Abstract
A key technical challenge in cartilage tissue engineering is the development of a noninvasive method for monitoring the composition, structure, and function of the tissue at different growth stages. Due to its noninvasive, three-dimensional imaging capabilities and the breadth of available contrast mechanisms, magnetic resonance imaging (MRI) techniques can be expected to play a leading role in assessing engineered cartilage. In this review, we describe the new MR-based tools (spectroscopy, imaging, and elastography) that can provide quantitative biomarkers for cartilage tissue development both in vitro and in vivo. Magnetic resonance spectroscopy can identify the changing molecular structure and alternations in the conformation of major macromolecules (collagen and proteoglycans) using parameters such as chemical shift, relaxation rates, and magnetic spin couplings. MRI provides high-resolution images whose contrast reflects developing tissue microstructure and porosity through changes in local relaxation times and the apparent diffusion coefficient. Magnetic resonance elastography uses low-frequency mechanical vibrations in conjunction with MRI to measure soft tissue mechanical properties (shear modulus and viscosity). When combined, these three techniques provide a noninvasive, multiscale window for characterizing cartilage tissue growth at all stages of tissue development, from the initial cell seeding of scaffolds to the development of the extracellular matrix during construct incubation, and finally, to the postimplantation assessment of tissue integration in animals and patients.
Introduction
Musculoskeletal disorders impact nearly one in three Americans annually, with injuries to cartilage accounting for a large fraction of these afflictions.1,2 In particular, damage to articular cartilage due to injuries in the knees, shoulders, and other joints comprises a major portion of these disorders.3–5 Osteoarthritis is a major cause of articular cartilage damage; one that is particularly evident in the aging population.6 Articular cartilage derives its load-bearing and viscoelastic properties from the tissue's extracellular matrix (ECM), which is composed primarily of type II collagen, proteoglycan, and water.7,8 Mature cartilage is an avascular tissue with a three-layered zonal structure and a few active chondrocytes, which results in a tissue with poor self-healing capabilities. The ideal treatment for cartilage injuries would be to re-establish a new articular cartilage at the injury site that functions under load in a manner identical to the asymptomatic joint.9 The repair should be accomplished via arthroscopic surgery and thereafter monitored noninvasively.
To develop a replacement for damaged or diseased cartilage, tissue engineers use a variety of approaches. These techniques combine biocompatible scaffolds (natural or synthetic), appropriate cell types, and cell signaling molecules to create a tissue that mimics the functional native tissue.10 Scaffolds are selected based on their ability to promote adhesion and proliferation of the desired cell types, although scaffold-free methods have also been reported in the literature.11,12 As a cell source, either chondrocytes harvested from cartilage or mesenchymal stem cells (MSCs) differentiating toward chondrocytes are used.13,14 Growth factors promoting chondrogenesis or biomimetic approaches are also used to promote chondrogenic differentiation of cells.13,15 Although much research progress has been reported in the past decade, the implantation of engineered cartilage tissue has not yet reached the orthopedic clinic. Scale up from animal studies and translation to the clinic requires noninvasive assessment of the growing tissue for an extended period of time (weeks to months). During this time, the researcher, physician, and the patient need specific information about all aspects of the ECM development and tissue integration.
New imaging tools are therefore needed to assess the success of current first-generation tissue regeneration techniques (microfracture, autographs, and autologous chondrocyte implantation), and to test the effectiveness of the emerging second-generation tissue-engineering methods (cells, scaffolds, and growth factor therapies). The imaging tools must be safe and ideally they should provide quantitative information at all stages of tissue growth, repair, and regeneration. Assessing the success of tissue engineering relies on our ability to observe cell growth, cellular differentiation, and tissue morphogenesis noninvasively and longitudinally. Different medical imaging modalities—optical, x-ray, and magnetic resonance—provide images with contrast that could be used to monitor the progress of growing engineered tissue. In addition, each technique has a characteristic resolution and a field of view that extends from the micron dimensions of a cell (for light microscopy) up to the length of the human femur (X-ray and magnetic resonance imaging [MRI]). These imaging tools are used at the bench, in the laboratory, and in the clinic: (1) to observe the cellular response to chemical growth factors and biomechanical forces, (2) to characterize the ability of scaffolds to support cell growth and differentiation, (3) to image the spatial organization of the ECM components, and ultimately, (4) to monitor the strength and integration of tissue implanted in animals and in humans. For example, in cartilage regeneration, tissue engineers must regulate and adjust many variables, from selecting the optimal cell-seeding density and timing for administration of growth factors, to monitoring in vivo development and integration of new tissue. A key aspect of this regeneration process is the feedback of information to the tissue engineer identifying the dynamics of tissue growth. Such feedback should be based on noninvasive imaging and available for the entire time period of tissue growth.
Presently, when tissue-engineered cartilage is being evaluated, the production of proteoglycan and type-II collagen are used as biomarkers for success. These ECM components are assessed by biochemical techniques that include quantitative glycosaminoglycan (GAG) and collagen assays, gene expression analyses using the quantitative real-time polymerase chain reaction, and histological staining.16–20 The mechanical properties are typically assessed by confined and unconfined compression or micro/nanoindentation.21–23 When scaffolds are implanted in animals, they are removed at different time points and analyzed ex vivo. Although informative and quantitative, such techniques for visualizing tissue growth are destructive, nonlocalized, and restricted to an individual sampling time. Another concern is that these biological techniques are usually performed after freezing or fixing of the tissue, which changes the tissue's properties, and therefore cannot provide de facto information about the dynamical processes and biomechanical properties in the in vivo environment.24–26 It is also noted that the analytical macromolecular content does not wholly represent the complex structural and functional aspects of cartilage, which are assessed by elasticity and molecular mobility in the tissue.27,28
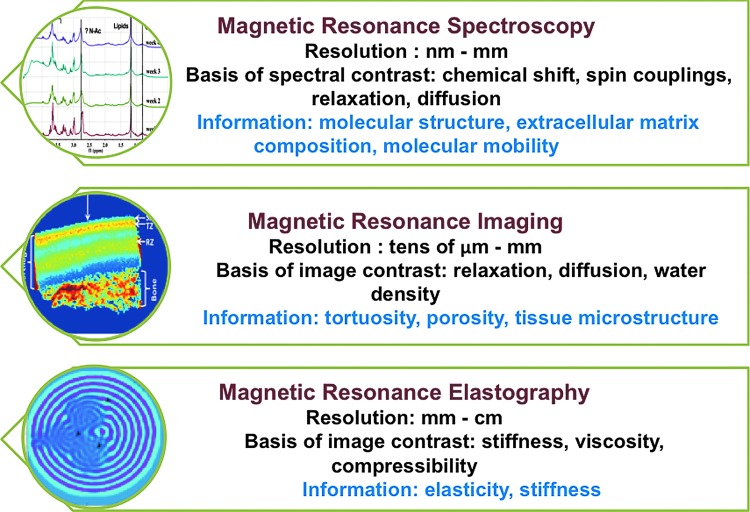
Magnetic resonance spectroscopy (MRS), MRI, and magnetic resonance elastography (MRE) are noninvasive methods that can provide biochemical, anatomical, and biomechanical measures of tissue. Figure 1 describes exemplary features of MR spectroscopy, imaging, and elastography techniques. MRS and imaging are well-established techniques that have been used to probe the structure and dynamics of biochemical changes in engineered tissues.28–42 MR elastography, while relatively new, is rapidly being employed to assess the mechanical properties of engineered tissues.30,43 Using MR spectroscopy, imaging, and elastography together, it is possible to create three-dimensional (3D) maps of chemical shifts, relaxation times, diffusion coefficient, spectral couplings, and tissue stiffness.44 These tissue parameters are strongly correlated with the physical, chemical, and mechanical properties of the growing tissue.
FIG. 1.
Key features of magnetic resonance spectroscopy, imaging, and elastography for characterizing biological tissues. Color images available online at www.liebertpub.com/teb
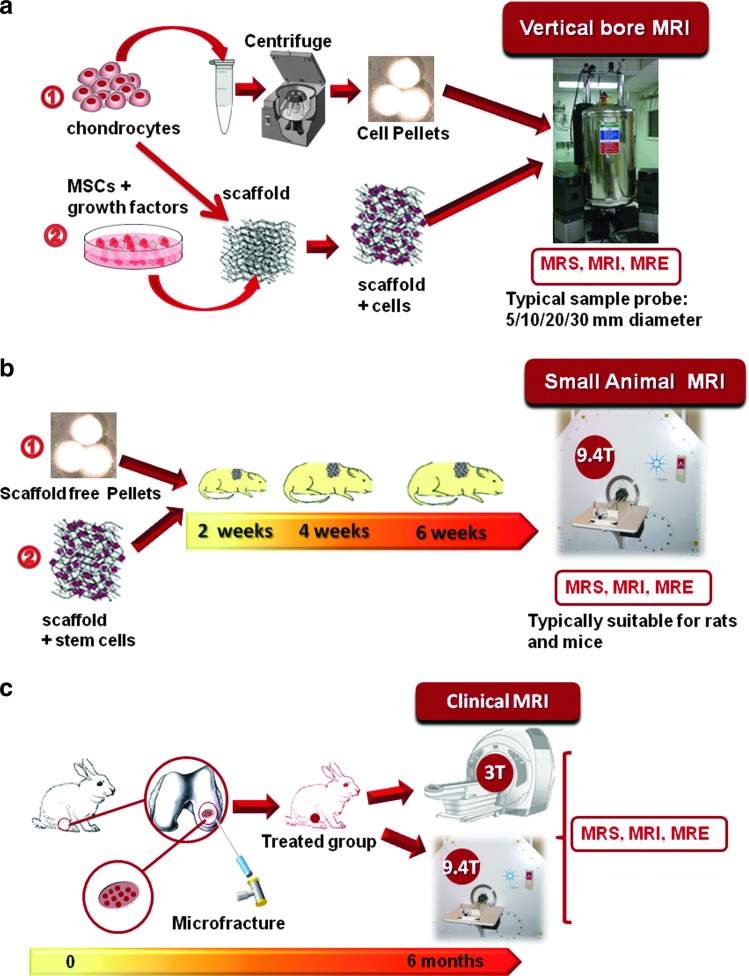
A proposed flowchart for the characterization of engineered tissue in vitro and in vivo using MR techniques is shown in Figure 2. Briefly, the chondrocytes or stem cells are expanded in a suitable scaffold-free or scaffold-based microenvironment with chondrogenic growth media used to ensure differentiation. The tissues are next grown in MR-compatible bioreactors or in a more traditional incubator under suitable culture conditions. The samples are taken for MRS, MRI, and MRE at the chosen time point and if grown in NMR-compatible bioreactors, they can be returned to the incubator for further growth. For in vivo studies, the implanted or regenerating tissue can be measured using MR techniques at the chosen time points on the same tissue constructs. Thus, it is possible to conduct a longitudinal study for the same tissue construct without destroying the tissue. Finally, quantitative MR-derived parametric maps (such as the chemical shift map, T2 and the apparent diffusion coefficient (ADC) map, tissue-stiffness map) can be created using a specific pulse protocol for each tissue. Such information is vital to tissue engineers so that they can regulate and optimize the tissue growth conditions.
FIG. 2.
