Abstract
It has recently been described that aging in C. elegans is accompanied by the progressive development of morphological changes in the nervous system. These include novel outgrowths from the cell body or axonal process, as well as blebbing and beading along the length of the axon. The formation of these structures is regulated by numerous molecular players including members of the well-conserved insulin/insulin growth factor-like (IGF)-1 signaling and mitogen-activated protein (MAP) kinase pathways. This review summarizes the recent literature on neuronal aging in C. elegans, including our own findings, which indicate a role for protein with tau-like repeats (PTL-1), the homolog of mammalian tau and MAP2/4, in maintaining neuronal integrity during aging.
Keywords: aging, neuronal aging, Caenorhabditis elegans, lifespan, protein with tau-like repeats
Cellular Features of Neuronal Aging in C. elegans
Caenorhabditis elegans is a useful model in which to study aging owing, in particular, to its relatively short lifespan. Like in humans, aging in worms is accompanied by physiological changes including progressive loss of mobility,1,2 sarcopenia and deterioration of other tissues1,3 and a decline in immune function.4 Human physiological aging (as differentiated from pathological aging) is additionally associated with subtle physical changes in the brain, such as neuronal restructuring and synaptic loss,5,6 and these changes have been linked to a progressive impairment of cognitive function.5,7 In light of these observations, it was somewhat surprising that initial studies did not detect any structural decline in the C. elegans nervous system with age.1 Recently, however, close examination of neuronal morphology in the nematode has revealed pronounced aging phenotypes such as aberrant outgrowths and beading along neuron processes, and age-associated synaptic deterioration has also been detected.8-10 The nematode model system therefore presents an opportunity to explore the mechanisms by which neuronal aging is regulated. In this model, it is also possible to dissect the relationship between neuronal aging and aging of the whole organism, by examining for example whether accelerated neuronal aging has repercussions for the entire organism.
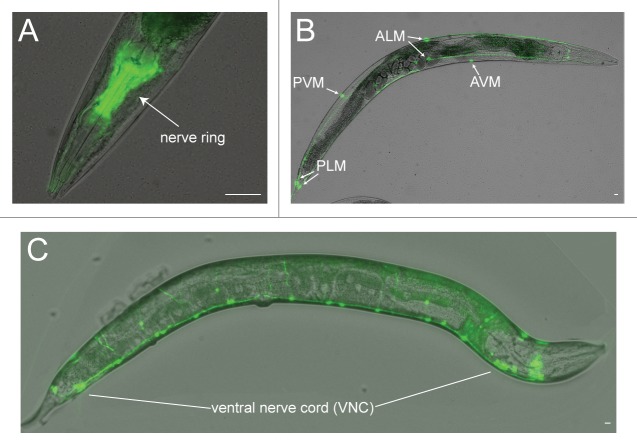
Such explorations will be greatly facilitated by the relative simplicity of the C. elegans nervous system, which consists of only 302 neurons. These neurons have been anatomically mapped and develop in a stereotypical manner,11,12 facilitating the study of age-related structural changes. To date, among the 302 neurons, age-associated changes have been identified in the mechanosensory “touch receptor” neurons, in the axons of cholinergic neurons in the ventral nerve cord (VNC), in the axons of GABAergic motor neurons in the ventral and dorsal nerve cord, and the nerve ring (Fig. 1). These changes include branching from the cell body and axon, as well as blebbing and beading along the axon.8-10 Representative images of these structures are shown in (Fig. 2). In neurons displaying such structures, nuclear DAPI staining appears intact even in severe cases, suggesting that these aged neurons are not undergoing apoptosis or necrosis.10
Figure 1. Fluorescence reporter lines enable the visualization of neurons in C. elegans. (A) Nerve ring, positioned between the anterior and terminal bulbs of the pharynx. The sIs11686(Pptl-1::gfp) reporter line is shown.54 (B) Touch receptor neurons, where cell bodies for the AVM, ALMs, PVM and PLMs are shown. The zdIs5(Pmec-4::gfp) reporter line is shown.55 (C) Ventral nerve cord GABAergic motor neurons. The oxIs12(Punc-47::gfp) reporter line is shown.56 Scale = 10 µm.
