Abstract
Cyclic AMP (cAMP) is a ubiquitous second messenger that regulates many proteins, most notably cAMP-dependent protein kinase (PKA). PKA holoenzymes (comprised of two catalytic (C) and two regulatory (R) subunits) regulate a wide variety of cellular processes, and its functional diversity is amplified by the presence of four R-subunit isoforms, RIα, RIβ, RIIα, and RIIβ. Although these isoforms all respond to cAMP, they are functionally non-redundant and exhibit different biochemical properties. In order to understand the functional differences between these isoforms, we screened cAMP derivatives for their ability to selectively activate RI and RII PKA holoenzymes using a fluorescence anisotropy assay. Our results indicate that RIα holoenzymes are selectively activated by C8-substituted analogs and RIIβ holoenzymes by N6-substituted analogs, where HE33 is the most prominent RII activator. We also solved the crystal structures of both RIα and RIIβ bound to HE33. The RIIβ structure shows the bulky aliphatic substituent of HE33 is fully encompassed by a pocket comprising of hydrophobic residues. RIα lacks this hydrophobic lining in Domain A and the side chains are displaced to accommodate the HE33 di-propyl groups. Comparison between cAMP-bound structures reveals that RIIβ, but not RIα, contains a cavity near the N6 site. This study suggests that the selective activation of RII over RI isoforms by N6 analogs is driven by the spatial and chemical constraints of Domain A and paves the way for the development of potent non-cyclic nucleotides activators to specifically target PKA iso-holoenyzmes.
Keywords: cAMP-dependent protein kinase, cyclic nucleotide analogs, isoform selectivity, fluorescence anisotropy, x-ray crystallography
INTRODUCTION
3′-5′-cyclic adenosine monophosphate (cAMP) is a second messenger that facilitates intracellular signal transduction upon extracellular stimulation. cAMP binds to and regulates the function of five major intracellular cAMP-binding proteins: cAMP-dependent protein kinase (Protein Kinase A or PKA), cAMP-activated guanine nucleotide-exchange factors (EPAC1 and EPAC2), phosphodiesterases, hyperpolarization-activated cyclic-nucleotide-modulated channels (HCN), and the catabolite gene activator protein (CAP). PKA is highly responsive to low levels of cAMP, making it one of the major receptors for this second messenger. PKA holoenzymes exist as an inactive tetrameric complex composed of two catalytic (C) subunits bound to two regulatory (R) subunits. Binding of cAMP to each of the two R-subunits releases the catalytic domains, thereby enabling downstream phosphorylation of PKA substrates.
PKA-mediated signaling is differentiated by the presence of four R-subunit isoforms (RIα, RIβ, RIIα, and RIIβ), each a product of different genes. Each isoform differs in their sub-cellular localization, abundance, affinity for the catalytic subunit, sensitivity to cAMP, and specificity for the scaffolding proteins A-Kinase Anchoring Proteins (AKAPs)(1). The relative cellular ratio between RI- and RII-subunits also plays a critical role in cell growth and differentiation(2). Deviations from this balance are correlated with several cancers (carney complex, breast, and ovarian), autoimmune disease (systemic lupus erythematosus (SLE)(3), and HIV(4)). For example, cancer cells express a significantly higher level of RI proteins than RII(5–7) and cancer cells that express more RIα display enhanced growth rates(8, 9). Furthermore, SLE patients display drastically reduced mRNA levels of RIα, RIβ, and RIIβ and SLE T-cells display altered levels of protein phosphorylation by the RIα and RIIβ holoenzymes (80% and 40%, respectively)(3, 10). Clearly, aberrant changes in the relative amounts of RI and RII protein and activity levels are correlated with diseased states, creating a pressing need to develop small molecules that can preferentially activate or inhibit one isoform over another.
Modulating PKA-specific isoform activity with small molecules has been particularly challenging since ATP competitors (such as H89 and staurosporine) inhibit other kinases and agents that increase cAMP levels in cells (such as forskolin) non-discriminately activate all PKA isoforms as well as the other cAMP-binding proteins (such as EPAC and HCN). Thus, for PKA, targeting the R-subunit cAMP-binding sites with cAMP analogs is an evident strategy for developing isoform-specific agonists of PKA activity.
In an effort to achieve isoform-selective activation, traditional methods initially focused on measuring the ability of cAMP analogs to bind isolated R-subunit isoforms. Comprehensive studies over several decades examined numerous cAMP analogs containing chemical substitutions on the phosphate moiety and the adenine and ribose rings(11). Activation profiles of R:C holoenzyme complexes using these analogs were not systematically measured, nor were these analogs thoroughly mapped against the crystal structures of RIα:cAMP2(12) and RIIβ:cAMP2(13) complexes. Moreover, recent crystal structures of PKA-Iα(14), PKA-IIα(15), and PKA-IIβ(16) holoenzymes reveal R-subunit cAMP binding pockets that differ drastically between the cAMP-bound and holoenzyme states. In order for cAMP analogs to effectively target an individual isoform, their selectivity must be established using R:C holoenzyme complexes, not only free R-subunits. Building on this foundation, the goals of this study are to broadly determine whether specific classes of cAMP analogs can activate individual PKA holoenzyme isoforms and to identify structural features that may govern such specificities.
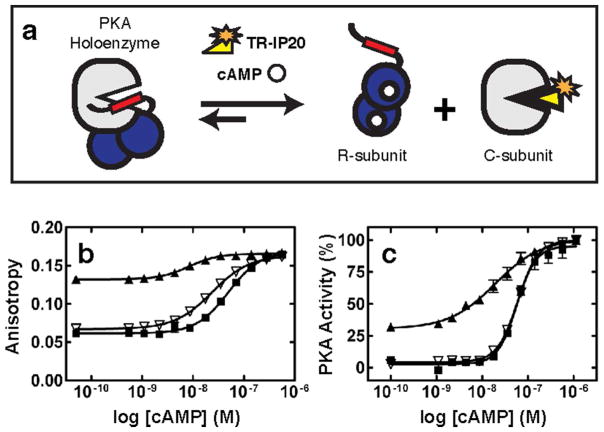
Measuring cAMP-mediated activation of PKA holoenzymes has not been readily amenable to a quick and easy high-throughput format. In the past, the dose-response activation of PKA holoenzymes by cAMP was characterized with assays that monitored through C-subunit catalytic activity (17). While accurate and sensitive, this method has not been performed in miniaturized microplate format, and thus not appropriate for the simultaneous measurement of many compounds. However, a method was developed to measure the cAMP-mediated activation of PKA holoenzymes in high throughput format (18). The integrity of the PKA holoenzyme complex upon addition of an allosteric activator (such as cAMP) can be measured by use of a high affinity fluorescently labeled peptide that competes with the regulatory subunit for binding to the catalytic subunit. Instead of measuring loss of PKA activity, the assay was developed to measure association and dissociation of the R- and C-subunits. This assay thereby enables measurement of agonists and antagonists and identifies binders that are not simply competitive with ATP, which almost all other known assays do. Binding of an agonist to a pre-formed holoenzyme complex induces complex dissociation and allows the labeled peptide to bind to the released catalytic subunit (Figure 1a). A fluorescence anisotropy change upon peptide binding to the catalytic subunit is used as a read-out for cAMP induced activation.