Schematic flowchart for in vitro (a) and in vivo (b, c) MR-based assessment of tissue-engineered cartilage. MR techniques are compatible with different tissue growth strategies for in vitro studies as shown in (a) as well as in vivo studies involving implanted engineered tissues (b) or tracking of cartilage regeneration, for example, using microfracture in rabbit knee cartilage (c) in small or large animal models. MR, magnetic resonance; MRI, magnetic resonance imaging; MRS, magnetic resonance spectroscopy; MRE, magnetic resonance elastography; MSCs, mesenchymal stem cells. Color images available online at www.liebertpub.com/teb
The sensitivity and specificity of MR techniques depend upon the choice of the magnetic field, nuclei, pulse sequence, and imaging gradients (for a review of the basic phenomena underlying MRI as applied to tissue engineering, see the earlier review by Xu et al.30). In this article, we will focus on the use of MRS, MRI, and MRE for assessing the growth of engineered cartilage tissue. We first discuss the principles of cartilage tissue engineering as practiced today. We then discuss recent advances using MR techniques to monitor cartilage tissue growth. We end with a discussion of the strengths and weaknesses of the individual techniques and the outlook for future developments.
Cartilage Tissue Engineering
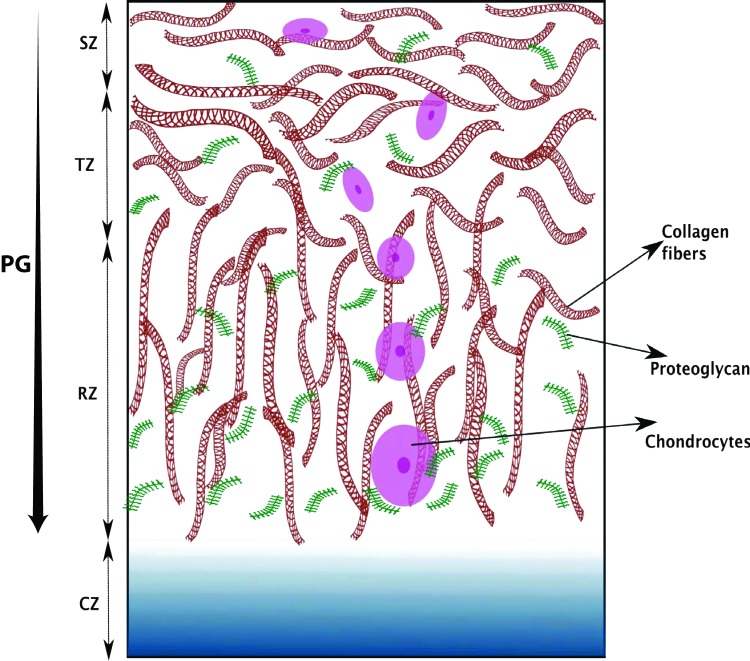
Articular cartilage has a three-layered zonal structure that is composed of chondrocytes and ECM components: proteoglycans, collagen, and synovial fluid. The collagen–proteoglycan network provides the mechanical strength needed for its load-bearing function. The synovial fluid provides nutrients to chondrocytes and is essential for maintaining a low-friction surface needed for articulation.45,46 These zones are named as the superficial zone (SZ), transitional zone (TZ), radial zone (RZ) with calcified zone (CZ) connecting to the bone as depicted in Figure 3.7,8,47 The cellular and ECM properties are depth dependent as shown in Figure 3. The natural cartilage's shear modulus is found to be of the order of ∼0.5–4 MPa and it changes with depth below the articular surface.48 It is recognized that both the collagen mesh network and mobility of proteoglycan aggregates contribute to the tissue stiffness and elasticity. The overall goal of cartilage tissue engineering is to mimic this natural structure using cells, scaffolds, growth factors, and mechanical conditions.13,49,50 Scaffolds composed of natural polymers such as, polysaccharide-based hydrogels (alginate beads, chitosan, or agarose) or protein-based hydrogels (collagen or fibrin) are now commonly used in cartilage tissue engineering.51–53 However, because of the ease with which synthetic polymer hydrogels can be functionalized and modified, polyethyleneglycol (PEG) and PEG-derived hydrogels or -fibrins are also making their way into cartilage tissue engineering.49,54,55 Unfortunately, both types of scaffolds yield products that fall short of the biochemical and mechanical properties of the native tissue. Thus, much research is currently underway to improve cartilage tissue-engineering techniques.10 Chondrocytes derived from natural sources or from MSCs differentiating toward chondrocytes are commonly used as a cell source in cartilage tissue engineering; however, there are unmet challenges in expanding and maintaining chondrocytes (e.g., the composition of the construct and the regulation of culture conditions need careful monitoring).13,14,50,56,57 In addition, chondrogenic growth factors play a key role in chondrocyte differentiation and cartilage generation.58 The most challenging aspect of cartilage tissue engineering is to replicate the natural compressive behavior of cartilage under mechanical loading conditions and its porous structure for cell nutrients. Mechanical stimulations in the form of dynamic compression and hydrostatic pressure have been found to be effective in promoting chondrogenic differentiation of stem cells and chondrocytes; however, results are mixed for other types of mechanical stimulation.53 Many of the early efforts in cartilage tissue engineering were focused on the production of proteoglycan and collagen as the measure of success; however, recent work attempts to achieve the more difficult goal of creating engineered cartilage with a three-layered zonal structure and the mechanical strength of native tissue.13,19,22,49,59,60 A recent work by Moutos et al. uses the 3D microweaving technique with controlled growth factor delivery to achieve a three-layered zonal structure and desired concentration of ECM molecules to create a native-like synthetic tissue.22 Another recent work also targets the development of a custom-shaped engineered tissue graft specifically designed for the cartilage defect in each patient.61
FIG. 3.
Illustration of zonal arrangement of proteoglycans, collagen, and chondrocytes in the hyaline articular cartilage. The picture does not correspond to the actual dimensions. The abbreviations in the figure with typical length scales are SZ-superficial zone (10%–20%); TZ-transitional zone (also called middle zone) (40%–60%); RZ-radial zone (also called deep zone) (30%–40%), and CZ-calcified zone as described in the text. The typical thickness of knee articular cartilage has been found to be ∼2 mm.47 As we move from the superficial to the deep zone, chondrocytes change in their shape and orientations, the collagen fiber orientation changes, and the amount of proteoglycans increase. PG, proteoglycan. Color images available online at www.liebertpub.com/teb
The future success of cartilage tissue-engineering hinges on the development of implants that have the biochemical, anatomical, and mechanical properties of native tissue. Thus, as engineered cartilage tissues are designed with increasing complexity, new noninvasive and volumetric monitoring imaging techniques will be needed to visualize tissue growth in vitro and in vivo. MR-based techniques, can address these needs by providing high-resolution multislice images, spectroscopic analysis of macromolecular composition and structure (cells, collagen, and proteoglycan), and elastography maps of tissue stiffness. All this information can be displayed in the form of quantitative maps of growing tissue that is acquired from living animals or humans in a noninvasive and longitudinal manner.
Magnetic Resonance Spectroscopy
MRS (or NMR spectroscopy as it is popularly known) records the radiofrequency signal whose amplitude and frequency are spectral fingerprints for the molecular structure and dynamics.62 Three biologically relevant NMR-active nuclear isotopes, proton (1H), carbon (13C), and sodium (23Na), have been studied in cartilage tissue engineering.63–65 These nuclei possess a nuclear magnetic moment with a nonzero spin quantum number. When placed in a strong static magnetic field (11.7 to 17.6 Tesla) and excited by an appropriate radiofrequency signal suitable for NMR-active nuclei (125 to 750 MHz), the nuclear spin states absorb and re-emit electromagnetic energy with a characteristic relaxation time scale depending upon the environment around the nuclei.62 Two time constants are commonly used in an NMR experiment for determining the molecular motion, the longitudinal relaxation time, T1, and the transverse relaxation time, T2.62 The complexity of NMR spectra results from the dependence of nuclear spin state energies on specific interactions (e.g., isotropic and anisotropic chemical shifts, through bond scalar interactions via J couplings and through space interactions (magnetic–magnetic dipole interaction), and electric quadrupolar interactions for the spin quantum number greater than one half).62 In a simple liquid, because of the rapid molecular tumbling, all anisotropic interactions average out leaving only the isotropic values of chemical shift and J-coupling. In solids, the strong anisotropic interactions, dipolar and quadrupolar coupling dominates the line shape of NMR spectra. As a consequence, the NMR lines for simple liquids are sharp and well resolved, while they are broad and featureless for solids. In most cases, biological tissues fall between these two extremes and multidimensional multinuclear spectroscopy must be used to resolve the nuclear sites and structural features.
The signal to noise ratio (SNR) for NMR spectra depends on the gyromagnetic ratio, γ, of the observed nuclei and the strength of nuclear interactions at the location of nuclei. Among the NMR-active nuclei, protons have the largest gyromagnetic ratio (42.58 MHz/Tesla), and thus, the highest SNR. The 13C nucleus, which has a lower gyromagnetic ratio (10.71 MHz/Tesla) or sensitivity than protons, has also been used in NMR for studying native and engineered cartilage. NMR spectroscopy with 13C has the advantage of giving a wider chemical shift range (∼220 ppm) as compared to protons (∼10 ppm), making the spectral assignment easier because of less overlap between different spectral groups. Special NMR spectroscopy techniques such as high-resolution magic angle spinning (HRMAS), cross-polarization magic angle spinning (CPMAS), and heteronuclear decoupling, which reduce line broadening significantly and enhance SNR, particularly for 13C NMR, have been used for NMR investigations of natural cartilage.27,28,66–69
Water is the most abundant molecule in biological tissues; therefore, the water signal dominates the proton NMR spectra of cartilage. The diffusion of water provides a window into tissue composition and dynamics. Using a long observation time in a diffusion NMR experiment, it has been shown that the typical mesh size for the collagen network in natural cartilage is on the order of 10 μm, which is rather large, and thus allows proteoglycan molecules to move freely and to exhibit a liquid-like motion with sharp resonances.27,28 Using 13C CPMAS, HRMAS, and pulsed field gradient diffusion NMR, Huster et al.27 concluded that the cartilage collagen undergoes a faster motion when compared with isolated collagen, whereas the motion of the chondroitin sulfate chain associated with proteoglycans is more restricted compared with the motion observed in a chondroitin sulfate solution. These investigators also found that the side chains of the triple helix structure of collagen are more flexible than the core chains.27 This unique information about molecular mobility, which in turn provides information about porosity noninvasively, is only available through NMR investigations.
To observe the proton signatures associated directly with the other ECM components, the water proton signal needs to be suppressed.70 The proton spectra associated with the macromolecules in the tissue, such as lipids, collagen, and proteoglycan aggregates, provide direct information about these macromolecules and they are important characteristics when investigating the development of engineered cartilage tissue. Using water suppressed proton NMR spectroscopy, Kotecha et al.71 have confirmed a previously published study72 showing that the chondrocyte pellet culture produces higher amounts of proteoglycans and collagen when compared with chondrocytes seeded in an alginate bead culture system. This article71 also reported that the residual dipolar coupling derived from proton double-quantum coherence spectroscopy was much smaller for the tissue grown as chondrocyte pellets and chondrocytes seeded in alginate beads when compared with the native tissue, which suggests a lack of preferred orientation in the collagen produced in engineered tissues.63,71
Scheidt et al.28 used 13C HRMAS NMR to study the site-specific motion of 13C nuclei in chondrocytes seeded in a collagen hydrogel for 3 weeks of culture time. They found that the T2 relaxation times of the carbon atoms in engineered tissue are shorter than those observed in the native tissue. Schulz et al.20,28 have used 13C magic angle spinning solid state NMR spectroscopy to quantify cell proliferation, metabolic activity, ECM composition, GAG mobility, and nanoarchitecture of engineered cartilage. Clearly, 13C NMR spectroscopy is a useful tool for probing cartilage tissue engineering, and it will play an important role in understanding the growth dynamics in vitro and in vivo. However, the 13C nuclei have a low natural abundance and a high value of dipolar coupling with protons. These factors increase the spectral line width and reduce SNR, which makes 13C MRS a challenge. Therefore, the emerging signal-enhancing techniques such as the dynamic nuclear polarization, low-power heteronuclear decoupling, the fast magic angle spinning, and paramagnetic signal enhancement will play an important role in extending the in vitro study of engineered cartilage using 13C NMR spectroscopy in the future.73–76 However, many of these new techniques cannot be performed in vivo, but they are still useful in adding to our understanding of the mechanism of tissue growth.
