Abstract
Pancreatic ductal adenocarcinoma (PDAC) is characterized by an abundant desmoplastic stroma. Interactions between cancer and stromal cells play a critical role in tumour invasion, metastasis and chemoresistance. Therefore, we hypothesized that gene expression profile of the stromal components of pancreatic carcinoma is different from chronic pancreatitis and reflects the interaction with the tumour. We investigated the gene expression of eleven stromal tissues from PDAC, nine from chronic pancreatitis and cell lines of stromal origin using the Affymetrix U133 GeneChip set. The tissue samples were microdissected, the RNA was extracted, amplified and labelled using a repetitive in vitro transcription protocol. Differentially expressed genes were identified and validated using quantitative RT-PCR and immuno-histochemistry. We found 255 genes to be overexpressed and 61 genes to be underexpressed within the stroma of pancreatic carcinoma compared to the stroma of chronic pancreatitis. Analysis of the involved signal transduction pathways revealed a number of genes associated with the Wnt pathway of which the differential expression of SFRP1 and WNT5a was confirmed using immunohistochemistry. Moreover, we could demonstrate that WNT5a expression was induced in fibroblasts during cocultivation with a pancreatic carcinoma cell line. The identified differences in the expression profile of stroma cells derived from tumour compared to cells of inflammatory origin suggest a specific response of the tissue surrounding malignant cells. The overexpression of WNT5a, a gene involved in the non canonical Wnt signalling and chondrocyte development might contribute to the strong desmoplastic reaction seen in pancreatic cancer.
Keywords: pancreatic ductal adenocarcinoma, chronic pancreatitis, expression profiling, microarray, stroma, microdissection, WNT5a
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the leading causes of cancer-related death today. In the United States it ranks fourth, accounting for approximately 30,000 deaths per year [1]. Apart from surgery there is no effective therapy, and even most patients who had undergone tumour resection die within the first 3 years after surgery [2]. The main reason for this poor prognosis is local recurrence and/or the occurrence of distant metastases. Whereas the genetic and epigenetic changes of tumour epithelia in general have been investigated in great detail in the last decades [3], the contribution of tumour stroma during the natural history is still incomplete [4]. Stromal cells might revert a tumourigenic phenotype [5], but participation of stromal tissue during metastatic processes has also been described [6]. The stromal compartment of a tumour might also harbour genetic changes indicating a coevolutionary process of tumour epithelium and stroma compartment [7]. In PDAC, it has been demonstrated that radiation of stromal fibroblasts increase the invasiveness of PDAC [8, 9]. Several mediators of stromal epithelial interaction have been described [10], of which TGF-β takes part in the development of fibrotic changes within the stroma of pancreatic tissue [11]. Only recently the relevance of the Wnt signalling pathway in mediating stromal–epithelial interactions of tumours has been described [12]. However, in contrast to many other solid tumours the canonical Wnt signal transduction pathway is rarely activated in PDAC, since translocation of β-catenin to the nucleus is seldom observed [13, 14]. Recently, various research groups have applied DNA-microarray technology to identify differentially expressed genes in PDAC for new diagnostic and therapeutic approaches [15–20]. Most of these studies were performed on whole tissue samples which contain different cell types and focused on the gene expression differences of the epithelial compartment of the tumour. Those expression profiles cannot be attributed to one cell type and thus, microdissection is the method of choice to generate a more precise picture of gene expression changes in a specific cell type. We have established microdissection and microarray-based gene expression profiling in PDAC epithelia cells [21], and we have now compared now gene expression profiles from tumour-associated stromal tissue with expression profiles from benign stromal tissue using the Affymetrix U133 GeneChip set. Since there are only very few stromal cells adjacent to normal pancreatic ducts, we analysed stromal cells from the fibrotic tissue of chronic pancreatitis a benign tissue remodelling process of the pancreas [22]. We identified genes whose expression levels differed significantly between stroma from PDAC and from chronic pancreatitis. Within this set of genes, we detected an over-representation of genes encoding Wnt signalling molecules within the PDAC stroma and we were able to validate the overexpression of WNT5a suggesting that the non-canonical Wnt signalling cascade is activated.
Material and methods
Patients and tissues
Freshly frozen tissue samples of PDACs (n = 11) were obtained from surgical specimens from patients who underwent surgery at the Department of Visceral-, Thoracic- and Vascular-Surgery, University Hospital Carl Gustav Carus, Technical University of Dresden and the Department of General Surgery, University of Kiel, Germany, between 1996 and 2003. Chronic pancreatitis specimens (n = 9) were obtained from patients who underwent pancreatic resection. Prior to surgery, all patients had signed an informed consent form that had been approved by the local ethics committee. Immediately after surgical removal the specimens were sectioned and microscopically evaluated. Suitable samples of tumour tissue or normal tissue were snap frozen in liquid nitrogen and stored at −80°C until further processing.
Microdissection
Microdissection was performed in the laboratory of the Institute of Pathology, University Schleswig-Holstein Campus Kiel, Kiel, Germany as already described [21]. In brief, frozen tissue specimens were cut into 10 μm sections and slides were immediately fixed in 70% ethanol. The sections were briefly stained with haematoxylin and eosin and coverslipped. Suitable areas for microdissection without visible neoplastic cell contamination were marked on these slides and served as a template for microdissection. The estimated cellularity was approx. 10,000–11,000 cells per microdissected sample and the purity of the dissections was approx. 90%. For comparison, we included the gene expression data from our previous study of microdissected tumour epithelia [21] and seven additional epithelia samples (two normal, five PDAC).
Cell culture
The cell lines CAPAN2, BxPC3, CAPAN1, ASPC1, COLO357, MiaPaCa2, Panc1, Panc89, PT45, PancTUI, Kif5, F13 [23] and immortalized primary stellate cells [24] were cultured in RPMI 1640 supplemented with 10% foetal bovine serum, 2 mM glutamine, non-essential amino acids (5 ml/l), penicillin (10,000 units/ml), and streptomycin (10 mg/ml) and passaged before they reached confluency. All cell culture materials were obtained from Invitrogen, Karlsruhe, Germany. 1.5 × 105 Panc89 and 1.5 × 105 Kif5 cells (human mesenchymal cells) were cocultured and separated by a 0.2 μm membrane (Becton Dickinson Labware, NJ, USA). After reaching confluence (48 hrs), the cells were trypsinized and transferred to new vessels to coculture cells for 60 additional hrs. Following trypsinization of the cells RNA isolation was performed using a Micro-to-Midi Total RNA Purification System (Invitrogen, Paisley, UK) according to the manufacturer's instruction.
