Proper temporal and spatial regulation of sumoylation during mitosis is critical for mitotic progression. The SUMO isopeptidases SENP1 and SENP2 localize to key mitotic structures, including kinetochores. Overexpression and RNAi studies demonstrate that SENP1 and SENP2 are important modulators of SUMO function in mitosis.
Abstract
Sumoylation of centromere, kinetochore, and other mitotic chromosome-associated proteins is essential for chromosome segregation. The mechanisms regulating spatial and temporal sumoylation of proteins in mitosis, however, are not well understood. Here we show that the small ubiquitin-related modifier (SUMO)–specific isopeptidases SENP1 and SENP2 are targeted to kinetochores in mitosis. SENP2 targeting occurs through a mechanism dependent on the Nup107-160 subcomplex of the nuclear pore complex and is modulated through interactions with karyopherin α. Overexpression of SENP2, but not other SUMO-specific isopeptidases, causes a defect in chromosome congression that depends on its precise kinetochore targeting. By altering SENP1 kinetochore associations, however, this effect on chromosome congression could be phenocopied. In contrast, RNA interference–mediated knockdown of SENP1 delays sister chromatid separation at metaphase, whereas SENP2 knockdown produces no detectable phenotypes. Our findings indicate that chromosome segregation depends on precise spatial and temporal control of sumoylation in mitosis and that SENP1 and SENP2 are important mediators of this control.
INTRODUCTION
Regulation of essential mitotic processes is achieved in large measure through the action of posttranslational protein modifications, including phosphorylation, ubiquitylation, and sumoylation. Phosphorylation has been particularly well studied, as some of the best-characterized regulators of kinetochore and microtubule interactions possess protein kinase activity, including the Aurora kinases and BUBR1 (Lens et al., 2010; Bolanos-Garcia and Blundell, 2011). Ubiquitylation also plays a number of well-established roles in controlling mitotic progression, in particular by facilitating proteasome-mediated degradation of proteins, including securin and the mitotic cyclins (Min and Lindon, 2012). Sumoylation represents a more recently discovered regulator of mitosis, and multiple studies revealed essential roles in controlling chromosome condensation and cohesion, kinetochore assembly and function, and spindle dynamics (Meluh and Koshland, 1995; Biggins et al., 2001; Fukagawa et al., 2001; Strunnikov et al., 2001; Bachant et al., 2002; Bylebyl et al., 2003; Stead et al., 2003; Dieckhoff et al., 2004; Azuma et al., 2005; Zhang et al., 2008; Baldwin et al., 2009; Mukhopadhyay et al., 2010). The molecular targets and mechanisms of action of sumoylation during mitosis, however, remain to be fully explored.
Small ubiquitin-related modifiers (SUMOs) are ∼100–amino acid proteins that, like ubiquitin, are covalently conjugated to lysine residues in substrate proteins (Johnson, 2004; Geiss-Friedlander and Melchior, 2007). Invertebrates express a single SUMO protein, whereas vertebrates express three predominant SUMO paralogues: SUMO-1, SUMO-2, and SUMO-3. SUMO-2 and SUMO-3 are highly related, sharing 96% sequence homology, and are therefore referred to collectively as SUMO-2/3. SUMO-1 shares only 45% similarity to SUMO-2/3. Biochemical and proteomic analyses identified distinct subsets of proteins that are modified uniquely by SUMO-1 or SUMO-2/3, indicating that SUMO paralogues may regulate unique biological processes and have distinct signaling properties (Vertegaal et al., 2006; Geiss-Friedlander and Melchior, 2007). Of particular interest, SUMO-1 and SUMO-2/3 are uniquely regulated and conjugated to distinct proteins during mitosis (Zhang et al., 2008). The molecular mechanisms regulating the spatial and temporal control of paralogue-selective modifications and functions, however, are also not well understood.
Sumoylation occurs through a three-step enzymatic cascade, which requires the concerted action of an ATP-dependent E1 activating enzyme (Aos1/Uba2 heterodimer), an E2 conjugating enzyme (Ubc9), and one of a number of SUMO-specific E3 ligases (Johnson, 2004; Gareau and Lima, 2010). Although regulation of substrate modification can occur at the level of conjugation, regulation at the level of desumoylation by SUMO-specific isopeptidases also plays an important role. Yeast express two major SUMO-specific isopeptidases, ubiquitin-like protease 1 (Ulp1) and Ulp2, whereas vertebrates express six enzymes, referred to as SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7 (Mukhopadhyay and Dasso, 2007). These enzymes possess conserved C-terminal catalytic domains and divergent N-terminal domains that determine subcellular localization and substrate selectivity (Nishida et al., 2000; Nishida and Yamada, 2008; Hang and Dasso, 2002; Zhang et al., 2002; Bailey and O'Hare, 2004; Di Bacco et al., 2006; Yun et al., 2008; Klein et al., 2009). In addition to being localized to distinct subcellular domains, the six vertebrate SENPs also exhibit differences in SUMO paralogue specificity. This specificity is determined by differences in both N-terminal and C-terminal domains (Mikolajczyk et al., 2007; Kolli et al., 2010).
Given the unique activities and localizations of the vertebrates SENPs, they represent potentially important spatial and temporal regulators of sumoylation in cells. Consistent with this, SENP2 and SENP6 have both been implicated in having important, but functionally distinct, roles in regulating sumoylation in mitosis. SENP6 affects kinetochore assembly by limiting sumoylation of the kinetochore-associated protein CENP-I and its degradation through the SUMO-targeted ubiquitin E3 ligase RNF4 (Mukhopadhyay et al., 2010). SENP2 is believed to regulate sumoylation of kinetochore-associated proteins necessary for the association of CENP-E with kinetochores. We previously demonstrated that the recruitment of CENP-E to kinetochores was dependent on its ability to interact noncovalently with SUMO2/3 and that SENP2 overexpression resulted in a loss of CENP-E from kinetochores (Zhang et al., 2008). How the substrate recognition and function of SENP2 and SENP6 are controlled spatially and temporally in mitosis is not understood but of great interest.
Here we present evidence that SENP1 and SENP2 are positioned to uniquely exert spatial and temporal control on sumoylation of proteins in mitosis. We show that SENP2 is distinct from SENP1 and SENP6 in its ability to cause a mitotic, prometaphase arrest when overexpressed in cultured mammalian cells. The ability of SENP2 to cause cell cycle arrest is due to a unique association with kinetochores during prophase that depends on interactions with the Nup107-160 subcomplex of the nuclear pore complex (NPC) and karyopherin α. We also find that SENP1 associates with the mitotic spindle and kinetochores in mitosis but has no effect on mitotic progression when overexpressed. In contrast to overexpression phenotypes, RNA interference (RNAi)–mediated knockdown of SENP1 prevents timely separation of sister chromatids at the metaphase-to-anaphase transition. Together our findings reveal critical and nonredundant roles for SENP1 and SENP2 in mitosis and demonstrate the importance that subcellular localization plays in defining SUMO mammalian isopeptidase function.
RESULTS
SENP2 overexpression uniquely induces prometaphase arrest
We previously demonstrated that overexpression of SENP2 in cultured mammalian cells results in a global reduction in sumoylation and a prometaphase arrest phenotype caused by defects in the targeting of CENP-E to kinetochores (Zhang et al., 2008). Because mammalian cells express multiple different SUMO-specific isopeptidases with distinct subcellular localizations and enzymatic activities, we sought to investigate whether overexpression studies could be used to identify functions for other SENPs in mitosis.