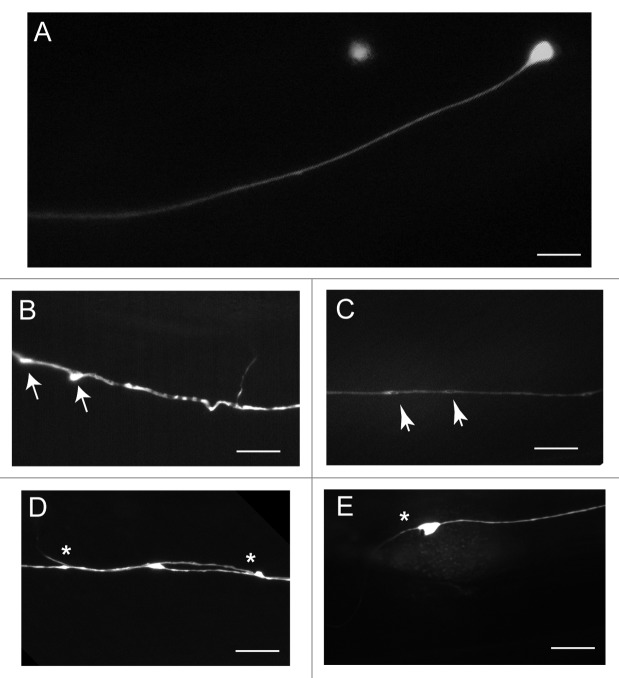
Figure 2. Touch receptor neurons develop abnormal structures in the cell body and axon in a progressive, age-dependent manner. Here, touch neurons are visualized using the zdIs5(Pmec-4::gfp) reporter line. (A) A “normal” touch receptor neuron (ALM). The cell body of AVM is out of focus. (B–E) Age-associated abnormal structures in ALM touch neurons. (B) Blebs (arrows) form along the process, creating a wavy appearance along the axon. (C) Beading (arrowheads) along the axon. (D) Branching (asterisks) along the axon. Some branches appear to develop from blebs. (E) Branching from the cell body. Scale = 10 µm.
Branching refers to novel extensions emanating from the neuronal cell body or along the axonal process. Branches have been visualized using several distinct fluorescent neuronal reporter transgenes8-10 and were also observed by immunostaining with an anti-acetylated α-tubulin antibody, revealing that the outgrowths contain acetylated microtubules.10 The branching process is dynamic, since protrusions have been observed to form and retract, and secondary branches may extend from existing branches.9,10
Blebs are defined as triangular-shaped protrusions from the processes.10 When several of these blebs form along the process, this can distort the structure of the axon, such that the axon adopts a wavy appearance.10 These wavy processes have been quantified in touch neurons and found to increase in frequency with age.9 In old animals, branches or even axon splitting can sometimes be observed at the sites of these blebs.10 Neither branches nor blebs co-localize with late endosome or lysosome markers,10 or with synaptic protein markers.9
Beading, or bubble-like lesions, refers to focal enlargements that occur along the length of the axonal process.9,10 These beads were observed in touch neurons, and have dark, fluorescence-free regions in the center of the structure.9,10
Complementing the work performed using transgenic strains expressing fluorescence reporter transgenes, examination of touch neurons in non-transgenic wild-type animals by electron microscopy (EM) also revealed these morphological changes. This indicates that the observed structures are not simply an artifact associated with fluorescence transgenes.9 In addition, EM studies revealed that some novel outgrowths developing from touch neuron processes co-localize with mitochondria at the branch points.9 These observations were confirmed by immunofluorescence assays showing co-localization of these branch points with mitochondria-specific GFP in touch neurons.9 It is unclear if mitochondria accumulate due to cytoskeletal changes at these points, or if the presence of mitochondria induces branching at these sites.
Another age-related effect found in the worm neurons is synaptic deterioration, such as a reduction in synaptic vesicles and size of the presynaptic terminal.9 Synaptic integrity is a well-established parameter of neuronal aging in mammals,13-15 and correlates well with memory and behavioral regression.16 Similarly, reduction in synaptic integrity is associated with impaired locomotion in aged C. elegans.9 Using EM studies, the number of synaptic vesicles in the nerve ring could be quantified, revealing that old adult worms have significantly fewer synaptic vesicles compared with young adult animals.9
Notably, different types of neurons showed differential susceptibility to age-related changes. For example, touch receptor neurons accumulate aberrant branching with age, whereas two of the nerve ring interneurons and some dopaminergic neurons remain intact.9 Moreover, different subsets of touch neurons also display differential severity of morphological defects, and even vary in the type of defects that can be observed.9,10 It is intriguing that this kind of aging heterogeneity exists even between neurons of the same type.
