Background: Intermediate filament (IF) proteins, including keratin 8 and GFAP, are regulated by serine phosphorylation, whereas phospho-tyrosine sites are poorly understood.
Results: Keratin 8 phospho-Tyr-267 is dephosphorylated by PTP1B and promotes insolubility and filament organization, as does the paralogous GFAP tyrosine.
Conclusion: Tyrosine phosphorylation is important for IF organization and dynamics.
Significance: These findings may provide a pathogenesis model for GFAP mutations in Alexander disease patients.
Keywords: Cytoskeleton, Intermediate Filaments, Phosphotyrosine, Post-translational Modification, Protein-tyrosine Phosphatase (Tyrosine Phosphatase), Alexander Disease, GFAP, Keratin, Tyrosine Phosphatase
Abstract
Post-translational modifications are important functional determinants for intermediate filament (IF) proteins. Phosphorylation of IF proteins regulates filament organization, solubility, and cell-protective functions. Most known IF protein phosphorylation sites are serines localized in the variable “head” and “tail” domain regions. By contrast, little is known about site-specific tyrosine phosphorylation or its implications on IF protein function. We used available proteomic data from large scale studies to narrow down potential phospho-tyrosine sites on the simple epithelial IF protein keratin 8 (K8). Validation of the predicted sites using a pan-phosphotyrosine and a site-specific antibody, which we generated, revealed that the highly conserved Tyr-267 in the K8 “rod” domain was basally phosphorylated. The charge at this site was critically important, as demonstrated by altered filament organization of site-directed mutants, Y267F and Y267D, the latter exhibiting significantly diminished solubility. Pharmacological inhibition of the protein-tyrosine phosphatase PTP1B increased K8 Tyr-267 phosphorylation, decreased solubility, and increased K8 filament bundling, whereas PTP1B overexpression had the opposite effects. Furthermore, there was significant co-localization between K8 and a “substrate-trapping” mutant of PTP1B (D181A). Because K8 Tyr-267 is conserved in many IFs (QYE motif), we tested the effect of the paralogous Tyr in glial fibrillary acidic protein (GFAP), which is mutated in Alexander disease (Y242D). Similar to K8, Y242D GFAP exhibited highly irregular filament organization and diminished solubility. Our results implicate the rod domain QYE motif tyrosine as an important determinant of IF assembly and solubility properties that can be dynamically modulated by phosphorylation.
Introduction
Keratin 8 (K8)2 is a cytoskeletal intermediate filament (IF) protein expressed in simple epithelial cells, including those in liver, intestine, and exocrine pancreas. As a major structural protein, K8 enables cells to withstand mechanical stress, a functional property shared by other IF proteins (1). Aside from keratins, human IF proteins also include the nuclear lamins, neurofilaments, glial fibrillary acidic protein (GFAP), vimentin, and desmin, among others (2). More than 70 human diseases, including neuropathies, myopathies, and digestive and skin diseases, are caused or aggravated by mutations in IF genes (3, 4). Understanding the regulation of the various IF proteins is a necessary step to a better appreciation for the mechanisms and potential targets for IF-related diseases.
With respect to K8, its roles in the maintenance of healthy digestive epithelia are tied to cellular signaling events associated with oxidative stress (5), apoptosis (5), protein aggregation (6), and mitochondrial function (7). Mechanistic ex vivo and in vivo studies involving transgenic mice have unequivocally demonstrated the importance of a properly functioning keratin cytoskeleton to the ability of simple epithelial cells to cope with stress (5, 8). In that regard, K8 post-translational modifications are critical modulators of its cellular functions.
K8 undergoes several post-translational modifications, including phosphorylation (9), sumoylation (10), acetylation (11), and transamidation (12), which take place across different segments of the K8 protein backbone. For example, the central and highly conserved α-helical coiled-coil “rod” domain contains the known sumoylation and acetylation sites, and it is flanked by the variable non-α-helical N-terminal “head” and C-terminal “tail” domains, which contain the known phosphorylation and transamidation sites on K8. There is accumulating evidence for cross-talk between the different types of K8 modifications, with phosphorylation playing a central role. For example, K8 acetylation modulates site-specific K8 phosphorylation (11), which in turn regulates K8 transamidation (12). Phosphorylation is also important in promoting K8 sumoylation (10), and may modulate keratin glycosylation (13).
From a functional standpoint, most of the cellular effects of K8 are tied to its phosphorylation status. In particular, serine phosphorylation of K8 at several sites results in filament reorganization and increased K8 solubility, as occurs during mitosis and cellular stress (14–16). For example, the highly abundant K8 becomes hyper-phosphorylated during stress and, in that regard, acts as a “phosphate sponge” for stress kinases (e.g. p38), which ultimately results in protection from apoptosis (17). Importantly, this mechanism appears to be compromised in the context of common human variants of K8 that predispose their carriers to liver disease (17). The biological relevance of phosphorylation is not unique to K8; the functions of other IF proteins, such as epidermal keratins, neurofilaments, and vimentin, for example, are critically modulated by phosphorylation under physiological and pathophysiological states (18–20). Therefore, understanding the nature and regulation of IF protein phosphorylation is critical as it may provide a mechanistic link between clinically relevant IF gene mutations and their disease manifestations.
All known phosphorylation sites on K8 are head and tail domain serine residues (Ser-24, Ser-74, Ser-432) (9). In contrast, site-specific characterization of phospho-tyrosine residues on K8 is currently lacking, although K8 has long been known to be a target for tyrosine phosphorylation in the presence of phosphatase inhibition by pervanadate (21). Other known, but poorly characterized, IF protein targets for tyrosine phosphorylation include K19, which becomes phosphorylated at Tyr-391 in the tail domain in the presence of Src kinase or pervanadate treatment (21, 22); vimentin, upon exposure of lymphoid cells to platelet-derived growth factor (23); and possibly peripherin (24). The elusive nature of tyrosine phosphorylation of K8, and IF proteins in general, is in part attributable to the low cellular abundance of phosphotyrosine relative to phosphoserine (25, 26).
