Summary
An assay has been developed in which the activity of an ubiquinol oxidase from Escherichia coli, cytochrome bo3 (cbo3), is determined as a function of the hydrophobic substrate ubiquinol-10 (UQ-10) in tethered lipid bilayers membranes (tBLMs). UQ-10 was added in situ, while the enzyme activity and the UQ-10 concentration in the membrane have been determined by cyclic voltammetry. Cbo3 is inhibited by UQ-10 at concentrations above 5-10 pmol/cm2, while product inhibition is absent. Cyclic voltammetry has also been used to characterise the effects of three inhibitors; Cyanide, inhibiting oxygen reduction; 2-n-Heptyl-4-hydroxyquinoline N-oxide (HQNO), inhibiting the quinone oxidation and Zn(II), thought to block the proton channels required for oxygen reduction and proton pumping activity. The electrochemical behaviour of cbo3 inhibited with HQNO and Zn(II) is almost identical, suggesting that Zn(II) ions inhibit the enzyme reduction by quinol, rather than oxygen reduction. This suggests that at Zn(II) concentration below 50 μM the proton release of cbo3 is inhibited, but not the proton uptake required to reduce oxygen to water.
Keywords: ubiquinol oxidase, cytochrome c oxidase, electrochemistry, tethered bilayer lipid membranes, enzyme mechanism
Introduction
Bacteria have highly diverse and highly branched respiratory chains [1] which consist of a range of enzymes which transfer electrons from many different substrates into a common pool of lipid soluble electron carriers, known collectively as quinones. There are a number of different quinones expressed in organisms including ubiquinone or coenzyme Q, menaquinone and plastoquinone. The composition of the quinone pool in the membrane is dependent on the species and growth conditions. Generally, anaerobic and gram positive bacteria tend to use menaquinones whereas aerobic gram negative bacteria and eukaryotic membranes are predominantly composed of ubiquinones.[2] Ubiquinone-8 is the predominant electron carrier in aerobically grown Escherichia coli.
In the E. coli aerobic respiratory pathway there are two terminal oxidases that catalyse the oxidation of ubiquinol to ubiquinone and reduce molecular oxygen to water. Expression of cytochrome bo3 (cbo3) is the favoured terminal oxidase under high oxygen conditions and cytochrome bd is expressed under microaerophilic conditions.[3, 4] Cbo3 is of particular interest as it is structurally related to the eukaryotic cytochrome c oxidase.[5] In the catalytic cycle of cbo3, ½O2 is reduced to H2O for every ubiquinol that is oxidised and a total of four protons are translocated across the membrane, two of which come from the oxidation of ubiquinol and reduction of ½O2. The remaining two are pumped through the protein in conjunction with the catalytic cycle.[6] Cbo3 contains two ubiquinol binding sites, a low affinity site (QL), which binds the substrate ubiquinol for catalysis and is in equilibrium with the free ubiquinone pool in the membrane, and a high affinity site (QH), which stabilises the semiquinone intermediate during catalysis and is not exchanged with the the UQ pool in the bilayer.[7] When the crystal structure of cbo3 was solved, no quinone was found in the QH but the structure enabled the authors to suggest a binding pocket.[5] This site was later confirmed and further refined by mutagenenis and labelling studies in the group of Gennis.[8-10]
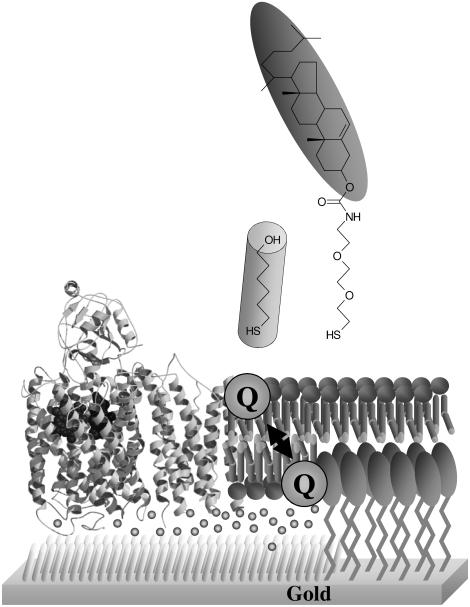
The characterisation of transmembrane redox enzymes can be problematic due to the atypical nature of their substrates. Analysis of enzyme kinetics with hydrophobic substrates can be complicated by incomplete knowledge of the true substrate concentration in the membrane.[11] Long chain quinones such as UQ-10 are extremely hydrophobic and rapidly aggregate in aqueous solution, so water soluble substrate analogues such as UQ-1, UQ-2 and duroquinol are commonly used to derive kinetic and mechanistic information. We have developed a membrane model in which the activity of cbo3 can be assayed under near native conditions in a tethered bilayer lipid membrane (tBLM, Figure 1).[12, 13] tBLMs are biomimetic surfaces that were first designed in the 90ies as a model membrane system to study a wide variety of membrane properties, ranging from the physical chemistry of lipid membranes, ion transport through ion channels to enzyme kinetics of membrane-bound oxidoreductases.[14-16]
Figure 1.
A schematic representation of the tBLM used in this work.