Genetic Aspects of Neuronal Aging in C. elegans
In addition to the cellular characteristics of neuronal aging in C. elegans that are reminiscent of age-associated neuronal changes in higher organisms, several conserved regulators of aging have also been identified.4,17-20
Insulin /IGF-1 signaling (IIS) pathway
The Insulin/IGF-1 signaling (IIS) pathway is one of the best-characterized regulators of aging in C. elegans. Reduction-of-function mutations in the DAF-2 insulin receptor result in a dramatic extension of lifespan.18 Activation of the DAF-2 receptor signals to downstream kinases to phosphorylate the forkhead box-O (FOXO) transcription factor DAF-16, which results in its exclusion from the nucleus and hence an inability to trigger the expression of genes associated with longevity. Thus, mutations in DAF-16 result in premature aging, and the lifespan extension observed in daf-2 mutants is dependent on DAF-16.18,21,22
The IIS pathway in C. elegans has been found to be involved in nervous system aging. daf-2 reduction-of-function mutants display delayed neuronal branching in touch receptor and cholinergic neurons.8-10 Furthermore, daf-16 appears to be required for the daf-2-mediated delay in appearance of neuronal defects, since daf-16;daf-2 double mutants display wild-type levels of branching in touch neurons.8 Interestingly, Toth and colleagues reported that the branching phenotypes observed in daf-2 and daf-16 mutant strains have subtle differences compared with aged wild-type animals. For example, ALM branching is rare in aged wild-type worms, but occurs at a 10% frequency in daf-2 mutant animals of the same stage.9 This indicates that the profile of neuronal aging phenotypes is different between wild-type animals and IIS mutants, although the reasons for this are unclear.
Like DAF-16, the heat shock factor (HSF)-1 transcription factor, is repressed by insulin signaling. HSF-1 functions with DAF-16 to regulate proteostasis and chaperone expression when active in response to heat stress.3 Reduction of HSF-1 activity results in a shortened lifespan23 as well as a significantly higher frequency of touch neuron defects compared with wild-type.9,10 Furthermore, hsf-1;daf-16 double mutants do not show enhancement of the accelerated onset of phenotypes observed in single mutants, suggesting that these transcription factors may largely act within the same pathway to regulate neuronal aging.10 Similarly, knockdown of HSF-1 by RNAi does not affect the lifespan of daf-16 mutants.23
Mechanosensory signal transduction
Mechanosensory-defective (or mec) mutants are defective in their response to gentle touch. Interestingly, some of these mutants also show lifespan phenotypes. Mutations in mechanosensory channel components MEC-2, MEC-4, MEC-6, MEC-10 and extracellular matrix (ECM) proteins MEC-5 and MEC-9 result in a shortened lifespan, whereas mutations in ECM protein MEC-1 and α-tubulin MEC-12 do not.10 However, these mutants all display a high frequency of neuronal defects at earlier ages compared with wild-type.10 Pan and colleagues suggest that defects in nerve attachment in certain mec-1 mutants may be responsible for the accelerated onset of defects observed in touch neurons. This is supported by the finding that animals carrying mutations in him-4 and fbl-1, which encode ECM proteins hemicentin24 and fibulin,25 are defective in nerve attachment and also display a high frequency of touch neuron defects at a young age.10
Some reports suggest that the ability of touch neurons to function correctly is correlated with healthy neuronal aging. Tank and colleagues found that animals that display high levels of branching in touch neurons are also generally less touch sensitive,8 although a similar experiment led by others did not display any significant correlation.9 In addition, a gain-of-function mutation in the neuronal SLO-1 hyperpolarising ion channel results in touch insensitivity. Analysis of this mutant demonstrates an accelerated onset of touch neuron defects.10 The involvement of synaptic activity is not limited to touch neurons. Mutations in unc-13 and unc-18 disrupt synaptic transmission and result in higher levels of beading in axons in the ventral and dorsal nerve cord at young stages, compared with wild type. Conversely, mutation of dgk-1 enhances synaptic activity and is associated with reduced beading in these same axons.10
MAPK signaling pathway
MAPK signaling pathways regulate many processes including cell proliferation, differentiation, survival and apoptosis. The neuronal c-Jun N-terminal kinase (jnk-1) is a positive modulator of DAF-16 and is involved in regulating longevity. Overexpression of jnk-1 extends lifespan in C. elegans, whereas loss of jnk-1 shortens lifespan.26 Loss of jnk-1 also results in a higher frequency of branching in both touch receptor and GABAergic neurons.8 JKK-1 and MEK-1 are stress-responsive kinases upstream of JNK-1 that both show strong similarity to the mammalian MAP kinase kinase MKK-7.27 Loss of mek-1 did not affect branching frequency, but loss of jkk-1 resulted in the acceleration of the branching phenotype.8 This suggests that a pathway involving JNK-1 and JKK-1, but not MEK-1, regulates neuronal aging in C. elegans.
