Abstract
P128 is a chimeric anti-staphylococcal protein having a catalytic domain from a Staphylococcus bacteriophage K tail associated structural protein and a cell wall targeting domain from the Staphylococcus bacteriocin-lysostaphin. In this study, we disclose additional properties of P128 and compared the same with lysostaphin. While lysostaphin was found to get inactivated by heat and was inactive on its parent strain S. simulans biovar staphylolyticus, P128 was thermostable and was lytic towards S. simulans biovar staphylolyticus demonstrating a difference in their mechanism of action. Selected mutation studies of the catalytic domain of P128 showed that arginine and cysteine, at 40th and 76th positions respectively, are critical for the staphylolytic activity of P128, although these amino acids are not conserved residues. In comparison to native P128, only the R40S mutant (P301) was catalytically active on zymogram gel and had a similar secondary structure, as assessed by circular dichroism analysis and in silico modeling with similar cell binding properties. Mutation of the arginine residue at 40th position of the P128 molecule caused dramatic reduction in the Vmax (∆OD600 [mg/min]) value (nearly 270 fold) and the recombinant lysostaphin also showed lesser Vmax value (nearly 1.5 fold) in comparison to the unmodified P128 protein. The kinetic parameters such as apparent Km (Km APP) and apparent Kcat (KcatAPP) of the native P128 protein also showed significant differences in comparison to the values observed for P301 and lysostaphin.
Keywords: Staphylococcus aureus, methicillin resistance, arginine mutation, disulfide bonds, in silico modeling, western blot
Introduction
Staphylococcus aureus is a common pathogen for both humans and animals, causing skin and soft tissue infections, pneumonia, endocarditis, meningitis and osteomyelitis, and serious nosocomial infections.1-3 The prevalence of multidrug-resistant strains of Staphylococcus aureus has increased worldwide, and their treatment has posed immense challenges.4 Consequently, alternatives to traditional antibiotics are being developed, such as bacteriophages,5,6 modified bacteriophages,7 and bacteriophage products.2,8
P128 is a Staphylococcus bacteriophage-derived anti-staphylococcal protein.2,9 This chimeric protein was constructed by fusing the tail-associated muralytic enzyme from a lytic bacteriophage (phage K)10 with the Src Homology (SH3b) staphylococcal cell wall-binding domain of lysostaphin, a bacteriocin with potential applications for treating multidrug-resistant staphylococci.11 P128 has been successfully tested on a panel of 3000 typed S. aureus strains including several methicillin-resistant S. aureus (MRSA) strains and clinically significant strains such as USA100, USA300, and USA400.2
The chimeric protein P128 contains a cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domain that is frequently found in proteins from bacteria, bacteriophages, archaea, and eukaryotes of the Trypanosomatidae family.12 The CHAP domain consists of 110 to 140 amino acids and is found in a wide range of protein architectures but is commonly associated with bacterial Src Homology (SH3) domains.13 Although many CHAP domain proteins have not been characterized, they are thought to function in peptidoglycan hydrolysis. CHAP proteins contain three highly conserved amino acid residues: invariant cysteine (Cys) and histidine (His) residues along with a polar residue such as asparagine (Asn), aspartic acid (Asp), or glutamic acid (Glu).14,15 These residues form part of the active site of the enzyme and are equivalent to the catalytic triad of papain-like thiol proteases.13
In our earlier studies, we have demonstrated the potentcy of an anti-staphylococcal molecule P128 against globally prevalent antibiotic resistant S. aureus isolates with potent efficacy against methicillin-resistant S. aureus in a rat nasal colonization model.2 This molecule has also been found to be stable in human serum, plasma and blood with no cytotoxic effect on eukaryotic cells such as Hep2 and Vero cells.16
In this paper, we have compared the properties of his-tagged lysostaphin, the bacteriocin produced by S. simulans biovar staphylolyticus, known to cleave the cross-linking pentaglycine bridges of the cell walls of staphylococci11,17,18 and the staphylolytic protein P128.
Site-directed mutagenesis of the conserved cysteine and histidine residues in the CHAP domain have confirmed their importance in the endopeptidase activity of the bacteriophage B30 lysin and within the CHAP domain of the LysWMY staphylococcal lysin.19,20 To gain more insights into the P128 molecule, we have performed systematic study on mutations of selected residues of the catalytic domain of this molecule and report their effect on its staphylolytic activity.
Results
Cloning, expression and purification of wild-type and mutant P128 proteins
The DNA and amino acid sequences of wild-type P128 and lysostaphin are shown in Figure S1. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed that all constructs expressed protein of expected sizes, where wild-type P128 and P128 mutants were of ~27 kDa and the recombinant lysostaphin was of ~32 kDa. The purified proteins were > 95% pure as judged by RP-HPLC and SDS-PAGE (data not shown).
Effect of lysostaphin and P128 on S. simulans biovar staphylolyticus
Lysostaphin at 50 µg and 100 µg protein did not show any lytic activity on S. simulans biovar staphylolyticus while P128 showed > 99.9% to 99.999% killing of the same cells under similar experimental conditions (Fig. 1).
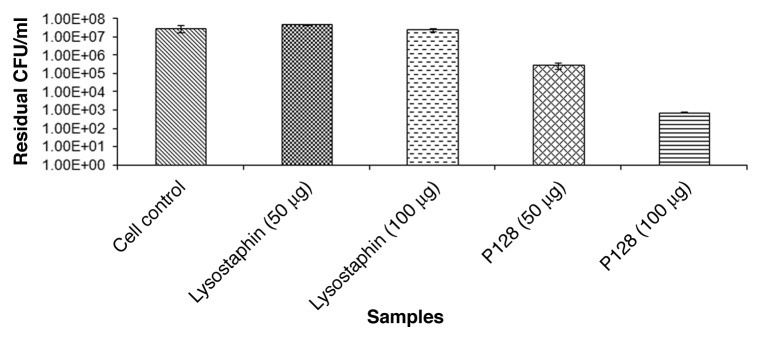
Figure 1. Effect of purified lysostaphin and P128 on S. simulans biovar staphylolyticus. Sample1: cell control; sample 2: cells +50 µg of lysostaphin; Sample 3: cells with 100 µg of lysostaphin; sample 4: cells with 50 µg of P128; sample 5: cells with 100 µg of P128. Detailed experimental conditions followed are described in Materials and Methods section.
Effect of heat on lysostaphin and P128
The results (Fig. 2) indicate that lysostaphin is less heat stable in comparison to P128. Nearly 7 log units reduction in CFU was seen with P128 while lysostaphin showed ~5 log unit reduction under the same experimental conditions when both the proteins were exposed to 37 °C for 1 h. While lysostaphin lost all its activity when exposed to 70 °C for 15 min, P128 retained its activity indicating its thermostable property.
