Abstract
Canonical Wnt signaling supports the pluripotency of embryonic stem cells (ESCs) but also promotes differentiation of early mammalian cell lineages. To explain these paradoxical observations, we explored the gene regulatory networks at play. Canonical Wnt signaling is intertwined with the pluripotency network comprising Nanog, Oct4, and Sox2 in mouse ESCs. In defined media supporting the derivation and propagation of ESCs, Tcf3 and β-catenin interact with Oct4; Tcf3 binds to Sox motif within Oct-Sox composite motifs that are also bound by Oct4-Sox2 complexes. Further, canonical Wnt signaling up-regulates the activity of the Pou5f1 distal enhancer via the Sox motif in ESCs. When viewed in the context of published studies on Tcf3 and β-catenin mutants, our findings suggest Tcf3 counters pluripotency by competition with Sox2 at these sites, and Tcf3 inhibition is blocked by β-catenin entry into this complex. Wnt pathway stimulation also triggers β-catenin association at regulatory elements with classic Lef/Tcf motifs associated with differentiation programs. The failure to activate these targets in the presence of a MEK/ERK inhibitor essential for ESC culture suggests MEK/ERK signaling and canonical Wnt signaling combine to promote ESC differentiation.
Keywords: Mouse embryonic stem cells, Wnt, β-catenin, 2i, pluripotency, differentiation
Introduction
A central question in all stem cell-based systems is how the balance of stem cell maintenance and commitment is regulated. Embryonic stem cells (ESCs) derived directly from the early mammalian embryo provide a particularly attractive model given their capacity for long-term propagation as stem cells under defined culture conditions and their potential to generate all cell types of the adult organism 1. The pluripotency of ESCs is dependent on a set of core transcriptional regulators, including Pou5f1/Oct4, Sox2, and Nanog (NOS) 2, 3. The co-expression of Oct4, Sox2, and Klf4, a member of a family of transcriptional regulators with redundant roles in ESC maintenance 4, is sufficient for a broad range of differentiated cell types to acquire a pluripotent state that closely resembles that of ESCs 5, 6. Direct analysis of the targets of these transcriptional regulators has demonstrated that core pluripotency factors co-occupy cis-regulatory elements near ESC specific genes, providing strong evidence for co-regulatory inputs into the pluripotency gene regulatory network, as well as mutual reinforcement of each factor’s own expression 2–4, 7, 8.
Several secreted factors are pivotal to maintaining ESC properties; their addition to culture medium replaces the requirement for serum and feeder cell support in the maintenance and propagation of ESCs 9–11. In particular, leukemia inhibitor factor (LIF) acts through Stat3 to maintain the pluripotency of mouse ESCs (mESCs), whereas bone morphogenetic protein (BMP) activity directed activation of inhibitor of DNA binding (Id) regulatory factors replaces serum requirements 9, 12.
Recent studies have identified two small molecule pathway modulators, PD0325901 (PD03) and CHIR99021 (CHIR), which substitute for LIF and BMP in defined ESC medium to enable the isolation and propagation of mouse ESCs 13, and for the first time ESCs from the rat 14. PD03 is an inhibitor of mitogen-activated protein kinase kinase (MEK) 15; MEK action lies downstream of several receptor tyrosine kinase-mediated signaling pathways 16 including the Fibroblast Growth Factor (FGF) pathway. FGF signaling is critical in establishing and maintaining trophectodermal (TE) precursors, the first differentiated cell lineage to be established by the totipotent mammalian embryo 17. CHIR inhibits glycogen synthase kinase-3 (GSK3); as GSK3β-directed phosphorylation and degradation of β-catenin suppresses canonical Wnt signaling, CHIR is a potent agonist of the Wnt signaling pathway 15, 18–20. In canonical Wnt signaling, the accumulation of cytoplasmic β-catenin enables its nuclear entry and complexing with members of the Lef/Tcf family of transcriptional regulators 21. In the absence of β-catenin, Lef/Tcf factors bind DNA directly at a consensus Lef/Tcf site, and recruit transducin-like enhancer of split (TLE) proteins to silence target gene activity. In contrast, their dimerization with β-catenin generates transcriptional activating complexes that bind to cis-regulatory modules activating target genes 22.