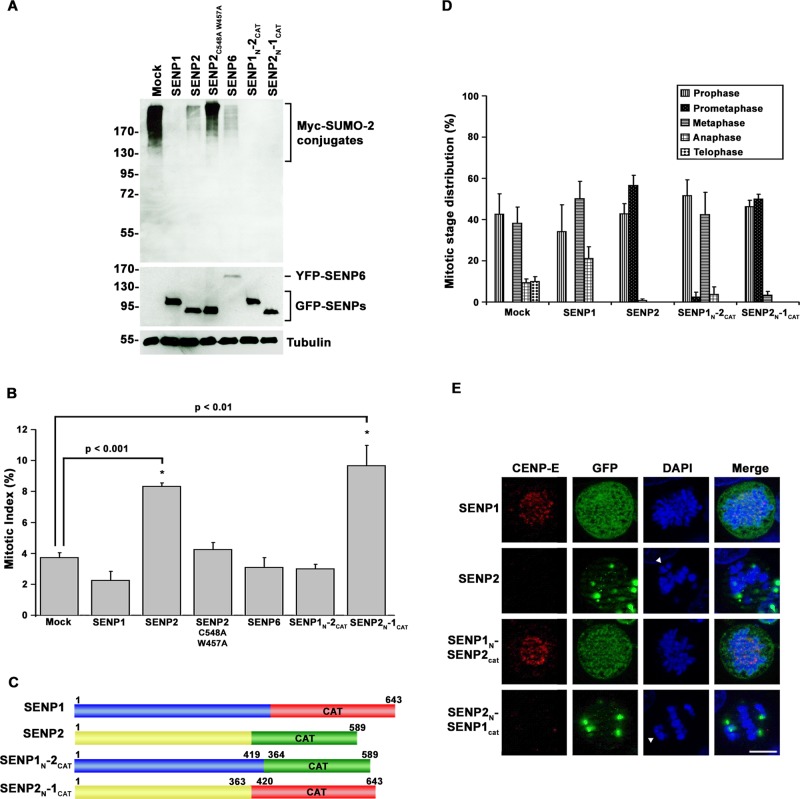
We transiently transfected constructs encoding green fluorescent protein (GFP)–SENP1, GFP-SENP2, or yellow fluorescent protein (YFP)–SENP6 together with Myc-tagged SUMO-2 into HeLa cells and first examined the effects of overexpressing these isopeptidases on sumoylation. SENP1 was chosen for comparison due to its close similarity to SENP2, and SENP6 due to previous evidence of mitotic functions (Mukhopadhyay et al., 2010). In comparison to control cells, overexpression of all three isopeptidases caused comparable global decreases in levels of high–molecular weight SUMO-2 conjugates (Figure 1A). To determine whether the suppression of sumoylation caused by SENP1, SENP2, or SENP6 overexpression resulted in similar prometaphase arrest phenotypes, we analyzed the cell cycle distribution of cells 48 h after transfection. As expected, we observed a reproducible twofold increase in the mitotic index of SENP2-overexpressing cells, with a majority of mitotic cells arresting specifically in prometaphase, with lagging chromosomes concentrated at the spindle poles (Figure 1, B, D, and E). In contrast, cells overexpressing SENP1 or SENP6 showed no noticeable defects in chromosome segregation, and the fraction of cells in mitosis was comparable to that of control cells (Figure 1, B, D, and E). Of note, the ability of SENP2 overexpression to cause a prometaphase arrest depended on suppression of SUMO-2 conjugate levels, as overexpression of a catalytically inactive mutant (SENP2C548A+W457A) did not affect levels of SUMO-2/3–modified proteins (Figure 1A) and had no effect on cell cycle progression (Figure 1B).
FIGURE 1:
SENP2 overexpression uniquely affects mitotic progression through mechanisms dependent on its N-terminal domain. (A) HeLa cells were cotransfected with constructs coding for Myc-tagged SUMO-2 and the indicated GFP-tagged SUMO isopeptidases or empty vector (Mock) as control. Cell lysates were analyzed by immunoblot analysis with antibodies specific for Myc, GFP, or tubulin. (B) HeLa cells were transfected with constructs coding for the indicated GFP-tagged SUMO isopeptidases or empty vector (Mock). The fraction of transfected cells in mitosis was determined by fluorescence microscopy 48 h after transfection. (C) Schematic diagram of SENP1, SENP2, and SENP1/2 chimeras. CAT, catalytic domain. (D) HeLa cells were transfected with constructs coding for the indicated SUMO isopeptidases or empty vector (Mock). The fraction of transfected cells present at each of the indicated stages of mitosis was determined by fluorescence microscopy 48 h after transfection. (E) HeLa cells were transfected with constructs coding for SENP1, SENP2, or the indicated chimeras. Cells were stained with CENP-E–specific antibodies and analyzed by immunofluorescence microscopy. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Arrowheads indicate unaligned chromosome pairs. Bar, 10 μm. Error bars represent SDs from three independent experiments.
SENP2-induced prometaphase arrest is determined by its N-terminal domain
The ability of SENP2 overexpression to uniquely affect mitotic chromosome segregation could be explained by distinct enzymatic activity, as determined by its C-terminal catalytic domain, or distinct subcellular localization or substrate specificity, as established by elements in its N-terminal domain (Mukhopadhyay and Dasso, 2007; Kolli et al., 2010; Goeres et al., 2011). To distinguish between these two possibilities, we generated expression constructs for chimeric SENP1 and SENP2 proteins. The N-terminal domain of SENP1 was fused to the catalytic domain of SENP2 to generate SENP1N-2CAT, and the N-terminal domain of SENP2 was fused to the catalytic domain of SENP1 to generate SENP2N-1CAT (Figure 1C). Constructs coding for these chimeric proteins were transfected into cells together with Myc-tagged SUMO-2, and their effects on sumoylation were monitored by immunoblot analysis. Overexpression of SENP1N-2CAT and SENP2N-1CAT both caused global decreases in high–molecular weight SUMO-2 conjugates similar to those obtained with overexpression of wild-type SENP1 and SENP2 (Figure 1A). To evaluate effects on cell cycle progression, we also analyzed transfected cells by fluorescence microscopy and quantified mitotic indices. Overexpression of SENP2N-1CAT caused arrest of cells in prometaphase, with chromosomes concentrated at the spindle poles, similar to the arrest observed with wild-type SENP2 (Figure 1, B, D, and E). Consistent with this effect, overexpression of both SENP2 and the SENP2N-1CAT chimera blocked recruitment of CENP-E to kinetochores, as previously reported for SENP2 (Zhang et al., 2008; Figure 1E). In contrast, and similar to wild-type SENP1, overexpression of the SENP1N-2CAT chimera had no noticeable effect on cell cycle progression, chromosome segregation, or CENP-E kinetochore targeting (Figure 1, B, D, and E). Thus the N-terminal domain of SENP2 contains unique determinants critical to its ability to affect CENP-E targeting and prometaphase arrest.
The N-terminal domains of SENP1 and SENP2 direct overlapping and unique subcellular localizations in interphase and mitosis
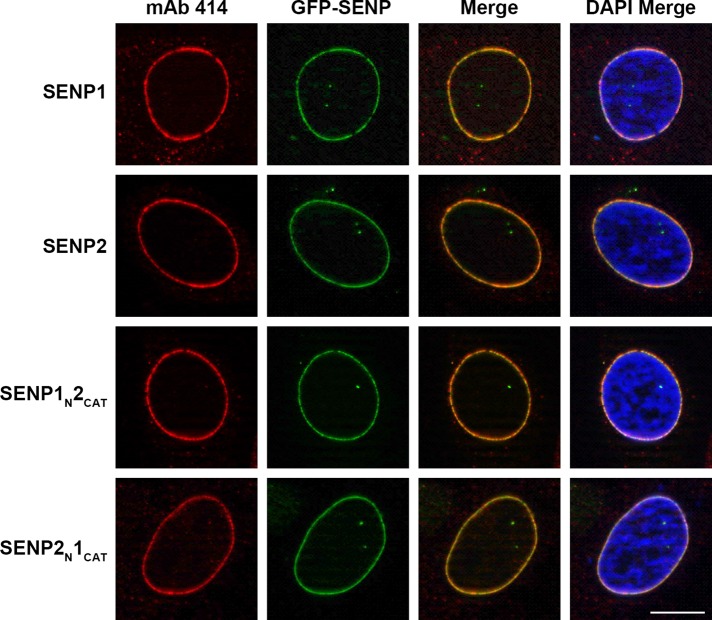
Because the N-terminal domains of SUMO-specific isopeptidases are important determinants of localization, we next examined the subcellular distributions of GFP-SENP1 and GFP-SENP2, as well as the chimeric proteins in interphase and mitosis. SENP1 and SENP2 were detected predominantly at the nuclear envelope and NPCs during interphase, as previously reported or suggested (Hang and Dasso, 2002; Zhang et al., 2002; Bailey and O'Hare, 2004; Chow et al., 2012; Figure 2). As predicted based on previous studies of SENP2 NPC targeting (Goeres et al., 2011), the localization of SENP2N-1CAT chimera was indistinguishable from that of wild-type SENP2 (Figure 2B). Similarly, the localization of the SENP1N-2CAT chimera was also indistinguishable from that of wild-type SENP1 (Figure 2B). Because the SENP2 catalytic domain alone contains no NPC-targeting signals (Goeres et al., 2011), this result is consistent with signals for NPC localization also residing within the N-terminal domain of SENP1.
