Abstract
Frequent genetic alterations of the components in the phosphoinositide 3-kinase (PI3K)/PTEN/AKT signaling pathway contribute greatly to breast cancer initiation and progression, which makes targeting this signaling pathway a promising therapeutic strategy for breast cancer treatment. In this study, we showed that in the presence of copper (Cu), disulfiram (DSF), a clinically used antialcoholism drug, could potently inhibit breast cancer cell growth regardless of the PIK3CA status. Surprisingly, the treatment with a mixture of DSF and copper (DSF-Cu) led to the decreased expression of PTEN protein and the activation of AKT in a dose- and time-dependent manner in different cell lines with or without PIK3CA mutations. Treatment of breast cancer cell lines with a combination of DSF-Cu and LY294002, a pan-PI3K inhibitor, resulted in the significant inhibition of cell growth when compared with either drug alone. In addition, the combined treatment of DSF and LY294002 significantly inhibited the growth of the breast tumor xenograft in nude mice induced by MDA-MB-231 cells expressing mutant PIK3CA-H1047R and PIK3CA-E545K, whereas neither DSF nor LY294002 alone could significantly retard tumor growth. Finally, the observed in vivo inhibitory effects are found associated with aberrant signaling alterations and apoptosis-inducing activities in tumor samples. Thus, our finding shows for the first time that treatment of breast cancer with DSF results in a novel feedback mechanism that activates AKT signaling. Our study also suggests that the combination of DSF and a PI3K inhibitor may offer a new combinational treatment model for breast cancer, particularly for those with PIK3CA mutations.
Introduction
The phosphoinositide 3-kinase (PI3K)/PTEN/AKT signaling pathway has been firmly established for its role in multiple cellular activities, including cell proliferation, survival, metabolism, cytoskeleton reorganization, and membrane trafficking (1–3). The abnormal activation of this signaling pathway leads to various diseases such as diabetes, autoimmunity, and cancer. PIK3CA, the gene encoding the α form of class IA PI3Ks, is amplified and overexpressed in an assortment of human cancers, including breast, ovarian, head and neck, urinary tract, cervical, and thyroid cancers (4–8). Recently, somatic mutations of PIK3CA were identified in various human cancers with different frequencies (9–13). Specifically, 20% to 30% of breast cancers were found to harbor PIK3CA mutations, making it one of the most frequent genetic alterations in breast cancer. The amplification and somatic mutation of PIK3CA ultimately leads to the activation of the PI3K/AKT signaling pathway, thereby making PIK3CA a promising target for breast cancer treatment. Although many groups have focused on designing specific PI3K inhibitors, some studies have shown that maximum efficacy and minimum side effects may result from combining the PI3K-AKT pathway inhibitor with other cancer therapeutics that have different mechanisms of action (14–17).
Accumulated experimental and clinical evidence supports the notion that the ubiquitin/proteasome-dependent pathway plays an essential role in the upregulation of proliferation, downregulation of apoptosis, and promotion of angiogenesis and development of drug resistance in human tumor cells (18–20). This implicates proteasome inhibitors as potentially novel anticancer drugs (21–24). Consistent with this idea, our group reported that disulfiram (DSF), a clinically used antialcoholism drug and copper-binding agent, is capable of binding copper to form a new complex (DSF-Cu), which inhibits the proteasome and induces apoptosis in breast cancer cell cultures and retards the growth of breast cancer xenografts (22). Although DSF has recently been tested in a phase I/II clinical trial for the treatment of metastatic melanoma and refractory solid tumors involving the liver, its exact mechanism of action has not been well characterized.
In this report, we investigated the effect of DSF-Cu on the AKT signaling pathway, a well-established survival signaling pathway in breast cancer. We found that DSF-Cu treatment on several breast cancer cell lines led to the downregulation of PTEN protein expression and activation of pAKT along with the induction of cell death. This observation prompted us to test the possible synergistic effects of DSF-Cu and the PI3K inhibitor, LY294002, in these breast cancer cell lines, as loss of PTEN and activation of AKT may make cells more dependent on the PI3K/AKT pathway, and consequently more sensitive to PI3K Inhibition. Indeed, our in vitro study showed that the combination of DSF-Cu and LY294002 suppressed the growth of various breast cancer cells more effectively than either treatment alone. In addition, the effect was more significant in breast cancer cells with PIK3CA mutations than in breast cancer cells with wild-type-PIK3CA. When given to mice with a human breast cancer cell line MDA-MB-231 expressing mutant PIK3CA, the combined treatment of DSF and LY294002 showed more significant tumor growth inhibition than either single treatment. Moreover, the in vivo inhibitory effects were found associated with signaling alterations, proteasome inhibition, and apoptosis activation, as shown by the accumulation of p27 and BAX proteins, poly ADP ribose polymerase (PARP) cleavage, and apoptotic nuclei formation detected by multiple assays using tumor samples. Taken together, the information from this study provides a basis for developing a promising therapeutic model in which a PI3K inhibitor and a proteasome inhibitor may be combined to effectively treat breast cancer.