This challenge has partially been overcome in recent years by the application of proteomic methodologies coupled with immune enrichment to identify phospho-tyrosine substrates (25). To that end, phospho-tyrosine peptides for most IF proteins have been identified in large scale proteomic studies (27–32). However, experimental confirmation and full characterization of any of these sites are presently lacking.
The availability of a pan-phosphotyrosine antibody and our generation of a site-specific antibody able to detect K8 phosphorylation allowed us to explore the hypothesis that tyrosine phosphorylation is an important determinant of K8 properties. In doing so, we identified a highly conserved tyrosine on K8 (Tyr-267) as a phosphorylation site that regulates filament organization and solubility and is a target for the protein-tyrosine phosphatase PTP1B. Furthermore, we showed that an Alexander disease mutation of the paralogous residue Tyr-242 in GFAP (Y242D) resulted in a highly insoluble protein with abnormal circular filament organization. These findings represent the first site-specific, functional characterization of tyrosine phosphorylation of any IF protein under basal conditions. The highly conserved nature of the K8 rod domain phospho-tyrosine residue suggests that this could be a mechanism for the regulation of the solubility properties and filament organization of other IF proteins.
EXPERIMENTAL PROCEDURES
Antibodies
We used the following antibodies: mouse pan-phosphotyrosine pY-100 (Cell Signaling), mouse anti-K8 (TS1) and mouse pan-actin (Thermo Scientific), rabbit anti-GFAP (DAKO), and mouse anti-HA (Covance). Generation of the p-Tyr-267 K8 antibody was done by a commercial source (AnaSpec) and involved immunizing two rabbits (A5276 and A5277), four times each, with the peptide VKAQ(pY)EDIAN-(LC)-C-NH2. The LC linker had the following sequence: NH2-CH2-CH2-CH2-CH2-CH2-COOH. Affinity purification was performed on a peptide column, and the purified antibody from rabbit A5277, which showed greater affinity and specificity, was used in the experiments from this study.
Site-directed Mutagenesis
The QuikChange site-directed mutagenesis kit (Stratagene) was used to generate mutants of K8 (in vector pcDNA3.1), GFAP (OriGene; in vector pCMV6-XL6), and HA-tagged PTP1B (Addgene plasmid 8601; in vector pJ3H). The wild-type (WT) and mutant constructs were confirmed by DNA sequencing.
Cell Cultures, Transfections, and Drug Treatment
BHK-21 (baby hamster kidney), NIH3T3 (mouse fibroblast), HT29 (human colon carcinoma), and HepG2 (human liver carcinoma) cells were obtained from American Type Culture Collection and cultured as recommended by the supplier. BHK-21 and HT29 cells were used primarily for biochemical analyses due to the high level of K8 expression after transfection or endogenously, respectively. NIH-3T3 and HepG2 cells were used primarily for immunofluorescence staining experiments to visualize filament organization after transfection (NIH-3T3) or endogenously (HepG2). Lipofectamine 2000 (for BHK-21 transfections) or Lipofectamine LTX (for NIH3T3 and HepG2 transfections) were used according to supplier (Invitrogen) instructions. Biochemical and immunofluorescence analyses were performed 18–24 h after transfection. Pharmacological inhibitors to epidermal growth factor receptor (EGFR) (Gefitinib) and PTP1B (TCS401) were purchased from Tocris, and PTP inhibitor XXII (PTPI-XXII) was purchased from EMD Millipore. All drug concentrations, specified in the figure legends, were chosen based on the reported IC50 values for their intended targets.
Immunoprecipitation, Immunoblot, and Immunofluorescence Analyses
Total cell lysates, detergent (Triton X-100)-soluble fractions, and high salt extracts (HSEs) were prepared and analyzed by immunoblotting as described previously (10). Immunoprecipitation was performed as described previously (11) using Dynabeads protein G (Invitrogen). For immunofluorescence analysis, cells were grown on 4-well poly-d-lysine-coated chamber slides and then processed and analyzed as described previously (11) using the specified antibodies.
Data Analysis
The pixel intensities of the scanned immunoblot images were estimated using Adobe Photoshop CS2 version 9.0. The graphs were presented, and the data were statistically analyzed using Prism 6 software (GraphPad Software).
RESULTS
K8 Is Phosphorylated on Conserved Rod Domain Residue Tyr-267
Upon surveying the literature on phosphotyrosine proteomic studies, we found that phospho-peptides of K8 (and its obligate binding partner K18) were reported in several independent studies that utilized different antibodies for immune enrichment (27–32). We validated one of the pan-phosphotyrosine antibodies used for immune enrichment (pY-100) (31) biochemically in a cell culture system to determine whether it can be used to detect dynamic changes in K8 phosphorylation. To do so, we tested the effects of epidermal growth factor (EGF) and blockade of its receptor (EGFR) in human colon carcinoma HT29 cells, which have endogenous expression of EGFR and are abundant in K8/K18. To enrich for K8/K18, we prepared HSEs, which contain mostly keratins, as seen by Coomassie Blue stain (Fig. 1A, bottom panel). Immunoblot analysis of the HSEs with the pY-100 antibody showed that transient (4 h) EGFR blockade using the reversible inhibitor gefitinib caused an increase in phosphorylation of both keratins in a dose-dependent manner (Fig. 1A, top panel). In contrast, treatment with EGF after gefitinib wash-out led to a decrease in K8 and K18 phosphorylation (Fig. 1B). These data show that the pY-100 antibody recognized phosphorylation sites on K8 and K18 that changed dynamically in response to modulation of the EGFR kinase pathway. Although the reported K18 phospho-peptides were primarily in the head domain (Tyr-13, Tyr-24, Tyr-36), highly conserved rod domain residues (Tyr-204, Tyr-267) were implicated as phosphorylation sites on K8 by the large scale proteomic studies (28–31). Using these predicted sites as a starting point, our next experiments were designed to study the consequences and regulation of conserved site-specific K8 tyrosine phosphorylation and to determine whether this process extends to other IF proteins.