FIGURE 2:
SENP1 and SENP2 association with NPCs is determined by N-terminal targeting signals. HeLa cells were transfected with constructs coding for GFP-tagged SENP1, SENP2, or the indicated chimeras, fixed with formaldehyde, and permeabilized with TNX-100. Cells were stained with monoclonal antibody (mAb) 414 to detect nuclear pore complexes and imaged by immunofluorescence microscopy. DNA was stained with DAPI. Bar, 10 μm.
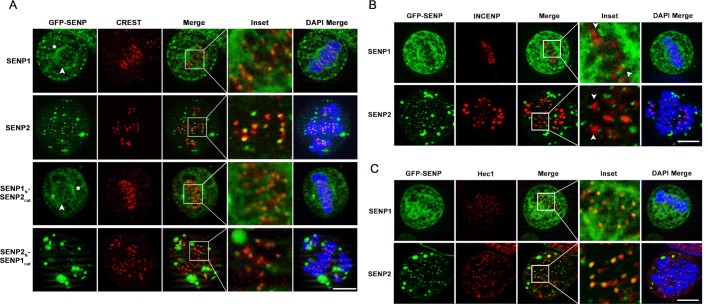
To evaluate localizations in mitosis, we labeled cells transfected with GFP-SENP1 and GFP-SENP2 with human CREST antibodies to mark the centromeres of mitotic chromosomes. SENP1 was detected at centrosomes and along spindle microtubules, as well as at foci partially colocalizing with CREST, indicative of kinetochore localization (Figure 3A). SENP2 was detected in nondescript aggregates but also at foci partially colocalizing with CREST. Unlike SENP1, SENP2 was not detected at centrosomes or on spindle microtubules (Figure 3A). Colocalization studies with INCENP and Hec1, markers for the inner centromere and outer kinetochore, respectively, further confirmed and narrowed the association of SENP1 and SENP2 to elements of the outer kinetochore (Figure 3, B and C). The SENP1N-2CAT and SENP2N-1CAT chimeras showed localization patterns in mitotic cells indistinguishable from those of wild-type SENP1 and SENP2, respectively (Figure 3A). These localization studies demonstrate that the N-terminal domains of SENP1 and SENP2 specify overlapping and distinct associations with NPCs in interphase and with elements of the mitotic spindle and kinetochore in mitosis.
FIGURE 3:
SENP1 and SENP2 are targeted to overlapping and distinct mitotic structures through their N-terminal domains. Constructs coding for GFP-tagged SENP1, SENP2, or SENP1/2 chimeras were transfected into HeLa cells. Cells expressing SENP1 or SENP1N2cat were permeabilized, fixed, and stained, where cells expressing SENP2 or SENP2N1cat cells were fixed, permeabilized, and stained. Cells were colabeled with (A) CREST anti-centromere antibodies (asterisks highlight centrosome staining, and arrowheads indicate spindle microtubule staining), (B) antibodies specific for the inner-centromere marker INCENP (arrowheads indicate centromeres flanked by kinetochore-associated SENP1 and SENP2 signals), and (C) antibodies specific for the outer-kinetochore marker Hec1. DNA was labeled with DAPI. Cells were imaged by immunofluorescence microscopy. Bar, 10 μm.
SENP1 and SENP2 both associate with the Nup107-160 subcomplex of the NPC but interact differentially with karyopherin α
The association of SENP2 with NPCs is mediated in part through interactions with the Nup107-160 subcomplex and karyopherins (Goeres et al., 2011). Both interactions also have the potential to affect targeting to kinetochores in mitosis (Wozniak et al., 2010). However, SENP2 interactions in mitotic cells have not previously been characterized, and interactions mediating SENP1 association with NPCs also have not been fully investigated. We therefore performed immunopurifications of GFP-tagged SENP1 and SENP2 from lysates prepared from synchronized cells arrested in mitosis, as well as from asynchronous cells. Interactions with karyopherin α3 and Nup107 were investigated by immunoblot analysis. Nup107 copurified with both SENP1 and SENP2 from both asynchronous and mitotic cell lysates, suggesting stable association of both isopeptidases with the Nup107-160 subcomplex throughout the cell cycle (Figure 4). In contrast, although karyopherin α3 copurified with SENP2 in immunopurifications from both asynchronous and mitotic extracts, interactions between SENP1 and karyopherin α3 were not detected under either condition. These overlapping and distinct interactions are consistent with the overlapping and distinct distributions of SENP1 and SENP2 in interphase and mitosis.
FIGURE 4:
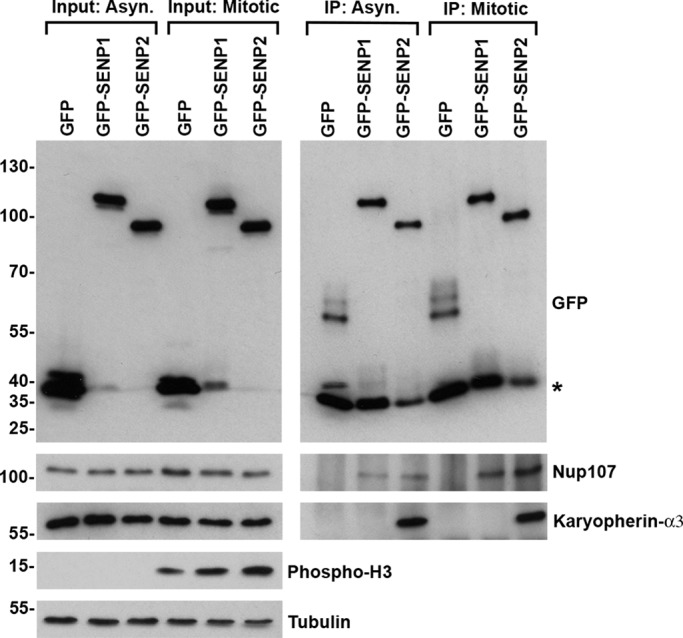
SENP1 and SENP2 interact with Nup107 but differentially associate with karyopherin α3. 293T cells were transfected with constructs coding for GFP, GFP-SENP1, or GFP-SENP2. Protein complexes were immunopurified, using GFP-specific antibodies, from lysates prepared from cells grown asynchronously or synchronized in mitosis by overnight incubation in the presence of nocodazole. Fractions of starting cell lysates (Input) and immunopurified protein complexes (IP) were analyzed by immunoblot analysis with antibodies specific for GFP, Nup107, karyopherin α3, tubulin, or phosphorylated histone H3. Asterisk denotes a contaminating band comigrating with GFP.
Two N-terminal elements of SENP2 specify kinetochore association and are required for prometaphase arrest
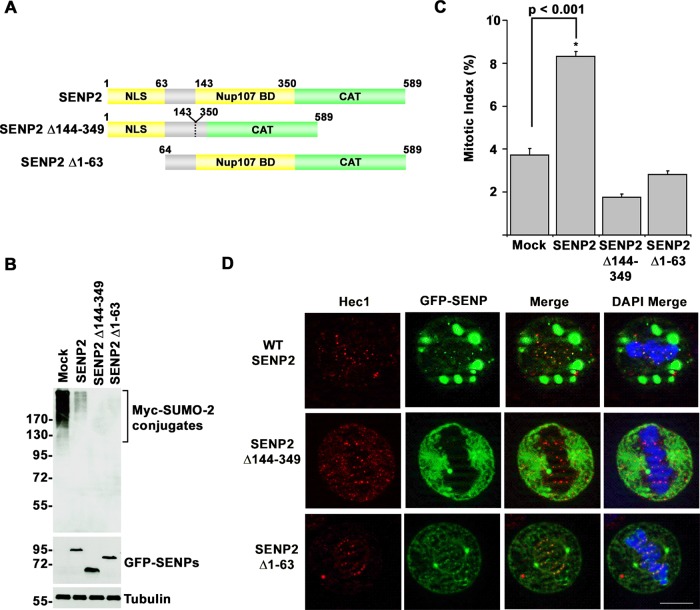
The Nup107-160 subcomplex of the NPC localizes to kinetochores in mitosis (Wozniak et al., 2010). To determine whether interactions with the Nup107-160 subcomplex facilitate SENP2 targeting to kinetochores and whether this targeting is required for prometaphase arrest, we transfected HeLa cells with a construct encoding GFP-tagged SENP2 Δ144-349 (Figure 5A). This SENP2 deletion mutant is defective in interactions with the Nup107-160 subcomplex (Goeres et al., 2011). Although overexpression of SENP2 Δ144-349 caused a decrease in high–molecular weight SUMO-2 conjugates similar to overexpression of wild-type SENP2 (Figure 5B), no detectable effect on cell cycle progression was observed based on quantitative analysis of mitotic indexes (Figure 5C). Consistent with a functional relationship between kinetochore localization and prometaphase arrest, SENP2 Δ144-349 exhibited a diffuse cytoplasmic distribution in mitotic cells devoid of kinetochore or spindle association (Figure 5D).