Materials and Methods
Cell culture and protease inhibitor experiment
Human breast cancer cell lines MDA-MB-157, MDA-MB-231, BT20, MCF7, T47D, and BT474 were obtained from the American Tissue Culture Collection and grown at 37°C in humidified 5% CO2 in DMEM (Invitrogen) supplemented with 10% fetal bovine serum. MDA-MB-157 and MDA-MB-231 are cells that express wild-type PIK3CA, whereas the MCF7 cell line contains a PIK3CA-E545K mutation. The T47D cell line contains a PIK3CA-H1047R mutation and the BT20 cell line contains two PIK3CA mutations (H1047R and P539R). All MDA-MB-157 and MDA-MB-231–derived cells were cultured with the same medium. The MCF10A and MCF10A-derived cell lines were cultured in serum free medium with insulin, hydrocortisone and epidermal growth factor as described in ref. 25. Caspase-3 inhibitor III was purchased from Calbiochem. Protease inhibitors N-acetyl-L-Leucyl-L-methioninal, trans-epoxysuccinyl-L-leucylamido-(4-guanidin) butane (E64), N-ethylmaleimide (NEM), phenylmethylsulfonyl fluoride (PMSF), Nα-p-tosyl-l-lysinechloromethyl ketone, and N-tosyl-l-phenylalanine chloromethyl ketone (TPCK) were obtained from Sigma. All these reagents were prepared according to the manufacturer’s instructions and were used to pretreat cells for 4 hours, followed by treatment with DSF-Cu for 6 hours. Protein lysates were then collected for the Western blot.
Construction of recombinant lentivirus and establishment of stable cells expressing mutant PIK3CA
The method was done as described (25).
Western blotting
For immunoblotting, we used the following primary antibodies: rabbit polyclonal antibodies against pAKT (Ser473), AKT, phosphorylated extracellular signal-regulated kinase (pERK), ERK, p-p70s6K, p70s6K, ubiquitin, PARP (Cell Signaling), p27 (Santa Cruz Biotechnology), and BAX (Sigma-Aldrich); and mouse monoclonal antibodies against V5 (Invitrogen), and β-actin (Sigma-Aldrich). A Western blot process was carried out as described (25).
Cell proliferation assay
Combinational treatment was performed by incubating cells with LY294002 for 2 days, adding DSF-Cu, and incubation for 1 additional day. After treatment, an MTT assay was performed as described in the protocol (Promega).
Human breast tumor xenograft experiments
Five-week-old female athymic nude mice were purchased from Taconic Research Animal Services and housed under pathogen-free conditions. Xenograft experiments were performed in compliance with the relevant laws and guidelines set forth by the Institutional Laboratory Animal Care and Use Committee of Wayne State University. MDA-MB-231/PIK3CA-H1047R and MDA-MB-231/PIK3CA-E545K cells (3 × 106) were injected s.c. into one flank of each mouse. Tumor size was measured with calipers every other day. Tumor volume (V) was determined by the equation V = (L × W2) × 0.5, in which L is the length and W is the width of the tumor. When xenografts reached volumes of ~200 mm3, the mice showing tumors were randomly assigned to a control group (n = 7), a DSF only group (n = 6), a PIK3CA inhibitor LY294002 only group (n = 6), and a combination group (DSF+LY294002; n = 6). These groups received the following treatments thrice weekly by s.c. injection on a different flank of the cell injection: solvent control (PBS/cremophor/DMSO/ethanol, 7.5:1.5:0.5:0.5), DSF (30 mg/kg), LY294002 (70 mg/kb), or DSF (30 mg/kg) plus LY294002 (70 mg/kb), respectively. The experiment was terminated and the mice were sacrificed when the control tumors reached ~1,000 mm3 (on day 29). The tumors were removed and photographed, and then used for multiple assays including Western blotting and immunohistochemistry (IHC).
H&E staining, IHC, and terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling assay
For H&E staining, paraffin-embedded sample slides were deparaffinized, hydrated, and then stained with hematoxylin for 1 minute. After rinsing, the slides were stained with eosin for 1 minute, rinsed, and sealed with coverslips using Per-mount. For immunohistochemical staining, mouse monoclonal anti-p27 antibody (1:40; Vector Laboratory) and rabbit polyclonal anti-BAX antibody (1:50; Zymed Labs, Inc.) were used. The terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) assay was performed using the ApopTag Peroxidase In situ Apoptosis Detection kit (Chemicon).
Statistical analysis
Evaluation of the synergism of two drugs DSF and LY294002 on cell proliferation for each cell line was carried out using a two-way ANOVA F test on the interaction of two factors: DSF concentration levels and presence/absence of LY294002. The evaluation of the synergism of DSF at a single concentration level and LY294002 on tumor burden in vivo was carried out using a balanced 2 × 2 factorial design with six replicates in each category. The drug effect of DSF + 10 μmol/L LY294002 is compared between mutated and wild-type cell lines using t test. All tests apply a significance level of 0.05 without adjustment for multiple tests. All the above-specified analyses were performed in the statistical software R.
