Abstract
Fusarium oxysporum is a devastating plant pathogen that oxidizes C18 fatty acids sequentially to jasmonates. The genome codes for putative dioxygenase (DOX)-cytochrome P450 (CYP) fusion proteins homologous to linoleate diol synthases (LDSs) and the allene oxide synthase (AOS) of Aspergillus terreus, e.g., FOXB_01332. Recombinant FOXB_01332 oxidized 18:2n-6 to 9S-hydroperoxy-10(E),12(Z)-octadecadienoic acid by hydrogen abstraction and antarafacial insertion of molecular oxygen and sequentially to an allene oxide, 9S(10)-epoxy-10,12(Z)-octadecadienoic acid, as judged from nonenzymatic hydrolysis products (α- and γ-ketols). The enzyme was therefore designated 9S-DOX-AOS. The 9S-DOX activity oxidized C18 and C20 fatty acids of the n-6 and n-3 series to hydroperoxides at the n-9 and n-7 positions, and the n-9 hydroperoxides could be sequentially transformed to allene oxides with only a few exceptions. The AOS activity was stereospecific for 9- and 11-hydroperoxides with S configurations. FOXB_01332 has acidic and alcoholic residues, Glu946-Val-Leu-Ser949, at positions of crucial Asn and Gln residues (Asn-Xaa-Xaa-Gln) of the AOS and LDS. Site-directed mutagenesis studies revealed that FOXB_01332 and AOS of A. terreus differ in catalytically important residues suggesting that AOS of A. terreus and F. oxysporum belong to different subfamilies. FOXB_01332 is the first linoleate 9-DOX with homology to animal heme peroxidases and the first 9-DOX-AOS fusion protein.
Keywords: cytochrome P450 class III, heme peroxidase, linoleate diol synthase, mutagenesis site-specific, oxylipin biosynthesis, oxygenation mechanism
Polyunsaturated fatty acids can be oxidized to biological mediators throughout the eukaryotic world by a large repertoire of oxygenating enzymes and synthases. In humans, the medically most important oxidative pathway is initiated by cyclooxygenases (COXs), which oxidize arachidonic acid to prostaglandin (PG)H2 (1). The latter is transformed by a series of specific synthases to PGs of the D, E, and F2α series and to prostacyclin and thromboxane A2 by specific cytochrome P450s (CYPs) (1, 2).
Jasmonic acid (JA) is a plant hormone and has a similar cyclopentane element as PGs (3, 4). JA is formed in plants from 18:3n-3 or 16:3n-3 in a series of sequential oxidations and transformations, which have been studied intensively (3, 5). The first step of JA biosynthesis is catalyzed by plant 13-lipoxygenases (LOXs), which form 13S-hydroperoxides. The latter are transformed to allene oxides by specific CYPs, and by allene oxide cyclases to the five carbon ring structure, 12-oxo-phytodienoic acid (3, 5). Allene oxides are also formed from 9S-hydroperoxides in plants (6, 7). JA is an important signal molecule in plant defense, growth, and development (5, 8), and it is also of commercial value in the perfume industry as a fragrance.
JA was first isolated and named from the essential oil of Jasminum grandiflorum in 1962, and the biosynthesis of JA was elucidated 22 years later by Vick and Zimmerman (8, 9). Fungal biosynthesis of JA and its derivatives was discovered in 1970 in Gibberella fujikuroi and Lasiodiplodia theobromae, and subsequently in Fusarium oxysporum and a few other species (10–13). In spite of prominent biosynthesis of JA by certain strains, the mechanism of fungal JA biosynthesis has not been determined. The analysis is complicated due to the highly variable capacity to form JA between different fungal isolates, and future work will be needed to reveal the regulation of JA biosynthesis. It is possible that the first steps of biosynthesis of the five-membered ring of JA occur by the same mechanism as in plants: oxidation and transformation of 18:3n-3 by 13S-LOX, allene oxide synthase (AOS), and allene oxide cyclase. This is supported by the identical absolute configuration of plant and fungal JA and its metabolites (11, 14), the detection of 12-oxo-phytodienoic acid as a metabolite of L. theobromae (15), and the recent characterization of 13S-LOX of F. oxysporum (16).
Aspergillus terreus does not form JA, but this fungus oxidizes 18:2n-6 sequentially to 9R-hydroperoxy-10(E),12(Z)-octadecadienoic acid (HPODE) and an allene oxide, 9R(10)-epoxy-11,(12Z)-octadecadienoic acid [9R(10)-EODE] (17). The AOS was recently identified in the CYP domain of a dioxygenase (DOX)-CYP fusion protein (ATEG_02036), whereas the origin of the 9R-DOX activity could not be determined (18).
The F. oxysporum species complex contains a wide range of plant pathogens and producers of a plethora of mycotoxins, and it comprises two of the top 10 fungal pathogens, F. oxysporum and Fusarium graminearum (19). Their genomes have been sequenced to facilitate research on their pathogenic mechanism and this information is available at the Fusarium Comparative Database. We hypothesized that F. oxysporum might produce allene oxides because its genome encodes homologous enzymes to the AOS of A. terreus. FOXB_01332 could be aligned to this AOS with high amino acid identity (∼50%, see below) (supplementary Fig. I).
The AOS of A. terreus belongs to the family of fungal DOX-CYP fusion proteins. The latter contain linoleate 8R-DOX or 10R-DOX in their N-terminal domains and 5,8- or 7,8-hydroperoxide isomerases or an AOS in the C-terminal CYP domains (18, 20–23); 9R-DOX activity could not be detected in the recombinant AOS of A. terreus. This fusion protein thus differs from the AOS of the coral Plexaura homomalla, which contains an N-terminal catalase-like domain with AOS activity and a C-terminal 8R-LOX domain (24).
The DOX domains of DOX-CYP fusion enzymes are homologous to COX, as illustrated in Fig. 1 by a comparison of the tetramer sequence including the tyrosine residue which catalyzes hydrogen abstraction and the proximal histidine heme ligand. These tetrameric sequences of ATEG_02036 and FOXB_01332 are identical, but differ at two positions from COX and at one from 5,8- and 7,8-linoleate diol synthase (LDS) with 8R-DOX activities (Fig. 1). Whether the evolutionary replacement of Trp with Phe in ATEG_02036 and FOXB_01332 supports DOX activities is unknown.
Fig. 1.

Comparison of partial sequences of DOX and CYP domains. A: Partial alignment of the DOX domains of COX-1 and -2, 7,8- and 5,8-LDS, ATEG_02036, and FOXB_01332 by ClustalW. The catalytically important Tyr residue and the proximal His ligand are in red, and differences to COX within the tetramer are marked blue. B: Partial alignment of CYP domains of 7,8- and 5,8-LDS by ClustalW. C: Partial alignment of ATEG_02036 and FOXB_01332 by ClustalW. The conserved motifs of aspergilli with catalytically important Asn and Gln residues are marked red along with the aligned residues of FOXB_01332 marked blue. 7,8-LDSgg, 7,8-LDS from Gaeumannomyces graminis; 5,8-LDSat, 5,8-LDS from A. terreus.
Comparison of the C-terminal sequences of ATEG_02036 and FOXB_01332 revealed an interesting difference. The Asn and Gln residues in the pentamer sequence (Ala-Asn-Xaa-Xaa-Gln) of 5,8-LDS and the AOS of A. terreus are of catalytic importance (18). These amide residues of 7,8-LDS and 5,8-LDS support the heterolytic cleavage of the dioxygen bonds of 8R-HPODE, and Asn964 of the AOS facilitates homolytic cleavage of the dioxygen bond of 9R-HPODE (18, 25). The ClustalW alignment suggests that Asn964 and Gln967 of the AOS of A. terreus are replaced by an acidic and an alcoholic residue, Glu946 and Ser949 respectively, in FOXB_01332 (Fig. 1). The latter sequence also contains two amide residues, Asn921 and Gln924, not far from the homologous position of Asn964 of ATEG_02036 (supplementary Fig. I).
The first objective of the present investigation was to express FOXB_01332 and to study its catalytic activity. Our experiments demonstrate that FOXB_01332 is a 9S-DOX-AOS fusion protein. The 9S-DOX motif, Tyr-Arg-Phe-His, occurs in a subset of fungal DOX-CYP fusion proteins (Fig. 1). The second objective was to determine whether the Phe residue in this sequence was essential for 9S-DOX activity. The third objective was to compare potentially important residues of the two fungal AOSs.