FIGURE 5:
N-terminal targeting elements in SENP2 specify mitotic arrest phenotypes. (A) Schematic diagram of SENP2 and targeting domain mutants. BD, binding domain; CAT, catalytic domain; NLS, nuclear localization signal. (B) HeLa cells were cotransfected with constructs coding for Myc-tagged SUMO-2 and wild type GFP-SENP2, the indicated GFP-tagged SENP2 mutants, or empty vector as control (Mock). Cell lysates were analyzed by immunoblot analysis with antibodies specific for Myc, GFP, or tubulin. (C) HeLa cells were transfected with constructs coding for wild-type GFP-SENP2, the indicated GFP-tagged SENP2 mutants, or empty vector (Mock). The fraction of transfected cells in mitosis was determined by fluorescence microscopy 48 h after transfection. (D) HeLa cells were transfected with constructs coding for wild-type GFP-SENP2 or the indicated GFP-tagged SENP2 mutants. Cells were permeabilized, fixed, and stained with Hec1-specific antibodies and analyzed by immunofluorescence microscopy. DNA was labeled with DAPI. Bar, 10 μm. Error bars represent SDs from three independent experiments.
A second NPC-targeting element in SENP2 consists of a nuclear localization signal (NLS) within the N-terminal 63 amino acids that mediates high-affinity interactions with karyopherin α (Goeres et al., 2011). To evaluate the role of karyopherin α binding in affecting SENP2 localization in mitosis, we next transfected HeLa cells with SENP2 Δ1-63, a deletion mutant lacking the N-terminal 63 amino acids (Figure 5A). Similar to wild-type SENP2, overexpression of SENP2 Δ1-63 resulted in global decreases in high–molecular weight SUMO-2 conjugates, as revealed by immunoblot analysis of whole-cell lysates (Figure 5B). Based on analysis of mitotic indices, however, overexpression of SENP2 Δ1-63 failed to produce a noticeable effect on cell cycle progression (Figure 5C). Surprisingly, analysis by fluorescence microscopy revealed that SENP2 Δ1-63 was detectable at kinetochores but also at centrosomes and along spindle microtubules, in a manner mirroring SENP1 localization (Figure 5D). These findings demonstrate that the association of SENP2 with kinetochores in mitosis requires interactions with the Nup107-160 subcomplex and that karyopherin α binding restricts SENP2 localization. Karyopherin α, however, was not detected at kinetochores in SENP2-overexpressing cells (Supplemental Figure S1). The results also demonstrate that the effect of SENP2 overexpression on mitotic progression correlates with restricted kinetochore localization.
Tethering SENP1 to kinetochores induces prometaphase arrest
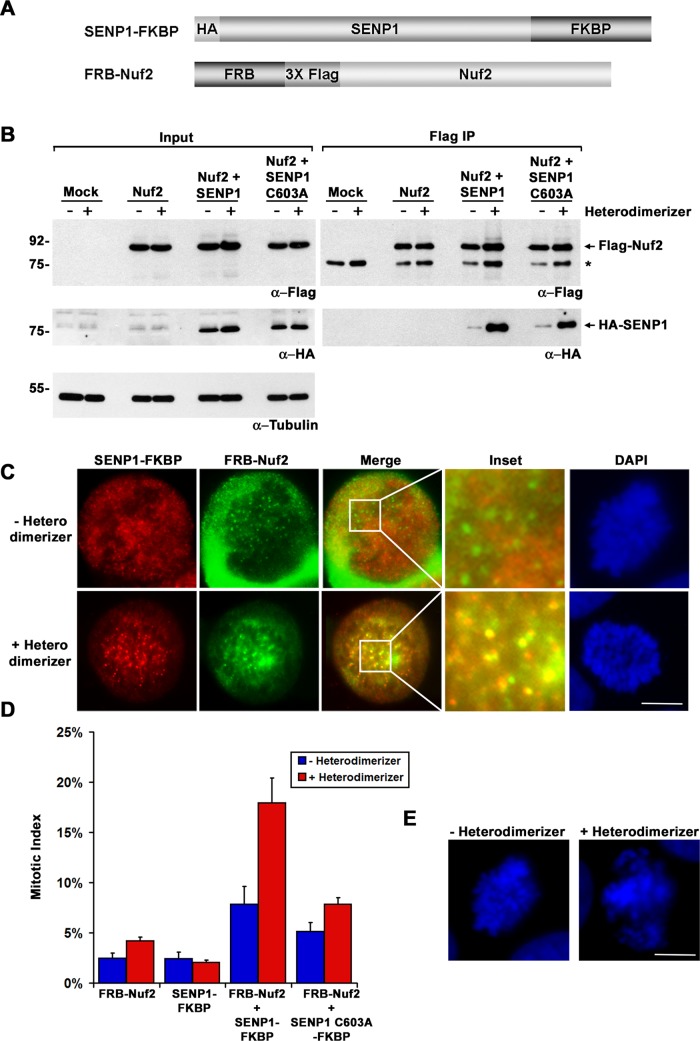
We hypothesized that a more stable association with kinetochores, or association with distinct kinetochore-associated proteins, could underlie the unique ability of SENP2 to affect mitotic progression. To test this hypothesis, we investigated the effects of artificially tethering SENP1 to kinetochores using the rapamycin-based heterodimerization system (Choi et al., 1996; Zhu et al., 2006). Specifically, we fused the ligand-binding domain of the FK506-binding protein (FKBP) to hemagglutinin (HA)-tagged SENP1 and the rapamycin-binding domain of the FKBP-rapamycin–associated protein (FRB) to FLAG-tagged Nuf2, an outer-kinetochore protein (Figure 6A). Cells were transiently transfected with a construct encoding FRB-FLAG-Nuf2 alone or together with a construct encoding HA-SENP1-FKBP. Transfected cells were then incubated in the presence or absence of the heterodimerizer, AP21967. To demonstrate effective AP21967-mediated heterodimerization, FRB-Nuf2 was immunopurified from transfected cell lysates using a FLAG-specific antibody, and immunoblot analysis was performed using an HA-specific antibody. As expected, copurification of SENP1-FKBP with FRB-Nuf2 depended on both coexpression and the presence of the AP21967 heterodimerizer. Minimal interaction between SENP1-FKBP and FRB-Nuf2 was detected in the absence of AP21967 (Figure 6B).
FIGURE 6:
SENP1 induces mitotic arrest when artificially tethered to kinetochores. (A) Schematic diagram of HA-tagged SENP1-FKBP and FLAG-tagged FRB-Nuf2 fusion proteins. (B) HeLa cells were transfected with empty vector, constructs coding for FRB-Nuf2, wild-type SENP1-FKBP, or catalytically inactive SENP1-FKBP (C603A), as indicated. Cells were cultured in the presence (+) or absence (–) of AP21967 heterodimerizer. FLAG-tagged Nuf2 was immunopurified from cell lysates, and immunoblot analysis was performed on starting cell lysates (Input) and immunopurified complexes using FLAG-, HA-, and tubulin-specific antibodies (IP). Asterisk denotes antibody heavy chain. (C) HeLa cells were cotransfected with constructs coding for HA-tagged SENP1-FKBP and FLAG-tagged FRB-Nuf2. Cells were cultured in the absence or presence of AP21967 heterodimerizer and analyzed by immunofluorescence microscopy using HA- and FLAG-specific antibodies. DNA was labeled with DAPI. Bar, 10 μm. (D) Cells were transfected with constructs coding for the indicated fusion proteins and cultured in the absence or presence of AP21967 heterodimerizer. Mitotic indexes were determined by fluorescence microscopy. Error bars denote SDs from three independent experiments. (E) Illustration of mitotic chromosomes observed in cells coexpressing FRB-Nuf2 and SENP1-FKBP and cultured in the absence or presence of AP21967 heterodimerizer.
To examine how heterodimerization with FRB-Nuf2 affects SENP1-FKBP localization, we examined the distributions of both fusion proteins in cotransfected cells cultured in the absence or presence of AP21967. Under the specific fixation and permeabilization conditions used, SENP1-FKBP was detected as a diffuse signal throughout mitotic cells in the absence of heterodimerizer, with no appreciable detection at kinetochores (Figure 6C). Under similar assay conditions, FRB-Nuf2 was detected in distinct foci that colocalized with the outer-kinetochore protein CENP-E (Figure 6C). When cells were cultured in the presence of heterodimerizer, SENP1-FKBP was detected in prominent foci throughout mitotic cells that colocalized with FRB-Nuf2, consistent with enhanced kinetochore localization (Figure 6C).
