 2009). The CRL5 complexes, comprised of Cul‐5, a suppressor of cytokine signaling (SOCS) family proteins, elongins B and C, and Rbx1, suppress JAK‐STAT signaling via degradation of JAK family proteins (Hilton, 1999). On the contrary, Cul‐5 has been identified as a possible tumor suppressor because its overexpression induces growth‐inhibition in breast cancer cells (Burnatowska‐Hledin et al., 2004; Johnson et al., 2007). As regards Cul‐7, to date, there have been no reports indicating neddylation of Cul‐7 in the CRL7 complexes. The Cul‐7‐based E3 ligase regulates insulin receptor substrate 1 (IRS1) and cyclin D1 as substrates for proteolytic degradation (Okabe et al., 2006; Xu et al., 2008). Interestingly, Cul‐7 contains two additional motifs, i.e. a DOC domain and a CPH domain, at the N‐terminal region distinct from other cullin family members (Grossberger et al., 1999; Kaustov et al., 2007). Cul‐7 and its homolog PARC are able to bind to p53 in the cytoplasm via a CPH domain and inhibit the p53 transactivation activity (Andrews et al., 2006; Kaustov et al., 2007).
As to target molecules of neddylation other than cullins, p53 has been shown to be modified with NEDD8 via a RING finger‐type E3 ligase MDM2, leading to the facilitated transactivation activity of p53 (Harper, 2004; Xirodimas et al., 2004). MDM2 also neddylates the proapoptotic protein TAp73 and thereby promotes its cytoplasmic localization to suppress the transactivation action (Watson et al., 2006). Moreover, MDM2 has been reported to be also involved in its self‐neddylation, which contributes to its stability (Watson et al., 2010; Xirodimas et al., 2004). In addition, breast cancer‐associated protein 3 (BCA3), which is highly expressed in breast and prostate cancers, has been identified as a NEDD8 substrate (Gao et al., 2006). BCA3 inhibits NF‐κB‐dependent transcription through its ability to bind to NF‐κB subunit p65 and the cyclin D1 promoter in a neddylation‐dependent manner.
3. Modulation of SCF E3 ligases for anticancer strategy
The SCF ubiquitin E3 ligases have been shown to be dysregulated in a wide range of cancers, resulting in unlimited cell‐proliferation and carcinogenesis via accumulation of their substrate proteins. Consequently, the modulation of these E3 ligases is attracting attention as a possible strategy for anticancer therapy. The components of SCF complexes (e.g. cullins, Skp1/2, F‐box proteins, and Rbx1/2) and regulators of neddylation are potential candidates in the therapeutic strategy.
3.1. Molecules involved in inhibition of SCF E3 ligases through deneddylation
Neddylation (conjugation of NEDD8 to substrate proteins) is catalyzed by three enzymes (E1–E3) in a multistep fashion. Recently, using high throughput screening, an adenosine sulfamate derivative MLN4924 was identified as a specific inhibitor of NAE (Soucy et al., 2009). Pharmaceutically, MLN4924 irreversibly forms a covalent adduct with NEDD8 via NAE that is involved in the first NEDD8 adenylation step. MLN4924 is a potent ATP‐competitive inhibitor that disrupts the thiolester bond between NEDD8 and Uba3, a subunit of NAE. Fundamentally, MLN4924‐mediated suppression of cullin neddylation has been shown to increase expression levels of CRL substrates (Figure 3). Moreover, recent studies revealed that MLN4924 increases the levels of CRL substrates such as IκBα to cause anticancer effects against acute myeloid leukemia (AML) both in vitro and in xenograft models (Milhollen et al., 2010; Swords et al., 2010). Clinical trials using MLN4924 are currently ongoing in anticancer therapy for patients with AML.
Figure 3.
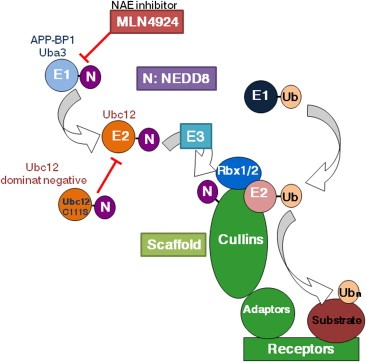
Modulation of NEDD8‐conjugation pathway by anticancer molecules.
Ubc12, an E2 NEDD8‐conjugation enzyme, is also a key molecule in the neddylation cascade. Activated NEDD8 is conjugated to the active site cysteine residue of Ubc12 via a thiolester bond. Finally, Ubc12 transfers NEDD8 to a lysine residue of the substrate protein for neddylation. Artificial Ubc12‐C111S with a substitution of Cys‐to‐Ser at the active site (Cys‐111) was shown to function as a dominant negative mutant against the endogenous wild‐type Ubc12, attributable to its covalent binding to NEDD8 (Wada et al., 2000) (Figure 3). This mutant Ubc12‐C111S has a forceful anti‐proliferative action on cancer cells (e.g. osteosarcoma, oral squamous cell carcinoma), concomitant with the instability of cellular morphology due to an actin cytoskeleton irregularity (Amir et al., 2002; Chairatvit and Ngamkitidechakul, 2007; Leck et al., 2010; Wada et al., 2000).