EXPERIMENTAL PROCEDURES
Materials
Unsaturated fatty acids (99%) were from Merck, Sigma, and Larodan, and were stored in 50–100 mM ethanol (−20°C). [13C18]18:2n-6 (98%) and 13-hydroxy-12-oxo-(9Z),(15Z)-octadecadienoic acid were from Larodan. 9-HPODE was prepared by photooxidation, purified by normal phase HPLC, and the S and R stereoisomers of 9-HPODE were partly separated on Reprosil Chiral NR-R (18, 26); the latter contained a few percent of the S stereoisomer. 9S-HPODE, 13S-HPODE, 13S-hydroperoxyoctadecatrienoic acid (HPOTrE), 13R-HPODE, and 13R-HPOTrE were prepared enzymatically [tomato LOX (27), soybean LOX-1, and 13R-MnLOX (28)] and purified by silica chromatography. The [11,11-2H2]18:2n-6 (>95%), [11S-2H]18:2n-6 (>98%), and [11R-2H]18:2n-6 (25%) were generous gifts of Dr. M. Hamberg (Karolinska Institute, Stockholm). The Champion pET Directional TOPO kit was from Invitrogen. Restriction enzymes and chemically competent Escherichia coli (NEB5α) were from New England BioLabs, Fermentas, and Invitrogen. The plasmid Midi kit was from Macherey-Nagel and the gel extraction kit and Pfu DNA polymerase were from Fermentas. Phusion DNA polymerase was from New England BioLabs. RNaseA and ampicillin were from Sigma. Primers were obtained from TIB MOLBIOL (Berlin, Germany). Sequencing was performed at Uppsala Genome Center (Rudbeck Laboratories, Uppsala University).
Expression in E. coli
The open reading frame of FOXB_01332 (GenBank, EGU88194) was purchased in pUC57 from GenScript (Hong Kong) and was subcloned to pET101D-TOPO vectors by PCR technology (forward primer: 5′-caccatgtcgttcaatgaaaag, reverse primer: 5′-atttcccaaaatctcatcctttc) following Invitrogen's instructions. The amplicon was ligated into pET101/D-TOPO and introduced into E. coli (BL21) by heat-shock transformation. Cells were grown to A600 0.6–0.8 in low salt Luria Bertani medium and protein expression was induced by 0.1 mM isopropyl β-D-1-thiogalactopyranoside. Cultures were grown for 5 h at room temperature with moderate shaking (∼100 rpm). Cells were harvested by centrifugation and sonicated (Bioruptor Next Gen, 10 × 30 s, 4°C) (23). At least three independent expressions were analyzed.
Site-directed mutagenesis
Site-directed mutagenesis was performed according to the Quick Change protocol (Stratagene) in pUC57 constructs (23). Amplicons were obtained from 10 ng of template by Pfu DNA polymerase (16 cycles) before digestion with Dpn1 (2 h, 37°C). Amplification of one distinct PCR product was confirmed by agarose gel electrophoresis before heat-shock transformation (NEB5α). Primers containing the designated replacements are listed in the supplementary material (supplementary Table I). All mutations were confirmed by sequencing before subcloning to pET101D-TOPO as described above.
Enzyme assays
Recombinant 9S-DOX-AOS of the bacterial lysate was incubated with 50–100 μM of fatty acids (16:3n-3, 18:1n-9, 18:1n-6, 18:2n-6, 18:3n-3, 18:3n-6, 18:4n-3, 20:2n-6, 20:3n-6, and 20:4n-6) for 30–40 min on ice. Mutants of 9S-DOX-AOS from triplicate transformations were incubated with 18:2n-6 in the same way. The reactions (0.3–0.5 ml) were terminated with ethanol (4 vol), and proteins were removed by centrifugation. The metabolites were extracted on octadecyl silica (SepPak/C18), evaporated to dryness, and dissolved in 40–50 μl of ethanol or 2% isopropyl alcohol in hexane. Five to ten microliters were subjected to LC-MS/MS analysis. Conversion of R and S stereoisomers of 9-HPODE and 13-HPOTrE, respectively, was assayed in the same way.
LC-MS/MS analysis of oxylipins
Reversed phase (RP)-HPLC with MS/MS analysis was performed with a Surveyor MS pump (Thermo Fisher) and an octadecyl silica column (5 μm, 2.1 × 150 mm; Phenomenex), which was usually eluted at 0.3–0.5 ml/min with methanol/water/acetic acid, 800/200/0.05 or 750/250/0.05. The effluent was subject to electrospray ionization in a linear ion trap mass spectrometer (LTQ, Thermo Fisher) with monitoring of carboxylate anions. The heated transfer capillary was set at 315°C; the ion isolation width at 1.5, 4, or 5 amu; the collision energy at 35 (arbitrary scale); and the tube lens varied between 90 and 120 V. PGF1α was infused for tuning. The mass spectra were recorded every second and consisted of an average of five microscans, each with a maximum injection time of 0.2 s. The mass spectra were routinely analyzed with selective ion monitoring of characteristic ions and also checked by subtraction of background spectra before and after the peaks of interest. These chromatograms were analyzed with the Xcalibur software with peak integration and manual selection of the integration set points.
Normal phase HPLC with MS/MS analysis was performed with a silicic acid column (5 μm, Kromasil 100 Si, 250 × 2 mm; Chrom Tech) using hexane/isopropyl alcohol/acetic acid, 98/2/0.01 or 97/3/0.01, at 0.3 ml/min (P2000, Thermo). The effluent was mixed with isopropyl alcohol/water, 60/40, at 0.2–0.3 ml/min from a second pump (Surveyor MS pump). The combined effluents were introduced by electrospray into the ion trap mass spectrometer (LTQ).
Steric analysis of 9-HODE by chiral phase (CP)-HPLC-MS/MS was performed with cellulose tribenzoate coated on silica gel (5 μm, 250 × 2 mm; Chiracel OB-H, Daicel Chemical Company) and the effluent (hexane/isopropyl alcohol/acetic acid, 95/5/0.01) was mixed with isopropyl alcohol/water as above. Steric analysis of hydroperoxyoctadecamonoenoic acid (HPOME) was performed with CP-HPLC-MS/MS analysis (8 μm, 250 × 2 mm, Reprosil Chiral NR; Dr. Maisch GmbH, Ammerbuch, Germany) (26). The stereoisomers of 9-hydroxy-10-oxo-(12Z)-octadecenoic acid were resolved and analyzed by CP-HPLC-MS/MS (5 μm, 2 × 250 mm; Reprosil Chiral AM) as described (17).
Determination of deuterium kinetic isotope effects
Deuterium kinetic isotope effects (D-KIEs) were estimated from parallel incubations of 9S-DOX-AOS with 100 μM [11,11-2H2]18:2n-6 and 100 μM [13C18]18:2n-6 (4°C, 30 min). The incubations were terminated with ethanol and combined. The metabolites were extracted (SepPak/C18), usually reduced by triphenylphosphine, and analyzed by LC-MS/MS. α-Ketols were the main products, and we estimated the D-KIEs from the conversion of [11,11-2H2]18:2n-6 to α-ketols with one or no deuterium label (the remaining deuterium at C-11 was partly lost from the α-ketol due to keto-enol tautomerism) in comparison with the formation of [13C18]-labeled α-ketol. We also estimated the D-KIEs from LC-MS/MS analysis of the ratio of [11-2H]9-HODE (m/z 296 → 171) and [13C18]9-HODE (m/z 313 → 180).
Bioinformatics
Proteins were aligned with the ClustalW algorithm (Lasergene, DNASTAR, Inc.). Phylogenetic trees were constructed with the MEGA5 software with bootstrap tests of the resulting nodes (29, 30). The distance within branches was based on the number of expected substitutions per amino acid position.
RESULTS
Bioinformatics
The genomes of the F. oxysporum complex, which are sequenced at the Broad Institute, were investigated for homologs of the AOS of A. terreus. FOXB_01322 of F. oxysporum Fo5176 (GenBank, EGU88194) could be aligned with ∼50% amino acid identity with the AOS of A. terreus (GenBank, AGH14485 or ATEG_02036). An alignment of the two sequences is shown in supplementary Fig. I. As expected, the NCBI database identified LDS- and CYP-like domains in FOXB_01332. We therefore decided to investigate FOXB_01322 for DOX and AOS activities.