RNA preparation and array hybridization
RNA preparation and array hybridization was performed as described earlier [21]. In brief, Poly A†-RNA from the microdissected surgical specimens and cell cultures was prepared using a PolyATtract 1000 kit (Promega, Heidelberg, Germany). For each sample the cDNA synthesis and repetitive in vitro transcription were performed three times. First-strand cDNA synthesis was initiated using the Affymetrix T7-oligo-dT promotor-primer combination at 0.1 mM. After second-strand cDNA synthesis the in vitro transcription was performed using Ambion's Megascript kit (Ambion, Huntington, UK), as recommended by the manufacturer. From the generated aRNA a new first-strand synthesis was initiated using 0.025 mM of a random hexamer as primer. After completion, the second-strand synthesis was performed using the Affymetrix T7-oligo-dT promotor-primer combination as primer at a concentration of 0.1 mM. A second in vitro transcription was performed and then the procedure was repeated one additional time. During the last in vitro transcription, biotin-labelled nucleotides were incorporated into the aRNA, as recommended by the Affymetrix protocol. Hybridization and detection of the labelled aRNA on the U133 A/B Affymetrix GeneChip set (Affymetrix, Santa Clara, CA, USA) were performed according to Affymetrix instructions.
Gene expression analysis
The U133 A/B Affymetrix GeneChip set utilized in this study consists of more than 44,000 probe sets. The Cel Files obtained from the Affymetrix MAS 5.0 software were loaded into dChip2006 (www.dchip.org), normalized and expression values were calculated using the PM/MM model [25]. To minimize the noise within the gene expression data set we used only the probe sets that displayed an intensity value of greater than 120 in more than 15% of the chips analysed and in which average intensity were below 4000. The cut-off of 120 for the intensity value was derived from the intensity values from the bacterial control probe sets for the Bacillus subtilis genes dapB, lys, pheA, thrC and trpE within the data set. Only 1% of those probe sets revealed intensity values above 120, thus limiting the probability of false positives due to random fluctuation of the background intensities. The expression values were exported and further explored using SAM (http://www-stat.stanford.edu/∼tibs/SAM/) [26] and Excel (Microsoft, Seattle, WA, USA). We scored genes as differentially expressed if they met the following criteria: a fold change >2 and a q-value ≤5%. Identification of probe sets expression overexpressed only in pancreatic tumour stroma was done using dChip (cut-off: fold change >4 and P-value < 0.05) by comparing the expression of tumour stroma samples (group 1) with chronic pan-creatitis stroma, tumour epithelia and normal epithelia samples (group 2).
Quantitative RT-PCR
The aRNA from the second amplification cycle, as stated above, was reversely transcribed into cDNA. 1 ng of cDNA was used for a TaqMan assay (Applied Biosystems, Weiterstadt, Germany). The genes were amplified with the TaqMan Universal PCR Master Mix according to the manufacturer's instructions, with an ABI PRISM 5700 Sequence Detection System using gene specific primers and probes. Gene expression was quantified by the comparative cT-Method, normalizing cT-values to a housekeeping gene (β-actin or G6PDH) and calculating the relative expression values [27] using the following primers: G6PDH forward: 5′-acgtgatgcagaaccac-ctactg; G6PDH reverse: 5′- acgacggctgcaaaagtggcg-3′; SFRP1 forward: 5′-gatgcaggaggctcaggtgat-3′; SFRP1 reverse: 5′-tgtcctgtgtatctgctggcaac-3′; WNT5a forward: 5′-taggcacgaaagcacaggtc-3′; WNT5a reverse: 5′-cacggcatctctctttcacca-3′; CXCL14 forward: 5′-ctgtgatggcgagacaaatg-3′; CXCL14 reverse: 5′-gttgggaacctcacatgctt-3′; β-actin forward: 5′-aatgc-tatcacctcccctgtgt-3′ and β-actin reverse: 5′-aagccaccccacttctctctaa-3′.
For the cocultivation experiments total RNA was prepared and the reverse transcription was performed using Superscript II reverse transcrip-tase (Invitrogen) and random hexamer primers (Applied Biosystems, Foster City, CA, USA) according to the manufacture's suggestions. The subsequent quantitative PCR reaction was conducted using SYBR green Super Mix (Bio-Rad Laboratories GmbH, Munich, Germany), gene specific primers and the following PCR conditions: 95°C, 5 min. (single step); 95°C, 30 sec.; 60°C, 45 sec.; 72°C, 70 sec. (40 cycles). The fluorescence was determined by Bio-Rad's Opticon PCR-System.
Immunohistochemistry
For immunohistochemistry, 5 μm sections were routinely prepared using sialinized slides (superfrost slides from Menzel Gl‰ser, Braunschweig, Germany). Immunohistochemistry for WNT5a and SFRP1 was performed using the streptavidin-biotin-peroxidase method as described previously [28]. Antigen retrieval was carried out in a microwave oven (250 W for 30 min. in a citrate solution pH 6.0). The primary antibodies used were a mouse poly-clonal antibody against the WNT5a protein (clone GT15034; 10 μg/ml; Neuromics, Northfield, MN and sc-23698, Santa Cruz, CA, USA), a polyclonal antiserum generated in rabbits with a synthesized peptide corresponding to amino acids 285-299 of the human SFRP1 protein (1:50) [29] and a goat antibody against the CXCL14 protein (AF866; 1:50; R&D Systems, Minneapolis, MN, USA). As negative control specimens were incubated without the primary antibody. Afterwards, the slides were briefly counterstained with haematoxylin and eosin. For the negative control, the primary antibody was omitted. The staining intensity was evaluated for each sample semi-quantitatively as absent, weak, moderate or strong by one pathologist (A.H. A.W. or G.K.) and by C.P. without knowledge of the histopathologic and molecular data. For the analysis of immunohistochemical staining of tumour and the adjacent normal tissue we employed the Wilcoxon Rank test.
Results
Gene expression analysis of stromal tissues
Using microdissection, stromal tissue adjacent to the tumour from 11 patients with PDAC and stromal tissue from nine patients with chronic pancreatitis were obtained and compared. The combination of fold change (cut-off: >2) and SAM q-value (cut-off: ≤5%) analysis yielded 331 differentially expressed probe sets from 316 genes. In total, 61 genes were under-expressed and 255 were overexpressed in the stromal tissue of PDAC (Table 1).
Table 1.