The COP9 signalosome (CSN) is a zinc metalloprotease complex comprising eight subunits (Deng et al., 2000). The CSN5 subunit (also known as Jab1) has the catalytic activity of cleavage at the isopeptide bonds between NEDD8 and cullins via the JAMM/MPN motif (Cope et al., 2002). Furthermore, CSN5 has been shown to be overexpressed in diverse cancer cells including breast, liver and pancreatic cancers (Adler et al., 2006; Berg et al., 2007; Kouvaraki et al., 2006) and play a crucial role in nuclear transportation and degradation of p27Kip1 (Tomoda et al., 1999, 2002). Importantly, knockdown of CSN5 induces defects in DNA repair response (Groisman et al., 2003) and also induces cell‐cycle arrest at multiple check‐points (Panattoni et al., 2008).
CAND1 interacts with un‐neddylated cullins to block the binding of NEDD8 and other adapter proteins (Goldenberg et al., 2004; Liu et al., 2002) (see Figure 2). Thus, CAND1 inhibits the assembly and activation of CRLs. In addition, CAND1 plays a key role in the ubiquitination activity of CRLs for diverse substrates. Specifically, it promotes the recruitment of F‐box proteins (Bosu and Kipreos, 2008; Dubiel, 2009). For instance, neddylated Cul‐1 was shown to be overexpressed in neuroendocrine lung cancer and be associated with downregulation of CAND1 (Salon et al., 2007). Moreover, it has recently been reported that CAND1 expression is suppressed by miR‐148a, which is one of human microRNAs (miRNAs), and that the knockdown of CAND1 promotes the proliferation of LNCaP cells (also known as a hormone‐sensitive prostate cancer cell line) (Murata et al., 2010).
3.2. NUB1 and NUB1L as potent tumor‐suppressor proteins
NUB1 is a NEDD8‐interacting protein composed of 601 amino acid residues with a calculated molecular mass of 69.1kDa. It is an interferon (IFN)‐inducible protein and predominantly localizes to the nucleus. NUB1L, a splicing variant of NUB1, possesses an insertion of 14 amino acids that codes for an additional ubiquitin‐associated (UBA) domain (Figure 4). Biologically, NUB1/NUB1L recruits NEDD8 and its conjugates to the proteasome for degradation and negatively regulates the NEDD8‐conjugation system (Kamitani et al., 2001; Kito et al., 2001; Tanaka et al., 2003; Tanji et al., 2005). Furthermore, NUB1 is expressed in some cancer cell lines, including rectal adenocarcinoma, neuroblastoma, malignant lymphoma, cervical adenocarcinoma, and RCC (Kito et al., 2001). Recently, NUB1 was shown to not only correlate with IFNα‐induced antimitogenic action, but also exert anticancer effects against RCC cells, concomitant with S‐phase transition during the cell cycle and apoptosis via accumulation of p27 and cyclin E (Hosono et al., 2010). Interestingly, overexpression of NUB1 strongly inhibits proliferation of IFNα‐resistant RCC cells (Hosono et al., 2010).
Figure 4.
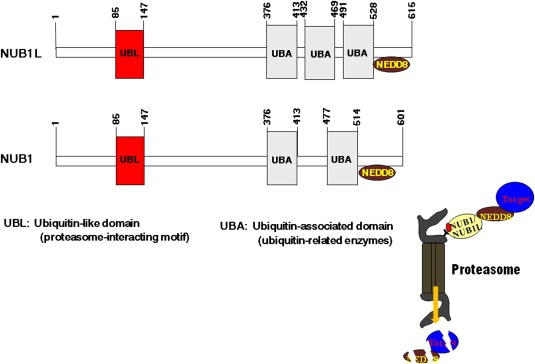
Domain structure of NUB1 and NUB1L.
4. Conclusion
New insights from experimental and clinical studies have revealed that negative regulation of the NEDD8‐conjugation pathway has a strong potential for cancer prevention. It is possible that this strategy is effective for numerous cancers in which components of SCF complexes are dysregulated, e.g. overexpression of Cul‐4A (Chen et al., 1998; Liu et al., 2009; Melchor et al., 2009), Skp‐2 (Kudo et al., 2005; Yang et al., 2002), and β‐TrCP (Belaidouni et al., 2005; Westbrook et al., 2008). MLN4924, a small inhibitor for SCF ligases, has recently been developed as a novel class of anticancer agent. It is distinct from agents used in conventional therapies. This compound is expected to exhibit better specificity for cancer cells and have reduced toxicity, compared to Bortezomib, which entirely blocks the ubiquitin–proteasome pathway as a proteasome inhibitor. In addition to MLN4924, the deneddylation‐related molecules (such as CSN and NUB1/NUB1L) are also attractive candidates for inhibition of the SCF ligase activity, because they are expected to achieve new strategies with high responsibility and tolerability in anticancer therapy.