Interestingly, the loss of genes strictly required for axon regeneration following laser axotomy, namely dlk-1, mkk-4 and pmk-3,28 does not affect the frequency of branching in touch neurons.8 This indicates that pathways involved in axon regeneration and age-related branching are distinct. Other C. elegans MAPK genes are also differentially involved in neuronal aging. MLK-1 is thought to activate the p38 homolog PMK-1, and loss of this protein results in accelerated branching in touch neurons. However, loss of NSY-1, orthologous to the human apoptosis signal-regulating kinases (ASKs),29 or SEK-1, a MAP kinase kinase able to activate both PMK-1 and JNK-1,30 did not affect neuronal aging.8
Longevity and aging effectors
The capacity of other factors that regulate lifespan independently of insulin-like signaling to influence neuronal aging has also been explored. For instance, the eat-2 mutant has impaired pharyngeal pumping and a DAF-16-independent lifespan extension that is attributed to caloric restriction.31 eat-2 mutants did not, however, show a delayed onset of neuronal defects.8,10
lmn-1 encodes a conserved nuclear lamin protein in C. elegans. Mutations in lamin genes in humans are associated with an aging disorder known as Hutchinson-Gilford Progeria Syndrome (HGPS),32 and in C. elegans result in a shortened lifespan.33 Transcript levels of lmn-1 are also reduced in adult worms compared with embryos.34 Interestingly, mutations in lmn-1 also result in a higher frequency of touch neuron defects in young adulthood.10
Respiration can also affect aging. Modest inhibition of respiration leads to an extension of lifespan in both invertebrates35,36 and vertebrates.37,38 clk-1 encodes a ubiquinone biosynthetic enzyme, and mutations in this gene result in reduced respiration and lifespan extension.36 clk-1 mutants also show delayed touch neuron branching.8 In contrast, mev-1 respiratory chain mutants are short-lived39 and display an accelerated onset of branching.8 These data suggest that factors that affect respiration rates also affect neuronal health.
Do regulators of neuronal aging act cell autonomously?
These reports indicate that many molecular players regulate neuronal aging in C. elegans. Although many of these genes are expressed in neurons, some, including daf-2 and daf-16, are also expressed in other tissues.40 Despite this potential complexity, Tank and colleagues found that neuronal re-expression of DAF-16 in daf-16;daf-2 double mutants was sufficient to rescue the delayed onset of neuronal defects observed in daf-2 single mutants, identifying neurons as the cellular focus of DAF-16-mediated control of neuronal aging.8 The converse experiment, using RNAi to knock down daf-16 solely in non-neuronal cells in a daf-2 mutant, resulted in a restoration to wild-type lifespan, but did not affect the delay in neuronal branching that is observed in daf-2 mutants.8 In contrast, Pan and colleagues found that re-expression of DAF-16 in all neurons or in body wall muscles in a daf-16 single mutant was not sufficient to rescue the increased accumulation of neuronal defects.10 It is possible that these contrasting observations could be due to differences in scoring parameters. In addition, Tank et al., observed that knock down of daf-2 in non-neuronal cells resulted in wild-type levels of branching in touch neurons despite these animals displaying an extended lifespan.8 The observations of Tank et al., suggest that daf-2 and daf-16 are able to regulate neuronal aging in a cell-autonomous manner. In addition, these observations indicate that, at least in the case of the IIS pathway, regulation of neuronal aging and whole organism lifespan can be decoupled from one another.
As previously mentioned, the IIS-regulated transcription factor HSF-1 was also reported to regulate neuronal aging.9,10 Interestingly, re-expression of HSF-1 solely in touch neurons was able to completely rescue this phenotype,9 indicating that, like DAF-16, HSF-1 can also act cell-autonomously to regulate neuronal aging.