The 100 most differentially expressed genes in PDAC stromal tissue
| Representative public ID | Affymetrix probeset ID | Fold change (TS/CP) | Gene symbol | Gene title |
|---|---|---|---|---|
| Up-regulated | ||||
| NM_005498 | 218261_at | 22.67 | AP1M2 | Adaptor-related protein complex 1, mu 2 subunit |
| NM_004887 | 218002_s_at | 13.25 | CXCL14 | Chemokine (C-X-C motif) ligand 14 |
| AI739132 | 229479_at | 8.97 | LOC646324 | Hypothetical LOC646324 |
| NM_001854 | 204320_at | 8.04 | COL11A1 | Collagen, type XI, α 1 |
| NM_005940 | 203878_s_at | 7.40 | MMP11 | Matrix metallopeptidase 11 (stromelysin 3) |
| NM_007036 | 208394_x_at | 6.55 | ESM1 | Endothelial cell-specific molecule 1 |
| BC006361 | 211050_x_at | 6.21 | DKFZP434B2016 | Similar to hypothetical protein LOC284701 |
| AF352728 | 221701_s_at | 6.09 | STRA6 | Stimulated by retinoic acid gene 6 homologue (mouse) |
| AU156710 | 227123_at | 5.68 | RAB3B | RAB3B, member RAS oncogene family |
| AB033025 | 212942_s_at | 5.25 | KIAA1199 | KIAA1199 |
| AI822137 | 230135_at | 5.17 | EST | CDNA FLJ42405 fis, clone ASTRO3000474 |
| BE327661 | 240649_at | 4.89 | EST | Transcribed locus |
| AA741296 | 238812_at | 4.69 | ZFAND6 | Zinc finger, AN1-type domain 6 |
| M86849 | 223278_at | 4.64 | GJB2 | Gap junction protein, β 2, 26 kD (connexin 26) |
| AW972824 | 244765_at | 4.63 | DUSP27 | Dual specificity phosphatase 27 (putative) |
| AK025453 | 228656_at | 4.33 | PROX1 | Prospero-related homeobox 1 |
| AW301806 | 217580_x_at | 4.33 | ARL6IP2 | ADP-ribosylation factor-like 6 interacting protein 2 |
| NM_006587 | 220356_at | 4.18 | CORIN | Corin, serine peptidase |
| AI376003 | 205941_s_at | 4.08 | COL10A1 | Collagen, type X, α 1(Schmid metaphyseal chondrodysplasia) |
| AF288571 | 221558_s_at | 3.90 | LEF1 | Lymphoid enhancer-binding factor 1 |
| AA382425 | 232109_at | 3.90 | UBXD3 | UBX domain containing 3 |
| T10030 | 228210_at | 3.87 | NXPH3 | Neurexophilin 3 |
| NM_003248 | 204776_at | 3.84 | THBS4 | Thrombospondin 4 |
| M57707 | 204188_s_at | 3.78 | RARG | Retinoic acid receptor, γ |
| NM_003326 | 207426_s_at | 3.76 | TNFSF4 | Tumour necrosis factor (ligand) superfamily, member 4 |
| AB051466 | 232327_at | 3.73 | THSD7B | Thrombospondin, type I, domain containing 7B |
| AL136559 | 223536_at | 3.61 | PSD2 | Pleckstrin and Sec7 domain containing 2 |
| NM_012261 | 219463_at | 3.60 | C20orf103 | Chromosome 20 open-reading frame 103 |
| M31159 | 210095_s_at | 3.50 | IGFBP3 | Insulin-like growth factor-binding protein 3 |
| AW296153 | 235540_at | 3.50 | EST | — |
| AI206039 | 231187_at | 3.41 | SLC28A1 | Solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| W74476 | 226997_at | 3.41 | EST | CDNA FLJ10196 fis, clone HEMBA1004776 |
| AB032931 | 223229_at | 3.34 | UBE2T | Ubiquitin-conjugating enzyme E2T (putative) |
| AW593143 | 205134_s_at | 3.32 | NUFIP1 | Nuclear fragile X mental retardation protein interacting protein 1 |
| NM_016931 | 219773_at | 3.30 | NOX4 | NADPH oxidase 4 |
| AI081522 | 244486_at | 3.29 | PINK1 | PTEN-induced putative kinase 1 |
| AB015329 | 215138_s_at | 3.27 | KIAA1026 | Kazrin |
| AI378647 | 230147_at | 3.26 | F2RL2 | Coagulation factor II (thrombin) receptor-like 2 |
| AW664964 | 230493_at | 3.21 | TMEM46 | Transmembrane protein 46 |
| AK024472 | 226530_at | 3.15 | BMF | Bcl2 modifying factor |
| NM_001845 | 211981_at | 3.14 | COL4A1 | Collagen, type IV, α 1 |
| BF476080 | 229779_at | 3.13 | COL4A4 | Collagen, type IV, α 4 |
| NM_021161 | 220727_at | 3.10 | KCNK10 | Potassium channel, subfamily K, member 10 |
| NM_002381 | 206091_at | 3.09 | MATN3 | Matrilin 3 |
| AI222435 | 230319_at | 3.07 | C4orf31 | Chromosome 4 open-reading frame 31 |
| U09716 | 203294_s_at | 3.04 | LMAN1 | Lectin, mannose-binding, 1 |
| U03115 | 217060_at | 3.01 | EST | T cell receptor b chain variable region (TCRB) mRNA, 5′ end |
| AI742057 | 226702_at | 2.99 | LOC129607 | Hypothetical protein LOC129607 |
| AA012950 | 239126_at | 2.98 | C19orf23 | Chromosome 19 open reading frame 23 |
| NM_003427 | 207494_s_at | 2.97 | ZNF76 | Zinc finger protein 76 (expressed in testis) |
| Down-regulated | ||||
| NM_019598 | 220782_x_at | 0.04 | KLK12 | Kallikrein-related peptidase 12 |
| NM_000273 | 206696_at | 0.05 | GPR143 | G protein-coupled receptor 143 |
| NM_005867 | 207258_at | 0.11 | DSCR4 | Down syndrome critical region gene 4 |
| AI090768 | 224808_s_at | 0.13 | C7orf20 | Chromosome 7 open reading frame 20 |
| NM_006461 | 203145_at | 0.14 | SPAG5 | Sperm-associated antigen 5 |
| AF105378 | 228206_at | 0.15 | HS3ST4 | Heparan sulphate (glucosamine) 3-O-sulfotransferase 4 |