Figure 1.
a) Schematic of the Ligand Regulated Competition (LiReC) Assay. b) Activation of PKA-RIα (black squares), PKA-RIIβ S112A (open triangles), and PKA-RIIβ (black triangles) holoenzymes by cAMP using the LiReC assay with TR-IP20 as a fluorescence probe. c) Activation of PKA-RIα (black squares), PKA-RIIβ S112A (open triangles), and PKA-RIIβ (black triangles) by cAMP using the catalytically coupled Cook assay.
Here, we modified and validated the previously described PKA activation assay to accommodate RIIβ holoenzymes and utilized the assay to analyze 21 cAMP analogs for their ability to activate RIα and RIIβ holoeznymes. Two striking trends emerged. First, EC50 values for RIα holoenzymes were lowest using cAMP compounds with C8 substitutions. Second, EC50 values for RIIβ holoenzymes were lowest using cAMP compounds with N6 substitutions. The most isoform selective compound tested was HE33, an N6 modified compound, activating RIIβ holoenzymes with a 9-fold lower EC50 than RIα. We also solved the structures of HE33 co-crystallized with both RIα and RIIβ. The structures illustrate how the Domain A pocket in RIIβ has spatially and electrostatically favorable properties that enable the stabilization of bulky and hydrophobic N6 substituents, whereas RIα does not. These results further support the proposed mechanisms of cAMP activation for each of the two different holoenzymes.
RESULTS AND DISCUSSION
Development of Ligand Regulated Competition (LiReC) Assay for Measuring PKA-RIα and PKA-RIIβ Activity
The LiReC assay was originally developed as a high-throughput agonist and antagonist screen for PKA-RIα holoenzymes using fluorescein-labeled IP20 as a probe (f-IP20, Protein Kinase Inhibitor peptide residues 5–25)(18). Under the same conditions, the LiReC assay could not be performed with PKA-RIIβ holoenzymes for several reasons. Not only were the fluorescence anisotropy (FA) measurements not consistent over time, FA values were also higher for the RIIβ/C-subunit/f-IP20 mixture than an equivalent mixture containing RIα (Figure 1b).
We suspected that RIIβ was being phosphorylated at serine 112 by the C-subunit in situ, therefore shifting the equilibrium towards a higher concentration of IP20:C complexes resulting in higher FA baselines. Phosphorylation of serine 112 in RIIβ has been shown to weaken the interaction with the catalytic subunit(19). Furthermore, previous experiments have shown that mutation of this serine to alanine resulted in a 40-fold increase in affinity between the R- and C-subunits(20). Since RIα is a pseudosubstrate (the P-site is occupied by an alanine), it cannot be phosphorylated therefore remains as a stable high affinity complex with the C-subunit over the course of the experiment. Since the basic principle of the LiRec assay relies on the competitive nature between the probe and R-subunit for the catalytic subunit, RIIβ with a wild-type P-site serine could not be used with the PKA-LiReC assay for high-throughput screening of cAMP analogs.
In order to minimize IP20:C-subunit complex formation in the absence of cAMP and maximize RIIβ:C complexes, we first altered the probe to decrease its affinity with the C-subunit. We exchanged the fluorophore from fluorescein to texas red (TR) and verified that TR-IP20 was still capable of monitoring cAMP-mediated activation of PKA-RIα. TR-IP20 concentration was fixed at 3 nM and serial dilutions of C-subunit determined the affinity for the labeled probe. The affinity between the C-subunit and TR-IP20 was measured to be 5.5 nM under these conditions. Results of cAMP-mediated activation of PKA holoenzymes using RIα with the TR-IP20 are consistent with experiments using f-IP20, (EC50 was 36 nM and 53 nM, respectively). In the following LiReC compound screening assays, 6 nM PKA and 3 nM TR-IP20 were used.
Next, we developed a suitable RIIβ construct for use in the LiReC assay. Based on our hypothesis that phosphorylation of RIIβ by the C-subunit was responsible for the poor FA response, we mutated serine 112 to alanine and measured EC50 values of cAMP-mediated activation of holoenzyme complexes. In the LiReC assay, RIα and RIIβ-S112A holoenzymes had comparable EC50 values (36 nM and 18 nM, respectively). (Figure 1b). Furthermore, the upper and lower anisotropy values were similar between the two complexes and the FA baselines of RIIβ:C complexes in the presence of TR-IP20 remained constant over a long period of time. To cross-validate our modified LiReC assay as an approach for measuring RIIβ holoenzyme activation, we also utilized the standard spectrophotometrically-based catalytic coupled assay(17). The catalytic coupled assay also resulted in similar EC50 values between RIα and RIIβ-S112A holoenzymes (57 nM and 60 nM, respectively) and a slightly lower EC50 value for wild-type RIIβ holoenzymes (19 nM) (Figure 1c).
