Abstract
Background
Hepatocellular carcinoma (HCC) in young subjects is rare but more devastating. We hypothesize that genes and etiological pathways are unique to young HCC (yHCC; ≤40 years old at diagnosis) patients. We therefore compared the gene expression profiles between yHCCs and HCCs from elderly patients.
Results
All 44 young HCCs (≤40 years old at the diagnosis; 23 cases in the training set while another 21 in the validation cohort) were positive for serum hepatitis B surface antigen (HBsAg), but negative for antibodies to hepatitis C virus (anti-HCV). All 48 elderly (>40 years old; 38 in the training set while another 10 in the validation cohort) HCC patients enrolled were also serum HBsAg positive and anti-HCV negative. Comparative genomics analysis was further performed for elucidating enriched or suppressed biological activities in different HCC subtypes.
The yHCC group showed more macroscopic venous invasions (60.9% vs. 10.5%, p < 0.001), fewer associated cirrhosis (17.4% vs. 63.2%, p < 0.001), and distinct profiles of expressed genes, especially those related to DNA replication and repair. yHCCs possessed increased embryonic stem cell (ESC) traits and were more dedifferentiated. A 309-gene signature was obtained from two training cohorts and validated in another independent data set. The ILF3 ESC gene, which was previously reported in poorly differentiated breast cancers and bladder carcinomas, was also present in yHCCs. Genes associated with HCC suppression, including AR and ADRA1A, were less abundant in yHCCs. ESC genes were also more enriched in advanced HCCs from elderly patients.
Conclusion
This study revealed the molecular makeup of yHCC and the link between ESC traits and HCC subtypes. Findings in elderly tumors, therefore, cannot be simply extrapolated to young patients, and yHCC should be treated differently.
Keywords: Young hepatocellular carcinoma, Embryonic stem cells, Dedifferentiation
Background
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide and chronic hepatitis B virus (HBV) infection is the most important cause of HCC in Taiwan [1,2]. Most HCC patients are diagnosed in old age with only a small portion of them younger than 40 years old [1,2]. Compared with the elderly HCC patients, young HCC (yHCC) cases (≤40 years of age) are more likely to be symptomatic at diagnosis and the HCC stage tends to be more advanced. Thus, there is a decreased chance of curative resection for the tumors in this group [3,4]. Although the presence of cirrhosis is less frequent in young patients [4], the time to yHCC recurrence after surgical resection was shorter and a one year survival rate was lower than those with elderly patients [4,5]. An aggressive clinical course and a poor prognosis have also been reported in children with HCC [6,7]. If yHCC patients survived longer than one year, their long-term survivals seemed to be better than those of elderly HCC patients due to fewer incidences of associated cirrhosis and relatively better liver function reserves [5]. High serum alpha-fetoprotein levels are more often found in yHCC patients [3,8]. HBV viral load is not a predictor in the development of HCC in young adults [9-11], in contrast, viral load and hepatic inflammatory activity were associated with late recurrence of HCC among elderly patients after resection of the primary HCC [12]. The aforementioned findings suggest that hepatocarcinogenesis in yHCCs is different from that in elderly patients. Yet the underlying mechanisms and the detail molecular portrait of yHCC remain unclear.
It has also been recognized that cancer cells, especially those of advanced and metastatic cancers, possess characteristics reminiscent of normal stem cells. The degree of stem cell gene reactivation or tumor cell dedifferentiation correlates with pivotal tumor features and prognosis [13,14]. A recent paper demonstrated by RT-qPCR, that the high expression levels of putative hepatic stem/progenitor cell biomarkers are related to tumor angiogenesis and a poor prognosis for HCC [15]. However, no similar study has addressed yHCC. Identifying genes involved in cancer progression and cell dedifferentiation offers another dimension to predict HCC recurrence, as well as providing novel therapeutic targets and prognosis markers.
Results
Clinical profiles, serological data, and histopathological findings for the HCCs from young and elderly patients enrolled in array analysis
The clinical profiles, serological data, and histopathological findings for young and elderly HCC patients in the training cohort are in Table 1. In 61 enrolled primary HBsAg positive HCC patients, 23 cases were yHCCs and 38 were elderly. Macroscopic venous invasion was more frequent (60.9% vs. 10.5%, p < 0.001), but accompanied cirrhosis was significantly fewer in younger subjects (17.4% vs. 63.2%, p < 0.001). Consistent with fewer cirrhotic patients in the younger group, the ICG-15 retention was lower (p = 0.0055) and the platelet counts tended to be higher (p = 0.087). There were no statistically significant differences in the remaining parameters between these two groups.
Table 1.