Analysis of ESC culture and embryonic development provide conflicting views of the role of β-catenin-dependent, canonical Wnt signaling on ESC cultures. Addition of recombinant Wnt3a, a Wnt ligand activating canonical Wnt signaling, together with LIF is reported to support ESC pluripotency in the absence of other factors 23–25. Further, CHIR-mediated stimulation of canonical Wnt signaling in the presence of PD03 blocks an intrinsic tendency of mouse ESCs to differentiate, enabling continued replication of ESCs in a pluripotent state 13. BIO, another GSK-3 inhibitor, has been reported to maintain ESCs via up-regulation of LIF 26 and enhance somatic cell reprogramming via cell-fusion through the accumulation of β-catenin 27. Wnt signaling also promotes reprogramming to induced pluripotent cells (iPSCs), substituting for c-Myc in the efficient propagation of iPSCs derived from mouse embryonic fibroblasts infected with Sox2, Oct4, and Klf4 28. The down-regulation of “stemness marker genes” in ESCs lacking functional β-catenin supports a role of canonical Wnt signaling in maintenance of pluripotency 29, although a study of an independently-generated β-catenin-deficient mES cell line reached a different conclusion 30.
At the DNA level, genome wide interaction studies of canonical Wnt signaling effectors have largely focused around transcription factor 7 like 1 (Tcf7l1, commonly known as Tcf3), a transcriptional component that is thought to predominantly repress Wnt-target genes. Tcf3 binding shows a strong intersection at sites co-bound by major pluripotency regulators 7, 31. Recent reports indicate that a loss of Tcf3 can substitute for CHIR in 2i, which is consistent with an inhibitory action of this member on the pluripotency program 32. CHIR-mediated stimulation of β-catenin activity is proposed to both abrogate Tcf3 repression on the pluripotency network through a transactivation independent mechanism and to promote pluripotency through an interaction with Oct4 32–34. Together these data provide evidence for a canonical Wnt/β-catenin pathway action in promoting the pluripotent state of stem cells.
Conversely, canonical Wnt signaling has also been shown to induce specification of TE and mesendoderm lineages 35–37, and mouse embryos lacking β-catenin, Wnt3, or two Wnt co-receptors, Lrp5 and Lrp6, arrest prior to gastrulation linking canonical Wnt signaling to axial specification and mesendodermal induction 38–43. The conflicting reports present a mechanistic paradox: how does β-catenin-dependent canonical Wnt signaling promotes both the stem cell state and the early commitment of pluripotent cells to specific cell lineages of the gastrulating embryo?
To address this question, we engineered a mESC line to produce a Biotin-FLAG epitope tagged form of β-catenin from the β-catenin (Ctnnb1) locus, and performed genome-wide chromatin immunoprecipitation (ChIP) in conjunction with high throughput sequencing (ChIP-seq) to directly address β-catenin target sites on canonical Wnt signaling activation in mouse ESCs. When these data are viewed in conjunction with extensive expression profiling of ESCs under pluripotency and differentiation promoting conditions, together with DNA binding studies of key pluripotency determinants and their complex formation with β-catenin, a mechanistic model emerges that can reconcile the opposing actions of canonical Wnt signaling discussed above.
Materials and methods
For full details see the supplementary methods.
Generation of ES cell lines
Ctnnb1-BioFLneo and Ctnnb1-BioFLneo; BirA knock-in ES cell lines generation are described in supplementary methods.
ESC culture
ESCs were cultured in serum+LIF complete media and 2i according to standard procedures (supplementary methods).
Immunofluorescence, Immunoblot, and Co-immunoprecipitation Assays
Immunofluorescence and immunoblot were performed according to standard techniques described in supplementary methods. Nuclear extracts were analyzed according to the manufacturer’s instructions for Nuclear Complex Co-IP Kit (Active Motif, 54001). Co-immunoprecipitation experiments were performed according to the manufacturer’s instructions for Protein A/G HP SpinTrap Buffer Kit (GE Healthcare, 28-9135-67).