Screening of commercial cAMP analogs for PKA activation
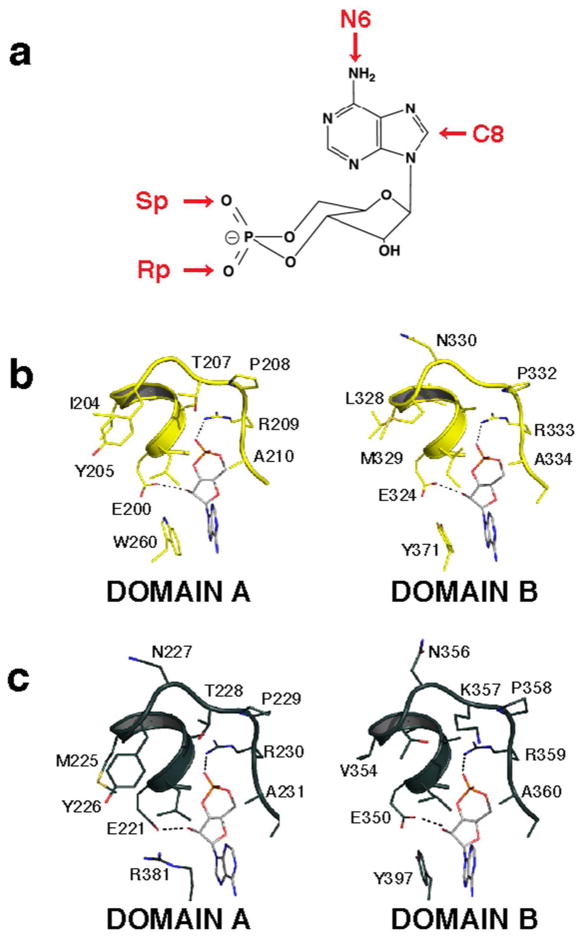
Next, we investigated whether PKA isozymes could be selectively activated with commercially available cAMP analogs. We selected a set of 21 cAMP derivatives (19 agonists and 2 antagonists) based on previous signaling pathway studies(21) and recommendations of some agonists as potent PKA isoform selective activators(11). The four main sites of modification on the cAMP molecule are shown in Figure 2a, where substitutions were made on the adenine ring or by replacing an oxygen atom with sulfur in the cyclic phosphate moiety.
Figure 2.
a) Sites of cAMP modifications. Detailed interactions between the phosphate binding cassette residues of RIα (b) and RIIβ (c) and cAMP.
The 21 analogs were tested for their ability to activate RIα or RIIβ-S112A holoenzymes using the modified LiReC HT assay. A summary of EC50 values is listed in Table 1. Results for the agonist experiments are grouped according to derivative type and their relative selectivity for the isoform class. The EC50 for cAMP activation of RIα was 35 nM and of RIIβ was 18 nM. Two significant trends were evident from the results. First, all analogues with substitutions on the 8-carbon position of the adenine ring preferentially activated RIα holoenzymes, with up to 5-fold selectivity. Second, all analogues with substitutions on the 6-nitrogen position preferentially activated RIIβ-S112A holoenzymes, with up to 9-fold selectivity. This preference held true for all compounds tested over IC50 ranges of 50 μM to 15 μM. Notably, replacement of exocyclic axial oxygen with sulfur in the cyclic phosphate moiety (Figure 2a) in 8-substituted Sp-CAMPS compounds induced a change in selectivity from type I to type II.
Table 1. Activation of PKA Isoforms by cAMP Analogs.
Activation of PKA RI and RII PKA were measured with the LiReC assay. EC50 results are grouped according to substitution type and selectivity. Fold selectivity is calculated based on selectivity based on preference for RIα or RIIβ holoenzymes.
| Compound | RIα EC50(nM) | RIIβ EC50 (nM) | RIIβ/RIα Fold Selectivity | |
|---|---|---|---|---|
|
| ||||
| cAMP | 36 | 18 | 2.0 | |
| cGMP | 15000 | 5080 | 3.0 | |
| RIα | 8-Cl cAMP | 58 | 275 | 0.2 |
| 8-CPT cAMP | 33 | 149 | 0.2 | |
| 8-PIP cAMP | 1020 | 3950 | 0.3 | |
| 8-HA cAMP | 358 | 1150 | 0.3 | |
| 8-Br cAMP | 81 | 184 | 0.4 | |
| 8-MA cAMP | 102 | 223 | 0.5 | |
| 8-AHA cAMP | 116 | 197 | 0.6 | |
| Sp cAMP | 1590 | 2270 | 0.7 | |
| RIIβ | HE33 | 414 | 45 | 9.2 |
| 6-MBC cAMP | 289 | 59 | 4.9 | |
| 6-Bnz cAMP | 170 | 35 | 4.9 | |
| 6-Phe cAMP | 92 | 20 | 4.6 | |
| Sp-5, 6-DCl BIMPS | 344 | 83 | 4.1 | |
| 6-MB cAMP | 140 | 39 | 3.6 | |
| Sp-8-CPT cAMP | 342 | 96 | 3.6 | |
| Sp-8-B cAMP | 1355 | 435 | 3.1 | |
| 5,6 DCl BIMPS | 270 | 129 | 2.1 | |
| Sp-8-PIP cAMP | 5048 | 2959 | 1.7 | |
| 2-OMe cAMP | 5666 | n.d. | n.d. | |
| 2-AEA cAMP | 8739 | 2652 | 3.3 | |
| 2-Cl cAMP | 42 | 45 | 0.9 | |
Structure of RIα(91–379) bound to HE33
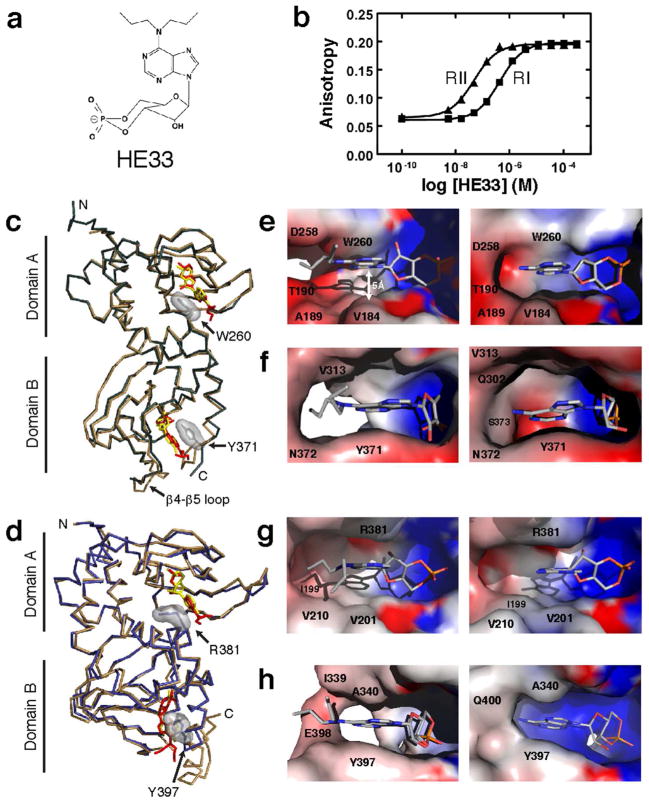
From our agonist studies, HE33, a N6 di-propyl substituted cAMP analog, exhibited the most selective activation between the RI and RII isoforms (Figure 3a,b). In our LiReC assay, HE33 activated RIIβ with an EC50 similar to cAMP (45 nM) and exhibited 9-fold selectivity over RIα (EC50 414 nM). To expand upon our FA analysis, we co-crystallized HE33 with both RIα and RIIβ with the aim of elucidating the molecular basis for this specificity.