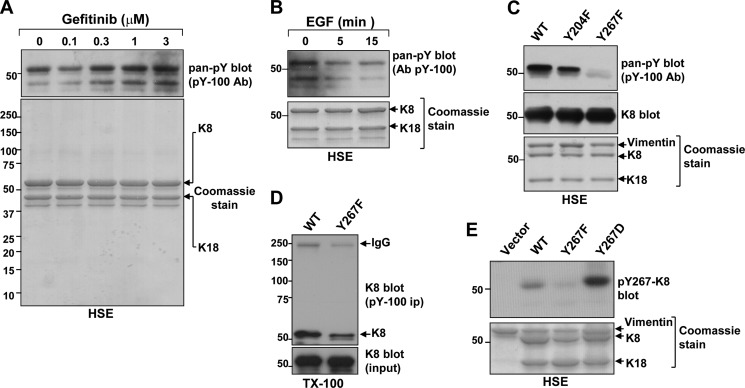
FIGURE 1.
Tyr-267 is a phosphorylated site on insoluble K8. A, pan-phosphotyrosine (pan-pY) immunoblot of HSEs from HT29 cells that were treated with DMSO vehicle (0) or the EGFR antagonist gefitinib at the designated concentrations for 4 h. Ab, antibody. B, Pan-pY immunoblot of HSEs from HT29 cells that were treated with 10 μm gefitinib for 4 h followed by drug wash-out and treatment with 20 ng/ml human recombinant EGF for 5 or 15 min. C, immunoblot comparison of HSEs from WT, Y204F K8-, and Y267F K8-expressing BHK-21 cells using the pan-pY antibody reveals that K8 Tyr-267 is a phosphorylated residue. K8 immunoblot and Coomassie Blue stain serve as loading controls. Note also the presence of endogenous vimentin and transfected WT K18, visible by Coomassie Blue stain. D, K8 immunoblot comparison of pan-pY immunoprecipitates (IP) of WT and Y267F K8-expressing BHK-21 cell Triton X-100 lysates (TX-100). E, validation of K8 Tyr-267 phosphorylation using a site-specific rabbit antibody. The antibody recognizes WT and the phospho-mimetic Y267D K8 mutant, but not the phosphorylation-deficient Y267F K8. HSEs of BHK-21 cells transfected with vector control or one of the three K8 constructs were analyzed by Coomassie Blue stain (as loading control) or immunoblotting with the purified p-Tyr-267 K8 antibody (pY267-K8).
We generated mutants of K8 Tyr-204 and Tyr-267 to nonphosphorylatable phenylalanines (Y204F and Y267F) and tested the ability of the pY-100 antibody to recognize the K8 mutants when compared with the WT K8 protein in an overexpression system using BHK-21 cells, which lack endogenous keratins but express vimentin. As shown in Fig. 1C, the Y267F mutation abolished K8 recognition by the pY-100 antibody in the insoluble HSE fraction. The detergent-soluble fraction of the BHK-21 lysates was also analyzed by immunoprecipitation with the pY-100 antibody followed by K8 immunoblot, and it showed the reduced, but not eliminated, presence of K8 tyrosine phosphorylation (Fig. 1D). Combined, these data indicate that Tyr-267 is a K8 phosphorylation site that is associated with the insoluble K8 pool.
To further study this modification, we generated an anti-peptide rabbit antibody against this phospho-epitope. The site-specific antibody (p-Tyr-267 K8) was validated in the overexpression system using HSEs of BHK-21 cells transfected with empty vector, WT, Y267F, or Y267D K8, the last being a phospho-mimetic mutation to rule out the possibility that the antibody recognized the unmodified tyrosine. As shown in Fig. 1E, the p-Tyr-267 K8 antibody recognized WT K8, and to a greater extent, Y267D K8, but not Y267F K8. Although the p-Tyr-267 K8 antibody detected overexpressed K8 by immunoblot of the HSE fraction, it was not suitable for immunocytochemistry and for immunoblot analysis of the detergent-soluble fraction, exhibiting a high degree of nonspecific binding in the latter case (not shown).
K8 Tyr-267 Is Important for Regulating Filament Organization and K8 Solubility
Because we were able to confirm Tyr-267 as a K8 phosphorylation site, we then investigated the effect of this residue on K8 properties. With respect to filament organization, the phosphorylation-deficient K8 Y267F mutant exhibited mostly short, perinuclear filaments, whereas the phospho-mimetic K8 Y267D mutant resulted in extensive bundling and filament aggregation when compared with WT K8 (Fig. 2A). Given the dramatic outcome on filament organization, we hypothesized that this site is important for modulating K8 solubility. Using HSE preparations, we found that the phospho-mimetic mutation (Y267D) led to a significant decrease in K8 solubility when compared with WT and Y267F K8 (Fig. 2, B and C).