| N53051 | 229378_at | 0.16 | STOX1 | Storkhead box 1 |
| NM_017748 | 218655_s_at | 0.17 | CCDC49 | Coiled-coil domain containing 49 |
| BE273906 | 238980_x_at | 0.18 | C17orf56 | Chromosome 17 open reading frame 56 |
| NM_002580 | 205815_at | 0.18 | REG3A | Regenerating islet-derived 3 alpha |
| AA419275 | 224970_at | 0.20 | NFIA | nuclear factor I/A |
| U05598 | 209699_x_at | 0.20 | AKR1C2 | Aldo-keto reductase family 1, member C2 |
| M25915 | 208792_s_at | 0.22 | CLU | Clusterin |
| BF439728 | 238317_x_at | 0.25 | RBMS1 | RNA binding motif, single-stranded interacting protein 1 |
| NM_001458 | 207876_s_at | 0.27 | FLNC | Filamin C, γ (actin-binding protein 280) |
| NM_001353 | 204151_x_at | 0.27 | AKR1C1 | Aldo-keto reductase family 1, member C1 |
| M13452 | 212089_at | 0.28 | LMNA | Lamin A/C |
| NM_006179 | 231785_at | 0.28 | NTF5 | Neurotrophin 5 (neurotrophin 4/5) |
| NM_003277 | 204482_at | 0.30 | CLDN5 | Claudin 5 (transmembrane protein deleted in velocardiofacial syndrome) |
| NM_001937 | 207977_s_at | 0.31 | DPT | Dermatopontin |
| AW157077 | 203197_s_at | 0.31 | C1orf123 | Chromosome 1 open reading frame 123 |
| AB056476 | 224339_s_at | 0.32 | ANGPTL1 | Angiopoietin-like 1 |
| BE646573 | 223217_s_at | 0.33 | NFKBIZ | Nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor |
| NM_002178 | 203851_at | 0.35 | IGFBP6 | Insulin-like growth factor-binding protein 6 |
| AA780381 | 215498_s_at | 0.35 | MAP2K3 | Mitogen-activated protein kinase kinase 3 |
| AA194312 | 227086_at | 0.36 | HIRA | HIR histone cell cycle regulation defective homologue A (S. cerevisiae) |
| AL353132 | 217021_at | 0.37 | CYB5A | Cytochrome b5 type A (microsomal) |
| NM_020190 | 218162_at | 0.37 | OLFML3 | Olfactomedin-like 3 |
| NM_001159 | 205083_at | 0.39 | AOX1 | Aldehyde oxidase 1 |
| NM_000504 | 205620_at | 0.39 | F10 | Coagulation factor X |
| AW779917 | 230003_at | 0.39 | EST | Transcribed locus |
| AV757441 | 238675_x_at | 0.40 | EST | — |
| AK022266 | 233817_at | 0.40 | NBPF10 | Neuroblastoma breakpoint family, member 10 |
| AF109161 | 209357_at | 0.40 | CITED2 | Cbp/p300-interacting transactivator |
| NM_014057 | 218730_s_at | 0.40 | OGN | Osteoglycin (osteoinductive factor, mimecan) |
| NM_006086 | 202154_x_at | 0.41 | TUBB3 | Tubulin, β 3 |
| NM_003012 | 202037_s_at | 0.42 | SFRP1 | secreted frizzled-related protein 1 |
| AW157094 | 209291_at | 0.42 | ID4 | Inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| AB028976 | 212845_at | 0.42 | SAMD4A | Sterile a motif domain containing 4A |
| AI830227 | 222065_s_at | 0.42 | FLII | Flightless I homologue (Drosophila) |
| M24317 | 209612_s_at | 0.43 | ADH1B | Alcohol dehydrogenase IB (class I), β polypeptide |
| AF043290 | 209447_at | 0.43 | SYNE1 | Spectrin repeat containing, nuclear envelope 1 |
| AI989530 | 227197_at | 0.45 | SGEF | Src homology 3 domain-containing guanine nucleotide exchange factor |
| NM_001449 | 201540_at | 0.45 | FHL1 | Four and a half LIM domains 1 |
| AK023795 | 222162_s_at | 0.45 | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| AB011538 | 203812_at | 0.46 | EST | CDNA clone IMAGE:5922621 |
| BE967532 | 203636_at | 0.46 | MID1 | Midline 1 (Opitz/BBB syndrome) |
| NM_001801 | 204154_at | 0.46 | CDO1 | Cysteine dioxygenase, type I |
| U36764 | 208756_at | 0.47 | EIF3S2 | Eukaryotic translation initiation factor 3, subunit 2 β, 36 kD |
| AL136694 | 223938_at | 0.47 | C1orf49 | Chromosome 1 open-reading frame 49 |
Gene selected for further validation are marked in bold and italic. TS, tumour stroma; CP, stroma of chronic pancreatitis. The complete table is available as supplemental data.
Hierarchical clustering of the stromal tissue samples using the 331 probe sets revealed two major clusters of stromal tissue of chronic pancreatitis and of PDAC (Fig. 1A).
Fig. 1.
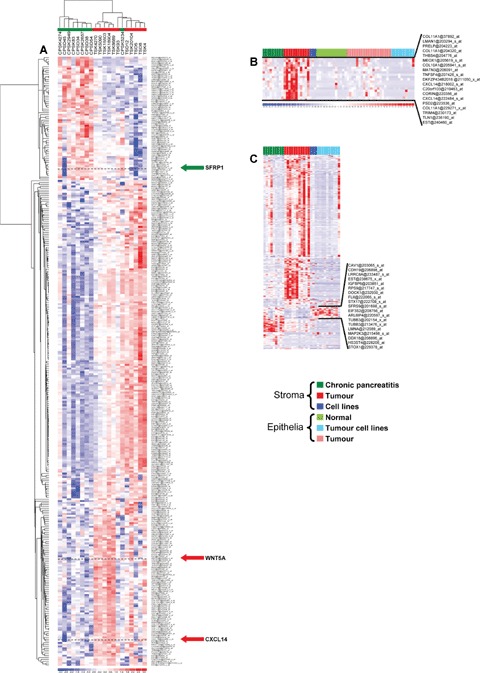
(A) Hierarchical clustering of the stromal tissue samples using the identified differentially expressed genes. (B) Hierarchical clustering of pancreatic tissues and cell lines using the 205 probe sets identified asdifferentially expressed between tumour stroma and all other primary tissues from the 331 probe set. Probe set within the black lines are highly expressed in tumour stroma and the genes of this cluster are depicted on the right. (C) Heatmap of the expression of the 331 probe sets in the data set. Probe sets within the black line are highly expressed in cell lines and are depicted on the right. The cell line PT45 is the last column to the right in Fig. 1B and C.