As outlined in Fig. 1 and the introduction, the short sequence with the proximal heme ligand and the important Tyr residue for 8R- and 10R-DOX catalysis were identical in FOXB_01332 and ATEG_02036. All ascomycetous sequences with over 50% identity to FOXB_01332/ATEG_02036 contained a Trp instead of the Phe in the Tyr-Arg-Phe-His motif of the DOX domains.
FOXB_01332 and ATEG_02036 differed in an interesting way in the CYP domain as mentioned above. The AOS and 5,8-LDS of A. terreus contain the conserved sequence Ala-Asn-Xaa-Xaa-Gln, and the amide residues, Asn and Gln, are important for catalysis (18). At the homologous positions of FOXB_01332, Asn964 and Gln967 are replaced with an acidic (Glu946) and an alcoholic residue (Ser949). There are several Asn residues in the putative I-helix of FOXB_01332, and one of them is present within a similar tetramer, Asn921-Val-Asp-Gln (supplementary Fig. I).
Oxidation of α-linolenic linoleic acids
Biosynthesis of 9S-hydroperoxides.
FOXB_01332 was expressed in E. coli and was found to possess both 9S-DOX and AOS activities and will therefore be referred to as 9S-DOX-AOS.
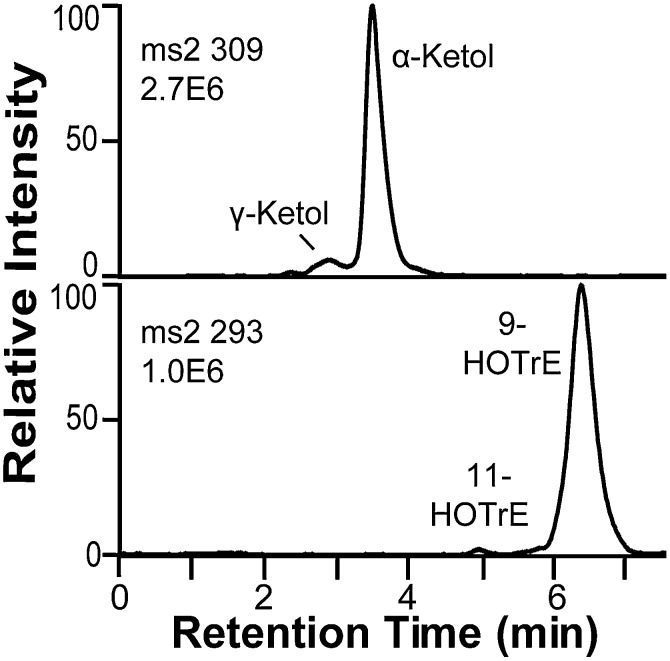
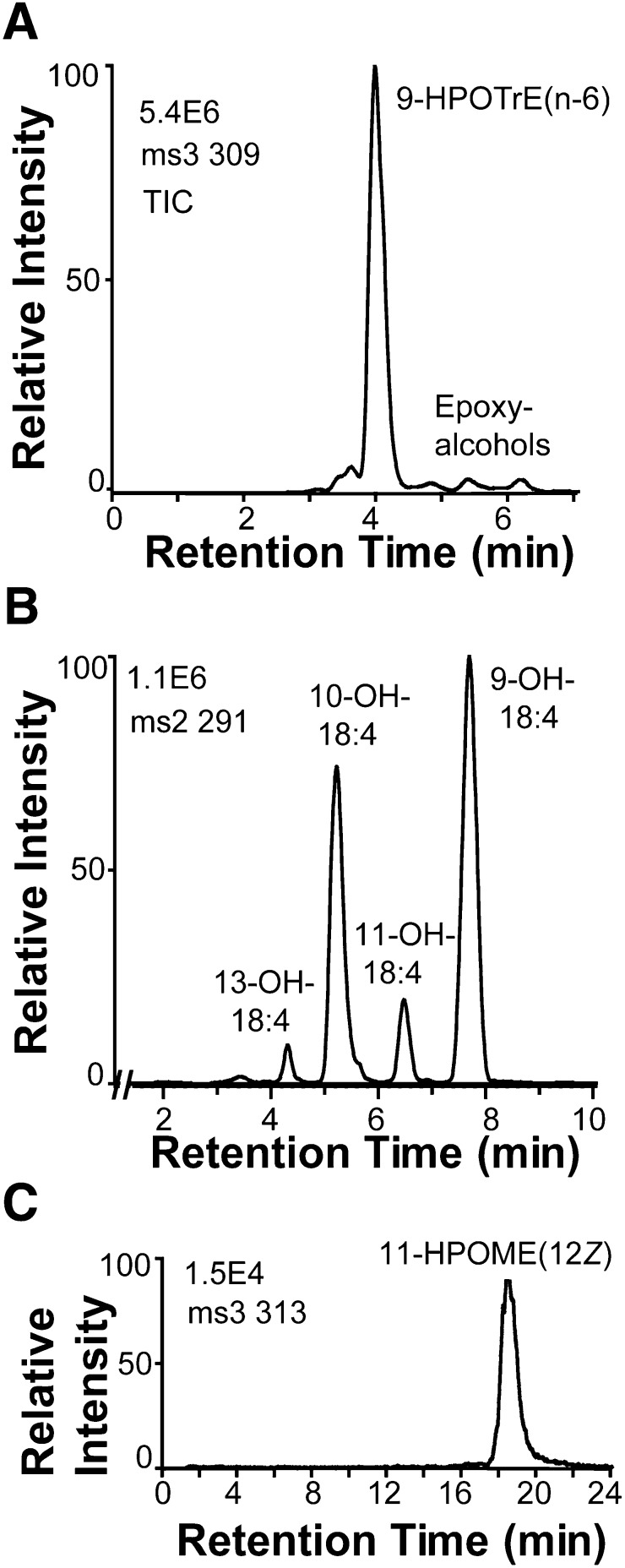
Oxidation of 18:3n-3 by 9S-DOX-AOS yielded 9-hydroxy-10(E),12(Z),15(Z)-octadecatrienoic acid (9-HOTrE), traces of 11-HOTrE, and two polar metabolites (Fig. 2). The MS3 spectra of 9-HPOTrE and 11-HPOTrE and the MS/MS spectra of the corresponding alcohols were as reported (31–33).
Fig. 2.
Oxidation of 18:3n-3 by recombinant FOXB_01332 and analysis of products by RP-HPLC-MS/MS. Top trace, MS2 analysis (m/z 309 → full scan) of α- and γ-ketols, likely formed from the allene oxide, 9S(10)-epoxy-10,12(Z),15(Z)-octadecatrienoic acid, by hydrolysis (see text for details). Bottom trace, MS2 analysis (m/z 293 → full scan) of HOTrEs. The hydroperoxides were reduced with triphenylphosphine before LC-MS analysis.
The 18:2n-6 was oxidized by 9S-DOX-AOS in the same way as 18:3n-3. Steric analysis by CP-HPLC-MS/MS showed that 95% 9S-HODE and 5% 9R-HODE were formed (Fig. 3A). Small amounts of the trans isomer of 9-HODE were also detected. The relatively low stereospecificity of 9S-DOX compared with 8R- and 10R-DOX is likely due to rapid transformation of 9S-HPODE by the AOS, which leads to the accumulation of autoxidation products.
Fig. 3.
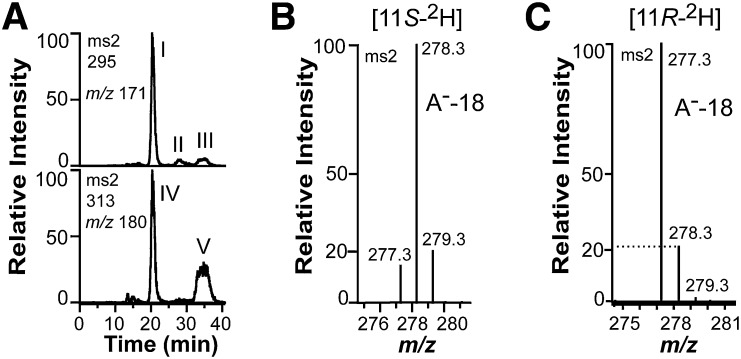
Oxidation of 18:2n-6, [11S-2H]18:2n-6, and [11,11-2H2]18:2n-6 by recombinant 9S-DOX-AOS (FOXB_01332). A: Steric analysis by CP-HPLC-MS/MS of the main hydroperoxide formed from 18:2n-6 by 9S-DOX-AOS after reduction to an alcohol (top trace). Small amounts of racemic [13C18]9-HODE were added to facilitate the analysis (bottom trace). The main biological product after reduction of hydroperoxides to alcohols, peak I, coeluted with the 9S stereoisomer of [13C18]9-HODE (peak IV) and not with [13C18]9R-HODE (peak V). Small amounts of 9R-HODE (peak III) and trans-trans 9-HODE (peak II) were also detected (top trace). B: Partial MS/MS spectrum of 9-HODE from oxidation of [11S-2H]18:2n-6 (95% 2H) by recombinant FOXB_01332. The deuterium label was retained as judged from the prominent signal at m/z 278 [296-18; loss of water from the carboxylate anion (A−)]. C: Partial MS/MS spectrum of 9-HODE from oxidation of [11R-2H]18:2n-6 (25% 2H) by the recombinant enzyme. The deuterium label was not retained as judged from the signals at m/z 277 and 278 [loss of water from the carboxylate anions (A−)]. The intensity of the latter ion was ∼20% of the intensity of m/z 277, and this is in agreement with the natural abundance of the 13C isotope (1.1% 13C at each of the 18 positions).