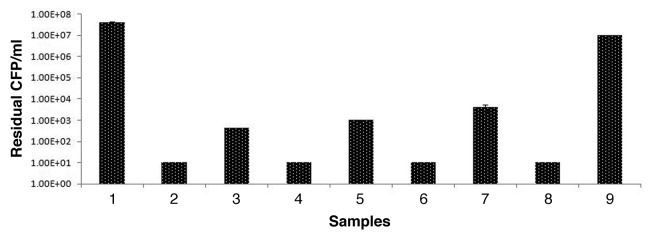
Figure 2. Effect of heat on lysostaphin and P128 preparation. Residual activity tested on S. carnosus cells by viability assay (CFU drop) in triplicates. Values are mean ± SD. Sample 1: cell control; sample 2: with P128 untreated; sample 3: with lysostaphin untreated; sample 4: with P128 after exposure to 37 °C, 15 min; sample 5: with lysostaphin after exposure to 37 °C, 15 min; sample 6: with P128 after exposure to 55 °C; sample 7; with lysostaphin after exposure to 55 °C for 15 min; sample 8: with P128 after exposure to 70 °C; sample 9; with lysostaphin after exposure to 70 °C for 15 min.
Effect of pH on activity of lysostaphin and P128
Both the lysostaphin and P128 proteins showed similar activity at all pHs tested (Fig. S2).
Rate of bactericidal activity of lysostaphin and P128
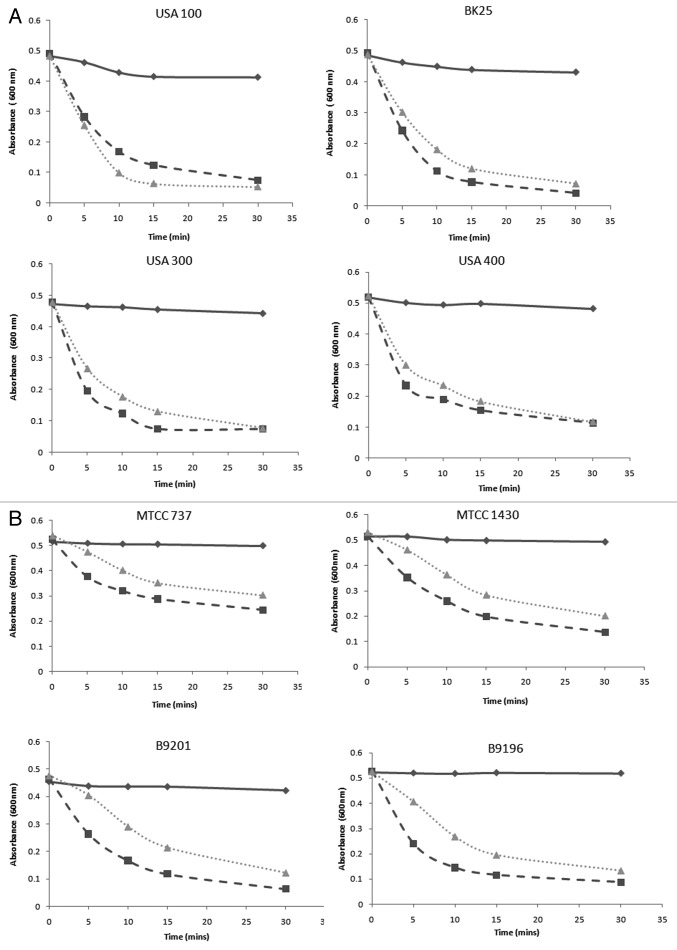
It is evident from Figure 3A and 3B that P128 showed higher rate of activity in comparison to lysostaphin for all the strains tested. For the MRSA strains, at the end of 10 min, P128 showed nearly 65–75% reduced turbidity while lysostaphin showed 60% turbidity reduction in almost all the cases. Similar results were seen for MSSA strains.
Figure 3. Effect of P128 and lysostaphin on selected Staphylococcus isolates by turbidity reduction assay. (A) Four MRSA isolates namely USA 100, USA 300, USA 400, and BK25 were grown in LB and taken for turbidity reduction assay with and without 6 µg each of lysostaphin and P128. Experiments were done in triplicates and values are mean ± SD. Control samples are represented by solid lines, P128 treated samples are represented by dashed lines while lysostaphin treated samples are represented by dotted lines. (B) Four MSSA isolates namely MTCC737, MTCC1430, B9201, and B9196 were grown in LB and taken for turbidity reduction assay with and without 6 µg each of lysostaphin and P128. Experiments were done in triplicates and values are mean ± SD. Control samples are represented by solid lines, P128 treated samples are represented by dashed lines while lysostaphin treated samples are represented by dotted lines.
From the studies described above, it is clear that P128 has better enzymatic properties in comparison to lysostaphin. To have better understanding of P128 at the molecular level, we modified selected residues of the P128 molecule and examined effect of such changes on its anti-staphylococcus activity and also tested P128 activity in presence of reducing agents such as DTT (5 mM) and thimerosal (a thiol protease inhibitor) at 0.001% for understanding the criticality of cysteine residues for the biological activity of P128 (Fig. S3).
Conserved residues in P128
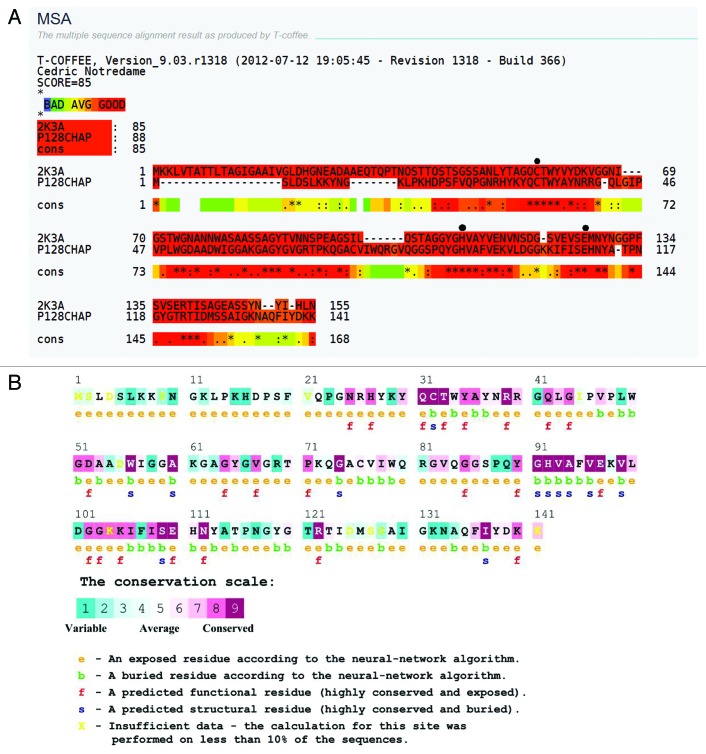
T-Coffee alignment of the P128 CHAP domain with that of the S. saprophyticus protein 2K3A shows conserved residues at positions Cys32, His92, and Glu110 (Fig. 4A). ConSeq results show an amino acid conservation score of 9.0 for the putative catalytic triad residues of Cys32, His92, and Glu110 and high scores for other residues that contribute to catalytic activity (e.g., Gln31, Thr33, Gly51, Gly91, and Asn112) (Fig. 4A). In addition, ConSeq results indicated several additional residues that appear to be conserved (Fig. 5B).