Competing interests
The authors have no conflicts of interest to declare.
Acknowledgments
This work was supported in part by National Institutes of Health Grants R01DK56298 and R01AG024497 (to T.K.).
Tanaka Tomoaki, Nakatani Tatsuya, Kamitani Tetsu, (2012), Inhibition of NEDD8‐conjugation pathway by novel molecules: Potential approaches to anticancer therapy, Molecular Oncology, 6, doi: 10.1016/j.molonc.2012.01.003.
Contributor Information
Tomoaki Tanaka, Email: tomoaki826@msic.med.osaka-cu.ac.jp.
Tatsuya Nakatani, Email: nakatani@med.osaka-cu.ac.jp.
Tetsu Kamitani, Email: tkamitani@georgiahealth.edu.
References
- Abida, W.M. , Nikolaev, A. , Zhao, W. , Zhang, W. , Gu, W. , 2007. FBXO11 promotes the neddylation of p53 and inhibits its transcriptional activity. J. Biol. Chem.. 282, 1797–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler, A.S. , Lin, M. , Horlings, H. , Nuyten, D.S. , van de Vijver, M.J. , Chang, H.Y. , 2006. Genetic regulators of large-scale transcriptional signatures in cancer. Nat. Genet.. 38, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir, R.E. , Iwai, K. , Ciechanover, A. , 2002. The NEDD8 pathway is essential for SCF(beta -TrCP)-mediated ubiquitination and processing of the NF-kappa B precursor p105. J. Biol. Chem.. 277, 23253–23259. [DOI] [PubMed] [Google Scholar]
- Andrews, P. , He, Y.J. , Xiong, Y. , 2006. Cytoplasmic localized ubiquitin ligase cullin 7 binds to p53 and promotes cell growth by antagonizing p53 function. Oncogene. 25, 4534–4548. [DOI] [PubMed] [Google Scholar]
- Angers, S. , Thorpe, C.J. , Biechele, T.L. , Goldenberg, S.J. , Zheng, N. , MacCoss, M.J. , Moon, R.T. , 2006. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol.. 8, 348–357. [DOI] [PubMed] [Google Scholar]
- Bawa-Khalfe, T. , Yeh, E.T. , 2010. SUMO losing balance: SUMO proteases disrupt SUMO homeostasis to facilitate cancer development and progression. Genes Cancer. 1, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaidouni, N. , Peuchmaur, M. , Perret, C. , Florentin, A. , Benarous, R. , Besnard-Guerin, C. , 2005. Overexpression of human beta TrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene. 24, 2271–2276. [DOI] [PubMed] [Google Scholar]
- Berg, J.P. , Zhou, Q. , Breuhahn, K. , Schirmacher, P. , Patil, M.A. , Chen, X. , Schafer, N. , Holler, T.T. , Fischer, H.P. , Buttner, R. , Gutgemann, I. , 2007. Inverse expression of Jun activation domain binding protein 1 and cell cycle inhibitor p27Kip1: influence on proliferation in hepatocellular carcinoma. Hum. Pathol.. 38, 1621–1627. [DOI] [PubMed] [Google Scholar]
- Bosu, D.R. , Kipreos, E.T. , 2008. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div.. 3, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnatowska-Hledin, M.A. , Kossoris, J.B. , Van Dort, C.J. , Shearer, R.L. , Zhao, P. , Murrey, D.A. , Abbott, J.L. , Kan, C.E. , Barney, C.C. , 2004. T47D breast cancer cell growth is inhibited by expression of VACM-1, a cul-5 gene. Biochem. Biophys. Res. Commun.. 319, 817–825. [DOI] [PubMed] [Google Scholar]
- Chairatvit, K. , Ngamkitidechakul, C. , 2007. Control of cell proliferation via elevated NEDD8 conjugation in oral squamous cell carcinoma. Mol. Cell Biochem.. 306, 163–169. [DOI] [PubMed] [Google Scholar]
- Chan, Y. , Yoon, J. , Wu, J.T. , Kim, H.J. , Pan, K.T. , Yim, J. , Chien, C.T. , 2008. DEN1 deneddylates non-cullin proteins in vivo. J. Cell Sci.. 121, 3218–3223. [DOI] [PubMed] [Google Scholar]
- Chen, L.C. , Manjeshwar, S. , Lu, Y. , Moore, D. , Ljung, B.M. , Kuo, W.L. , Dairkee, S.H. , Wernick, M. , Collins, C. , Smith, H.S. , 1998. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res.. 58, 3677–3683. [PubMed] [Google Scholar]
- Ciechanover, A. , 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J.. 17, 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover, A. , Schwartz, A.L. , 1998. The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. U.S.A.. 95, 2727–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope, G.A. , Suh, G.S. , Aravind, L. , Schwarz, S.E. , Zipursky, S.L. , Koonin, E.V. , Deshaies, R.J. , 2002. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 298, 608–611. [DOI] [PubMed] [Google Scholar]