Oxidation of [11S-2H]18:2n-6 and [11R-2H]18:2n-6 revealed that the proS hydrogen was retained in 9S-HODE (Fig. 3B) and that the proR hydrogen was lost (Fig. 3C). This is in agreement with antarafacial hydrogen abstraction and oxygenation.
Non-enzymatic hydrolysis of allene oxides to ketols.
The MS/MS spectrum of the major polar metabolite of 18:2n-6 was identical to the α-ketol 9-hydroxy-10-oxo-12(Z)-octadecenoic acid (17). This MS/MS spectrum and the corresponding MS/MS spectrum of 9-hydroxy-10-oxo-12(Z),15(Z)-octadecadienoic acid showed characteristic ions with even mass at m/z 200. The MS3 spectra (m/z 309 → 291 → full scan and m/z 311 → 293 → full scan, respectively) of these metabolites also contain an informative ion at m/z 263 (291-28, loss of CO) and m/z 265, respectively (18). The MS/MS spectrum of the minor polar product of 18:2n-6 was similar to that of the γ-ketol, 13-hydroxy-10-oxo-11(E)-octadecenoic acid (17). In analogy, the MS/MS spectrum of the minor polar product of 18:3n-3 was consistent with 9-hydroxy-10-oxo-12(Z),15(Z)-octadecadienoic acid.
Product profile over time and chromatographic reproducibility.
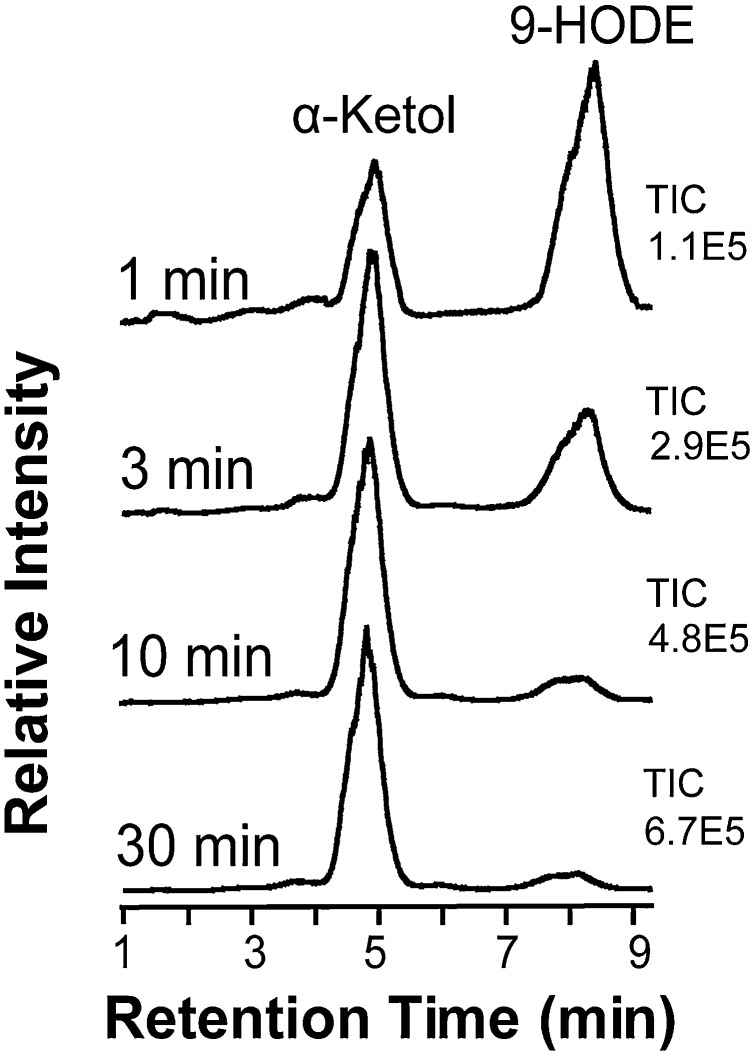
Analysis of the total ion current during RP-HPLC-MS/MS analysis of metabolites formed from 18:2n-6 by 9S-DOX-AOS showed that 9-HODE was the main product. The ion current averaged 67% at short incubation time (1 min), whereas the α-ketol was the main product after 3 min (63%) and dominated at later time points (10 min, 87%; 30 min, 91%), as illustrated in Fig. 4. The corresponding numbers of the total ion current of a second experiment were 73% 9-HPODE at 1 min, and 61, 89, and 94% α-ketol at the later time points. Ten replicate analyses of the same sample by RP-HPLC-MS/MS showed that the product profile was reproducible, as the relative amount of the α-ketol in this sample averaged 72.4% with a standard deviation of 1.55%.
Fig. 4.
Time profile of products formed from 18:2n-6 by 9S-DOX-AOS. 9S-DOX-AOS was incubated for 1, 3, 10, and 30 min, and the products were analyzed by RP-HPLC-MS/MS after reduction of hydroperoxides to alcohols by triphenylphosphine. The chromatograms show the total ion current (TIC), which is normalized to the height of the largest peak, and the absolute numbers are given to the right of each chromatogram.
Steric analysis of 9-hydroxy-10-oxo-12(Z)-octadecenoic acid.
These ketols are likely formed by hydrolysis of allene oxides, 9S(10)-epoxy-10,12(Z),15(Z)-octadecatrienoic acid and 9S(10)-epoxy-10,12(Z)-octadecadienoic acid [9S(10)-EODE], respectively (17, 18). This was supported by CP-HPLC-MS/MS analysis of 9-hydroxy-10-oxo-12(Z)-octadecenoic acid (Fig. 5A). The latter mainly consisted of the 9R-hydroxy isomer, which is in agreement with nonenzymatic hydrolysis of 9S(10)-EODE formed by potato stolons (17).
Fig. 5.
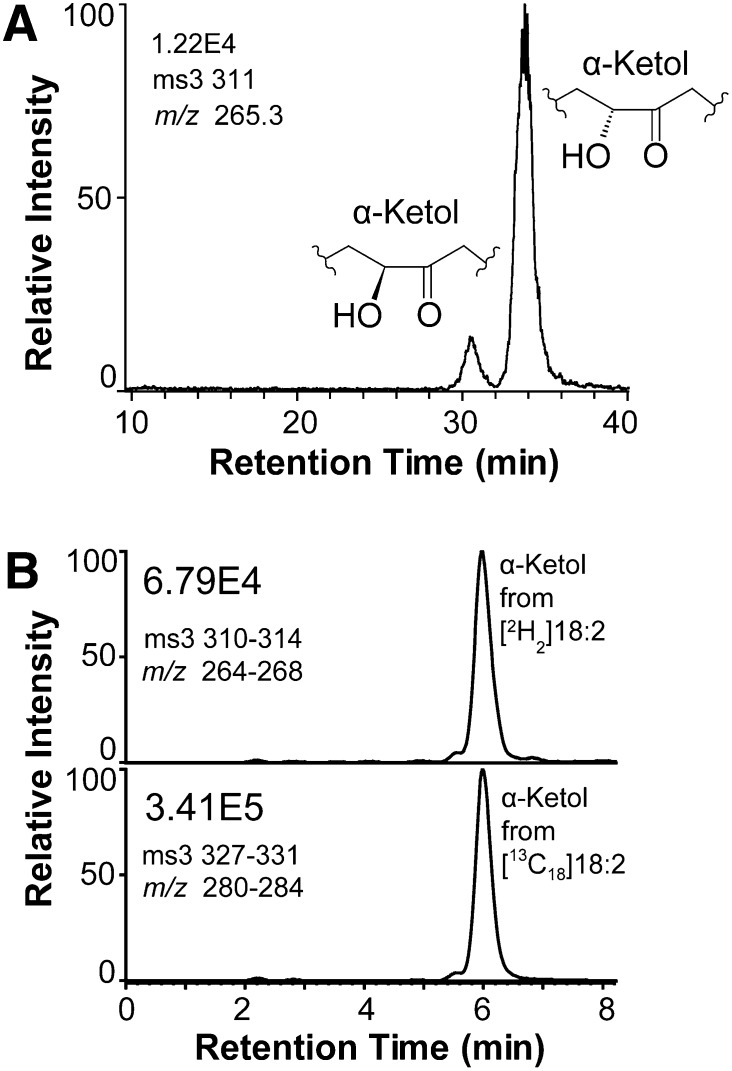
CP-HPLC analysis of the α-ketols and estimation of the D-KIE of 9S-DOX-AOS. A: Steric analysis of the α-ketols, which are formed in different isomeric amounts due to steric factors during nonenzymatic hydrolysis of 9S(10)-EODE (17). B: Apparent D-KIE from the formation of the α-ketol from [11,11-2H2]18:2n-6 and from [13C18]18:2n-6. The 9S-DOX activity abstracts the proR hydrogen at C-11, whereas the α-ketol may lose the remaining deuterium label at C-11 due to keto-enol tautomerism. The D-KIE was therefore estimated by MS3 with a mass window of four during the first and second selection, as indicated in the figure.
