Abstract
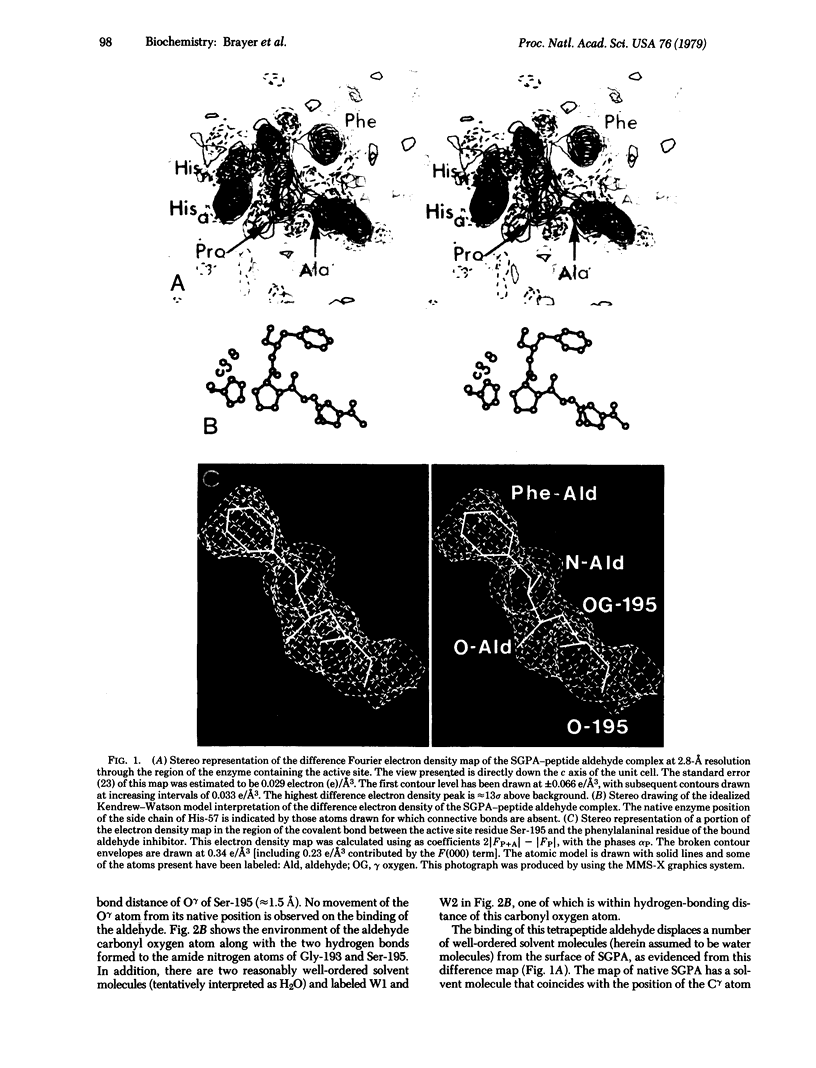
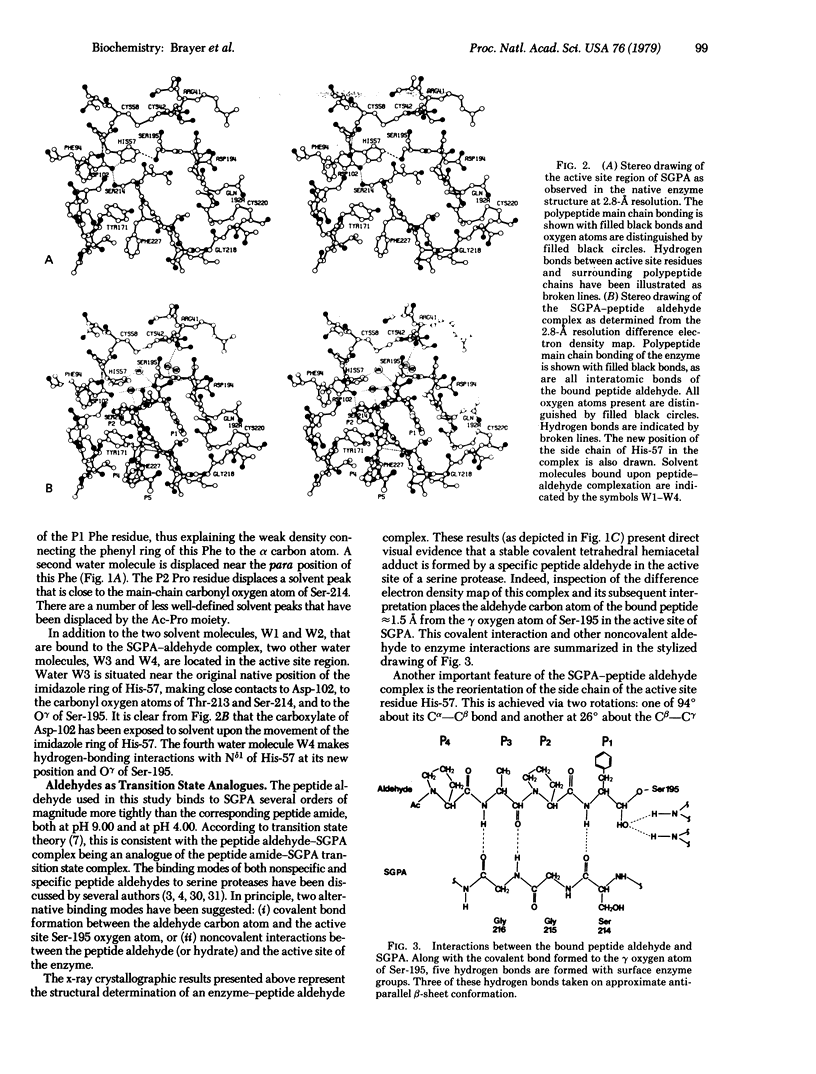
X-ray crystallographic data show that a specific tetrapeptide aldehyde inhibitor (N-acetylprolylalanylprolylphenylalaninal) forms a stable, covalent, tetrahedral addition complex with the serine protease, SGPA, from Streptomyces griseus. Earlier proposals, based on kinetic measurements, for the covalent nature of such linkages are confirmed, and the difference electron density map of this aldehyde inhibitor indicates that a major conformational change of the histidyl-57 side chain occurs on inhibitor binding.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer C. A., Löfqvist B. Studies of the heterogeneity of Streptomyces griseus protease. Isolation and characterization of an alkaline serine protease from commercial pronase-P derived from Streptomyces griseus K1. Acta Chem Scand. 1973;27(9):3147–3166. doi: 10.3891/acta.chem.scand.27-3147. [DOI] [PubMed] [Google Scholar]
- Bauer C. A., Pettersson G. Effect of pH on the catalytic activity of Streptomyces griseus protease 3. Eur J Biochem. 1974 Jun 15;45(2):469–472. doi: 10.1111/j.1432-1033.1974.tb03571.x. [DOI] [PubMed] [Google Scholar]
- Bauer C. A., Thompson R. C., Blout E. R. The active centers of Streptomyces griseus protease 3 and alpha-chymotrypsin: enzyme-substrate interactions remote from the scissile bond. Biochemistry. 1976 Mar 23;15(6):1291–1295. doi: 10.1021/bi00651a019. [DOI] [PubMed] [Google Scholar]
- Brayer G. D., Delbaere L. T., James M. N. Molecular structure of crystalline Streptomyces griseus protease A at 2.8 A resolution. I. Crystallization, data collection and structural analysis. J Mol Biol. 1978 Sep 5;124(1):243–259. doi: 10.1016/0022-2836(78)90158-4. [DOI] [PubMed] [Google Scholar]
- Brayer G. D., Delbaere L. T., James M. N. Molecular structure of crystalline Streptomyces griseus protease A at 2.8 A resolution. II. Molecular conformation, comparison with alpha-chymotrypsin and active-site geometry. J Mol Biol. 1978 Sep 5;124(1):261–283. doi: 10.1016/0022-2836(78)90159-6. [DOI] [PubMed] [Google Scholar]
- Breaux E. J., Bender M. L. The binding of specific and non-specific aldehyde substrate analogs to alpha-chymotrypsin. FEBS Lett. 1975 Aug 1;56(1):81–84. doi: 10.1016/0014-5793(75)80116-5. [DOI] [PubMed] [Google Scholar]
- Codding P. W., Delbaere L. T., Hayakawa K., Hutcheon W. L., James M. N., Jurásek L. The 4.5 Angstrom resolution structure of a bacterial serine protease from Streptomyces griseus. Can J Biochem. 1974 Mar;52(3):208–220. doi: 10.1139/o74-034. [DOI] [PubMed] [Google Scholar]