The general principle of a tBLM is based on a surface that is modified with synthetic lipids, which consist of a lipid headgroup, a linker moiety and a group that binds to the surface. In our system the lipid headgroup is a cholesterol, the linker consist of three repeats of ethyleneglycol and the surface-binding group is a thiol (Figure 1, top). When these modified surfaces are incubated with lipid vesicles, the vesicles spread onto the modified surfaces and cover the synthetic lipids with phospholipid layer and thus creating a bilayer. In our system, the coverage of the synthetic cholesterol linker is reduced by mixing it with 6-mercaptohexanol. Because the two thiol molecules phase separate on the gold surface, cholesterol-free regions are created with space for integral membrane proteins (Figure 1, bottom).[17] A particularly important feature of this membrane model is that the native, hydrophobic quinones can be incorporated in the bilayer.
We have previously shown that tBLMs can be used to study the enzyme kinetics of quinone converting enzymes like cbo3 using voltammetric methods.[13, 18, 19] In this method, ubiquinone in the tBLM is electrochemically reduced by the electrode and after diffusion to cbo3 ubiquinol is catalytically oxidised, creating a redox loop. By measuring the electron consumption at the surface (the electrode current), the enzyme activity is thus established. In the past we have incorporated cbo3 by assembling the tBLM with proteoliposomes that were prepared with purified enzyme.[18] More recently, we have prepared tBLMs from total inner membrane extract from E. coli.[13, 19] The advantage of latter system is that no complex or harsh purification procedures are used and that the protein of interest is permanently retained within its natural membrane environment.
We have continued this previous work by developing an assay to determine enzymes kinetics as a function of UQ-10 concentration in the membrane. By adding UQ-10 in situ, higher concentration of UQ-10 can be reached than we reported previously.[13] We have also carried out inhibition studies to interpret and understand mechanisms and behaviour of quinone converting enzymes. We present voltammetric evidence of different modes of inhibition from three distinct inhibitors which are known to bind and to different sites of cbo3.
Materials and Methods
Materials
EO3-cholesteryl was made as previously described[20]. 6-Mercaptohexanol (Fluka) was used without further purification. All solvents were HPLC grade (Fisher) and used as received. 20 mM MOPS buffer with 30 mM Na2SO4 adjusted to pH 7.4 was used for the preparation of membrane extracts and all electrochemistry experiments. For inhibition studies, ubiquinone-10 (Sigma) was made up as 1mg/ml stock solution in chloroform and stored at −20° C. E. coli polar extract (Avanti) was stored in 5 mg dry aliquots stored under nitrogen at −20° C. NaCN and ZnSO4 were added from a aqueous stock solution of 5 and 10mM, respectively. 2-n-Heptyl-4-hydroxyquinoline N-oxide (HQNO, Alexis biochemicals) was made up as a stock solution of 20mM in methanol and stored at −20° C. For in situ titration of UQ-10, UQ-10 was made up as stock solutions of 500nM, 5μM, 50μM, 500μM and 5mM in dimethyl sulfoxide (DMSO).
E. coli inner membrane purification
E. coli inner membranes were prepared from strain GO105/pJRhisA in which the cbo3 protein is overexpressed and no cytochrome bd is present[21]. E. coli was grown to mid-log phase at 37° C with shaking in LB medium supplemented with 500 μM CuSO4 and 100 μg/ml carbenicillin. E. coli cells were harvested from the growth medium by centrifugation at 12000 g for 30 minutes and cell paste was frozen at −20° C overnight. Thawed E. coli cell paste was resuspended in 20 mM MOPS 30mM Na2SO4 buffer at approximately 30 mL buffer per 10 g cell paste and passed through a cell disrupter two times at 35000 psi. Cell debris was removed by centrifugation at 12000 g for 30 minutes. The supernatant containing the membrane fraction was centrifuged at 131000 g for 2 hours and the membrane pellet was resuspended in 25% w/w sucrose-MOPS/Na2SO4 buffer. A 30% w/w to 55% w/w sucrose gradient with centrifugation at 131000 g for 16 hours with no deceleration or breaking was used to separate the inner membrane from the outer membrane. The inner membrane fraction was removed from the sucrose gradient and washed several times with buffer by centrifugation at 131000 g for 2 hours. The protein concentration of the inner membrane preparation was determined using a Schaffner-Weissman assay[22]. Inner membrane vesicles were resuspended in buffer and stored in 5 mg/ml protein aliquots at −80° C.
Mixed Vesicle Preparation
For the inhibition studies 1% w/w ubiquinol-10 solution was added to E. coli polar extract lipids and dried under nitrogen to form a multilammelar film on the sides of a glass vial. The lipid/UQ-10 film was resuspended in buffer (5 mg/ml) by vortexing. Lipid vesicles were formed by extrusion through a track etched membrane (200nm, Avanti), mixed with inner membrane vesicles (10% v/v) and subjected to three rounds of freeze-thawing before passing through a track etched membrane another 11 times. For membranes used in the in situ UQ-10 titration, mixed vesicles were prepared as described but no UQ-10 was added to the E. coli polar extract lipids.
Electrodes and tethered membrane formation
Template-stripped gold (TSG) surfaces were formed as previously described[23]. Briefly, 150 nm of gold was evaporated onto a clean, polished silicon wafer. 12 mm by 12 mm clean glass microscope slides were glued to the gold surface using Epo-tek 377 and cured for 2 hours at 120° C. Once cooled, the glass slides could be removed from the silicon wafer to expose the template-stripped gold surface ready for the formation of the self-assembled monolayer (SAM).
SAMs were formed by incubating a freshly exposed TSG slide in 0.11 mM EO3-cholesteryl and 0.89 mM 6-mercaptohexanol in propanol for 16 hours. This forms a 60%/40% EO3-cholesteryl/6-mercaptohexanol area ratio on the surface which was checked with impedance spectroscopy before each experiment[17]. The slides were rinsed with propanol and methanol and dried under nitrogen before being incorporated into the electrochemical cell.