Demographic data in relation to age of the training cohort HCC patients undergoing surgical resection
|
All patients |
Age ≦40 |
Age >40 |
p* | |
|---|---|---|---|---|
| (n = 61) | (n = 23) | (n = 38) | ||
|
Patient demographics | ||||
| Age (years) (median; 25 and 75 percentiles) (range) |
47.5; 36.5-60.2 (26-75.7) |
35.5; 30.8-37.0 (26-39.5) |
56.8; 50.6-64.3 (40.8-75.7) |
<0.001 |
| Sex (M:F) |
54:7 |
19:4 |
35:3 |
0.409 |
| Albumin (g/dL) (median; 25 and 75 percentiles) |
4.0; 3.8-4.4 |
4.2; 3.9-4.4 |
4.0; 3.7-4.3 |
0.129 |
| Total bilirubin (mg/dL) (median; 25 and 75 percentiles) |
0.80; 0.6-1.2 |
0.7; 0.5-1.1 |
0.9; 0.6-1.2 |
0.688 |
| ALT (U/L) (median; 25 and 75 percentiles) |
41.0; 30.5-57.0 |
46.0; 35.0-61.0 |
38.0; 29.8-57.0 |
0.946 |
| AST (U/L) (median; 25 and 75 percentiles) |
40.0; 27.0-66.0 |
42.0; 27.0-65.0 |
39.5; 27.0-67.5 |
0.871 |
| Platelet (/mm3) (median; 25 and 75 percentiles) |
182000; 139000-224500 |
204000; 171000-264000 |
167000; 131750-2017250 |
0.087 |
| ICG-15R (%) (retention rate) (median; 25 and 75 percentiles) |
8; 5.3-12.8 |
6; 4-8 |
10; 6-14.3 |
0.0055 |
| Child-Pugh A/B (%) |
59/2 (96.7%/3.3%) |
22/1 (95.7%/4.4%) |
37/1 (97.4%/2.6%) |
1.000 |
|
Tumor characteristics | ||||
| Tumor size (cm) (median; 25 and 75 percentiles) |
4.8; 3.4-8.7 |
5.0; 4.0-11.0 |
4.4; 3.2-6.4 |
0.523 |
| Multinodularity (%) |
28 (45.9%) |
9 (39.1%) |
19 (50.0%) |
0.440 |
| AFP (ng/ml) (median; 25 and 75 percentiles) |
190; 15-1541 |
316; 18.4-5499 |
169.5; 12.9-728.5 |
0.742 |
| Macroscopic venous invasion (%) |
18 (29.5%) |
14 (60.9%) |
4 (10.5%) |
<0.001 |
| Daughter nodule (%) |
32 (62.8%) |
11 (55.0%) |
21 (67.7%) |
0.389 |
|
Histopathological findings | ||||
| Cirrhosis in non-tumor part (yes/no) (%) |
33/28 (54.1%/45.9%) |
4/19 (17.4%/82.6%) |
24/14 (63.2%/36.8%) |
<0.001 |
| Edmondson grading (I or II/ III or IV) (%) |
33/24 (57.9%/42.1%) |
11/12 (47.8%/52.2%) |
22/12 (64.7%/35.3%) |
0.276 |
| Microscopic venous invasion (%) | 43 (70.5%) | 17 (73.9%) | 26 (68.4%) | 0.775 |
ICG-15R: indocyanine green retention rate at 15 minutes;
*p value: comparison between younger and elderly HCC patients.
Molecular signatures of yHCCs
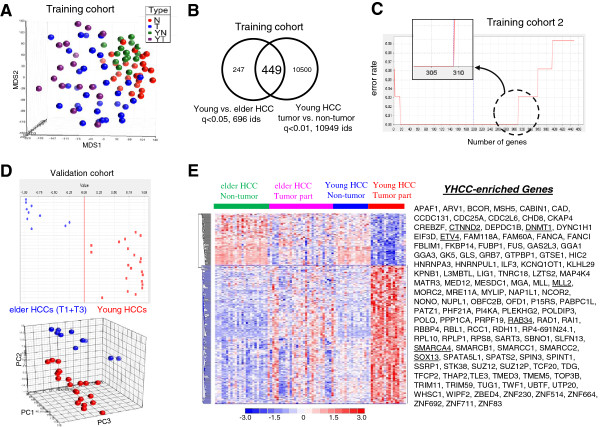
Data analysis steps were summarized in Additional file 1: Figure S1 online. To explore the molecular mechanisms governing the diverse clinical behaviors of the different HCCs, we delineated gene expression profiles of 48 primary HCC samples, as well as those of 39 non-cancerous tissues, from the above 61 patients as a training data set. A multidimensional scaling (MDS) plot using the whole transcriptome showed that the mRNA profiles of normal and cancerous tissues were different, while tumors of different age groups were similar (Figure 1A). We compared tumor samples to non-tumor counterparts for minimizing stromal and myometrial contamination. A total of 449 probe sets were differentially expressed between young and elderly HCCs (positive false discovery rate (pFDR) q < 0.05), as well as between tumor and non-tumor tissues of yHCC patients (Figure 1B).
Figure 1.
Distinct gene expression patterns in HCC from young or elderly patients. (A) A multidimensional scaling (MDS) plot using the whole transcriptome illustrates the mRNA profiles of normal and cancerous tissues. (B) A Venn diagram summarizing genes differentially expressed between the HCC tumor tissues of young and elderly patients, or between tumor and non-tumor yHCC samples. (C) Advanced signature training using a machine learning strategy and an independent testing elderly HCC data set. When probe sets were ranked by signal-to-noise ratios (weights), the top 309 features was the largest panel to give the lowest error rate (i.e., a best classification effect; upper panel). (D) The discrimination ability of the 309-probeset signature on the validation data set. The prediction strength plot (upper) and the PCA plot (lower) show the discriminating power of the identified 309 probe sets in separating young and elderly patients in the validation cohort. (E) A heat map shows the 309 probes sets differentiating young and elderly HCCs in the training data set 1, as well as discriminating tumor and non-tumor tissues. Columns represent tumor samples; rows represent probe sets. Genes in red: increased; in blue: decreased. Genes underlined: discussed in the text.
The discrimination ability of these 449 probe sets were further trained by performing supervised machine learning that combined weighted voting algorithm and leave-one-out cross validation (LOOCV) [16], on the 2nd external data set (downloaded from the Expression Project for Oncology (expO)). An error rate of 9.4% (2 out of 16 yHCCs and 1 out of 16 elderly HCCs in the validation set; P < 0.001 by permutation test) was found (Figure 1C). The top 309 features (ranked by the weighted value of each probe set [16]) form a largest panel to have the best discrimination ability than that of the 449-probeset signature (error rate 0 vs. 9.4%; Figure 1C, upper panel). The discrimination ability of these 309 probe sets was evaluated on an independent testing data set that included another 21 yHCCs and 10 Taiwanese elderly HCCs (4 were at T1 stage and the remaining 6 were at T3 stage by 6th edition American Joint Committee on Cancer (AJCC)/International Union Against Cancer (UICC) staging system [17,18]). Prediction strength (PS; Figure 1D, upper) and principle component analysis (PCA; Figure 1D, lower) plots showed that these 309 probe sets distinguished young and elderly HCCs well.
The distribution of these 309 probe sets among sample groups were examined by hierarchical clustering. The differences in gene expression profiles between elderly and yHCC were more striking in tumor parts as compared to those in non-tumor parts (Figure 1E). A heat map for these genes indicated the unique gene expression levels in yHCC, with 225 probe sets being predominantly up in yHCCs (Table 2) while another 84 being down (Figure 1E). Many of yHCC-enriched genes, such as CTNND2 (delta 2 catenin), RAB34 (a member of the RAS oncogene family), SOX13 (SRY (sex determining region Y)-box 13), ETV4 (ets variant gene 4), DNMT1 (DNA cytosine-5-methyltransferase 1), TLE3 (transducin-like enhancer of split 3), MLL (myeloid/lymphoid or mixed-lineage leukemia), and MLL2, have been associated with tumor malignancy and poor patient outcomes in HCC or other cancers (Figure 1E, underlined). These consistent findings support the reliability of our gene list. Genes down-regulated in yHCC (i.e. more abundant in elderly HCCs) are shown in Additional file 2: Table S1.