- DIXON M. The determination of enzyme inhibitor constants. Biochem J. 1953 Aug;55(1):170–171. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbaere L. T., Hutcheon W. L., James M. N., Thiessen W. E. Tertiary structural differences between microbial serine proteases and pancreatic serine enzymes. Nature. 1975 Oct 30;257(5529):758–763. doi: 10.1038/257758a0. [DOI] [PubMed] [Google Scholar]
- Diamond R. Real-space refinement of the structure of hen egg-white lysozyme. J Mol Biol. 1974 Jan 25;82(3):371–391. doi: 10.1016/0022-2836(74)90598-1. [DOI] [PubMed] [Google Scholar]
- Gertler A. Inhibition of Streptomyces griseus protease B by peptide chloromethyl ketones: partial mapping of the binding site and identification of the reactive residue. FEBS Lett. 1974 Jul 1;43(1):81–85. doi: 10.1016/0014-5793(74)81110-5. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G., Kar D., Momii R. K. NMR evidence against covalent attachment of an aldehyde 'transition-state' analogue to alpha-chymotrypsin. Biochem Biophys Res Commun. 1976 Nov 8;73(1):105–111. doi: 10.1016/0006-291x(76)90503-9. [DOI] [PubMed] [Google Scholar]
- Henderson P. J. A linear equation that describes the steady-state kinetics of enzymes and subcellular particles interacting with tightly bound inhibitors. Biochem J. 1972 Apr;127(2):321–333. doi: 10.1042/bj1270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A., Tokawa K., Shimizu B. Peptide aldehydes inhibiting chymotrypsin. Biochem Biophys Res Commun. 1972 Oct 17;49(2):343–349. doi: 10.1016/0006-291x(72)90416-0. [DOI] [PubMed] [Google Scholar]
- James M. N., Delbaere L. T., Brayer G. D. Amino acid sequence alignment of bacterial and mammalian pancreatic serine proteases based on topological equivalences. Can J Biochem. 1978 Jun;56(6):396–402. doi: 10.1139/o78-062. [DOI] [PubMed] [Google Scholar]
- Johnson P., Smillie L. B. The amino acid sequence and predicted structure of Streptomyces griseus protease A. FEBS Lett. 1974 Oct 1;47(1):1–6. doi: 10.1016/0014-5793(74)80412-6. [DOI] [PubMed] [Google Scholar]
- Jurásek J., Johnson P., Olafson R. W., Smillie L. B. An improved fractionation system for pronase on CM-sephadex. Can J Biochem. 1971 Nov;49(11):1195–1201. doi: 10.1139/o71-171. [DOI] [PubMed] [Google Scholar]
- KURTZ A. N., NIEMANN C. Use of the pH-stat in kinetic studies of reactions whose products are capable of functioning as buffers. Biochemistry. 1962 Mar;1:238–243. doi: 10.1021/bi00908a007. [DOI] [PubMed] [Google Scholar]
- Lienhard G. E. Enzymatic catalysis and transition-state theory. Science. 1973 Apr 15;180(4082):149–154. doi: 10.1126/science.180.4082.149. [DOI] [PubMed] [Google Scholar]
