Abstract
Background and Aims LFY
homologues encode transcription factors that regulate the transition from vegetative to reproductive growth in flowering plants and have been shown to control inflorescence patterning in model species. This study investigated the expression patterns of LFY homologues within the diverse inflorescence types (head-like, umbel-like and inflorescences with elongated internodes) in closely related lineages in the dogwood genus (Cornus s.l.). The study sought to determine whether LFY homologues in Cornus species are expressed during floral and inflorescence development and if the pattern of expression is consistent with a function in regulating floral development and inflorescence architectures in the genus.
Methods
Total RNAs were extracted using the CTAB method and the first-strand cDNA was synthesized using the SuperScript III first-strand synthesis system kit (Invitrogen). Expression of CorLFY was investigated by RT–PCR and RNA in situ hybridization. Phylogenetic analyses were conducted using the maximum likelihood methods implemented in RAxML-HPC v7.2.8.
Key Results
cDNA clones of LFY homologues (designated CorLFY) were isolated from six Cornus species bearing different types of inflorescence. CorLFY cDNAs were predicted to encode proteins of approximately 375 amino acids. The detection of CorLFY expression patterns using in situ RNA hybridization demonstrated the expression of CorLFY within the inflorescence meristems, inflorescence branch meristems, floral meristems and developing floral organ primordia. PCR analyses for cDNA libraries derived from reverse transcription of total RNAs showed that CorLFY was also expressed during the late-stage development of flowers and inflorescences, as well as in bracts and developing leaves. Consistent differences in the CorLFY expression patterns were not detected among the distinct inflorescence types.
Conclusions
The results suggest a role for CorLFY genes during floral and inflorescence development in dogwoods. However, the failure to detect expression differences between the inflorescence types in the Cornus species analysed suggests that the evolutionary shift between major inflorescence types in the genus is not controlled by dramatic alterations in the levels of CorLFY gene transcript accumulation. However, due to spatial, temporal and quantitative limitations of the expression data, it cannot be ruled out that subtle differences in the level or location of CorLFY transcripts may underlie the different inflorescence architectures that are observed across these species. Alternatively, differences in CorLFY protein function or the expression or function of other regulators (e.g. TFL1 and UFO homologues) may support the divergent developmental trajectories.
Keywords: Cornus, dogwood, inflorescence evolution, LFY homologues, CorLFY expression, RT–PCR, in situ hybridization
INTRODUCTION
FLORICAULA/LEAFY (FLO/LFY) homologues are transcription factors regulating the transition from vegetative growth to reproductive growth in flowering plants. They were first identified in the analysis of two mutants, floricaula in Antirrhinum majus (Coen et al., 1990) and lfy in Arabidopsis thaliana (Schultz and Haughn, 1991; Weigel et al., 1992). FLO/LFY homologues have since been identified and studied in many lineages of land plants (e.g. Mouradov et al., 1998; Ahearn et al., 2001; Chujo et al., 2003; Dornelas and Rodriguez, 2005; Maizel et al., 2005; Sliwinski et al., 2006, summarized in Benlloch et al., 2007; Bosch et al., 2008; Hamès et al., 2008). In land plants, LFY is known to encode a protein with highly conserved N-terminal and C-terminal domains that are connected by a more variable interdomain region (Maizel et al., 2005). The C-terminal contains a DNA-binding domain that is structurally related to helix–turn–helix domains (Hamès et al., 2008), while the N-terminal encodes a homodimerization domain (Siriwardana and Lamb, 2012), and the interdomain region is of unknown function. Unlike many plant developmental regulators that exist as multigene families (Riechmann and Ratcliffe, 2000; Martinez-Castilla and Alvarez-Buylla, 2003; Shiu et al., 2005), LFY exists as a single-copy gene in most angiosperms that have been investigated (Southerton et al., 1998; Frohlich and Parker, 2000; Yoon and Baum, 2004), although two or more copies of LFY homologues have been reported in some angiosperm lineages, including Ionopsidium acaule (violet cress; Shu et al., 2000), Nicotiana (tobacco; Ahearn et al., 2001), Zea mays (maize; Bomblies et al., 2003), Rosaceae (Maloideae; Wada et al., 2002; Esumi et al., 2005), Fabaceae (Caesalpinoideae, Archambault and Bruneaua, 2004), Lamiales (Aagaard et al., 2005), Alliaceae (garlic, Rotem et al., 2007) and Idahoa scapigera (Brassicaceae, Sliwinski et al., 2007).
The role of the LFY gene in controlling floral meristem identity (Weigel et al., 1992; Weigel and Meyerowitz, 1993; Weigel and Nilsson, 1995; Blázquez et al., 1997; Nilsson et al., 1998) and floral organ initiation and patterning (Weigel et al., 1992; Parcy et al., 1998; Chae et al., 2008; Hamès et al., 2008), which was initially reported in Arabidopsis and Antirrhinum, has been supported by gene expression and/or functional data from a number of other plants, including both monocots [Z. mays, Bomblies et al., 2003; Allium sativum (garlic), Rotem et al., 2007; Oryza sativa (rice), Rao et al., 2008] and dicots [Pisum sativum (pea), Hofer et al., 1997; Petunia, Souer et al., 1998, 2008; Solanum lycopersicum (tomato), Molinero-Rosales et al., 1999; Ionopsidium acaule (violet cress), Shu et al., 2000; Vitis vinifera (grapevine), Carmona et al., 2002; Eschscholzia californica (California poppy), Busch and Gleissberg, 2003, Wreath et al., 2013; Hevea brasiliensis (rubber tree), Dornelas and Rodriguez, 2005; Idahoa, Sliwinski et al., 2007; and Populus, An et al., 2011]. A recent study has demonstrated that LEAFY (LFY) stimulates flower development and the formation of floral primordia via control of auxin response pathways (Li et al., 2013). Furthermore, several studies have supported a role for LFY in controlling inflorescence architectures. For instance, constitutive expression of LFY resulted in a solitary terminal flower in Nicotiana (Ahearn et al., 2001; Koes, 2008); ectopic expression of LFY caused internodal compression of inflorescences in Arabidopsis (Sliwinski et al., 2007) and Malus domestica (apple; Flachowsky et al., 2010); changes in the activity or expression of LFY were associated with the origins of rosette flowering in Idahoa, Ionopsidium and Leavenworthia (Brassicaceae) (Shu et al., 2000; Bosch et al., 2008); and LFY activity repressed pedicel elongation and orientation in Arabidopsis, contributing to the variation of inflorescence architecture in the species (Yamaguchi et al., 2012). In maize, the LFY homologue (ZFL) promoted spikelet formation (Ikeda-Kawakatsu et al., 2012), while double mutants of LFY homologues (zfl1 and zfl2) exhibited reduced tassel branching (Bomblies et al., 2003). In rice, RNAi knockdowns of the LFY homologue, RFL, severely decreased panicle branching, while overexpression of RFL resulted in small panicles (Rao et al., 2008) due to the suppression of spikelet meristem formation (Ikeda-Kawakatsu et al., 2012). In wheat, the expression pattern of the LFY orthologue (WFL) was associated with spikelet formation (Shitsukawa et al., 2006). These data seem to suggest contrasting effects of LFY activity on inflorescence size between dicots and monocots (reduction in dicots but enlargement in monocots).
In the current study, we investigated the expression of LFY homologues during floral and inflorescence development of several dogwood species (Cornus, s.l.) in an effort to detect differences in LFY expression that might explain the evolutionary divergence in this genus of determinate umbel-like and head-like inflorescences from inflorescences with elongated internodes. The dogwood genus consists of four closely related lineages with similar inflorescence branching patterns (Feng et al., 2011), but differing in their aspect (Fig. 1). All inflorescences have cymous lateral branches and possess a terminal flower (the thyrsoids of Endress, 2010; Feng et al., 2011). Species within the blue- or white-fruited lineage (BW) produce determinate, elongated, large compound inflorescences. Within the cornelian cherry (CC) lineage, the inflorescences are determinate and umbel-like. Species within the dwarf dogwood lineage (DW) bear depauperate, condensed inflorescences with up to four lateral dichasia with very short, but evident inflorescence branches and pedicels (the minidichasia of Feng et al., 2011). Those within the big-bracted dogwood lineage (BB) produce flowers in head-like structures (Harris, 1999; Harris and Harris, 2001) (Fig. 1).
Fig. 1.
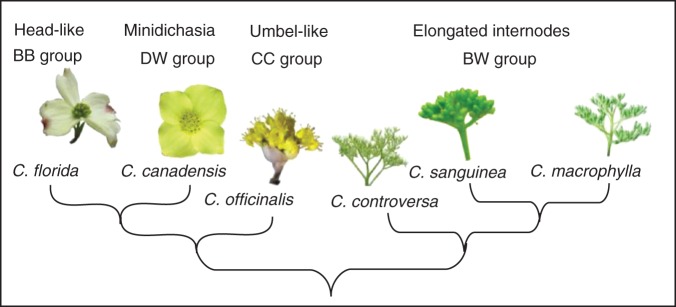
Four major clades of Cornus showing their phylogenetic relationships (from Xiang et al., 2008) and inflorescence types (modified from Xiang et al., 2008 and Feng et al., 2011). BB, big-bracted; DW, dwarf dogwood; CC, cornelian cherry; BW, blue- or white-fruited.