Table 2.
ESC genes overexpressed in yHCC patients (q < 0.05, Young HCC vs. elder HCC)
| Probe set ID | UniGene ID | Gene title | Gene symbol | Location | Folds |
|---|---|---|---|---|---|
| 219010_at |
Hs.518997 |
chromosome 1 open reading frame 10 |
C1orf106 |
chr1q32.1 |
3.05 |
| 202715_at |
Hs.377010 |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
CAD |
chr2p22-p21 |
1.68 |
| 1555772_a_at |
Hs.437705 |
cell division cycle 25 homolog A (S. pombe) |
*CDC25A |
chr3p21 |
1.89 |
| 226980_at |
Hs.482233 |
DEP domain containing 1B |
DEPDC1B |
chr5q12.1 |
2.08 |
| 201697_s_at |
Hs.202672 |
DNA (cytosine-5-)-methyltransferase 1 |
*DNMT1 |
chr19p13.2 |
1.74 |
| 229115_at |
Hs.649497 |
dynein, cytoplasmic 1, heavy chain 1 |
*DYNC1H1 |
chr14q32.3-qter |
1.78 |
| 200005_at |
Hs.55682 |
eukaryotic translation initiation factor 3, subunit D |
EIF3D |
chr22q13.1 |
1.55 |
| 1554576_a_at |
Hs.434059 |
ets variant gene 4 (E1A enhancer binding protein, E1AF) |
ETV4 |
chr17q21 |
2.02 |
| 220060_s_at |
family with sequence similarity 222, member B |
FAM222B |
chr17q11.2 |
1.56 |
|
| 213008_at |
Hs.513126 |
Fanconi anemia, complementation group I |
*FANCI |
chr15q26.1 |
1.78 |
| 219390_at |
Hs.571333 |
FK506 binding protein 14, 22 kDa |
FKBP14 |
chr7p15.1 |
1.72 |
| 223079_s_at |
Hs.116448 |
glutaminase |
GLS |
chr2q32-q34 |
2.26 |
| 215942_s_at |
Hs.386189 |
G-2 and S-phase expressed 1 |
*GTSE1 |
chr22q13.2-q13.3 |
1.94 |
| 242890_at |
Hs.655830 |
Helicase, lymphoid-specific |
*HELLS |
chr10q24.2 |
2.47 |
| 212966_at |
Hs.632767 |
hypermethylated in cancer 2 |
*HIC2 |
chr22q11.21 |
2.71 |
| 208930_s_at |
Hs.465885 |
interleukin enhancer binding factor 3, 90 kDa |
*ILF3 |
chr19p13.2 |
2.32 |
| 208974_x_at |
Hs.532793 |
karyopherin (importin) beta 1 |
KPNB1 |
chr17q21.32 |
1.82 |
| 202726_at |
Hs.1770 |
ligase I, DNA, ATP-dependent |
*LIG1 |
chr19q13.2-q13.3 |
1.82 |
| 65588_at |
Hs.400876 |
lncRNA LOC388796 |
LOC388796 |
65588_at |
1.68 |
| 224473_x_at |
Hs.523221 |
leucine zipper, putative tumor suppressor 2 |
*LZTS2 |
chr10q24 |
1.61 |
| 64432_at |
Hs.333120 |
MAPKAPK5 antisense RNA 1 |
MAPKAPK5-AS1 |
chr12q24.12 |
1.61 |
| 242260_at |
Hs.268939 |
Matrin 3 |
MATR3 |
chr5q31.2 |
2.35 |
| 235409_at |
Hs.187569 |
MAX gene associated |
MGA |
chr15q14 |
1.79 |
| 228097_at |
Hs.484738 |
myosin regulatory light chain interacting protein |
MYLIP |
chr6p23-p22.3 |
1.64 |
| 208752_x_at |
Hs.524599 |
nucleosome assembly protein 1-like 1 |
NAP1L1 |
chr12q21.2 |
1.66 |
| 214107_x_at |
Hs.740414 |
aminopeptidase puromycin sensitive |
NPEPPS |
chr17q21 |
1.85 |
| 200057_s_at |
Hs.533282 |
non-POU domain containing, octamer-binding |
*NONO |
chrXq13.1 |
1.52 |
| 228566_at |
Hs.464912 |
Cyclin-dependent kinase 2B-inhibitor-related protein |
P15RS |
chr18q12.2 |
1.51 |
| 203103_s_at |
Hs.502705 |
PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae) |
*PRPF19 |
chr11q12.2 |
1.49 |
| 1555630_a_at |
Hs.301853 |
RAB34, member RAS oncogene family |
RAB34 |
chr17q11.2 |
3.14 |
| 206499_s_at |
Hs.469723 |
regulator of chromosome condensation 1 |
*RCC1 |
chr1p36.1 |
1.59 |
| 200858_s_at |
Hs.512675 |
ribosomal protein S8 |
RPS8 |
chr1p34.1-p32 |
1.36 |
| 209127_s_at |
Hs.584842 |
squamous cell carcinoma antigen recognized by T cells 3 |
SART3 |
chr12q24.1 |
2.03 |
| 214728_x_at |
Hs.327527 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, a4 |
*SMARCA4 |
chr19p13.2 |
1.61 |
| 201072_s_at |
Hs.476179 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, c1 |
SMARCC1 |
chr3p23-p21 |
1.89 |
| 228990_at |
Hs.632377 |
small nucleolar RNA host gene 12 (non-protein coding) |
SNHG12 |
chr1p35.3 |
1.86 |
| 218324_s_at |
Hs.654826 |
spermatogenesis associated, serine-rich 2 |
SPATS2 |
chr12q13.12 |
1.84 |
| 202826_at |
Hs.233950 |
serine peptidase inhibitor, Kunitz type 1 |
SPINT1 (HAI-1) |
chr15q15.1 |
3.36 |
| 200956_s_at |
Hs.523680 |
structure specific recognition protein 1 |
*SSRP1 |
chr11q12 |
1.78 |
| 207627_s_at |
Hs.48849 |
transcription factor CP2 |
TFCP2 |
chr12q13 |
1.66 |
| 212770_at |
Hs.709205 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
TLE3 |
chr15q22 |
1.51 |
| 208837_at |
Hs.513058 |
transmembrane emp24 protein transport domain containing 3 |
TMED3 |
chr15q24-q25 |
2.77 |
| 238797_at |
Hs.13543 |
tripartite motif-containing 11 |
TRIM11 |
chr1q42.13 |
1.51 |
| 235476_at |
Hs.212957 |
tripartite motif-containing 59 |
TRIM59 |
chr3q26.1 |
2.00 |
| 209053_s_at |
Hs.113876 |
Wolf-Hirschhorn syndrome candidate 1 |
WHSC1 |
chr4p16.3 |
1.97 |
| 204799_at |
Hs.475208 |
zinc finger, BED-type containing 4 |
ZBED4 |
chr22q13.33 |
1.72 |
| 228988_at | Hs.326801 | zinc finger protein 711 | ZNF711 | chrXq21.1-q21.2 | 3.40 |
*Genes discussed in the text.