To form tethered bilayer lipid membranes (tBLMs), mixed vesicles were added to the SAM surface at a final concentration of 0.5 mg/ml in the presence of 10 mM CaCl2 and incubated for 2 hours. The surface was then rinsed several times with buffer and 1 mM EDTA to remove any traces of calcium ions in the cell.
Electrochemistry
Electrochemical measurements were carried out in a glass electrochemical cell which holds 2 mL of buffer and was thermostatted at 20° C. A saturated calomel electrode (SCE) or saturated Ag/AgCl reference electrode was used as the reference electrode (Radiometer Analytical) and a platinum wire was used as the counter electrode. All potentials are quoted versus the Standard Hydrogen Electrode (SHE). The gold-SAM working electrode was secured at the open base of the electrochemical cell in a PTFE electrode holder, exposing an electrode area of 0.25 cm2. The electrochemical cell was housed in a Faraday cage to minimise electrical noise and argon was used to purge the cell of oxygen. For the in situ UQ-10 titration, the experimental apparatus was stored and assembled inside a nitrogen filled glovebox (MBraun MB 150 B-G) where the O2 levels were <1 ppm and air equilibrated buffer was added to the electrochemical cell to give an oxygen concentration of 100 μM, well in excess of the KM. Electrochemical measurements were recorded using an Autolab (Ecochemie) electrochemical analyser with a PGSTAT30 potentiostat and a FRA2 frequency analyser. All cyclic voltammetry experiments were carried out by holding the potential at 0.4 V for 5 seconds and cycling to −0.4 V and back at a scan rate of 0.01 V/s.
For the in situ UQ-10 titration, the UQ-10 concentration within the bilayer was monitored using cyclic voltammetry at 1V/s scan rate. At this high scan rate, the catalytic activity of the enzyme in the tBLM is outrun and a clear peak is observed due to the UQ-10 reduction. The area underneath the peak, determined by the software ‘Utils’, kindly provided by Dr. H.A. Heering, was used to calculate the UQ-10 concentration in the tBLM.
For analysis of the inhibition data, the baseline of the CV was determined from the fully inhibited trace (using the ‘Utils’ software) and subsequently subtracted from all traces. To determine the IC50 values, the relative current at −0.35V vs SHE was taken from the baseline corrected data, although no significant difference was observed between IC50 values determined using baseline corrected data and the raw data.
Results
In situ titration of UQ-10
Several organic solvents were tried to titrate UQ-10 in the electrochemical cell, including ethanol and acetone. DMSO was found to be the most suitable solvent for in situ addition as UQ-10 is soluble in DMSO up to concentrations of 5 mM, DMSO easily mixes with water and is non-volatile. The main drawback is that DMSO has been suggested to disrupt and permeablise cell membranes.[24] However, we did not observe and significant disruption to the tBLM (only a small decrease in double layer capacitance was observed between 0 and 40% DMSO). Finally, no difference in the cbo3 activity was observed with cyclic voltammetry when adding small amounts of DMSO (< 2%).
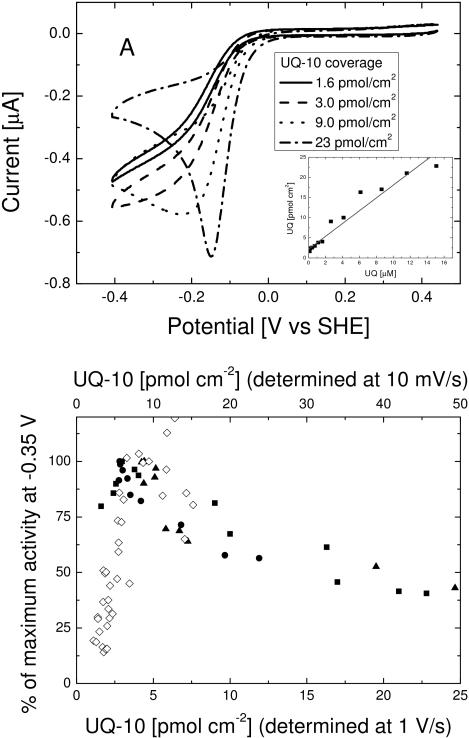
Figure 2a shows cyclic voltammograms (10 mV/s) of the tBLM at increasing amounts of UQ-10. DMSO content stayed below 10% (v/v) throughout the titration. As is clear from Figure 2a, the UQ-10 addition significantly changes the shape of the cyclic voltammograms. At low concentration, the shape remains largely unchanged, but the current increases. At high concentration, the wave converts into a peak shaped voltammogram, with the current at low potential (< −0.3 V vs SHE) decreasing. The peak is partly due to large amount of UQ-10 in the membrane, thus becoming more prominent relative to the catalytic wave. Analysis of the peak area presented in the Discussion section shows that this peak signal cannot be explained by UQ-10 reduction alone.
Figure 2.
(Top) Cyclic voltammograms of a tBLM containing cbo3 at increasing UQ-10 coverage (determined separately using cyclic voltammograms at 1 V/s). Electrode area is ~ 0.25 cm2 (Top, Insert) The UQ-10 coverage (determined using cyclic voltammograms at 1 V/s) as a function of UQ-10 added to the electrolyte from a DMSO stock solution.