Figure 4. Conserved residues in P128 CHAP. (A) T-Coffee alignment of 2K3A and P128 CHAP domains. Highly conserved cysteine (C), histidine (H), and glutamic acid (E) residues in both proteins are identified as black dots. (B) ConSeq results for P128 CHAP protein. Residues and corresponding conservation scores are shown with color codes.
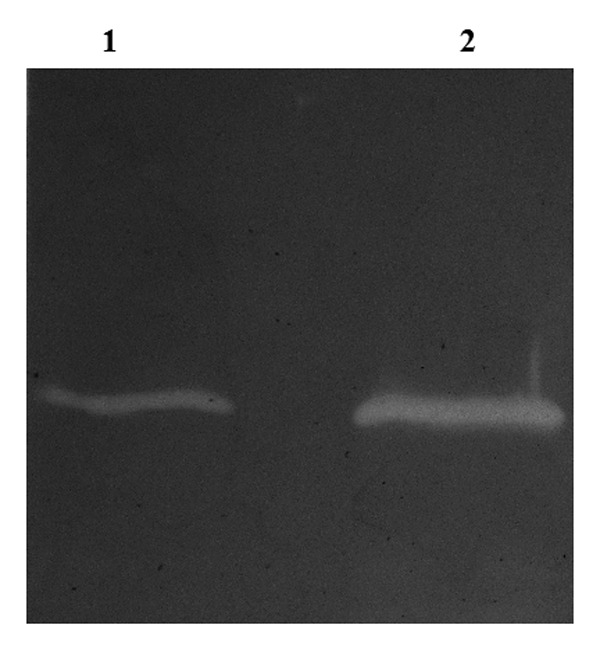
Figure 5. Zymogram analysis of P128 and P301 proteins. Zymogram was carried out with autoclaved cells of S. aureus RN4220 and 1 µg purified proteins. Lane 1: P301, lane 2: P128.
Bactericidal activity and zymogram analysis
The viability reduction activity values of wild-type and mutant P128 proteins is shown in Table 1. It is clear that the mutations of C32S, H92S, and E110D abolished staphylolytic activity of P128 since these amino acids form the catalytic triad of the CHAP domain (Table 1). It was also interesting to see that mutation of the other cysteine residue C76S resulted in loss of staphylolytic activity of P128, indicating the requirement of free cysteine for antistaphylococcal activity of P128. Surprisingly, mutation of a non-conserved residue like arginine at 40th position (R40S) also abolished P128 activity, as determined by the viability assay, although it retained the catalytic activity by zymogram assay (Table 1),
Table 1. Details of P128 and its mutant variants and their activity profiles.
| Construct | Point of mutation | Primers used for carrying out SDMa | Viability reduction assayb | Activity on Zymogramc |
|---|---|---|---|---|
| pGDC 173 |
Native P128 |
Nil |
>99.9% killing |
Active |
| pGDC 175 |
Cys32 to Ser32 |
FP 5′-CATTATAAGT ATCAGAGCAC ATGGTATGC-3′ RP 5′-GCATACCATG TGCTCTGATA CTTATAATG-3′ |
Inactive |
Inactive |
| pGDC 276 |
His92 to Thr92 |
FP 5′-AGCCCACAAT ATGGTACCGT AGCGTTTGTA GAG-3′ RP 5′-CTCTACAAAG CGTACGGTAC CATATTGTGG GCT-3′ |
Inactive |
Inactive |
| pGDC 187 |
Glu110 to Asp110 |
FP 5′-ATATTTATCT CTGACCATAA CTATGCTACC-3′ RP 5′-GGTAGCATAG TTATGGTCAG AGATAAATAT-3′ |
Inactive |
Inactive |
| pGDC 171 |
Cys76 to Ser76 |
FP 5′-CCTAAACAAG GTGCTAGCGT TATATGGCAA AGAGG-3′ RP 5′-CCTCTTTGCC ATATAACGCT AGCACCTTGT TTAGG-3′ |
Inactive |
Inactive |
| pGDC 301 | Arg40 to Ser40 | FP 5′-TGGTATGCTT ATAATTCTAG AGGTCAATTA GGC-3′ RP 5′-GCCTAATTGA CCTCTAGAAT TATAAGCATA CCA-3′ |
Inactive | Active |
a Sequence in bold refer to the point of mutation. bAssay performed with crude protein preparation. cZymogram gels with S. aureus RN4220 autoclaved cells.
As shown in Figure 5, wild-type P128 and the R40S mutant (P301) showed activity in the zymogram gel containing staphylococcus cells. Since no other P128 mutants showed zymogram activity, studies were restricted to P128 and the R40S mutant alone.
Binding, western blot, and CD analysis
Of all the protein variants tested, only P301 and P128 proteins showed activity on zymogram,, hence all subsequent assays were carried out only on these two proteins.
The negligible staphylolytic activity of P301 in vitro suggested possibility of its reduced binding to the bacterial cell wall. However, the P128 western blot analysis showed that both P128 and P301 proteins were able to bind to Staphylococcus (Fig. S4) with similar efficiencies. This indicated that the binding domains of both P128 and P301 maintain their original conformation which is substantiated by similar secondary composition by CD spectra analysis (data not shown). The antibody used for the P128 western blot studies showed signal only with the P128 protein (Fig. S5, lane 2) and did not light up any of the E. coli host proteins (Fig. S5, lane 1) showing the specificity of the antibody used.
In silico modeling
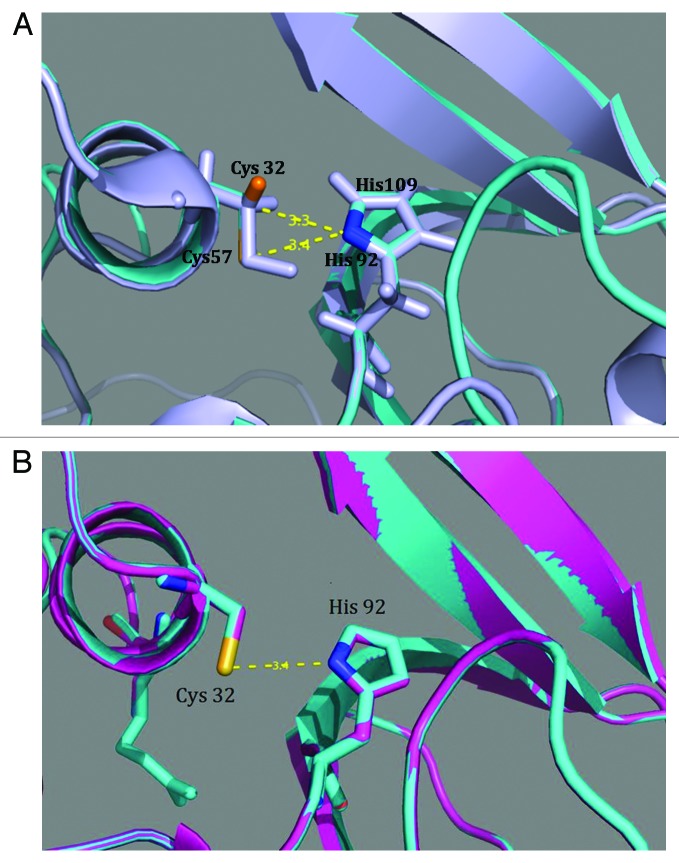
The superimposed models for 2K3A (lavender) and the 16-kDA catalytic domain of P128 (cyan) show the highly conserved cysteine and histidine residues in the active site; dotted lines indicate the distance in Angstrom units (Fig. 6A). The distance between cysteine and histidine residues is the same for P128 CHAP and P301 (Fig. 6B).
