Abstract
We recently demonstrated that the phytotoxin cichorine is produced by Aspergillus nidulans. Through a set of targeted deletions, we have found a cluster of seven genes that are required for its biosynthesis. Two of the deletions yielded molecules that give information about the biosynthesis of this metabolite.
Introduction
Filamentous fungi produce a variety of low-molecular weight secondary metabolites, many of which have been proven to possess remarkable biological activities. They include the cholesterol-lowering agent lovastatin and the immunosuppressant cyclosporine.1 It came as a surprise, then, that the analysis of recently sequenced fungal genomes suggested that the number of metabolites obtained in laboratory conditions is still far fewer than the number of putative secondary metabolite genes.2-5
This realization has prompted efforts to activate these silent genes through various approaches, including epigenetic modification,6 promoter exchange with an inducible promoter,7 controlled expression of a gene coding for a pathway-specific transcription factor,8 and co-cultivation with bacteria.9 A different strategy, however, is to alter the media conditions in which the fungus is cultivated, as different environmental cues may switch on heretofore unactivated pathways.10 For the filamentous fungus Aspergillus nidulans, this approach has been key to the isolation of aspoquinolones,11 aspernidine A/B,12 and F9775 A and B.13
With this approach we obtained the polyketide cichorine from the cultivation of Aspergillus nidulans on YES (Yeast Extract Sucrose) plates at 37°C for 5 days, during the course of our work with prenylated xanthones from this species.14 Cichorine is a phytotoxin active against knapweed, corn, and soybeans.15-16 It was isolated from Aspergillus silvaticus as well as Alternaria cichorii, which produces foliar blight in the important pest Russian knapweed.15 It had not before been observed in A. nidulans. The compact, functionalized isoindolin-1-one framework was an attractive target for total synthesis.17-18 Compounds featuring this framework have been shown to possess antimicrobial,19 anti-HIV,20 and antitumor properties.21
Because the genome of A. nidulans has been sequenced and relatively well-annotated,2 and because a straightforward gene targeting system has been developed for this species,22-23 we could proceed to efficiently investigate the genes that are responsible for the formation of cichorine. The presence of an aromatic group in cichorine indicated that it is a product of a nonreduced polyketide synthase (NR-PKS). Targeted gene deletions revealed that AN6448.4Δ alone failed to produce any detectable amount of cichorine. (We use the gene nomenclature of the Central Aspergillus Data Repository, CADRE, http://www.cadre-genomes.org.uk/, and the Aspergillus genome database, http://www.aspgd.org/.)
In our current investigation, we endeavored to identify other genes that are responsible for cichorine formation anticipating that intermediate compounds accumulating as a result of interruption of the pathway would help us understand how cichorine is synthesized.
Results and discussion
Genes required for the synthesis of a particular secondary metabolite tend to cluster together in the genome in fungi. For this reason we deleted genes both upstream and downstream of the identified PKS gene, AN6448.4 (Table 1). All strains carried the deletion of stcJ, a gene responsible for the carcinogenic secondary metabolite sterigmatocystin.24 Eliminating stcJ may facilitate isolation of other metabolites and also free up precursors to boost their production. The strains also carried nkuAΔ to improve frequencies of correct gene targeting.22
Table 1.