(Bottom) (Closed symbols) Normalised activity of cbo3 as a function of UQ-10 coverage for three different experiments (squares, triangles and circles). The UQ-10 coverage (bottom axis) is determined by determination of the UQ-10 peak area from voltammograms measured at 1 V/s. (Open symbols) Comparison with previously published data[13] in which UQ-10 is cosolubilised in vesicles prior to tBLM formation and the UQ-10 coverage (top axis) is determined at 10 mV/s after inhibition of cbo3 with NaCN. The top and bottom axis have different scales due to observed differences in UQ-10 coverage when determined at 1 V/s and 10 mV/s (see text).
The reduction in current at low potential indicates that cbo3 is inhibited by UQ-10, a property we could only tentatively conclude previously due to scatter in the data[13], but is unambiguous from the in situ titrations with UQ-10. Finally, Figure 2a shows that the onset of the wave occurs at higher potential when the UQ-10 concentration is increased. This is not due to the DMSO, as no change in the CV is seen upon addition of DMSO alone. The reason for this shift is explained in the Discussion section.
Using fast-scan voltammetry, the relationship between the concentration of UQ-10 in the electrolyte and the concentration determined in the bilayer can be determined. At 1 V/s, the voltammogram outruns catalysis and even at low UQ-10 concentrations, the wave transforms to a peak corresponding to the UQ-10 reduction in the membrane. The area under the peak is used to determine the UQ-10 coverage and the relation between UQ-10 added to the electrolyte (in DMSO) and QU-10 incorporation in the membrane is shown in the insert of Figure 2a. This relation is approximately linear, although the slope varies between experiments. The quinone content in the membrane determined at 1 V/s is observed to be lower than that at 10 mV/s (after inhibition of the enzyme with NaCN). Approximately 50% of the UQ-10 signal is lost when the scan rate is increased from 10 mV/s to 1 V/s. Previously, we have reported that the diffusion of UQ-10 across the tethered membrane is slow with a ‘flip’ time between 0.05 and 1 s−1.[12] We thus assume that the loss of signal in the fast-scan voltammetry is due to the diffusion kinetics of the UQ-10 in the membrane.
The relative activity of cbo3 with different UQ-10 concentrations in a tBLM at −0.35 V vs SHE on the reverse scan of the CV is shown in Figure 2b (filled symbols). Correcting for the loss in apparent UQ-10 coverage between 10 mV/s and 1 V/s, the data fits alongside data we reported previously[13], in which UQ-10 was added to the lipid vesicles prior to tBLM formation. In this latter approach, the UQ-10 content of the membrane was determined at 10 mV/s after inhibiting the enzyme with NaCN.
Inhibition of cbo3
Besides the in situ titration of UQ-10, the interaction between UQ-10 and cbo3 has also been studied using inhibitors. Note that the cyclic voltammogram contains direct information about this interaction, as at each potential the enzyme activity (or current) is related to the quinol/quinone ratio, which, when using slow scan rates, is in equilibrium throughout the scan.
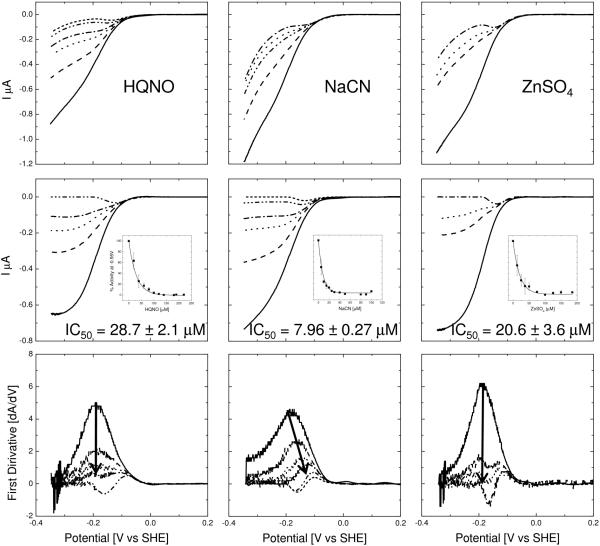
Cyanide is a well known and potent inhibitor of heme-copper oxidases. It binds to the binuclear centre in cbo3 and prevents the reduction of oxygen to water. Figure 3, middle, shows the forward part of the CVs collected after addition of NaCN. With increasing NaCN concentration the size of the catalytic UQ wave decreases in magnitude and then transforms to a peak shape. Closer examination reveals that the addition of NaCN reduces the maximum current without changing the onset of the wave. This results in a shift of the midpoint of the wave, which is more clearly visible in the first derivative shown at the bottom of Figure 3. This behaviour is identical (albeit reversed) to the behaviour we have previously indentified when activating anaerobic membrane samples with increasing oxygen substrate concentrations[13], consistent with the fact the cyanide inhibits oxygen reduction. At high concentrations (>50 μM) of NaCN, the enzyme is completely inhibited and no further reduction of the peak is observed. The IC50 of NaCN with cbo3 in the native-like tBLM is 7.96 ± 0.27 μM.
Figure 3.
(Top) Cyclic voltammograms for cytochrome bo3 inhibited with (NaCN) at 0μM, 5μM, 10μM, 20μM, 30μM and 90μM, (HQNO) at 0μM, 40μM, 60μM, 80μM and 190μM and (ZnSO4) at 0μM, 25μM, 50μM and 125μM. The QU-10 concentration in the membrane is between 5 and 10 pmol/cm2. The electrode area is 0.25 cm2 (Middle) Baseline corrected voltammograms of the data shown in Top. The baseline was taken from the cyclic voltrammogram measured at the highest concentration of inhibitor. (Middle, insert) Extracted data for IC50 determination. Error bars are standard deviations determined from at least three experiments. (Bottom) First derivatives of the data shown in Middle.