Developmental studies using scanning electron microscopy and histology have shown that the structural differences among these inflorescence types are largely determined during early development of inflorescences and are due to variations in the pattern, number and elongation of inflorescence branch meristems. Inflorescence branch meristems (IBMs) are generated in all four types during early development, but in umbel-like and head-like inflorescences the inflorescence branches and the rachis supporting the central inflorescence meristem (IM) do not elongate during development (for detailed differences among the four types, see Feng et al., 2011). The observation that the generation of new inflorescence meristems ceases in all four inflorescence types when the central inflorescence meristem transitions into a floral meristem (as evidenced by floral organ formation) clearly indicates that these four inflorescence types are all determinate (Feng et al., 2011). The inflorescence buds are preformed in the fall and expand in the following spring (Feng et al., 2011). Previous studies have indicated that the determinate umbel-like and head-like inflorescences in Cornus evolved in parallel from elongated, branched inflorescences (Xiang and Thomas, 2008; Feng et al., 2011).
The primary goal of this study was to identify orthologues of Arabidopsis LFY from the different lineages of dogwood (Cornus) species (hereafter CorLFY) and examine their expression patterns during floral and inflorescence development in the genus. We also hoped that a comparison of the expression patterns of the LFY genes among these species would provide insight into the potential role of these genes in the evolution of umbel-like and head-like inflorescences in Cornus.
MATERIALS AND METHODS
Sampling and cDNA preparation
Six species of Cornus representing the four major clades of the genus were included in the study. These were C. officinalis (CC group), C. florida (BB group) and C. canadensis (DW group) and C. macrophylla, C. sanguinea and C. controversa (BW group). Inflorescence buds were collected from plants growing on the NCSU campus and JC Raulston Arboretum for all species except C. canadensis, which were grown in the NCSU Phytotron and introduced from several wild populations in West Virginia and New Hampshire.
Inflorescence buds in early and late developmental stages were collected from these species for investigation of CorLFY expression. The early stages (Stages I–IV in Feng et al., 2011) span from the initiation of the inflorescence meristems to the formation of floral organ primordia. These early developmental events occur in the fall season. The late stage (Stage V in Feng et al., 2011) represents the subsequent development (or maturation) of the inflorescence and flowers in the following spring. Inflorescence buds of the late development stage were further divided into three different developmental phases: (1) bud unopened; (2) inflorescence bud opened, flower buds enlarging and bracts expanding; and (3) flower buds open and bracts fully expanded and white (in C. canadensis and C. florida). Table 1 provides the dates of inflorescence sample collections. Samples of developing young leaves were also collected from each species for analysis as a comparison with inflorescence buds. All samples of buds and leaves were stored in RNAlater solution (Ambion) immediately after their removal from the living plants to stabilize the RNA. Total RNA was extracted from bracts, flowers and young leaves using the CTAB method (Chang et al., 1993). For buds in early developmental stages it was not possible to separate bracts from the flowers manually. The RNA samples obtained were used as the template for first-strand cDNA synthesis using the SuperScript III First-Strand Synthesis System kit and the supplied oligo(dT) primer (Invitrogen, Carlsbad, CA, USA). The cDNAs were used for cloning CorLFY and the gene expression analyses described below.
Table 1.
Dates (month/day) samples were collected for RT–PCR analysis
| Cornus species | Stage 0 | Stage 1 | Stage 2 | Stage 3 |
|---|---|---|---|---|
| C. florida | 7/19 | 2/16 | 3/30 | 4/7 |
| C. canadensis | 0·8–1·5 cm | >1·5 cm | ||
| C. officinalis | 4/27 | 6/8 | 10/20 | 2/16 |
| C. macrophylla | 7/27 | 4/7 | 4/20 | 5/27 |
| C. sanguinea | 7/11 | 3/16 | 3/30 | 5/27 |
| C. controversa | 7/11 | 7/27 | 3/16 | 4/20 |
Stage 0, formation of IM, IBM, FM; Stage 1, inflorescence bud unopened; Stage 2, bud open, flower buds and inflorescence bracts (if present) expanding, except in C. officinalis, in which buds are expanding but bracts are not fully expanded; Stage 3, flower mature and bracts fully expanded. For C. canadensis grown in the phytotron, developmental stages were identified by morphology (size of bud) as plants in a growth chamber do not grow in synchrony with external seasons.
CorLFY gene cloning
The cDNA from early-stage inflorescences was used to isolate CorLFY homologues using PCR with degenerate primers. The cDNA sequences of CorLFY were obtained by amplification and sequencing of three overlapping fragments (I, II and III) (see Table 2 for the primer sequences). Degenerate primers were first designed based on LFY-like sequences of other flowering plants from GenBank to amplify fragments I and II using PCR (Table 2). The 3′ end of CorLFY (fragment III) was amplified in two steps. First, an oligo-dT primer (Table 2) containing a unique adaptor sequence was used for synthesis of the first strand. The products were then used as the templates for PCR with the forward primer lfyF2-3-2 and the reverse primer lfy3′, which were designed from the sequence of 3′RT. Unless specified, the Tm value for PCR was set at two degrees higher than the average value of the Tm of forward and reverse primers. PCR products were cloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA). For each Cornus species, at least five to ten positive clones from PCR products of each fragment were sequenced. Sequencing reactions were performed using the M13 primers and DNA sequencing kit (Applied Biosystems, Warrington, UK). Sequences obtained for the three fragments were assembled manually to build the cDNA sequence based on overlapping regions of approximately 100 base pairs (bp), which showed no polymorphisms among clones in all regions, including the overlapping regions. The complete cDNA sequence for Cornus canadensis was also amplified as a single fragment using PCR with primers (P5LFY_COcaN GAGGGATTGTAATTGTGTTGC and P3LFY_COca CGTACTACAGATGATATAAGG) to confirm the sequence assembled from fragments. The cDNA sequences of CorLFY were aligned and translated into amino acid sequences using the translation tool implemented in Geneious v5·1 (http://www.geneious.com/; Drummond et al., 2010). The N and C domains were identified in the amino acid sequences of CorLFY by comparison with that in Arabidopsis (see Results and Fig. 2). Identity of CorLFY was confirmed by a BLAST search using the amino acid sequence from Cornus as the query sequence and the non-redundant protein database on GenBank as well as by phylogenetic analyses of the CorLFY sequences with other LFY homologous sequences from Genbank.
Table 2.
Primers designed for CorLFY cloning, RT-PCR and probe synthesis for in situ hybridization in Cornus
| PCR |
Forward primer (5′–3′) | Reverse primer (5′–3′) | |
|---|---|---|---|
| Gene cloning | I | lfyL2 TTCWCGGCSAGYTTRTTCAAGTGGG | lfyR3-1 ACGTAGTGYCKCATYTTS GGCTTGTTKATGT |
| II | lfyF2-3-2 GAGGTKGCRCGYGGRAAGAAGAACGG | Cornus_3R1_yg CAGAGCTGGCGGAGCTTGGTNGGG | |
| RT | 3′ RT CCGGATCCTCTAGAGCGGCCGC(T)17 | ||
| III | lfyF2-3-2 GAGGTKGCRCGYGGRAAGAAGAACGG | lfy3′ CCGGATCCTCTAGAGCGGCCGC | |
| RT–PCR | P-I, Tm 60 °C | PF C. florida TCTATGAGCAGTGCCGTGATTTCTTG | PR C. florida CGTAGTGTCGCATT TTGGGCTTGTTA |
| PF C. canadensis TCTACGAGCAGTGCCGTGATTTCTTG | PR C. canadensis CGTAGTGCCGC ATCTTGGGCTTGTTA | ||
| PF C. officinalis TCTACGAGCAGTGCCGTGATTTCTTG | PR C. officinalis CGTAGTGCCGC ATTTTGGGCTTGTTA | ||
| PF C. macrophylla TCTACGAGCAGTGCCGTGATTTCTTG | PR C. macrophylla CGTAGTGCCG CATTTTGGGCTTGTTA | ||
| PF C. sanguinea TCTACGAACAGTGCCGTGATTTCTTG | PR C. sanguinea CGTAGTGTCGCA TCTTAGGCTTGTTA | ||
| PF C. controversa TCTACGAGCAGTGCCGTGATTTCTTG | PR C. controversa CGTAGTGTCGC ATTTTGGGCTTGTTA | ||
| 26S, Tm 55 °C | 12F GTCCTAAGATGAGCTC | 2782R GGTAACTTTTCTGACACCTC | |
| In situ probe | Tm 55 °C | lfyL2 TTCWCGGCSAGYTTRTTCAAGTGGG | lfyR1 CAACGCMCTTGATGCACWCT |
Fig. 2.