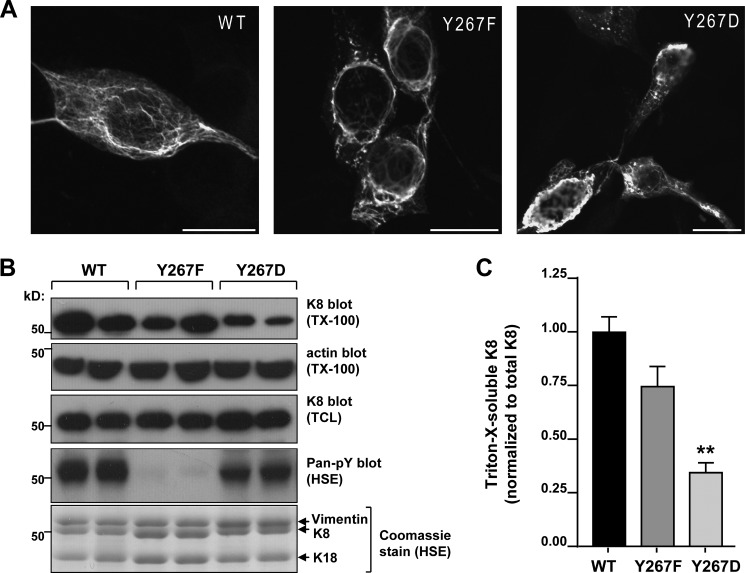
FIGURE 2.
Tyr-267 is important for K8 filament organization and solubility. A, immunofluorescence staining analysis of K8/K18-transfected NIH-3T3 cells reveals a partially disrupted filament network in the Y267F K8 mutant and abnormal filament aggregation in the Y267D K8 mutant. Scale bars = 20 μm. B, immunoblot analysis of Triton X-100-soluble fractions (TX-100) of BHK-21 cells expressing WT, Y267F, and Y267D K8 shows diminished solubility of the phospho-mimetic Y267D K8. Actin blot (Triton X-100 fraction) and K8 blot (total cell lysate (TCL)) served as loading controls. Pan-pY immunoblot and corresponding Coomassie Blue stain confirm the phosphorylation status of K8. Each condition was assayed in duplicate transfections. C, densitometric analysis (average of three independent experiments) shows significant difference in the solubility of Y267D K8 relative to WT (p < 0.01; one-way analysis of variance).
Abnormal Filaments and Decreased Solubility in Alexander Disease GFAP Mutant Affecting Conserved Tyr-242
We next asked whether this residue is important for regulating the properties of other IF proteins. In addition to being conserved across all type I and type II IF proteins (with the exception of K40), K8 Tyr-267 is also conserved across all type III, and most type IV, IFs (Fig. 3A). Importantly, the paralogous residue in GFAP (Tyr-242) is mutated (Y242D) in Alexander disease (33), an early-onset neurodegenerative disease that is associated with seizures, developmental delays, loss of psychomotor function, and a life expectancy of 5–10 years (34, 35). Because Alexander disease Tyr-242 mutation in GFAP is also to a negatively charged aspartic acid residue, we probed whether this mutation paralleled the effects on K8 with respect to alterations in filament organization and solubility. As shown in Fig. 3B, the Y242D GFAP mutant appeared as numerous abnormal circular deposits in the majority of transfected cells (60.5 ± 5.3%), in stark contrast to WT GFAP, which formed a normal-appearing filament network. Furthermore, biochemical analysis revealed that the solubility of Y242D was dramatically less in comparison with WT GFAP (Fig. 3, C and D). These data demonstrate that the K8 phospho-residue Tyr-267 and the corresponding conserved site in GFAP (Tyr-242) are critical for regulating IF organization and solubility, which may carry significance to other IF proteins and their disease mechanisms.
FIGURE 3.
Alexander disease-associated GFAP mutant at conserved Tyr-242 is highly insoluble and exhibits abnormal filament organization. A, sequence alignment of K8 with other human IFs (type III and type IV) showing the conserved nature of K8 Tyr-267 and the surrounding motif, QYE. NFL, neurofilament. B, immunofluorescence staining analysis of GFAP-transfected NIH-3T3 cells reveals a dramatically abnormal circular filament phenotype of Alexander disease mutant Y242D GFAP. Scale bars = 20 μm. C, immunoblot analysis of the Triton X-100-soluble fractions (TX-100) and total cell lysates (TCL) of NIH-3T3 cells transfected with WT or Y242D GFAP. Actin blot serves as loading control for the Triton X-100 fraction. D, densitometric quantification of the soluble fraction of WT and Y242D GFAP (***, p < 0.001; unpaired t test) from panel C.
The Nonreceptor Tyrosine Phosphatase PTP1B Regulates K8 Phosphorylation and Solubility
We next sought to identify potential regulators of K8 Tyr-267 phosphorylation status. Because our results indicated that an inability of K8 to become dephosphorylated at Tyr-267, as mimicked by the negative charge of the Y267D mutation, limits K8 solubility, we investigated the regulation of K8 Tyr-267 phosphorylation by tyrosine phosphatases. After an initial screen (not shown) using several tyrosine phosphatase inhibitors with either broad specificity (pervanadate, BVT948) or phosphatase selectivity (NSC 87877, NSC 95397, and TCS 401, which target SHP1/2, CDC25, and PTP1B, respectively), we focused on the nonreceptor tyrosine phosphatase PTP1B. The reason for selecting PTP1B was our finding of enhanced K8 tyrosine phosphorylation using two structurally and mechanistically distinct small molecule inhibitors of this enzyme: TCS401 and PTP inhibitor XXII (Fig. 4, A and B). We used the latter inhibitor in all subsequent experiments and refer to it as PTPI-XXII. In line with its ability to increase K8 tyrosine phosphorylation, PTPI-XXII decreased K8 solubility in a dose-dependent manner (Fig. 4C). These data are consistent with the decreased solubility of the phospho-mimetic Y267D K8 mutant (Fig. 2). The decrease in K8 solubility coincided with an increase in the number of cells that contained K8/K18 filament bundles following PTPI-XXII treatment (Fig. 4, D and E). However, the effect on filament organization was not as dramatic when compared with the Y242D K8 mutant, indicating that the cellular context is important for the observed effects on filament organization.
FIGURE 4.