- Deng, X.W. , Dubiel, W. , Wei, N. , Hofmann, K. , Mundt, K. , Colicelli, J. , Kato, J. , Naumann, M. , Segal, D. , Seeger, M. , Carr, A. , Glickman, M. , Chamovitz, D.A. , 2000. Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development. Trends Genet.. 16, 202–203. [DOI] [PubMed] [Google Scholar]
- Dubiel, W. , 2009. Resolving the CSN and CAND1 paradoxes. Mol. Cell. 35, 547–549. [DOI] [PubMed] [Google Scholar]
- Furukawa, M. , He, Y.J. , Borchers, C. , Xiong, Y. , 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol.. 5, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Furukawa, M. , Xiong, Y. , 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell Biol.. 25, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F. , Cheng, J. , Shi, T. , Yeh, E.T. , 2006. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat. Cell Biol.. 8, 1171–1177. [DOI] [PubMed] [Google Scholar]
- Goldenberg, S.J. , Cascio, T.C. , Shumway, S.D. , Garbutt, K.C. , Liu, J. , Xiong, Y. , Zheng, N. , 2004. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 119, 517–528. [DOI] [PubMed] [Google Scholar]
- Gong, L. , Yeh, E.T. , 1999. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem.. 274, 12036–12042. [DOI] [PubMed] [Google Scholar]
- Gong, L. , Kamitani, T. , Millas, S. , Yeh, E.T. , 2000. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J. Biol. Chem.. 275, 14212–14216. [DOI] [PubMed] [Google Scholar]
- Groisman, R. , Polanowska, J. , Kuraoka, I. , Sawada, J. , Saijo, M. , Drapkin, R. , Kisselev, A.F. , Tanaka, K. , Nakatani, Y. , 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 113, 357–367. [DOI] [PubMed] [Google Scholar]
- Grossberger, R. , Gieffers, C. , Zachariae, W. , Podtelejnikov, A.V. , Schleiffer, A. , Nasmyth, K. , Mann, M. , Peters, J.M. , 1999. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J. Biol. Chem.. 274, 14500–14507. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro, D. , Pagano, M. , 2004. Oncogenic aberrations of cullin-dependent ubiquitin ligases. Oncogene. 23, 2037–2049. [DOI] [PubMed] [Google Scholar]
- Haas, A.L. , Ahrens, P. , Bright, P.M. , Ankel, H. , 1987. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem.. 262, 11315–11323. [PubMed] [Google Scholar]
- Harper, J.W. , 2004. Neddylating the guardian; Mdm2 catalyzed conjugation of Nedd8 to p53. Cell. 118, 2–4. [DOI] [PubMed] [Google Scholar]
- Hershko, A. , 2005. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ.. 12, 1191–1197. [DOI] [PubMed] [Google Scholar]
- Hershko, A. , Ciechanover, A. , 1998. The ubiquitin system. Annu. Rev. Biochem.. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hilton, D.J. , 1999. Negative regulators of cytokine signal transduction. Cell Mol. Life Sci.. 55, 1568–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T. , Osaka, F. , Chiba, T. , Miyamoto, C. , Okabayashi, K. , Shimbara, N. , Kato, S. , Tanaka, K. , 1999. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 18, 6829–6834. [DOI] [PubMed] [Google Scholar]
- Hosono, T. , Tanaka, T. , Tanji, K. , Nakatani, T. , Kamitani, T. , 2010. NUB1, an interferon-inducible protein, mediates anti-proliferative actions and apoptosis in renal cell carcinoma cells through cell-cycle regulation. Br. J. Cancer. 102, 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. , McCall, C.M. , Ohta, T. , Xiong, Y. , 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol.. 6, 1003–1009. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Zacharek, S. , He, Y.J. , Lee, H. , Shumway, S. , Duronio, R.J. , Xiong, Y. , 2008. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev.. 22, 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D.T. , Paydar, A. , Zhuang, M. , Waddell, M.B. , Holton, J.M. , Schulman, B.A. , 2005. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol. Cell. 17, 341–350. [DOI] [PubMed] [Google Scholar]
- Ichimura, Y. , Kirisako, T. , Takao, T. , Satomi, Y. , Shimonishi, Y. , Ishihara, N. , Mizushima, N. , Tanida, I. , Kominami, E. , Ohsumi, M. , Noda, T. , Ohsumi, Y. , 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408, 488–492. [DOI] [PubMed] [Google Scholar]