Results
The effect of DSF-Cu on breast epithelial cells with or without PIK3CA mutation
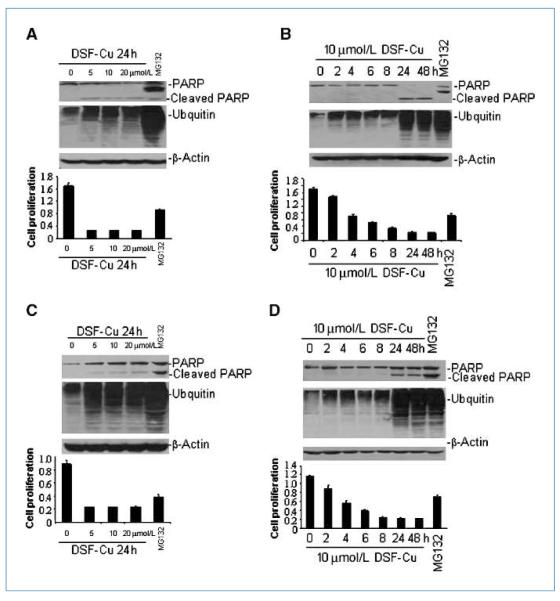
In an effort to investigate whether DSF-Cu has different effects on breast cancer cell lines with different PIK3CA status, we treated BT20 cells (with P539R and H1047R mutations) and MDA-MB-231 cells (with wt-PIK3CA) with various doses of DSF-Cu under various time courses. There was no observable difference in the dose- or time-dependent effects of DSF-Cu treatment on proliferation in these two cell lines. In addition, DSF-Cu treatment induced significant accumulation of ubquitinated proteins and PARP cleavage in both breast cancer cell lines (Fig. 1). Interestingly, treatment with the authentic proteasome inhibitor MG132, which was used as a control, generated a p85 PARP cleavage fragment in both cell lines, whereas DSF-Cu treatment resulted in a p85 fragment in MDA-MB-231 cells and a p65 fragment in BT20 cells (Fig. 1). Taken together, these data suggests that inhibition of cell proliferation induced by DSF-Cu is a common phenomenon in breast cancer cell lines regardless of PIK3CA status.
Figure 1.
The effect of DSF-Cu on breast cancer cell lines with or without PIK3CA mutations. A and C, the effect of different doses of DSF-Cu (0–20 μm) on BT20 cells (A) and MDA-MB-231 cells (C) for 24 h. B and D, the effect of different time courses (0–48 h) of 10 μmol/L DSF treatment on BT20 cells (B) and MDA2-MB-231 cells (D). For all the figures, top, Western blot showing PARP cleavage; middle, the accumulation of ubiquitinated proteins (UB-PRS); bottom, cell viability after different time courses or doses for DSF-Cu treatment. The proteasome inhibitor MG132 is used as a positive control.
DSF-Cu treatment results in PTEN protein decrease and AKT activation
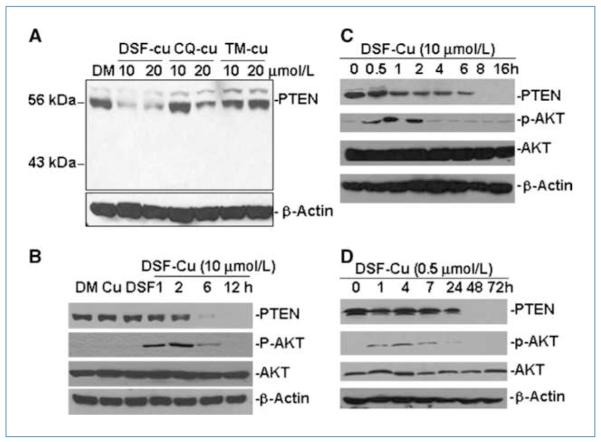
We next investigated the effect of DSF-Cu on the AKT signaling pathway, which plays a critical role in cell survival and proliferation. We first treated MDA-MB-231 cells with a mixture containing copper and either DSF, clioquinol (CQ), or tetrathiomolybdate (TM). CQ is a therapeutic agent for Alzheimer’s disease and has antiproteasomal, androgen receptor–suppressive, proapoptotic, and antitumorigenic activities in human breast and prostate cancer cells in the presence of copper (26, 27). TM is a strong copper chelator currently being tested in clinical trials (28, 29) and was used as a control here. To our surprise, we found that treatment with 10 or 20 μmol/L DSF-Cu or 20 μmol/L CQ-Cu mixtures could significantly inhibit PTEN protein expression, whereas TM-Cu at both concentrations had no effect (Fig. 2A).
Figure 2.
The effects of DSF-Cu on PTEN and AKT signaling in breast cancer cell lines with or without PIK3CA mutation. A, a Western blot showing the effects of different doses of DSF-Cu, CQ-Cu, and TM-Cu on PTEN protein expression in MDA-MB-231 cell. β-Actin was used as a protein loading control. B, kinetic effect of DSF-Cu (10 μmol/L) on PTEN and AKT signaling in MDA-MB-231 cells. MDA-MB-231 cells were treated with DMSO (DM), 10 μmol/L CuCl2 (Cu), DSF, or DSF-Cu mixture for the indicated number of hours. C and D, kinetic effects of high (10 μmol/L; C) versus low doses (0.5 μmol/L; D) of DSF-Cu on PTEN and AKT signaling in BT20 cell. BT20 cells were treated with DSF-Cu for the indicated number of hours. For B, C, and D, Western blots were performed with a PTEN, a p-AKT, and a total AKT antibody. β-Actin was used as a protein loading control.
A kinetic experiment using MDA-MB-231 cells showed a time-dependent decrease in PTEN protein expression with DSF-Cu treatment, which correlated with an initial increase in pAKT levels that decreased at later time points. The total AKT signal was not changed in this treatment (Fig. 2B). To confirm that this phenomenon was not cell line specific, we treated BT20 cells that harbor PIK3CA mutations with 10 μmol/L DSF-Cu and showed a significant time-dependent decrease in PTEN protein expression in these cells as well. Again, p-AKT was abruptly increased with DSF-Cu treatment (0.5–2 h) and then decreased to normal levels (after 2 h; Fig. 2C). A similar phenomenon was also observed with a low level of DSF-Cu treatment (0.5 μmol/L DSF) in BT20 cells, although the decrease of PTEN protein expression was postponed until after 24 hours of treatment and pAKT activation did not last longer than 24 hours (Fig. 2D).