Figure 3.
Comparison of interactions between HE33 and RIα or RIIβ. a) Chemical structure of HE33. b) LiReC Assay results of HE33 activation of RIα and RIIβ. c) Crystal structure of RIα:cAMP2 (tan) and RIα:HE332 (green). Surface rendering of the cyclic nucleotide binding pocket in Domain A (e) and Domain B (f), with HE33 (left) and cAMP (right). d) Crystal structure of RIIβ:cAMP2 (tan) and RIIβ:HE332 (blue). Surface rendering of the nucleotide binding pocket in Domain A (g) and Domain B (h), with HE33 (left) and cAMP (right).
Previously solved crystal structures of both RIα and RIIβ bound to cAMP(12, 13) laid the groundwork for the structural analysis of our HE33-bound complexes. Although each of the four regulatory subunits differs in sequence and function, they all share the same domain architecture. Each regulatory subunit contains an N-terminal dimerization domain that binds to scaffolding proteins, a flexible linker region that contains the inhibitor site, and two tandem cyclic nucleotide binding domains, Domain A and B. Both crystal structures of RIα and RIIβ bound to cAMP were solved using R-subunit constructs containing only the inhibitor site and cAMP-binding domains (residues 91–379 for RIα and residues 112–416 for RIIβ). These crystal structures show that both RIα and RIIβ share a similar overall globular shape, where the two cyclic nucleotide-binding domains come together at a common interface. Despite these similarities, the molecular details for each isoform are clearly distinct and each will be discussed in turn.
The crystal structure of RIα(91–379) bound to 2 molecules of HE33 was solved to 2.95 Å resolution using the RIα:Sp-cAMP2 structure (PDB code 1NE6) as a molecular replacement probe. The complex crystallized in the P6522 space group with one molecule in the asymmetric unit (Table 2). The overall conformation of RIα in its HE33-bound state is similar to the previous cAMP-bound conformation, where the protein forms a compact globular shape with a kinked αB/C helix connecting the two modules (Figure 4a). The backbones between the cAMP- and HE33-bound structures had a high degree of alignment, with the exception of the β4-β5 loop in Domain B and the C-terminal tail (Figure 3c).
Table 2.
X-ray Diffraction Data and Refinement Statistics.
| Crystal | RIα:HE332 | RIIβ:HE332 |
|---|---|---|
|
| ||
| Space Group | P6522 | P41212 |
| Unit cell dimensions (Å) | a=b=89.8, c=185.1 | a=b=54.5, c=200.0 |
| Number of crystals | 1 | 1 |
| Wavelength (Å) | 1.0 | 1.0 |
| Mosaicity (°) | 0.85 | 0.63 |
| Resolution (Å) | 50–2.95 | 50–2.5 |
| Total observations | 90832 | 400152 |
| Unique observations | 9906 (476) | 15916 (1249) |
| Average redundancy | 9.2 (7.6) | 25.1 (19.1) |
| Rsym (%) | 7.4 (41.7) | 8.0 (32.5) |
| Completeness (%) | 99.7 (99.4) | 95.1 (88.4) |
| <I>/<σ> | 40.6 (3.3) | 39.3 (5.5) |
| No. molecules per asymmetric unit | 1 | 1 |
| Number of protein atoms | 267 | 271 |
| Solvent content (%) | none | 71 |
| RMSD bond lengths (Å) | 0.01 | 0.008 |
| RMSD bond angles (°) | 1.42 | 1.40 |
| Rwork | 21.8 | 23.8 |
| Rfree | 28.6 | 27.8 |
| Ramachandran angles most favored (%) | 85.0 | 82.9 |
| Disallowed (%) | none | none |
Values in parentheses are for highest-resolution shell.
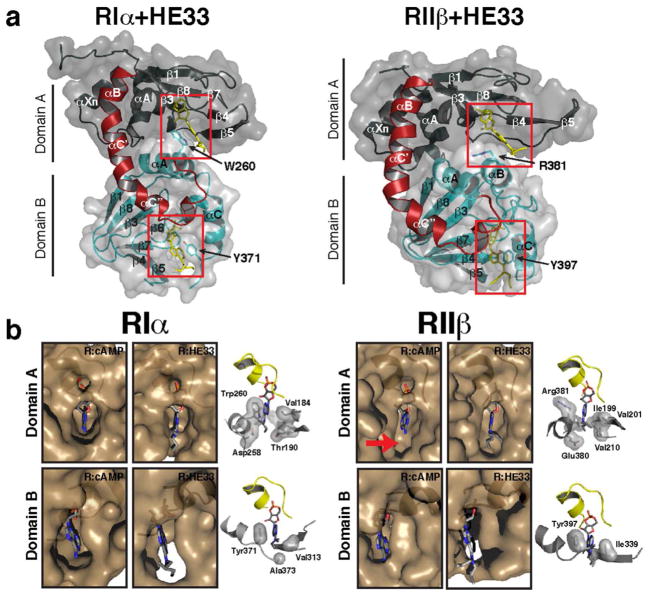
Figure 4.
Comparison of cyclic nucleotide binding pockets between RIα and RIIβ in cAMP-bound and HE33-bound bound states. a) Full view crystal structures of RIα-HE33 and RIIβ-HE33. b) Zoomed view of RIα and RIIβ nucleotide binding pockets in the presence of cAMP and HE33.
HE33 binds to the nucleotide-binding pockets of RIα in the same orientation as cAMP. In the cAMP-bound structure, a hydrophobic residue packs against the adenine ring of cAMP in each of the domains (Trp260 for Domain A and Tyr371 for Domain B) and studies have shown that both residues are critical players in facilitating cAMP-mediated activation of holoenzyme complexes(14). In the HE33 structure, these same residues also stack against the adenine ring of the analog (Figure 3c). In addition, both ribose rings from cAMP and HE33 form the same hydrogen bond network with polar amino acids in RIα. Specifically, in both Domains A and B, the exocylic and 2′ oxygen form hydrogen bonds with a glutamate and arginine residue in the phosphate binding cassette, respectively (Glu200 and Arg209 in Domain A and Glu324 and Arg333 in Domain B, Figure 2b).