To characterize the effects of artificially tethering SENP1 to the outer kinetochore, we next analyzed the cell cycle distribution of cotransfected cells cultured in the absence or presence of heterodimerizer (Figure 6D). Expression of either FRB-Nuf2 or SENP1-FKBP alone did not significantly affect cell cycle progression, irrespective of the presence or absence of AP21967. Coexpression of both fusion proteins caused a twofold to threefold increase in the mitotic index when cells were cultured in the absence of the heterodimerizer, possibly reflecting the low level of basal interaction detected by immunopurification (Figure 6, B and D). Of most significance, however, a fivefold to sixfold increase in the mitotic index was detected in cells coexpressing FRB-Nuf2 and SENP1-FKBP and cultured in the presence of AP21967. Of importance, this effect on cell cycle progression depended on SUMO deconjugation, as heterodimerization of a catalytically inactive SENP1 mutant (SENP1 C603A) with FRB-Nuf2 had significantly reduced effects on the mitotic index relative to effects observed in the absence of heterodimerizer (Figure 6, B and D). Similar to the phenotype observed with SENP2 overexpression, tethering SENP1 to Nuf2 at the outer kinetochore resulted in the accumulation of prometaphase-arrested cells, many of which exhibited lagging chromosomes present at the spindle poles (Figure 6E). These results support a model in which the inhibitory effects of SENP2 overexpression on mitotic progression are mediated through precisely localized deconjugation of SUMO-modified proteins at kinetochores.
RNAi depletion reveals a critical function for SENP1 in mitosis
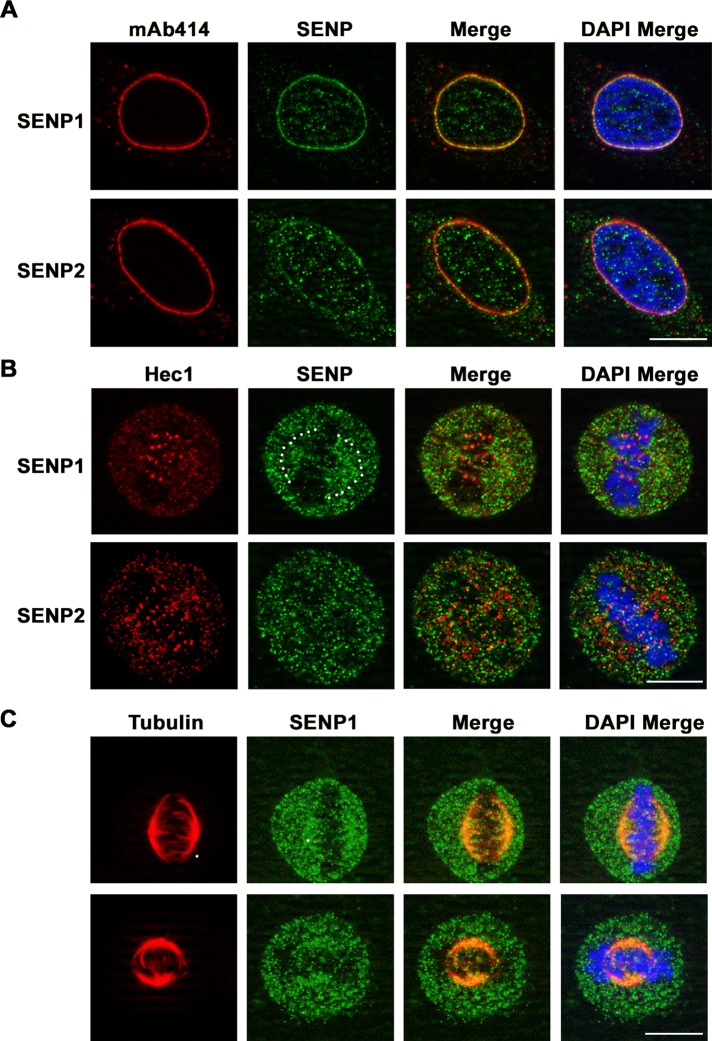
The findings outlined earlier involved characterization of exogenously expressed SENP1 and SENP2. To evaluate endogenous proteins, we performed immunofluorescence microscopy with SENP1- and SENP2-specific antibodies. As previously reported, SENP2-specific antibodies weakly stained the nucleoplasm and cytoplasm and were concentrated at the nuclear envelope in interphase cells (Figure 7A; Goeres et al., 2011). To evaluate colocalization with kinetochores in mitosis, we colabeled cells with antibodies specific for Hec1. Although SENP2 antibodies labeled foci associated with condensed mitotic chromosomes, we were unable to definitively localize endogenous SENP2 to kinetochores due to limitations of the antibody and SENP2 expression levels (Figure 7B). Large GFP-SENP2 foci observed in interphase and mitotic cells were not observed when analyzing endogenous SENP2. Consistent with GFP-SENP1 localization, antibodies specific for SENP1 labeled predominantly the nuclear envelope in interphase cells, with a slight nucleoplasmic signal also being detected (Figure 7A). In mitotic cells, SENP1 was detected as diffuse puncta throughout cells, with additional concentrations observed at mitotic spindles (Figure 7, B and C). As with SENP2, definitive localization at kinetochores was not possible.
FIGURE 7:
Immunofluorescence microscopy analysis of endogenous SENP1 and SENP2. HeLa cells were fixed, permeabilized, stained, and analyzed by immunofluorescence microscopy. DNA was labeled with DAPI. Cells were colabeled with (A) SENP1- or SENP2-specific antibodies and mAb 414 to detect nuclear pore complexes in interphase, (B) SENP1- or SENP2-specific antibodies and Hec1-specific antibodies in mitosis, and (C) SENP1- and tubulin-specific antibodies in mitosis. Bar, 10 μm.
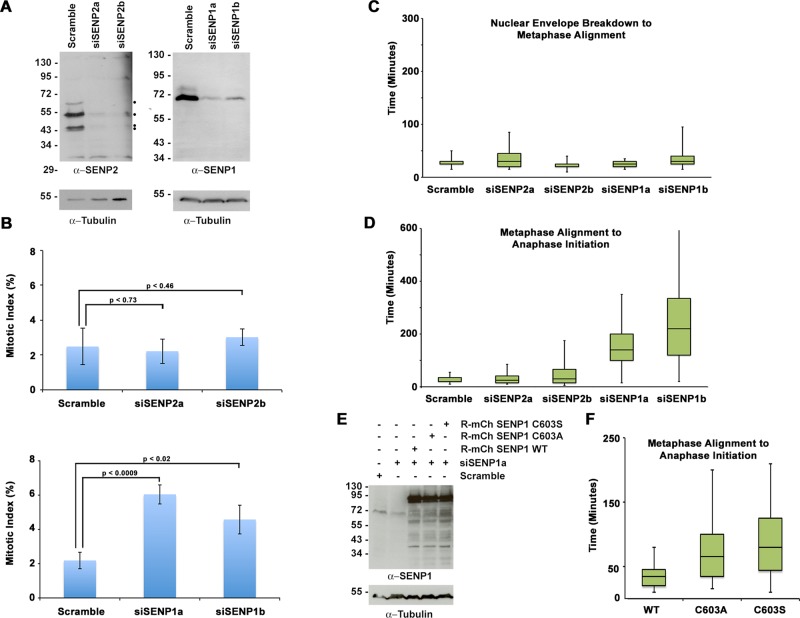
To further explore the functions of endogenous SENP1 and SENP2 in mitosis, we finally turned to RNAi to knock down protein expression in HeLa cells. In HeLa cells, SENP2 is present as multiple isoforms ranging from 45 to 60 kDa that are believed to arise from alternative pre-mRNA splicing (Goeres et al., 2011). Each of these SENP2 isoforms was reduced by >90% using two independent small interfering RNAs (siRNAs; Figure 8A). SENP1-specific antibodies detected a predominant band of ∼70 kDa, which was also reduced by >90% using two independent siRNAs (Figure 8A). The effects of SENP1 or SENP2 depletion on cell cycle progression were analyzed by fluorescence microscopy and quantitative assessment of mitotic indices. This analysis revealed no detectable effect on cell cycle progression in SENP2-depleted cells (Figure 8B). In SENP1-depleted cells, a reproducible twofold to threefold increase in mitotic index was observed relative to control cells using both independent siRNAs (Figure 8B).