A. nidulans strains used in this study
| Strain | Genotype | Source |
|---|---|---|
|
stcJ
Δ
LO2026 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB | 6 |
|
AN11921Δ
LO5000-LO5002 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;AN11921::AfpyrG | This study |
|
cicA
Δ
LO4517-LO4519 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;cicA::AfpyrG | This study |
|
cicB
Δ
LO4482-LO4484 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;cicB::AfpyrG | This study |
|
cicC
Δ
LO4487-LO4488 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;cicC::AfpyrG | This study |
|
cicD
Δ
LO4492, LO4494 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;cicD::AfpyrG | This study |
|
cicE
Δ
LO4497-LO4499 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;cicE::AfpyrG | This study |
|
cicF
Δ
LO4503-LO4505 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;cicF::AfpyrG | This study |
|
cicG
Δ
LO4568-LO4569 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;cicG::AfpyrG | This study |
|
cicH
Δ
LO4507-LO4508 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;cicH::AfpyrG | This study |
|
AN6450Δ
LO4975-LO4977 |
pyrG89; pyroA4, nkuA::argB; riboB2, stcJ::AfriboB;AN6450::AfpyrG | This study |
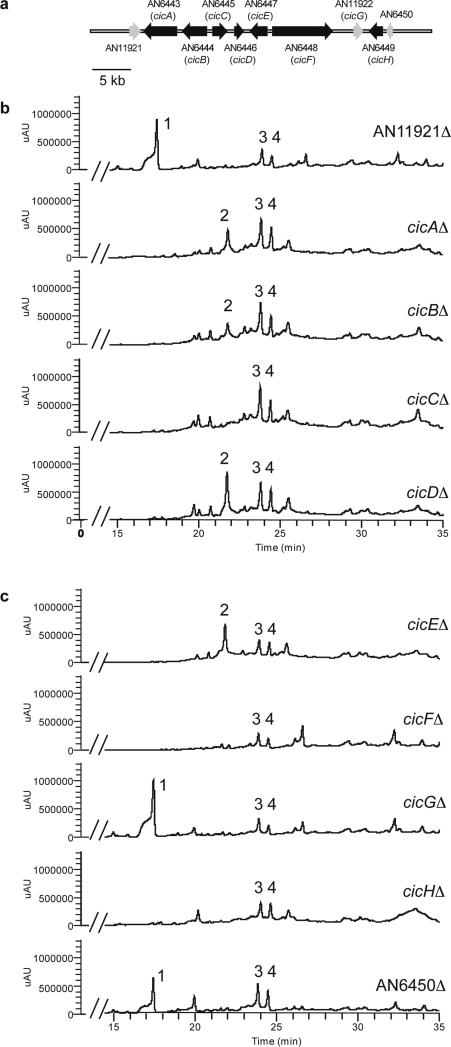
All strains were cultivated in the same cultivation medium that yields cichorine. (Cichorine has also been generated via cultivation in Raulin-Thom media.25) All deletions were verified by diagnostic PCR (see Experimental section). The result was the elimination of cichorine in strains ranging from AN6443.4Δ to AN6449.4Δ, with the exception of AN11922.4Δ (Figure 1). Table 2 lists the putative functions of the genes within the cluster and the ones immediately outside it. We now designate these genes as cicA-cicH. Aside from the PKS gene, the cluster contains a transporter (cicA) and transcriptional activator gene (cicD), and also four tailoring genes (cicB, cicC, cicE, and cicH).
Figure 1.
(A) Organization of the genes surrounding the PKS cicF involved in cichorine biosynthesis. Genes shown in black are required for cichorine biosynthesis, whereas those in gray are not. (B) HPLC extracts of strains AN11921Δ to cicDΔ as detected by UV absorbance at 254 nm. (C) The extracts (as in B) of cicEΔ to AN6450Δ. (D) Structures of cichorine (1), asperthecin (2), austinol (3), and dihydroaustinol (4). The biochemical origins of 2-4, differing from cichorine, have been previously determined.27-28
Table 2.
Putative functions of the genes within the cichorine cluster, as indicated from BLAST searches.