Figure 6. In silico modeling. (A) Superimposition of highly conserved Cys-His active site residues of 2K3A (lavender) used for homology modeling and in silico model of P128 CHAP (cyan). Conserved residues Cys57 and His117 (2K3A) and Cys32 and His92 (P128 CHAP) are labeled along with distance measurements. Broken lines with labels indicate distance (in Å) between residues. (B) Superimposition of in silico model of wild-type P128 CHAP (cyan) and the arginine 40 mutant P301 (magenta) showing the same distance between cysteine and histidine residues (in Å). Figures were generated using PyMOL.
Kinetic characterization studies
Table 2 shows the kinetic characterization values of native P128, and its R40S mutant P301 and lysostaphin. While the Vmax (∆OD600 [mg/min]) of the native P128 was found to be 29365 milliunits/min/mg, the R40S mutant P301 showed a Vmax value of 108 milliunits/min/mg while lysostaphin showed a value of 19800 milliunits/min/mg. The apparent Km (KmAPP) of the P128 native protein was 34 ng /ml while it was 9260 ng /ml for the R40S mutant P301 and lysostaphin showed an KmAPP of 50 ng /ml. KcatAPP of the native P128 was 0.074 moles while it was 20.5 n moles for the P301 protein and 0.12 n moles for lysostaphin. This data indicates that the arginine mutation causes at least 270 fold reduction in P128 activity in comparison to the unmodified P128 protein and P128 has better activity properties in comparison to lysostaphin under the experimental conditions described.
Table 2. Kinetic values for native P128, Arginine mutant of P128 (P301) and recombinant lysostaphin.
| Protein samples |
Vmax (∆OD600 [mg/min]) | KmAPP (ng/ml) | KcatAPP (molecules/sec) | KcatAPP (nmoles/s) |
|---|---|---|---|---|
| P128 |
29365 milliunits/min/mg |
34.00 |
4.4 × 1013 |
0.074 nmoles |
| P301 |
108 milliunits/min/mg |
9260.00 |
12.0 × 1016 |
20.50 nmoles |
| lysostaphin | 19800 milliunits/min/mg | 50.00 | 7.2 × 1013 | 0.120 nmoles |
Experiments were done in triplicates. Michaelis-Menten parameters are reported ± SD.
Discussion
A significant number of reports on lysostaphin resistant variants have been reported since its in-vitro use21-25 and endopeptidase resistance gene (epr) is the contributory factor for the observed lysostaphin resistance.26,27 The observations of P128 having lytic activity toward S. simulans biovar staphylolyticus indicates that the epr resistant gene does not affect P128 activity demonstrating a difference in the mechanism of action between these two staphylolytic proteins.
The observations of higher thermal stability of P128 over lysostaphin will have significant bearing in various biotechnological applications since thermostable proteins have longer survival times, better reaction kinetics and diminished microbial contamination.28 Since proteins of increased stability have longer serum half lives and are more resistant to mutations than the protein from which they are derived,27,29 our present observations of higher thermostability of P128 assumes critical importance. Reports on thermostable nature of a wide number of bacteriophage virion associated peptidoglycan hydrolysases also exists.30 Higher thermostability of proteins have been attributed to greater hydrophobicity, increased helical content and increased polar surface area.31 Additional work on P128 in these lines needs to be performed for deciphering reasons for its thermostable property. Interestingly enough, lysostaphin from the extracellular broth of S. simulans biovar staphylolyticus is also reported to be heat-labile.17
It has previously been reported that although deletion of the cell wall targeting domain of lysostaphin does not interfere with its endopeptidase activity, it abolishes its ability to distinguish between S. aureus and S. simulans biovar staphylolyticus.32 The present observations of P128 attacking S. simulans biovar staphylolyticus directly imply that the catalytic domain of P128 is more potent in comparison to the catalytic domain of the lysostaphin molecule. Also, the observations of higher rate of activity of P128 over lysostaphin in both MRSA and MSSA isolates along with better kinetic values of P128 (like Vmax, Km APP, Kcat APP) over lysostaphin supports our claims that the P128 has a potent catalytic domain in comparison to lysostaphin. Although, methicillin resistance does not give rise to resistance to lysostaphin per se, Sabath et al.33 observed that methicillin-resistant clones from heterogenously resistant staphylococcus strains were more resistant to lysostaphin due to variations in cell wall structure, antibiotic susceptibility pattern, and other toxin encoding genes in the host.34 Such variations do not seem to affect P128 since P128 is active on lysostaphin and methicillin resistant isolates.
The CHAP domain is present in a large number of enzymes that cleave peptidoglycan.13 Our results show that Cys26, His92, and Glu110, which form the catalytic triad of other CHAP proteins,15 are also essential for P128 activity. We also observed that Cys76 mutation results in complete loss of P128 activity, indicating the critical nature of this residue, even though sequence analysis do not indicate this as a conserved residue. This finding is consistent with that of Pritchard et al.19 who showed that modification of non-conserved cysteine residues in the CHAP domain of the B30 endolysin reduces activity. Interestingly, P128 activity was not affected by DTT while its activity was totally inhibited by thimerosal indicating the essentiality of both the cysteines of P128 for its lytic activity and implying the absence of a disulfide linkage in this molecule.
CD measurements in the far UV region play an important role in complementing the higher resolution structural approaches of X-ray crystallography and nuclear magnetic resonance. Also, secondary and tertiary structural characteristics of a range of mutant proteins can be assessed very rapidly by CD.35 Our results on similar CD spectra for both the native P128 and its R40S mutant (P301) reflect similar secondary composition. Also, similar property of binding of P128 and P301 to S. aureus cells is consistent with the observations of Sakurada et al.,36 who reported that the anti-staphylococcal protein lysostaphin binds to cells that are resistant to its staphylolytic action.
Zymography is a method to detect cell wall hydrolase activity on dead cells impregnated in the SDS-PAGE.37 Our observations of P301 demonstrating activity only on zymogram gel with no CFU reduction are similar to those of Sass and Bierbaum38 and Raltson et al.39 for a phage endolysin. It is tempting to speculate that the zymogram positive reaction with the P301 protein could be due to the extended time of exposure (more than 16 h) of the protein to the impregnated substrate while the insignificant activity of P301 in the viability assay (CFU drop) (Table 1) could be attributed to less exposure of the P301 protein to the live cells (1 h) during the viability assay experimental conditions. Conversely, Rodriguez et al.40 has described bacteriophage-derived tail enzymes that exhibit lytic activity against S. aureus Sa9 cells by viability assay but no activity on zymogram gels.