Pharmacologic inhibition of the protein-tyrosine phosphatase PTP1B increases K8 phosphorylation, reduces K8 solubility, and increases filament bundling. A, immunoblot analysis for K8 tyrosine phosphorylation in HSEs of HT29 cells treated for 6 h with the general tyrosine phosphatase inhibitor BVT948 (30 μm) or the PTP1B-selective inhibitor TCS401 (6 μm). Veh, vehicle; Ab, antibody. B, immunoblot analysis for K8 tyrosine phosphorylation in HSEs of BHK-21 cells transfected with WT or Y267F K8 and treated with DMSO vehicle or the PTP1B-selective inhibitor PTPI-XXII (30 μm) for 1 h. Vim., vimentin. C, decreased solubility of K8 in PTPI-XXII-treated (2 h) HepG2 cells. Actin blot serves as a loading control. D, increased K8/K18 filament bundling in PTPI-XXII-treated (4 h) HepG2 cells. Scale bars = 20 μm. E, quantification of filament bundling (n = 70 cells per condition) shows an increase in the PTPI-XXII-treated group (*, p < 0.05; unpaired t test).
A Decrease in Filamentous and Insoluble K8 Is Mediated by PTP1B Dephosphorylation of K8 Tyr-267
As an alternative to the pharmacological approach to examine the role of PTP1B on K8 phosphorylation, we turned to an overexpression system using HA-tagged PTP1B. After transient overexpression into HepG2 cells, we noted a partial K8-PTP1B co-localization with a more diffuse filament staining pattern in the PTP1B-expressing cells (Fig. 5A and inset). In an effort to stabilize a potential K8-PTP1B interaction without significantly affecting filament organization, we co-transfected a “substrate-trapping” PTP1B mutant (D181A) into HepG2 cells. Mutation of the PTP1B invariant catalytic acid (Asp-181), which protonates the tyrosyl leaving group of the substrate, to an Ala results in an enzyme that retains its ability to bind tightly to cellular substrates, but is markedly attenuated in enzymatic activity (36). As shown in Fig. 5B, there was significant co-localization between D181A PTP1B and K8 filaments, which also appeared more filamentous when compared with WT PTP1B-expressing cells. Biochemical analysis also revealed an increased association between K8 and D181A PTP1B. (Fig. 5C). Furthermore, in cells expressing WT PTP1B, the levels of K8 protein in the HSE fraction were significantly diminished (Fig. 5D). To further ascertain that K8 is a substrate of PTP1B, we assessed K8 phosphorylation at Tyr-267 in the presence of PTP1B overexpression and in the absence or presence of PTPI-XXII. PTP1B overexpression significantly diminished K8 phosphorylation, and this was partially reversed by PTPI-XXII (Fig. 5E), which demonstrates that PTP1B is a K8 phosphatase that dephosphorylates K8 Tyr-267.
FIGURE 5.
Overexpression of WT and a substrate-trapping D181A mutant of PTP1B demonstrates that PTP1B is a K8 phosphatase that regulates K8 filament organization and solubility. A, immunofluorescence staining of endogenous K8 (red) and overexpressed WT PTP1B (green) in HepG2 cells showing partial co-localization and altered filament organization in the presence of PTP1B overexpression. Inset, comparison of PTP1B-transfected and untransfected cells B, immunofluorescence staining of endogenous K8 (red) and overexpressed D181A PTP1B (green) in HepG2 cells showing significant co-localization. Merged images in A and B show DAPI co-staining, and the areas demarcated by white boxes represent the magnified panels at the bottom. Scale bars = 10 μm. C, co-immunoprecipitation analysis of BHK-21 Triton X-100 lysates after triple transfection with K8, K18, and either WT PTP1B or D181A PTP1B, demonstrating increased association between K8 and D181A PTP1B. Each blot shows two nonconsecutive lanes of the same, equally exposed membrane. D, co-expression of WT PTP1B with WT K8/K18 in BHK-21 cells (24 h of transfection) results in a significant decrease in insoluble K8 (HSE fraction), as demonstrated by immunoblot and Coomassie Blue stain. TX-100, Triton X-100 fraction. E, K8 phosphotyrosine blot of HSEs from K8/K18-transfected BHK-21 cells that were also co-transfected with vector or WT PTP1B (simultaneous 18 h transfection) and treated with either vehicle or the PTP1B-selective inhibitor PTPI-XXII (30 μm) for 1 h prior to lysis. PTP1B expression decreases solubility and p-Tyr-267 K8 (K8-pY267), and this is partially reversed by PTPI-XXII.
DISCUSSION
Although serine phosphorylation is known to be a critical modulator of IF cytoskeletal dynamics, there is very little known regarding the role of site-specific tyrosine phosphorylation. Despite the lack of molecular insight, there is substantial evidence for tyrosine kinase signaling-mediated regulation of IF properties. For example, there is a significant reorganization of K8/K18 filaments within minutes of EGFR stimulation, characterized by enlargement of the peripheral keratin network, which uses focal adhesions as nucleation sites (37). In the present study, we demonstrated that EGFR activation is a negative regulator of K8/K18 tyrosine phosphorylation in the insoluble compartment. Because K8 phosphorylation at Tyr-267 decreases K8 solubility, our findings align with a recent study showing that longer EGFR activation (1 h) increases the levels of soluble K17 (38).
The specific kinase signaling pathways that lead to K8 Tyr-267 phosphorylation remain to be identified, but it is likely that this process will be highly cell- and tissue-dependent. The detection of K8 phospho-tyrosine peptides in lung cancer was associated with those tissues and cell lines that had high expression of the proto-oncogene tyrosine kinase ROS (31). ROS is an orphan receptor tyrosine kinase that controls cell survival, growth, differentiation, and proliferation pathways via its ability to form oncogenic fusion proteins (39). ROS fusions are common in cholangiocarcinoma (28), a highly aggressive form of hepatocellular carcinoma with poorly understood molecular mechanisms. Based on proteomic analysis, K8 is also phosphorylated at Tyr-267 in human cholangiocarcinoma tissues (28), which provides a possible disease context to be addressed in future studies.