D-KIE of 9S-DOX-AOS.
The D-KIE during the sequential oxidation to 9-HPODE and α-ketols was estimated by comparison of the rate of oxidation of [11,11-2H2]18:2n-6 and [13C18]18:2n-6 as illustrated in Fig. 5B. The D-KIE for formation of α-ketols ranged between 3 and 5, suggesting little or no hydrogen tunneling in the sequential biosynthesis of the 9-hydroperoxide and the allene oxide/α-ketol. Analysis of the formation rate of the 9-hydroperoxide from deuterated and 13C-labeled 18:2n-6 yielded similar results. We conclude that 9S-DOX catalyzes hydrogen abstraction with a small D-KIE in analogy with other DOX of the animal peroxidase gene family.
Transformation of hydroperoxides by AOS
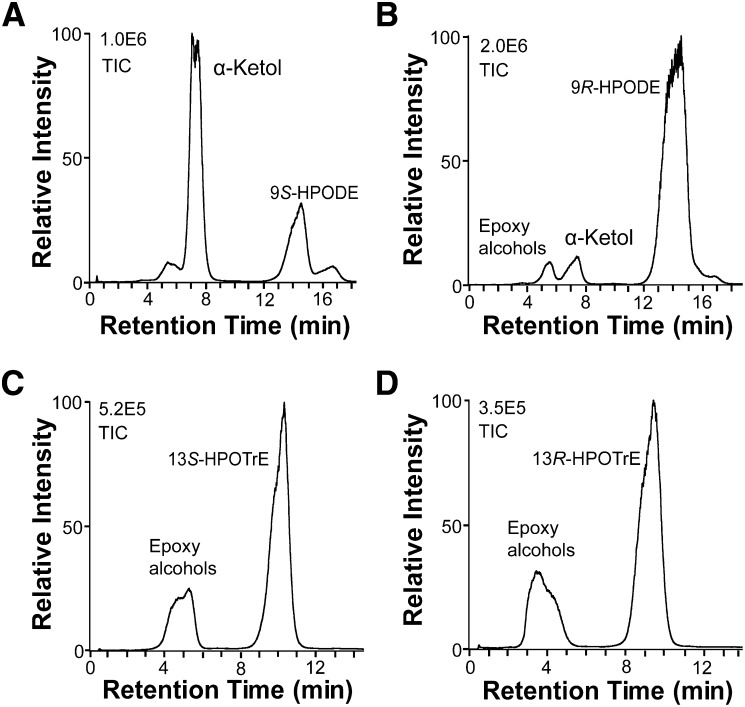
9S-DOX-AOS converted 9S-HPODE to α- and γ-ketols, likely formed by hydrolysis of 9S(10)-EODE as discussed above, along with small amounts of epoxy alcohols (Fig. 6A). The 9R-HPODE preparation contains a few percent 9S-HPODE, and only the latter was apparently transformed to an α-ketol by 9S-DOX-AOS (Fig. 6B). 13S-HPODE, 13S-HPOTrE, 13R-HPODE, and 13R-HPOTrE were not transformed to detectable amounts of allene oxides/α-ketols, but variable amounts of epoxy alcohols accumulated (Fig. 6). We compared MS3 spectra of the epoxy alcohols formed from 13S-HPOTrE for ions in the MS3 spectrum of authentic 13-hydroxy-12-oxo-(9Z),(15Z)-octadecadienoic acid (α-ketol), but could not detect significant amounts of this α-ketol. The epoxy alcohols could be formed by heme of the DOX and the CYP domains, and 13R- and 13S-HPODE were transformed to 11-hydroxy-12(13)-epoxy- and 9-hydroxy-12(13)-epoxy-octadecadienoic acids in almost equal relative amounts (cf. supplementary Fig. II).
Fig. 6.
LC-MS analysis of the transformation of hydroperoxides by 9S-DOX-AOS. A: 9S-HPODE was transformed to α- and γ-ketols. B: The 9R-HPODE preparation, which contained small amounts of 9S-HPODE, was only transformed to small amounts of the α-ketol. C, D: 13S-HPOTrE and 13R-HPOTrE were mainly converted to epoxy alcohols. α-Ketols could not be detected in (C) and (D). TIC, total ion current.
Oxidation of other fatty acids
The 18:3n-6 was oxidized at C-9, but this hydroperoxide was not transformed by the AOS activity (Fig. 7A). The 18:4n-3 was oxidized by hydrogen abstraction at C-11 and C-8 with oxygen insertion mainly at C-9, C-10, and C-11 (Fig. 7B). The 9-hydroperoxide of 18:4n-3 was partly transformed by the AOS activity with hydrolysis to an α-ketol as judged from the MS/MS and MS3 spectra with similar fragmentation to the α-ketols described above.
Fig. 7.
LC-MS analysis of the oxidation of 18:3n-6, 18:4n-3, and 18:1n-6 by 9S-DOX-AOS. A: The 18:3n-6 was transformed to the 9-hydroperoxide (9-HPOTrE) as the main product and to traces of epoxy alcohols. B: The 18:4n-3 was oxidized mainly at C-9, C-10, and C-11, and relatively small amounts of α-ketol were also detected (data not shown). C: The 18:1n-6 was oxidized at C-11 and resolved by CP-HPLC to one major isomer and this hydroperoxide was likely the 11R stereoisomer of 11-HPOME; oxygen is presumably inserted at C-11 from the same direction as in the biosynthesis of 9S-HPODE, but designated 11R-HPOME due to the Cahn-Ingold-Prelog priority rules. The metabolites were separated by normal phase HPLC. TIC, total ion current.
The 18:1n-9 and 18:1n-6 were oxidized at C-11. CP-HPLC revealed that 11-HPOME(9Z) was almost racemic, whereas 11-HPOME(12Z) consisted of one stereoisomer (Fig. 7C), likely 11R-HPOME, based on the antarafacial hydrogen abstraction and oxygen insertion during oxidation of 18:2n-6 discussed above.
We next examined the effect of chain elongation to C20 and shortening to C16. The 20:2n-6 and 20:3n-6 were oxidized at C-11, and these hydroperoxides were almost completely further transformed by the AOS activity followed by nonenzymatic hydrolysis to α-ketols (supplementary Fig. III). The 20:4n-6 was oxidized at C-11, but 11-hydroperoxy-5(Z),8(Z),12(E),14(Z)-eicosatetraenoic acid accumulated and it was much less efficiently transformed by the AOS activity in comparison with 11-hydroperoxy-8(Z),12(E),14(Z)-eicosatrienoic acid and 11-hydroperoxy-12(E),14(Z)-eicosadienoic acid (supplementary Fig. III).
The 16:3n-3 was a poor substrate and was mainly oxidized by hydrogen abstraction at C-12 with formation of 10-hydro(pero)xy-7(Z),11(E),13(Z)-hexadecatrienoic acid without further transformation by the AOS activity (data not shown). This metabolite is also formed by L. theobromae (34).