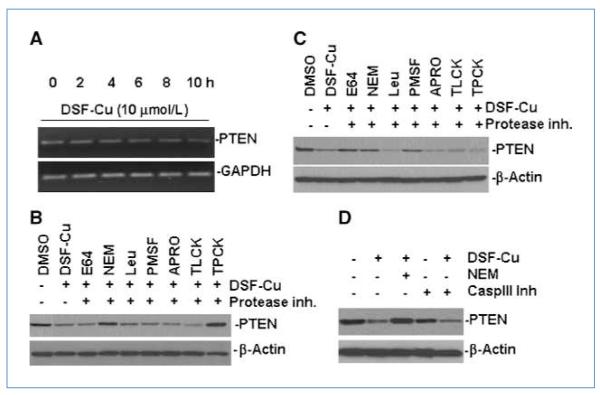
We next sought to explore the mechanism underlying the decrease in expression of PTEN upon the treatment of DSF-Cu. Reverse transcription-PCR analysis showed no significant change in PTEN mRNA level after DSF-Cu treatment in BT20 cells (Fig. 3A). These data suggest that protein stability rather than transcriptional regulation is the major mechanism responsible for the decreased PTEN protein levels in response to DSF-Cu treatment. We then examined the effects of different protease inhibitors on the decreased expression of PTEN protein induced by DSF-Cu in cultured cells. As shown in Fig. 3B, decreased PTEN expression induced by DSF-Cu treatment was attenuated in BT20 cells pretreated with two cysteine protease inhibitors NEM and TPCK. However, when MDA-MB-231 cells were tested, three protease inhibitors, E64, NEM, and PMSF, showed effect in blocking PTEN degradation induced by DSF-Cu (Fig. 3C). In contrast, as shown in Fig. 3D, a caspase-3 inhibitor could not rescue the PTEN degradation in BT20 cells treated with DSF-Cu. Therefore, caspase-3 does not play a critical role in PTEN degradation in DSF-Cu–treated cells.
Figure 3.
The mechanism of PTEN deregulation upon DSF-Cu treatment. A, the expression level of PTEN mRNA in BT20 cells after DSF-Cu treatment. Cells were treated with 10 μmol/L DSF-Cu for 0 to 10 h. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a RNA loading control. B and C, Western blot shows PTEN protein level after pretreatment with different protease inhibitor for 4 h, followed by DSF-Cu treatment for 6 h in BT20 cells (B) and MDA-MB-231 cells (C). D, caspase-3 inhibitor has no effect in preventing PTEN protein degradation upon DSF-Cu treatment. Different treatments were performed as indicated. Caspase-3 inhibitor (150 μmol/L) was pretreated for 4 h.
To investigate whether other authentic proteasome inhibitors could also cause a decrease in PTEN protein expression, we used MG132 and PS341, two well-recognized proteasome inhibitors, on breast cancer cell lines with different dose and time courses. The result showed that both MG132 and PS341 treatments led to the activation of pAKT and apparent decrease of PTEN protein in BT20 cells. The effects were less apparent in MDA-MB-231 cells (Supplementary Data S1). These data suggest that AKT activation and PTEN protein decrease may be common to all the proteasome inhibitors.
Combined treatment of LY294002 and DSF-Cu inhibits cell growth in vitro
AKT is a major cell survival molecule and activation of pAKT is a common cellular response to proapoptotic signaling. PTEN antagonizes PI3K and negatively regulates the AKT signaling pathway (30–32). Suppression of PTEN expression in breast cancer cells leads to AKT activation and confers sensitivity to inhibitors of mammalian target of rapamycin and AKT (33). In addition, breast cancer cell lines with PIK3CA mutation are more sensitive to PI3K inhibitor than cells with wt-PIK3CA, as evidenced by several reports including our own using newly established cell lines (Supplementary Data S2–4).
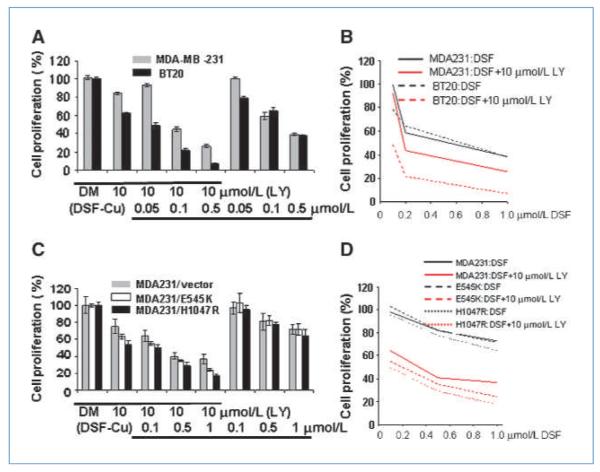
Considering that DSF-Cu treatment leads to multiple biological effects such as decreasing PTEN and activating AKT, along with its cell death induction function, we investigated whether the combined treatment of DSF-Cu with LY294002 would show an additive or a synergistic effect in killing breast cancer cells and also whether this effect would become more significant in cells with PIK3CA mutation. First, we performed in vitro cell proliferation assays on BT20, MCF7, T47D, and MDA-MB-231, four cell lines with different PIK3CA mutation statuses. The combination of DSF-Cu and LY294002 treatment led to a more significant decrease in cell proliferation than each single treatment alone in all four cell lines (Fig. 4A). Specifically, the effect in BT20 cells was more significant than in MDA-MB-231 cells at three concentrations (P = 0.007, 0.001, and <0.001). MCF7 and T47D cells had very similar effect in response to the combinational treatment (Supplementary Data S5). Statistical analysis indicated that synergism of the two drug treatments was found in all four cell lines (Supplementary Data S5; Fig. 4B). In addition, we performed a similar treatment on a set of isogenic cell models: MDAMB-231/vector cells, MDA-MB-231/PIK3CA-H1047R, and MDA-MB-231/PIK3CA-E545K (Fig. 4C). We found that combined treatment led to increased proliferation inhibition compared with each single treatment alone in all three cell lines. The effect was more significant in MDA-MB-231/PIK3CA-H1047R and MDA-MB-231/PIK3CA-E545K cells than in MDA-MB-231/vector cells with 10 μmol/L LY294002 plus 1 μmol/L DSF-Cu (Fig. 4C), although no significant synergism was found with indicated treatment conditions (Fig. 4D). Moreover, the effect of MDA-MB-231/PIK3CA-H1047R and MDA-MB-231/PIK3CA-E545K cells responding to the combinational treatment is similar (Fig. 4C). We also observed similar results in MDA-MB-157 and MCF10A-derived model cells (Supplementary Data S6).