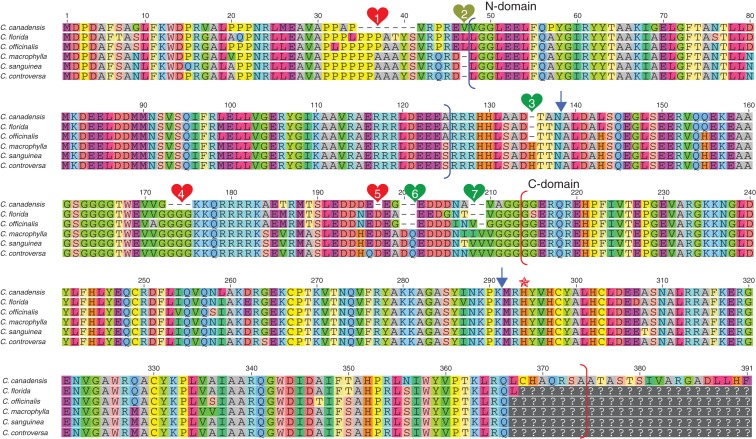
Aligned amino acid sequences of CorLFY (C. florida, C. canadensis, C. officinalis, C. macrophylla, C. sanguinea and C. controversa). Identical amino acids are shown in the same colour. Composite numbering with gaps (dashes) is indicated at the top. Unless specified, composite numbering is used in descriptions. N-terminal domain: sites 48–126, marked by black brackets; C-terminal domain: sites 214–374, marked by red brackets. Two blue arrows indicate the boundary between exons 1 and 2 and the boundary between exons 2 and 3, respectively. The seven numbers (1–7) shown within coloured heart symbols indicate the seven indels. Red heart symbols represent deletions in C. canadensis, the yellow heart symbol represents a deletion in the BW group (C. macrophylla, C. sanguinea and C. controversa) and green heart symbols represent deletions shared by the DW (C. canadensis), BB (C. florida) and CC (C. officinalis) groups and by the DW (C. canadensis) and BB (C. florida) groups. The red star indicates the conserved histidine residue (His at site 294 of CorLFY) in the C-terminal domain, which was predicted to play an essential role in DNA-binding activity (Maizel et al., 2005). The complete coding region was obtained only in C. canadensis; question marks indicate missing data in the other species.
Analyses of gene expression using PCR
Expression of CorLFY in early and late development of inflorescences and young leaves was investigated by PCR using the cDNA libraries derived from total RNA samples. Species-specific PCR primers (P-I) in CorLFY binding to exons II and III, respectively, were designed and used to amplify a fragment of 150 bp spanning exons II and III. The primer sequences are listed in Table 2. The amplicons from each species and each type of organ (i.e. leaf, bract and flower) were obtained by 30 cycles of PCR and sequenced to confirm their identity. The PCR analyses were repeated at least once for the same cDNA samples with each pair of primers for all six species. For the three species with involucral bracts (bracts appearing to subtend the entire inflorescence) (C. florida, C. canadensis and C. officinalis), PCR analyses were further repeated using different cDNA samples from at least one additional biological replicate. For all PCR analyses, we were able to use 26S rDNA as the internal control. Although 26S rDNA is usually not expected to be present in the cDNA libraries prepared using oligo(T) due to the lack of a poly(A) tail in 26S rRNA molecules, polyadenylation of 26S rRNA was found in Nicotiana shoots (Lewandowska et al., 2007) and reverse transcription of 18S rRNA with poly(dT)18 and other homopolymers was successful in several diverse flowering plant species representing bryophytes, ferns, monocots and eudicots (Bogdanović et al., 2013). Our sequencing results for the PCR products amplified using 26S rDNA primers confirmed the amplicon as a part of the gene. PCR controls using an RNA sample in an amount similar to that of the cDNA samples with the species-specific primers or 26S rDNA primers did not amplify a band.
Analyses of gene expression using RNA in situ hybridization
RNA in situ hybridization was used to examine the spatial expression pattern of CorLFY during early development of inflorescences and flowers. Young inflorescence buds at Stages I–IV were collected in summer and autumn from the six species. The sampled inflorescence buds were fixed in formaldehyde for at least 8 h at 4 °C and then dehydrated in a series of cold ethanol concentrations, permeated with an analytical grade xylene (Fisher, Fair Lawn, NJ, USA) series and embedded in Paraplast Plus (Fisherbrand, Houston, TX, USA) for sectioning, following Feng et al. (2012).
Before in situ hybridization, RNA probes were transcribed in vitro following the methods described in Franks et al. (2002) using the primers listed in Table 2. The probes were approximately 400 bp long and were derived from the first exon of CorLFY. The in situ hybridization protocol followed that of Feng et al. (2012), which was modified from Franks et al. (2002), Kim et al. (2005) and Souer et al. (2008). The tissues were sectioned at a thickness of 8 μm using a microtome and mounted on slides. The slides were treated with proteinase K (0·5 ng μl−1) at 37 °C for 30 min and hybridized with species-specific probes (0·8–1·5 ng μl−1) at 60 °C overnight. Slides were washed twice in 0·2 × SSC at 65 °C with agitation. Signals were detected using Western blue (Promega, Madison, WI, USA). The in situ experiments for each probe were carried out at least twice with several biological replicates (different inflorescence buds) in each experiment. Both positive and negative controls were included in each experiment. We used the antisense probes of CorAP3 from Feng et al. (2012) as positive control (data not shown). A sense strand probe was used as a negative control.
Gene and protein sequence analyses
We performed phylogenetic analyses to determine the evolutionary relationships of CorLFY genes and identify evolutionary changes in gene sequences that may be correlated with the gene expression pattern and inflorescence architectures. The phylogenetic analyses were conducted for both the cDNA sequences and the amino acid sequences of CorLFY with and without inclusion of some LFY-like sequences from the asterid and rosid clades (Table 3). These sequences were randomly selected among the completed cDNA pool of LFY in GenBank. The sequences were first downloaded from NCBI, then translated to the predicted amino acid sequences and aligned in Geneious v5·1 (http://www.geneious.com/; Drummond et al., 2010), followed by manual adjustment. The Cornus LFY sequences were manually added to the matrix. The final matrices, with and without inclusion of some LFY-like sequences from the asterid and rosid clades respectively, contained 1320/1101 bp, 440/367 amino acids (gaps included) and 19/6 taxa. The cDNA sequence matrices were further adjusted based on the protein sequence alignments. Phylogenetic analyses were conducted using the maximum likelihood (ML) methods implemented in RAxML-HPC v7.2.8 (available at http://www.phylo.org/; Miller et al., 2010). The ML analyses for cDNA matrices were performed using the GTR model with a gamma distribution for site variation, all parameter values as ‘estimated’ and rapid bootstrapping of 500 replicates. The JTT model was selected as the best model in the ML analyses of the protein sequences. Trees from ML analyses were rooted using the six rosid species and viewed and edited using MEGA 4.0.2 (Tamura et al., 2007). Analyses including only CorLFY were not rooted.
Table 3.
Taxon sampling for phylogenetic analysis
| Lineage | Order | Family | Species |
|---|---|---|---|
| Rosids | Cornales | Cornaceae | Cornus florida |
| Cornales | Cornaceae | Cornus canadensis | |
| Cornales | Cornaceae | Cornus officinalis | |
| Cornales | Cornaceae | Cornus macrophylla | |
| Cornales | Cornaceae | Cornus sanguinea | |
| Cornales | Cornaceae | Cornus controversa | |
| Ericales | Ericaceae | Impatiens balsamina | |
| Lamiales | Phrymaceae | Mimulus lewisii | |
| Lamiales | Phrymaceae | Mimulus guttatus | |
| Lamiales | Plantaginaceae | Antirrhinum majus | |
| Solanales | Solanaceae | Nicotiana tabacum | |
| Asterales | Asteraceae | Helianthus annuus | |
| Asterales | Asteraceae | Chrysanthemum morifolium | |
| Solanales | Solanaceae | Solanum tuberosum | |
| Lamiales | Scrophulariaceae | Buddleja davidii | |
| Asterids | Fagales | Fagaceae | Castanea mollissima |
| Fagales | Juglandaceae | Juglans regia | |
| Rosales | Rosaceae | Cydonia oblonga | |
| Rosales | Rosaceae | Pyrus communis |
RESULTS
Sequence characteristics of LFY homologues in Cornus
The full-length cDNA sequence of CorLFY was obtained in C. canadensis (C. canaLFY) and cloned from products of single PCR amplification. For the other five species (C. florida, C. officinalis, C. macrophylla, C. sanguinea and C. controversa), approximately 70 bp of the 3′ end of the cDNA coding sequences were missing (C. floLFY, C. offiLFY, C. macroLFY, C. sanLFY and C. conLFY) due to failure to obtain the 3′ end of the coding sequence in PCR products using the 3′ RACE approach. Sequencing of multiple clones of PCR products from different species (see Materials and methods) did not detect paralogous sequences (no polymorphisms among clones were observed), indicating that CorLFY is likely to be a single-copy gene in Cornus. In C. canadensis, the CorLFY cDNA coding sequence was 1125 bp long and predicted to encode a protein of 375 amino acids that shared extensive sequence similarity with the Arabidopsis LFY protein. Three exons were predicted based on sequence similarity and conserved splicing sites in LFY of Arabidopsis and confirmed by sequencing of genomic DNA in C. florida. The sizes of the three predicted exons are shown in Table 4. The two conserved domains present in other LFY-like proteins were also found in CorLFY, with the N-terminal domain spanning from amino acid 48 to amino acid 126 and the C-terminal domain from site 214 to site 374 (Fig. 2). In addition, CorLFY was characterized by a proline-rich region (amino acid residues 1–47) and a highly acidic region (amino acid residues 80–88) within the N-terminal domain (amino acid residues 1–138), and another highly acidic region (amino acid residues 192–205) in the second exon (amino acid residues 139 to site 291) (Fig. 2). Thirteen of the 47 amino acid sites were prolines in the proline-rich region. Compared with the conserved N- and C-terminal domains, the proline-rich region and highly acidic region in the interdomain were more variable among Cornus species and contained seven indels (designated indels 1, 2, 3, 4, 5, 6 and 7 in Fig. 2). Three of the seven indels (1, 4 and 5) occurred uniquely in C. canadensis (DW group). Three other indels (3, 6 and 7) were shared by C. canadensis (DW group), C. florida (BB group) and C. officinalis (CC group). Finally, indel 2 was a deletion of one amino acid (site 47) shared by the three species of the BW group (C. macrophylla, C. sanguinea and C. controversa).