- Jackson, S. , Xiong, Y. , 2009. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci.. 34, 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath, S. , Barlogie, B. , Berenson, J.R. , Singhal, S. , Alexanian, R. , Srkalovic, G. , Orlowski, R.Z. , Richardson, P.G. , Anderson, J. , Nix, D. , Esseltine, D.L. , Anderson, K.C. , 2005. Bortezomib in recurrent and/or refractory multiple myeloma. Initial clinical experience in patients with impaired renal function. Cancer. 103, 1195–1200. [DOI] [PubMed] [Google Scholar]
- Johnson, A.E. , Le, I.P. , Buchwalter, A. , Burnatowska-Hledin, M.A. , 2007. Estrogen-dependent growth and estrogen receptor (ER)-alpha concentration in T47D breast cancer cells are inhibited by VACM-1, a cul 5 gene. Mol. Cell Biochem.. 301, 13–20. [DOI] [PubMed] [Google Scholar]
- Kaelin, W.G. , Ratcliffe, P.J. , 2008. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 30, 393–402. [DOI] [PubMed] [Google Scholar]
- Kamitani, T. , Kito, K. , Fukuda-Kamitani, T. , Yeh, E.T. , 2001. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J. Biol. Chem.. 276, 46655–46660. [DOI] [PubMed] [Google Scholar]
- Kamitani, T. , Kito, K. , Nguyen, H.P. , Yeh, E.T. , 1997. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem.. 272, 28557–28562. [DOI] [PubMed] [Google Scholar]
- Kamura, T. , Conrad, M.N. , Yan, Q. , Conaway, R.C. , Conaway, J.W. , 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev.. 13, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura, T. , Koepp, D.M. , Conrad, M.N. , Skowyra, D. , Moreland, R.J. , Iliopoulos, O. , Lane, W.S. , Kaelin, W.G. , Elledge, S.J. , Conaway, R.C. , Harper, J.W. , Conaway, J.W. , 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 284, 657–661. [DOI] [PubMed] [Google Scholar]
- Kapetanaki, M.G. , Guerrero-Santoro, J. , Bisi, D.C. , Hsieh, C.L. , Rapic-Otrin, V. , Levine, A.S. , 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. U.S.A.. 103, 2588–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaustov, L. , Lukin, J. , Lemak, A. , Duan, S. , Ho, M. , Doherty, R. , Penn, L.Z. , Arrowsmith, C.H. , 2007. The conserved CPH domains of Cul7 and PARC are protein-protein interaction modules that bind the tetramerization domain of p53. J. Biol. Chem.. 282, 11300–11307. [DOI] [PubMed] [Google Scholar]
- Kipreos, E.T. , Lander, L.E. , Wing, J.P. , He, W.W. , Hedgecock, E.M. , 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 85, 829–839. [DOI] [PubMed] [Google Scholar]
- Kito, K. , Yeh, E.T. , Kamitani, T. , 2001. NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J. Biol. Chem.. 276, 20603–20609. [DOI] [PubMed] [Google Scholar]
- Kouvaraki, M.A. , Korapati, A.L. , Rassidakis, G.Z. , Tian, L. , Zhang, Q. , Chiao, P. , Ho, L. , Evans, D.B. , Claret, F.X. , 2006. Potential role of Jun activation domain-binding protein 1 as a negative regulator of p27kip1 in pancreatic adenocarcinoma. Cancer Res.. 66, 8581–8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo, Y. , Kitajima, S. , Ogawa, I. , Kitagawa, M. , Miyauchi, M. , Takata, T. , 2005. Small interfering RNA targeting of S phase kinase-interacting protein 2 inhibits cell growth of oral cancer cells by inhibiting p27 degradation. Mol. Cancer Ther.. 4, 471–476. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Tomooka, Y. , Noda, M. , 1992. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun.. 185, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Kurz, T. , Chou, Y.C. , Willems, A.R. , Meyer-Schaller, N. , Hecht, M.L. , Tyers, M. , Peter, M. , Sicheri, F. , 2008. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell. 29, 23–35. [DOI] [PubMed] [Google Scholar]
- Kurz, T. , Ozlu, N. , Rudolf, F. , O'Rourke, S.M. , Luke, B. , Hofmann, K. , Hyman, A.A. , Bowerman, B. , Peter, M. , 2005. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae . Nature. 435, 1257–1261. [DOI] [PubMed] [Google Scholar]
- Laney, J.D. , Hochstrasser, M. , 1999. Substrate targeting in the ubiquitin system. Cell. 97, 427–430. [DOI] [PubMed] [Google Scholar]
- Leck, Y.C. , Choo, Y.Y. , Tan, C.Y. , Smith, P.G. , Hagen, T. , 2010. Biochemical and cellular effects of inhibiting Nedd8 conjugation. Biochem. Biophys. Res. Commun.. 398, 588–593. [DOI] [PubMed] [Google Scholar]