Figure 4.
Combinational effects of DSF-Cu and LY294002 (LY) on breast cancer cell lines in vitro. A, the effect of combined treatment on BT20 and MDA-MB-231 cells. Three treatments were performed: (1) LY294002 at 10 μmol/L; (2) DSF-Cu at 0.05, 0.1, or 0.5 μmol/L; and (3) LY294002 plus DSF-Cu combination. DM, DMSO treatment as a negative control. B, scatter plot of cell proliferation (%) versus dose concentrations (μmol/L). Percent proliferations (%) were calculated as the ratios to the controls. Straight lines connect the mean cell proliferations. Synergism is indicated in both cell lines BT20 (P = 0.0014) and MDA-MB-231 (P = 0.0495) by ANOVA test. C, MDA-MB-231/vector, MDA-MB-231/PIK3CA-E545K, and MDA-MB-231/PIK3CA-H1047R cells were used to test the treatment regimens. Combined treatments (10 μmol/L LY294002 plus 1 μmol/L DSF) showed more significant inhibition of cell proliferation than a single treatment in all cell lines (all P < 0.002). D, the drug effects of combined treatment (10 μmol/L LY294002 plus DSF) among three cell lines are significant. The P values of one way ANOVA test at three concentration levels of DSF are 0.003, 0.019, and <0.001, respectively.
Combined treatment of LY294002 and DSF synergistically inhibits tumorigenesis of breast cancer cells in vivo
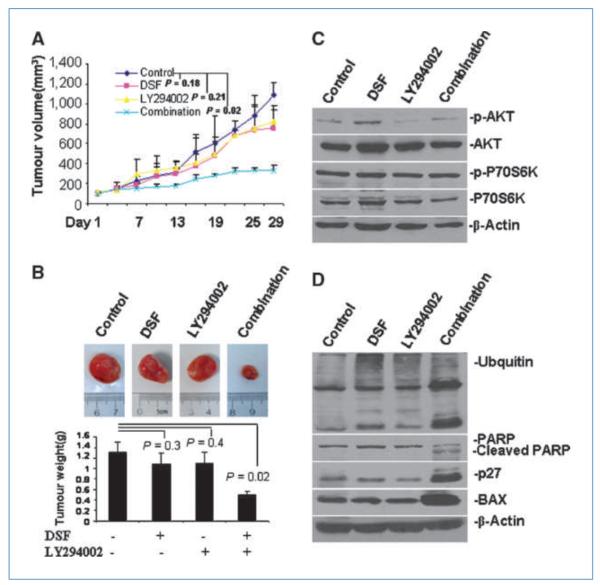
To further test our hypothesis in vivo, four different treatments were delivered to a xenograft mouse model created by s.c. injection with MDA-MB-231/PIK3CA H1047R cells. We found that combined treatment, as well as treatment with DSF or LY294002 alone, inhibited tumor growth by 74.5% (P = 0.02), 35% (P = 0.18), and 27% (P = 0.21), respectively, compared with the solvent control (Fig. 5A). Furthermore, tumor burden in the DSF and LY294002 single-treatment groups was reduced by 25% (P = 0.3) and 16.7% (P = 0.4), respectively, whereas in the combined treatment group, tumor burden was reduced by 58.3% (P = 0.02), compared with the control group (Fig. 5B). Two-way ANOVA test was performed and showed synergism of the two drugs DSF and LY294002 on tumor burden in vivo (P = 0.002; Supplementary Data S7). Similarly, in contrast to either single treatment, only the combined treatment could significantly retard the growth of the xenograft tumor induced by the MDA-MB-231/PIK3CA-E545K cells (Supplementary Data S8). Taken together, the in vivo data are consistent with in vitro data and support the hypothesis that the combined treatment of DSF and LY294002 has a more significant effect in targeting breast cancer cells. In addition, the mutation in the helical and kinase domain had no significant difference in response to the combined treatment.
Figure 5.
Combinational effects of DSF-Cu and LY294002 on the tumorigenesis of MDA-MB-231/PIK3CA-H1047R mutant cells in vivo. A, tumor growth chart showing the effect of the different treatments in vivo. Points, mean of tumor volume in each experimental group; bars, SD. B, comparison of final tumor weights after four different treatments in vivo. Top, representative pictures of one tumor group with four different treatments. Bottom, final weights after four treatments. Columns, mean; bars, SD. C, representative Western blot analysis of tumor tissue extract with antibodies against pAKT, AKT, p70S6K, pP70S6K, and β-actin. D, representative Western blots analysis of tumor tissue extract with antibodies against p27, BAX, PARP, ubiquitin, and β-actin.