PTL-1 Regulates Neuronal Aging and Lifespan
Protein with tau-like repeats (PTL-1) is the sole C. elegans homolog of tau and MAP2/4, which are members of the mammalian family of microtubule-associated proteins (MAPs).41,42 Tau is pathologically important in several neurodegenerative diseases, as the progressive accumulation of tau and the formation of insoluble aggregates is associated with disease progression (reviewed in refs. 43–46). PTL-1 is expressed in neurons, with particularly high levels detected in the touch receptor neurons. It regulates microtubule assembly in vitro41,42 and in vivo regulates kinesin/dynein motility, physically associating with UNC-104 (kinesin-3).47 These latter observations are mirrored in the mammalian system where tau impairs kinesin-dependent anterograde transport by pathologically interacting with kinesin adaptor molecules such as JIP1.48,49 We have recently demonstrated a role for PTL-1 in the regulation of neuronal aging and lifespan of C. elegans.50
In our study, we investigated the effect of mutations in PTL-1 on the touch receptor and VNC GABAergic neurons. Two mutant strains of ptl-1 are available: ptl-1(ok621) is a null allele51 whereas ptl-1(tm543) putatively generates a truncated protein lacking the microtubule-binding repeat region of PTL-1. We observed branching and beading in ptl-1 mutants at a higher frequency in younger stages compared with wild-type controls. This was observed in both touch receptor and GABAergic neurons, indicating PTL-1 is likely to be a general regulator of neuronal aging. By imaging ALM touch neurons in populations of animals over time, we found that cell body branching often appears first with the highest prevalence, followed by axon beading and axon branching later in adulthood. In addition, we observed the heterogeneous sensitivity toward aging within the six touch receptor neurons. Posterior touch neurons had an accelerated onset and more severe aging defects compared with the anterior touch neurons. We were able to rescue the appearance of these defects to wild-type rates by re-expressing PTL-1 in the null mutant under the regulation of the ptl-1 promoter. Interestingly, increasing the gene dosage of PTL-1 by stable integration of a PTL-1 transgene also resulted in a higher frequency of abnormal neuronal structures in young transgenic animals compared with wild-type.50
We also observed that worms lacking PTL-1 showed a 37% shortening of lifespan compared with wild-type controls.50 It therefore appears that PTL-1 is a neuronal-specific protein41,42 that regulates whole organism aging.50 There are several examples in which the manipulation of neurons alone affects lifespan. In C. elegans, neuron-specific expression of daf-2 or age-1 (homologous to phosphoinositide 3-kinase) was able to rescue lifespan extension in daf-2 or age-1 mutants, respectively.8,52 In addition, direct ablation of some olfactory and gustatory neurons affects lifespan.53 These examples highlight the importance of neurons and neuronal gene function in the regulation of lifespan. PTL-1 activity in neurons may therefore contribute to wild-type longevity.
Conclusions
C. elegans is a powerful model to study aging, due to its well-characterized anatomical structures, short lifespan and ease of maintenance. Recent reports have described progressive morphological changes in C. elegans neurons,8-10 and these phenotypes can be used as parameters to track neuronal aging in the worm. Although the specific molecular mechanisms governing the formation of these novel outgrowths are yet to be defined, neuronal aging clearly involves conserved pathways such as insulin- and MAPK-regulated signaling. We have also observed premature neuronal aging in worms lacking the homolog of tau, a protein that in mammals is known to be involved in neurodegenerative disorders. Future studies on neuronal aging could investigate the molecular mechanisms that influence age-associated structural changes, as well as the interplay between neuronal health and whole organismal lifespan. The use of C. elegans to investigate neuronal aging provides important information on physiological aging of the nervous system as well as insights into age-associated pathology.