| Gene designation | Putative function | CADRE/AspGD annotation | Broad annotation |
|---|---|---|---|
| AN11921 | Amino acid transporter | AN11921.4 | ANID_11921.1 |
| cicA | ABC transporter | AN6443.4 | ANID_6443.1 |
| cicB | Conserved hypothetical protein | AN6444.4 | ANID_6444.1 |
| cicC | Oxidoreductase | AN6445.4 | ANID_6445.1 |
| cicD | Regulatory protein | AN6446.4 | ANID_6446.1 |
| cicE | O-methyltransferase | AN6447.4 | ANID_6447.1 |
| cicF | Polyketide synthase | AN6448.4 | ANID_6448.1 |
| cicG | Conserved hypothetical protein | AN11922.4 | ANID_11922.1 |
| cicH | Cytochrome P450 | AN6449.4 | ANID_6449.1 |
| AN6450 | Ribosomal protein | AN6450.4 | ANID_6450.1 |
Initially, we were unable to detect any obvious biosynthetic intermediates from the knockout strains. This was perhaps to be expected for the transporter, transcriptional activator, and backbone PKS genes, but it was reasonable to predict that at least some of the tailoring genes would yield intermediates upon deletion. Some polyketides feature a carboxylic acid motif or otherwise become negatively charged in aqueous media. It stands to reason, then, that these molecules will not partition into the organic layer during extraction. In these cases, it is necessary to neutralize the molecules with concentrated acid. This strategy was necessary for the acquisition of the polyketides orsellinic acid and F9775 A and B in A. nidulans.13
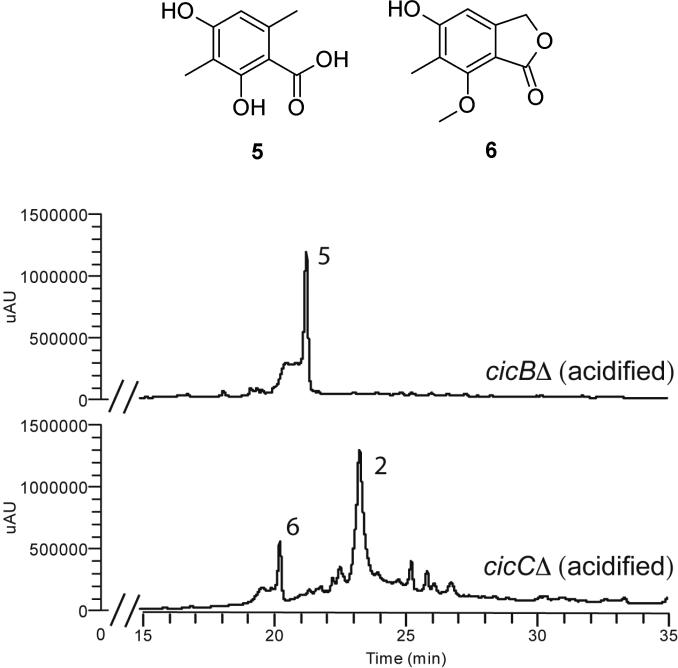
Acidifying the aqueous media allowed us to detect one compound from the cicBΔ strain and an additional compound from the cicCΔ strain. To determine their structures, we scaled up the cultivation and purified the metabolites using silica gel chromatography followed by preparative HPLC. Chemical structures were determined by 1H and 13C NMR, and the spectra were also compared with literature data.
The cicBΔ intermediate is a simple polyketide, 5, 3-methylorsellinic acid (Figure 2). Interestingly, this is the same molecule that we have recently acquired through the upregulation of the PKS gene, cicF. The cicCΔ intermediate, nidulol, 6, has been isolated from Aspergillus and Alternaria species, including a strain of A. nidulans, and it was found to be mildly cytotoxic toward human epidermoid carcinoma KB cells.26
Figure 2.
3-methylorsellinic acid (5) and nidulol (6), intermediates acquired from acidified extracts of cichorine deletant strains cicBΔ and cicCΔ, respectively.