Table 4.
Variation in sizes (bp) of the three predicted exons for CorLFY from different species
| Species | First exon | Second exon | Third exon* | Accession number |
|---|---|---|---|---|
| Cornus canadensis L. f. | 414 | 347 | 364 | KC332279 |
| Cornus florida L. | 435 | 359 | 294+ ? | KC332280 |
| Cornus officinalis Seib. & Zucc. | 435 | 362 | 290+ ? | KC332275 |
| Cornus macrophylla Wall. | 435 | 365 | 290+ ? | KC332278 |
| Cornus sanguinea L. | 435 | 371 | 290+ ? | KC332276 |
| Cornus controversa Hemsl. | 435 | 371 | 290+ ? | KC332277 |
| Amino acid sites in Fig. 2 | 1–138 | 139–291 | 292–391 |
* Values followed by ‘?’ are estimated and may range up to approximately 70 bp larger.
Expression of CorLFY detected by PCR analysis
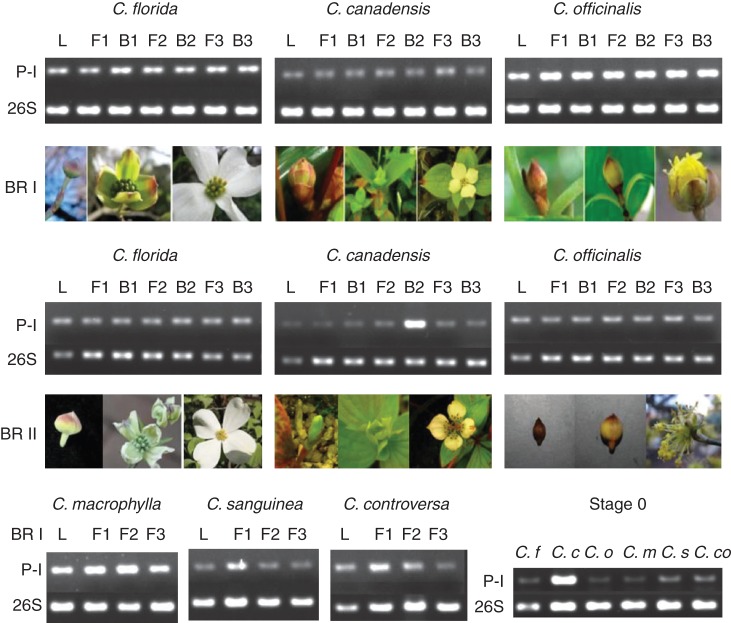
PCR using the species-specific primer sets (P-I) for the cDNA libraries derived from total RNAs resulted in a single DNA amplicon approximately150 bp in length in all six species during early and late inflorescence development (Fig. 3). The P-I primer pairs span the junction between exons 2 and 3 in the CorLFY homologues. The P-I primer pairs detected expression in young leaves, developing bracts, flowers and stage 0 inflorescence samples by PCR of 30 cycles (Fig. 3). However, due to the limited number of biological replicates and the non-quantitative nature of our assay we cannot rule out the possibility of differences in the expression levels of CorLFY homologues that we did not detect by the PCR analyses, either between species or within species, of different tissue or developmental samples.
Fig. 3.
Expression of CorLFY in late developmental stages of Cornus bracts and flowers and young leaves detected by RT–PCR. P-I, RT-PCR using six different pairs of species-specific primers developed for each Cornus species. BR I and BR II indicate two biological replicates; BR II included only the three species with involucral bracts (C. florida, C. canadensis and C. officinalis). For description of the three developmental stages, refer to Table 1. Inflorescence images represent the three stages analysed and are modified from Feng et al. 2012. L, leaf; B1–B3, three late developmental stages of bracts representing unopened bracts, expanding bracts and whitened bracts, respectively; F1–F3: three late developmental stages of flowers associated with the three bract developmental stages. 26S rDNA was used as the internal control. The cDNA concentrations used for PCR were the same as those for 26S, both of which were run at 30 cycles.
Expression of CorLFY detected by RNA in situ hybridization
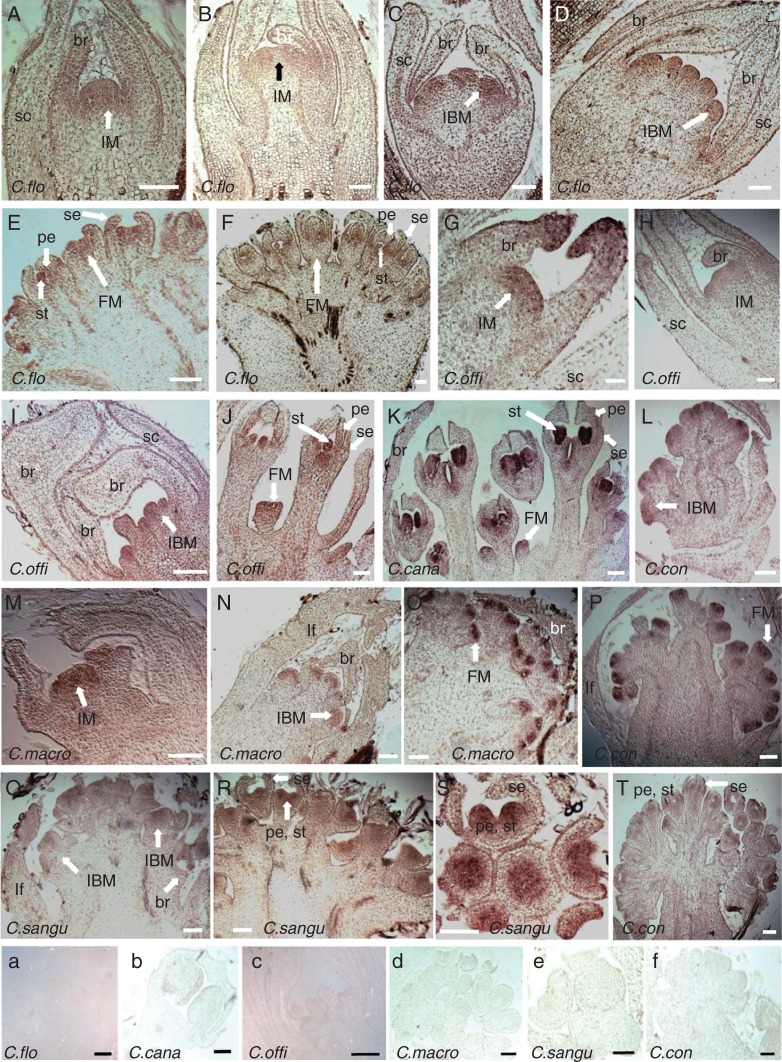
Our in situ hybridization experiments detected expression of CorLFY in the developing inflorescence meristem (IM in Fig. 4A, B, G, K), inflorescence branch meristem (IBM in Fig. 4C, D, H, L, N, Q), in the floral meristem (FM in Fig. 4I, J, M, O) and during early floral organogenesis (Fig. 4E, F, I, J, P, R) of all inflorescence types except the depauperate inflorescences of C. canadensis. Due to lack of materials, we were only able to observe expression in floral meristems and floral organ primordia of C. canadensis (Fig. 4K). For the three BW species, one or two of these stages are also missing due to lack of material (i.e. floral organ stage for C. macrophylla, IM stage for C. sanguinea, and IM and FM for C. controversa; the stages not shown in Fig. 4). Furthermore, we could not examine CorLFY expression at the earlier stages (i.e. when apical meristems are still vegetative) due to difficulty obtaining these samples. The in situ hybridization results obtained showed no apparent differences in the expression patterns of CorLFY among the head-like, umbel-like inflorescences and elongated inflorescence types found in the species studied. Sense-strand negative control probes were used to estimate the levels of non-specific background signal (Fig. 4A–F).