Acknowledgments
H.R.N. is supported by a University of Sydney Re-entry Fellowship, J.G. by the Estate of Dr Clem Jones AO, and grants from the Australian Research Council and the National Health and Medical Research Council of Australia. The authors thank members of the Nicholas lab and Götz lab for helpful discussions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/25288
References
- 1.Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–14. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 2.Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2004;101:8084–9. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–12. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youngman MJ, Rogers ZN, Kim DH. A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002082. doi: 10.1371/journal.pgen.1002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- 6.Hedden T, Gabrieli JD. Insights into the ageing mind: a view from cognitive neuroscience. Nat Rev Neurosci. 2004;5:87–96. doi: 10.1038/nrn1323. [DOI] [PubMed] [Google Scholar]
- 7.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–35. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tank EM, Rodgers KE, Kenyon C. Spontaneous age-related neurite branching in Caenorhabditis elegans. J Neurosci. 2011;31:9279–88. doi: 10.1523/JNEUROSCI.6606-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toth ML, Melentijevic I, Shah L, Bhatia A, Lu K, Talwar A, et al. Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J Neurosci. 2012;32:8778–90. doi: 10.1523/JNEUROSCI.1494-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan CL, Peng CY, Chen CH, McIntire S. Genetic analysis of age-dependent defects of the Caenorhabditis elegans touch receptor neurons. Proc Natl Acad Sci USA. 2011;108:9274–9. doi: 10.1073/pnas.1011711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall DH, Russell RL. The posterior nervous system of the nematode Caenorhabditis elegans: serial reconstruction of identified neurons and complete pattern of synaptic interactions. J Neurosci. 1991;11:1–22. doi: 10.1523/JNEUROSCI.11-01-00001.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 13.Bertoni-Freddari C, Fattoretti P, Paoloni R, Caselli U, Galeazzi L, MeierRuge W. Synaptic structural dynamics and aging. Gerontologia. 1996;42:170–80. doi: 10.1159/000213789. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto A. Synaptic changes in the perineal motoneurons of aged male rats. J Comp Neurol. 1998;400:103–9. doi: 10.1002/(SICI)1096-9861(19981012)400:1<103::AID-CNE7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 15.Geinisman Y, de Toledo-Morrell L, Morrell F. Loss of perforated synapses in the dentate gyrus: morphological substrate of memory deficit in aged rats. Proc Natl Acad Sci USA. 1986;83:3027–31. doi: 10.1073/pnas.83.9.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geinisman Y, Detoledo-Morrell L, Morrell F, Heller RE. Hippocampal markers of age-related memory dysfunction: behavioral, electrophysiological and morphological perspectives. Prog Neurobiol. 1995;45:223–52. doi: 10.1016/0301-0082(94)00047-L. [DOI] [PubMed] [Google Scholar]
- 17.Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/S0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- 18.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 19.Gaglia MM, Jeong DE, Ryu EA, Lee D, Kenyon C, Lee SJ. Genes that act downstream of sensory neurons to influence longevity, dauer formation, and pathogen responses in Caenorhabditis elegans. PLoS Genet. 2012;8:e1003133. doi: 10.1371/journal.pgen.1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alper S, McElwee MK, Apfeld J, Lackford B, Freedman JH, Schwartz DA. The Caenorhabditis elegans germ line regulates distinct signaling pathways to control lifespan and innate immunity. J Biol Chem. 2010;285:1822–8. doi: 10.1074/jbc.M109.057323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–9. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 22.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–83. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2003;15:657–64. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel BE, Hedgecock EM. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development. 2001;128:883–94. doi: 10.1242/dev.128.6.883. [DOI] [PubMed] [Google Scholar]
- 25.Kubota Y, Nagata K, Sugimoto A, Nishiwaki K. Tissue architecture in the Caenorhabditis elegans gonad depends on interactions among fibulin-1, type IV collagen and the ADAMTS extracellular protease. Genetics. 2012;190:1379–88. doi: 10.1534/genetics.111.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci USA. 2005;102:4494–9. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villanueva A, Lozano J, Morales A, Lin X, Deng X, Hengartner MO, et al. jkk-1 and mek-1 regulate body movement coordination and response to heavy metals through jnk-1 in Caenorhabditis elegans. EMBO J. 2001;20:5114–28. doi: 10.1093/emboj/20.18.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–6. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wes PD, Bargmann CI. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 2001;410:698–701. doi: 10.1038/35070581. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka-Hino M, Sagasti A, Hisamoto N, Kawasaki M, Nakano S, Ninomiya-Tsuji J, et al. SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 2002;3:56–62. doi: 10.1093/embo-reports/kvf001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raizen DM, Lee RY, Avery L. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics. 1995;141:1365–82. doi: 10.1093/genetics/141.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prokocimer M, Barkan R, Gruenbaum Y. Hutchinson-Gilford progeria syndrome through the lens of transcription. Aging Cell. 2013 doi: 10.1111/acel.12070. In press. In press. [DOI] [PubMed] [Google Scholar]