ChIP, ChIP-seq and ChIP-qPCR
ChIP was performed according to Vokes et al., 2007 (supplementary methods). The construction of ChIP-seq libraries was performed with ChIP-seq DNA Sample Prep Kit (Illumina, IP-102-1001) according to manufacturer’s instruction and sequenced on Genome Analyzer II (Illumina) machine. qPCR was performed with Biorad iQ™ SYBR ® Green Supermix (#170-8880). Fold enrichment was calculated by normalizing ChIP sample against input, and target region against control region as follows. ΔCt=Ct(ChIP)-Ct(input); ΔΔCt=ΔCt(target region)-ΔCt(control region); Fold enrichment=2−ΔΔCt
Expression profiling
Microarrays were conducted in triplicate using Affymetrix GeneChip mouse Gene 1.0 ST array (Affymetrix, 901169) according to the manufacturer’s instructions, and data normalized by RMA in R (supplementary methods).
EMSA
Nuclear extracts were isolated from 293T cells overexpressing Pou5f1, Sox2, or Tcf7l1 as described 44. Diogoxigenin-labeled probes were constructed using the DIG Gel Shift Kit (Roche, 3353591). Binding reaction and subsequent EMSA was performed according to the manufacture’s instruction.
Bioinformatic data analysis
See supplementary methods for complete description of data analysis.
Accession numbers
Sequencing and microarray data have been deposited to GEO with accession number GSE43597.
Results
Genome-wide profiling of the canonical Wnt regulatory network in mESCs
To take advantage of in vivo biotinylation and FLAG-tag technologies in analyzing canonical Wnt signaling in mESCs, we generated a Ctnnb1-Biotin-3xFLAG knock-in ESC line (Ctnnb1-BioFLneo ESC) using gene-targeting strategies 45, 46. The modified allele places a carboxyl-terminal epitope tag on β-catenin comprising three tandem copies of a FLAG (3xFLAG) epitope 47 and a short peptide that serves as a substrate for in vivo biotinylation in cells expressing the Escherichia coli Biotin ligase, BirA 46, 48, 49 (Figure S1A). Correct targeting of the modified Ctnnb1 knocked-in allele was confirmed by long-range PCR (Figure S1B). Ctnnb1-BioFLneo ESCs were then engineered to stably express BirA to ensure biotinylation of β-catenin-BioFL proteins (Ctnnb1-BioFLneo; BirA ESC).
We confirmed the integrity, specificity and activity of the allele through the following observations. First, production and localization of β-catenin-BioFL protein was comparable to that of the wild-type protein (Figure S1C and S1D). Second, biotinylated β-catenin-BioFL proteins were detected using streptavidin conjugated reagents in Ctnnb1-BioFLneo; BirA ESCs, but not in Ctnnb1-BioFLneo ESCs (Figure S1D). Finally, biotinylated β-catenin-BioFL appeared to function normally; mice homozygous for Ctnnb1-BioFL alleles carrying BirA ligase are viable with no apparent abnormalities (SO and APM, manuscript in preparation). As a control cell line for subsequent analyses, we also generated a BirA-expressing ESC line (BirA ESC; Figure S1E).
To better understand roles of canonical Wnt signaling in ESC biology, we set out to identify genomic targets of β-catenin, applying ChIP-seq to Ctnnb1-BioFLneo; BirA ESCs. Ctnnb1-BioFLneo; BirA ESCs were cultured on feeder cells in standard condition with serum+LIF complete media (abbreviation: CM), and treated with CHIR (CM+CHIR) for 16 hours, then subjected to a series of ChIP-seq procedures. β-catenin-DNA complexes were pulled down using anti-FLAG antibody (FLAG-ChIP) or streptavidin (Biotin-ChIP) in parallel. DNA obtained from each ChIP procedure was independently sequenced. We also repeated FLAG-ChIP on Ctnnb1-BioFLneo; BirA ESCs without CHIR treatment.