Despite these similarities, there are critical differences between the two crystal structures. Figure 4b illustrates the spatial occupancy of both cAMP and HE33 within each nucleotide-binding pocket. In Domain A, RIα envelops the entire cAMP molecule, creating an induced-fit cavity. The computed electrostatic potential in the nucleotide-binding pocket emphasizes the preferential polar binding sites at each end of the cAMP molecule (Figure 3e, right). In the RIα:HE332 structure, the pocket is expanded to accommodate the bulky di-propyl chemical group at the N6 position. This expansion specifically stems from side chain movements of residues Thr190 and Asp258 as well as β4 and β5 movements away from the nucleotide relative to the conformation observed in the cAMP-bound state. The hydrophobicity of the N6-substituent likely facilitates the structural rearrangement in order to relieve both steric and electrostatic constraints.
In Domain B, the RIα nucleotide-binding pocket again encapsulates the cAMP molecule (Figure 3f and 4b). In the HE33-bound state, the nucleotide-binding pocket is expanded to accommodate the di-propyl substituent on HE33. In contrast to Domain A where only a few side chain atoms are rearranged, electron density was not observed in large sections of Domain B. These regions include the β4-β5 loop (residues 303–310) and the C-terminal tail (residues 373–379). The absence of electron density in these regions suggests that HE33 is not particularly stable in Domain B, in contrast to Domain A.
Structure of RIIβ(108–402) bound to HE33
The crystal structure of RIIβ (108–402) bound to 2 molecules of HE33 was solved to 2.5 Å resolution using the RIIβ:cAMP2 structure as a molecular replacement probe (PDB code 1CX4). The complex crystallized in the P41212 space group with one molecule in the asymmetric unit (Table 2). The electron density from the two HE33 molecules is clearly observed (Figure S1). As with the RIα:HE332 structure, the protein forms a compact globular shape with a kinked αB/C helix connecting the two modules (Figure 4a), and the main chain atoms between the RIIβ:HE332 and RIIβ:cAMP2 complexes align very well (Figure 3d). The two hydrophobic capping residues in RIIβ (Arg381 for Domain A and Tyr397 for Domain B) also stack against the adenine ring in both HE33-bound and cAMP-bound structures.
In Domain A, RIIβ has a large pocket near the N6 position of cAMP that is absent in RIα (Figure 4b, arrow). This pocket is lined with a hydrophobic environment created by aliphatic side chains in the β4 and β5 sheets of Domain A and the αB helix of Domain B, primarily Ile199, Val201, Val210, Gln377, Glu380, and the capping residue Arg381 (Figure 3g and 4b). In the HE33-bound state, the space that was present in the cAMP-bound state is now occupied by the HE33 N6 alkyl substituent. In fact, the dipropyl groups of HE33 are supported in the same hydrophobic environment observed in the cAMP-bound state. Irrespective of whether cAMP or HE33 is bound, the size and shape of this pocket remained largely unperturbed.
In Domain B, electron density in the nucleotide binding pocket is not well formed in the RIIβ:HE33 structure compared to the cAMP-bound structure and electron density for several amino acids was not observed, similar to RIα. Despite the disordered regions in Domain B, clear electron density was still observed for the hydrophobic capping residue, Tyr397. Figure 3c and 3d illustrate how Tyr397, like Arg381 in Domain A, packs against the HE33 adenine ring.
Structural insights elucidating the preferential activation of RIIβ over RIα holoenzymes with HE33
The overall goal of this study was to determine whether one could obtain selective dissociation, hence activation, between Type I and Type II PKA holoenzymes with cAMP analogs. Results from the LiReC activation assays demonstrated a clear trend where N6-substituted compounds were selective for RIIβ holoenzymes, and C8-substituted compounds were selective for RIα holoenzymes (Table 1). Previous experiments measuring the binding affinities between cAMP derivatives and Domain A of free R-subunits using 3H-cAMP competition methods also exhibited similar trends to our activation assays(11). In Domain A, C8-substituted compounds bound to RIα with the highest affinity, while the N6 substituted compounds bound to RIIβ with the highest affinity. In Domain B, there was no clear correlation between N6 and C8-substitutions for RI- and RII-subunit binding. The correlation between the Domain-specific binding affinity data and our holoenzyme activation data together suggests that Domain A may play a more important role in the activation of PKA holoenzymes.
Our structural studies provide key insights towards understanding why N6-substituted cAMP analogs are more suitable to bind RIIβ, but not RIα. Based on the RIIβ:cAMP2 crystal structure, RIIβ is predisposed to accommodate N6-substituents in Domain A due to a large cavity near the N6-position (Figure 4b). Our RIIβ:HE332 structure confirms this hypothesis where the additional aliphatic groups on the N6 atom in HE33 fill the vacant space with minimal structural perturbation. The HE33 di-propyl groups are also surrounded in a hydrophobic environment provided by the β4 and β5 sheets of Domain A and the αB helix of Domain B. These interactions may stabilize HE33 within RIIβ such that activation of holoenzyme complexes occurs at levels similar to cAMP. 6-Bnz, 6-Phe, and 6-MB cAMP derivatives are also potent RIIβ activators, suggesting that a hydrophobic substituent is also necessary for high affinity binding and activation of RIIβ holoenzymes.
In contrast, there are no equivalent hydrophobic residues in RIα to stabilize interactions of HE33 in Domain A. Although HE33 still binds to RIα, the nucleotide-binding site in Domain A is more open and lacks the hydrophobic network around the N6 substituent as observed in RIIβ. In the RIα cAMP-bound structure, hydrophilic residues line the region where HE33 di-propyl groups would be located. This loss of hydrophobic interactions and increase in hydrophilic blocking in the αA helix of RIα may explain the increased EC50 for activation of RIα by HE33.
In addition to structural differences between RIα:HE332 and RIIβ:HE332 complexes, the model of PKA activation by cAMP differs between RI and RII complexes. Mutagenesis studies revealed cAMP-induced activation as a cooperative and ordered process for RIα, where cAMP must first bind to Domain B before Domain A is accessible. Binding of cAMP to Domain A, in turn, initiates the release of active C-subunit(22). In contrast, activation of RIIβ holoenzyme by cAMP is not an ordered process. Instead, binding of cAMP to either Domain A or Domain B activates PKA(23).