Identifcation of genes overexpressed in tumour stroma
To identifiy genes highly expressed in pancreatic tumour stroma, we compared the expression of the 331 probe sets between pancreatic tumour stroma and other primary pancreatic tissues excluding pancreatic cell lines. 19 of the 331 probe sets representing 16 genes (including CXCL14, COL10A1 and COL11A1) were up-regulated by a fold change of at least 4 (Fig. 1B).
Signal transduction pathway analysis of differentially expressed genes
Using the FatiGO† tool in conjunction with the KEGG database from the Babelomics web server, we were able to assign 45 out of the 316 differentially expressed genes to a KEGG pathway (Fig. 2) [30]. Interestingly, five overexpressed and one underexpressed genes were associated with the general Wnt signal transduction cascade indicating an activation of the pathway in PDAC stroma. Moreover, the KEGG pathway analysis showed that three genes of the Mitogen-activated protein kinases (MAPK) signalling pathway were underexpressed and one was overexpressed in PDAC stroma (Fig. 2A and B).
Fig. 2.
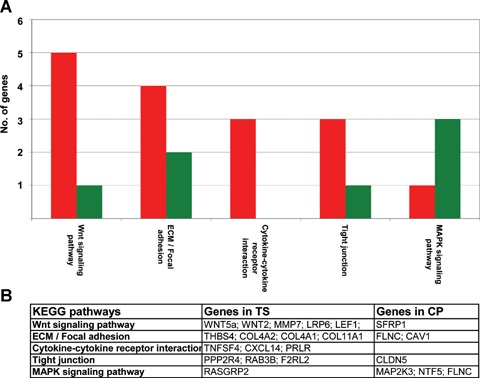
(A) Association of the differentially expressed genes with KEGG signal transduction pathways. Green: genes underexpressed; red: genes overex-pressed in pancreatic ductal adenocarcinoma (PDAC) stroma compared to stroma from chronic pancreatitis. (B) Genes identified in the KEGG pathways, TS, tumour stroma; CP, stroma of chronic pancreatitis.
Validation of differential expression of WNT5a, CXCL14 and SFRP1
We choose to validate the differential expression of CXCL14, SFRP1 and WNT5a using quantitative RT-PCR and immunohistochemistry. The selection of those three genes was based on their assignment to important signal transduction pathways and on their differential expression. We could show that all genes, the underexpressed SFRP1, the only lightly overexpressed WNT5a and the highly overex-pressed CXCL14 are differentially expressed in the stroma of PDACs compared to the stromal tissue of chronic pancreatitis. Quantitative RT-PCR showed that SFRP1 is down-regulated by a fold change of 5.27, whereas WNT5a is up-regulated by a fold change of 2.06 and CXCL14 is up-regulated by a fold change of 13.25 in the stroma of PDACs (Fig. 3). Using immunohistochemistry, we analysed stroma tissue from PDAC, peritumoural chronic pancreatitis or peritumour-al benign stromal tissue for the expression of WNT5a, CXCL14 and SFRP1. For WNT5a, we detected an up-regulation in 92% of the cases (47/51, P < 0.01, Fig. 4A and B). Analysis of the stromal tissue surrounding normal ducts revealed only rarely positive cells for WNT5a. Recently, it has been described that WNT5a is regulated by CUTL1 in PDAC [31]. Analysis of the CUTL1 expression in our gene expression data revealed mean arbitrary intensity units for CUTL1 in tumour stroma of 251 and 252 in chronic pancreatitis stroma indicating that CUTL1 is expressed in pancreatic stroma tissue, but not overexpressed. For CXCL14, an overexpression could be observed in 54% of the cases of PDAC-associated stroma compared to peritu-moural chronic pancreatitis tissue (7/13 Fig. 4C and D) and for SFRP1, we observed a loss of protein in stromal PDAC tissue in 65% of the cases analysed (24/37, Fig. 4E and F).
Fig. 3.
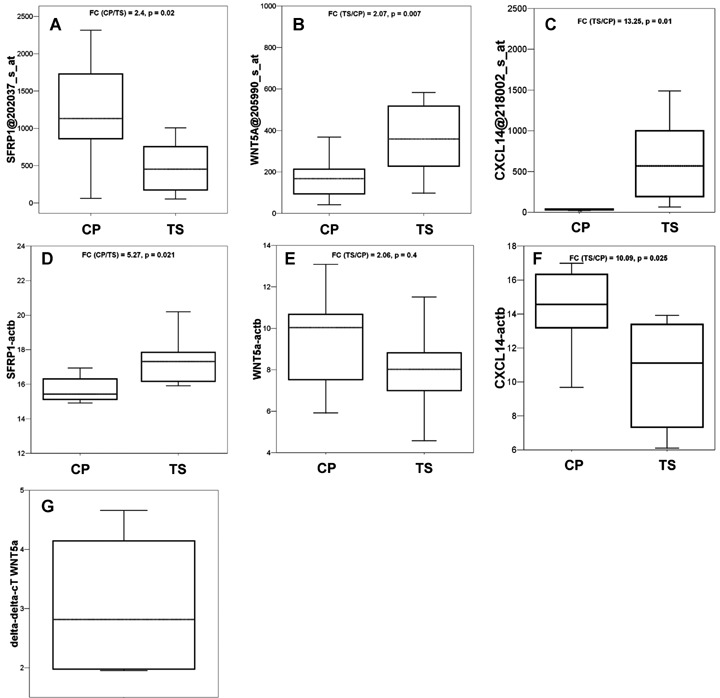
Boxplots of the distribution of expression of SFRP1, WNT5a and CXCL14 in stroma of PDAC and chronic pancreatitis. (A–C) Arbitrary intensity units of the probe sets from the Affymetrix U133 GeneChips; (D–F): Δ-cT values from the quantitative PCR, data were calculated as Δ-cT values using β-actin values for normalization, therefore lower values indicate higher expression of a given gene in the samples; (G) Relative expression of WNT5a in KIF5 stromal cells cocultured with Panc89 cells. WNT5a is induced in fibroblasts by a median of 2.82-fold if WNT5a expression is normalized to G6PDH expression and referred to the expression of WNT5a in KIF5 cells grown in monolayers; dotted lines: median; FC, fold change; TS, tumour stroma; CP, stroma of chronic pan-creatitis. P-values were calculated using the t-test.
Fig. 4.