FIGURE 8:
Analysis of endogenous SENP1 and SENP2 by siRNA knockdown. (A) HeLa cells were transfected with a control scramble siRNA, two independent SENP2-specific siRNAs, or two independent SENP1-specific siRNAs. Cell lysates were analyzed by immunoblot analysis with SENP1- or SENP2-specific antibodies and anti-tubulin antibodies as indicated. Dots indicate multiple SENP2 isoforms. (B) HeLa cells were transfected with a control scramble siRNA, two independent SENP2-specific siRNAs, or two independent SENP1-specific siRNAs, and mitotic indices were determined by fluorescence microscopy analysis of DAPI-stained cells 48 h after transfection. Error bars equal the SDs from three independent experiments. (C, D) YFP-H2B–expressing HeLa cells were transfected with control scramble, SENP1, or SENP2 siRNAs and imaged by live-cell fluorescence microscopy starting 48 h after transfection. Time from nuclear envelope breakdown to metaphase alignment and metaphase alignment to anaphase onset was quantified for ≥65 cells from at least three independent experiments for each condition. Outliers, defined as being 1.5× the interquartile range above or below the data set minimum or maximum, are not shown. (E) YFP-H2B–expressing HeLa cells were cotransfected with control scramble or SENP1a siRNA and siRNA-resistant constructs of mCherry-SENP1 WT or the catalytic mutants C603A and C603S. Cells lysates were analyzed by immunoblot analysis with SENP1- or tubulin-specific antibodies. (F) YFP-H2B–expressing HeLa cells were cotransfected with control scramble or SENP1a siRNA and siRNA-resistant constructs of mCherry-SENP1 WT or the catalytic mutants C603A and C603S. Time from metaphase alignment to anaphase onset was measured by live-cell fluorescence microscopy starting 48 h after transfection and quantified for ≥35 transfected cells from three independent experiments for each condition. Outliers, defined as being 1.5× the interquartile range above or below the data set minimum or maximum, are not shown.
To characterize effects on mitotic progression in greater detail, we repeated SENP1 and SENP2 depletions in HeLa cells expressing YFP-tagged histone H2B. We used live-cell imaging to measure the time between nuclear envelope breakdown (NEBD) and metaphase alignment, as well as metaphase alignment to initiation of anaphase (Figure 8, C and D). Consistent with the mitotic index assessment, SENP2-depleted cells did not exhibit an increase in the time spent in mitosis compared with the control cells (Figure 8, C and D). In contrast, although SENP1-depleted cells progressed from NEBD to metaphase normally, they exhibited a clear delay in anaphase onset after metaphase alignment (Figure 8, C and D). SENP1-depleted cells ultimately completed cell division despite prolonged times in metaphase, demonstrating that the SENP1-depletion conditions resulted in a mitotic delay but not an arrest. To validate that this effect was SENP1 specific, we cotransfected cells with siRNAs and constructs coding for siRNA-resistant forms of wild-type or catalytically inactive (C603A or C603S) mCherry-SENP1 and repeated the live-cell image analysis. Each of the mCherry-SENP1 proteins was expressed at comparable levels, whereas endogenous SENP1 expression was clearly reduced (Figure 8E). As predicted, wild-type SENP1 expression restored normal metaphase-to-anaphase kinetics, whereas the catalytically inactive forms of SENP1 failed to do so (Figure 8F). These results demonstrate that SENP1 and its isopeptidase activity are critical for a timely metaphase-to-anaphase transition. Collectively the findings reveal that SENP1 and SENP2 depletions produce distinct results from SENP1 and SENP2 overexpression and reveal a critical, nonredundant role for SENP1 in chromosome segregation.
DISCUSSION
Sumoylation is essential for chromosome segregation in organisms ranging from yeast to humans (Dasso, 2008; Wan et al., 2012). This requirement is related in part to spatial and temporal regulation of kinetochore assembly, including the association of CENP-E and the CENP-H/I/K complex with kinetochores during prophase in mammalian cells (Zhang et al., 2008; Mukhopadhyay et al., 2010). In this study we provided evidence that the SUMO-specific isopeptidases SENP1 and SENP2 are positioned to affect spatial and temporal control of sumoylation through unique associations with kinetochores, spindle microtubules, and centrosomes. Consistent with roles in affecting spatial and temporal control of sumoylation in mitosis, manipulating the expression levels of SENP1 or SENP2 induced defects in chromosome congression in prometaphase or sister chromatid separation at metaphase. Of importance, observed overexpression phenotypes correlated with the precise subcellular localizations of SENP1 and SENP2, demonstrating the crucial connection between isopeptidase targeting and biological function.
Our studies to elucidate the localizations of SENP1 and SENP2 in mitosis relied largely on analysis of exogenously expressed GFP-tagged proteins. A number of lines of evidence indicated that the mitotic localizations that we observed with GFP-SENP1 and GFP-SENP2 accurately reflect the localizations of the endogenous proteins. First, both proteins were found to associate with the Nup107-160 subcomplex, whose localizations to spindle microtubules and kinetochores in mitosis are well established. In the case of SENP2, we confirmed that its association with kinetochores depended on interactions with the Nup107-160 subcomplex through analysis of SENP2 Δ144-349 mutant. Although we were unable to obtain definitive evidence for endogenous SENP2 at kinetochores using immunofluorescence microscopy, this likely reflected the combination of low SENP2 expression levels and the relatively small fraction of the Nup107-160 subcomplex that is targeted to kinetochores (Wozniak et al., 2010). In contrast to SENP2, our immunofluorescence analysis revealed that a fraction of SENP1 is enriched at the mitotic spindle, consistent with the observed localization of GFP-SENP1. Determining whether SENP1 localization depends on the Nup107-160 complex, however, will require additional studies.
Previous studies established that signals in the N-terminal domains of mammalian SENPs specify their subcellular localizations (Hang and Dasso, 2002; Zhang et al., 2002; Bailey and O'Hare, 2004; Di Bacco et al., 2006; Goeres et al., 2011). Consistent with this, we found, using chimeric fusion proteins, that the N-terminal domains of SENP1 and SENP2 determine their unique associations with NPCs, kinetochores, and spindle microtubules. The N-terminal domain of SENP2 contains multiple elements that mediate association with NPCs during interphase, including an element that binds the Nup107-160 subcomplex of the NPC and an element that binds karyopherins (Goeres et al., 2011). Our findings indicate that both of these elements also contribute to the association of SENP2 with kinetochores in mitosis. SENP1 also associates with the Nup107-160 subcomplex but is distinct from SENP2, in that stable associations with karyopherin α were not detected.
The Nup107-160 subcomplex redistributes from NPCs to spindle poles, microtubules, and the outer-kinetochore plate in mitosis, consistent with a role in mediating SENP1 and SENP2 localizations to these structures (Loiodice et al., 2004; Orjalo et al., 2006; Zuccolo et al., 2007; Wozniak et al., 2010). The Nup107-160 subcomplex plays important roles in controlling mitotic events, including chromosome segregation, by affecting the distribution of Aurora B and other chromosome passenger complex (CPC) proteins (Platani et al., 2009). Of note, several members of the CPC are sumoylated, including Aurora B and Borealin (Klein et al., 2009; Fernandez-Miranda et al., 2010; Ban et al., 2011). Our findings that SENP1 and SENP2 are both associated with the Nup107-160 subcomplex suggest the intriguing possibility that its effects on CPC distribution may be related in part to control of CPC sumoylation.
Although SENP1 localized to the mitotic spindle and kinetochores, the distribution of SENP2 was distinct in its more restricted localization to kinetochores. The restricted localization of SENP2 depended on interactions with the Nup107-160 subcomplex but also on interactions mediated by N-terminal residues that include a functional NLS. This NLS mediates high-affinity interactions with karyopherin α and, in particular, RanGTP-insensitive interactions with karyopherin α3 (Goeres et al., 2011). Deletion of the N-terminal NLS caused SENP2 to localize to spindle microtubules, as well as to kinetochores, a distribution comparable to the localization of the Nup107-160 subcomplex and SENP1. This change in localization could be explained by higher levels of soluble SENP2 Δ1-63 relative to full-length SENP2. However, detection of spindle staining for both proteins was unaffected by expression levels. Based on current knowledge, it is therefore hypothesized that both SENP1 and SENP2 are targeted to spindle microtubules and kinetochores through interactions with the Nup107-160 subcomplex and that interactions with karyopherin α function to further stabilize or restrict SENP2 localization at kinetochores. Further studies are required to test this hypothesis.