Use of kinetic parameters like Km APP, Vmax, Kcat APP for assessing activity of native and mutated antibacterial agents like lysozyme is well documented.41 The observations of differences in catalytic properties of the native P128 and P301 reported in this article support such literature reports. The observation of significantly lesser apparent Km value for the native P128 in comparison to its R40S mutant P301 indicates higher affinity of P128 for the substrate in comparison to the R40S mutant. The observations of drastic reduction in the activity of the P128 protein after arginine 40 substitution indicates that residues in CHAP domain, conserved or non-conserved, could play critical role in the activity of such antimicrobial agents. The higher apparent Km value for lysostaphin in comparison to P128 indicates higher potency of the P128 over lysostaphin and this could be attributed to the potent activity of the catalytic domain of P128 over lysostaphin since these two molecules have similar cell wall binding domains.
In general, coagulase-negative staphylococci were found to be relatively less susceptible to the lytic action of lysostaphin than coagulase-positive staphylococci.42 In the coagulase-negative staphylococcal species namely Staphylococcus simulans and Staphylococcus capitis, incorporation of serine residues into the third and fifth position of the interpeptide bridge, through the activities of the lif and epr genes in S. simulans and S. capitis, respectively, is believed to be responsible for the natural low susceptibility to lysostaphin of these species.21,43 However, Koehl et al.44 report that the tested staphylococcus strains did not show any difference in the pentaglycine composition but were different in endogenous autolysin activity and such endogenous autolysins have been implicated in conferring sensitivity to lysostaphin on whole cells. It is interesting to see that susceptibility of both MRSA and MSSA are similar for P128 and lysostaphin while the MSSA strains showed better susceptibility to P128 than lysostaphin. The present observation of P128 having better activity in MSSA strains possibly suggests that the autolysins do not affect the activity of P128.
Studies described in this paper reveal binding site locations and identify residues that are likely to take part in the endopeptidase reaction of P128 with the peptidoglycan residues of Staphylococcus aureus cells. It is expected that the present work will provide a better understanding of the molecular mechanisms involved in catalysis of P128 and would assist us for predicting catalytic residues in P128 in future which would be achieved by site-directed mutagenesis, kinetic analyses of analogs and synthetic substrates. Work on further sequential mutations of the P128 molecule for achieving a higher and better stable P128 molecule is in progress.
Materials and Methods
Chemicals, strains, and plasmids
All chemicals and customized oligonucleotides were procured from Sigma-Aldrich Co. The P128 polyclonal antibody was obtained from Abexome Biosciences, and the restriction endonucleases and E. coli ER2566 were purchased from New England Biolabs and Bangalore Genei Pvt. Ltd. The Quick-Change Lightning site-directed mutagenesis (SDM) kit was purchased from Stratagene (210518). The S. aureus strain COL was a kind gift from Dr. Barry Kreiswirth (Public Health Research Institute, Newark, NJ USA), and S. aureus RN4220 was a kind gift from Dr Richard Novick (Skirball Institute, New York, NY USA). MRSA strains (USA100, USA300, USA400, and BK25) were obtained from Dr Barry Kreiswirth (Public Health Research Institute Center, NJ USA) while MSSA isolates MTCC 737 and MTCC 1430 were procured from Microbial Type Culture collection Centre, IMTECH, Chandigarh, India and the other two MSSA isolates namely B9201 and B9196 were collected from various hospitals in and around Bangalore, India.
The BioTrace nitrocellulose blotting membrane was purchased from Pall Corporation (P/N 66485) while the Ni-NTA agarose was from Qiagen (30230). The Escherichia coli strain DH5α was purchased from Bangalore Genei Pvt. Ltd., and the pET26b vector used for cloning P128 and its variants was purchased from Merck AG (69862-3) while pRG5 plasmid was procured from ATCC (ATCC® 67076™). pRSETA plasmid was procured from Invitrogen.
Construction of lysostaphin expression cassette
The mature peptide of the lysostaphin gene was amplified using pRG5 plasmid DNA as a template PCR. The forward primer (5′-CGCGGATCCC AGGTTGTACA GAATGCTGCA ACACATGAAC ATTCAGC-3′) and the reverse primer (5′-CGCGGATCAC TTTATAGTTC CCCAAAGAAC ACC-3′) were used for the PCR conditions that included an initial denaturation step of 95 °C for 5 min, followed by 8 cycles of 95 °C for 30 s, 53 °C for 30 s, and 72 °C for 1 min and 22 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min with a final extension at 72 °C for 7 min. The amplified PCR product was digested with the BamHI and ligated pRSETA vector at the same site. Recombinant clones were screened by PCR and confirmed by DNA sequencing.
Purification of recombinant His-lysostaphin
The induced cell pellet of E. coli ER2566 cells with the plasmid containing the lysostaphin gene under the transcriptional control of the T7 promoter (0.26 L culture equivalent to 43.4 g wet weight) was resuspended and lysed in 400 ml of 0.1 M Tris base solution containing 0.5 M NaCl and 0.1 mM EDTA and sonicated for 20 min. Post lysis, the pH of the crude lysate was adjusted to 8.0 with 1N HCl and imidazole and polyethylene imine (PEI) were added at 10 mM and 0.1% concentration respectively.
The clarified material obtained upon centrifugation of the crude lysate at 15,880 x g for 20 min was loaded onto a Ni2+–NTA affinity column pre-equilibrated with equilibration buffer (0.1 M Tris.HCl, pH 8.0 with 0.5 M NaCl, 0.1 mM EDTA and 0.01M imidazole). Five bed volumes of the wash buffer (0.1 M Tris. HCl, pH 8.0 containing 0.5 M NaCl, 0.1 mM EDTA, and 0.02 M imidazole) and bound proteins was eluted by applying imidazole gradient of 0.02 M to 0.3M imidazole in 0.1 M Tris. HCl, pH 8.0 containing 0.5 M NaCl, and 0.1 mM EDTA. All the fractions were analyzed on 12% SDS-PAGE and the fractions of interest were pooled and dialyzed against 100 volumes of 0.05 M Tris. HCl, pH 7.5 at 4 °C for 16 h followed by filter sterilization using 0.2 µM syringe filter.
Cloning, expression, and purification of wild-type and mutant P128 proteins
P128 cloning and expression details have been described elsewhere.2 The wild-type P128 plasmid was used as a template to generate mutants by SDM, which were sequenced before use in expression studies. The wild-type and mutant P128 proteins were purified by ammonium sulfate (20–50% saturation) precipitation followed by ion-exchange column (Merck-Millipore, CM52) at pH 8.0. Comparison studies of P128 was done in detail only with its R40S mutant, (R40S) designated as P301, since only this mutant showed catalytic activity other than P128.
Bactericidal activity assay
The Staphylococcus aureus strain COL, is a methicillin resistant staphylococcus aureus isolate that expresses several proteases and toxins.45 This strain was grown in Luria-Bertani (LB) broth until A600 nm reached 1.0 and then diluted in fresh LB broth to obtain 1 × 108 cells /ml. Aliquots of the cell suspension (100 μl, 107 cells) were treated with 100 μl of crude or purified wild-type or mutant protein (10 μg /ml) and incubated at 37 °C for 60 min with shaking at 200 rpm. Viable cells were enumerated as colony-forming units (CFUs) by serial dilution and plating on LB agar plates.