Coordinated functional module changes in yHCCs
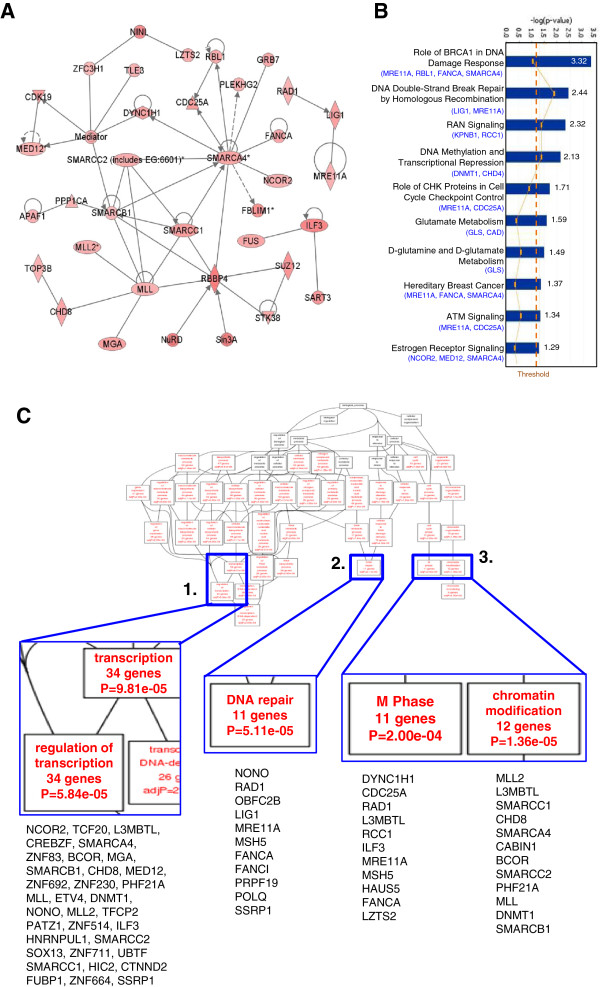
To understand how genes enriched in yHCC are related to each other, as well as to spot the more critical yHCC genes, we performed systems biology analysis. A major genetic network contains known cancer-related or pro-proliferating genes, including CDC25A, CDK19, FUS (fused in sarcoma), TLE3, and ILF3 (interleukin enhancer binding factor 3) was formed (Figure 2A). Central to the network, there were hub genes (genes with higher connectivity to others), including MLL, SMARCA4, SMARCB1, SMARCC1, and RBBP4 (retinoblastoma binding protein 4) (Figure 2A).
Figure 2.
Interaction network and gene set enrichment analyses as frameworks for interpreting yHCC biology. (A) A genetic network composed of multiple yHCC genes. This network is displayed graphically as nodes (gene products) and edges (biological relationships between nodes) mapped by the Ingenuity Pathway Analysis (IPA) tool. The intensity of the node color indicates the degree of upregulation. (B) Canonical pathway analysis. Genes that are more abundant in yHCC were subjected to IPA search. (C) Altered biological modules in yHCCs. 282 probe sets that are more abundant in yHCC were subjected to Gene Ontology database search. The number of genes, gene symbols, and p values for each category that are significantly enriched are listed (p < 0.05).
To understand better how gene expression profiles correlate with pathogenesis and tumor phenotypes, signature probe sets were subjected into canonical pathways and functional group analysis using the Ingenuity Pathway Analysis (IPA) and Gene Ontology (GO) databases, respectively. The most significant canonical pathway mapped is the “BRCA1 in DNA Damage Response” pathway (Figure 2B). Other predominant pathways were DNA double-strand break repair, DNA methylation and transcriptional repression and ATM Signaling (Figure 2B). Consistent with the unique expression profile of yHCCs, the genes involved in the regulation of transcription were enriched in yHCCs (p = 5.84*10e-05; Figure 2C, panel 1). Genes involved in chromatin modification are also unique in yHCCs (p = 1.36*10e-5; Figure 2C, panel 3). Other related predominant GO processes included those pertaining to DNA repair (p = 5.11*10e-5) and M phase cell cycle (p = 2.00*10e-4) (Figure 2C, panels 2–3).
Increased embryonic stem cell (ESC) traits in HCCs, especially those from young patients
Stemness genes are known to contribute largely in tumorigenesis and disease progression [13,14]. For narrowing down key genes and obtaining more insights in yHCC pathogenesis, the above 309 probe sets were used to compare the relationships between HCCs and ESC. Transcriptome distances were measured by calculating the average linkage distances. Compared with non-tumor tissues, HCCs of different age categories were closer to ESCs (Figure 3A), suggesting the re-expression of ESC genes is a characteristic feature during tumorigenesis. The closest correlation between ESC and yHCC was observed, indicating the level of ESC gene re-expression was inversely correlated with patient age (Figure 3A).
Figure 3.