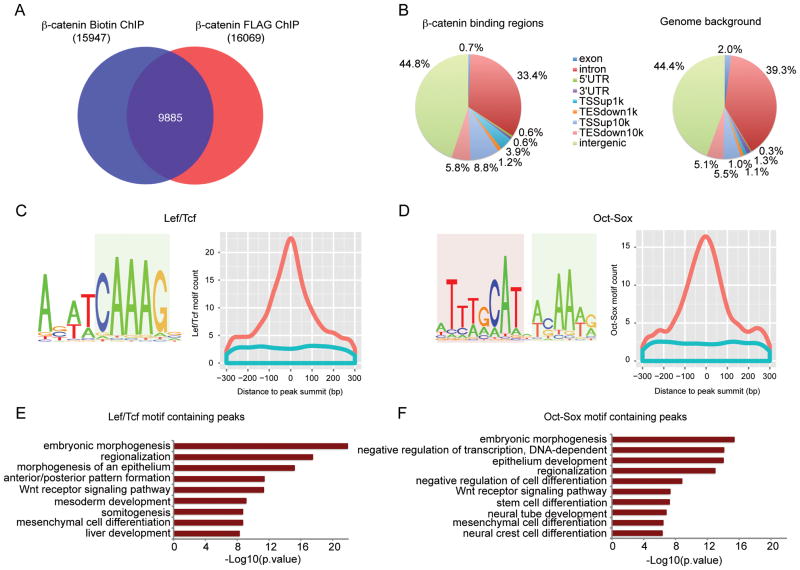
We obtained 15947 and 16069 binding regions for Biotin-ChIP and FLAG-ChIP replicates, respectively, from ChIP-seq of CHIR treated Ctnnb1-BioFLneo; BirA ESCs (Figure S2A–C; see supplementary methods for peak calling program and criteria). In contrast, only a small number of regions were bound by β-catenin in CM without CHIR (data not shown), suggesting only background levels of endogenous canonical Wnt signaling in CM supplemented feeder-supported cultures. The intersection of the two CHIR-dependent data sets identified 9885 regions (62.0% in Biotin-ChIP and 61.5% in FLAG-ChIP) in common (pearson correlation=0.89, Figure S2C). Shared peaks have a higher intensity and peak ranking than ChIP regions unique to a single dataset suggesting that the intersection represents the most robust set of bone-fide interaction sites (Figure S2D). This independently validated intersection formed the foundation for subsequent analysis (Figure 1A) and representative peaks associated with pluripotency sustaining transcriptional components were validated by qPCR (Figure S2E). All binding sites were annotated relative to ref-seq gene predictions (Figure 1B and Table S1). When compared across the genome, β-catenin associated regions show enrichment within 10kb of the transcriptional start site (TSS), and a relative depletion in intronic and exonic regions (Figure 1B). Approximately 16% of all annotated genes are associated with β-catenin binding in CHIR treated mESCs using a ‘gene’ definition as the region 10 kb upstream of the TSS plus the gene body.
Figure 1. Genome-wide mapping of β-catenin binding regions in mESCs cultured in CM.
(A) Venn diagram showing the overlap between β-catenin Biotin ChIP-seq and FLAG ChIP-seq peaks.
(B) Genome-wide distribution of β-catenin binding regions relative to mouse genes compared with random control region genomic distribution. Binding regions were annotated as exon, introns, 5′ un-translated region (5′ UTR), 3′ UTR, within 0–1 kb upstream of TSS (TSSup1k), within 1–10 kb upstream of TSS (TSSup10k), within 0–1 kb downstream of TES (TESdown1k), within 1–10kb downstream of TES (TESdown10k), or > 10kb away from the nearest genes (intergenic).
(C) (D) Top enriched motifs recovered from de novo motif analysis of β-catenin binding regions. Left panels show motif logos. HMG box motif is highlighted in light blue, and POU family motif in light red. Right panels show histogram of motifs ± 300bp around peak summit of β-catenin (orange) or matched control peak (blue).