Fig. 4.
Expression pattern of CorLFY in early developmental stages of Cornus inflorescences detected by in situ RNA hybridization. (A–F) Expression of CorLFY in developing inflorescence primordia (IM, IBM, FM, br, se, pe, st) of C. florida. (G–J) Expression of CorLFY in developing inflorescence primordia (IM, IBM, FM, br, se, pe, st) of C. officinalis. (K) Expression of CorLFY in developing inflorescence primordia (FM, se, pe, st) of C. canadensis. (M–O) Expression of CorLFY in developing inflorescence primordia (IM, IBM, FM) meristems of C. macrophylla. (L, P, T) Expression of CorLFY in developing inflorescence meristems (IBM, FM, se, pe, st) of C. controversa. (Q–S) Expression of CorLFY in developing inflorescence primordia (IBM, FM, br, se, pe, st) of C. sanguinea. (a–f) Negative control of CorLFY expression in the six Cornus species: (a) inflorescence meristem of C. florida, comparable to (E); (b) young flower bud of C. canadensis, comparable to one of those in (K); (c) inflorescence branch meristems, comparable to stage in (I); (d) young flower bud of C. macrophylla, comparable to stage in (O); (e) young flower bud of C. sanguinea, comparable to stage in (S); (f) floral meristem of C. controversa, comparable to stage in (P). VM, vegetative meristem; IM, inflorescence meristem; IBM, inflorescence branch meristem; FM, flower meristem; C. flo, C. florida; C. cana, C. canadensis; C. offi, C. officinalis; C. macro, C. macrophylla; C. sangu, C. sanguinea; C. con, C. controversa; lf, leaf; br, bract; se, sepal; pe, petal; st, stamen; sc, scale. Scale bars = 100 μm.
Gene genealogy and evolutionary changes
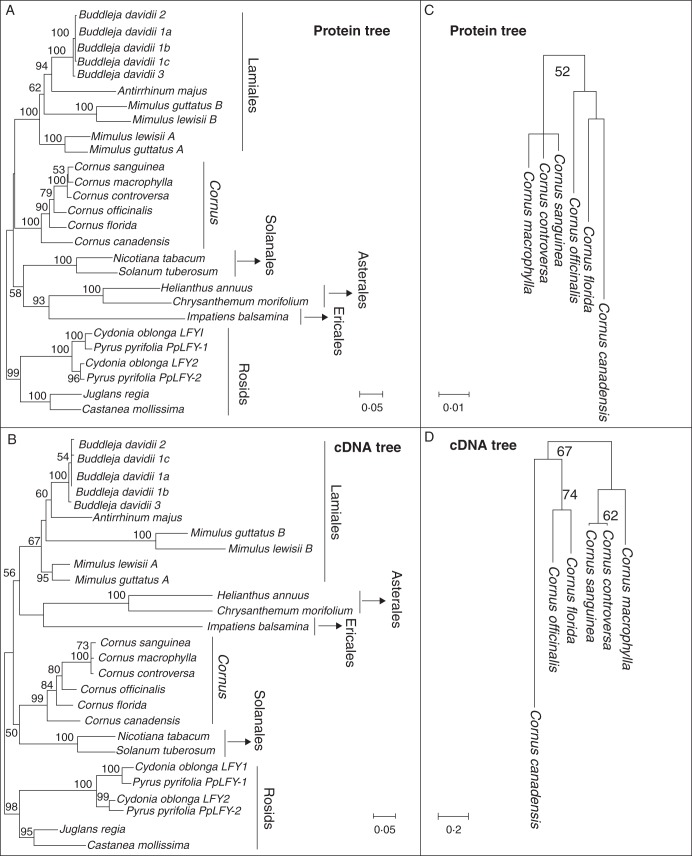
In rooted analysis of cDNA and protein sequences including Cornus and other taxa, the CorLFY sequences formed a well-supported monophyletic group closely allied with LFY homologues from Solanales and Lamiales, respectively (Fig. 5A, B). The LFY-like genes from Solanaceae, Lamiales, Asteraceae and Rosids formed monophyletic clades, but relationships among these clades and Cornus remained unclear (weakly supported) (Fig. 5A, B). However, within the Cornus clade, relationships among the BB, DW, CC and BW lineages were different from those found in phylogenetic studies of Cornus using several molecular markers from plastic and nuclear genomes that included many to all species (Fan and Xiang 2003; Xiang et al., 2006, 2008; Xiang and Thomas, 2008), which showed (BW(CC(BB, DW))). The CorLFY cDNA and protein phylogenies, in contrast, showed (DW(BB(CC,BW))) (Fig. 5A, B). This incongruence could be an artefact of the distant outgroup rooting, ambiguity in alignment between Cornus and the divergent outgroup taxa in variable portions of the interdomain region, or due to other unknown reasons. The sister of Cornus, Alangiaceae, and all other families of Cornales were missing in the phylogenetic analyses due to lack of data. Unknown factors in the molecular evolution of LFY may also contribute to this conflict, e.g. selection and functional constraint. A number of amino acid sites support a closer relationships of BB, CC and DW, as found in previous phylogenetic studies, but a few amino acid changes were shared by the BB and CC species, which bear head-like and umbel-like inflorescences, respectively (i.e. site 37 in indel 1 and site 47 in indel 2; Fig. 2). These changes support a closer sister relationship between BB and CC and are in agreement with the lack of elongation of IM and IBMs in these two lineages, but a closer relationship between lineages BB and CC is in conflict with the previous phylogeny, which showed a closer relationship between BB and DW.
Fig. 5.
Phylogram of LFY orthologues resulting from ML analyses of cDNA sequences and amino acid sequences. Numbers at nodes are bootstrap supports from ML analyses. Only bootstrap support ≥50 % is shown (italicized numbers). (A, B) Rooted trees for (A) proteins and (B)cDNA. (C, D) Unrooted trees for (C) proteins and (D) cDNA.
Results of the unrooted phylogenetic analyses of cDNA and protein sequences including only CorLFY were consistent with the observed pattern of protein sequence variation described above (Fig. 5C, D). The alignment of amino acid sequences of CorLFY also showed that variation at a number of sites is correlated with the divergence of lineages with different inflorescence types (Fig. 2). Three deletions, of sites 35–41, 173–175 and 197, and 10 other unique substitutions were observed in C. canadensis of the DW group (four small minidichasia). Eight, six and 15 substitutions were observed for C. florida (head-like inflorescence), C. officinalis (umbel-like inflorescence) and the BW group with thyrsoids with elongated internodes, respectively. These substitutions can be observed in the aligned matrix shown in Fig. 2.
DISCUSSION
Expression of CorLFY during inflorescence development and the evolution of Cornus inflorescence architectures
Flowering plants exhibit a diverse array of inflorescence architectures. Despite the importance of the inflorescence in angiosperm reproduction and evolution, little progress has been made in understanding the developmental-genetic bases of inflorescence evolution. This is largely because the genetic programme controlling inflorescence development is complicated and remains unknown in most species. Nonetheless, available data have supported key roles of LFY and TFL1 in structuring racemes and cymes (Souer et al., 1998, 2008; Molinero-Rosales et al., 1999; Ahearn et al., 2001; Jack, 2004; Conti and Bradley, 2007; Prusinkiewicz et al., 2007; Koes, 2008; Thouet et al., 2012).
By in situ hybridization and PCR on cDNA clones, we detected expression of CorLFY mRNA in the IM, IBMs, FMs and developing floral organs in head-like inflorescences, umbel-like inflorescences and elongated inflorescences of different Cornus species examined (Figs 3 and 4). This expression of CorLFY is consistent with the expectation that CorLFY plays a role in the specification of inflorescence and floral meristem identity in Cornus. However, our experiments did not show any differences in the levels or patterns of CorLFY expression between the three different inflorescence types in Cornus that might have helped us to define the molecular mechanisms that underlie the divergent morphologies. A lower level of CorLFY activity in IM and emerging IBMs might be expected in the species with more highly branched inflorescence architectures and elongated inflorescence branches, as has been demonstrated in petunia and tobacco (Souer et al., 1998; Amaya et al., 1999; Ahearn et al., 2001). Thus, one might predict that lower levels of CorLFY activity would be functioning in the IM and IBMs of the BW species (e.g. C. macrophylla, C. controversa and C. sanguinea) producing elongated inflorescences and more highly branched inflorescences, relative to those species producing head-like or umbel-like inflorescences (e.g. C. florida and C. officinalis). Further experiments are needed to test this hypothesis. Although the nature of our experiments did not allow us to observe differences in the CorLFY expression patterns in these different inflorescence types, it remains possible that quantitative differences in expression level exist among the species. It is also possible that the CorLFY gene products are regulated at a post-transcriptional level, and differences in CorLFY sequences among the Cornus species may play a role at this level. Additionally, the differential development of these inflorescence types could also be attributed to functions of TFL1 or other proteins (e.g. UFO homologues).