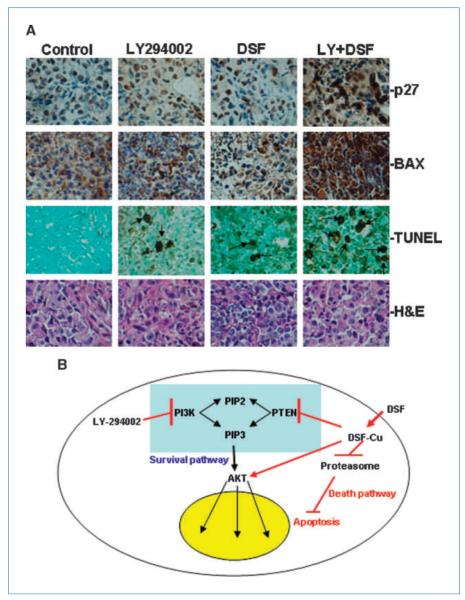
To further investigate whether the observed inhibitory effects of the combined treatments and single treatments are associated with signaling alterations or apoptosis induction, tumor samples from xenograft mice were subjected to Western blotting and IHC assays. Western blots using the tumor extracts showed that DSF treatment slightly increased pAKT, whereas LY294002 treatment decreased pAKT and the combined treatment led to a relatively low-level of pAKT compared with control. No significant changes in p70S6K or p-p70 S6K were observed (Fig. 5C). In addition, combined treatment led to significant PARP cleavage compared with single treatment alone or control. Subtle accumulation of ubiquitinated protein was also observed in DSF-, LY294002-, and combination-treated tumors (Fig. 5D). An increase in the levels of the proteasome-targeting proteins p27 and BAX were also observed in the combination-treated sample (Fig. 5D) and confirmed by IHC (Fig. 6A), indicating proteasomal inhibition in these tumor samples. Moreover, apoptosis in tissues derived from xenografts that received the combined treatment occurred about thrice more frequently than in tissues derived from xenografts treated with DSF or LY294002 alone as shown by the TUNEL assay (Fig. 6A; positive control was shown in Supplementary Data S9). Apoptosis induction in tumor samples was further observed by the H&E staining results. In contrast, apoptosis occurrence was not apparent in the control xenograft tumors (Fig. 6A). Overall, the combined treatment of DSF and LY294002 showed significant inhibition of breast tumor growth in nude mice, and this inhibition was associated with aberrant signaling alterations, proteasome inhibition, and induction of apoptosis.
Figure 6.
Alterations of apoptosis signaling in xenograft tumor samples and illustration of potential mechanism. A, IHC assays and TUNEL assay using mouse tumor samples. Tumors were collected after 29 d of treatment and the prepared tissue slides were analyzed with IHC using anti-p27 and anti-BAX antibodies, TUNEL assay, and H&E staining assay. P27 showed positive nucleus staining and BAX stained positively in the cytosol; arrows, stronger TUNEL-positive nuclei as well as apoptotic condensed nuclei. Top, the origins of all four-tumor samples. Magnification for all pictures, 10 × 40. B, illustration of a potential mechanism explaining the effects of current combinational treatment strategy, using the feedback loop induced by DSF-Cu treatment.
Discussion
Although DSF is clinically used as an antialcoholism drug, our group, along with several others, reported that DSF could be used as an anticancer drug (22). One possible mechanism is that DSF may interact with elevated levels of endogenous copper in tumor tissues (34–36) and form DSF-Cu complexes with active antitumor properties, specifically proteasomal inhibition and apoptotic induction. Consistent with previous observations, our current study found that DSF treatment induced an accumulation of many ubiquitinated proteins in different cell lines regardless of the PIK3CA status. We also showed that a low dose of DSF-Cu could induce the accumulation of ubiquitinated proteins and activate AKT, although it did not significantly inhibit tumor growth in vivo. This observation prompted us to speculate that a combination of low-dose DSF with reagents harboring different mechanisms could result in high therapeutic effects and low toxicity in vivo.
In this study, we showed that DSF-Cu treatment decreased the expression of PTEN protein (but not RNA) levels in different breast cancer cell lines in a time- and dose-dependent manner. Therefore, inhibition of protein synthesis and/or increase of protein degradation could be the mechanisms underlying this serendipitous observation. However, because decreased expression of PTEN protein upon DSF-Cu treatment was observed in the very early hours, we thus focused on the protein degradation mechanism. Our study showed that different protease inhibitors could reverse the PTEN degradation induced by DSF-Cu in two different cell lines and the cysteine protease inhibitor NEM commonly works in two different cell lines. This result not only confirmed our hypothesis but also is consistent with several previous reports that protease expression could be regulated by a feedback mechanism in response to some proteasome inhibitors (37, 38). Therefore, in our study, the DSF treatment might lead to the upregulation of some protease activity, which in turn may lead to the degradation of the PTEN protein.
In the past several years, numerous studies have identified a high frequency of PIK3CA somatic mutations in cancer cell lines and tumor samples, and have clarified the oncogenic properties of mutant PIK3CA in cancer tumorigenesis (39, 40). More importantly, 80% of the PIK3CA somatic mutations were identified in two hotspots, which are separately located in the helical and kinase domains of the PIK3CA molecule. Several recent studies suggest that the mutations in the helical and kinase domains trigger a gain of function through different mechanisms and speculate the contrasting roles of p85 and Ras-GTP in these two mutations (41–43). In addition, these two mutations were also reported to be associated with different prognoses for disease-free survival (44). However, the results of several previous studies using different mutant model cell lines indicated that H1047R and E545K, two mutations located separately in the kinase and helical domains, have very similar response to a pan-PI3K inhibitor LY294002 (13, 25, 45). The in vitro and in vivo assay results of the current study further support this notion, suggesting that the inhibition of the ATP binding site of PI3Ks is a feasible strategy in targeting cancer cells with different PIK3CA mutations.