- Li, W. , Kong, A.N. , 2009. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog.. 48, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan, J. , Imbert, G. , Wirbelauer, C. , Gstaiger, M. , Krek, W. , 1999. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev.. 13, 1822–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Furukawa, M. , Matsumoto, T. , Xiong, Y. , 2002. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell. 10, 1511–1518. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Lee, S. , Zhang, J. , Peters, S.B. , Hannah, J. , Zhang, Y. , Yin, Y. , Koff, A. , Ma, L. , Zhou, P. , 2009. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell. 34, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.C. , Pan, J. , Zhang, C. , Fan, W. , Collinge, M. , Bender, J.R. , Weissman, S.M. , 1999. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc. Natl. Acad. Sci. U.S.A.. 96, 4313–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, K.R. , Haas, A.L. , 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem.. 267, 7806–7813. [PubMed] [Google Scholar]
- Lyapina, S. , Cope, G. , Shevchenko, A. , Serino, G. , Tsuge, T. , Zhou, C. , Wolf, D.A. , Wei, N. , Deshaies, R.J. , 2001. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Melchor, L. , Saucedo-Cuevas, L.P. , Munoz-Repeto, I. , Rodriguez-Pinilla, S.M. , Honrado, E. , Campoverde, A. , Palacios, J. , Nathanson, K.L. , Garcia, M.J. , Benitez, J. , 2009. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res.. 11, R86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza, H.M. , Shen, L.N. , Botting, C. , Lewis, A. , Chen, J. , Ink, B. , Hay, R.T. , 2003. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem.. 278, 25637–25643. [DOI] [PubMed] [Google Scholar]
- Milhollen, M.A. , Traore, T. , Adams-Duffy, J. , Thomas, M.P. , Berger, A.J. , Dang, L. , Dick, L.R. , Garnsey, J.J. , Koenig, E. , Langston, S.P. , Manfredi, M. , Narayanan, U. , Rolfe, M. , Staudt, L.M. , Soucy, T.A. , Yu, J. , Zhang, J. , Bolen, J.B. , Smith, P.G. , 2010. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 116, 1515–1523. [DOI] [PubMed] [Google Scholar]
- Mizushima, N. , Noda, T. , Yoshimori, T. , Tanaka, Y. , Ishii, T. , George, M.D. , Klionsky, D.J. , Ohsumi, M. , Ohsumi, Y. , 1998. A protein conjugation system essential for autophagy. Nature. 395, 395–398. [DOI] [PubMed] [Google Scholar]
- Morimoto, M. , Nishida, T. , Honda, R. , Yasuda, H. , 2000. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1). Biochem. Biophys. Res. Commun.. 270, 1093–1096. [DOI] [PubMed] [Google Scholar]
- Morimoto, M. , Nishida, T. , Nagayama, Y. , Yasuda, H. , 2003. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem. Biophys. Res. Commun.. 301, 392–398. [DOI] [PubMed] [Google Scholar]
- Murata, T. , Takayama, K. , Katayama, S. , Urano, T. , Horie-Inoue, K. , Ikeda, K. , Takahashi, S. , Kawazu, C. , Hasegawa, A. , Ouchi, Y. , Homma, Y. , Hayashizaki, Y. , Inoue, S. , 2010. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis.. 13, 356–361. [DOI] [PubMed] [Google Scholar]
- Okabe, H. , Lee, S.H. , Phuchareon, J. , Albertson, D.G. , McCormick, F. , Tetsu, O. , 2006. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS One. 1, e128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka, F. , Kawasaki, H. , Aida, N. , Saeki, M. , Chiba, T. , Kawashima, S. , Tanaka, K. , Kato, S. , 1998. A new NEDD8-ligating system for cullin-4A. Genes Dev.. 12, 2263–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved, S. , Mosesson, Y. , Zwang, Y. , Santonico, E. , Shtiegman, K. , Marmor, M.D. , Kochupurakkal, B.S. , Katz, M. , Lavi, S. , Cesareni, G. , Yarden, Y. , 2006. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem.. 281, 21640–21651. [DOI] [PubMed] [Google Scholar]
- Pagano, M. , 1997. Cell cycle regulation by the ubiquitin pathway. FASEB J.. 11, 1067–1075. [DOI] [PubMed] [Google Scholar]
- Panattoni, M. , Sanvito, F. , Basso, V. , Doglioni, C. , Casorati, G. , Montini, E. , Bender, J.R. , Mondino, A. , Pardi, R. , 2008. Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J. Exp. Med.. 205, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard, L. , Willis, J.H. , Willems, A. , Johnson, J.L. , Srayko, M. , Kurz, T. , Glaser, S. , Mains, P.E. , Tyers, M. , Bowerman, B. , Peter, M. , 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 425, 311–316. [DOI] [PubMed] [Google Scholar]