Cbo3 was also studied with the substrate analogue 2-n-Heptyl-4-hydroxyquinoline N-oxide (HQNO), which is a napthoquinone analogue of the semiquinone intermediate. As with the NaCN data, a transition is observed from wave to peak upon increasing HQNO concentration. However, in contrast to NaCN, the midpoints of the wave remain unaltered. In the baseline corrected voltammograms, an interesting double wave feature becomes apparent at high HQNO concentration, which is due to the superposition of the quinol reduction peak and the catalytic wave of reduced magnitude. As with NaCN we find that HQNO can completely and fully inhibit the enzyme. The IC50 for HQNO is 28.7 ± 2.1 μM.
Zinc is known to be an inhibitor of a wide variety of bioenergetic enzymes including cytochrome c oxidase[25, 26], mitochondrial complex I[27] and cbo3[3]. As with the HQNO, the voltammograms for partially inhibited cbo3 shows that the midpoint of the wave remains unaltered, ultimately resulting in the double wave feature. The IC50 for cbo3 with ZnSO4 is 20.6 ± 3.6 μM. We note that the inhibition with ZnSO4 showed some variability in the results, suggesting that the detailed structural properties of the tBLM influences the Zn(II) enzyme interaction.
Discussion
Substrate inhibition
Quinones are hydrophobic electron carriers and an essential component of respiratory chains in all organisms to mediate electron transfer between individual protein complexes. A significant problem in being able to fully understand and interpret the roles of quinones in biological processes is the lack of appropriate, physiologically relevant assays available. Currently, the most widespread methods are spectrophotometric assays using short, water soluble quinone analogues as long chain hydrophobic quinones can aggregate and form micelles which increase the turbidity of the assay solution and interfere with accurate measurements[28]. Another issue is the determination of accurate and absolute quinone concentrations partitioned into the cell membrane. By using model bilayers tethered to an electrode the activity of quinones with transmembrane enzymes can be easily and quickly monitored[29]. With this approach quinone concentrations within the bilayer may be rapidly and accurately determined using fast scan voltammetric methods and the quinone concentration can be changed in situ by the addition of ubiquinone solubilised in DMSO. The in situ UQ-10 addition data presented here corroborates our previously published data in that cbo3 is inhibited by UQ-10. The shape of the voltammogram at high UQ-10 content (apparent coverage of 23 pmol/cm2 at 1 V/s corresponds to ~46 pmol/cm2 at 10 mV/s, Figure 2a) shows a peak shape upon lowering the potential in which the area underneath the peak far exceeds the quinone content of membrane. This behaviour can be explained by assuming substrate inhibition, but not product inhibition. At high potential all UQ-10 is oxidised, thus not inhibiting cbo3. In the initial stages of the catalytic wave, some UQ-10 is reduced and substrate generated, but not yet to an extent that the enzyme is inhibited. Upon reducing the potential further, more substrate is generated, inhibiting the enzyme and thereby reducing the current, resulting in a peak shaped voltammogram. Besides the catalytic wave, this peak shape is further enhanced by the large amount of UQ-10 in the membrane as this induces a UQ-10 reduction peak in itself. Modelling studies shown in the supporting information support this hypothesis. When both the product and the substrate inhibits cbo3, the modelling shows the expected behaviour that the area underneath the peak is equal or less than the UQ-10 coverage, in contrast with our data in which the area is larger than the UQ-10 coverage. However, when only substrate inhibits cbo3 the model indicates that the peak area can exceed that of the UQ-10 coverage.
The modelling studies also confirm that by increasing the UQ-10 in the tBLM, the onset of the catalytic wave shifts to more positive potentials. The reason for this is complex, but can approximately be explained by noting that at a given potential, a similar fraction of UQ-10 is in its reduced state. At higher coverage of UQ-10, this will lead to larger substrate concentration and thus a higher turn-over of the enzyme.
Substrate inhibition (without product inhibition) has been observed previously by Musser et al. [30] for cbo3 using water-soluble quinone analogues, UQ-1 and UQ-2. Musser et al. could not excluded that the observed kinetic properties were due to the high concentrations of UQ-1 and UQ-2 used in the essays, which resulted in non-ideal solution conditions. Although the data presented here confirm the substrate inhibition, it can still not be excluded that the substrate inhibition is due to some non-ideal behaviour of the quinones in the membrane such as aggregation of ubiquinone or disruption of the lipid bilayer. However, no indication of non-ideal behaviour was observed and the more likely explanation is that the enzyme is subjected to substrate inhibition. It is possible that a second ubiquinol binds close or at the active site (QL) during turn-over, disrupting the catalytic reaction at this site. However, the fact that the inhibition is specific to ubiquinol (and not ubiquinone) makes this less likely. An alternative model was previously suggested; at high concentrations of ubiquinol, the quinone in high affinity ubiquinone site, QH, is replaced by an ubiquinol, making it impossible for the QH site to be reduced to the semiquinone intermediate that is required during turnover.[30, 31] The location of the QH site close to the surface of the enzyme[5, 8] suggest it could be possible that the ubiquinone is replaced. However, the quinone is known to be tightly bound and not easily removed from the QH site, making it less likely that replacement can occur at a fast rate. An alternatively explanation is that at high ubiquinol concentration in the membrane, ubiquinol binds close to the QH site in an a-specific manner such that the ubiquinone in the QH site is reduced to a ubiquinol. In this model, the inhibition would also depend on the redox state of the ubiquinone pool or, in other words, inhibition would only take place when most of the ubiquinol is in the reduced state.