Sodium (23Na) is the second most abundant NMR-active nucleus with a gyromagnetic ratio of 11.26 MHz/Tesla. It has been widely used to study cartilage.77–79 Sodium binds to the negatively charged proteoglycans, and therefore, its concentration in cartilage is twice (∼240–300 mM) as high as that in the tissue fluid (∼150 mM).77 The GAG component of proteoglycans is directly correlated with the sodium signal intensity in an MRS/MRI experiment. In the early stages of osteoarthritis, the loss of proteoglycans reduces the local sodium ion concentration and this change can be measured quantitatively by using sodium NMR/MRI.77,80,81 The potential of sodium MRI to monitor the proteoglycan concentration in engineered cartilage was demonstrated by Novotny et al.41 They used a 4.7 T research scanner to generate sodium maps of tissue samples showing an average concentration range of 260–278 mM after 8 weeks of incubation. However, more studies of this type need to be completed.
Sodium is a spin 3/2 quadrupolar nucleus and when this quadrupolar interaction is not averaged out by rapid motion (for example, sodium ions in biological tissues), double- and triple-quantum coherence spectra with biexponential relaxation times are observed, which provides an insight into the dynamics and preferred orientation of collagen fibers (order) in the tissue.63,77,82 The sodium signal intensity and its relaxation properties have also been used to quantify proteoglycans in normal and diseased or damaged cartilage, where it correlates directly with the amount of proteoglycans.77,83–85 However, in engineered cartilage, sodium NMR has been primarily used for quantification of the order or preferred orientation of collagen fibers.15,63,86 For example, Keinan-Adamsky et al.86 have used multiquantum coherence spectroscopy in pig articular cartilage samples from birth to 39 months of age to obtain the average quadrupolar coupling, which provides information about collagen fiber orientation order. Using 1H T2 MRI and 23Na double- and triple-quantum filtered NMR spectroscopy, they concluded that the collagen fibril order and the density of the collagen fibers increased from birth to maturation in the pig articular cartilage. Kotecha et al.63 have used 23Na triple-quantum coherence spectroscopy and 1H double-quantum coherence spectroscopy to study tissue-engineered cartilage over a 4-week culture period in chondrocytes seeded in alginate beads, scaffold-free chondrocyte pellets, and in MSCs seeded in collagen/chitosan-based scaffolds. They concluded from this study that the tissue-engineered cartilage collagen lacks order or preferred orientation and consists of randomly oriented collagen fibers. They also found that the scaffold plays an important and non-negligible role in the NMR relaxation properties of sodium ions. The chondrocytes seeded in alginate bead culture had shorter relaxation times as compared to the chondrocyte pellets and MSCs seeded in collagen/chitosan scaffolds—presumably because of the calcium ion network in the alginate beads. In the future, additional studies of these parameters with different scaffolds and cell sources can be used to optimize tissue growth strategy.
Multidimensional solid-state NMR spectroscopy is a standard tool for the study of the structure and interactions at the molecular level for membrane proteins and amyloid fibrils,75,87 and it can also be used as a tool to probe the structure and dynamics of tissue formation. Such information would be useful for tissue engineers who do not have access to these tools and rely on empirical approaches to make progress in assessing tissue growth strategies.
Magnetic Resonance Imaging
MRI generates 3D images of soft tissues with a submillimeter resolution.88 Image contrast can be tuned to the water content, NMR relaxation times (T1 or T2), or the ADC.30 Native cartilage consists of 75% water (wet tissue weight) permeating a complex matrix of macromolecules (largely type II collagen and proteoglycans). The structural and morphological changes that arise in the development of engineered tissue become visible in T1- or T2-weighted images after as few as two weeks of incubation.30 The ADC also decreases by a factor of three or more as the motion of water becomes more hindered and restricted by the synthesis of macromolecules.89 Thus, we can expect MRI to be sensitive to changes in the tissue microenvironment during the different developmental stages observed in cartilage tissue engineering.
In developing tissue, the changing porosity and tortuosity partitions water into multiple compartments (e.g., water bound to proteins or trapped in cells and organelles), which restricts the exchange and mixing of the free and bound water fractions on the millisecond time scales used in MRI data acquisition. The dynamics of these processes are known to correlate with the ECM components in developing cartilage, and they can be measured in terms of the T1rho relaxation times,41 sodium MRI,41 and magnetization transfer (MT).33,42,90 Much of the current work using MRI to monitor tissue-engineered cartilage is focused on these more specific measures of tissue complexity.40
Yin et al.,89 for example, found that the relaxation times and diffusion coefficient (T1, T1rho, ADC, and T2) all decreased (T1 by 18%, T2 by 42%, and ADC by 26%) in a 4-week study of chondrocyte pellets (a scaffold-free tissue-engineering model). However, in a 4-week study of chondrocytes grown in an agarose gel, Miyata et al.,35 found that while the longitudinal relaxation times (T1) and apparent diffusion coefficient (ADC) decreased, the transverse relaxation time (T2) increased. These results demonstrate that MRI analysis is able to detect unexpected changes in the tissue microenvironment through measurements, in this case, of the T2 relaxation times.
In recent high SNR studies of native articular cartilage, the T1 relaxation time was found to be highly correlated with the proteoglycan content, the ADC and T1rho parameters were found to reflect the total amount of collagen and proteoglycans, and the T2 relaxation time was associated with both the amount of collagen and the orientation of collagen fibers.91,92 However, despite this interrelationship between MRI parameters and cartilage components, a study by Greco and Spencer93 of engineered cartilage grown in a hollow fiber bioreactor system showed excellent correlation over 4 weeks of incubation time between the T1, T2, ADC parameters and both proteoglycans and collagen content, while the MT ratio was found to be better correlated with the amount of collagen. It is likely that during engineered tissue growth, both the T1 and T2 relaxation times would decrease due to accumulation of both proteoglycans and collagen. It is possible to separate the contributions of water, proteoglycans and collagen content to the observed T2 data of engineered cartilage using multiexponential analysis,38,39,94 but such experiments require a very high SNR, hence, use of signals from the entire sample, not from individual image pixels. Nevertheless, using this technique, Reiter et al. have found that the proteoglycan component in cartilage tissue taken from young and mature bovine nasal cartilage was highly correlated with T2 and with the biochemical analysis.94 Using sodium and T1rho MRI, Novotny et al.41 have shown that culture of high-density porcine chondrocytes produced a similar amount of proteoglycans in 8 weeks as found in natural cartilage. In addition, Li et al.33 have shown a statistically significant correlation between MT parameters (bound water fraction and cross-relaxation rates) and the GAG content-engineered cartilage tissue for the period of 3 weeks. Finally, Irrechukwu et al.39 used both T2 and MT to monitor the stimulatory effects of pulsed low-intensity ultrasound on the development of ECM components (proteoglygans and collagen) after 3 and 5 weeks of treatment.
In addition to quantifying the amount of proteoglycans and collagen present in developing cartilage tissue, tracking cell differentiation and proliferation is also important for optimizing strategies for cartilage tissue engineering. As discussed in the Cartilage Tissue Engineering section, chondrocytes and MSCs are commonly used as a cell source for cartilage tissue engineering and regeneration. One way to observe these cells is to label them with superparamagnetic iron oxide (SPIO) particles. SPIO particles were used for tracking chondrocytes grown in agarose by Ramaswamy et al.95 for an incubation time of 6 weeks. In another study, the T2 values of SPIO-labeled cells were found to decrease with the increasing cell number, which can be used as a signal contrast standard for labeled cells.96 SPIO-labeled chondrocytes were also shown to maintain their phenotype and to continue the production of cartilage-specific ECM components in hollow-fiber bioreactor experiments or when grown in alginate beads.95–97 It was also confirmed that the chondrocyte differentiation of human bone marrow-derived MSCs seeded in porcine osteochondral plugs was not affected by SPIO labeling in vitro and this strategy can be used for MRI cell tracking in cartilage tissue engineering effectively.98 Chen et al.99 observed optimal low-dosage SPIO-labeled cells in a minipig model and found that the labeled cells could be tracked for at least 12 weeks using T2-weighted MR imaging. Recently, there have also been attempts to visualize cell clusters without SPIO labeling with a resolution of 40 μm3 by optimizing the MRI data acquisition conditions, which opens up new avenues for tissue-engineered cartilage tracking as well.100 It is anticipated that by optimized MRI for cell tracking, future MR imaging of cell numbers and distribution in engineered cartilage tissue may be possible.
Magnetic Resonance Elastography
MRE uses MRI to visualize shear wave motion in tissue perturbed by low-frequency (50–5000 Hz) mechanical vibrations.101 From the recorded motion sensitive phase data, images of the tissue mechanical properties (the shear modulus and viscosity) can be reconstructed.102 Changes in the mechanical properties of articular cartilage provide a direct indication of its integrity.103 In general, the mechanical properties of cartilage can be classified by its tensile, compressive, and shear properties.21 While the compressive properties are commonly assessed using ex vivo samples by the mechanical application of uniaxial tensile or compression forces,28,48 the shear properties, which are derived from volume-conserving deformations, can be observed ex vivo using shear loading or by using ultrasonic or MR elastography.104 The tensile properties of SZs are found to be important in cartilage health21; however, MR elastography methods are not suited for such studies.
The relationship between transverse and axial strain is given by the Poisson ratio, which links the shear and compression properties of materials (elastic, bulk, and shear moduli). The tissue property commonly assessed in MRE is represented by the complex (shear) modulus G, which is defined at a specific frequency (ω), as the ratio of shear stress and shear strain. The real part of G, the storage modulus G′, is a measure of the ability of a material to store energy. The imaginary part of G′′, the loss modulus characterizes the dissipation of mechanical energy. The complex modulus therefore is a frequency-dependent quantity, G(ω). For biological tissues, there is a consensus that power law models are the most suitable for characterizing the observed frequency behavior (dispersion) of G(ω).105,106 In these models, the frequency variation of G(ω) involves terms raised to fractional powers (typically described by α, where 0<α<1).107 A rheological model exhibiting an α power law dispersion is the so-called springpot (or single fractional element).108 The springpot's stress/strain behavior is intermediate between the spring (α=0) and the dashpot (α=1), and can be visualized as a fractal arrangement of a large number of individual elastic springs and viscous dashpots. Schiessel and Blumen109 have shown that the power α depends on the geometrical arrangement and the relative values of the basic elements. In dynamic elastography, the wave velocity and the damping coefficient are directly related to the complex modulus G.110 The stiffness, a property often specified in elastographic studies, is defined as the squared wave velocity multiplied with the volume density of a material. In the elastic case, the loss modulus is assumed to be zero, and as a consequence, the stiffness is frequency independent and identical to the storage modulus G′. Hence, in this simple case, G′ can be determined if the wave velocity (frequency times wavelength) and the density are known, or measured using MRI.