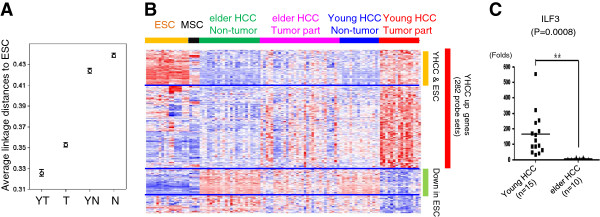
ESC traits in yHCCs. (A) Relationships between ESC, HCCs of different ages, and non-tumor tissues. Average linkage distances between tissues and ESC were calculated using the filtered 309 probe sets. The confidence limits as shown represent the standard error. YT and T: cancerous HCC samples from young and elderly patients, respectively. YN and N: non-tumor samples. (B) A heat map showing shared genes between yHCC and ESC. (C) Validation of ILF3 array data by real-time RT-PCR. The mean expression levels of the target genes were compared to the GAPDH control. **: p < 0.01 by t-test.
The distributions of these 309 probe sets among sample groups were shown using a heat map (Figure 3B). Among genes enriched in yHCC, a subgroup of genes was also abundant in stem cells, especially in ESC (Figure 3B). Table 2 shows ESC genes overexpressed in YHCC patients. Among them, 9 genes were involved in cell cycle control (CDC25A, DYNC1H1, FANCI, GTSE1, HELLS, ILF3, LIG1, LZTS2, and RCC1; p = 1.3*10e-3, gene enrichment analysis was done based on the GO database), 5 genes in DNA repair (FANCI, PRPF19, LIG1, NONO, and SSRP1; p = 8.3*10e-3), and 2 genes in blastocyst growth (PRPF19 and SMARCA4; p = .031) (Table 2, genes with asterisks). Intriguingly, ILF3 is among the ‘Core 9’ ESC transcription regulators that were highly expressed in poorly differentiated breast cancers, glioblastomas, and bladder carcinomas (13). The differential expression of ILF3 between young and elderly HCCs was verified by RT-qPCR (Figure 3C).
Decreasing hepatic differentiation program in yHCCs and during disease progression in elderly HCCs
We hypothesized that yHCCs also forfeited genes associated with liver differentiation and thereby were more dedifferentiated and malignant. Liver precursor characteristics were examined in the yHCC samples by comparing the relationships between HCC subgroups and liver progenitor cells (derived from the H9 ESC line [19]). An inverse correlation between the hepatogenesis process with patient ages was observed (Figure 4A, left panel; the direction of ESC hepatogenesis is indicated by a green arrow). Such impressions were strengthened by calculating the transcriptome distances between the sample groups (Figure 4A, right panel). Among the 309 yHCC genes, 15 genes were more abundant in differentiated liver progenitor cells (day 20; Additional file 3: Figure S2 online). These 15 genes, which are also downregulated in yHCCs, hold the potentials of being novel tumor suppressor genes in yHCCs.
Figure 4.
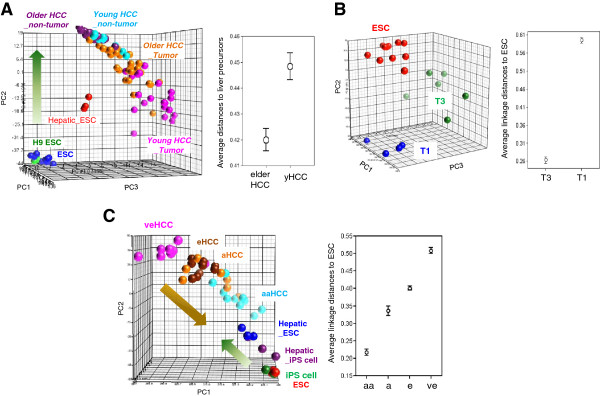
Decreased hepatogenesis characters in yHCCs but increased ESC traits in advanced HCCs. (A) Forfeiting of liver differentiation program in yHCCs. (Left) A PCA plot using genes differentiating the H9 ESCs and day 20 hepatic differentiated progenies (q < 10-4). The transcriptome drift directions during hepatic differentiation are indicated by an arrow. Hepatic_ESC: day 20 (d20) hepatic differentiated progenies. (right) Relationships between liver precursors and HCCs of different age groups. (B) Relationships between ESC and HCC of different histopathological stages. (Left) A PCA plot based on 977 probe sets genes distinguishing early (T1) and late (T3) HCCs from elderly patients. (Right) Relationships between ESC and T1/T3 HCCs. (C) Dedifferentiation-like transcriptome reprogramming during the progression of HCV-related HCC. (Left) A PCA plot using genes differentiating very early (ve) and advanced (a) HCC (q < 0.01, 1700 probe sets). The orange arrow represents the transcriptome drift direction during HCC progression. eHCC, early HCC; aHCC, advanced HCC; Hepatic_iPS cells: day 20 hepatic differentiated progenies of iPS cells. (Right) Relationships between ESC and HCC at different clinical stages.
The above observation inspired us to hypothesize further that the forfeiting of hepatogenesis traits may have also occurred during disease progression in HCCs of the same age group. We examined the associations between ESC gene patterns and clinical stage. Early (T1) and late (T3) HCCs [18] used in the validation cohort were applied to compare the relationships with ES cells and the advanced T3 cases were closer to ES cells (Figure 4B). Such relationships were validated by evaluating another independent serum anti-HCV positive HCC data set [20]. This data set included four neoplastic stages (very early HCC to very advanced metastatic tumors) from patients with HCV infection [20]. When the relationships between the different pathological HCC subgroups and pluripotent stem cells (including ESCs and induced pluripotent stem cells (iPS cells) [19]) were compared, an increased stemness that accurately reflected the pathological progression of the disease was again observed (Figure 4C). A dedifferentiation-like transcriptome drift (indicated by an orange arrow, Figure 4C) was anti-correlated with the hepatic differentiation program of pluripotent stem cells (indicated by a green arrow), indicating a dedifferentiation status during the progression of HCV-related HCC.
Discussion
This study explored the gene expression profile of yHCC. We found the age difference between HCC patients is mirrored in their gene expression profiles. A similar observation has been reported for other cancers: there was a clear segregation of the pediatric and adult germ cell tumors [21], and pediatric glioblastomas also have a characteristic transcriptome profile different from that of adult tumors [22,23]. The outcomes of melanoma in the younger and the elderly populations were also different and these 2 patient groups express distinct microRNA profiles [24]. Thus, age difference between patients with the same disease can be mirrored in their gene expression profiles. Patients of different ages but with the same tumor should be treated in different ways.