In petunia and tomato, the expression of floral meristem identity was found to be regulated via transcription of DOUBLE TOP (DOT)/ANANTHA, a homologue of UNUSUAL FLORAL ORGANS (UFO) rather than by the transcription of LFY (Souer et al., 2008). DOT was found to activate ALF (homologue of LFY) by a post-translational mechanism in petunia (Souer et al., 2008). The study by Souer et al. (2008) suggested that divergent expression patterns of LFY and UFO homologues supported differential spatiotemporal control of floral identity in these distinct species. The expression data we present here suggest that either (1) the expression of CorLFY at the transcription level does not play a key role in regulating the evolutionary shift from elongated, highly branched inflorescences to condensed head-like and umbel-like inflorescences in Cornus, or (2) the transcription level of CorLFY does play a role but at a quantitative level that was not detected in this study. The divergence in spatiotemporal control of floral identity in different dogwood lineages may also involve differences in the activity of the CorLFY protein or the expression or activities of CorTFL1 and CorUFO. We hope to test this in the future using the genetic transformation system we have established in C. canadensis (Liu et al., 2013).
Sequence characterization and evolution of CorLFY
LFY homologues exist as two copies in gymnosperm species (Mouradov et al., 1998; Mellerowicz et al., 1998; Frohlich and Parker, 2000) and as a single copy in most angiosperms (Coen et al., 1990; Weigel et al., 1992; Souer et al., 1998; Molinero-Rosales et al., 1999; Kyozuka et al., 1998; Rottmann et al., 2000; Carmona et al., 2002; An et al., 2011). The fact that two or more copies of LFY homologues found in a number of distantly related angiosperm lineages, such as Ionopsidium acaule (violet cress; Shu et al., 2000) and Idaho ascapigera (Sliwinski et al., 2007) (Brassicaceae), Nicotiana (Ahearn et al., 2001) (Solanaceae), several fruit tree species of Rosaceae (Wada et al., 2002; Esumi et al., 2005), Allium sativum (Rotem et al., 2007) (Alliaceae) and Zea mays (Bomblies et al., 2003) (Poaceae), demonstrates that duplication of LFY occurred multiple times during angiosperm evolution. Our sequencing of cloned PCR products (>20 clones from the three fragments) from degenerate primers (designed from aligned sequences of asterids) failed to detect any paralogous sequences within a species, indicating that CorLFY likely exists in Cornus as a single copy, as it does in most angiosperms.
Like most LFY homologues found in other angiosperm species, the six CorLFY genes were predicted to have three exons and the predicted proteins have two conserved regions, an N-terminal domain and a C-terminal domain (Fig. 2). The predicted amino acid sequences of CorLFY share some other motifs with previously characterized LFY proteins, including a proline-rich region and a highly acidic region in the second exon (Fig. 2). The proline-rich region is well conserved in LFY homologues of angiosperms but is not well conserved in those of gymnosperms (Frohlich and Meyerowitz, 1997; Carmona et al., 2002). However, the proline-rich motif was later found to be absent from FLO/LFY homologues of some angiosperm species, such as Malus domestica (apple; Wada et al., 2002) and Eucalyptus (Dornelas et al., 2004). In Eucalyptus, homologues lacking the proline-rich region are able to restore the wild type phenotype of lfy mutants, which suggests that the proline-rich region may not be functionally significant in this species (Dornelas et al., 2004). Maizel et al. (2005) have demonstrated that the C-terminal domain of LFY functions as a DNA-binding domain. In the present study, we also observed a conserved histidine residue in the C-terminal domain of CorLFY (His at site 294; Fig. 2) previously shown to be important for LFY DNA-binding activity (Maizel et al., 2005). A number of amino acid changes were observed in the C domain (Fig. 2). It is unclear whether these changes may affect the role of the protein in its regulatory function on inflorescence development in the different dogwood lineages. In addition to the proline-rich and DNA-binding domains described above, there is a highly acidic region in the N-terminal domain of the first exon of CorLFY. Our alignment of CorLFY with previously identified LFY-like proteins confirmed that the N-terminal domain is conserved throughout angiosperm LFY homologues (data not shown).
Among the six CorLFY sequences, many changes in the gene and protein sequences were found, with the C. canadensis sequences showing the greatest number of changes (Fig. 2). These changes were dispersed in different regions, with the indels restricted to the interdomain and proline-rich regions, while amino acid substitutions were observed in all regions.
Expression of CorLFY in vegetative organs and in flowers
Expression of LFY homologues in the vegetative shoot apical meristem and leaf primordia has previously been reported in many angiosperms, such as Nicotiana tabacum (Kelly et al., 1995), Arabidopsis (Blázquez et al., 1997; Hempel et al., 1997), Impatiens sp. (Pouteau et al., 1997), Solanum lycopersicum (Molinero-Rosales et al., 1999), Ionopsidium acaule (Shu et al., 2000), Populus (Rottmann et al., 2000), Malus domestica (Wada et al., 2002), Carica papaya (Yu et al., 2005), Idahoa (Sliwinski et al., 2007), Silene (Allnutt et al., 2007), Oryza sativa (Rao et al., 2008) and Dendranthema (Ma et al., 2008), which suggests a role of LFY in leaf organogenesis. However, the exact mechanism is not yet completely understood. Li et al. (2013) demonstrated that LFY stimulates the formation of floral and leaf primordia through auxin-regulated pathways. Expression of LFY homologues in developing leaves has also been reported in many investigated angiosperm species (Kelly et al., 1995; Bradley et al., 1997; Blázquez et al., 1997; Hofer et al., 1997; Pouteau et al., 1997; Molinero-Rosales et al., 1999; Rottmann et al., 2000; Souer et al., 1998; Southerton et al., 1998; Walton et al., 2001; Carmona et al., 2002; Dong et al., 2005; Ma et al., 2008), including Cornus in this study (Fig. 3). In pea and tomato plants, lfy mutants showed changes in leaf morphology, confirming the function of LFY homologues in leaf development (Hofer et al., 1997; Molinero-Rosales et al., 1999). Interestingly, expression of LFY homologues in vegetative meristems was not detected in the snapdragon (Coen et al., 1990), rubber tree (Dornelas and Rodriguez, 2005) and Populus tomentosa (An et al., 2011). However, the lack of expression of LFY homologues in vegetative meristems of rubber tree was attributed to a season-dependent mechanism that regulates its expression (Dornelas and Rodriguez, 2005). Seasonally dependent expression of LFY homologues has also been reported in other woody species, such as kiwifruit, grape and apple (Walton et al., 2001; Carmona et al., 2002; Wada et al., 2002; Almada et al., 2009). Therefore, LFY homologues may have a role in leaf development that is largely conserved across angiosperm lineages. It has been suggested that LFY orthologues have a general role in regulating indeterminacy in lateral primordia derived from apical meristems, and that this role may reflect an ancestral function of the gene (Hofer et al., 1997).
LFY-like genes have been found to play key roles in the transition from inflorescence meristem to floral meristem in a diversity of plants (Weigel et al., 1992; Weigel and Nilsson, 1995; Blázquez et al., 1997; Nilsson et al., 1998; Molinero-Rosales et al., 1999; Pidkowich et al., 1999; Peña et al., 2001; Wada et al., 2002; Bomblies et al., 2003; Kim et al., 2005; Zhang et al., 2010). In our study, expression in floral meristems and during floral organogenesis was observed for all species studied (Figs 3 and 4). These results and the expression of CorLFY throughout the development of inflorescences are consistent with the expectation that activity of CorLFY is likely required in floral and inflorescence development in Cornus, as has been suggested in other plants.
ACKNOWLEDGEMENTS
We thank the J. C. Raulston Arboretum for providing living plants for experiments, the phytotron at North Carolina State University for caring for cultured plants and D. E. Boufford for correction of the English and assistance in collecting C. canadensis from the field. We thank April Wynn for assistance with the in situ hybridization experiments. The study benefitted from a CAS/SAFEA International Partnership Program for Creative Research Teams. The research was supported by a National Science Foundation grant (IOS-10 24629) and Chinese Education Ministry [China Scholarship Council, Grant No. (2010) 3006, to J. L.].