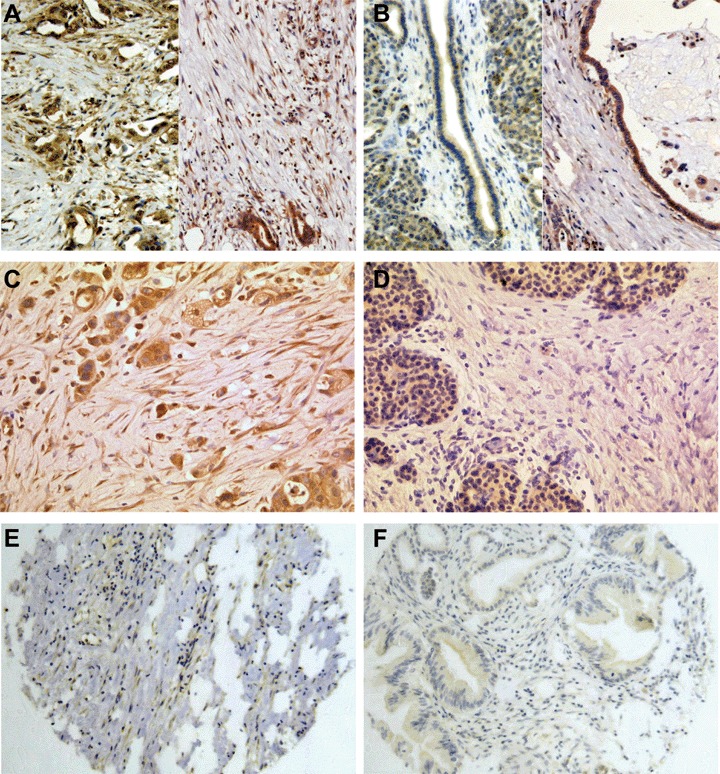
Immunohistochemistry of Wnt pathway associated genes. (A) Strong expression of WNT5a in PDAC stroma cell and (B) lack of expression of WNT5a in stromal tissue adjacent to normal ducts in two different samples. The surrounding acinar cells of the pancreas show staining for WNT5a, whereas the normal duct epithelia show only a faint stain and staining is absent within the stromal cells; (C) CXCL14 expression in PDAC epithelia and adjacent stromal cells. (D) Lack of expression of CXCL14 in a stroma of chronic pancreatitis. (E) Expression of SFRP1 in the stroma of chronic pancreatitis and blood vessels; (F) Loss of SFRP1 expression in the stroma of a well-differentiated PDAC. (A–F) 200×.
Induction of WNT5a by cocultivation
We also analysed three cell lines from stromal origin for their expression of the identified 331 probe sets. The stromal cell lines F13, Kif5 and Stellate, as well as the cell lines derived from PDAC tissues except PT45 expressed most of the genes in only low amounts under standard cultivation conditions (Fig. 1C). However, cocultivation of the human foreskin fibrob-last cell line Kif5 with the pancreas cancer cell line Panc89 resulted in a median 2.82-fold induction of WNT5a RNA in the fibroblasts. This indicates that WNT5a expression might be induced by soluble factors derived from pancreatic tumours, since the two cell lines were separated by a membrane with 0.2 μm pore size (Fig. 3G).
Discussion
A strong desmoplastic reaction is typical for PDACs, but may also be found in other carcinomas, such as colon or breast carcinoma [32–34]. Since tumour desmoplasia seems to have an association with tumour progression, analysis of stromal gene expression may increase our understanding of this process. Using the Affymetrix U133 GeneChip set we performed the first whole genome gene expression analysis of microdissected cells from PDAC stroma and chronic pancreatitis stroma. Thus the tumour-specific stromal reaction can be discriminated from the severe, but benign reaction represented by chronic pancreatitis. We identified 316 differentially expressed genes represented by 331 Affymetrix probe sets, of which 255 were up-regulated and 61 genes down-regulated in the stromal tissue of PDAC compared to chronic pancreatitis stroma. Within the set of genes up-regulated in PDAC stromal tissue, we identified MMP11 and CXCL14 which has been shown to be activated in stromal tissue other carcinomas [35–37]. Despite the importance of the stromal tissue for tumour development and maintenance, few gene expression studies on stromal tissue have been performed. This might be due to the fact that microdissection is a prerequisite for profiling stromal tissue. We compared our data with the data set published by Binkley et al. and found an overlap of six genes (COL10A1, COL4A2, IFI30, IFI6, POSTN and UCP2) [38]. The reason for this small overlap might be found in the different types of analysis. Binkley et al. used a bioin-formatic approach to identify stromal gene expression by subtracting epithelial gene expression data from their data of pancreatic cancer gene expression; whereas, we analysed the gene expression of tumours stroma directly. The different GeneChip formats might also be another cause for the small overlap, we observed as Binkley et al. analysed only 6800 human genes, whereas we analysed of the whole human transcriptome (∼30,000 genes). Of the 61 genes underex-pressed in PDAC stroma, some are highly expressed in normal epithelia. This might be owed to spurious epithelial cells within the stroma of chronic pancreatitis which cannot be eliminated by manual microdissection. However, the immunohistochemical validation of WNT5a, SFRP1 and CXCL14 clearly demonstrates that the genes identified by our approach are differentially expressed between PDAC and chronic pancreatitis stroma.
We classified the genes according to their involvement in signal transduction pathways and found a high proportion of Wnt pathway members as dysregulated suggesting that this pathway plays a role in stroma–tumour interaction in PDACs. Interestingly, we found a down-regulation of SFRP1 in PDAC stroma. As it has been shown that SFRP1 expression is lost by hypermethylation in the majority of cancers and PDACs [39, 40], this is the first report of the down-regulation of SFRP1 in primary stromal tissue. We may speculate that SFRP1 down-regulation in PDAC stroma is linked to hypermethylation of the SFRP1 gene in PDAC stroma as observed in cancer epithelia. SFRP1 is an inhibitor of canonical Wnt action and it is also capable of transducing signals without the participation of Wnt molecules via Frizzled receptors [41]. As WNT5a is known to be able to inactivate the canonical pathway, our observed overexpression of WNT5a in the stromal and the epithelial compartment of PDAC might be a reason for the dormancy of the canonical Wnt pathway [42]. WNT5a has been shown to activate MMP7 and to enhance the invasiveness of breast cancer cells via the non-canonical JNK pathway which might also occur in PDAC stroma, since we observed a MMP7 overexpression in those cells [43]. Interestingly, in PDAC cells WNT5a might also promote growth and invasion [31] indicating that the stromal compartment might contribute to these processes. However, despite the overexpression of WNT5a in PDAC stromal tissue we did not observe an overexpression of CUTL1 suggesting that also other signalling pathways might contribute to the transcriptional activation of WNT5a and may act via soluble factors as the induction of WNT5a expression in fibroblasts demonstrated. These factors are most likely proteins of the hedgehog family, which are overexpressed in PDAC and known to regulate WNT5a expression [44, 45]. WNT5a has also been characterized as a modulator of chondrocyte development and associated with chondrocyte hyper-trophy [46]. Chondrocyte hypertrophy is also associated with the expression of COL10A1 [47] which we and others identified to be overexpressed in PDAC stroma [38]. Therefore, WNT5a overexpres-sion in tumour stroma might result in overexpression of COL10A1. COL10A1 belongs to the type of network-forming collagens; whereas, COL11A1 which we also observed as overexpressed in PDAC stroma is a fibrillar collagen [48]. The overexpression of these two types of collagen might have profound effects on the density of the extracellular matrix surrounding the PDAC cells and might contribute to the callous form of tumours seen in PDAC. Gene expression profiling of microdissected tissues has been shown feasible and generates a complete picture of changes not only in the tumour epithelia [49], but also as shown here in the stromal tissue surrounding the tumour. The differential expression we have observed in our data might therefore lead to a better understanding of the generation of desmoplasia in PDAC.