Due to the structural similarity between the isoforms of PI3Ks, a specific inhibitor targeting PIK3CA is still unavailable and, as a result, most developed PI3K inhibitors are pan-PI3K inhibitors. Meanwhile, accumulating evidence has shown that various other PI3K isoforms (e.g., β, δ, and γ) are also involved in tumorigenesis (46–49). Therefore, a pan-PI3K isoform inhibitor may offer enhanced therapeutic benefits compared with an isoform-specific inhibitor. This notion has been further supported by a recent report that shows that although p110α activation is required to sustain the proliferation of established PIK3CA-mutant tumors, PTEN-deficient tumors depend instead on p110β signaling (50). In this regard, a pan-PI3K inhibitor is expected to have the broadest clinical application, as it will target the growth of both PIK3CA mutations and PTEN-null cancers. In our current study, DSF treatment led to decreased PTEN protein and activation of AKT in both cell types with and without PIK3CA mutations. Therefore, we expect that the combination of a pan-PI3K inhibitor with DSF would be more effective than an isoform-specific inhibitor.
Many newly identified drugs tested in research or preclinical settings are common apoptosis inducers and the mechanisms of these drugs on cells are not entirely clarified. This drawback limits their transition from research to clinical applications. In our current study, we show for the first time that DSF-Cu treatment on breast cancer cell lines leads to a decrease in the PTEN protein level and an increase in the pAKT level. Both changes are believed to ultimately lead to the activation of the AKT signaling pathway. Therefore, DSF seems to have two mechanisms of action. On one hand, DSF-Cu inhibits the proteasome, which in turn triggers the death pathway and induces apoptosis. On the other hand, DSF-Cu also activates the PI3K/PTEN/AKT survival signaling pathway (Fig. 6B). Consistent with this observation, our study showed that the PI3K inhibitor, LY294002, combined with DSF treatment, not only effectively inhibited cell proliferation in vitro, but also significantly decreased the tumor formation in vivo. Taken together, our current observations reinforce the notion that effective therapy could result from combinations of therapeutic agents with different functional mechanisms. More significantly, our results suggest that a potential proteasome inhibitor and a pan PI3K inhibitor could be a novel and promising combined treatment option for breast cancer therapy.
Supplementary Material
Acknowledgments
We thank Drs. David Gorski, Julie Boerner, Zengquan Yang, Ramsi Hadad, and Craig Giroux for their insightful discussions and technical suggestions; Dr. Seema Sethi for the assistance with the IHC analysis; Dr. Youming Xie for providing the PS341; and Dr. Cecilia Speyer and Elizabeth A. Katz for editing our manuscript.
Grant Support Susan G. Komen grant KG080465 (G. Wu), a pilot grant from Karmanos Cancer Institute, Wayne State University (G. Wu), and supporting program for young university teachers (2006jql141zd) and Natural Science Key Foundation (KJ2010A299) from Department of Education, Anhui Province, People’s Republic of China (H. Zhang).
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–9. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 2.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 3.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 4.Knuutila S, Bjorkqvist AM, Autio K, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152:1107–23. [PMC free article] [PubMed] [Google Scholar]
- 5.Ma YY, Wei SJ, Lin YC, et al. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–44. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 6.Racz A, Brass N, Heckel D, Pahl S, Remberger K, Meese E. Expression analysis of genes at 3q26-q27 involved in frequent amplification in squamous cell lung carcinoma. Eur J Cancer. 1999;35:641–6. doi: 10.1016/s0959-8049(98)00419-5. [DOI] [PubMed] [Google Scholar]
- 7.Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 8.Wu G, Mambo E, Guo Z, et al. Uncommon mutation, but common amplifications, of the PIK3CA gene in thyroid tumors. J Clin Endocrinol Metab. 2005;90:4688–93. doi: 10.1210/jc.2004-2281. [DOI] [PubMed] [Google Scholar]
- 9.Bachman KE, Argani P, Samuels Y, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–5. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 10.Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–81. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 11.Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–9. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 12.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, Xing M, Mambo E, et al. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005;7:R609–16. doi: 10.1186/bcr1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan QW, Specht KM, Zhang C, Goldenberg DD, Shokat KM, Weiss WA. Combinatorial efficacy achieved through two-point blockade within a signaling pathway-a chemical genetic approach. Cancer Res. 2003;63:8930–8. [PubMed] [Google Scholar]
- 15.Hu L, Hofmann J, Lu Y, Mills GB, Jaffe RB. Inhibition of phosphatidylinositol 3′-kinase increases efficacy of paclitaxel in in vitro and in vivo ovarian cancer models. Cancer Res. 2002;62:1087–92. [PubMed] [Google Scholar]
- 16.Klejman A, Rushen L, Morrione A, Slupianek A, Skorski T. Phosphatidylinositol-3 kinase inhibitors enhance the anti-leukemia effect of STI571. Oncogene. 2002;21:5868–76. doi: 10.1038/sj.onc.1205724. [DOI] [PubMed] [Google Scholar]
- 17.Weisberg E, Banerji L, Wright RD, et al. Potentiation of antileukemic therapies by the dual PI3K/PDK-1 inhibitor, BAG956: effects on BCR-ABL- and mutant FLT3-expressing cells. Blood. 2008;111:3723–34. doi: 10.1182/blood-2007-09-114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almond JB, Cohen GM. The proteasome: a novel target for cancer chemotherapy. Leukemia. 2002;16:433–43. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- 19.Dou QP, Li B. Proteasome inhibitors as potential novel anticancer agents. Drug Resist Updat. 1999;2:215–23. doi: 10.1054/drup.1999.0095. [DOI] [PubMed] [Google Scholar]