We conclude that the DOX activity appears to have a relatively broad substrate specificity for unsaturated C18 to C20 fatty acids, whereas the AOS activity appears to be specific for 9S- and 11S-hydroperoxides of C18 and C20 fatty acids with the notable exception of 9S-HPOTrE(n-6) (Fig. 7A).
Site-directed mutagenesis of 9S-DOX-AOS
Site-directed mutagenesis of 9S-DOX.
The conserved Tyr-(His/Arg)-Trp-His motif contains the catalytic Tyr residue, which is oxidized by the heme to a radical that performs the hydrogen abstraction of LDS and COX (Fig. 1). ATEG_02036 and FOXB_01332 retain the Tyr residue but have a Phe residue instead of a Trp in this sequence. To assess whether this residue could be of catalytic importance, we replaced Phe416 of FOXB_01332 with a Trp residue. The Phe416Trp mutant oxidized 18:2n-6 to 9-HPODE with formation of the α-ketol as the main product (>90%), but an increased formation of other hydroperoxides could not be detected. We conclude that the replacement of Phe416 with Trp does not affect the product profile of FOXB_01332.
Site-directed mutagenesis of the AOS.
The LDS and AOS of A. terreus contain the conserved sequence Ala-Asn-Xaa-Xaa-Gln in their CYP domains (Fig. 1). These Asn and Gln residues were shown to be crucial for the hydroperoxide isomerase activities of 5,8- and 7,8-LDS, respectively, and the Asn residue for the AOS activity (18, 25). At the homologs positions according to ClustalW alignments, 9S-DOX-AOS contains an acidic and an alcoholic residue (Glu946 and Ser949, respectively) (Fig. 1). In order to determine whether these residues were critical for the AOS activity, we replaced them with Val and Ala residues, respectively. The mutant Glu946Val oxidized 18:2n-6 as the native enzyme with formation of the α-ketol as the main product after 30 min, as judged from the total ion current (range 87–93%; n = 3). The Ser949Ala mutant also formed α-ketol as the native enzyme. This acid-alcohol pair thus lacks catalytic importance for the AOS activity.
The putative I-helix of the AOS domain contains several Asn residues, one of which is present in the sequence Asn921-Val-Asp-Gln (supplementary Fig. I). Asn residues appear to be critical for all AOSs identified so far (18, 20, 35, 36). We therefore examined whether replacement of Asn921 with Val could influence the AOS activity of FOXB_01332 in analogy with Asn964 of AOS of A. terreus. The Asn921Val mutant oxidized 18:2n-6 with formation of 9-HPODE, α-ketols, and γ-ketols as the native 9S-DOX-AOS (data not shown).
DISCUSSION
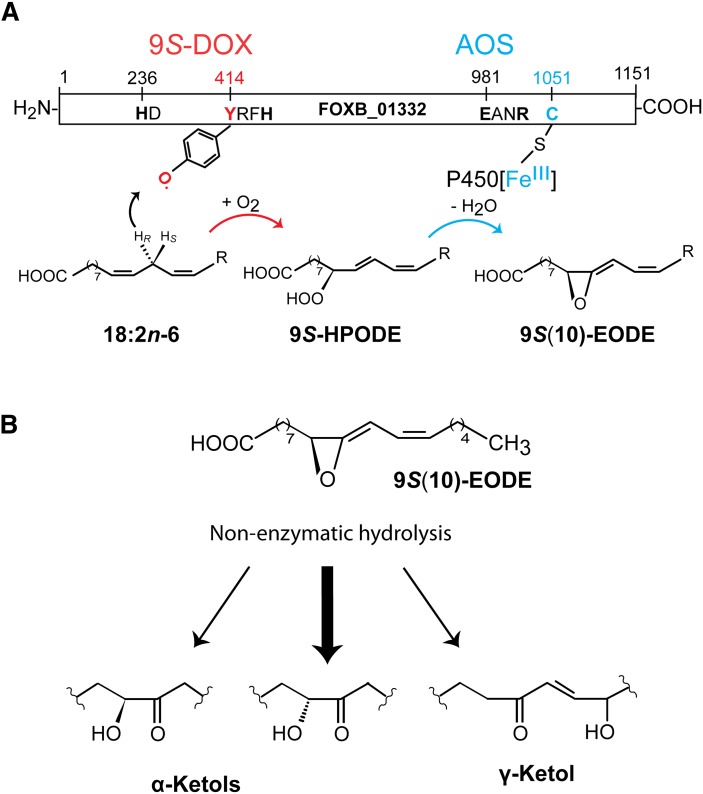
We have expressed the first 9S-DOX, and it belongs to the animal heme peroxidase gene family. This enzyme was found to be fused with a CYP domain, which encodes an AOS. Our report demonstrates a novel route to allene oxides without the need of a LOX. An overview of the catalytic domains and the reaction mechanism of 9S-DOX-AOS are shown in Fig. 8A.
Fig. 8.
Overview of 9S-DOX-AOS domains, the biosynthesis of 9S-HPODE and 9S(10)-EODE from 18:2n-6, and the nonenzymatic hydrolysis of 9S(10)-EODE. A: Overview of the 9S-DOX and AOS domains. The former contains the distal heme ligand, His236 (marked HD) and the catalytic domain with Tyr414. The latter is likely oxidized to a tyrosyl radical, which abstracts the proR hydrogen at C-11 and molecular oxygen is inserted at C-9. The AOS domain contains the salt bridge (Glu981 and Arg984) and the heme thiolate ligand (Cys1051). CYP[FeIII] catalyzes homolytic scission of the oxygen bond with formation of CYP[FeIVOH]. The latter desaturates the 9(10)-epoxide intermediate with a radical at C-11 and forms the double bond between C-10 and C-11. B: Nonenzymatic hydrolysis of 9S(10)-EODE yields α-ketols and small amounts of γ-ketols. The relative amounts of the two α-ketols differ during nonenzymatic hydrolysis of 9S(10)-EODE due to steric hindrance at C-9.
Biological relevance
The discovery of the 9S-DOX-AOS has biological implications. It extends the family of fungal DOX-CYP fusion enzymes, which now consists of LDS, 10R-DOX, AOS, and 9S-DOX-AOS. The two AOSs form different allene oxides, 9S(10)-EODE and 9R(10)-epoxy-11,(12Z)-octadecadienoic acid, and they are not homologous to plant AOSs of the CYP74 family (35). The two AOSs also differ in the active site as the position of the catalytically important Asn941 residue for homolytic cleavage of 9R-HPODE is not conserved in the homologous position of 9S-DOX-AOS (18). The fact that F. oxysporum has the capacity to form allene oxides from 9S-HPODE in analogy with plants suggests that these allene oxides may have biological functions, but little is yet known about the biological role of AOS other than in JA biosynthesis (8), and whether additional AOSs occur in pathogens of the Fusarium complex.
9S-DOX of F. oxysporum and 9R-DOX of A. terreus
The 9S-DOX activity of FOXB_01332 with 18:2n-6 and 18:3n-3 as substrates shows many similarities to 9S-LOX of plants, and it may therefore be mistaken as a LOX. 9S-DOX and 9-LOX catalyze antarafacial hydrogen abstraction and oxygenation (27, 37). A detailed analysis revealed two characteristic differences between hydrogen abstraction by a tyrosyl radical and by a LOX metal center with a catalytic base, e.g., Fe3+OH−. First, the D-KIE of 9S-DOX-AOS was in the same order of magnitude as reported for COX (38), and at least one order of magnitude less than the D-KIE of sLOX-1 (39). Second, 9S-DOX oxidizes 18:1n-6 by hydrogen abstraction at C-11 in analogy with the efficient oxidation of 18:1n-9 at C-8 by 8R-DOX of LDS, whereas monounsaturated fatty acids are poor substrates of LOX (26, 40, 41).
A. terreus and L. theobromae possess prominent 9R-DOX activities, which are linked to AOS (17, 34). The 9R-DOX activity of A. terreus retains a broad substrate specificity and a D-KIE of the same magnitude as 9S-DOX, but it catalyzes suprafacial hydrogen abstraction and oxygenation (17, 18).
The sequence homology between the DOX domains of FOXB_01332 and ATEG_02036 is intriguing. It raises two questions. Could the lack of 9R-DOX activity of recombinant ATEG_02036 be due to cloning or expression artifacts? If this is the case, what is the structural basis of supra- or antarafacial oxygen insertion? 9R-DOX could also be unrelated to ATEG_02036 and 9S-DOX, but the low D-KIE of 9R-DOX is consistent with other DOXs of the animal peroxidase gene family (38, 42).