Of importance, the mitotic arrest phenotype observed upon SENP2 overexpression depended on the more restricted, karyopherin α–dependent localization of SENP2. We interpret this finding as an indication that the restricted localization of SENP2 at kinetochores affects its substrate selectivity and thereby acts to distinguish SENP2 from SENP1. By facilitating kinetochore association, karyopherin α could affect SENP2 substrate selectivity by enhancing its local concentration at kinetochores. Alternatively, karyopherin α could function to target SENP2 to a distinct subdomain of the kinetochore or more directly facilitate association with specific SUMO-modified proteins. Our finding that SENP1 could be detected at kinetochores when overexpressed but exerted no mitotic defects unless artificially tethered to Nuf2 is consistent with the interpretation that the precise localizations or concentrations of SENP1 and SENP2 dictate their substrate selectivity and function. Identification of the SUMO-modified proteins recognized by SENP1 and SENP2 in mitosis will help to better clarify how localization, concentration, or other parameters affect substrate specificity.
How karyopherin α affects the association of SENP2 with kinetochores is unclear, but karyopherins in general have a number of established roles in affecting protein localization during mitosis (Mosammaparast and Pemberton, 2004). Of particular interest, the relative distribution of hKid between spindle microtubules and chromosome arms is determined by interactions with karyopherin α and β. In a manner reminiscent of our findings for SENP2, interactions with karyopherin α and β function to restrict hKid localization to spindle microtubules while promoting interactions with chromosome arms (Tahara et al., 2008). Also of potential relevance, studies in budding yeast demonstrated a mitotic role for the karyopherin Kap121p in the transport of Ulp1 from NPCs to the septin ring (Makhnevych et al., 2007). Further studies are required to determine the precise molecular details of how karyopherins affect protein targeting in mitosis, including the targeting and dynamic association of SENP2 with kinetochores.
In addition to providing evidence of mitotic roles for SENP1 and SENP2 in mitosis using protein overexpression studies, we also made important observations in cells depleted of SENP1 or SENP2. We observed a reproducible twofold to threefold increase in the mitotic index upon depletion of endogenous SENP1. Our time-lapse microscopy analysis demonstrated that this increase is due to a delay in the separation of sister chromatids after normal alignment at the metaphase plate. Furthermore, we demonstrated that this effect is due to the loss of SENP1 isopeptidase activity, as only a wild-type siRNA-resistant SENP1 rescued this phenotype. Although further studies are required to define the precise defect, a delay in the metaphase-to-anaphase transition may be related to improper desumoylation of a protein involved in the spindle assembly checkpoint or dissolution of sister chromatid cohesion. Intriguingly, gene-knockout studies in chicken DT40 cells revealed a requirement for SENP1 in the maintenance of sister chromatid cohesion in the presence of microtubule-destabilizing agents (Era et al., 2012). Although our SENP1 RNAi studies revealed no defects in cohesion maintenance, differences in findings could be explained by the unique experimental conditions, including the presence and absence of drug treatments, transient knockdown versus knockout, and human HeLa cells versus chicken DT40 cells. In contrast to SENP1, time-lapse microscopy did not identify any obvious mitotic defects in SENP2-depleted cells, which could be explained in a number of ways. First, SENP2 may be redundant with other SENPs, including SENP3, which affects desumoylation of Borealin (Klein et al., 2009). Alternatively, hypersumoylation of mitotic proteins resulting from SENP2 depletion may have limited functional consequences in contrast to hyposumoylation resulting from SENP2 overexpression.
Our evaluations of the overexpression and knockdown phenotypes of SENP1 and SENP2 reveal critical roles for both sumoylation and desumoylation at multiple points during mitosis. Our findings also highlight the importance that subcellular localization plays in defining the activities of SENP1 and SENP2 in mitosis and their unique and nonredundant functions. Ultimately, the identification of SENP1 and SENP2 mitotic substrates will be essential to more fully understand the role that these enzymes play in regulating chromosome segregation and, more generally, how sumoylation controls this vital cellular process.
MATERIALS AND METHODS
Antibodies
SENP2 and GFP rabbit polyclonal antibodies were produced as described previously (Goeres et al., 2011). SENP1 antibody, a gift from Mary Dasso (National Institutes of Health, Bethesda, MD), was generated by injecting rabbits with GST-SENP1 (273–449) as previously described (Chow et al., 2012). Antibodies were affinity purified using appropriate antigens and standard protocols.
Remaining antibodies were obtained from the following sources: anti-Nup107 antibody was kindly provided by Joseph Glavy (Charles V. Schaefer, Jr., School of Engineering & Science, Stevens Institute of Technology, Hoboken, NJ); CREST human autoantibodies were a generous gift from Ted Salmon (University of North Carolina, Chapel Hill, NC); anti–karyopherin α3 was provided by Stephen Adams (Northwestern University, Chicago, IL); anti–CENP-E (Active Motif, Carlsbad, CA); anti-GFP (Clontech, Mountain View, CA); anti-INCENP (Active Motif); anti-HEC1 (BD Biosciences, San Jose, CA); anti-tubulin (Sigma-Aldrich, St. Louis, MO); anti-Myc (Santa Cruz Biotechnology, Santa Cruz, CA); anti-HA (Santa Cruz Biotechnology); anti-FLAG M2 (Sigma-Aldrich); mAb414 (Abcam, Cambridge, MA); and anti-phospho-histone H3 (Ser-10; EMD Millipore, Billerica, MA).
Plasmid constructs
SENP2 cDNA and GFP-tagged expression constructs were obtained as previously described (Zhang et al., 2002; Goeres et al., 2011). GFP-SENP1 and YFP-SENP6 vectors were a gift from Mary Dasso. To generate SENP1 and SENP2 chimeric protein expression constructs, we used site-directed mutagenesis to create restriction sites at the junction between the N-terminal domain and the catalytic domain of both SENP1 and SENP2. The catalytic domain of one isopeptidase was PCR amplified and ligated into this restriction site to replace the original catalytic domain. FLAG-Nuf2 was subcloned into pC4-RHE vector for mammalian expression (Ariad Pharmaceuticals, Cambridge, MA). SENP1 was subcloned into the pC4EN-F1 vector (Ariad Pharmaceuticals). SENP1 was subcloned into a pmCherry-C2 vector, and the siRNA resistant clones and catalytic mutants were generated using site-directed mutagenesis.
Cells, cell culture, transfection, and RNA interference
HeLa cells stably expressing YFP–histone H2B were a gift from Andrew Holland (Johns Hopkins University School of Medicine, Baltimore, MD). HeLa or 293T cells were maintained at 37°C in DMEM supplemented with 10% fetal bovine serum, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 8.0), and 1% penicillin-streptomycin. Cells were transfected with the indicated plasmids at a confluency of 40–50% using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. For RNAi, cells were grown to 40–50% confluency and then transfected using RNAiMax (Invitrogen). siRNA oligos were used at a final concentration of 20 nM. siRNA oligos included the following: scramble control, 5′-CUUCCUCUCUUUCUCUCCCUUGUGA-3′; SENP2 oligo(a), 5′-AUAUCUGGAUUCUAUGGGAUU-3′; SENP2 oligo(b), 5′-GAAAGAGAGAAGUACCGAAtt-3′; SENP1 oligo(a), 5′- UCCUUUACACCUGUCUCGAUGUCUU-3′; and SENP1 oligo(b), 5′-GCAAAUGGCCAAUGG AGAAAUUCUA-3′. Cells were harvested for immunoblotting, immunofluorescence microscopy, or time-lapse microscopy 48 h after transfection. For cotransfection of siRNA oligos and siRNA-resistant forms of SENP1, cells were grown to 40–50% confluency and transfected using Lipofectamine 2000 using a final concentration of siRNA oligo of 20 nM.
Heterodimerization
For heterodimerization experiments, cells were transfected with indicated plasmids and cultured in the presence or absence of 250 nM AP21967 for 48 h (Ariad Pharmaceuticals) before analysis by immunofluorescence microscopy. For immunopurification, cells were lysed in buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM sodium chloride, 1% Triton-X 100, 1% sodium deoxycholate, 2 mM EDTA, 1 μg/ml leupeptin and pepstatin A, 20 μg/ml aprotinin, and 2 mM phenylmethylsulfonyl fluoride (PMSF). Lysates were placed on ice for 5 min and then sonicated for 30 s and centrifuged at 16,000 × g for 15 min. Lysates were incubated with M2 FLAG agarose beads (Sigma-Aldrich) for 4 h at 4°C, and then beads were washed six times in phosphate-buffered saline (PBS), and bound proteins were eluted directly in SDS-sample buffer.