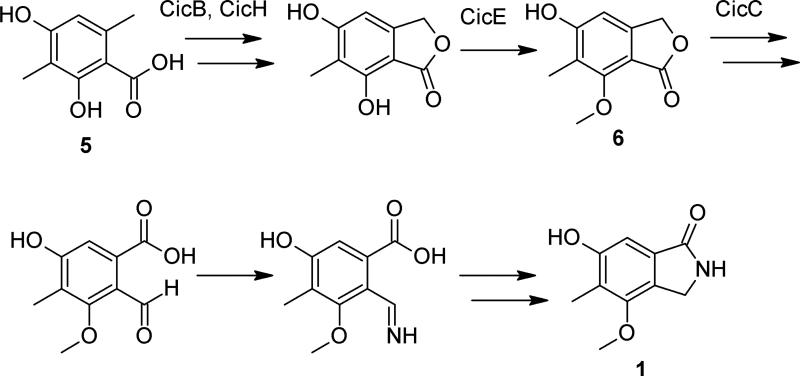
The deletants and the intermediates allow a better understanding of cichorine biosynthesis. Because upregulation of the backbone PKS gene cicF and deletion of cicB yield the same molecule, it is reasonable to suspect that the first step in cichorine biosynthesis is the generation of 5, followed by formation of the next (unidentified) intermediate catalyzed by the gene product of cicB. Interestingly, the cytochrome P450 monoxygenase gene cicH is homologous to mpaD in Penicillium brevicompactum, which is predicted to be catalyze the lactonization of the well-known immunosuppressant mycophenolic acid. Therefore, cicH may play a similar ring-closing role for cichorine.
The O-methyltransferase gene, cicE, is plausibly the responsible gene for the methylation of one of the phenol groups. Functionalized, two-ring 6 from the cicCΔ strain may be a later-stage intermediate, but lactone to lactam formation must still occur in order to generate cichorine. Other genes would be required for this conversion, and these genes are not found within this cluster. Based on this information, we have outlined a hypothetical biosynthesis as depicted in Figure 3. Although the entire set of genes pertaining to a particular fungal secondary metabolite may fit in one single cluster, it is being increasingly found in A. nidulans that such genes may be split into at least two distinct genomic loci, demonstrated with the prenylated xanthones14 and the terpenes27 from A. nidulans, and possibly with F9775 A and B.13
Figure 3.
Proposed biosynthesis of cichorine (1).
Conclusion
In summary we have shown that genes from a distinct cluster are required to generate cichorine in A. nidulans. The cluster contains the PKS gene, one regulatory and transporter gene each, and four genes involved in the tailoring of the polyketide backbone. We have also acquired two intermediates, improving our understanding of the biosynthesis of this isoindolin-1-one-based molecule.
Experimental
Generation of fusion PCR fragments A. nidulans protoplasting, and transformation
The gene deletions were performed using established gene targeting procedures.23 Two ~1000 base pair fragments upstream and downstream of every targeted gene were amplified from A. nidulans genomic DNA using PCR. Primers used in this study are listed in Table S1. The two amplified flanking sequences and an A. fumigatus pyrG selectable marker cassette were fused together by PCR using nested primers. A. nidulans strains in this study are listed in Table 1. Protoplast generation and transformation were utilized as previously described.23 The strain LO2026 carrying a deletion of the stcJ gene that eliminates sterigmatocystin production was used as the recipient strain. Diagnostic PCR of the deletant strains was performed employing the external primers from the first round of PCR. The difference in the size between the gene replaced by the selective marker and the native gene enabled us to determine if the transformants carried correct gene replacements. For further verification, diagnostic PCR was performed two more times, with one of the external primers and a primer located inside the marker gene, then the other external primer and an internal primer. In these cases, the deletants yielded the PCR product of the expected size whereas no product was present in non-deletants.
Fermentation and LC/MS analysis
YES medium was prepared by combining 20 g yeast extract, 120 g sucrose, 20 g agar, and 2 mL trace element solution in 1 L H2O. For the LC/MS screening experiments, spores of LO2026 (the control strain) and three strains of each gene deletant were individually inoculated (1 × 107 spores) onto 10 × 150 mm petri dishes which contained YES agar, and they were cultivated for five days at 37°C. Following, the agar was split into ~2 cm2 pieces, and the material was placed into 250 mL Ehrlenmeyer flasks and covered with methnol. The flasks were placed in a sonicator (Branson, Model 5510) for an hour. The methanol was then decanted. The pieces were covered with 1:1 methanol:dichloromethane, followed by another hour of sonication. The combined organic solvents were removed in vacuo, then partitioned between H2O (25 mL) and ethyl acetate (25 mL × 2). The combined ethyl acetate layers were evaporated. The dried, crude material was dissolved at a concentration of 20 mg/mL in DMSO, and then a sample was diluted 5-fold in methanol for LC/MS analysis.