The stiffness of articular cartilage reflects the mesh of its collagen-rich fibril network and the distribution of proteoglycans in this mesh.48,111 From the Poisson ratio and the bulk modulus of natural cartilage—derived in ex vivo non-MRE mechanical compression tests48—we can assess the static shear stiffness, which is nonuniform, and increases moving from the superficial into the deeper regions, (changes on the order of ∼0.5–4.0 MPa). Engineered protocartilage tissue is much less stiff, with stiffness values that range from MRE derived ∼3 kPa for chondrocytes encased in alginate beads112 up to nanoindentation derived 0.5 MPa for engineered tissue augmented with the polyglycolic acid yarn to mimic collagen fibers in zonal artificial cartilage.22 The stiffness also changes over the entire tissue growth phase as cells proliferate, differentiate, generate ECM components, and as the scaffold degrades. Since cartilage is always undergoing mechanical loads, information about its stiffness is critical for selecting the best scaffold for each application, as well as for monitoring the development of the engineered tissue, both in vitro and in vivo. Current mechanical measurement techniques, such as uniaxial compression tests, while useful for showing stiffness changes over time,28 are destructive for tissue-engineered cartilage and cannot provide a complete volumetric map of the tissue's mechanical properties.
MRE can provide quantitative maps of tissue's mechanical properties. In an MRE experiment, external vibrations in the audio range are introduced into the target tissue and snapshots of the resulting mechanical wave are encoded in the phase of the MR signal using motion-sensitized MR pulse sequences.101 The mechanical properties of the tissue are determined by applying inversion techniques to recover tissue stiffness from the wave images. Treating tissue as a viscoelastic material, direct inversion of the wave equation102 can be performed to reconstruct images of the complex modulus G(ω). While providing both, the storage and the loss modulus, this approach is very sensitive to noise and requires the assumption of an infinite medium. A more stable inversion can be achieved using the local frequency estimation (LFE).113 From the LFE-derived wavelengths, an image of the stiffness can be calculated, but at the cost of not being able to determine the loss modulus. In specimens with finite geometries (e.g., thin plates and articular cartilage) and with minimal damping, wave reflections occur, which make it necessary to apply finite element-based inversion algorithms114 for the determination of G(ω). Fortunately, independent of the inversion technique used, the frequency-independent tissue parameters can be found by fitting the dispersion relations of selected rheological models to the experimental G(ω).
While not yet widely used in the study of cartilage, MRE was shown to be sensitive to the grade of liver fibrosis115,116 and it has revealed abnormal mechanical properties in the brains of patients suffering from neurodegenerative diseases.117–119 In these soft tissues, the macroscopic mechanical behavior (stiffness) has highly correlated changes in the tissue composition and structure identified by histology on a microscopic scale. Therefore, it is reasonable to expect that changes in the structure and composition of engineered cartilage during growth will alter the MRE-derived stiffness. A few preliminary MRE studies on engineered tissue constructs have already been conducted.29,43 This work applied line-profile MRE to osteogenic and adipogenic tissue constructs embedded in gelatin sponges for 4 weeks and found that, while the stiffness increased in osteogenic tissues it decreased in adipose tissues,43 reflecting the stiffening of the developing bone and the softening of the prototype adipose tissue.
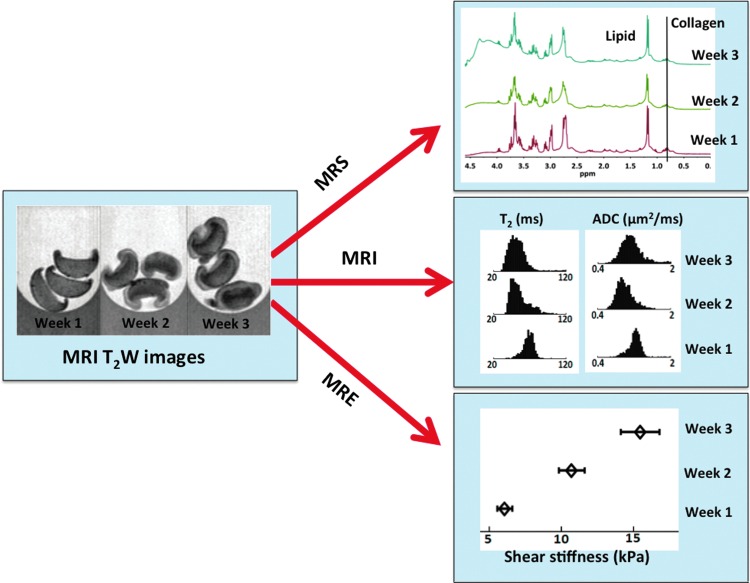
MRE of articular cartilage is challenging because of large (MPa) values of the tissue stiffness and the small dimensions (∼2-mm thickness). This requires the application of mechanical excitation frequencies in the kHz range. The main issue with high-frequency excitation is the strong damping of shear waves. Using a 5-kHz excitation frequency, Lopez et al.120 were able to measure the mean shear stiffness of 8-mm-thick natural bovine hyaline cartilage plugs and found it to be on the order of 2 MPa. In this study, they also reported a decrease in tissue stiffness following selective enzymatic degradation, which released the proteoglycans from the cartilage. To compensate for the strong damping at kHz frequencies, Yasar et al.121 have developed a novel geometric focusing approach, in which, the energy absorption is compensated for by the concentration of the wave energy on gradually decreasing wave fronts. This technique has been successfully applied to determine the mechanical properties of alginate beads suspended in the agarose gel.112 In a different MRE study, Yin Z. et al.122 measured the tissue stiffness of chondrocyte pellets (scaffold-free model) over 4 weeks of culture time and found that as proteoglycans and collagen content increases, the tissue stiffness also increases by threefold (see Fig. 4). To apply such MRE techniques in vivo, and in stiffer materials, such as fully developed cartilage, advances are needed in MRI hardware as well as for MRE, more efficient mechanical actuators (particularly at high excitation frequencies), faster MRE pulse sequences, and gradient coils with faster rise times.
FIG. 4.
An example of MR assessment of engineered cartilage tissue development using MR spectroscopy, imaging, and elastography measurements. Here we followed the growth of scaffold-free chondrocyte pellets over 3 weeks of culture time. Further details are listed in the text in appropriate sections. Color images available online at www.liebertpub.com/teb
The MRE studies, described above, focus on shear mechanical properties of tissue. A very recent work reports that the compressive properties also correlate with physiological changes in the human brain.123 Hirsch et al. claim that the MRE-derived volumetric strain, expressed as the divergence of the 3D displacement field, is related to the dilation and compression of fluid pores. As such, the volumetric strain might also be suitable for monitoring the growth of the porous scaffold of engineered cartilage.
In Vivo MR Techniques for Monitoring of Cartilage Tissue Engineering and Repair
The MRS, MRI, and MRE techniques for assessing cartilage tissue engineering described in this review have emphasized their use for in vitro and ex vivo studies; however, the techniques in most cases can be adopted for in vivo cartilage regeneration studies with some modifications to improve the RF coil filling factor and SNR.124 MRI is currently used to study the cartilage regeneration and restoration in animal models and in human patients, and there have been attempts to incorporate MRS information as well.125–127
In vivo studies using bioreactors (incubators) and animal models are important for validating new approaches of tissue engineering. MR compatible bioreactors allow the investigator to continuously monitor tissue regeneration over a period of days to weeks, which is important for optimizing growth conditions and viewing developing tissue structures. Recent 7-week studies of chondrocytes growing in an MR-compatible hollow cylinder bioreactor tracked the development of new cartilage-like tissue using T2 relaxation times and SPIO nanoparticles.93,95 Alternatively, in vivo imaging of small animal models (e.g., SCID mice, rats, and rabbits) can be performed weekly to test the efficacy of tissue-engineering strategies, often using narrow bore (20–30 cm) high field (4.7–11 Tesla) animal MRI systems. Large animals (e.g., dogs, goats, pigs, and horses) have also been used for cartilage repair studies,127 but they present challenges as clinical MR scanners must be used in most cases. The advantage of using large animal models is the relatively thick cartilage of these animals and the clear ability to translate findings into clinical protocols. Small animal cartilage MRI in rats and mice is feasible,128 however, because of the small size of cartilage, its use is often limited to the high field systems. Small animals can, of course, be used as a bioreactor in MRI studies designed to observe tissue growth and if successful, the implanted cartilage tissue and its interaction with the host tissue can be periodically monitored.
The application of multiparametric MRI (3D relaxation time and diffusion coefficient maps) for assessing cartilage regeneration after treatment or surgery in large animal models is under active investigation.129–132 For example, in studies of the repair of osteochondral defects in rabbits, Kim et al. did not find any difference after 6 weeks between the T2 values of the control and the repair,133 while Ramaswamy et al. did report a difference after 5 weeks.134 For an excellent review of cartilage repair in large animal models with emphasis on assessing the cartilage microstructure during repair using as relaxation times (T1 and T2), see the recent article by Trattnig et al.135 MRI is also used clinically to monitor cartilage repair and regeneration procedures not involving tissue-engineered constructs, but these studies are beyond the scope of this article.
Outlook and Future Prospects
In Figure 4, we present an example of how MR spectroscopy, imaging, and elastography techniques have been used to monitor tissue growth in chondrocyte pellets (a scaffold-free tissue-engineering model) over a period of 3 weeks. This study has the advantages of the combined modality approach in which, we visualize tissue growth and collect quantitative data on each pixel (or regions of interest) without the need for contrast agents, tissue sampling, or sample fixation. In addition, these techniques can provide information about collagen fibril orientation, tissue stiffness and porosity not readily available using other methods. We expect that these techniques will play a major role in monitoring cartilage tissue engineering in the future.
In Table 1, we provide a summary of selected MR parameters that are correlated with changes in the tissue composition and structure. This table can serve as a guide to tissue engineers when selecting techniques for characterizing engineered cartilage and it provides an estimate of the range of information MRS, MRI, and MRE can provide.
Table 1:
Summary of Current MR Techniques Used to Characterize Tissue Engineered Cartilage and Their Correlation with Different Components, Such as Cell Density, Extra Cellular Matrix Components, Proteoglycans and Collagen of Engineered Tissue
| Current Methods | Parameter | Macromolecule probed | Nature of correlation |
|---|---|---|---|
| Magnetic Resonance Spectroscopy (MRS) (1H, 13C, 23Na) | Chemical shift | Proteoglycans, Collagen, Cell density and phenotype | Quantitative |
| Line integral and broadening | Proteoglycans, Collagen | Quantitative | |
| Site specific relaxation rates | Proteoglycans (HRMAS), Collagen (CPMAS) | Site specific motion | |
| Multiple-quantum coherence spectroscopy | Proteoglycans, Collagen order | Molecular motion and tissue anisotropy | |
| Magnetic Resonance Imaging (MRI) | Image contrast | Proteoglycans, Collagen, Cell density for SPIO labeled cells | Non-specific, qualitative |
| T1 relaxation time | Proteoglycans, Tissue Water | Primarily proteoglycans, but all ECM component contribute | |
| T2 relaxation time | Proteoglycans, Collagen, Tissue water | Depends on the scaffold/cells, Primarily collagen but all ECM component contribute | |
| Apparent Diffusion Coefficient (ADC) | Proteoglycans, Collagen, Tissue Water | For short observation time - water content and for long observation time - ECM component contribute | |
| Magnetization Transfer Ratio (MT) | Proteoglycans, Collagen, Tissue Water | Primarily proteoglycans, both ECM component can contribute depending upon experimental conditions | |
| T1rho relaxation time | Proteoglycans, Collagen | Primarily proteoglycans, both ECM component can contribute depending upon experimental conditions | |
| Magnetic Resonance Elastography (MRE) | Tissue stiffness | Proteoglycans, Collagen, Tissue water | All ECM components contribute |
| Shear Elasticity (Storage Modulus) | Proteoglycans, Collagen, Tissue water | Primarily Collagen | |
| Shear Viscosity (Loss Modulus) | Proteoglycans, Collagen, Tissue Water | Primarily Tissue Water content |
ECM, extracellular matrix.