- 20.Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–58. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 21.Adams J. Development of the proteasome inhibitor PS-341. Oncologist. 2002;7:9–16. doi: 10.1634/theoncologist.7-1-9. [DOI] [PubMed] [Google Scholar]
- 22.Chen D, Cui QC, Yang H, Dou QP. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006;66:10425–33. doi: 10.1158/0008-5472.CAN-06-2126. [DOI] [PubMed] [Google Scholar]
- 23.Dou QP, Goldfarb RH. Bortezomib (millennium pharmaceuticals) IDrugs. 2002;5:828–34. [PubMed] [Google Scholar]
- 24.Milacic V, Chen D, Ronconi L, Landis-Piwowar KR, Fregona D, Dou QP. A novel anticancer gold(III) dithiocarbamate compound inhibits the activity of a purified 20S proteasome and 26S proteasome in human breast cancer cell cultures and xenografts. Cancer Res. 2006;66:10478–86. doi: 10.1158/0008-5472.CAN-06-3017. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Liu G, Dziubinski M, Yang Z, Ethier SP, Wu G. Comprehensive analysis of oncogenic effects of PIK3CA mutations in human mammary epithelial cells. Breast Cancer Res Treat. 2008;112:217–27. doi: 10.1007/s10549-007-9847-6. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Cui QC, Yang H, et al. Clioquinol, a therapeutic agent for Alzheimer’s disease, has proteasome-inhibitory, androgen receptor-suppressing, apoptosis-inducing, and antitumor activities in human prostate cancer cells and xenografts. Cancer Res. 2007;67:1636–44. doi: 10.1158/0008-5472.CAN-06-3546. [DOI] [PubMed] [Google Scholar]
- 27.Daniel KG, Chen D, Orlu S, Cui QC, Miller FR, Dou QP. Clioquinol and pyrrolidine dithiocarbamate complex with copper to form protea-some inhibitors and apoptosis inducers in human breast cancer cells. Breast Cancer Res. 2005;7:R897–908. doi: 10.1186/bcr1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brewer GJ. The use of copper-lowering therapy with tetrathiomolyb-date in medicine. Expert Opin Investig Drugs. 2009;18:89–97. doi: 10.1517/13543780802621859. [DOI] [PubMed] [Google Scholar]
- 29.Henry NL, Dunn R, Merjaver S, et al. Phase II trial of copper depletion with tetrathiomolybdate as an antiangiogenesis strategy in patients with hormone-refractory prostate cancer. Oncology. 2006;71:168–75. doi: 10.1159/000106066. [DOI] [PubMed] [Google Scholar]
- 30.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci U S A. 1999;96:4240–5. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fine B, Hodakoski C, Koujak S, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–5. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons R. Human cancer, PTEN and the PI-3 kinase pathway. Semin Cell Dev Biol. 2004;15:171–6. doi: 10.1016/j.semcdb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 33.Steelman LS, Navolanic PM, Sokolosky ML, et al. Suppression of PTEN function increases breast cancer chemotherapeutic drug resistance while conferring sensitivity to mTOR inhibitors. Oncogene. 2008;27:4086–95. doi: 10.1038/onc.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habib FK, Dembinski TC, Stitch SR. The zinc and copper content of blood leucocytes and plasma from patients with benign and malignant prostates. Clin Chim Acta. 1980;104:329–35. doi: 10.1016/0009-8981(80)90390-3. [DOI] [PubMed] [Google Scholar]
- 35.Nayak SB, Bhat VR, Upadhyay D, Udupa SL. Copper and ceruloplasmin status in serum of prostate and colon cancer patients. Indian J Physiol Pharmacol. 2003;47:108–10. [PubMed] [Google Scholar]
- 36.Rizk SL, Sky-Peck HH. Comparison between concentrations of trace elements in normal and neoplastic human breast tissue. Cancer Res. 1984;44:5390–4. [PubMed] [Google Scholar]
- 37.Lee CS, Tee LY, Warmke T, et al. A proteasomal stress response: pre-treatment with proteasome inhibitors increases proteasome activity and reduces neuronal vulnerability to oxidative injury. J Neurochem. 2004;91:996–1006. doi: 10.1111/j.1471-4159.2004.02813.x. [DOI] [PubMed] [Google Scholar]
- 38.Meiners S, Heyken D, Weller A, et al. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem. 2003;278:21517–25. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- 39.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–9. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang CH, Mandelker D, Schmidt-Kittler O, et al. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science. 2007;318:1744–8. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 42.Miled N, Yan Y, Hon WC, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–42. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 43.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110α of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–7. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbareschi M, Buttitta F, Felicioni L, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–9. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 45.Samuels Y, Diaz LA, Jr., Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–73. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 46.Ciraolo E, Iezzi M, Marone R, et al. Phosphoinositide 3-kinase p110β activity: key role in metabolism and mammary gland cancer but not development. Sci Signal. 2008;1:ra3. doi: 10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia S, Liu Z, Zhang S, et al. Essential roles of PI(3)K-p110β in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–9. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110β, -γ, and -δ isoforms of class I phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2006;103:1289–94. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110α and p110β phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–8. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wee S, Wiederschain D, Maira SM, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–62. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.