In light of these biochemical models of cAMP-induced PKA activation, our structural analysis of HE33 bound to both RIα and RIIβ may explain how selectivity of N6 analogues for RIIβ is achieved. HE33 is able to bind both free RIα and RIIβ proteins, as observed from the crystal structures. However, our data also show major structural differences in Domain B of RIα between the cAMP and HE33-bound conformations. RIα completely encloses cAMP in Domain B, whereas HE33 induces local conformational changes such that the nucleotide-binding pocket becomes completely opened. In the context of the activation process, if HE33 cannot be stabilized in Domain B, Domain A cannot be accessed, therefore attenuating C-subunit dissociation and activation. This could explain why there is a 10-fold increase in EC50 for HE33 relative to cAMP. For RIIβ, our structural analysis shows that HE33 forms well-ordered interactions in Domain A, stabilized by a number of hydrophobic interactions. The space around the N6 pocket also provides extra room for bulky N6 substituents, thereby reducing the potential for steric clashes. Since activation of RIIβ holoenzymes does not require ordered and sequential cAMP-binding events, the binding of N6-cAMP derivatives in Domain A would be sufficient to activate the RIIβ isoforms. Taken together, our biochemical and structural data suggests that N6-substituted analogs are well suited to bind Domain A of RIIβ-subunits.
Our studies provide the initial steps towards the development of isoform-specific cAMP analogs for drug therapies. The crystal structure of HE33 bound to RIIβ will enable rational design and development of analogues with higher selectivity for RIIβ, certainly with opportunities around modifying the N6 position with larger hydrophobic groups. It may also be possible to increase selectivity by discovering cAMP analogs that can be additionally stabilized by the hydrophilic side chains of the αB helix to increase binding to RIIβ. With these studies, future work can be done to optimize the chemical substituents for even greater selectivity of PKA isoforms.
CONCLUDING REMARKS
To determine a possible mechanism for why cAMP compounds with N6-subustituents preferentially activate RII-subunits, we selected to co-crystallize the most selective agonist for RIIβ, HE33, with both RIα and RIIβ-subunits. HE33 has a di-propyl alkyl group at the N6 position. We solved the structure of both RIα and RIIβ bound to HE-33. Comparison between RIIβ:HE332 and RIIβ:cAMP2 demonstrated that a space near the N6 position in the cAMP structure is occupied by the HE33 N6 alkyl substituent, surrounded in a hydrophobic environment. Conversely, RIα lacks this hydrophobic environment and binding of HE-33 results in a more open pocket. In Domain B, both RIα and RIIβ display opened pockets. In light of the different biochemical mechanisms of cAMP-induced activation between RI and RII subunits, these structural studies explain why selectivity of N6 analogues for RIIβ is achieved.
This structure-activity relationship derived from a pre-existing set of cAMP analogs suggests the potential to diverge into highly selective activation. Understanding the structural basis of pathways in biology that are non-redundantly regulated allows rational approaches to analog discovery, and is an important step in generalizing the approach.
METHODS
Protein preparation
The catalytic subunit was expressed and purified in E. coli BL21 (DE3) cells (Novagen) as described previously(24). Both RIα and RIIβ proteins were expressed and purified as described previously(25). The RIIβ (108–402) S112A mutant was generated using site-directed mutagenesis, then expressed and purified using the wild-type protocol. N-terminal deletions of RIα and RIIβ subunits were used in this study in order to reduce the tendency for proteolytic cleavage and aggregation. Additionally, RIIβ was truncated by 14 residues at the C-terminus to reduce proteolytic cleavage(26).
Synthesis of Texas red-labeled IP20
To synthesize TR-IP20, 1 mg of Texas red-X succinimidyl ester and 2 mg of IP20 were incubated in 1 mL of PBS/DMF (20:80) overnight at 4 °C with gentle agitation. The fluorescent peptide was purified by C18 reverse-phase HPLC.
High Throughput Screening of PKA activation with cAMP analogs
cAMP stocks for assays were dissolved in H2O at 9 mM. All cAMP analogs were dissolved in DMSO. Assays were performed in 384 well Nunc flat black bottom plates. Assay mixtures contained 50 mM HEPES, 0.005% (v/v) Triton X-100, 2 mM ATP, 10 mM MgCl2, 2 mM DTT, and 6.25% (v/v) DMSO, 3 nM TR-IP20, and 6 nM PKA holoenzyme. 75 μL of this assay mix and 5 μL of cAMP compounds (concentrations ranging from 0.25 nM and 1 μM) were combined into each well. Fluorescence readings were taken on a GenesisPro plate reader (Tecan) at 570/20 nm excitation and 630/20 nm emission using a 590 nm dichromic mirror. Measurements were taken periodically over approximately 2 hours following addition of compound. A G-factor of 1.0011 was used for the TR fluorophore. All data were fit to a sigmoidal dose-response curve and EC50 values were calculated using GraphPad Prism 5 software (GraphPad).
cAMP Activation of PKA using a Catalytic Coupled Assay
PKA activity was measured using a modified protocol of the standard spectrophotometric method described by Cook et al.(17). The assay was performed in 96-well clear bottom untreated Costar plates (Corning, cat no. 3631) where 5 nM PKA holoenzyme, 25 mM HEPES (pH 7), 75 mM KCl, 10 mM MgCl2, 1 mM ATP, 1 mM phosphoenolpyruvate, 15 units/ml lactate dehydrogenase, 7 u/ml pyruvate kinase, and 0.2 mM NADH was incubated with various concentrations of cAMP for 20 minutes at room temperature. Each reaction was initiated with 0.2 M Kemptide substrate peptide (LRRASLG) using a multi-channel pipette. The activity of free C-subunit was monitored spectrophotometrically by a change in absorbance at 340 nm over time due to oxidation of NADH. Data were fit with GraphPad Prism 5 software (GraphPad) to determine the apparent activation constant, EC50. Substituted cAMP analogues were sourced from BioLog. HE33 was prepared by an optimized procedure originally reported by Kataoka et al.(27).
Co-crystallization of RIα(91–379) and HE33
Purified RIα was dialyzed into 50 mM MES (pH 5.8), 200 mM NaCl, 2 mM EDTA, 2 mM EGTA, 2 mM TCEP-HCl, and 10% (v/v) glycerol. RIα was incubated with a ten-fold excess of HE33 on ice for 2 hours. Excess compound was removed by buffer exchange in an Ultra-4 (Amicon) centrifugal device.
RIα(91-379):HE33 crystals were obtained with the hanging drop method in 6.3% (w/v) PEG 3350, 0.074 M sodium malonate (pH 7.0) after 3 weeks of growth at 25°C. Crystals were harvested, transferred to mother liquor containing 20% (v/v) glycerol, and flash-frozen in liquid nitrogen.