AOS of F. oxysporum and A. terreus
The AOS activity of FOXB_01332 converted 9S-hydroperoxides of C18 fatty acids as well as 11S-hydroperoxides of C20 fatty acids. Importantly, 9R-, 13S-, and 13R-hydroperoxides were not transformed. It is therefore unlikely that this AOS is involved in the biosynthesis of allene oxides as precursors of JA.
9S-HPODE undergoes homolytic cleavage by the AOS. This likely occurs with formation of CYP[FeIVOH] and an intermediate, 9S(10R)-epoxy-12(Z)-octadecenoic acid, with a radical at C-11, followed by biosynthesis of an allene oxide by electron transfer from C-11 to the metal center and proton loss, as described for plant AOS (35, 43). Allene oxides are unstable in aqueous solutions with nonenzymatic hydrolysis to α- and γ-ketols as illustrated in Fig. 8B, but allene oxides can be isolated and purified at low temperatures (7, 44).
We expected the catalytically important Asn964 of AOS of A. terreus to align with an important region of 9S-DOX-AOS. The acid-alcohol pair in the sequence Gly-Xaa-(Glu/Asp)-(Ser/Thr) of CYP hydroxylases participates in cleavage of molecular oxygen (45). The acidic (Glu946) and alcoholic (Ser949) residues of FOXB_01332 at the homologous position of the amide residues in Ala-Asn-Xaa-Xaa-Gln of LDS and AOS of A. terreus appeared to lack catalytic importance (cf. Fig. 1). Replacement of these Glu and Ser residues with nonpolar amino acids did not alter the AOS activity.
Asn residues facilitate the AOS activities of plant and fungal AOS (18, 20, 35, 36). The putative I-helix of 9S-DOX-AOS contains four Asn and four Glu residues (supplementary Fig. I). One of them, Asn921, is present in the sequence Asn-Asn921-Val-Asp-Gln. Replacement of Asn921 with Val did not alter the AOS activity. Additional work is needed to determine whether any of the other Asn residues facilitate the homolytic cleavage of 9S-HPODE.
Homologs to DOX-AOS
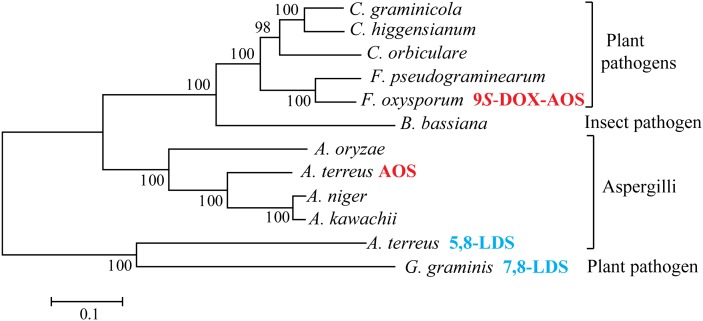
Our results raise the question whether the two first steps of fungal JA biosynthesis are catalyzed by separate 13S-LOX and AOS as in plants or by putative DOX-AOS fusion proteins. Sequence analysis of ascomycetous fungi revealed several proteins with at least 50% amino acid identity to 9S-DOX-AOS and AOS of A. terreus (Fig. 9). All these proteins contained the Tyr-Arg-Phe-His motif in the DOX domains and several consensus sequences of animal heme peroxidases, whereas the CYP domains contained relatively few conserved sequences in addition to the Glu-Xaa-Xaa-Arg salt bridge motif and the catalytic loop with the heme thiolate ligand [Gly-Xaa-His-Glu-Cys-Xaa-(Ala/Gly)].
Fig. 9.
Phylogenetic tree of protein sequences of ascomycetes with at least 50% sequence identity to AOS of A. terreus and/or 9S-DOX-AOS of F. oxysporum. The phylogenetic tree (MEGA5) is based on the alignment of selected protein sequences from the ten listed organisms and, for reference, 7,8-LDS of G. graminis and 5,8-LDS of A. terreus (marked in blue). In addition to LDS, the only nonorphan proteins are 9S-DOX-AOS of F. oxysporum and AOS of A. terreus (marked in red). The horizontal bar indicates an estimation of the substitution rate per amino acid position (29). The GenBank ID numbers of the proteins are: EFQ27323, Colletotrichum graminicola; CCF39565, Colletotrichum higgensianum; ENH82400, Colletotrichum orbiculare; EKJ79444, Fusarium pseudograminearum; EGU88194 (9S-DOX-AOS), F. oxysporum; EJP69120 (CYP6003A1), Beauveria bassiana; BAE60972, Aspergillus oryzae; AGH14485 (AOS), A. terreus; EHA25900, Aspergillus niger; GAA91201, Aspergillus kawachii; AGA95448 (5,8-LDS), A. terreus; AAD49559 (7,8-LDS), Gaeumannomyces graminis.
The two fungal AOS domains also lack characteristic motifs in support of homolytic cleavage of 9-HPODE other than the thiolate heme ligand. It might therefore be difficult to identify novel fungal AOS by bioinformatics alone. Nevertheless, synthesized open reading frames of candidate genes can be used for protein expression. This may provide important information on DOX-CYP proteins and JA biosynthesis by the F. oxysporum complex.
CONCLUSIONS
A 9S-DOX and an AOS are present as a fusion protein of F. oxysporum. 9S-DOX is homologous to LDS, 10R-DOX, and other animal heme peroxidases, and the AOS is a CYP with homology to the AOS of A. terreus. The homolytic cleavage of 9R- and 9S-HPODE by the heme thiolate group of these AOSs is facilitated by different amino acids, and these enzymes may therefore belong to separate DOX-AOS subfamilies.
Supplementary Material
Acknowledgments
The authors thank Professor Mats Hamberg, the Karolinska Institute, for the generous gift of stereospecifically deuterated linoleic acids.
Footnotes
Abbreviations:
- AOS
- allene oxide synthase
- COX
- cyclooxygenase
- CP
- chiral phase
- CYP
- cytochrome P450
- D-KIE
- deuterium kinetic isotope effect
- DOX
- dioxygenase
- HOTrE
- hydroxy-10(E),12(Z),15(Z)-octadecatrienoic acid
- HPODE
- hydroperoxy-10(E),12(Z)-octadecadienoic acid
- HPOME
- hydroperoxyoctadecamonoenoic acid
- HPOTrE
- hydroperoxyoctadecatrienoic acid
- JA
- jasmonic acid
- LDS
- linoleate diol synthase
- LOX
- lipoxygenase
- 9S(10)-EODE
- 9S(10)-epoxy-10,12(Z)-octadecadienoic acid
- P450
- cytochrome P450
- PG
- prostaglandin
- RP
- reversed phase
This work was supported by the Swedish Research Council (03X-06523), the Knut and Alice Wallenberg Foundation (KAW 2004.123), and Uppsala University.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures and one table.