- Podust, V.N. , Brownell, J.E. , Gladysheva, T.B. , Luo, R.S. , Wang, C. , Coggins, M.B. , Pierce, J.W. , Lightcap, E.S. , Chau, V. , 2000. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl. Acad. Sci. U.S.A.. 97, 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut, G. , Peter, M. , 2008. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep.. 9, 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao-Naik, C. , delaCruz, W. , Laplaza, J.M. , Tan, S. , Callis, J. , Fisher, A.J. , 1998. The rub family of ubiquitin-like proteins. Crystal structure of Arabidopsis rub1 and expression of multiple rubs in Arabidopsis. J. Biol. Chem.. 273, 34976–34982. [DOI] [PubMed] [Google Scholar]
- Read, M.A. , Brownell, J.E. , Gladysheva, T.B. , Hottelet, M. , Parent, L.A. , Coggins, M.B. , Pierce, J.W. , Podust, V.N. , Luo, R.S. , Chau, V. , Palombella, V.J. , 2000. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol. Cell Biol.. 20, 2326–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, P.G. , Sonneveld, P. , Schuster, M.W. , Irwin, D. , Stadtmauer, E.A. , Facon, T. , Harousseau, J.L. , Ben-Yehuda, D. , Lonial, S. , Goldschmidt, H. , Reece, D. , San-Miguel, J.F. , Blade, J. , Boccadoro, M. , Cavenagh, J. , Dalton, W.S. , Boral, A.L. , Esseltine, D.L. , Porter, J.B. , Schenkein, D. , Anderson, K.C. , 2005. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N. Engl. J. Med.. 352, 2487–2498. [DOI] [PubMed] [Google Scholar]
- Salon, C. , Brambilla, E. , Brambilla, C. , Lantuejoul, S. , Gazzeri, S. , Eymin, B. , 2007. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J. Pathol.. 213, 303–310. [DOI] [PubMed] [Google Scholar]
- San Miguel, J.F. , Schlag, R. , Khuageva, N.K. , Dimopoulos, M.A. , Shpilberg, O. , Kropff, M. , Spicka, I. , Petrucci, M.T. , Palumbo, A. , Samoilova, O.S. , Dmoszynska, A. , Abdulkadyrov, K.M. , Schots, R. , Jiang, B. , Mateos, M.V. , Anderson, K.C. , Esseltine, D.L. , Liu, K. , Cakana, A. , van de Velde, H. , Richardson, P.G. , 2008. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N. Engl. J. Med.. 359, 906–917. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C. , Serino, G. , Callis, J. , Crosby, W.L. , Lyapina, S. , Deshaies, R.J. , Gray, W.M. , Estelle, M. , Deng, X.W. , 2001. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Scott, D.C. , Monda, J.K. , Grace, C.R. , Duda, D.M. , Kriwacki, R.W. , Kurz, T. , Schulman, B.A. , 2010. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell. 39, 784–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, T. , Ohta, T. , Tong, K.I. , Kokubu, A. , Odogawa, R. , Tsuta, K. , Asamura, H. , Yamamoto, M. , Hirohashi, S. , 2008. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. U.S.A.. 105, 13568–13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, J.D. , Gurian-West, M. , Clurman, B. , Roberts, J.M. , 1999. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev.. 13, 2375–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar, J.R. , D'Angiolella, V. , Pagan, J.K. , Pagano, M. , 2009. SnapShot: F Box Proteins II. Cell. 137, 1358 1358.e1 [DOI] [PubMed] [Google Scholar]
- Soucy, T.A. , Smith, P.G. , Milhollen, M.A. , Berger, A.J. , Gavin, J.M. , Adhikari, S. , Brownell, J.E. , Burke, K.E. , Cardin, D.P. , Critchley, S. , Cullis, C.A. , Doucette, A. , Garnsey, J.J. , Gaulin, J.L. , Gershman, R.E. , Lublinsky, A.R. , McDonald, A. , Mizutani, H. , Narayanan, U. , Olhava, E.J. , Peluso, S. , Rezaei, M. , Sintchak, M.D. , Talreja, T. , Thomas, M.P. , Traore, T. , Vyskocil, S. , Weatherhead, G.S. , Yu, J. , Zhang, J. , Dick, L.R. , Claiborne, C.F. , Rolfe, M. , Bolen, J.B. , Langston, S.P. , 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 458, 732–736. [DOI] [PubMed] [Google Scholar]
- Stebbins, C.E. , Kaelin, W.G. , Pavletich, N.P. , 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 284, 455–461. [DOI] [PubMed] [Google Scholar]
- Sugasawa, K. , Okuda, Y. , Saijo, M. , Nishi, R. , Matsuda, N. , Chu, G. , Mori, T. , Iwai, S. , Tanaka, K. , Hanaoka, F. , 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 121, 387–400. [DOI] [PubMed] [Google Scholar]
- Swords, R.T. , Kelly, K.R. , Smith, P.G. , Garnsey, J.J. , Mahalingam, D. , Medina, E. , Oberheu, K. , Padmanabhan, S. , O'Dwyer, M. , Nawrocki, S.T. , Giles, F.J. , Carew, J.S. , 2010. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 115, 3796–3800. [DOI] [PubMed] [Google Scholar]