X-ray diffraction data was collected at The Advanced Light Source beamline 8.2.1 (Lawrence Berkeley National Laboratory, Berkeley, CA). Diffraction data were integrated and scaled with HKL2000(28). The space group was unambiguously determined to be P6522. Initial phases were generated by molecular replacement using the program phaser(29) and the coordinates for the RIα:Sp-cAMP2 complex (PDB code 1NE6)(30) as a search model. The two Sp-cAMP molecules were removed from the PDB file to prevent model bias. One molecule was found in the asymmetric unit (RFZ=9.0, RFZ=37.8) corresponding to a solvent content of 63.5%. All ambiguous main chains and side chains manually rebuilt using Coot(31), followed by iterative cycles of structure refinement using REFMAC in the CCP4 suite(32, 33). TLS refinement(34) was implemented for each lobe. Simulated annealing via the program Phenix(35, 36) was used in the final stages of refinement because geometric restraints were too loose with REFMAC. The final model contained R-subunit residues 107–372 bound to two HE-33 molecules. All figures were made using PyMol(37) (DeLano Scientific).
Co-crystallization of RIIβ(108-402) and HE33
The strategy used to co-crystallize RIIβ and HE33 was adapted from co-crystallization of RIIβ bound to cAMP(13). Briefly, initial protein samples were eluted from a Superdex 75 (GE bioscience) column in 50 mM MES, 200 mM NaCl, 2 mM EDTA, 2 mM EGTA, and 5 mM DTT. The protein sample was incubated with ten-fold excess HE33 overnight at 4°C. Excess compound was removed by buffer exchange in an Amicon Ultra-4 centrifugal device.
Crystallization trials were set-up using the Oryx crystallization robot (Douglas Instruments) in modified microbatch mode. Drop size of 0.4 μL at 50% (v/v) protein was used, and drops were covered with Al’s Oil (Hampton Research) directly following mixing of reagents. The final crystallization condition was obtained with the hanging drop method consisting of 20% (w/v) PEG 4000, 80 mM Bis-Tris 6.0, and 50 mM MgCl2. Crystals were harvested, transferred to mother liquor containing 10% (v/v) glycerol, and flash-frozen in liquid nitrogen.
Two data sets were collected at the Advanced Light Sources (ALS) at Lawrence Livermore National Lab beamline 8.2.2. Data collected at 2.5 Å was chosen for processing and analysis. Initial phases were obtained using the structure of cAMP bound to RIIβ (PDB code 1CX4) as a search model using the CCP4 package program PHASER. Initial density maps were obtained and the model of RIIβ-cAMP was fit into the density for RIIβ-HE33. Initial refinement of the RIIβ-cAMP model into the RIIβ-HE33 density map was performed using the REFMAC5 program in CCP4.
Electrostatic Potential Calculations
Pdb coordinates were converted to pqr format at http://biophysics.cs.vt.edu/H++. Electrostatic potentials were generated in pymol with the Apbs plugin(38).
Supplementary Material
Electron density of HE33 in RIIβ-HE33 crystal structures. The 2Fo-FC map is rendered at 1σ. Domain A is shown on the left and domain B is shown on the right.
Acknowledgments
We thank M. Deal (University of California, San Diego) for providing the C-subunit protein used in these studies and A. Kornev for helpful discussions and assistance with structural rendering. This work was funded in part by GM34921 to S.S.T, NIH Training Grant T32 CA0095523 and National Research Service Award Training Grant to S.H.B. and NIH Training Grant T32 GM08326 and AHA (Western States Affiliate) 07015019Y to C.Y.C.
Abbreviations
- cAMP
cyclic adenosine monophosphate
- PKA
protein kinase A or cAMP-dependent protein kinase
- C-subunit
catalytic subunit
- R-subunit
regulatory subunit
- AKAP
a kinase anchoring protein
- EPAC
cAMP-activated guanine nucleotide-exchange factor
- HNC
hyperpolarization-activated cyclic-nucleotide-modulated channel
- PBC
phosphate binding cassette
Footnotes
Accession Codes. PDB coordinates for the RIα(91–379):HE332 structure is 4JV4. PDB coordinates for the RIIβ(108–402):HE332 structure is 4JVA.
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nature reviews. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 2.Neary CL, Nesterova M, Cho YS, Cheadle C, Becker KG, Cho-Chung YS. Protein kinase A isozyme switching: eliciting differential cAMP signaling and tumor reversion. Oncogene. 2004;23:8847–8856. doi: 10.1038/sj.onc.1208165. [DOI] [PubMed] [Google Scholar]
- 3.Kammer GM, Khan IU, Malemud CJ. Deficient type I protein kinase A isozyme activity in systemic lupus erythematosus T lymphocytes. The Journal of clinical investigation. 1994;94:422–430. doi: 10.1172/JCI117340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skalhegg BS, Funderud A, Henanger HH, Hafte TT, Larsen AC, Kvissel AK, Eikvar S, Orstavik S. Protein kinase A (PKA)--a potential target for therapeutic intervention of dysfunctional immune cells. Current drug targets. 2005;6:655–664. doi: 10.2174/1389450054863644. [DOI] [PubMed] [Google Scholar]
- 5.Meoli E, Bossis I, Cazabat L, Mavrakis M, Horvath A, Stergiopoulos S, Shiferaw ML, Fumey G, Perlemoine K, Muchow M, Robinson-White A, Weinberg F, Nesterova M, Patronas Y, Groussin L, Bertherat J, Stratakis CA. Protein kinase A effects of an expressed PRKAR1A mutation associated with aggressive tumors. Cancer research. 2008;68:3133–3141. doi: 10.1158/0008-5472.CAN-08-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller WR. Regulatory subunits of PKA and breast cancer. Annals of the New York Academy of Sciences. 2002;968:37–48. doi: 10.1111/j.1749-6632.2002.tb04325.x. [DOI] [PubMed] [Google Scholar]
- 7.Cheadle C, Nesterova M, Watkins T, Barnes KC, Hall JC, Rosen A, Becker KG, Cho-Chung YS. Regulatory subunits of PKA define an axis of cellular proliferation/differentiation in ovarian cancer cells. BMC medical genomics. 2008;1:43. doi: 10.1186/1755-8794-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDaid HM, Cairns MT, Atkinson RJ, McAleer S, Harkin DP, Gilmore P, Johnston PG. Increased expression of the RIalpha subunit of the cAMP-dependent protein kinase A is associated with advanced stage ovarian cancer. British journal of cancer. 1999;79:933–939. doi: 10.1038/sj.bjc.6690149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho-Chung YS. Role of cyclic AMP receptor proteins in growth, differentiation, and suppression of malignancy: new approaches to therapy. Cancer research. 1990;50:7093–7100. [PubMed] [Google Scholar]