- Poulos T. L., Alden R. A., Freer S. T., Birktoft J. J., Kraut J. Polypeptide halomethyl ketones bind to serine proteases as analogs of the tetrahedral intermediate. X-ray crystallographic comparison of lysine- and phenylalanine-polypeptide chloromethyl ketone-inhibited subtilisin. J Biol Chem. 1976 Feb 25;251(4):1097–1103. [PubMed] [Google Scholar]
- Richards F. M. The matching of physical models to three-dimensional electron-density maps: a simple optical device. J Mol Biol. 1968 Oct 14;37(1):225–230. doi: 10.1016/0022-2836(68)90085-5. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Alden R. A., Birktoft J. J., Kraut J., Powers J. C., Wilcox P. E. An x-ray crystallographic study of the binding of peptide chloromethyl ketone inhibitors to subtilisin BPN'. Biochemistry. 1972 Jun 20;11(13):2439–2449. doi: 10.1021/bi00763a009. [DOI] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967 Apr 20;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Schultz R. M., Cheerva A. C. The binding of a non-specific "transition state analogue" to alpha-chymotrypsin. FEBS Lett. 1975 Jan 15;50(1):47–49. doi: 10.1016/0014-5793(75)81037-4. [DOI] [PubMed] [Google Scholar]
- Segal D. M., Powers J. C., Cohen G. H., Davies D. R., Wilcox P. E. Substrate binding site in bovine chymotrypsin A-gamma. A crystallographic study using peptide chloromethyl ketones as site-specific inhibitors. Biochemistry. 1971 Sep 28;10(20):3728–3738. doi: 10.1021/bi00796a014. [DOI] [PubMed] [Google Scholar]
- Tatsuta K., Mikami N., Fujimoto K., Umezawa S., Umezawa H. The structure of chymostatin, a chymotrypsin inhibitor. J Antibiot (Tokyo) 1973 Nov;26(11):625–646. doi: 10.7164/antibiotics.26.625. [DOI] [PubMed] [Google Scholar]
- Thompson R. C. Peptide aldehydes: potent inhibitors of serine and cysteine proteases. Methods Enzymol. 1977;46:220–225. doi: 10.1016/s0076-6879(77)46023-3. [DOI] [PubMed] [Google Scholar]
- Thompson R. C. Use of peptide aldehydes to generate transition-state analogs of elastase. Biochemistry. 1973 Jan 2;12(1):47–51. doi: 10.1021/bi00725a009. [DOI] [PubMed] [Google Scholar]
- Umezawa H. Structures and activities of protease inhibitors of microbial origin. Methods Enzymol. 1976;45:678–695. doi: 10.1016/s0076-6879(76)45058-9. [DOI] [PubMed] [Google Scholar]
- Westerik J. O., Wolfenden R. Aldehydes as inhibitors of papain. J Biol Chem. 1972 Dec 25;247(24):8195–8197. [PubMed] [Google Scholar]
- Wolfenden R. Transition state analog inhibitors and enzyme catalysis. Annu Rev Biophys Bioeng. 1976;5:271–306. doi: 10.1146/annurev.bb.05.060176.001415. [DOI] [PubMed] [Google Scholar]