Inhibitor studies
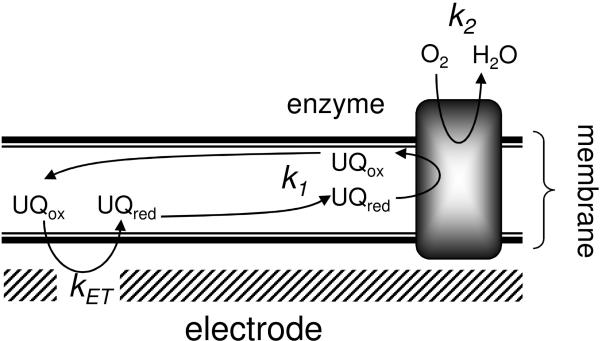
Two distinct voltammetric behaviours are observed depending on the type of compound used to inhibit cbo3. We hypothesise this is due to the inhibition of the two active sites in cbo3; the quinol oxidation site and the oxygen reducing site (with rates k1 and k2, respectively, see Figure 4). NaCN, which induces a shift in the wave’s midpoints (Figure 3), binds to the binuclear centre in the active site, thereby directly reducing the oxygen reduction reaction (rate k2 in Figure 4). The other inhibitors, HQNO and ZnSO4, have a distinct response and do not cause a shift in the midpoint of the catalytic wave.
Figure 4.
Schematic representation of the tBLM and the rates that influence the currents measured with cyclic voltammetry.
The effect of these inhibitors on the voltammograms can be explained by taking into account that the total current is dependent on both k1 and k2 with the slowest reaction being the rate limiting step. The second important feature is that the rate k1 is dependent on the redox state of the ubiquinone pool, which in turn is dependent on the applied potential. Thus, at high potential (the onset of the wave), little ubiquinol is reduced and thus k1 is low and rate limiting. At low potential (the plateau region), all ubiquinol is reduced and the activity is either limited by k1 or k2. Cyanide inhibits the oxygen reducing site and thus lowers the rate k2. As a result, by increasing the cyanide concentration, k2 will become rate limiting. The lower the rate of k2, the higher the potential at which it becomes rate limiting (as the rate k1 is dependent on potential). Increasing cyanide thus changes the shape as well as the magnitude of the wave.
In contrast, HQNO reduces the rate k1 and if this is done in a truly competitive manner, rate k1 will still be dependent on the oxidation state of the quinone pool and thus the wave shape will remain unaltered although the magnitude decreases. HQNO is indeed thought to inhibit cbo3 competitively by binding at the QL site in the enzyme[32, 33] although kinetic analysis suggests that HQNO may actually be an uncompetitive[30] or non competitive[34] inhibitor, possibly also interacting with the QH site. The IC50 value (~21 μM) determined here is considerably higher than previously determined values (0.3 μM) [18]. We propose that this difference is due to the different assay conditions used, changing the nature of the quinol substrate and ‘effective’ quinol and inhibitor concentration in the lipid environment. Although during the cyclic voltammogram, the concentration of substrate varies, differentiating between non competitive and uncompetitive inhibition based on the cyclic voltammetry would only be possible if the quinol oxidation is irreversible (if the reduction potential of cbo3 is much lower than UQ-10). In contrast, the fact that the midpoint of the catalytic wave remains unaltered upon addition of HQNO can only be explained if cbo3 is in redox equilibrium with the quinol/quinone pool in the membrane and the reduction of cbo3 is not significantly different from that of UQ-10. If this was not case, the onset of the catalytic wave would remain unchanged upon addition of HQNO as in this situation the enzyme activity during the scan would only be limited by the ubiquinol concentration and not influenced by ubiquinone (or the ratio of ubiquinol/ubiquinone). Modelling studies (Supporting Information) indeed confirm that the data can only be simulated if a model is assumed in which cbo3 is in redox equilibrium with the UQ-10 pool.
Zinc and other selective divalent cations, like cadmium, nickel and cobalt[35, 36] have been shown to inhibit heme copper oxidases such as cbo3 and cytochrome c oxidase. At low concentration, the cations are thought to inhibit the enzymes by blocking the release of protons[25, 36]. The voltammograms after inhibition with HQNO and ZnSO4 showed a very similar behaviour contrasting that of NaCN. Especially at the intermediate inhibitor concentrations, both inhibitors induce a double wave effect in the voltammogram. These results thus confirm that Zn(II) inhibition is not affecting proton uptake to supply protons for oxygen reduction, but mainly inhibits enzyme reduction by quinol or the coupled release of protons (from quinol oxidation). This latter observation might be influenced by the enzyme’s orientation on the surface. If the enzyme is oriented with the N-side towards the surface, Zn(II) ions can only bind to the P-side, corresponding to the impairment of proton release as shown for cytochrome c oxidase.[25]
Supplementary Material
Acknowledgement
E .coli strain GO105/pJRhisA was kindly provided by Dr. R. B. Gennis (Univ. Illinois). We also thank Moazur Rahman for help with using the cell disruptor and Stefanie Jourdan for help with the French pressure cell. The production of membranes was assisted by equipment funded by the Welcome Trust and technical help funded by the European membrane consortium (EMeP). Finally, this work was funded by a BBSRC grant (BB/D008131/1).