Properties and characterization studies of purified recombinant lysostaphin and P128
The recombinant lysostaphin and P128 purified proteins were tested for their activity profiles by viability assay, turbidity reduction assay, thermostability studies and rate of killing on four randomly chosen MRSA and MSSA isolates. The protocols followed for various methods are described below.
Effect of heat on lysostaphin and P128
Ten micrograms proteins of purified lysostaphin and P128 were treated at 37, 55, and 70 °C in a water bath for 1 h. The samples were brought to room temperature by cooling on ice and then used for bactericidal assay as reported earlier.3
Effect of pH on activity of lysostaphin and P128
S. aureus ATCC 25923 cells46 (1 × 109 cells /ml) grown in LB broth was taken for this study. Nearly 2 × 108 cells in buffers of different pHs (5.0–9.0) were taken in a 1 ml cuvette after which 5 µg of purified P128 and lysostaphin was added separately and the absorbance was measured at 5 min intervals. For comparing activity at various pHs, the activity achieved at pH 7.0 was taken as 100% and a graph of % activity versus pH was plotted. Values are reported as average ± SD. For pH 5.0 and 6.0, 50 mM sodium acetate was used, for pH 7.0 and 8.0, 50 mM Tris.HCl was used while for pH 9.0, 50 mM glycine-NaOH was used. Since lysostaphin is known to be stable with 150 mM sodium chloride,47 we included 150 mM sodium chloride in all the buffers for the present assay.
Effect of lysostaphin and P128 on S. simulans biovar staphylolyticus
Staphylococcus simulans biovar staphylolyticus is known to secrete the glycylglycine endopeptidase called lysostaphin.48 The post-exponential-phase cultures of this strain is resistant to the action of lysostaphin, due to a plasmid based resistance factor called endopeptidase resistance gene epr11,49 and we wished to evaluate if P128 activity is affected by such a resistant factor.
Rate of activity of lysostaphin and P128
To determine the rate of activity of lysostaphin and P128, we randomly chose four MRSA and four MSSA isolates from our staphylococcus library and followed activity of both the proteins (with 6 µg protein) by turbidity reduction assay as described earlier.3
Conserved residues in P128
The CHAP domain is reported to contain a catalytic triad of cysteine, histidine, and a polar residue such as glutamic acid. Rossi et al.15 have identified a Cys-His-Glu-Asn proteolytic relay in a CHAP domain protein from S. saprophyticus by structural analysis.
While conserved residues in the CHAP domain were identified by comparing P128 with 2K3A (a CHAP domain protein from Staphylococcus saprophyticus) using the T-Coffee multiple sequence alignment program (http://www.tcoffee.org),50,51 functionally and structurally important residues in the 16-kDa catalytic domain of P128 containing the CHAP domain was analyzed using the ConSeq Server (http://conseq.tau.ac.il) as done by Berezin et al.52 Homologs were identified using PSI-BLAST53 and repeated for three iterations with a cutoff E-value of 0.0001 and unique sequences used for calculations.
Site-directed mutagenesis
The oligonucleotides used to generate mutations are shown in Table 1. PCR conditions included initial denaturation at 95 °C for 30 s, followed by 18 cycles of denaturation step at 95 °C for 30 s and annealing at 68 °C for 1 min, with a final extension at 68 °C for 5 min. PCR products were digested with DpnI and introduced into competent cells of E. coli Top10. Clones were screened by restriction digestion, and confirmed by DNA sequencing.
Circular dichroism spectra
To evaluate changes in secondary structure, wild-type P128 and the R40S mutant P301 were purified and subjected to circular dichroism (CD) spectroscopy. Far UV CD spectra were recorded at 25 °C from 260 to 195 nm using a JASCO J-715 spectropolarimeter and a quartz cuvette with 2-mm path length. Proteins were measured at 0.25 mg /ml in 20 mM sodium acetate buffer (pH 6.0). The K2d server (http://www.embl-heidelberg de andrade k2d) was used to estimate the percentages of protein secondary structure.54 The spectra represent scans after correction for buffer baseline and were reported as mean residue ellipticity (θ).
In silico modeling
The automated protein structure homology-modeling server SWISS MODEL (http://swissmodel.expasy.org/) was used for in silico model building.55 The modeled residue range was 20 to 139 based on template 2K3A. The template has 30% sequence identity to the 16-kDa catalytic domain of P128 and is therefore suitable for homology modeling. The modeling figures were generated using PyMOL.56
Zymogram studies
Zymography were performed as described by Lepeuple et al.57 where protein samples (crude or purified) were separated by 12% SDS-PAGE on gels containing 0.2% autoclaved S. aureus RN4220 cells. After electrophoresis, the zymograms were washed for 30 min with distilled water at room temperature followed by transfer to a buffer (25 mM Tris.HCl, pH 7.5 with 0.1% Triton X-100) for 16 h at 37 °C for renaturation. The gels were further stained with 0.1% methylene blue and 0.001% KOH for 2 h at room temperature and the muralytic activity was detected as a clear zone against the dark blue background of methylene blue.
Binding studies and western blot analysis of P128 and P301
Staphylococcus aureus strain RN4220 was grown overnight in LB medium at 37 °C and the following day, mid-log-phase cultures of RN4220 of 10 OD600 were used. Suitable volumes of the bacterial suspensions (1 × 1010 CFU /ml) were mixed with 10 µg reaction of the purified P128 protein and P301 proteins and incubated for 10 min on ice. Proteins bound to staphylococci was collected by centrifugation for 5 min at 16,000 × g, and supernatant with unbound protein was removed. Bacterial sediments were suspended in sample buffer, and were analyzed by 10% SDS-polyacrylamide gel electrophoresis. The abundances of the proteins were assessed by western blot using polyclonal anti-P128 antibody.
The proteins from SDS-PAGE were transferred to a nitrocellulose blotting membrane and blocked overnight in 1× TBS Tween containing 3% bovine serum albumin. After washing with 1× TBS-Tween, the membrane was incubated with anti-P128 antibody (1: 12,00,000) followed by three washes with 1× TBS-Tween. After incubatation with the secondary goat anti-rabbit alkaline phosphatase conjugate (1:500), the blot was developed using BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) solution. Suitable controls were run for determining specificity of the P128 antibody used.
Kinetic characterization studies
The catalytic properties of native P128, P301 and recombinant lysostaphin was examined by turbidity reduction assay with S. carnosus cells since S. carnosus is a GRAS organism and is protease free and is the most susceptible strain for lysozymes.58 The assay was performed in a total reaction volume of 250 µl with S. carnosus cell suspension (1 OD600) and suitable amounts of all the test proteins (2 µg /ml) in a microplate format in Spectramax Plus 381 (Molecular Devices). The reaction was monitored for 30 min and readings were taken at an interval of 1 min using Softmax Pro 5.0 version. Vmax, apparent Km (Km APP) and apparent Kcat (Kcat APP) values were calculated based on the △OD obtained. While Vmax was defined as the maximum rate of reaction (∆A600) obtained for 1 mg of protein per min following the protocol described by Scanlon et al.,59 APP was defined as that amount of protein that is required to bring a fall in the absorbance of cell suspension at 600 nm by 0.001 units per min under specified conditions of pH 7.0 (25 mM Tris. HCl + 150 mM sodium chloride) at 37 °C.58 Kcat APP is the turnover number of the enzyme defined as number of protein molecules used per second for fall in the OD of cell suspension by 0.001 (1 milliunit).