Assessment of tissue engineering using MR-based technology is promising; however, its use by tissue engineers is not yet wide spread. The factors inhibiting more wide spread use of MR techniques for monitoring and assessing cartilage tissue engineering laboratories are varied: the availability of an MR scanner or spectrometer near to the tissue engineering laboratory, the need to establish a team with MR specialists, the expense, and the use of often long time data acquisition methods. The MR community is actively working to address many of these concerns and to bring the technology closer to tissue-engineering groups. A bench-top MRI scanner developed by the Mäder group for characterization of scaffolds is one such example.31 In the clinical setting, gadolinium-enhanced MRI could prove to be useful; therefore, the correlation of proteoglycan with a fixed charged density derived from gadolinium-enhanced MRI for cartilage tissue-engineering constructs is encouraging.36 In addition, as sodium MRI becomes more feasible (with the advent of more high field clinical MR scanners), it may be possible to derive fixed charge density information without the use of a contrast agent.136 MRS has many untapped advantages and provides quantitative information about engineered tissues, however, it is time-consuming and the SNR for useful MR techniques such as water suppressed MRS, 13C and 23Na MRS and multiple-quantum coherence MR is low. As discussed in MRS sections, the development of new signal-enhancing techniques will help resolve some of these issues in the future. MRS can also be used to understand the dynamics and structure of engineered tissues and to monitor tissue formation. MRI is currently used for monitoring the cartilage repair and regeneration and the growth of engineered cartilage. MRI provides parametric images, which can be directly correlated with the tissue composition and microstructure. The main shortcoming of MRI at the moment seems to be the lack of standards for assessing relaxation parameters and efforts are underway to resolve this issue.137,138 MR elastography, while relatively new in the field of tissue engineering is gaining popularity because it is able to directly measure the mechanical properties of the tissue. New techniques are under development to increase the dynamic range and to improve the resolution, as well as to extend the method to include compression as well as shear moduli. We expect that in the near future, all the MR-based techniques discussed above will augment the current characterization methods so as to provide quantitative information of cartilage tissue growth at all stages, from cell seeding to postimplantation in animals and in the clinic.
Acknowledgments
This study was supported by NIH/NIBIB R01 grant EB007537. The authors wish to thank Dr. Anne George (University of Illinois at Chicago, Oral Biology), Dr. Thomas M. Schmid (Rush University, Biochemistry), Dr. Thomas J. Royston (University of Illinois at Chicago, Bioengineering), Dr. Sriram Ravindran (University of Illinois at Chicago, Oral Biology), Dr. Vincent Wang (Rush University, Orthopedic Surgery), and Dr. Brian Roman (University of Chicago, Radiology) for their useful discussions and insights in cartilage tissue engineering, repair and regeneration and its analytical characterization.
Disclosure Statement
No competing financial interests exist.
References
- 1.Prevalence, most common causes of disability among adults—united states, 2005. MMWR Morb Mortal Wkly Rep. 2009;58:421. [PubMed] [Google Scholar]
- 2.Bernstein J. King T. Lawry G.V. Musculoskeletal medicine educational reform in the bone and joint decade. Arthrit Rheum-Arthr. 2007;57:1109. doi: 10.1002/art.23001. [DOI] [PubMed] [Google Scholar]
- 3.Centers for disease control, prevention, arthritis foundation: A national public health agenda for osteoarthritis. 2010. www.cdc.gov/arthritis/docs/OAagenda.pdf www.cdc.gov/arthritis/docs/OAagenda.pdf
- 4.Lawrence R.C. Felson D.T. Helmick C.G. Arnold L.M. Choi H. Deyo R.A. Gabriel S. Hirsch R. Hochberg M.C. Hunder G.G. Jordan J.M. Katz J.N. Kremers H.M. Wolfe F. Workgrp N.A.D. Estimates of the prevalence of arthritis and other rheumatic conditions in the united states. Arthritis Rheum. 2008;58:26. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy L. Helmick C.G. The impact of osteoarthritis in the united states: a population-health perspective a population-based review of the fourth most common cause of hospitalization in u.S. Adults (reprinted from Am J Nur 112, s13, 2012) Orthop Nurs. 2012;31:85. doi: 10.1097/NOR.0b013e31824fcd42. [DOI] [PubMed] [Google Scholar]
- 6.Tchetina E.V. Developmental mechanisms in articular cartilage degradation in osteoarthritis. Arthritis. 2011;2011:683970. doi: 10.1155/2011/683970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckwalter J.A. Mankin H.J. Articular cartilage: degeneration and osteoarthritis, repair, regeneration, and transplantation. Instr Course Lect. 1998;47:487. [PubMed] [Google Scholar]
- 8.Buckwalter J.A. Mankin H.J. Articular cartilage: Tissue design and chondrocyte-matrix interactions. Instr Course Lect. 1998;47:477. [PubMed] [Google Scholar]
- 9.Hunziker E.B. Articular cartilage repair: basic science and clinical progress. A review of the current status and prospects. Osteoarthritis Cartilage. 2002;10:432. doi: 10.1053/joca.2002.0801. [DOI] [PubMed] [Google Scholar]
- 10.Yoon H.H. Bhang S.H. Shin J.Y. Shin J. Kim B.S. Enhanced cartilage formation via three-dimensional cell engineering of human adipose-derived stem cells. Tissue Eng Part A. 2012;18:1949. doi: 10.1089/ten.tea.2011.0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheuk Y.-C. Wong M.W.-N. Lee K.-M. Fu S.-C. Use of allogeneic scaffold-free chondrocyte pellet in repair of osteochondral defect in a rabbit model. J Orthopaed Res. 2011;29:1343. doi: 10.1002/jor.21339. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z. Mccaffery J.M. Spencer R.G. Francomano C.A. Hyaline cartilage engineered by chondrocytes in pellet culture: Histological, immunohistochemical and ultrastructural analysis in comparison with cartilage explants. J Anat. 2004;205:229. doi: 10.1111/j.0021-8782.2004.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein T.J. Malda J. Sah R.L. Hutmacher D.W. Tissue engineering of articular cartilage with biomimetic zones. Tissue Eng Part B Rev. 2009;15:143. doi: 10.1089/ten.teb.2008.0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meretoja V.V. Dahlin R.L. Kasper F.K. Mikos A.G. Enhanced chondrogenesis in co-cultures with articular chondrocytes and mesenchymal stem cells. Biomaterials. 2012;33:6362. doi: 10.1016/j.biomaterials.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotecha M. Ravindran S. Vaidyanathan A. George A. Magin R.L. Chaarcterization of ECM embedded biomimitic scaffolds for cartilage tissue engineering using sodium triple qunatum coherence spectroscopy. Proceedings of the International Society for Magnetic Resonance in Medicine (ISMRM) 20th Annual Meeting and Exhibition; Melbourne, Australia. May 5–11;2012 ; Abstract no. 1703. [Google Scholar]
- 16.Petit B. Masuda K. Dsouza A.L. Otten L. Pietryla D. Hartmann D.J. Morris N.P. Uebelhart D. Schmid T.M. Thonar E.J.M.A. Characterization of crosslinked collagens synthesized by mature articular chondrocytes cultured in alginate beads: comparison of two distinct matrix compartments. Exp Cell Res. 1996;225:151. doi: 10.1006/excr.1996.0166. [DOI] [PubMed] [Google Scholar]
- 17.Chandrasekhar S. Esterman M.A. Hoffman H.A. Microdetermination of proteoglycans and glycosaminoglycans in the presence of guanidine-hydrochloride. Anal Biochem. 1987;161:103. doi: 10.1016/0003-2697(87)90658-0. [DOI] [PubMed] [Google Scholar]
- 18.Chubinskaya S. Huch K. Schulze M. Otten L. Aydelotte M.B. Cole A.A. Gene expression by human articular chondrocytes cultured in alginate beads. J Histochem Cytochem. 2001;49:1211. doi: 10.1177/002215540104901003. [DOI] [PubMed] [Google Scholar]
- 19.Hoemann C.D. Molecular and biochemical assays of cartilage components. Methods Mol Med. 2004;101:127. doi: 10.1385/1-59259-821-8:127. [DOI] [PubMed] [Google Scholar]
- 20.Schulz R. Hohle S. Zernia G. Zscharnack M. Schiller J. Bader A. Arnold K. Huster D. Analysis of extracellular matrix production in artificial cartilage constructs by histology, immunocytochemistry, mass spectrometry, and NMR spectroscopy. J Nanosci Nanotechnol. 2006;6:2368. doi: 10.1166/jnn.2006.506. [DOI] [PubMed] [Google Scholar]
- 21.Little C.J. Bawolin N.K. Chen X. Mechanical properties of natural cartilage and tissue-engineered constructs. Tissue Eng Part B Rev. 2011;17:213. doi: 10.1089/ten.TEB.2010.0572. [DOI] [PubMed] [Google Scholar]
- 22.Moutos F.T. Freed L.E. Guilak F. A biomimetic three-dimensional woven composite scaffold for functional tissue engineering of cartilage. Nat Mater. 2007;6:162. doi: 10.1038/nmat1822. [DOI] [PubMed] [Google Scholar]
- 23.Bryant S.J. Davis-Arehart K.A. Luo N. Shoemaker R.K. Arthur J.A. Anseth K.S. Synthesis and characterization of photopolymerized multifunctional hydrogels: water-soluble poly(vinyl alcohol) and chondroitin sulfate macromers for chondrocyte encapsulation. Macromolecules. 2004;37:6726. [Google Scholar]
- 24.Purea A. Webb A.G. Reversible and irreversible effects of chemical fixation on the NMR properties of single cells. Magn Reson Med. 2006;56:927. doi: 10.1002/mrm.21018. [DOI] [PubMed] [Google Scholar]
- 25.Fishbein K.W. Gluzband Y.A. Kaku M. Ambia-Sobhan H. Shapses S.A. Yamauchi M. Spencer R.G. Effects of formalin fixation and collagen cross-linking on t2 and magnetization transfer in bovine nasal cartilage. Magn Reson Med. 2007;57:1000. doi: 10.1002/mrm.21216. [DOI] [PubMed] [Google Scholar]