Footnotes
This document is the Accepted Manuscript version of a Published Work that appeared in the final form in the BBA - Bioenergetics, copyright Elsevier after peer review and technical editing by the publisher. To access the final edited and published work see http://dx.doi.org/10.1016/j.bbabio.2010.01.012.
References
- [1].Anraku Y. Bacterial Electron-Transport Chains. Annu. Rev. Biochem. 1988;57:101–132. doi: 10.1146/annurev.bi.57.070188.000533. [DOI] [PubMed] [Google Scholar]
- [2].Soballe B, Poole RK. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology-(UK) 1999;145:1817–1830. doi: 10.1099/13500872-145-8-1817. [DOI] [PubMed] [Google Scholar]
- [3].Kita K, Konishi K, Anraku Y. Terminal oxidases of Escherichia coli aerobic respiratory chain .1. Purification and properties of cytochrome b562o complex from cells in the early exponential phase of aerobic growth. J. Biol. Chem. 1984;259:3368–3374. [PubMed] [Google Scholar]
- [4].Kita K, Konishi K, Anraku Y. Terminal oxidases of Escherichia coli aerobic respiratory chain .2. Purification and properties of cytochrome b558d complex from cells grown with limited oxygen and evidence of branched electron-carrying systems. J. Biol. Chem. 1984;259:3375–3381. [PubMed] [Google Scholar]
- [5].Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikstrom M. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat. Struct. Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- [6].Ek MS, Brzezinski P. Oxidation of ubiquinol by cytochrome bo3 from Escherichia coli: Kinetics of electron and proton transfer. Biochemistry. 1997;36:5425–5431. doi: 10.1021/bi962478e. [DOI] [PubMed] [Google Scholar]
- [7].Sato-Watanabe M, Mogi T, Ogura T, Kitagawa T, Miyoshi H, Iwamura H, Anraku Y. Identification of a novel quinone-binding site in the cytochrome bo complex from Escherichia coli. J. Biol. Chem. 1994;269:28908–28912. [PubMed] [Google Scholar]
- [8].Lin MT, Samoilova RI, Gennis RB, Dikanov SA. Identification of the Nitrogen Donor Hydrogen Bonded with the Semiquinone at the QH Site of the Cytochrome bo3 from Escherichia coli. J. Am. Chem. Soc. 2008;130:15768. doi: 10.1021/ja805906a. + [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yap LL, Samoilova RI, Gennis RB, Dikanov SA. Characterization of mutants that change the hydrogen bonding of the semiquinone radical at the QH site of the cytochrome bo3 from Escherichia coli. J. Biol. Chem. 2007;282:8777–8785. doi: 10.1074/jbc.M611595200. [DOI] [PubMed] [Google Scholar]
- [10].Yap LL, Samoilova RI, Gennis RB, Dikanov SA. Characterization of the exchangeable protons in the immediate vicinity of the semiquinone radical at the QH site of the cytochrome bo3 from Escherichia coli. J. Biol. Chem. 2006;281:16879–16887. doi: 10.1074/jbc.M602544200. [DOI] [PubMed] [Google Scholar]
- [11].Fato R, Castelluccio C, Palmer G, Lenaz G. A simple method for the determination of the kinetic constants of membrane enzymes utilizing hydrophobic substrates - Ubiquinol cytochrome c reductase. Biochimica Et Biophysica Acta. 1988;932:216–222. doi: 10.1016/0005-2728(88)90158-2. [DOI] [PubMed] [Google Scholar]
- [12].Jeuken LJC, Weiss SA, Henderson PJF, Evans SD, Bushby RJ. Impedance spectroscopy of bacterial membranes: Coenzyme-Q diffusion in a finite diffusion layer. Anal. Chem. 2008;80:9084–9090. doi: 10.1021/ac8015856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Weiss SA, Bushby RJ, Evans SD, Henderson PJF, Jeuken LJC. Characterization of cytochrome bo3 activity in a native-like surface-tethered membrane. Biochem. J. 2009;417:555–560. doi: 10.1042/BJ20081345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Janshoff A, Steinem C. Transport across artificial membranes - an analytical perspective. Anal. Bioanal. Chem. 2006;385:433–451. doi: 10.1007/s00216-006-0305-9. [DOI] [PubMed] [Google Scholar]
- [15].Jeuken LJC. Electrodes for integral membrane enzymes. Natural Product Reports. 2009;26:1234–1240. doi: 10.1039/b903252e. [DOI] [PubMed] [Google Scholar]
- [16].Knoll W, Naumann R, Friedrich M, Robertson JWF, Losche M, Heinrich F, McGillivray DJ, Schuster B, Gufler PC, Pum D, Sleytr UB. Solid supported lipid membranes: New concepts for the biomimetic functionalization of solid surfaces. Biointerphases. 2008;3:FA125–FA135. doi: 10.1116/1.2913612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jeuken LJC, Daskalakis NN, Han XJ, Sheikh K, Erbe A, Bushby RJ, Evans SD. Phase separation in mixed self-assembled monolayers and its effect on biomimetic membranes. Sensors And Actuators B-Chemical. 2007;124:501–509. [Google Scholar]