Gender disparity is a well known phenotype in HCC, and animal studies suggest that it may be due to the stimulatory effects of androgen and the protective effects of estrogen (see reviews [25,26]). Estrogen can protect hepatocytes from malignant transformation [27]. Intriguingly, both the androgen receptor (AR) and estrogen receptor 1 (ESR1) sex hormone receptors are down-regulated in yHCCs (Additional file 2: Table S1 & not shown). Genes involved in estrogen receptor signaling are also enriched in the yHCC signature (Figure 2B). Since all of our yHCC patients were sexually matured (the youngest case is a 26-year old female; Table 1), our data indicates an original and a unique pathogenesis mechanism in yHCCs.
HCC with stemness-related marker expression has recently been proposed to be a new and more aggressive subtype of HCC [28,29]. It is important that a suitable marker panel is developed to facilitate the diagnosis of this devastating HCC subtype. RT-qPCR analysis on elderly HCCs demonstrated that the high expression levels of 7 putative hepatic stem/progenitor cell biomarkers (including keratin 19 (K19), ABCG2, CD44, Nestin, CD133, EPCAM and OV6), is related to tumor angiogenesis and a poor prognosis for the HCC [15,28]. Recently, a stemness-related marker, CK19, was found well correlated with clinicopathologic features of tumor aggressiveness, vascular invasion, and poor differentiation in elderly HCCs [30]. No similar study has been addressed on yHCCs. Identifying genes involved in both cancer progression and cell dedifferentiation will offer another dimension to pathogenesis mechanisms, as well as providing novel therapeutic targets and prognosis markers. ILF3 (NF90) is one of the shared top genes between ESC and yHCC. LIF3 is among the ‘Core 9’ ESC genes highly re-expressed in advanced and poorly differentiated tumors [13] and is a prognostic factor in non-small cell lung cancer [31]. Another ESC gene overexpressed in yHCCs is DNMT1 and is responsible for the maintenance of DNA methylation patterns during replication. Inhibitors of this enzyme may potentially lead to DNA hypomethylation and re-expression of tumor suppressor genes [32]. Also, SOX13 contributes to control Wnt/TCF activity [33], crucial in HCC pathogenesis and cancer stem cell renewal [34]. Targeting these genes or pathways may restrain invasion by yHCC.
In addition to stemness genes, we also filtrated out 15 differentiation-related genes from in yHCCs. Eleven of these genes, including GSTK1 (glutathione S-transferase kappa 1) and SAR1B (SAR1 gene homolog B), are within the top 50 most down-regulated genes in yHCC patients (Additional file 2: Table S1; labeled with asterisks in Additional file 3: Figure S2). The repressed transcript levels and increased gene expression patterns during ESC hepatogenesis implied that these genes might function as novel tumor suppressor genes (TSGs). GSTK1 belongs to the glutathione S-transferase (GST) gene family that are critical for detoxification via conjugation of reduced glutathione (GSH) with numerous substrates such as pharmaceuticals and environmental pollutants [35]. GSTP1, another member of the GST family, has recently been identified to be a novel TSG for elderly HCCs, and the methylation frequency in GSTP1 is associated with HCC occurrence [36]. Roles of GSTK1 in yHCCs tumorigenesis and prognosis, as well as in ESC hepatogenesis, are awaited to be elucidated in the future.
Conclusion
This study revealed the molecular makeup of yHCC and the link between ESC traits and HCC subtypes. Therefore, molecular mechanisms in elderly HCC patients cannot be simply extrapolated to younger patients. Our results also helped to identify transcriptional programs that can be used as potential therapeutic targets for various HCC subgroups.
Methods
Patient profiles and microarray expression data sets
Data analysis and RNA isolation details were summarized in Additional file 4: Supplementary Materials and Methods online. The diagnosis of all the HCC patients had been tissue-verified by pathological examination of the surgically removed HCC and neighboring liver tissue. All 44 young HCCs (≤40 years old at the diagnosis; 23 cases in the training set while another 21 in the validation cohort) were positive for serum hepatitis B surface antigen (HBsAg), but negative for antibodies to hepatitis C virus (anti-HCV). All 48 elderly (>40 years old; 38 in the training set while another 10 in the validation cohort) HCC patients enrolled were also serum HBsAg positive and anti-HCV negative. The HCC samples used in this study were the original tumors obtained from the first operations of patients. The current study complies with the Helsinki Declaration. Informed consents for taking small part of the resected HCC and the surrounding non-tumor liver specimens for study were obtained from patients. The tissue sample analysis was approved by the Institutional Review Board of Taipei Veterans General Hospital (VGHIRB No.: 97-09-17A), Taiwan.
Fresh HCC tissues and non-tumor counter parts that had been removed during surgery were snap frozen and kept in liquid nitrogen for RNA extraction. All array data were deposited into the NCBI Gene expression omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database [37] with the accession number GSE45436 (see Additional file 1: Figure S1; training set 1 GSE45267, training set 2 GSE45434, and validation set GSE45435).
The embryonic stem cell (ESC) array data had been published previously [38]. HCV (+) HCC array data were downloaded from the GEO database (accession number GSE6764) [20]. Array data of the induced pluripotent stem cells (iPS cells) and ESCs, as well as their hepatic differentiated progenies, were from GEO dataset GSE14897 [19]. The second batch of elderly HCCs of the training data set were downloaded from the Expression Project for Oncology (expO) of the International Genomics Consortium (http://www.intgen.org/, accession number GSE2109 in the GEO database).
Abbreviations
HCC: Hepatocellular carcinoma; yHCC: Young HCC; ESC: Embryonic stem cells; HBV: Hepatitis B virus; HBsAg: Hepatitis B surface antigen; HCV: Hepatitis C virus; GEO: Gene expression omnibus; iPS: Induced pluripotent stem cells; FDR: False discovery rate; PCA: Principle component analysis; PS: Prediction strength; IPA: Ingenuity pathway analysis; GO: Gene Ontology; LOOCV: Leave-one-out-cross validation.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HW Wang and JC Wu conceived the study and identified its value to research. HW Wang and JC Wu designed the analysis approach. GY Chau, CW Su, and JC Wu collected tumor samples and carried out clinicopathologic feature analysis. HW Wang, SY Huang, TH Hsieh and CY Tung carried out the implementation of data analysis and wetlab experiments. HW Wang and JC Wu provided biological guidance during the analysis process. The manuscript was written by HW Wang and then revised by JC Wu. All authors read and approved the final manuscript.