Several other oncogenic tyrosine kinases have been shown to directly or indirectly associate with and affect the IF cytoskeleton. For example, in cells expressing constitutively active Src kinase, K19 becomes phosphorylated at Tyr-391, which then preferentially partitions into the soluble compartment (22). Furthermore, the interaction between Src and K19 appears to act as a critical facilitator of EGF-stimulated oncogenesis because it is necessary for the activation of Src by EGF-stimulated tissue transglutaminase (40). On the other hand, a phosphorylation-independent association between Src and K6, which is induced in keratinocytes during wound repair, inhibits Src kinase activity, resulting in attenuated keratinocyte migration (41).
Aside from keratins, vimentin IFs are also responsive to tyrosine kinase signaling. As shown recently, cell death induced by inhibition of Jak2 kinase is accompanied by a breakdown and cleavage of the vimentin IF network (42), which carries potential significance to diseases associated with aberrant Jak2 signaling such as leukemia, lymphoma, and myeloma. Furthermore, enhanced tyrosine kinase-mediated Rac activation promotes the collapse of the GFAP filament network into the perinuclear regions of human schwannoma cells (43). This process is thought to be linked to the ability of schwannoma cells to exhibit enhanced integrin-dependent adhesion and motility (43). It is not clear whether vimentin or GFAP phosphorylation, per se, played a role in the above mechanisms. However, based on large scale proteomic studies, both proteins are predicted to be phosphorylated at the highly conserved Tyr-276 in vimentin (28) and Tyr-242 in GFAP (31), which, as we demonstrated here, is critically important for GFAP filament organization and solubility.
The mutation of the Tyr-242 residue in GFAP to the negatively charged aspartic acid, and the resultant phenotype of the GFAP filaments, are significant from a disease and mechanistic standpoint. This residue is very close to the most commonly mutated GFAP site (Arg-239) in Alexander disease (35), suggesting that Arg-239 GFAP mutations may potentially affect Tyr-242 GFAP phosphorylation and thereby disturb filament organization. In fact, the striking circular filament phenotype that we observe with the Y242D GFAP mutant is somewhat reminiscent of what has previously been shown with an R239C GFAP mutant, which also had decreased solubility (44). Furthermore, a characteristic feature of astrocytes in Alexander disease is the presence of GFAP aggregates called Rosenthal fibers (34). Whether site-specific GFAP tyrosine phosphorylation controls Rosenthal fiber formation and contributes to the pathogenesis of Alexander disease remains to be determined.
Another relevant aspect of our study is the finding that the protein-tyrosine phosphatase PTP1B dephosphorylates K8. PTP1B is the founding member of a family of tissue-specific protein-tyrosine phosphatases (PTPs) that are the products of over 100 human genes (45). A major function of PTP1B is to negatively regulate receptor and nonreceptor tyrosine kinases by catalyzing their dephosphorylation in an endocytosis-dependent manner (45). PTP1B is considered a potential drug target for obesity, diabetes, and cancer (46) because mice lacking PTP1B have increased insulin sensitivity and are resistant to diet-induced obesity (47) and mammary tumorigenesis (48).
The C terminus of PTP1B contains a hydrophobic region that targets it to the endoplasmic reticulum (45), with the catalytic domain facing the cytoplasm where it has the potential to affect the phosphorylation of many different substrates and to affect various cell functions, including, for example, actin cytoskeleton dynamics (49). A previous study integrated proteome-wide evidence using a Bayesian model to predict substrates of PTP1B (50). The model was highly effective in predicting actual PTP1B substrates, such as EGFR, but in addition, it predicted K8 as a very high ranking substrate with a score near that of EGFR (0.897 for K8 versus 0.926 for EGFR) (50). Our present findings demonstrate that PTP1B is a bona fide K8 phosphatase and raise the possibility that PTP1B may be an important regulator of K8 function under various types of epithelial cell stress, including in the liver, which is a major site of PTP1B action (51).
Acknowledgments
We thank Jessica Leonard and Peter Altshuler for technical assistance with the experiments in this study.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK52951 (to M. B.O.) and K01 DK093776 (to N. T. S.). This work was also supported by a Department of Veterans Affairs Merit Review Award (to M. B. O.).
This article was selected as a Paper of the Week.