LITERATURE CITED
- Aagaard J, Willis J, Phillips P. Duplication of floral regulatory genes in the Lamiales. American Journal of Botany. 2005;92:1284–1293. doi: 10.3732/ajb.92.8.1284. [DOI] [PubMed] [Google Scholar]
- Ahearn KP, Johnson HA, Weigel D, Wagner DR. NFL1, a Nicotiana tabacum LEAFY-like gene, controls meristem initiation and floral structure. Plant Cell Physiology. 2001;42:1130–1139. doi: 10.1093/pcp/pce143. [DOI] [PubMed] [Google Scholar]
- Allnutt GV, Rogers HJ, Francis D, Herbert RJ. A LEAFY-like gene in the long-day plant, Silene coeli-rosa is dramatically up-regulated in evoked shoot apical meristems but does not complement the Arabidopsis lfy mutant. Journal of Experimental Botany. 2007;58:2249–2259. doi: 10.1093/jxb/erm090. [DOI] [PubMed] [Google Scholar]
- Almada R, Cabrera N, Casaretto J, Ruiz-Lara S, Villanueva EG. VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Reports. 2009;28:1193–1203. doi: 10.1007/s00299-009-0720-4. [DOI] [PubMed] [Google Scholar]
- Amaya I, Ratcliffe OJ, Bradley DJ. Expression of the CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell. 1999;11:1405–1417. doi: 10.1105/tpc.11.8.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An XM, Wang DM, Wang ZL, et al. Isolation of a LEAFY homolog from Populus tomentosa: expression of PtLFY in P. tomentosa floral buds and PtLFY-IR-mediated gene silencing in tobacco (Nicotiana tabacum) Plant Cell Reports. 2011;30:89–100. doi: 10.1007/s00299-010-0947-0. [DOI] [PubMed] [Google Scholar]
- Archambault A, Bruneaua A. Phylogenetic utility of the LEAFY/FLORICAULA gene in the Caesalpinioideae (Leguminosae): gene duplication and a novel insertion. Systematic Botany. 2004;29:609–626. [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueño F. Floral initiation and inflorescence architecture: a comparative view. Annals of Botany. 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- Bogdanović MD, Dragićević MB, Tanić NT, et al. Reverse transcription of 18S rRNA with Poly(dT)18 and other homopolymers. Plant Molecular Biology Reporter. 2013;31:55–63. [Google Scholar]
- Bomblies K, Wang RL, Ambrose BA, Schmidt RJ, Meeley RB, Doebley J. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development. 2003;130:2385–2395. doi: 10.1242/dev.00457. [DOI] [PubMed] [Google Scholar]
- Bosch JA, Heo K, Sliwinski MK, Baum DA. An exploration of LEAFY expression in independent evolutionary origins of rosette flowering in Brassicaceae. American Journal of Botany. 2008;95:286–293. doi: 10.3732/ajb.95.3.286. [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- Busch A, Gleissberg S. EcFLO, a FLORICAULA-like gene from Eschscholzia californica is expressed during organogenesis at the vegetative shoot apex. Planta. 2003;217:841–848. doi: 10.1007/s00425-003-1046-z. [DOI] [PubMed] [Google Scholar]
- Carmona MJ, Cubas P, Martinez-Zapater JM. VFL, the grapevine FLORICAULA/LEAFY ortholog, is expressed in meristematic regions independently of their fate. Plant Physiology. 2002;130:68–77. doi: 10.1104/pp.002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Tan QK, Hill TA, Irish VF. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135:1235–1245. doi: 10.1242/dev.015842. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Chujo A, Zhang Z, Kishino H, Shimamoto K, Kyozuka J. Partial conservation of LFY function between rice and Arabidopsis. Plant Cell Physiology. 2003;44:1311–1319. doi: 10.1093/pcp/pcg155. [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. Floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Conti L, Bradley D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell. 2007;19:767–778. doi: 10.1105/tpc.106.049767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong ZC, Zhao Z, Liu CW, et al. Floral patterning in Lotus japonicus. Plant Physiology. 2005;137:1272–1282. doi: 10.1104/pp.104.054288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornelas MC, Amaral WAN, Rodriguez APM. EgLFY, the Eucalyptus grandis homolog of the Arabidopsis gene LEAFY is expressed in reproductive and vegetative tissues. Brazilian Journal of Plant Physiology. 2004;16:105–114. [Google Scholar]
- Dornelas MC, Rodriguez AP. The rubber tree (Hevea brasiliensis Muell. Arg.) homologue of the LEAFY/FLORICAULA gene is preferentially expressed in both male and female floral meristems. Journal of Experimental Botany. 2005;56:1965–1974. doi: 10.1093/jxb/eri194. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, et al. Geneious. v5. 1. Auckland, New Zealand: Biomatters Ltd; 2010. [Google Scholar]
- Endress PK. Disentangling confusions in inflorescence morphology: patterns and diversity of reproductive shoot ramification in angiosperms. Journal of Systematics and Evolution. 2010;48:225–239. [Google Scholar]
- Esumi T, Tao R, Yonemori K. Isolation of LEAFY and TERMINAL FLOWER 1 homologues from six fruit tree species in the subfamily Maloideae of the Rosaceae. Sexual Plant Reproduction. 2005;17:277–287. [Google Scholar]
- Fan CZ, Xiang Q-Y. Phylogenetic analyses of Cornales based on 26S rDNA and combined 26S rDNA-matK-rbcL sequence data. American Journal of Botany. 2003;90:1357–1372. doi: 10.3732/ajb.90.9.1357. [DOI] [PubMed] [Google Scholar]
- Feng CM, Xiang QY, Franks RG. Phylogeny-based developmental analyses illuminate evolution of inflorescence architectures in dogwoods (Cornus s. l., Cornaceae) New Phytologist. 2011;191:850–869. doi: 10.1111/j.1469-8137.2011.03716.x. [DOI] [PubMed] [Google Scholar]
- Feng CM, Liu X, Yu Y, Xie DY, Franks RG, Xiang QY. Evolution of bract development and B-class MADS box gene expression in petaloid bracts of Cornus s. l. (Cornaceae) New Phytologist. 2012;196:631–643. doi: 10.1111/j.1469-8137.2012.04255.x. [DOI] [PubMed] [Google Scholar]
- Flachowsky H, Hattasch C, Hofer M, Peil A, Hanke MV. Overexpression of LEAFY in apple leads to a columnar phenotype with shorter internodes. Planta. 2010;231:251–263. doi: 10.1007/s00425-009-1041-0. [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang CX, Levin JZ, Liu ZC. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development. 2002;129:253–263. doi: 10.1242/dev.129.1.253. [DOI] [PubMed] [Google Scholar]
- Frohlich MW, Meyerowitz EM. The search for flower homeotic gene homologs in basal angiosperms and Gnetales: a potential new source of data on the evolutionary origin of flowers. International Journal of Plant Sciences. 1997;158:131–142. [Google Scholar]
- Frohlich MW, Parker DS. The mostly male theory of flower evolution origins: from genes to fossils. Systematic Botany. 2000;25:155–170. [Google Scholar]
- Hamès C, Ptchelkine D, Grimm C, et al. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO Journal. 2008;27:2628–2637. doi: 10.1038/emboj.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EM. Capitula in the Asteridae: a widespread and varied phenomenon. Botanical Review. 1999;65:348–369. [Google Scholar]
- Harris JG, Harris MW. Plant identification terminology. Spring Lake, UT: Spring Lake Publishing; 2001. [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, et al. Floral determination and expression of floral regulatory genes in Arabidopsis. Development. 1997;124:3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, et al. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Current Biology. 1997;7:581–587. doi: 10.1016/s0960-9822(06)00257-0. [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh JI, Nagato Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant Journal. 2012;69:168–180. doi: 10.1111/j.1365-313X.2011.04781.x. [DOI] [PubMed] [Google Scholar]
- Jack T. Molecular and genetic mechanism of floral control. Plant Cell. 2004;16:S1–S17. doi: 10.1105/tpc.017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Bonnlander MB, Meeks-Wagner DR. NFL, the tobacco homolog of FLORICAULA and LFY, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell. 1995;7:225–234. doi: 10.1105/tpc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Koh J, Ma H, et al. Sequence and expression studies of A-, B-, and E-class MADS-box homologues in Eupomatia (Eupomatiaceae): support for the bracteate origin of the calyptra. International Journal of Plant Sciences. 2005;166:185–198. [Google Scholar]
- Koes R. Evolution and development of virtual inflorescences. Trends in Plant Science. 2008;13:1–3. doi: 10.1016/j.tplants.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K. Down-regulation of RFL, the FLO/LFY homolog of rice accompanied with panicle branch initiation. Proceedings of the National Academy of Sciences of the USA. 1998;95:1979–1982. doi: 10.1073/pnas.95.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska M, Borcz B, Kamińska J, Wawrzyński A, Sirkoet A. Polyadenylation and decay of 26S rRNA as part of Nicotiana tabacum response to cadmium. Acta Biochemica Polonica. 2007;54:747–755. [PubMed] [Google Scholar]