- 26.Puchtler H. Meloan S.N. On the chemistry of formaldehyde fixation and its effects on immunohistochemical reactions. Histochemistry. 1985;82:201. doi: 10.1007/BF00501395. [DOI] [PubMed] [Google Scholar]
- 27.Huster D. Schillar J. Naji L. Kaufmann J. Arnold K. NMR studies of cartilage—dynamics, diffusion, degradation. Lect Notes Phys. 2004;634:465. [Google Scholar]
- 28.Scheidt H.A. Schibur S. Magalhaes A. De Azevedo E.R. Bonagamba T.J. Pascui O. Schulz R. Reichert D. Huster D. The mobility of chondroitin sulfate in articular and artificial cartilage characterized by c-13 magic-angle spinning NMR spectroscopy. Biopolymers. 2010;93:520. doi: 10.1002/bip.21386. [DOI] [PubMed] [Google Scholar]
- 29.Xu H.H. Othman S.F. Hong L. Peptan I.A. Magin R.L. Magnetic resonance microscopy for monitoring osteogenesis in tissue-engineered construct in vitro. Phys Med Biol. 2006;51:719. doi: 10.1088/0031-9155/51/3/016. [DOI] [PubMed] [Google Scholar]
- 30.Xu H.H. Othman S.F. Magin R.L. Monitoring tissue engineering using magnetic resonance imaging. J Biosci Bioeng. 2008;106:515. doi: 10.1263/jbb.106.515. [DOI] [PubMed] [Google Scholar]
- 31.Nitzsche H. Metz H. Lochmann A. Bernstein A. Hause G. Groth T. Mader K. Characterization of scaffolds for tissue engineering by benchtop-magnetic resonance imaging. Tissue Eng Part C Methods. 2009;15:513. doi: 10.1089/ten.TEC.2008.0488. [DOI] [PubMed] [Google Scholar]
- 32.Nitzsche H. Noack A. Lochmann A. Oliveira C. Besheer A. Metz H. Bernstein A. Hause G. Groth T. Mader K. Scaffold properties and interaction with cells investigated by magnetic resonance methods. Tissue Eng Part A. 2008;14:907. [Google Scholar]
- 33.Li W. Hong L. Hu L. Magin R.L. Magnetization transfer imaging provides a quantitative measure of chondrogenic differentiation and tissue development. Tissue Eng Part C Methods. 2010;16:1407. doi: 10.1089/ten.tec.2009.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Constantinidis I. Simpson N.E. Grant S.C. Blackband S.J. Long R.C. Sambanis A. Non-invasive monitoring of tissue-engineered pancreatic constructs by NMR techniques. Adv Exp Med Biol. 2006;585:261. doi: 10.1007/978-0-387-34133-0_18. [DOI] [PubMed] [Google Scholar]
- 35.Miyata S. Numano T. Homma K. Tateishi T. Ushida T. Feasibility of noninvasive evaluation of biophysical properties of tissue-engineered cartilage by using quantitative MRI. J Biomech. 2007;40:2990. doi: 10.1016/j.jbiomech.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Miyata S. Homma K. Numano T. Tateishi T. Ushida T. Evaluation of negative fixed-charge density in tissue-engineered cartilage by quantitative MRI and relationship with biomechanical properties. J Biomech Eng. 2010;132:071014. doi: 10.1115/1.4001369. [DOI] [PubMed] [Google Scholar]
- 37.Potter K. Butler J.J. Adams C. Fishbein K.W. Mcfarland E.W. Horton W.E. Spencer R.G. Cartilage formation in a hollow fiber bioreactor studied by proton magnetic resonance microscopy. Matrix Biol. 1998;17:513. doi: 10.1016/s0945-053x(98)90099-3. [DOI] [PubMed] [Google Scholar]
- 38.Reiter D.A. Irrechukwu O. Lin P.-C. Moghadam S. Thaer S.V. Pleshko N. Spencer R.G. Improved MR-based characterization of engineered cartilage using multiexponential t2 relaxation and multivariate analysis. NMR Biomed. 2012;25:476. doi: 10.1002/nbm.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irrechukwu O.N. Reiter D.A. Lin P.C. Roque R.A. Fishbein K.W. Spencer R.G. Characterization of engineered cartilage constructs using multiexponential t-2 relaxation analysis and support vector regression. Tissue Eng Part C Methods. 2012;18:433. doi: 10.1089/ten.tec.2011.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irrechukwu O.N. Lin P.C. Fritton K. Doty S. Pleshko N. Spencer R.G. Magnetic resonance studies of macromolecular content in engineered cartilage treated with pulsed low-intensity ultrasound. Tissue Eng Part A. 2011;17:407. doi: 10.1089/ten.tea.2010.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novotny J.E. Turka C.M. Jeong C. Wheaton A.J. Li C. Presedo A. Richardson D.W. Reddy R. Dodge G.R. Biomechanical and magnetic resonance characteristics of a cartilage-like equivalent generated in a suspension culture. Tissue Eng. 2006;12:2755. doi: 10.1089/ten.2006.12.2755. [DOI] [PubMed] [Google Scholar]
- 42.Nugent A.E. Reiter D.A. Fishbein K.W. Mcburney D.L. Murray T. Bartusik D. Ramaswamy S. Spencer R.G. Horton W.E., Jr. Characterization of ex vivo-generated bovine and human cartilage by immunohistochemical, biochemical, and magnetic resonance imaging analyses. Tissue Eng Part A. 2010;16:2183. doi: 10.1089/ten.tea.2009.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Othman S.F. Curtis E.T. Plautz S.A. Pannier A.K. Butler S.D. Xu H.H. MR elastography monitoring of tissue-engineered constructs. NMR Biomed. 2012;25:452. doi: 10.1002/nbm.1663. [DOI] [PubMed] [Google Scholar]
- 44.Kotecha M. Yin Z. Yasar T.K. Klatt D. Royston T.J. Magin R.L. Functional Imaging for Regenerative Medicine Workshop; Gaithersburg, MD: May 31, 2012. High field magnetic resonance spectroscopy, imaging and elastography for cartilage tissue engineering. June 1. [Google Scholar]
- 45.Kuettner K.E. Aydelotte M.B. Thonar E.J. Articular cartilage matrix and structure: a minireview. J Rheumatol Suppl. 1991;27:46. [PubMed] [Google Scholar]
- 46.Jay G.D. Torres J.R. Warman M.L. Laderer M.C. Breuer K.S. The role of lubricin in the mechanical behavior of synovial fluid. Proc Natl Acad Sci. 2007;104:6194. doi: 10.1073/pnas.0608558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shepherd D.E. Seedhom B.B. Thickness of human articular cartilage in joints of the lower limb. Ann Rheum Dis. 1999;58:27. doi: 10.1136/ard.58.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S.S. Falcovitz Y.H. Schneiderman R. Maroudas A. Sah R.L. Depth-dependent compressive properties of normal aged human femoral head articular cartilage: relationship to fixed charge density. Osteoarthritis Cartilage. 2001;9:561. doi: 10.1053/joca.2001.0424. [DOI] [PubMed] [Google Scholar]
- 49.Klein T.J. Rizzi S.C. Reichert J.C. Georgi N. Malda J. Schuurman W. Crawford R.W. Hutmacher D.W. Strategies for zonal cartilage repair using hydrogels. Macromol Biosci. 2009;9:1049. doi: 10.1002/mabi.200900176. [DOI] [PubMed] [Google Scholar]
- 50.Schrobback K. Malda J. Crawford R.W. Upton Z. Leavesley D.I. Klein T.J. Effects of oxygen on zonal marker expression in human articular chondrocytes. Tissue Eng Part A. 2012;18:920. doi: 10.1089/ten.TEA.2011.0088. [DOI] [PubMed] [Google Scholar]
- 51.Ceuninck F. Lesur C. Pastoureau P. Caliez A. Sabatini M. Culture of chondrocytes in alginate beads. Methods Mol Med. 2004;100:15. doi: 10.1385/1-59259-810-2:015. [DOI] [PubMed] [Google Scholar]
- 52.Spiller K.L. Maher S.A. Lowman A.M. Hydrogels for the repair of articular cartilage defects. Tissue Eng Part B Rev. 2011;17:281. doi: 10.1089/ten.teb.2011.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kock L. Van Donkelaar C.C. Ito K. Tissue engineering of functional articular cartilage: the current status. Cell Tissue Res. 2012;347:613. doi: 10.1007/s00441-011-1243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddi A.H. Becerra J. Andrades J.A. Nanomaterials and hydrogel scaffolds for articular cartilage regeneration. Tissue Eng Part B Rev. 2011;17:301. doi: 10.1089/ten.TEB.2011.0141. [DOI] [PubMed] [Google Scholar]
- 55.Becerra J. Andrades J.A. Guerado E. Zamora-Navas P. Lopez-Puertas J.M. Reddi A.H. Articular cartilage: Structure and regeneration. Tissue Eng Part B Rev. 2010;16:617. doi: 10.1089/ten.TEB.2010.0191. [DOI] [PubMed] [Google Scholar]
- 56.Schrobback K. Klein T.J. Schuetz M. Upton Z. Leavesley D.I. Malda J. Adult human articular chondrocytes in a microcarrier-based culture system: expansion and redifferentiation. J Orthop Res. 2011;29:539. doi: 10.1002/jor.21264. [DOI] [PubMed] [Google Scholar]
- 57.Yoon D.M. Hawkins E.C. Francke-Carroll S. Fisher J.P. Effect of construct properties on encapsulated chondrocyte expression of insulin-like growth factor-1. Biomaterials. 2007;28:299. doi: 10.1016/j.biomaterials.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 58.Yoon D.M. Fisher J.P. Chondrocyte signaling and artificial matrices for articular cartilage engineering. Adv Exp Med Biol. 2006;585:67. doi: 10.1007/978-0-387-34133-0_5. [DOI] [PubMed] [Google Scholar]
- 59.Spiller K.L. Liu Y. Holloway J.L. Maher S.A. Cao Y.L. Liu W. Zhou G.D. Lowman A.M. A novel method for the direct fabrication of growth factor-loaded microspheres within porous nondegradable hydrogels: controlled release for cartilage tissue engineering. J Control Release. 2012;157:39. doi: 10.1016/j.jconrel.2011.09.057. [DOI] [PubMed] [Google Scholar]
- 60.Harley B.A. Lynn A.K. Wissner-Gross Z. Bonfield W. Yannas I.V. Gibson L.J. Design of a multiphase osteochondral scaffold iii: Fabrication of layered scaffolds with continuous interfaces. J Biomed Mater Res Part A. 2010;92:1078. doi: 10.1002/jbm.a.32387. [DOI] [PubMed] [Google Scholar]
- 61.Koo S. Hargreaves B.A. Gold G.E. Dragoo J.L. Fabrication of custom-shaped grafts for cartilage regeneration. Int J Artif Organs. 2010;33:731. [PMC free article] [PubMed] [Google Scholar]
- 62.Abragam A. Principles of Nuclear Magnetism. NY: Oxford University Press; 2002. [Google Scholar]
- 63.Kotecha M. Ravindran S. Schmid T.M. Vaidyanathan A. George A. Magin R.L. Application of sodium triple-quantum coherence NMR spectroscopy for the study of growth dynamics in cartilage tissue engineering. NMR Biomed. 2013;26:709. doi: 10.1002/nbm.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huster D. Zernia G. Hohle S. Schulz R. Zscharnack M. Schiller J. Bader A. Arnold K. Investigation of the dynamics of the macromolecules in articular and tissue engineered cartilage by NMR spectroscopy. Biophys J. 2005;88:519a. [Google Scholar]