- Tanaka, T. , Kawashima, H. , Yeh, E.T. , Kamitani, T. , 2003. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J. Biol. Chem.. 278, 32905–32913. [DOI] [PubMed] [Google Scholar]
- Tanji, K. , Tanaka, T. , Kamitani, T. , 2005. Interaction of NUB1 with the proteasome subunit S5a. Biochem. Biophys. Res. Commun.. 337, 116–120. [DOI] [PubMed] [Google Scholar]
- Tomoda, K. , Kubota, Y. , Arata, Y. , Mori, S. , Maeda, M. , Tanaka, T. , Yoshida, M. , Yoneda-Kato, N. , Kato, J.Y. , 2002. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J. Biol. Chem.. 277, 2302–2310. [DOI] [PubMed] [Google Scholar]
- Tomoda, K. , Kubota, Y. , Kato, J. , 1999. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 398, 160–165. [DOI] [PubMed] [Google Scholar]
- Wada, H. , Yeh, E.T. , Kamitani, T. , 1999. Identification of NEDD8-conjugation site in human cullin-2. Biochem. Biophys. Res. Commun.. 257, 100–105. [DOI] [PubMed] [Google Scholar]
- Wada, H. , Yeh, E.T. , Kamitani, T. , 1999. The von Hippel-Lindau tumor suppressor gene product promotes, but is not essential for, NEDD8 conjugation to cullin-2. J. Biol. Chem.. 274, 36025–36029. [DOI] [PubMed] [Google Scholar]
- Wada, H. , Yeh, E.T. , Kamitani, T. , 2000. A dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8 conjugation in vivo. J. Biol. Chem.. 275, 17008–17015. [DOI] [PubMed] [Google Scholar]
- Walden, H. , Podgorski, M.S. , Huang, D.T. , Miller, D.W. , Howard, R.J. , Minor, D.L. , Holton, J.M. , Schulman, B.A. , 2003. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell. 12, 1427–1437. [DOI] [PubMed] [Google Scholar]
- Watson, I.R. , Blanch, A. , Lin, D.C. , Ohh, M. , Irwin, M.S. , 2006. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J. Biol. Chem.. 281, 34096–34103. [DOI] [PubMed] [Google Scholar]
- Watson, I.R. , Li, B.K. , Roche, O. , Blanch, A. , Ohh, M. , Irwin, M.S. , 2010. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene. 29, 297–304. [DOI] [PubMed] [Google Scholar]
- Westbrook, T.F. , Hu, G. , Ang, X.L. , Mulligan, P. , Pavlova, N.N. , Liang, A. , Leng, Y. , Maehr, R. , Shi, Y. , Harper, J.W. , Elledge, S.J. , 2008. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 452, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby, F.G. , Xia, G. , Pickart, C.M. , Hill, C.P. , 1998. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem.. 273, 34983–34991. [DOI] [PubMed] [Google Scholar]
- Willam, C. , Nicholls, L.G. , Ratcliffe, P.J. , Pugh, C.W. , Maxwell, P.H. , 2004. The prolyl hydroxylase enzymes that act as oxygen sensors regulating destruction of hypoxia-inducible factor alpha. Adv. Enzyme Regul.. 44, 75–92. [DOI] [PubMed] [Google Scholar]
- Xirodimas, D.P. , Saville, M.K. , Bourdon, J.C. , Hay, R.T. , Lane, D.P. , 2004. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 118, 83–97. [DOI] [PubMed] [Google Scholar]
- Xu, X. , Sarikas, A. , Dias-Santagata, D.C. , Dolios, G. , Lafontant, P.J. , Tsai, S.C. , Zhu, W. , Nakajima, H. , Nakajima, H.O. , Field, L.J. , Wang, R. , Pan, Z.Q. , 2008. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol. Cell. 30, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G. , Ayala, G. , De Marzo, A. , Tian, W. , Frolov, A. , Wheeler, T.M. , Thompson, T.C. , Harper, J.W. , 2002. Elevated Skp2 protein expression in human prostate cancer: association with loss of the cyclin-dependent kinase inhibitor p27 and PTEN and with reduced recurrence-free survival. Clin. Cancer Res.. 8, 3419–3426. [PubMed] [Google Scholar]
- Yang, X. , Zhou, J. , Sun, L. , Wei, Z. , Gao, J. , Gong, W. , Xu, R.M. , Rao, Z. , Liu, Y. , 2007. Structural basis for the function of DCN-1 in protein neddylation. J. Biol. Chem.. 282, 24490–24494. [DOI] [PubMed] [Google Scholar]
- Yeh, E.T. , Gong, L. , Kamitani, T. , 2000. Ubiquitin-like proteins: new wines in new bottles. Gene. 248, 1–14. [DOI] [PubMed] [Google Scholar]
- Yen, H.C. , Elledge, S.J. , 2008. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 322, 923–929. [DOI] [PubMed] [Google Scholar]
- Zhong, W. , Feng, H. , Santiago, F.E. , Kipreos, E.T. , 2003. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 423, 885–889. [DOI] [PubMed] [Google Scholar]