GFP-SENP immunopurifications
For GFP-SENP immunopurifications, rabbit anti-GFP antibodies were immobilized on Protein-A Plus agarose beads (Thermo Scientific, Rockford, IL) for 1 h and cross-linked with disuccinimidyl suberate for 30 min. Beads were washed through a series of four buffers including 50 mM Tris (pH 7.5), 100 mM glycine (pH 3.0), PBS, and lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM dithiothreitol, 0.05% sodium deoxycholate). 293T cells were transfected with the indicated plasmids for ∼36 h, treated with or without 0.1 μg/ml nocodazole overnight, and then harvested 48 h after transfection. Cells were lysed in lysis buffer supplemented with 1 mM PMSF, 5 μg/ml pepstatin A, 5 μg/ml leupeptin, and 10 mM N- ethylmaleimide, sonicated, and centrifuged 16,000 × g for 20 min at 4ºC. Protein lysates were quantified using a bicinchoninic acid protocol (Thermo Scientific) to normalize protein inputs. Antibody-bound beads were incubated with cell lysates for 5 h at 4ºC and washed six times with lysis buffer, and proteins were eluted directly in SDS-sample buffer.
Immunoblotting
Immunoblot analysis was performed using enzyme-linked chemiluminescence ECL-Prime reagent (GE Healthcare, Silver Spring, MD).
Immunofluorescence microscopy and live-cell imaging
HeLa cells were cultured on glass coverslips. Unless otherwise stated, cells were fixed in 2% formaldehyde for 30 min and permeabilized in 0.2% Triton-X 100 for 7 min at room temperature. For colocalization with kinetochore proteins, cells were fixed in 3.5% paraformaldehyde in PBS for 7 min and permeabilized in 0.5% Triton-X 100 in PBS for 20 min at room temperature. Localization of GFP-SENP1 was examined by preextracting in 20 μg/ml digitonin in buffer containing 200 mM HEPES (pH 6.5), 110 mM potassium acetate, 20 mM magnesium acetate, 1 μg/ml leupeptin and pepstatin A, 20 μg/ml aprotinin, and 1 mM PMSF for 15 min at room temperature and then fixing in 2% formaldehyde in PBS for 30 min. Immunostaining was carried out as previously described (Goeres et al., 2011) using secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Life Technologies, Grand Island, NY), unless otherwise noted. Images were acquired using a Zeiss Observer Z1 fluorescence microscope with a Zeiss Plan-Apochromat 63× objective (numerical aperture 1.40) and Apotome VH optical sectioning grid (Carl Zeiss, Jena, Germany). Images were obtained using a Zeiss AxioCam MRm camera and processed using AxioVision Software Release 4.8.2.
For live-cell imaging, cells were cultured in Lab-Tek Chambered #1.0 Borosilicate Coverglass slides (Nunc, Rochester, NY), transfected, and then imaged 48 h posttransfection. For imaging, cells were maintained in culture medium at 37ºC with 5% CO2 on a Zeiss Observer Z1 fluorescence microscope fitted with an incubation chamber. Images were acquired using a Zeiss EC Plan-Neofluar 40× objective (numerical aperture 1.3) every 5 min for 15 h with a Zeiss AxioCam MRm camera.
Supplementary Material
Acknowledgments
We thank all the members of the Matunis lab for helpful discussions and suggestions during the course of these studies. We also acknowledge Mary Dasso and her laboratory for generously providing SENP1 antibodies and plasmids and for helpful discussions. This work was supported through National Institutes of Health Grant GM060980 (to M.J.M.) and the Alfred Sommer Scholar's Program (to C.C.P.).
Abbreviations used:
- CPC
chromosome passenger complex
- FKBP
FK506-binding protein
- FRB
FKBP-rapamycin–associated protein
- NPC
nuclear pore complex
- SUMO
small ubiquitin-related modifier
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0230) on September 18, 2013.
*These authors contributed equally.
†Present address: Laboratory of Biochemistry and Genetics, NIDDK, National Institutes of Health, 8 Center Dr., Bethesda, MD 20892.
REFERENCES
- Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Bailey D, O'Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3406–3419. doi: 10.4161/cc.8.20.9911. [DOI] [PubMed] [Google Scholar]
- Ban R, Nishida T, Urano T. Mitotic kinase Aurora-B is regulated by SUMO-2/3 conjugation/deconjugation during mitosis. Genes. 2011;16:652–669. doi: 10.1111/j.1365-2443.2011.01521.x. [DOI] [PubMed] [Google Scholar]
- Biggins S, Bhalla N, Chang A, Smith DL, Murray AW. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 2001;159:453–470. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia VM, Blundell TL. BUB1 and BUBR1: multifaceted kinases of the cell cycle. Trends Biochem Sci. 2011;36:141–150. doi: 10.1016/j.tibs.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Chow KH, Elgort S, Dasso M, Ullman KS. Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus. 2012;3:349–358. doi: 10.4161/nucl.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 2008;3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckhoff P, Bolte M, Sancak Y, Braus GH, Irniger S. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol Microbiol. 2004;51:1375–1387. doi: 10.1046/j.1365-2958.2003.03910.x. [DOI] [PubMed] [Google Scholar]
- Era S, Abe T, Arakawa H, Kobayashi S, Szakal B, Yoshikawa Y, Motegi A, Takeda S, Branzei D. The SUMO protease SENP1 is required for cohesion maintenance and mitotic arrest following spindle poison treatment. Biochem Biophys Res Commun. 2012;426:310–316. doi: 10.1016/j.bbrc.2012.08.066. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miranda G, Perez de Castro I, Carmena M, Aguirre-Portoles C, Ruchaud S, Fant X, Montoya G, Earnshaw WC, Malumbres M. SUMOylation modulates the function of Aurora-B kinase. J Cell Sci. 2010;123:2823–2833. doi: 10.1242/jcs.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Regnier V, Ikemura T. Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells. Nucleic Acids Res. 2001;29:3796–3803. doi: 10.1093/nar/29.18.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Goeres J, Chan PK, Mukhopadhyay D, Zhang H, Raught B, Matunis MJ. The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107-160 nucleoporin subcomplex. Mol Biol Cell. 2011;22:4868–4882. doi: 10.1091/mbc.E10-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Klein UR, Haindl M, Nigg EA, Muller S. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell. 2009;20:410–418. doi: 10.1091/mbc.E08-05-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli N, Mikolajczyk J, Drag M, Mukhopadhyay D, Moffatt N, Dasso M, Salvesen G, Wilkinson KD. Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem J. 2010;430:335–344. doi: 10.1042/BJ20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T, Ptak C, Lusk CP, Aitchison JD, Wozniak RW. The role of karyopherins in the regulated sumoylation of septins. J Cell Biol. 2007;177:39–49. doi: 10.1083/jcb.200608066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J Biol Chem. 2007;282:26217–26224. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- Min M, Lindon C. Substrate targeting by the ubiquitin-proteasome system in mitosis. Semin Cell Dev Biol. 2012;23:482–491. doi: 10.1016/j.semcdb.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Arnaoutov A, Dasso M. The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol. 2010;188:681–692. doi: 10.1083/jcb.200909008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Nishida T, Tanaka H, Yasuda H. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem. 2000;267:6423–6427. doi: 10.1046/j.1432-1327.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- Nishida T, Yamada Y. SMT3IP1, a nucleolar SUMO-specific protease, deconjugates SUMO-2 from nucleolar and cytoplasmic nucleophosmin. Biochem Biophys Res Commun. 2008;374:382–387. doi: 10.1016/j.bbrc.2008.07.047. [DOI] [PubMed] [Google Scholar]
- Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M, Santarella-Mellwig R, Posch M, Walczak R, Swedlow JR, Mattaj IW. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol Biol Cell. 2009;20:5260–5275. doi: 10.1091/mbc.E09-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Aravind L, Koonin EV. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 2001;158:95–107. doi: 10.1093/genetics/158.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara K, et al. Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J Cell Biol. 2008;180:493–506. doi: 10.1083/jcb.200708003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- Wan J, Subramonian D, Zhang XD. SUMOylation in control of accurate chromosome segregation during mitosis. Curr Protein Pept Sci. 2012;13:467–481. doi: 10.2174/138920312802430563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67:2215–2230. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol. 2008;183:589–595. doi: 10.1083/jcb.200807185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhang H, Matunis MJ. SUMO modification through rapamycin-mediated heterodimerization reveals a dual role for Ubc9 in targeting RanGAP1 to nuclear pore complexes. Exp Cell Res. 2006;312:1042–1049. doi: 10.1016/j.yexcr.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Zuccolo M, et al. The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.