Acknowledgments
This study was supported by the Deutsche Krebshilfe (70-2937-SaI). The complete list of the differentially expressed genes is available as supplemental data. The complete data set is available at ArrayExpress E-MEXP-950 and E-MEXP-1121.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Lohr M. Is it possible to survive pancreatic cancer? Nat Clin Pract Gastroenterol Hepatol. 2006;3:236–7. doi: 10.1038/ncpgasthep0469. [DOI] [PubMed] [Google Scholar]
- 3.Moore PS, Beghelli S, Zamboni G, Scarpa A. Genetic abnormalities in pancreatic cancer. Mol Cancer. 2003;2:7. doi: 10.1186/1476-4598-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu GC, Kimmelman AC, Hezel AF, Depinho RA. Stromal biology of pancreatic cancer. J Cell Biochem. 2007;101:887–907. doi: 10.1002/jcb.21209. [DOI] [PubMed] [Google Scholar]
- 5.Kenny PA, Bissell MJ. Tumor reversion: correction of malignant behavior by microenvironmental cues. Int J Cancer. 2003;107:688–95. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tlsty TD. Stromal cells can contribute oncogenic signals. Semin Cancer Biol. 2001;11:97–104. doi: 10.1006/scbi.2000.0361. [DOI] [PubMed] [Google Scholar]
- 7.Fukino K, Shen L, Matsumoto S, Morrison CD, Mutter GL, Eng C. Combined total genome loss of heterozygosity scan of breast cancer stroma and epithelium reveals multiplicity of stromal targets. Cancer Res. 2004;64:7231–6. doi: 10.1158/0008-5472.CAN-04-2866. [DOI] [PubMed] [Google Scholar]
- 8.Ohuchida K, Mizumoto K, Murakami M, Qian LW, Sato N, Nagai E, Matsumoto K, Nakamura T, Tanaka M. Radiation to stromal fibroblasts increases invasiveness of pancreatic cancer cells through tumorstromal interactions. Cancer Res. 2004;64:3215–22. doi: 10.1158/0008-5472.can-03-2464. [DOI] [PubMed] [Google Scholar]
- 9.Skrzydlewska E, Sulkowska M, Koda M, Sulkowski S. Proteolytic-antiproteolytic balance and its regulation in carcinogenesis. World J Gastroenterol. 2005;11:1251–66. doi: 10.3748/wjg.v11.i9.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Micke P, Ostman A. Tumour-stroma interaction: cancer-associated fibroblasts as novel targets in anti-cancer therapy? Lung cancer. 2004;45(Suppl 2):S163–75. doi: 10.1016/j.lungcan.2004.07.977. [DOI] [PubMed] [Google Scholar]
- 11.Lohr M, Schmidt C, Ringel J, Kluth M, Muller P, Nizze H, Jesnowski R. Transforming growth factor-beta1 induces desmoplasia in an experimental model of human pancreatic carcinoma. Cancer Res. 2001;61:550–5. [PubMed] [Google Scholar]
- 12.Katoh M, Katoh M. STAT3-induced WNT5A signaling loop in embryonic stem cells, adult normal tissues, chronic persistent inflammation, rheumatoid arthritis and cancer. Int J Mol Med. 2007;19:273–8. [PubMed] [Google Scholar]
- 13.Wong SC, Lo ES, Lee KC, Chan JK, Hsiao WL. Prognostic and diagnostic significance of beta-catenin nuclear immunos-taining in colorectal cancer. Clin Cancer Res. 2004;10:1401–8. doi: 10.1158/1078-0432.ccr-0157-03. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe I, Hasebe T, Sasaki S, Konishi M, Inoue K, Nakagohri T, Oda T, Mukai K, Kinoshita T. Advanced pancreatic ductal cancer: fibrotic focus and beta-catenin expression correlate with outcome. Pancreas. 2003;26:326–33. doi: 10.1097/00006676-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Grutzmann R, Boriss H, Ammerpohl O, Luttges J, Kalthoff H, Schackert H, Kloppel G, Saeger H, Pilarsky C. Meta-analysis of microarray data on pancreatic cancer defines a set of commonly dysregulated genes. Oncogene. 2005;24:5079–88. doi: 10.1038/sj.onc.1208696. [DOI] [PubMed] [Google Scholar]
- 16.Brandt R, Grutzmann R, Bauer A, Jesnowski R, Ringel J, Lohr M, Pilarsky C, Hoheisel JD. DNA microarray analysis of pancreatic malignancies. Pancreatology. 2004;4:587–97. doi: 10.1159/000082241. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T, Fidler IJ, Coombes KR. Gene expression profile of metastatic human pancreatic cancer cells depends on the organ microenvironment. Cancer Res. 2007;67:139–48. doi: 10.1158/0008-5472.CAN-06-2563. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura T, Furukawa Y, Nakagawa H, Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N, Miyamoto M, Hirano S, Kondo S, Katoh H, Nakamura Y, Katagiri T. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. 2004;23:2385–400. doi: 10.1038/sj.onc.1207392. [DOI] [PubMed] [Google Scholar]
- 19.Buchholz M, Braun M, Heidenblut A, Kestler HA, Kloppel G, Schmiegel W, Hahn SA, Luttges J, Gress TM. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–36. doi: 10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- 20.Missiaglia E, Blaveri E, Terris B, Wang YH, Costello E, Neoptolemos JP, Crnogorac-Jurcevic T, Lemoine NR. Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int J Cancer. 2004;112:100–12. doi: 10.1002/ijc.20376. [DOI] [PubMed] [Google Scholar]
- 21.Grutzmann R, Pilarsky C, Ammerpohl O, Luttges J, Bohme A, Sipos B, Foerder M, Alldinger I, Jahnke B, Schackert HK, Kalthoff H, Kremer B, Kloppel G, Saeger HD. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia. 2004;6:611–22. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloppel G. Chronic pancreatitis of alcoholic and nonalcoholic origin. Semin Diagn Pathol. 2004;21:227–36. doi: 10.1053/j.semdp.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Kapischke M, Prinz K, Tepel J, Tensfeldt J, Schulz T. Precoating of alloplastic materials with living human fibroblasts–a feasibility study. Surg Endosc. 2005;19:791–7. doi: 10.1007/s00464-004-9222-1. [DOI] [PubMed] [Google Scholar]