Supplementary Material
Acknowledgements
We would like to thank the Department of Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India, for carrying out the CD analysis. Authors are also indebted to Dr J Ramachandran, Chairman and CEO, GangaGen Inc., USA, for his constant support and encouragement. Preliminary work related to thermostability studies of lysostaphin and P128 performed by Ms Thejashree Sarapalle and Ms Veena Gopalakrishna is gratefully acknowledged. Authors also thank Mr Murali Durgaiah for carrying out the effect of thimerosal on P128 activity.
Glossary
Abbreviations:
- CD
circular dichroism
- CFU
colony forming units
- CHAP
Cysteine, histidine dependent amino peptidase/hydrolase
- GRAS
Generally regarded as safe
- MRSA
Methicillin resistant Staphylococcus aureus
- MSSA
Methicillin sensitive Staphylococcus aureus
- SDM
Site-Directed Mutagenesis
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TBS
Tris buffered saline
Citation: Saravanan S, Paul V, George S, Sundarrajan S, Kumar N, Hebbur M, Kumar N, Veena A, Maheshwari U, Appaiah C, et al. Properties and mutation studies of a bacteriophage-derived chimeric recombinant staphylolytic protein P128: Comparison to recombinant lysostaphin. Bacteriophage 2013; 3:e26564; 10.4161/bact.26564
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/bacteriophage/article/26564
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/26564
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–32. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Paul VD, Rajagopalan SS, Sundarrajan S, George SE, Asrani JY, Pillai R, Chikkamadaiah R, Durgaiah M, Sriram B, Padmanabhan S. A novel bacteriophage Tail-Associated Muralytic Enzyme (TAME) from Phage K and its development into a potent antistaphylococcal protein. BMC Microbiol. 2011;11:226. doi: 10.1186/1471-2180-11-226. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vipra AA, Desai SN, Roy P, Patil R, Raj JM, Narasimhaswamy N, Paul VD, Chikkamadaiah R, Sriram B. Anti-staphylococcal activity of bacteriophage derived chimeric protein P128. BMC Microbiol. 2012;12:1. doi: 10.1186/1471-2180-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans HL, Sawyer RG. Cycling chemotherapy: a promising approach to reducing the morbidity and mortality of nosocomial infections. Drugs Today (Barc) 2003;39:733–8. doi: 10.1358/dot.2003.39.9.799480. [DOI] [PubMed] [Google Scholar]
- 5.Anderson B, Rashid MH, Carter C, Pasternack G, Rajanna C, Revazishvili T, Dean T, Senecal A, Sulakvelidze A. Enumeration of bacteriophage particles: Comparative analysis of the traditional plaque assay and real-time QPCR- and nanosight-based assays. Bacteriophage. 2011;1:86–93. doi: 10.4161/bact.1.2.15456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sulakvelidze A, Kutter E. Bacteriophage therapy in humans. In: Kutter E, Sulakvelidze A. (eds.) Bacteriophages: Biology and Application, Boca Raton, FL, CRC Press, 2005: 381-436. [Google Scholar]
- 7.Paul VD, Sundarrajan S, Rajagopalan SS, Hariharan S, Kempashanaiah N, Padmanabhan S, Sriram B, Ramachandran J. Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. BMC Microbiol. 2011;11:195. doi: 10.1186/1471-2180-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastagia M, Euler C, Chahales P, Fuentes-Duculan J, Krueger JG, Fischetti VA. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob Agents Chemother. 2011;55:738–44. doi: 10.1128/AAC.00890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padmanabhan S, Paul VD, Saravanan SR, Bharathi S. Phage derived antimicrobial activities. United States Patent No. US 8,202,516 B2 USA, 2012.
- 10.O’Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J Bacteriol. 2004;186:2862–71. doi: 10.1128/JB.186.9.2862-2871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastos MCF, Coutinho BG, Coelho MLV. Lysostaphin: A Staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals (Ott) 2010;3:1139–61. doi: 10.3390/ph3041139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigden DJ, Jedrzejas MJ, Galperin MY. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends Biochem Sci. 2003;28:230–4. doi: 10.1016/S0968-0004(03)00062-8. [DOI] [PubMed] [Google Scholar]
- 13.Bateman A, Rawlings ND. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci. 2003;28:234–7. doi: 10.1016/S0968-0004(03)00061-6. [DOI] [PubMed] [Google Scholar]
- 14.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi P, Aramini JM, Xiao R, Chen CX, Nwosu C, Owens LA, Maglaqui M, Nair R, Fischer M, Acton TB, et al. Structural elucidation of the Cys-His-Glu-Asn proteolytic relay in the secreted CHAP domain enzyme from the human pathogen Staphylococcus saprophyticus. Proteins. 2009;74:515–9. doi: 10.1002/prot.22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George SE, Chikkamadaiah R, Durgaiah M, Joshi AA, Thankappan UP, Madhusudhana SN, Sriram B. Biochemical characterization and evaluation of cytotoxicity of antistaphylococcal chimeric protein P128. BMC Res Notes. 2012;5:280. doi: 10.1186/1756-0500-5-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schindler CA, Schuhardt VT. Lysostaphin: A new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci U S A. 1964;51:414–21. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindler CA, Schuhardt VT. Purification and properties of lysostaphin--a lytic agent for S. aureus. Biochim Biophys Acta. 1965;97:242–50. doi: 10.1016/0304-4165(65)90088-7. [DOI] [PubMed] [Google Scholar]
- 19.Pritchard DG, Dong S, Baker JR, Engler JA. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology. 2004;150:2079–87. doi: 10.1099/mic.0.27063-0. [DOI] [PubMed] [Google Scholar]
- 20.Yokoi KJ, Kawahigashi N, Uchida M, Sugahara K, Shinohara M, Kawasaki K, Nakamura S, Taketo A, Kodaira K. The two-component cell lysis genes holWMY and lysWMY of the Staphylococcus warneri M phage varphiWMY: cloning, sequencing, expression, and mutational analysis in Escherichia coli. Gene. 2005;351:97–108. doi: 10.1016/j.gene.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Boyle-Vavra S, Carey RB, Daum RS. Development of vancomycin and lysostaphin resistance in a methicillin-resistant Staphylococcus aureus isolate. J Antimicrob Chemother. 2001;48:617–25. doi: 10.1093/jac/48.5.617. [DOI] [PubMed] [Google Scholar]
- 22.Climo MW, Ehlert K, Archer GL. Mechanism and suppression of lysostaphin resistance in oxacillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1431–7. doi: 10.1128/AAC.45.5.1431-1437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Climo MW, Patron RL, Goldstein BP, Archer GL. Lysostaphin treatment of experimental methicillin-resistant Staphylococcus aureus aortic valve endocarditis. Antimicrob Agents Chemother. 1998;42:1355–60. doi: 10.1128/aac.42.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strandén AM, Ehlert K, Labischinski H, Berger-Bächi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover FC, Biddle JW, Lancaster MV. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg Infect Dis. 2001;7:327–32. doi: 10.3201/eid0702.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heath LS, Heath HE, Sloan GL. Plasmid-encoded lysostaphin endopeptidase gene of Staphylococcus simulans biovar staphylolyticus. FEMS Microbiol Lett. 1987;44:129–33. doi: 10.1111/j.1574-6968.1987.tb02256.x. [DOI] [Google Scholar]