Supplementary Material
A workflow summarizing experimental design and the filtration of yHCC genes.
Top 50 genes down-regulated in yHCC patients (q < 0.05, Young HCC vs. elder HCC).
Differentiation-related yHCC genes. A Venn diagram illustrates that 83 ESC hepatogenesis-related yHCC genes are also present in the 309 yHCC genes. A heat map based on these 83 genes is shown. Array data of H9 ESC and differentiated liver precursor cells (day 20, d20) [19] were from GEO dataset GSE14897. *: genes discussed in the text.
Supplementary Materials and Methods.
Contributor Information
Hsei-Wei Wang, Email: hwwang@ym.edu.tw.
Tsung-Han Hsieh, Email: smallbeard2003@hotmail.com.
SSu-Yi Huang, Email: silmariola@gmail.com.
Gar-Yang Chau, Email: gychau@vghtpe.gov.tw.
Chien-Yi Tung, Email: cytung@ym.edu.tw.
Chien-Wei Su, Email: cwsu2@vghtpe.gov.tw.
Jaw-Ching Wu, Email: jcwu@vghtpe.gov.tw.
Acknowledgements
The authors acknowledge the efforts of IGC and expO, and the array services provided by Microarray & Gene Expression Analysis Core Facility, which is supported by the National Research Program for Genomic Medicine, National Science Council (NSC), of the National Yang-Ming University VGH Genome Research Center (VYMGC). The authors also acknowledge the Taiwan Liver Cancer Network for providing the validating set of HCC RNAs.
This work is supported by grants from Taipei Veterans General Hospital (TVGH V97ER2-016 & Center of Excellence for Cancer Research at TVGH, DOH102-TD-C-111-007), National Science Council (NSC98-3112-B-010-017, NSC99-3112-B-010-017, and NSC100-2325-B-010-003), and Yang-Ming University (Ministry of Education, Aim for the Top University Plan). This work is also support in part by the UST-UCSD International Center for Excellence in Advanced Bioengineering sponsored by the Taiwan NSC I-RiCE Program under grant number NSC101-2911-I-009-101.
References
- Hsu CY, Huang YH, Hsia CY, Su CW, Lin HC, Loong CC, Chiou YY, Chiang JH, Lee PC, Huo TI. et al. A new prognostic model for hepatocellular carcinoma based on total tumor volume: the Taipei integrated scoring system. J Hepatol. 2010;53(1):108–117. doi: 10.1016/j.jhep.2010.01.038. [DOI] [PubMed] [Google Scholar]
- Chen CH, Su WW, Yang SS, Chang TT, Cheng KS, Lin HH, Wu SS, Lee CM, Changchien CS, Chen CJ. et al. Long-term trends and geographic variations in the survival of patients with hepatocellular carcinoma: analysis of 11,312 patients in Taiwan. J Gastroenterol Hepatol. 2006;21(10):1561–1566. doi: 10.1111/j.1440-1746.2006.04425.x. [DOI] [PubMed] [Google Scholar]
- Lam CM, Chan AO, Ho P, Ng IO, Lo CM, Liu CL, Poon RT, Fan ST. Different presentation of hepatitis B-related hepatocellular carcinoma in a cohort of 1863 young and old patients–implications for screening. Aliment Pharmacol Ther. 2004;19(7):771–777. doi: 10.1111/j.1365-2036.2004.01912.x. [DOI] [PubMed] [Google Scholar]
- Cho SJ, Yoon JH, Hwang SS, Lee HS. Do young hepatocellular carcinoma patients with relatively good liver function have poorer outcomes than elderly patients? J Gastroenterol Hepatol. 2007;22(8):1226–1231. doi: 10.1111/j.1440-1746.2007.04914.x. [DOI] [PubMed] [Google Scholar]
- Chen CH, Chang TT, Cheng KS, Su WW, Yang SS, Lin HH, Wu SS, Lee CM, Changchien CS, Chen CJ. et al. Do young hepatocellular carcinoma patients have worse prognosis? The paradox of age as a prognostic factor in the survival of hepatocellular carcinoma patients. Liver Int. 2006;26(7):766–773. doi: 10.1111/j.1478-3231.2006.01309.x. [DOI] [PubMed] [Google Scholar]
- Ni YH, Chang MH, Hsu HY, Hsu HC, Chen CC, Chen WJ, Lee CY. Hepatocellular carcinoma in childhood: clinical manifestations and prognosis. Cancer. 1991;68(8):1737–1741. doi: 10.1002/1097-0142(19911015)68:8<1737::AID-CNCR2820680815>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Kakizaki S, Sohara N, Sato K, Takagi H, Arai H, Abe T, Katakai K, Kojima A, Matsuzaki Y. et al. Hepatocellular carcinoma in young adults: the clinical characteristics, prognosis, and findings of a patient survival analysis. Dig Dis Sci. 2007;52(4):1103–1107. doi: 10.1007/s10620-006-9578-2. [DOI] [PubMed] [Google Scholar]
- Sezaki H, Kobayashi M, Hosaka T, Someya T, Akuta N, Suzuki F, Tsubota A, Suzuki Y, Saitoh S, Arase Y. et al. Hepatocellular carcinoma in noncirrhotic young adult patients with chronic hepatitis B viral infection. J Gastroenterol. 2004;39(6):550–556. doi: 10.1007/s00535-003-1341-2. [DOI] [PubMed] [Google Scholar]
- Tsai FC, Liu CJ, Chen CL, Chen PJ, Lai MY, Kao JH, Chen DS. Lower serum viral loads in young patients with hepatitis-B-virus-related hepatocellular carcinoma. J Viral Hepat. 2007;14(3):153–160. doi: 10.1111/j.1365-2893.2006.00780.x. [DOI] [PubMed] [Google Scholar]
- Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, Wang LY, Lu SN, You SL, Chen DS. et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100(16):1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295(1):65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- Wu JC, Huang YH, Chau GY, Su CW, Lai CR, Lee PC, Huo TI, Sheen IJ, Lee SD, Lui WY. Risk factors for early and late recurrence in hepatitis B-related hepatocellular carcinoma. J Hepatol. 2009;51(5):890–897. doi: 10.1016/j.jhep.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SJ, Wang TY, Tsai CY, Hu TF, Chang MD, Wang HW. Increased epithelial stem cell traits in advanced endometrial endometrioid carcinoma. BMC Genomics. 2009;10:613. doi: 10.1186/1471-2164-10-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XR, Xu Y, Yu B, Zhou J, Qiu SJ, Shi GM, Zhang BH, Wu WZ, Shi YH, Wu B. et al. High expression levels of putative hepatic stem/progenitor cell biomarkers related to tumour angiogenesis and poor prognosis of hepatocellular carcinoma. Gut. 2010;59(7):953–962. doi: 10.1136/gut.2008.176271. [DOI] [PubMed] [Google Scholar]