- [18].Jeuken LJC, Connell SD, Henderson PJF, Gennis RB, Evans SD, Bushby RJ. Redox enzymes in tethered membranes. J. Am. Chem. Soc. 2006;128:1711–1716. doi: 10.1021/ja056972u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Weiss SA, Jeuken LJC. Electrodes modified with lipid membranes to study quinone oxidoreductases. Biochem. Soc. Trans. 2009;37:707–712. doi: 10.1042/BST0370707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Boden N, Bushby RJ, Clarkson S, Evans SD, Knowles PF, Marsh A. The design and synthesis of simple molecular tethers for binding biomembranes to a gold surface. Tetrahedron. 1997;53:10939–10952. [Google Scholar]
- [21].Rumbley JN, Nickels EF, Gennis RB. One-step purification of histidine-tagged cytochrome bo3 from Escherichia coli and demonstration that associated quinone is not required for the structural integrity of the oxidase. Biochim. Biophys. Acta-Protein Struct. Molec. Enzym. 1997;1340:131–142. doi: 10.1016/s0167-4838(97)00036-8. [DOI] [PubMed] [Google Scholar]
- [22].Schaffner W, Weissman C. Rapid, sensitive, and specific method for determination of protein in dilute solution. Analytical Biochemistry. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- [23].Jeuken LJC, Bushby RJ, Evans SD. Proton transport into a tethered bilayer lipid membrane. Electrochemistry Communications. 2007;9:610–614. [Google Scholar]
- [24].Gurtovenko AA, Anwar J. Modulating the structure and properties of cell membranes: The molecular mechanism of action of dimethyl sulfoxide. J. Phys. Chem. B. 2007;111:10453–10460. doi: 10.1021/jp073113e. [DOI] [PubMed] [Google Scholar]
- [25].Faxen K, Salomonsson L, Adelroth P, Brzezinski P. Inhibition of proton pumping by zinc ions during specific reaction steps in cytochrome c oxidase. Biochim. Biophys. Acta-Bioenerg. 2006;1757:388–394. doi: 10.1016/j.bbabio.2006.05.010. [DOI] [PubMed] [Google Scholar]
- [26].Qin L, Mills DA, Hiser C, Murphree A, Garavito RM, Ferguson-Miller S, Hosler J. Crystallographic location and mutational analysis of Zn and Cd inhibitory sites and role of lipidic carboxylates in rescuing proton path mutants in cytochrome c oxidase. Biochemistry. 2007;46:6239–6248. doi: 10.1021/bi700173w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sharpley MS, Hirst J. The inhibition of mitochondrial complex I (NADH : ubiquinone oxidoreductase) by Zn2+ J. Biol. Chem. 2006;281:34803–34809. doi: 10.1074/jbc.M607389200. [DOI] [PubMed] [Google Scholar]
- [28].Lenaz G, Degli Esposti M. Physical properties of ubiquinones in model systems and membranes. In: Lenaz G, editor. Coenzyme Q Biochemistry, Bioenergetics and Clinical Applications of Ubiquinone. Wiley; 1985. [Google Scholar]
- [29].Marchal D, Pantigny J, Laval JM, Moiroux J, Bourdillon C. Rate constants in two dimensions of electron transfer between pyruvate oxidase, a membrane enzyme, and ubiquinone (coenzyme Q(8)), its water-insoluble electron carrier. Biochemistry. 2001;40:1248–1256. doi: 10.1021/bi002325y. [DOI] [PubMed] [Google Scholar]
- [30].Musser SM, Stowell MHB, Lee HK, Rumbley JN, Chan SI. Uncompetitive substrate inhibition and noncompetitive inhibition by 5-n-undecyl-6-hydroxy-4,7-dioxobenzothiazole (UHDBT) and 2-n-Nonyl-4-hydroxyquinoline-N-oxide (NQNO) is observed for the cytochrome bo3 complex: Implications for a Q(H-2)-loop proton translocation mechanism. Biochemistry. 1997;36:894–902. doi: 10.1021/bi961723r. [DOI] [PubMed] [Google Scholar]
- [31].Kobayashi K, Tagawa S, Mogi T. Transient formation of ubisemiquinone radical and subsequent electron transfer process in the Escherichia coli cytochrome bo. Biochemistry. 2000;39:15620–15625. doi: 10.1021/bi0014094. [DOI] [PubMed] [Google Scholar]
- [32].Welter R, Gu LQ, Yu CA, Rumbley J, Gennis RB. Identification of the substrate-binding site in Escherichia coli cytochrome bo ubiquinol oxidase. Biophys. J. 1994;66:A367–A367. [PubMed] [Google Scholar]
- [33].Meunier B, Madgwick SA, Reil E, Oettmeier W, Rich PR. New inhibitors of the quinol oxidation sites of bacterial cytochromes bo and bd. Biochemistry. 1995;34:1076–1083. doi: 10.1021/bi00003a044. [DOI] [PubMed] [Google Scholar]
- [34].Sato-Watanabe M, Mogi T, Miyoshi H, Iwamura H, Matsushita K, Adachi O, Anraku Y. Structure-function studies on the ubiquinol oxidation site of the cytochrome bo complex from Escherichia coli using p-benzoquinones and substituted phenols. J. Biol. Chem. 1994;269:28899–28907. [PubMed] [Google Scholar]
- [35].Kita K, Konishi K, Anraku Y. Purification and properties of 2 terminal oxidase complexes of Escherichia coli aerobic respiratory chain. Methods In Enzymology. 1986;126:94–113. doi: 10.1016/s0076-6879(86)26012-7. [DOI] [PubMed] [Google Scholar]
- [36].Mills DA, Schmidt B, Hiser C, Westley E, Ferguson-Miller S. Membrane potential-controlled inhibition of cytochrome c oxidase by zinc. J. Biol. Chem. 2002;277:14894–14901. doi: 10.1074/jbc.M111922200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.