- 27.Heath HE, Heath LS, Nitterauer JD, Rose KE, Sloan GL. Plasmid-encoded lysostaphin endopeptidase resistance of Staphylococcus simulans biovar staphylolyticus. Biochem Biophys Res Commun. 1989;160:1106–9. doi: 10.1016/S0006-291X(89)80117-2. [DOI] [PubMed] [Google Scholar]
- 28.Bruins ME, Janssen AEM, Boom RM. Thermozymes and their applications: a review of recent literature and patents. Appl Biochem Biotechnol. 2001;90:155–86. doi: 10.1385/ABAB:90:2:155. [DOI] [PubMed] [Google Scholar]
- 29.Kiss C, Temirov J, Chasteen L, Waldo GS, Bradbury ARM. Directed evolution of an extremely stable fluorescent protein. Protein engg. design and selection. Protein Eng. 2009;22:313–23. doi: 10.1093/protein/gzp006. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-Rubio L, Martínez B, Donovan DM, Rodríguez A, García P. Bacteriophage virion-associated peptidoglycan hydrolases: potential new enzybiotics. Critical Reviews Microbiol. 2012;Early Online:1–8. doi: 10.3109/1040841X.2012.723675. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Tsai CJ, Nussinov R. Factors enhancing protein thermostability. Protein Eng. 2000;13:179–91. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- 32.Baba T, Schneewind O. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996;15:4789–97. [PMC free article] [PubMed] [Google Scholar]
- 33.Sabath LD, Leaf CD, Gerstein DA, Finland M. Altered cell walls of Staphylococcus aureus resistant to methicillin. Nature. 1970;225:1074–1074. doi: 10.1038/2251074a0. [DOI] [PubMed] [Google Scholar]
- 34.Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol. 2006;44:108–18. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751:119–39. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Sakurada J, Murai M, Zhijun L, Usui A, Seki K, Kobayashi K, Sumi Y, Jitsukawa H, Masuda S. Efficient adsorption of lysostaphin on bacterial cells of lysostaphin-resistant Staphylococcus aureus mutant. Microbiol Immunol. 1993;37:29–34. doi: 10.1111/j.1348-0421.1993.tb03175.x. [DOI] [PubMed] [Google Scholar]
- 37.Prado Acosta M, Mercedes Palomino M, Allievi MC, Sanchez Rivas C, Ruzal SM. Murein hydrolase activity in the surface layer of Lactobacillus acidophilus ATCC 4356. Appl Environ Microbiol. 2008;74:7824–7. doi: 10.1128/AEM.01712-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sass P, Bierbaum G. Lytic activity of recombinant bacteriophage phi11 and phi12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl Environ Microbiol. 2007;73:347–52. doi: 10.1128/AEM.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ralston DJ, Baer BS, Lieberman M, Krueger AP. Virolysin: a virus-induced lysin from staphylococcal phage lysates. Proc Soc Exp Biol Med. 1955;89:502–7. doi: 10.3181/00379727-89-21859. [DOI] [PubMed] [Google Scholar]
- 40.Rodríguez L, Martínez B, Zhou Y, Rodríguez A, Donovan DM, García P. Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88. BMC Microbiol. 2011;11:138. doi: 10.1186/1471-2180-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YC, Yang D. Determination of lysozyme activities in a microplate format. Anal Biochem. 2002;310:223–4. doi: 10.1016/S0003-2697(02)00320-2. [DOI] [PubMed] [Google Scholar]
- 42.Zygmunt WA, Browder HP, Tavormina PA. Susceptibility of coagulase-negative staphylococci to lysostaphin and other antibiotics. Appl Microbiol. 1968;16:1168–73. doi: 10.1128/am.16.8.1168-1173.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daum RS, Gupta S, Sabbagh R, Milewski WM. Characterization of Staphylococcus aureus isolates with decreased susceptibility to vancomycin and teicoplanin: isolation and purification of a constitutively produced protein associated with decreased susceptibility. J Infect Dis. 1992;166:1066–72. doi: 10.1093/infdis/166.5.1066. [DOI] [PubMed] [Google Scholar]
- 44.Koehl JL, Muthaiyan A, Jayaswal RK, Ehlert K, Labischinski H, Wilkinson BJ. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48:3749–57. doi: 10.1128/AAC.48.10.3749-3757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravipaty S, Reilly JP. Comprehensive characterization of methicillin-resistant Staphylococcus aureus subsp. aureus COL secretome by two-dimensional liquid chromatography and mass spectrometry. Mol Cell Proteomics. 2010;9:1898–919. doi: 10.1074/mcp.M900494-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemaire S, Tulkens PM, Van Bambeke F. Contrasting effects of acidic pH on the extracellular and intracellular activities of the anti-gram-positive fluoroquinolones moxifloxacin and delafloxacin against Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:649–58. doi: 10.1128/AAC.01201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabala I, Jonsson IM, Tarkowski A, Bochtler M. Anti-staphylococcal activities of lysostaphin and LytM catalytic domain. BMC Microbiol. 2012;12:97. doi: 10.1186/1471-2180-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Browder HP, Zygmunt WA, Young JR, Tavormina PA. Lysostaphin: enzymatic mode of action. Biochem Biophys Res Commun. 1965;19:383–9. doi: 10.1016/0006-291X(65)90473-0. [DOI] [PubMed] [Google Scholar]
- 49.DeHart HP, Heath HE, Heath LS, LeBlanc PA, Sloan GL. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl Environ Microbiol. 1995;61:1475–9. doi: 10.1128/aem.61.4.1475-1479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39(Web Server issue):W13–7. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fenton M, Cooney JC, Ross RP, Sleator RD, McAuliffe O, O’Mahony J, Coffey A. In silico modeling of the staphylococcal bacteriophage-derived peptidase CHAP(K) Bacteriophage. 2011;1:198–206. doi: 10.4161/bact.1.4.18245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–4. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 53.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrade MA, Chacón P, Merelo JJ, Morán F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–90. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 55.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 56.De Lano WL. The PyMOL Molecular Graphics System, De Lano Scientific, San Carlos, CA, USA, 2002. [Google Scholar]
- 57.Lepeuple AS, Chapot-Chartier MP, Van Gemert E. Analysis of the bacteriolytic enzymes of the autolytic lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: identification of a prophage-encoded enzyme. Appl Environ Microbiol. 1998;64:4142–8. doi: 10.1128/aem.64.11.4142-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bera A, Biswas R, Herbert S, Götz F. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect Immun. 2006;74:4598–604. doi: 10.1128/IAI.00301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scanlon TC, Teneback CC, Gill A, Bement JL, Weiner JA, Lamppa JW, Leclair LW, Griswold KE. Enhanced antimicrobial activity of engineered human lysozyme. ACS Chem Biol. 2010;5:809–18. doi: 10.1021/cb1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.