- 24.Jesnowski R, Furst D, Ringel J, Chen Y, Schrodel A, Kleeff J, Kolb A, Schareck WD, Lohr M. Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: deactivation is induced by matrigel and N-acetylcysteine. Lab Invest. 2005;85:1276–91. doi: 10.1038/labinvest.3700329. [DOI] [PubMed] [Google Scholar]
- 25.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle RM. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–33. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- 28.Stoehr R, Wissmann C, Suzuki H, Knuechel R, Krieg RC, Klopocki E, Dahl E, Wild P, Blaszyk H, Sauter G, Simon R, Schmitt R, Zaak D, Hofstaedter F, Rosenthal A, Baylin SB, Pilarsky C, Hartmann A. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Lab Invest. 2004;84:465–78. doi: 10.1038/labinvest.3700068. [DOI] [PubMed] [Google Scholar]
- 29.Klopocki E, Kristiansen G, Wild PJ, Klaman I, Castanos-Velez E, Singer G, Stohr R, Simon R, Sauter G, Leibiger H, Essers L, Weber B, Hermann K, Rosenthal A, Hartmann A, Dahl E. Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol. 2004;25:641–9. [PubMed] [Google Scholar]
- 30.Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–4. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ripka S, Konig A, Buchholz M, Wagner M, Sipos B, Kloppel G, Downward J, Gress T, Michl P. WNT5A – target of CUTL1 and potent modulator of tumor cell migration and invasion in pancreatic cancer. Carcinogenesis. 2007;28:1178–87. doi: 10.1093/carcin/bgl255. [DOI] [PubMed] [Google Scholar]
- 32.Kim JB, Stein R, O’Hare MJ. Tumourstromal interactions in breast cancer: the role of stroma in tumourigenesis. Tumour Biol. 2005;26:173–85. doi: 10.1159/000086950. [DOI] [PubMed] [Google Scholar]
- 33.Bilalovic N, Vranic S, Serdarevic F, Foco F. The role of the stroma in carcinogenesis. Bosn J Basic Med Sci. 2006;6:33–8. doi: 10.17305/bjbms.2006.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung LW, Baseman A, Assikis V, Zhau HE. Molecular insights into prostate cancer progression: the missing link of tumor microenvironment. J Urol. 2005;173:10–20. doi: 10.1097/01.ju.0000141582.15218.10. [DOI] [PubMed] [Google Scholar]
- 35.Basset P, Bellocq JP, Lefebvre O, Noel A, Chenard MP, Wolf C, Anglard P, Rio MC. Stromelysin-3: a paradigm for stromaderived factors implicated in carcinoma progression. Crit Rev Oncol Hematol. 1997;26:43–53. doi: 10.1016/s1040-8428(97)00010-3. [DOI] [PubMed] [Google Scholar]
- 36.Ricci F, Kern SE, Hruban RH, Iacobuzio-Donahue CA. Stromal responses to carcinomas of the pancreas: juxtatumoral gene expression conforms to the infiltrating pattern and not the biologic subtype. Cancer Biol Ther. 2005;4:302–7. doi: 10.4161/cbt.4.3.1501. [DOI] [PubMed] [Google Scholar]
- 37.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Binkley CE, Zhang L, Greenson JK, Giordano TJ, Kuick R, Misek D, Hanash S, Logsdon CD, Simeone DM. The molecular basis of pancreatic fibrosis: common stromal gene expression in chronic pan-creatitis and pancreatic adenocarcinoma. Pancreas. 2004;29:254–63. doi: 10.1097/00006676-200411000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, Galm O, Camara O, Durst M, Kristiansen G, Huszka C, Knuchel R, Dahl E. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25:3479–88. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 40.Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–42. [PubMed] [Google Scholar]
- 41.Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, Trousse F, Dwivedy A, Holt C, Bovolenta P. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005;8:1301–9. doi: 10.1038/nn1547. [DOI] [PubMed] [Google Scholar]
- 42.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:570–82. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pukrop T, Klemm F, Hagemann T, Gradl D, Schulz M, Siemes S, Trumper L, Binder C. Wnt5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc Natl Acad Sci USA. 2006;103:5454–9. doi: 10.1073/pnas.0509703103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kayed H, Kleeff J, Osman T, Keleg S, Buchler MW, Friess H. Hedgehog signaling in the normal and diseased pancreas. Pancreas. 2006;32:119–29. doi: 10.1097/01.mpa.0000202937.55460.0c. [DOI] [PubMed] [Google Scholar]
- 45.Reddy S, Andl T, Bagasra A, Lu MM, Epstein DJ, Morrisey EE, Millar SE. Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech Dev. 2001;107:69–82. doi: 10.1016/s0925-4773(01)00452-x. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Topol L, Lee H, Wu J. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development. 2003;130:1003–15. doi: 10.1242/dev.00324. [DOI] [PubMed] [Google Scholar]
- 47.Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol. 2003;162:833–42. doi: 10.1083/jcb.200211089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer H, Stenling R, Rubio C, Lindblom A. Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis. 2001;22:875–8. doi: 10.1093/carcin/22.6.875. [DOI] [PubMed] [Google Scholar]
- 49.Kristiansen G, Jacob J, Buckendahl AC, Grutzmann R, Alldinger I, Sipos B, Kloppel G, Bahra M, Langrehr JM, Neuhaus P, Dietel M, Pilarsky C. Peroxisome proliferator-activated receptor gamma is highly expressed in pancreatic cancer and is associated with shorter overall survival times. Clin Cancer Res. 2006;12:6444–51. doi: 10.1158/1078-0432.CCR-06-0834. [DOI] [PubMed] [Google Scholar]