LC/MS was performed using a ThermoFinnigan LCQ Advantage ion trap mass spectrometer with an RP C18 column (Alltech Prevail; 2.1 × 100 mm with a 3 μm particle size) at a flow rate of 125 μL/min and monitored by a UV detector at 254 nm. The solvent gradient was 95% MeCN-H2O (solvent B) in 5% MeCN-H2O (solvent A) both containing 0.05% formic acid: 0% B from 0 to 5 min, 0 to 100% B from 5 to 35 min, 100% B from 35 to 40 min, 100% B to 0% B from 40 to 45 min, and reequilibration with 0% B from 45 to 50 min.
Isolation of metabolites
The LO2026 (stcJΔ), LO3387 (cicBΔ), and LO3337 (cicCΔ) strains were each cultivated in 25 × 150 mm petri dishes containing YES medium for 5 days at 37°C. As with LC/MS analysis, the agar was chopped and sonicated in methanol, then 1:1 methanol:dichloromethane. The organic material was evaporated and extracted 4x with an equal volume of ethyl acetate. For LO3387 and LO3337, the aqueous layer was acidified with concentrated HCl to a pH of 2. The crude material was subjected to silica gel column chromatography, using 98:2 dichloromethane:methanol as the eluent for 1 and 5, and 100% dichloromethane for 6. The materials were further separated by preparative HPLC [Phenomenex Luna 5 μm C18 (2), 250 × 21.2 mm] with a flow rate of 5.0 mL/min and measured by a UV detector at 250 nm. See Supplementary Information for more details about isolation.
Cichorine (1)
white powder; 1H NMR and 13C NMR data (DMSO-d6), in good agreement with the published data.17,181H NMR (DMSO-d6): δ = 2.04 (3H, s), 3.83 (3H, s), 4.39 (2H, s), 6.81 (1H, s), 7.40 (1H, br s), 8.42 (1H, s), 9.76 (1H, s); 13C NMR (DMSO-d6): δ= 10.1, 43.9, 59.5, 103.7, 119.7, 123.8, 132.7, 154.2, 157.1, 170.6. For UV-Vis and ESIMS spectra, see Fig. S1. m/z (M+H) = 194.4; calculated = 194.1.
3-methylorsellinic acid (5)
white powder; 1H NMR and 13C NMR data (acetone-d6), in good agreement with the published data.291H NMR (acetone-d6): δ = 2.03 (3H, s), 2.48 (3H, s), 6.35 (1H, s); 13C NMR (acetone-d6): δ= 7.4, 23.6, 107.4, 108.6, 110.6, 140.5, 160.3, 174.2. For UV-Vis and ESIMS spectra, see Fig. S1. m/z (M-H) = 181.2; calculated = 181.1.
Nidulol (6)
white powder; 1H NMR and 13C NMR data (acetone-d6), in good agreement with the published data.301H NMR (acetone-d6): δ = 2.11 (3H, s), 3.98 (3H, s), 5.15 (2H, s), 6.87 (1H, s); 13C NMR (acetone-d6): δ= 8.0, 61.3, 68.5, 103.6, 108.2, 118.1, 148.8, 158.4, 163.3, 168.6. For UV-Vis and ESIMS spectra, see Fig. S1. m/z (M-H) = 193.6; calculated = 193.1.
Supplementary Material
Acknowledgements
The work was supported by grant PO1GM084077 from the National Institute of General Medical Sciences to BRO and CCCW and by the Kansas University Endowment Fund to BRO.