- 33.Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, et al. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:16690–5. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–95. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Copeland JM, Cho J, Lo T, Jr., Hur JH, Bahadorani S, Arabyan T, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–8. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Kayser EB, Sedensky MM, Morgan PG, Hoppel CL. Mitochondrial oxidative phosphorylation is defective in the long-lived mutant clk-1. J Biol Chem. 2004;279:54479–86. doi: 10.1074/jbc.M403066200. [DOI] [PubMed] [Google Scholar]
- 37.Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/- mice. J Biol Chem. 2008;283:26217–27. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dell’Agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, et al. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–44. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 39.Ishii N, Takahashi K, Tomita S, Keino T, Honda S, Yoshino K, et al. A methyl viologen-sensitive mutant of the nematode Caenorhabditis elegans. Mutat Res. 1990;237:165–71. doi: 10.1016/0921-8734(90)90022-J. [DOI] [PubMed] [Google Scholar]
- 40.Kimura KD, Riddle DL, Ruvkun G. The C. elegans DAF-2 insulin-like receptor is abundantly expressed in the nervous system and regulated by nutritional status. Cold Spring Harb Symp Quant Biol. 2011;76:113–20. doi: 10.1101/sqb.2011.76.010660. [DOI] [PubMed] [Google Scholar]
- 41.McDermott JB, Aamodt S, Aamodt E. ptl-1, a Caenorhabditis elegans gene whose products are homologous to the tau microtubule-associated proteins. Biochemistry. 1996;35:9415–23. doi: 10.1021/bi952646n. [DOI] [PubMed] [Google Scholar]
- 42.Goedert M, Baur CP, Ahringer J, Jakes R, Hasegawa M, Spillantini MG, et al. PTL-1, a microtubule-associated protein with tau-like repeats from the nematode Caenorhabditis elegans. J Cell Sci. 1996;109:2661–72. doi: 10.1242/jcs.109.11.2661. [DOI] [PubMed] [Google Scholar]
- 43.Lee G, Leugers CJ. Tau and tauopathies. Prog Mol Biol Transl Sci. 2012;107:263–93. doi: 10.1016/B978-0-12-385883-2.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris M, Maeda S, Vossel K, Mucke L. The many faces of tau. Neuron. 2011;70:410–26. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Götz J, Lim YA, Ke YD, Eckert A, Ittner LM. Dissecting toxicity of tau and beta-amyloid. Neurodegener Dis. 2010;7:10–2. doi: 10.1159/000283475. [DOI] [PubMed] [Google Scholar]
- 46.Götz J, Ittner LM, Fändrich M, Schonrock N. Is tau aggregation toxic or protective: a sensible question in the absence of sensitive methods? J Alzheimers Dis. 2008;14:423–9. doi: 10.3233/jad-2008-14410. [DOI] [PubMed] [Google Scholar]
- 47.Tien NW, Wu GH, Hsu CC, Chang CY, Wagner OI. Tau/PTL-1 associates with kinesin-3 KIF1A/UNC-104 and affects the motor’s motility characteristics in C. elegans neurons. Neurobiol Dis. 2011;43:495–506. doi: 10.1016/j.nbd.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 48.Ittner LM, Ke YD, Götz J. Phosphorylated Tau interacts with c-Jun N-terminal kinase-interacting protein 1 (JIP1) in Alzheimer disease. J Biol Chem. 2009;284:20909–16. doi: 10.1074/jbc.M109.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–9. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chew YL, Fan X, Götz J, Nicholas HR. PTL-1 regulates neuronal integrity and lifespan in C. elegans. J Cell Sci. 2013;126:2079–91. doi: 10.1242/jcs.jcs124404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon P, Hingula L, Krasny ML, Swienckowski JL, Pokrywka NJ, Raley-Susman KM. The invertebrate microtubule-associated protein PTL-1 functions in mechanosensation and development in Caenorhabditis elegans. Dev Genes Evol. 2008;218:541–51. doi: 10.1007/s00427-008-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–50. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 53.Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/S0896-6273(03)00816-X. [DOI] [PubMed] [Google Scholar]
- 54.McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol. 2003;68:159–70. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 55.Clark SG, Chiu C. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development. 2003;130:3781–94. doi: 10.1242/dev.00571. [DOI] [PubMed] [Google Scholar]
- 56.McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389:870–6. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]