- Li W, Zhou Y, Liu X, Yu P, Cohen JD, Meyerowitz EM. LEAFY controls auxin response pathways in floral primordium formation. Science Signaling. 2013;6 doi: 10.1126/scisignal.2003937. pra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Feng CM, Franks R, Qu R, Xie DY, Xiang QY. Plant regeneration and genetic transformation of C. canadensis: a non-model plant appropriate for investigation of flower development in Cornus (Cornaceae) Plant Cell Report. 2013;32:77–87. doi: 10.1007/s00299-012-1341-x. [DOI] [PubMed] [Google Scholar]
- Ma YP, Fang XH, Chen F, Dai SL. DFL, a FLORICAULA/LEAFY homologue gene from Dendranthema lavandulifolium is expressed both in the vegetative and reproductive tissues. Plant Cell Reports. 2008;27:647–654. doi: 10.1007/s00299-007-0489-2. [DOI] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, et al. The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science. 2005;308:260. doi: 10.1126/science.1108229. [DOI] [PubMed] [Google Scholar]
- Martinez-Castilla LP, Alvarez-Buylla ER. Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proceedings of the National Academy of Sciences of the USA. 2003;100:13407–13412. doi: 10.1073/pnas.1835864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Horgan K, Walden A, Coker A, Walter C. Pinus radiata homologue of FLORICAULA and LEAFY is expressed in buds containing vegetative shoot and undifferentiated male cone primordia. Planta. 1998;206:619–629. doi: 10.1007/s004250050440. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010. New Orleans, LA. 2010:1–8. [Google Scholar]
- Molinero-Rosales N, Jamilena M, Zurita S, Gomez P, Capel J, Lozano R. FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant Journal. 1999;20:685–693. doi: 10.1046/j.1365-313x.1999.00641.x. [DOI] [PubMed] [Google Scholar]
- Mouradov A, Sawbridge T, Hamdorf B, et al. Genetic engineering of reproductive sterility in Pinus radiata. Acta Horticulturae. 1998;461:417–423. [Google Scholar]
- Nilsson O, Lee I, Blázquez MA, Weigel D. Flowering time genes modulate the response to LEAFY activity. Genetics. 1998;150:403–410. doi: 10.1093/genetics/150.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. A genetic framework for floral patterning. Nature. 1998;395:561–566. doi: 10.1038/26903. [DOI] [PubMed] [Google Scholar]
- Peña L, Martín-Trillo M, Juárez J, Pina JA, Navarro L, Martínez-Zapater JM. Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nature Biotechnology. 2001;19:263–267. doi: 10.1038/85719. [DOI] [PubMed] [Google Scholar]
- Pidkowich MS, Klenz JE, Haughn GW. The making of a flower: control of floral meristem identity in Arabidopsis. Trends in Plant Science. 1999;4:64–70. doi: 10.1016/s1360-1385(98)01369-7. [DOI] [PubMed] [Google Scholar]
- Pouteau S, Nicholls D, Tooke F, Coen E, Battey N. The induction and maintenance of flowering in Impatiens. Development. 1997;124:3343–3351. doi: 10.1242/dev.124.17.3343. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- Rao NN, Prasad K, Kumar PR, Vijayraghavan U. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proceedings of the National Academy of Sciences of the USA. 2008;105:3646–3651. doi: 10.1073/pnas.0709059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Current Opinion in Plant Biology. 2000;3:423–434. doi: 10.1016/s1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- Rotem N, Shemesh E, Peretz Y, et al. Reproductive development and phenotypic differences in garlic are associated with expression and splicing of LEAFY homologue gaLFY. Journal of Experimental Botany. 2007;58:1133–1141. doi: 10.1093/jxb/erl272. [DOI] [PubMed] [Google Scholar]
- Rottmann WH, Meilan R, Sheppard LA, et al. Diverse effects of overexpression of Leafy and PTLF, a poplar (Populus) homolog of Leafy/Floricaula, in transgenic poplar and Arabidopsis. Plant Journal. 2000;22:235–246. doi: 10.1046/j.1365-313x.2000.00734.x. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell. 1991;3:771–781. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N, Takagishi A, Ikari C, Takumi S, Murai K. WFL, a wheat FLORICAULA/LEAFY ortholog, is associated with spikelet formation as lateral branch of the inflorescence meristem. Genes & Genetic Systems. 2006;81:13–20. doi: 10.1266/ggs.81.13. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Shih MC, Li WH. Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiology. 2005;139:18. doi: 10.1104/pp.105.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu G, Amaral W, Hileman LC, Baum DA. LEAFY and the evolution of rosette flowering in violet cress (Ionopsidium acaule, Brassicaceae) American Journal of Botany. 2000;87:634–641. [PubMed] [Google Scholar]
- Siriwardana NS, Lamb RS. The poetry of reproduction: the role of LEAFY in Arabidopsis thaliana flower formation. International Journal of Developmental Biology. 2012;56:207–221. doi: 10.1387/ijdb.113450ns. [DOI] [PubMed] [Google Scholar]
- Sliwinski MK, White MA, Maizel A, Weigel D, Baum DA. Evolutionary divergence of LFY function in the mustards Arabidopsis thaliana and Leavenworthia crassa. Plant Molecular Biology. 2006;62:279–289. doi: 10.1007/s11103-006-9020-3. [DOI] [PubMed] [Google Scholar]
- Sliwinski MK, Bosch JA, Yoon HS, Balthazar M, Baum DA. The role of two LEAFY paralogs from Idahoa scapigera (Brassicaceae) in the evolution of a derived plant architecture. Plant Journal. 2007;51:211–219. doi: 10.1111/j.1365-313X.2007.03148.x. [DOI] [PubMed] [Google Scholar]
- Souer E, van der Krol A, Kloos D, et al. Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development. 1998;125:733–742. doi: 10.1242/dev.125.4.733. [DOI] [PubMed] [Google Scholar]
- Souer E, Rebocho AB, Bliek M, Kusters E, de Bruin RA, Koes R. Patterning of inflorescences and flowers by the F-Box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell. 2008;20:2033–2048. doi: 10.1105/tpc.108.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southerton SG, Strauss SH, Olive MR, et al. Eucalyptus has a functional equivalent of the Arabidopsis floral meristem identity gene LEAFY. Plant Molecular Biology. 1998;37:897–910. doi: 10.1023/a:1006056014079. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4·0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thouet J, Quinet M, Lutts S, Kinet JM, Périlleux C. Repression of floral meristem fate is crucial in shaping tomato inflorescence. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031096. e31096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Cao QF, Kotoda N, Soejima J, Masuda T. Apple has two orthologues of FLORICAULA/LEAFY involved in flowering. Plant Molecular Biology. 2002;49:567–577. doi: 10.1023/a:1015544207121. [DOI] [PubMed] [Google Scholar]
- Walton EF, Podivinsky E, Wu RM. Bimodal pattern of floral gene expression over the two seasons that kiwifruit flowers develop. Physiologia Plantarum. 2001;111:396–404. doi: 10.1034/j.1399-3054.2001.1110318.x. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. Activation of floral homeotic genes in Arabidopsis. Science. 1993;261:1723–1726. doi: 10.1126/science.261.5129.1723. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Wreath S, Bartholmes C, Hidalgo O, Scholz A, Gleissberg S. Silencing of EcFLO, a FLORICAULA/LEAFY gene of the California poppy (Eschscholzia californica), affects flower specification in a perigynous flower context. International Journal of Plant Sciences. 2013;174:139–153. [Google Scholar]
- Xiang Q-Y, Thomas DT. Tracking character evolution and biogeographic history through time in Cornaceae – does choice of methods matter? Journal of Systematics and Evolution. 2008;46:349–374. [Google Scholar]
- Xiang Q-Y, Thomas DT, Zhang W-H, Manchester SR, Murrell Z. Species level phylogeny of the dogwood genus Cornus (Cornaceae) based on molecular and morphological evidence– implication in taxonomy and Tertiary intercontinental migration. Taxon. 2006;55:9–30. [Google Scholar]
- Xiang Q-Y, Thorne JL, Seo T, Zhang W, Thomas DT, Ricklefs RE. Rates of nucleotide substitution in Cornaceae (Cornales) – pattern of variation and underlying causal factors. Molecular Phylogenetics and Evolution. 2008;49:327–342. doi: 10.1016/j.ympev.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Yamaguchi A, Abe M, Wagner D, Komeda Y. LEAFY controls Arabidopsis pedicel length and orientation by affecting adaxial-abaxial cell fate. Plant Journal. 2012;69:844–856. doi: 10.1111/j.1365-313X.2011.04836.x. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Baum DA. Transgenic study of parallelism in plant morphological evolution. Proceedings of the National Academy of Sciences of the USA. 2004;101:6524–6529. doi: 10.1073/pnas.0401824101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QY, Moore PH, Albert HH, Roader AHK, Ming R. Cloning and characterization of a FLORICAULA/LEAFY ortholog, PFL, in polygamous papaya. Cell Research. 2005;15:576–584. doi: 10.1038/sj.cr.7290327. [DOI] [PubMed] [Google Scholar]
- Zhang JX, Wu KL, Zeng SJ, Duan J, Tian LN. Characterization and expression analysis of PhalLFY, a homologue in Phalaenopsis of FLORICAULA/LEAFY genes. Scientia Horticulturae. 2010;124:482–489. [Google Scholar]