References
- 1.Cox RJ. Org. Biomol. Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 2.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 3.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto KI, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu JJ, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 4.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafton A, Latge JP, Li WX, Lord A, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, de Cordoba SR, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, de Aldana CRV, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 5.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, van Dijck PWM, van Dijk A, Dijkhuizen L, Driessen AJM, d'Enfert C, Geysens S, Goosen C, Groot GSP, de Groot PWJ, Guillemette T, Henrissat B, Herweijer M, van den Hombergh J, van den Hondel C, van der Heijden R, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel M, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij N, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun JB, Ussery D, Varga J, Vervecken W, de Vondervoort P, Wedler H, Wosten HAB, Zeng AP, van Ooyen AJJ, Visser J, Stam H. Nat. Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 6.Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Nat. Chem. Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CCC. J. Am. Chem. Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Nat. Chem. Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 9.Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bode HB, Bethe B, Hofs R, Zeeck A. Chembiochem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Scherlach K, Hertweck C. Org. Biomol. Chem. 2006;4:3517–3520. doi: 10.1039/b607011f. [DOI] [PubMed] [Google Scholar]
- 12.Scherlach K, Schuemann J, Dahse HM, Hertweck C. J. Antibiot. 2010;63:375–377. doi: 10.1038/ja.2010.46. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Oakley CE, Bok JW, Keller N, Oakley BR, Wang CCC. Mol. Biosyst. 2010;6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, Wang CCC, Oakley BR. J. Am. Chem. Soc. 2011;133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stierle A, Hershenhorn J, Strobel G. Phytochemistry. 1993;32:1145–1149. [Google Scholar]
- 16.Fujita M, Yamada M, Nakajima S, Kawai K, Nagai M. Chem. Pharm. Bull. 1984;32:2622–2627. doi: 10.1248/cpb.32.2622. [DOI] [PubMed] [Google Scholar]
- 17.Moreau A, Couture A, Deniau E, Grandclaudon P, Lebrun S. Org. Biomol. Chem. 2005;3:2305–2309. doi: 10.1039/b504602e. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Valdez G, Olguin-Uribe S, Millan-Ortiz A, Gamez-Montano R, Miranda LD. Tetrahedron. 2011;67:2693–2701. [Google Scholar]
- 19.Breytenbach JC, van Dyk S, van den Heever I, Allin SM, Hodkinson CC, Northfield CJ, Page MI. Bioorg. Med. Chem. Lett. 2000;10:1629–1631. doi: 10.1016/s0960-894x(00)00306-1. [DOI] [PubMed] [Google Scholar]
- 20.Zhao XZ, Maddali K, Marchand C, Pommier Y, Burke TR. Bioorg. Med. Chem. 2009;17:5318–5324. doi: 10.1016/j.bmc.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardcastle IR, Ahmed SU, Atkins H, Farnie G, Golding BT, Griffin RJ, Guyenne S, Hutton C, Kallblad P, Kemp SJ, Kitching MS, Newell DR, Norbedo S, Northen JS, Reid RJ, Saravanan K, Willems HMG, Lunec J. J. Med. Chem. 2006;49:6209–6221. doi: 10.1021/jm0601194. [DOI] [PubMed] [Google Scholar]
- 22.Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Nat. Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 24.Brown DW, Adams TH, Keller NP. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14873–14877. doi: 10.1073/pnas.93.25.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawahara N, Nozawa K, Nakajima S, Udagawa SI, Kawai KI. Chem. Pharm. Bull. 1988;36:398–400. doi: 10.1248/cpb.36.398. [DOI] [PubMed] [Google Scholar]
- 26.Phuwapraisirisan P, Rangsan J, Siripong P, Tip-pyang S. Nat. Prod. Res. 2009;23:1063–1071. doi: 10.1080/14786410802265415. [DOI] [PubMed] [Google Scholar]
- 27.Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CCC, Oakley BR. Appl. Environ. Microbiol. 2008;74:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, Chiang YM, Oakley BR, Wang CCC. J. Am. Chem. Soc. 2012;134:4709–4720. doi: 10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishitoba Y, Nishimura H, Nishiyama T, Mizutani J. Phytochemistry. 1987;26:3181–3185. [Google Scholar]
- 30.Achenbach H, Muhlenfeld A, Brillinger GU. Liebigs. Ann. Chem. 1985;1985:1596–1628. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.