- K
- keratin
- EGFR
- epidermal growth factor receptor
- GFAP
- glial fibrillary acidic protein
- HSE
- high salt extract
- IF
- intermediate filament
- PTP1B
- protein-tyrosine phosphatase 1B
- PTPI-XXII
- PTP inhibitor XXII
- pY
- phospho-tyrosine
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Herrmann H., Strelkov S. V., Burkhard P., Aebi U. (2009) Intermediate filaments: primary determinants of cell architecture and plasticity. J. Clin. Invest. 119, 1772–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eriksson J. E., Dechat T., Grin B., Helfand B., Mendez M., Pallari H. M., Goldman R. D. (2009) Introducing intermediate filaments: from discovery to disease. J. Clin. Invest. 119, 1763–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Omary M. B. (2009) “IF-pathies”: a broad spectrum of intermediate filament-associated diseases. J. Clin. Invest. 119, 1756–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pan X., Hobbs R. P., Coulombe P. A. (2013) The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr. Opin. Cell Biol. 25, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toivola D. M., Strnad P., Habtezion A., Omary M. B. (2010) Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 20, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ku N. O., Strnad P., Zhong B. H., Tao G. Z., Omary M. B. (2007) Keratins let liver live: Mutations predispose to liver disease and crosslinking generates Mallory-Denk bodies. Hepatology 46, 1639–1649 [DOI] [PubMed] [Google Scholar]
- 7. Toivola D. M., Tao G. Z., Habtezion A., Liao J., Omary M. B. (2005) Cellular integrity plus: organelle-related and protein-targeting functions of intermediate filaments. Trends Cell Biol. 15, 608–617 [DOI] [PubMed] [Google Scholar]
- 8. Omary M. B., Ku N. O., Strnad P., Hanada S. (2009) Toward unraveling the complexity of simple epithelial keratins in human disease. J. Clin. Invest. 119, 1794–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Omary M. B., Ku N. O., Tao G. Z., Toivola D. M., Liao J. (2006) “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem. Sci. 31, 383–394 [DOI] [PubMed] [Google Scholar]
- 10. Snider N. T., Weerasinghe S. V., Iñiguez-Lluhí J. A., Herrmann H., Omary M. B. (2011) Keratin hypersumoylation alters filament dynamics and is a marker for human liver disease and keratin mutation. J. Biol. Chem. 286, 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snider N. T., Leonard J. M., Kwan R., Griggs N. W., Rui L., Omary M. B. (2013) Glucose and SIRT2 reciprocally mediate the regulation of keratin 8 by lysine acetylation. J. Cell Biol. 200, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwan R., Hanada S., Harada M., Strnad P., Li D. H., Omary M. B. (2012) Keratin 8 phosphorylation regulates its transamidation and hepatocyte Mallory-Denk body formation. FASEB J. 26, 2318–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ku N. O., Toivola D. M., Strnad P., Omary M. B. (2010) Cytoskeletal keratin glycosylation protects epithelial tissue from injury. Nat. Cell Biol. 12, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toivola D. M., Zhou Q., English L. S., Omary M. B. (2002) Type II keratins are phosphorylated on a unique motif during stress and mitosis in tissues and cultured cells. Mol. Biol. Cell 13, 1857–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liao J., Ku N. O., Omary M. B. (1997) Stress, apoptosis, and mitosis induce phosphorylation of human keratin 8 at Ser-73 in tissues and cultured cells. J. Biol. Chem. 272, 17565–17573 [DOI] [PubMed] [Google Scholar]
- 16. Ridge K. M., Linz L., Flitney F. W., Kuczmarski E. R., Chou Y. H., Omary M. B., Sznajder J. I., Goldman R. D. (2005) Keratin 8 phosphorylation by protein kinase C δ regulates shear stress-mediated disassembly of keratin intermediate filaments in alveolar epithelial cells. J. Biol. Chem. 280, 30400–30405 [DOI] [PubMed] [Google Scholar]
- 17. Ku N. O., Omary M. B. (2006) A disease- and phosphorylation-related nonmechanical function for keratin 8. J. Cell Biol. 174, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sihag R. K., Inagaki M., Yamaguchi T., Shea T. B., Pant H. C. (2007) Role of phosphorylation on the structural dynamics and function of types III and IV intermediate filaments. Exp. Cell Res. 313, 2098–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ivaska J., Pallari H. M., Nevo J., Eriksson J. E. (2007) Novel functions of vimentin in cell adhesion, migration, and signaling. Exp. Cell Res. 313, 2050–2062 [DOI] [PubMed] [Google Scholar]
- 20. Pan X., Kane L. A., Van Eyk J. E., Coulombe P. A. (2011) Type I keratin 17 protein is phosphorylated on serine 44 by p90 ribosomal protein S6 kinase 1 (RSK1) in a growth- and stress-dependent fashion. J. Biol. Chem. 286, 42403–42413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng L., Zhou X., Liao J., Omary M. B. (1999) Pervanadate-mediated tyrosine phosphorylation of keratins 8 and 19 via a p38 mitogen-activated protein kinase-dependent pathway. J. Cell Sci. 112, 2081–2090 [DOI] [PubMed] [Google Scholar]
- 22. Zhou Q., Snider N. T., Liao J., Li D. H., Hong A., Ku N. O., Cartwright C. A., Omary M. B. (2010) Characterization of in vivo keratin 19 phosphorylation on tyrosine-391. PLoS One 5, e13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valgeirsdóttir S., Claesson-Welsh L., Bongcam-Rudloff E., Hellman U., Westermark B., Heldin C. H. (1998) PDGF induces reorganization of vimentin filaments. J. Cell Sci. 111, 1973–1980 [DOI] [PubMed] [Google Scholar]
- 24. Angelastro J. M., Ho C. L., Frappier T., Liem R. K., Greene L. A. (1998) Peripherin is tyrosine-phosphorylated at its carboxyl-terminal tyrosine. J. Neurochem. 70, 540–549 [DOI] [PubMed] [Google Scholar]
- 25. Ding S. J., Qian W. J., Smith R. D. (2007) Quantitative proteomic approaches for studying phosphotyrosine signaling. Expert Rev. Proteomics 4, 13–23 [DOI] [PubMed] [Google Scholar]