- 65.Huster D. Naji L. Schiller J. Arnold K. Dynamics of the biopolymers in articular cartilage studied by magic angle spinning NMR. Appl Magn Reson. 2004;27:471. [Google Scholar]
- 66.Schiller J. Huster D. Fuchs B. Naji L. Kaufmann J. Arnold K. Evaluation of cartilage composition and degradation by high-resolution magic-angle spinning nuclear magnetic resonance. Methods Mol Med. 2004;101:267. doi: 10.1385/1-59259-821-8:267. [DOI] [PubMed] [Google Scholar]
- 67.Schiller J. Naji L. Huster D. Kaufmann J. Arnold K. 1h and 13c HR-MAS NMR investigations on native and enzymatically digested bovine nasal cartilage. MAGMA. 2001;13:19. doi: 10.1007/BF02668647. [DOI] [PubMed] [Google Scholar]
- 68.Naji L. Kaufmann J. Huster D. Schiller J. Arnold K. 13C NMR relaxation studies on cartilage and cartilage components. Carbohydr Res. 2000;327:439. doi: 10.1016/s0008-6215(00)00064-1. [DOI] [PubMed] [Google Scholar]
- 69.Ling W. Regatte R.R. Schweitzer M.E. Jerschow A. Characterization of bovine patellar cartilage by NMR. NMR Biomed. 2008;21:289. doi: 10.1002/nbm.1193. [DOI] [PubMed] [Google Scholar]
- 70.Lam B. Simpson A.J. Direct (1)h NMR spectroscopy of dissolved organic matter in natural waters. Analyst. 2008;133:263. doi: 10.1039/b713457f. [DOI] [PubMed] [Google Scholar]
- 71.Kotecha M. Schmid T.M. Magin R.L. Proton NMR characterization of chondrocyte re-differentiation in alginate beads and pellet culture; Annual Meeting of the Biomedical Engineering Society; Atlanta, Georgia. Oct. 2012. Abstract no. P-A-326. [Google Scholar]
- 72.Bernstein P. Dong M. Corbeil D. Gelinsky M. Gunther K.P. Fickert S. Pellet culture elicits superior chondrogenic redifferentiation than alginate-based systems. Biotechnol Progr. 2009;25:1146. doi: 10.1002/btpr.186. [DOI] [PubMed] [Google Scholar]
- 73.Kotecha M. Wickramasinghe N.P. Ishii Y. Efficient low-power heteronuclear decoupling in c-13 high-resolution solid-state NMR under fast magic angle spinning. Magn Reson Chem. 2007;45:S221. doi: 10.1002/mrc.2151. [DOI] [PubMed] [Google Scholar]
- 74.Hall D.A. Maus D.C. Gerfen G.J. Inati S.J. Becerra L.R. Dahlquist F.W. Griffin R.G. Polarization-enhanced NMR spectroscopy of biomolecules in frozen solution. Science. 1997;276:930. doi: 10.1126/science.276.5314.930. [DOI] [PubMed] [Google Scholar]
- 75.Wickramasinghe N.P. Parthasarathy S. Jones C.R. Bhardwaj C. Long F. Kotecha M. Mehboob S. Fung L.W. Past J. Samoson A. Ishii Y. Nanomole-scale protein solid-state NMR by breaking intrinsic 1ht1 boundaries. Nat Methods. 2009;6:215. doi: 10.1038/nmeth.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wickramasinghe N.P. Kotecha M. Samoson A. Past J. Ishii Y. Sensitivity enhancement in (13)c solid-state NMR of protein microcrystals by use of paramagnetic metal ions for optimizing (1)h t(1) relaxation. J Magn Reson. 2007;184:350. doi: 10.1016/j.jmr.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borthakur A. Mellon E. Niyogi S. Witschey W. Kneeland J.B. Reddy R. Sodium and t-1 rho MRI for molecular and diagnostic imaging of articular cartilage. NMR Biomed. 2006;19:781. doi: 10.1002/nbm.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borthakur A. Hancu I. Boada F.E. Shen G.X. Shapiro E.M. Reddy R. In vivo triple quantum filtered twisted projection sodium MRI of human articular cartilage. J Magn Reson. 1999;141:286. doi: 10.1006/jmre.1999.1923. [DOI] [PubMed] [Google Scholar]
- 79.Jerschow A. Ling W. Navon G. Regatte R.R. Phys 281-23na and cest MRI for assessing osteoarthritis and disc disorders. Abstr Pap Am Chem Soc. 2008;236 [Google Scholar]
- 80.Borthakur A. Shapiro E.M. Beers J. Kudchodkar S. Kneeland J.B. Reddy R. Sensitivity of MRI to proteoglycan depletion in cartilage: Comparison of sodium and proton MRI. Osteoarthr Cartilage. 2000;8:288. doi: 10.1053/joca.1999.0303. [DOI] [PubMed] [Google Scholar]
- 81.Wheaton A.J. Borthakur A. Shapiro E.M. Regatte R.R. Akella S.V.S. Kneeland J.B. Reddy R. Proteoglycan loss in human knee cartilage: quantitation with sodium MR imaging—feasibility study. Radiology. 2004;231:900. doi: 10.1148/radiol.2313030521. [DOI] [PubMed] [Google Scholar]
- 82.Navon G. Shinar H. Eliav U. Seo Y. Multiquantum filters and order in tissues. NMR Biomed. 2001;14:112. doi: 10.1002/nbm.687. [DOI] [PubMed] [Google Scholar]
- 83.Reddy R. Insko E.K. Noyszewski E.A. Dandora R. Kneeland J.B. Leigh J.S. Sodium MRI of human articular cartilage in vivo. Magn Reson Med. 1998;39:697. doi: 10.1002/mrm.1910390505. [DOI] [PubMed] [Google Scholar]
- 84.Shapiro E.M. Borthakur A. Dandora R. Kriss A. Leigh J.S. Reddy R. Sodium visibility and quantitation in intact bovine articular cartilage using high field na-23 MRI and mrs. J Magn Reson. 2000;142:24. doi: 10.1006/jmre.1999.1932. [DOI] [PubMed] [Google Scholar]
- 85.Shapiro E.M. Borthakur A. Gougoutas A. Reddy R. Na-23 MRI accurately measures fixed charge density in articular cartilage. Magn Reson Med. 2002;47:284. doi: 10.1002/mrm.10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keinan-Adamsky K. Shinar H. Navon G. Multinuclear NMR and MRI studies of the maturation of pig articular cartilage. Magn Reson Med. 2006;55:532. doi: 10.1002/mrm.20775. [DOI] [PubMed] [Google Scholar]
- 87.Hu K.N. Tycko R. What can solid state NMR contribute to our understanding of protein folding? Biophys Chem. 2010;151:10. doi: 10.1016/j.bpc.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haacke E.M. Brown R.W. Thompson M.R. Venkatesan R. Magnetic Resonance Imaging: Physical Principles and Sequence Design. NewYork: Wiley-Liss; 1999. [Google Scholar]
- 89.Yin Z. Schmid T.M. Madsen L. Kotecha M. Magin R.L. Monitoring the formation of tissue-engineered cartilage in scaffold-free pellet culture using magnetic resonance imaging. Proceedings of the International Society for Magnetic Resonance in Medicine (ISMRM) 20th Annual Meeting and Exhibition; Melbourne, Australia. May 5–11;2012 ; Abstract no. 1396. [Google Scholar]
- 90.Welsch G.H. Trattnig S. Scheffler K. Szomonanyi P. Quirbach S. Marlovits S. Domayer S. Bieri O. Mamisch T.C. Magnetization transfer contrast and T2 mapping in the evaluation of cartilage repair tissue with 3T MRI. J Magn Reson Imaging. 2008;28:979. doi: 10.1002/jmri.21516. [DOI] [PubMed] [Google Scholar]
- 91.Crema M.D. Roemer F.W. Marra M.D. Burstein D. Gold G.E. Eckstein F. Baum T. Mosher T.J. Carrino J.A. Guermazi A. Articular cartilage in the knee: Current MR imaging techniques and applications in clinical practice and research. Radiographics. 2011;31:37. doi: 10.1148/rg.311105084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Akella S.V. Regatte R.R. Gougoutas A.J. Borthakur A. Shapiro E.M. Kneeland J.B. Leigh J.S. Reddy R. Proteoglycan-induced changes in t1rho-relaxation of articular cartilage at 4t. Magn Reson Med. 2001;46:419. doi: 10.1002/mrm.1208. [DOI] [PubMed] [Google Scholar]
- 93.Greco J.B. Spencer R.G. Cartilage growth in magnetic resonance microscopy compatible hollow-fiber bioreactors. In: Chaudhuri J., editor; Al-Rubeai M., editor. Bioreactors for Tissue Engineering: Principles, Design and Operation. J Kluwer; Netherlands: Springer; 2005. pp. 135–163. [Google Scholar]
- 94.Reiter D.A. Roque R.A. Lin P.C. Irrechukwu O. Doty S. Longo D.L. Pleshko N. Spencer R.G. Mapping proteoglycan-bound water in cartilage: Improved specificity of matrix assessment using multiexponential transverse relaxation analysis. Magn Reson Med. 2011;65:377. doi: 10.1002/mrm.22673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramaswamy S. Greco J.B. Uluer M.C. Zhang Z.J. Zhang Z.L. Fishbein K.W. Spencer R.G. Magnetic resonance imaging of chondrocytes labeled with superparamagnetic iron oxide nanoparticles in tissue-engineered cartilage. Tissue Eng Part A. 2009;15:3899. doi: 10.1089/ten.tea.2008.0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu G. Xia C. Wang Z. Lv F. Gao F. Gong Q. Song B. Ai H. Gu Z. Magnetic resonance imaging probes for labeling of chondrocyte cells. J Mater Sci. 2011;22:601. doi: 10.1007/s10856-010-4227-x. [DOI] [PubMed] [Google Scholar]
- 97.Farrell E. Wielopolski P. Pavljasevic P. Kops N. Weinans H. Bernsen M.R. Van Osch G.J.V.M. Cell labelling with superparamagnetic iron oxide has no effect on chondrocyte behaviour. Osteoarthritis Cartilage. 2009;17:961. doi: 10.1016/j.joca.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 98.Van Buul G.M. Kotek G. Wielopolski P.A. Farrell E. Bos P.K. Weinans H. Grohnert A.U. Jahr H. Verhaar J.A. Krestin G.P. Van Osch G.J. Bernsen M.R. Clinically translatable cell tracking and quantification by MRI in cartilage repair using superparamagnetic iron oxides. PLoS One. 2011;6:e17001. doi: 10.1371/journal.pone.0017001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen J. Wang F. Zhang Y. Jin X. Zhang L. Feng Y. Lin X. Yang L. In vivo tracking of superparamagnetic iron oxide nanoparticle labeled chondrocytes in large animal model. Ann Biomed Eng. 2012;40:2568. doi: 10.1007/s10439-012-0621-5. [DOI] [PubMed] [Google Scholar]
- 100.Abarrategi A. Fernandez-Valle M.E. Desmet T. Castejon D. Civantos A. Moreno-Vicente C. Ramos V. Sanz-Casado J.V. Martinez-Vazquez F.J. Dubruel P. Miranda P. Lopez-Lacomba J.L. Label-free magnetic resonance imaging to locate live cells in three-dimensional porous scaffolds. J R Soc Interface. 2012;9:2321. doi: 10.1098/rsif.2012.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Muthupillai R. Lomas D.J. Rossman P.J. Greenleaf J.F. Manduca A. Ehman R.L. Magnetic resonance elastography by direct visualization of propagating acoustic strain waves. Science. 1995;269:1854. doi: 10.1126/science.7569924. [DOI] [PubMed] [Google Scholar]