REFERENCES
- 1.Smith W. L., Urade Y., Jakobsson P. J. 2011. Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem. Rev. 111: 5821–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ullrich V., Brugger R. 1994. Prostacyclin and thromboxane synthase: new aspects of hemethiolate catalysis. Angew. Chem. Int. Ed. Engl. 33: 1911–1919 [Google Scholar]
- 3.Wasternack C., Kombrink E. 2010. Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 5: 63–77 [DOI] [PubMed] [Google Scholar]
- 4.Hamberg M., Gardner H. W. 1992. Oxylipin pathway to jasmonates: biochemistry and biological significance. Biochim. Biophys. Acta. 1165: 1–18 [DOI] [PubMed] [Google Scholar]
- 5.Gfeller A., Dubugnon L., Liechti R., Farmer E. E. 2010. Jasmonate biochemical pathway. Sci. Signal. 3: cm3. [DOI] [PubMed] [Google Scholar]
- 6.Hamberg M. 2000. New cyclopentenone fatty acids formed from linoleic and linolenic acids in potato. Lipids. 35: 353–363 [DOI] [PubMed] [Google Scholar]
- 7.Brash A. R., Boeglin W. E., Stec D. F., Voehler M., Schneider C., Cha J. K. 2013. Isolation and characterization of two geometric allene oxide isomers synthesized from 9S-hydroperoxylinoleic acid by cytochrome P450 CYP74C3: stereochemical assignment of natural fatty acid allene oxides. J. Biol. Chem. 288: 20797–20806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wasternack C. 2007. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.). 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vick B. A., Zimmerman D. C. 1984. Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 75: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross B. E., Webster G. R. B. 1970. New metabolites of Gibberella fujikuroi. Part XV. N-jasmonoyl- and N-dihydrojasmonoyl-isoleucine. J. Chem. Soc. C. 1970: 1839–1842 [DOI] [PubMed] [Google Scholar]
- 11.Aldridge D. C., Galt S., Giles D., Turner W. B. 1971. Metabolites of Lasiodiplodia theobromae. J. Chem. Soc. C. 1971: 1623–1627 [Google Scholar]
- 12.Miersch O., Bohlmann H., Wasternack C. 1999. Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry. 50: 517–523 [Google Scholar]
- 13.Miersch O., Günther T., Fritsche W., Sembdner G. 1993. Jasmonates from different fungal species. Nat. Prod. Lett. 2: 293–299 [Google Scholar]
- 14.Miersch O., Preiss A., Sembdner G., Schreiber K. 1987. (+)-7-Iso-jasmonic acid and related compounds from Botryodiplodia theobromae. Phytochemistry. 26: 1037–1039 [Google Scholar]
- 15.Tsukada K., Takahashi K., Nabeta K. 2010. Biosynthesis of jasmonic acid in a plant pathogenic fungus, Lasiodiplodia theobromae. Phytochemistry. 71: 2019–2023 [DOI] [PubMed] [Google Scholar]
- 16.Brodhun F., Cristobal-Sarramian A., Zabel S., Newie J., Hamberg M., Feussner I. 2013. An iron 13S-lipoxygenase with an α-linolenic acid specific hydroperoxidase activity from Fusarium oxysporum. PLoS ONE. 8: e64919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jernerén F., Hoffmann I., Oliw E. H. 2010. Linoleate 9R-dioxygenase and allene oxide synthase activities of Aspergillus terreus. Arch. Biochem. Biophys. 495: 67–73 [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann I., Jernerén F., Oliw E. H. 2013. Expression of fusion proteins of Aspergillus terreus reveals a novel allene oxide synthase. J. Biol. Chem. 288: 11459–11469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean R., Van Kan J. A., Pretorius Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., Rudd J. J., Dickman M., Kahmann R., Ellis J., et al. 2012. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee D. S., Nioche P., Hamberg M., Raman C. S. 2008. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature. 455: 363–368 [DOI] [PubMed] [Google Scholar]
- 21.Garscha U., Jernerén F., Chung D., Keller N. P., Hamberg M., Oliw E. H. 2007. Identification of dioxygenases required for Aspergillus development. Studies of products, stereochemistry, and the reaction mechanism. J. Biol. Chem. 282: 34707–34718 [DOI] [PubMed] [Google Scholar]
- 22.Brodhun F., Göbel C., Hornung E., Feussner I. 2009. Identification of PpoA from Aspergillus nidulans as a fusion protein of a fatty acid heme dioxygenase/peroxidase and a cytochrome P450. J. Biol. Chem. 284: 11792–11805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann I., Jernerén F., Garscha U., Oliw E. H. 2011. Expression of 5,8-LDS of Aspergillus fumigatus and its dioxygenase domain. A comparison with 7,8-LDS, 10-dioxygenase, and cyclooxygenase. Arch. Biochem. Biophys. 506: 216–222 [DOI] [PubMed] [Google Scholar]
- 24.Oldham M. L., Brash A. R., Newcomer M. E. 2005. The structure of coral allene oxide synthase reveals a catalase adapted for metabolism of a fatty acid hydroperoxide. Proc. Natl. Acad. Sci. USA. 102: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann I., Oliw E. H. 2013. 7,8- and 5,8-linoleate diol synthases support the heterolytic scission of oxygen-oxygen bonds by different amide residues. Arch. Biochem. Biophys. 539: 87–91 [DOI] [PubMed] [Google Scholar]
- 26.Oliw E. H., Wennman A., Hoffmann I., Garscha U., Hamberg M., Jernerén F. 2011. Stereoselective oxidation of regioisomeric octadecenoic acids by fatty acid dioxygenases. J. Lipid Res. 52: 1995–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthew J. A., Chan H. W., Galliard T. 1977. A simple method for the preparation of pure 9-D-hydroperoxide of linoleic acid and methyl linoleate based on the positional specificity of lipoxygenase in tomato fruit. Lipids. 12: 324–326 [DOI] [PubMed] [Google Scholar]
- 28.Su C., Oliw E. H. 1998. Manganese lipoxygenase. Purification and characterization. J. Biol. Chem. 273: 13072–13079 [DOI] [PubMed] [Google Scholar]
- 29.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall B. G. 2013. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30: 1229–1235 [DOI] [PubMed] [Google Scholar]
- 31.MacMillan D. K., Murphy R. C. 1995. Analysis of lipid hydroperoxides and long-chain conjugated keto acids by negative ion electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 6: 1190–1201 [DOI] [PubMed] [Google Scholar]
- 32.Oliw E. H., Garscha U., Nilsson T., Cristea M. 2006. Payne rearrangement during analysis of epoxyalcohols of linoleic and alpha-linolenic acids by normal phase liquid chromatography with tandem mass spectrometry. Anal. Biochem. 354: 111–126 [DOI] [PubMed] [Google Scholar]
- 33.Oliw E. H., Su C., Skogström T., Benthin G. 1998. Analysis of novel hydroperoxides and other metabolites of oleic, linoleic, and linolenic acids by liquid chromatography-mass spectrometry with ion trap MSn. Lipids. 33: 843–852 [DOI] [PubMed] [Google Scholar]
- 34.Jernerén F., Eng F., Hamberg M., Oliw E. H. 2012. Linolenate 9R-dioxygenase and allene oxide synthase activities of Lasiodiplodia theobromae. Lipids. 47: 65–73 [DOI] [PubMed] [Google Scholar]
- 35.Brash A. R. 2009. Mechanistic aspects of CYP74 allene oxide synthases and related cytochrome P450 enzymes. Phytochemistry. 70: 1522–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao B., Boeglin W. E., Brash A. R. 2008. Role of the conserved distal heme asparagine of coral allene oxide synthase (Asn137) and human catalase (Asn148): mutations affect the rate but not the essential chemistry of the enzymatic transformations. Arch. Biochem. Biophys. 477: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider C., Pratt D. A., Porter N. A., Brash A. R. 2007. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem. Biol. 14: 473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann I., Hamberg M., Lindh R., Oliw E. H. 2012. Novel insights into cyclooxygenases, linoleate diol synthases, and lipoxygenases from deuterium kinetic isotope effects and oxidation of substrate analogs. Biochim. Biophys. Acta. 1821: 1508–1517 [DOI] [PubMed] [Google Scholar]
- 39.Rickert K. W., Klinman J. P. 1999. Nature of hydrogen transfer in soybean lipoxygenase 1: separation of primary and secondary isotope effects. Biochemistry. 38: 12218–12228 [DOI] [PubMed] [Google Scholar]
- 40.Clapp C. H., Senchak S. E., Stover T. J., Potter T. C., Findeis P. M., Novak M. J. 2001. Soybean lipoxygenase-mediated oxygenation of monounsaturated fatty acids to enones. J. Am. Chem. Soc. 123: 747–748 [DOI] [PubMed] [Google Scholar]
- 41.Oliw E. H., Jernerén F., Hoffmann I., Sahlin M., Garscha U. 2011. Manganese lipoxygenase oxidizes bis-allylic hydroperoxides and octadecenoic acids by different mechanisms. Biochim. Biophys. Acta. 1811: 138–147 [DOI] [PubMed] [Google Scholar]
- 42.Wu G., Lü J. M., van der Donk W. A., Kulmacz R. J., Tsai A. L. 2011. Cyclooxygenase reaction mechanism of prostaglandin H synthase from deuterium kinetic isotope effects. J. Inorg. Biochem. 105: 382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L., Chang Z., Pan Z., Fu Z. Q., Wang X. 2008. Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc. Natl. Acad. Sci. USA. 105: 13883–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brash A. R., Baertschi S. W., Ingram C. D., Harris T. M. 1988. Isolation and characterization of natural allene oxides: unstable intermediates in the metabolism of lipid hydroperoxides. Proc. Natl. Acad. Sci. USA. 85: 3382–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poulos T. L. 2005. Structural biology of heme monooxygenases. Biochem. Biophys. Res. Commun. 338: 337–345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.