- 26. Hunter T. (2009) Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 21, 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Artemenko K. A., Bergström Lind S., Elfineh L., Mayrhofer C., Zubarev R. A., Bergquist J., Pettersson U. (2011) Optimization of immunoaffinity enrichment and detection: toward a comprehensive characterization of the phosphotyrosine proteome of K562 cells by liquid chromatography-mass spectrometry. Analyst 136, 1971–1978 [DOI] [PubMed] [Google Scholar]
- 28. Gu T. L., Deng X., Huang F., Tucker M., Crosby K., Rimkunas V., Wang Y., Deng G., Zhu L., Tan Z., Hu Y., Wu C., Nardone J., MacNeill J., Ren J., Reeves C., Innocenti G., Norris B., Yuan J., Yu J., Haack H., Shen B., Peng C., Li H., Zhou X., Liu X., Rush J., Comb M. J. (2011) Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One 6, e15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo A., Villén J., Kornhauser J., Lee K. A., Stokes M. P., Rikova K., Possemato A., Nardone J., Innocenti G., Wetzel R., Wang Y., MacNeill J., Mitchell J., Gygi S. P., Rush J., Polakiewicz R. D., Comb M. J. (2008) Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. U.S.A. 105, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moritz A., Li Y., Guo A., Villén J., Wang Y., MacNeill J., Kornhauser J., Sprott K., Zhou J., Possemato A., Ren J. M., Hornbeck P., Cantley L. C., Gygi S. P., Rush J., Comb M. J. (2010) Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci. Signal. 3, ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 32. Bergström Lind S., Artemenko K. A., Elfineh L., Mayrhofer C., Zubarev R. A., Bergquist J., Pettersson U. (2011) Toward a comprehensive characterization of the phosphotyrosine proteome. Cell. Signal. 23, 1387–1395 [DOI] [PubMed] [Google Scholar]
- 33. Gorospe J. R., Naidu S., Johnson A. B., Puri V., Raymond G. V., Jenkins S. D., Pedersen R. C., Lewis D., Knowles P., Fernandez R., De Vivo D., van der Knaap M. S., Messing A., Brenner M., Hoffman E. P. (2002) Molecular findings in symptomatic and pre-symptomatic Alexander disease patients. Neurology 58, 1494–1500 [DOI] [PubMed] [Google Scholar]
- 34. Liem R. K., Messing A. (2009) Dysfunctions of neuronal and glial intermediate filaments in disease. J. Clin. Invest. 119, 1814–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quinlan R. A., Brenner M., Goldman J. E., Messing A. (2007) GFAP and its role in Alexander disease. Exp. Cell Res. 313, 2077–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flint A. J., Tiganis T., Barford D., Tonks N. K. (1997) Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 94, 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Felkl M., Tomas K., Smid M., Mattes J., Windoffer R., Leube R. E. (2012) Monitoring the cytoskeletal EGF response in live gastric carcinoma cells. PLoS One 7, e45280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chung B. M., Murray C. I., Van Eyk J. E., Coulombe P. A. (2012) Identification of novel interaction between annexin A2 and keratin 17: evidence for reciprocal regulation. J. Biol. Chem. 287, 7573–7581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charest A., Wilker E. W., McLaughlin M. E., Lane K., Gowda R., Coven S., McMahon K., Kovach S., Feng Y., Yaffe M. B., Jacks T., Housman D. (2006) ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 66, 7473–7481 [DOI] [PubMed] [Google Scholar]
- 40. Li B., Antonyak M. A., Druso J. E., Cheng L., Nikitin A. Y., Cerione R. A. (2010) EGF potentiated oncogenesis requires a tissue transglutaminase-dependent signaling pathway leading to Src activation. Proc. Natl. Acad. Sci. U.S.A. 107, 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rotty J. D., Coulombe P. A. (2012) A wound-induced keratin inhibits Src activity during keratinocyte migration and tissue repair. J. Cell Biol. 197, 381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Majumder A., Kirabo A., Karrupiah K., Tsuda S., Caldwell-Busby J., Cardounel A. J., Keseru G. M., Sayeski P. P. (2011) Cell death induced by the Jak2 inhibitor, G6, correlates with cleavage of vimentin filaments. Biochemistry 50, 7774–7786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Utermark T., Schubert S. J., Hanemann C. O. (2005) Rearrangements of the intermediate filament GFAP in primary human schwannoma cells. Neurobiol. Dis. 19, 1–9 [DOI] [PubMed] [Google Scholar]
- 44. Hsiao V. C., Tian R., Long H., Der Perng M., Brenner M., Quinlan R. A., Goldman J. E. (2005) Alexander-disease mutation of GFAP causes filament disorganization and decreased solubility of GFAP. J. Cell Sci. 118, 2057–2065 [DOI] [PubMed] [Google Scholar]
- 45. Stuible M., Tremblay M. L. (2010) In control at the ER: PTP1B and the down-regulation of RTKs by dephosphorylation and endocytosis. Trends Cell Biol. 20, 672–679 [DOI] [PubMed] [Google Scholar]
- 46. Yip S. C., Saha S., Chernoff J. (2010) PTP1B: a double agent in metabolism and oncogenesis. Trends Biochem. Sci. 35, 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elchebly M., Payette P., Michaliszyn E., Cromlish W., Collins S., Loy A. L., Normandin D., Cheng A., Himms-Hagen J., Chan C. C., Ramachandran C., Gresser M. J., Tremblay M. L., Kennedy B. P. (1999) Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548 [DOI] [PubMed] [Google Scholar]
- 48. Julien S. G., Dubé N., Read M., Penney J., Paquet M., Han Y., Kennedy B. P., Muller W. J., Tremblay M. L. (2007) Protein tyrosine phosphatase 1B deficiency or inhibition delays ErbB2-induced mammary tumorigenesis and protects from lung metastasis. Nat. Genet. 39, 338–346 [DOI] [PubMed] [Google Scholar]
- 49. Stuible M., Dubé N., Tremblay M. L. (2008) PTP1B regulates cortactin tyrosine phosphorylation by targeting Tyr446. J. Biol. Chem. 283, 15740–15746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ferrari E., Tinti M., Costa S., Corallino S., Nardozza A. P., Chatraryamontri A., Ceol A., Cesareni G., Castagnoli L. (2011) Identification of new substrates of the protein-tyrosine phosphatase PTP1B by Bayesian integration of proteome evidence. J. Biol. Chem. 286, 4173–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haj F. G., Zabolotny J. M., Kim Y. B., Kahn B. B., Neel B. G. (2005) Liver-specific protein-tyrosine phosphatase 1B (PTP1B) re-expression alters glucose homeostasis of PTP1B−/− mice. J. Biol. Chem. 280, 15038–15046 [DOI] [PubMed] [Google Scholar]