- 10.Khan IU, Laxminarayana D, Kammer GM. Protein kinase A RI beta subunit deficiency in lupus T lymphocytes: bypassing a block in RI beta translation reconstitutes protein kinase A activity and augments IL-2 production. J Immunol. 2001;166:7600–7605. doi: 10.4049/jimmunol.166.12.7600. [DOI] [PubMed] [Google Scholar]
- 11.Ogreid D, Ekanger R, Suva RH, Miller JP, Doskeland SO. Comparison of the two classes of binding sites (A and B) of type I and type II cyclic-AMP-dependent protein kinases by using cyclic nucleotide analogs. European journal of biochemistry / FEBS. 1989;181:19–31. doi: 10.1111/j.1432-1033.1989.tb14689.x. [DOI] [PubMed] [Google Scholar]
- 12.Su Y, Dostmann WRG, Herberg FW, Durick K, Xuong NH, Ten Eyck LF, Taylor SS, Varughese KI. Regulatory (RIa) Subunit of Protein Kinase A: Structure of Deletion Mutant with cAMP Binding Domains. Science. 1995;269:807–819. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- 13.Diller TC, Madhusudan Xuong NH, Taylor SS. Molecular basis for regulatory subunit diversity in cAMP-dependent protein kinase: crystal structure of the type II beta regulatory subunit. Structure. 2001;9:73–82. doi: 10.1016/s0969-2126(00)00556-6. [DOI] [PubMed] [Google Scholar]
- 14.Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Brown SH, von Daake S, Taylor SS. PKA type IIalpha holoenzyme reveals a combinatorial strategy for isoform diversity. Science. 2007;318:274–279. doi: 10.1126/science.1146447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown SH, Wu J, Kim C, Alberto K, Taylor SS. Novel Isoform-specific Interfaces Revealed by PKA RIIbeta Holoenzyme Structures. Journal of molecular biology. 2009 doi: 10.1016/j.jmb.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook PF, Neville ME, Jr, Vrana KE, Hartl FT, Roskoski R., Jr Adenosine cyclic 3′,5′-monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry. 1982;21:5794–5799. doi: 10.1021/bi00266a011. [DOI] [PubMed] [Google Scholar]
- 18.Saldanha SA, Kaler G, Cottam HB, Abagyan R, Taylor SS. Assay principle for modulators of protein-protein interactions and its application to non-ATP-competitive ligands targeting protein kinase A. Anal Chem. 2006;78:8265–8272. doi: 10.1021/ac061104g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosen OM, Erlichman J. Reversible Autophosphorylation of a Cyclic 3′:5′-AMP-Dependent Protein Kinase from Bovine Cardiac Muscle. J Biol Chem. 1975;250:7788–7794. [PubMed] [Google Scholar]
- 20.Diskar M, Zenn HM, Kaupisch A, Prinz A, Herberg FW. Molecular basis for isoform-specific autoregulation of protein kinase A. Cellular signalling. 2007;19:2024–2034. doi: 10.1016/j.cellsig.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Poppe H, Rybalkin SD, Rehmann H, Hinds TR, Tang XB, Christensen AE, Schwede F, Genieser HG, Bos JL, Doskeland SO, Beavo JA, Butt E. Cyclic nucleotide analogs as probes of signaling pathways. Nature methods. 2008;5:277–278. doi: 10.1038/nmeth0408-277. [DOI] [PubMed] [Google Scholar]
- 22.Herberg FW, Taylor SS, Dostmann WR. Active Site Mutations Define the Pathway for Cooperative Activation of cAMP-dependent Protein Kinase. Biochemistry. 1996;35:2934–2942. doi: 10.1021/bi951647c. [DOI] [PubMed] [Google Scholar]
- 23.Zawadzki KM, Taylor SS. cAMP-dependent protein kinase regulatory subunit type IIbeta: active site mutations define an isoform-specific network for allosteric signaling by cAMP. J Biol Chem. 2004;279:7029–7036. doi: 10.1074/jbc.M310804200. [DOI] [PubMed] [Google Scholar]
- 24.Herberg FW, Bell SM, Taylor SS. Expression of the Catalytic Subunit of cAMP-dependent Protein Kinase in E. coli: Multiple Isozymes Reflect Different Phosphorylation States. Protein Engineering. 1993;6:771–777. doi: 10.1093/protein/6.7.771. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Brown S, Xuong NH, Taylor SS. RIalpha subunit of PKA: a cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B. Structure. 2004;12:1057–1065. doi: 10.1016/j.str.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 26.Anand GS, Hotchko M, Brown SH, Ten Eyck LF, Komives EA, Taylor SS. R-subunit isoform specificity in protein kinase A: distinct features of protein interfaces in PKA types I and II by amide H/2H exchange mass spectrometry. Journal of molecular biology. 2007;374:487–499. doi: 10.1016/j.jmb.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kataoka S, Yamaji N, Kato M, Kawada T, Imai S. Studies on the synthesis of compounds related to adenosine 3′,5′-cyclic phosphate. VII. Synthesis and cardiac effects of N6,N6-dialkyl adenosine 3′,5′-cyclic phosphates. Chem Pharm Bull (Tokyo) 1990;38:3147–3154. doi: 10.1248/cpb.38.3147. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski ZaMW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276:Macromolecular Crystallography, part A. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta crystallographica. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Jones JM, Nguyen-Huu X, Ten Eyck LF, Taylor SS. Crystal structures of RIalpha subunit of cyclic adenosine 5′-monophosphate (cAMP)-dependent protein kinase complexed with (Rp)-adenosine 3′,5′-cyclic monophosphothioate and (Sp)-adenosine 3′,5′-cyclic monophosphothioate, the phosphothioate analogues of cAMP. Biochemistry. 2004;43:6620–6629. doi: 10.1021/bi0302503. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.The CCP4 suite: programs for protein crystallography. Acta crystallographica. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta crystallographica. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC. Recent developments in the PHENIX software for automated crystallographic structure determination. Journal of synchrotron radiation. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta crystallographica. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1 2010 [Google Scholar]
- 38.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electron density of HE33 in RIIβ-HE33 crystal structures. The 2Fo-FC map is rendered at 1σ. Domain A is shown on the left and domain B is shown on the right.