- Jen CH, Yang TP, Tung CY, Su SH, Lin CH, Hsu MT, Wang HW. Signature Evaluation Tool (SET): a Java-based tool to evaluate and visualize the sample discrimination abilities of gene expression signatures. BMC Bioinforma. 2008;9(1):58. doi: 10.1186/1471-2105-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27(42):5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- Greene FLPD, Fleming ID, Fritz A, Balch CM, Haller DG, Morrow M. AJCC cancer staging manual. 6. Chicago: Springer; 2002. p. 435. [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51(1):297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J. et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- Palmer RD, Barbosa-Morais NL, Gooding EL, Muralidhar B, Thornton CM, Pett MR, Roberts I, Schneider DT, Thorne N, Tavare S. et al. Pediatric malignant germ cell tumors show characteristic transcriptome profiles. Cancer Res. 2008;68(11):4239–4247. doi: 10.1158/0008-5472.CAN-07-5560. [DOI] [PubMed] [Google Scholar]
- Faury D, Nantel A, Dunn SE, Guiot MC, Haque T, Hauser P, Garami M, Bognar L, Hanzely Z, Liberski PP. et al. Molecular profiling identifies prognostic subgroups of pediatric glioblastoma and shows increased YB-1 expression in tumors. J Clin Oncol. 2007;25(10):1196–1208. doi: 10.1200/JCO.2006.07.8626. [DOI] [PubMed] [Google Scholar]
- Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, Bax DA, Coyle B, Barrow J, Hargrave D. et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukic DM, Rao UN, Kelly L, Skaf JS, Drogowski LM, Kirkwood JM, Panelli MC. Microrna profiling analysis of differences between the melanoma of young adults and older adults. J Transl Med. 2010;8:27. doi: 10.1186/1479-5876-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SH, Chen PJ. Gender disparity of hepatocellular carcinoma: the roles of sex hormones. Oncology. 2010;78(Suppl 1):172–179. doi: 10.1159/000315247. [DOI] [PubMed] [Google Scholar]
- Giannitrapani L, Soresi M, La Spada E, Cervello M, D’Alessandro N, Montalto G. Sex hormones and risk of liver tumor. Ann N Y Acad Sci. 2006;1089:228–236. doi: 10.1196/annals.1386.044. [DOI] [PubMed] [Google Scholar]
- Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317(5834):121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- Teufel A, Galle PR. Collecting evidence for a stem cell hypothesis in HCC. Gut. 2010;59(7):870–871. doi: 10.1136/gut.2009.206672. [DOI] [PubMed] [Google Scholar]
- Rountree CB, Mishra L, Willenbring H. Stem cells in liver diseases and cancer: recent advances on the path to new therapies. Hepatology. 2012;55(1):298–306. doi: 10.1002/hep.24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee JE, Cho JY, Yoo JE, Choi JS, Park YN. Human hepatocellular carcinomas with “Stemness”-related marker expression: keratin 19 expression and a poor prognosis. Hepatology. 2011;54(5):1707–1717. doi: 10.1002/hep.24559. [DOI] [PubMed] [Google Scholar]
- Guo NL, Wan YW, Tosun K, Lin H, Msiska Z, Flynn DC, Remick SC, Vallyathan V, Dowlati A, Shi X. et al. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Cancer Res. 2008;14(24):8213–8220. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson LR, Tatham AL, Lin Z, Denny WA. Epigenetic regulation of gene expression as an anticancer drug target. Curr Cancer Drug Targets. 2011;11(2):199–212. doi: 10.2174/156800911794328510. [DOI] [PubMed] [Google Scholar]
- Marfil V, Moya M, Pierreux CE, Castell JV, Lemaigre FP, Real FX, Bort R. Interaction between Hhex and SOX13 modulates Wnt/TCF activity. J Biol Chem. 2010;285(8):5726–5737. doi: 10.1074/jbc.M109.046649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Mishra L. Cancer stem cells and hepatocellular carcinoma. Cancer Biol Ther. 2009;8(18):1691–1698. doi: 10.4161/cbt.8.18.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Vasiliou V. Analysis of the glutathione S-transferase (GST) gene family. Hum Genomics. 2004;1(6):460–464. doi: 10.1186/1479-7364-1-6-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N, Kudo M, Nagasaka T, Ikai I, Goel A. Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma. Hepatology. 2012;56(3):994–1003. doi: 10.1002/hep.25706. [DOI] [PubMed] [Google Scholar]
- Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TS, Hsieh JY, Wu YH, Jen CH, Tsuang YH, Chiou SH, Partanen J, Anderson H, Jaatinen T, Yu YH. et al. Functional network reconstruction reveals somatic stemness genetic maps and dedifferentiation-like transcriptome reprogramming induced by GATA2. Stem Cells. 2008;26(5):1186–1201. doi: 10.1634/stemcells.2007-0821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A workflow summarizing experimental design and the filtration of yHCC genes.
Top 50 genes down-regulated in yHCC patients (q < 0.05, Young HCC vs. elder HCC).
Differentiation-related yHCC genes. A Venn diagram illustrates that 83 ESC hepatogenesis-related yHCC genes are also present in the 309 yHCC genes. A heat map based on these 83 genes is shown. Array data of H9 ESC and differentiated liver precursor cells (day 20, d20) [19] were from GEO dataset GSE14897. *: genes discussed in the text.
Supplementary Materials and Methods.