- 102.Manduca A. Oliphant T.E. Dresner M.A. Mahowald J.L. Kruse S.A. Amromin E. Felmlee J.P. Greenleaf J.F. Ehman R.L. Magnetic resonance elastography: Non-invasive mapping of tissue elasticity. Med Image Anal. 2001;5:237. doi: 10.1016/s1361-8415(00)00039-6. [DOI] [PubMed] [Google Scholar]
- 103.Kempson G.E. Spivey C.J. Swanson S.A.V. Freeman M.A.R. Patterns of cartilage stiffness on normal and degenerate human femoral heads. J Biomech. 1971;4:597. doi: 10.1016/0021-9290(71)90049-2. [DOI] [PubMed] [Google Scholar]
- 104.Glaser K.J. Manduca A. Ehman R.L. Review of MR elastography applications and recent developments. J Magn Reson Imaging. 2012;36:757. doi: 10.1002/jmri.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sack I. Beierbach B. Wuerfel J. Klatt D. Hamhaber U. Papazoglou S. Martus P. Braun J. The impact of aging and gender on brain viscoelasticity. NeuroImage. 2009;46:652. doi: 10.1016/j.neuroimage.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 106.Sinkus R. Siegmann K. Xydeas T. Tanter M. Claussen C. Fink M. Mr elastography of breast lesions: understanding the solid/liquid duality can improve the specificity of contrast-enhanced MR mammography. Magn Reson Med. 2007;58:1135. doi: 10.1002/mrm.21404. [DOI] [PubMed] [Google Scholar]
- 107.Magin R.L. Fractional Calculus in Bioengineering. Begell House, Inc.; Redding CT: Begell House: 2006. [Google Scholar]
- 108.Schiessel H. Metzler R. Blumen A. Nonnenmacher T.F. Generalized viscoelastic models: their fractional equations with solutions. J Phys A-Math Gen. 1995;28:6567. [Google Scholar]
- 109.Schiessel H. Blumen A. Mesoscopic pictures of the sol-gel transition—ladder models and fractal networks. Macromolecules. 1995;28:4013. [Google Scholar]
- 110.Klatt D. Hamhaber U. Asbach P. Braun J. Sack I. Noninvasive assessment of the rheological behavior of human organs using multifrequency MR elastography: a study of brain and liver viscoelasticity. Phys Med Biol. 2007;52:7281. doi: 10.1088/0031-9155/52/24/006. [DOI] [PubMed] [Google Scholar]
- 111.Zeytin S. Konduk B.A. Ipek M. Bindal C. Ucisik A.H. An evaluation of human articular cartilage on femoral head. Mater Sci Eng C. 2002;20:219. [Google Scholar]
- 112.Yin Z. Yasar T.K. Magin R.L. Measurement of in vitro local shear stiffness of alginate beads using microscopic MR elastography; Abstract presented at the annual meeting of Biomedical Engineering Society; Atlanta, Georgia. 2012. Abstract no. OP-1–19. [Google Scholar]
- 113.Knutsson H. Westin C.F. Granlund G. Local multiscale frequency and bandwidth estimation. Image Processing, 1994 Proceedings ICIP-94, IEEE International Conference; 1994. p. 36. [Google Scholar]
- 114.Perrinez P.R. Kennedy F.E. Van Houten E.E.W. Weaver J.B. Paulsen K.D. Magnetic resonance poroelastography: an algorithm for estimating the mechanical properties of fluid-saturated soft tissues. IEEE Trans Med Imaging. 2010;29:746. doi: 10.1109/TMI.2009.2035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Asbach P. Klatt D. Schlosser B. Biermer M. Muche M. Rieger A. Loddenkemper C. Somasundaram R. Berg T. Hamm B. Braun J. Sack I. Viscoelasticity-based staging of hepatic fibrosis with multifrequency MR elastography. Radiology. 2010;257:80. doi: 10.1148/radiol.10092489. [DOI] [PubMed] [Google Scholar]
- 116.Yin M. Talwalkar J.A. Glaser K.J. Manduca A. Grimm R.C. Rossman P.J. Fidler J.L. Ehman R.L. Assessment of hepatic fibrosis with magnetic resonance elastography. Clin Gastroenterol Hepatol. 2007;5:1207. doi: 10.1016/j.cgh.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Freimann F.B. Streitberger K.J. Klatt D. Lin K. Mclaughlin J. Braun J. Sprung C. Sack I. Alteration of brain viscoelasticity after shunt treatment in normal pressure hydrocephalus. Neuroradiology. 2012;54:189. doi: 10.1007/s00234-011-0871-1. [DOI] [PubMed] [Google Scholar]
- 118.Wuerfel J. Paul F. Beierbach B. Hamhaber U. Klatt D. Papazoglou S. Zipp F. Martus P. Braun J. Sack I. MR-elastography reveals degradation of tissue integrity in multiple sclerosis. NeuroImage. 2010;49:2520. doi: 10.1016/j.neuroimage.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 119.Murphy M.C. Huston J. Jack C.R. Glaser K.J. Manduca A. Felmlee J.P. Ehman R.L. Decreased brain stiffness in alzheimer's disease determined by magnetic resonance elastography. J Magn Reson Imaging. 2011;34:494. doi: 10.1002/jmri.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lopez O. Amrami K.K. Manduca A. Ehman R.L. Characterization of the dynamic shear properties of hyaline cartilage using high-frequency dynamic MR elastography. Magn Reson Med. 2008;59:356. doi: 10.1002/mrm.21474. [DOI] [PubMed] [Google Scholar]
- 121.Yasar T.K. Royston T.J. Magin R.L. Wideband MR elastography for viscoelasticity model identification. Magn Reson Med. 2012 doi: 10.1002/mrm.24495. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yin Z. Yifie L. Magin R.L. Evaluation of Tissue Engineered Cartilage using Microscopic Magnetic Resonance Elastography; 54th Experimental Nuclear Magnetic Resonance Conference (ENC); Apr 14–19;2013 ; Pacific Grove, CA. [Google Scholar]
- 123.Hirsch S. Klatt D. Freimann F.B. Scheel M. Braun J. Sack I. Divergence MRI of the human brain: in vivo measurement of volumetric strain induced by arterial pulsation and harmonic waves. Magn Reson Med. 2012 doi: 10.1002/mrm.24499. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 124.Wang Y.X. In vivo magnetic resonance imaging of animal models of knee osteoarthritis. Lab Anim. 2008;42:246. doi: 10.1258/la.2007.06041e. [DOI] [PubMed] [Google Scholar]
- 125.Pinney J.R. Taylor C. Doan R. Burghardt A.J. Li X. Kim H.T. Benjamin Ma C. Majumdar S. Imaging longitudinal changes in articular cartilage and bone following doxycycline treatment in a rabbit anterior cruciate ligament transection model of osteoarthritis. Magn Reson Imaging. 2012;30:271. doi: 10.1016/j.mri.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gold G.E. Pauly J.M. Macovski A. Herfkens R.J. Mr spectroscopic imaging of collagen: Tendons and knee menisci. Magn Reson Med. 1995;34:647. doi: 10.1002/mrm.1910340502. [DOI] [PubMed] [Google Scholar]
- 127.Chu C.R. Szczodry M. Bruno S. Animal models for cartilage regeneration and repair. Tissue Eng Part B Rev. 2010;16:105. doi: 10.1089/ten.teb.2009.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beuf O. Jaillon F. Saint-Jalmes H. Small-animal MRI: Signal-to-noise ratio comparison at 7 and 1.5 t with multiple-animal acquisition strategies. MAGMA. 2006;19:202. doi: 10.1007/s10334-006-0048-9. [DOI] [PubMed] [Google Scholar]
- 129.Kwack K.S. Cho J.H. Kim M.M. Yoon C.S. Yoon Y.S. Choi J.W. Kwon J.W. Min B.H. Sun J.S. Kim S.Y. Comparison study of intraarticular and intravenous gadolinium-enhanced magnetic resonance imaging of cartilage in a canine model. Acta Radiol. 2008;49:65. doi: 10.1080/02841850701552934. [DOI] [PubMed] [Google Scholar]
- 130.Kwack K.S. Min B.H. Cho J.H. Kim J.M. Yoon S.H. Kim S.Y. T2 relaxation time mapping of proximal tibiofibular cartilage by 3-tesla magnetic resonance imaging. Acta Radiol. 2009;50:1049. doi: 10.3109/02841850903215918. [DOI] [PubMed] [Google Scholar]
- 131.Simon T.M. Aberman H.M. Cartilage regeneration and repair testing in a surrogate large animal model. Tissue Eng Part B Rev. 2010;16:65. doi: 10.1089/ten.TEB.2009.0304. [DOI] [PubMed] [Google Scholar]
- 132.Wang V.M. Yin Z. Lee A. Van Thiel G. Karas V. Hussey K. Shewman E. Magin R.L. Cole B. Orthopedic Research Society; San Antonio, TXno: Jan 26–29, 2013. Analysis of cartilage regeneration in a rabbit glenohumeral joint model following microfracture and autologous matrix induced chondrogenesis using high-field (11.7 tesla) MRI. [Google Scholar]
- 133.Kim M. Foo L.F. Uggen C. Lyman S. Ryaby J.T. Moynihan D.P. Grande D.A. Potter H.G. Pleshko N. Evaluation of early osteochondral defect repair in a rabbit model utilizing fourier transform-infrared imaging spectroscopy, magnetic resonance imaging, and quantitative t2 mapping. Tissue Eng Part C Methods. 2010;16:355. doi: 10.1089/ten.tec.2009.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ramaswamy S. Gurkan L. Sharma B. Cascio B. Fishbein K.W. Spencer R.G. Assessment of tissue repair in full thickness chondral defects in the rabbit using magnetic resonance imaging transverse relaxation measurements. J Biomed Mater Res B. 2008;86B:375. doi: 10.1002/jbm.b.31030. [DOI] [PubMed] [Google Scholar]
- 135.Trattnig S. Winalski C.S. Marlovits S. Jurvelin J.S. Welsch G.H. Potter H.G. Magnetic resonance imaging of cartilage repair: A review. Cartilage. 2011;2:5. doi: 10.1177/1947603509360209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gold G.E. Chen C.A. Koo S. Hargreaves B.A. Bangerter N.K. Recent advances in MRI of articular cartilage. AJR Am J Roentgenol. 2009;193:628. doi: 10.2214/AJR.09.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Newbould R.D. Miller S.R. Toms L.D. Swann P. Tielbeek J.A. Gold G.E. Strachan R.K. Taylor P.C. Matthews P.M. Brown A.P. T2* measurement of the knee articular cartilage in osteoarthritis at 3t. J Magn Reson Imaging. 2012;35:1422. doi: 10.1002/jmri.23598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Newbould R.D. Miller S.R. Tielbeek J.A.W. Toms L.D. Rao A.W. Gold G.E. Strachan R.K. Taylor P.C. Matthews P.M. Brown A.P. Reproducibility of sodium MRI measures of articular cartilage of the knee in osteoarthritis. Osteoarthr Cartilage. 2012;20:29. doi: 10.1016/j.joca.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]