(E) (F) GO terms enriched for β-catenin peaks containing Lef/Tcf motif (E) or Oct-Sox motif (F) using GREAT. The –log10 of the raw binomial p-value is reported.
Motif analysis was performed on 400 base-pair regions centered on the peak summit of β-catenin association to identify statistically enriched DNA motifs within the data set (Figure S2F). As expected, the DNA target site for Lef/Tcf factors, the DNA-binding partner for β-catenin, was highly enriched in the dataset: 35.0% of all β-catenin peaks predicted a Lef/Tcf site, versus 11.1% in matched control regions (two-proportion z test, p-value < 1e-350). Strikingly, an Oct-Sox composite motif was also highly enriched (26.1% of all β-catenin peaks, p-value < 1e-324) and like Lef/Tcf predictions, centered on the predicted peak of β-catenin binding (Figure 1C and 1D). We also identified motifs that matched binding sites for Klf4, Zic, Esrrb, E2a, and AP-2 (Figure S2F). The distribution of these suggests enrichment in the region but a less direct association with β-catenin binding (data not shown). Importantly, the enrichment of Lef/Tcf sites provides strong support for the quality of the data set, while the enrichment of Oct-Sox motifs near β-catenin peak summits suggests an interplay between β-catenin and the pluripotency circuit on canonical Wnt signaling stimulation. Interestingly, gene ontology (GO) analysis using Genomic Regions Enrichment of Annotations Tool (GREAT)50 revealed that while embryogenesis-related and Wnt receptor signaling pathway-related genes were both enriched in Lef/Tcf motif-containing and Oct-Sox motif-containing β-catenin peaks, the former category was also enriched in mesoderm development-related term, and the latter stem cell- and neural-related terms (Figure 1E and 1F).
Analysis of β-catenin, Tcf3, Sox2, Oct4, and Nanog interactions at target genes points to distinct enhancer modules mediating the actions of canonical Wnt signaling
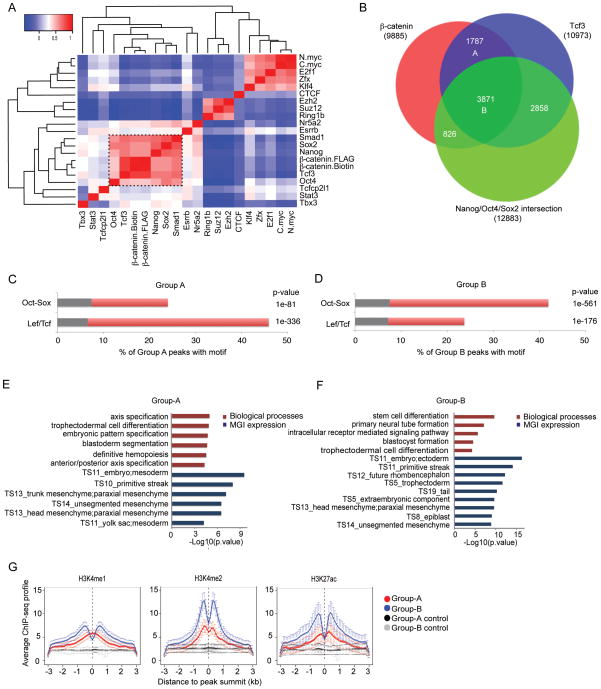
The recovery of Oct-Sox motif within β-catenin binding regions prompted us to compare the β-catenin data with previously-published ChIP-seq data for 19 TFs associated with maintenance of pluripotency, induction of iPS cells, and Wnt action (Figure S3A): the core pluripotency factors Nanog/Oct4/Sox2 (NOS, see Figure S3B and S3C for the comparison of two independent datasets) 7, 8; Smad1/Stat3, effectors of key mESCs signaling pathways 8; Tcfcp2l1/Tbx3/Klf4/C-myc/N-myc/Zfx, reprogramming factors important for self-renewal 8, 51; Ring1b/Ezh2/Suz12, components of polycomb repressive complexes (PRC) 8, 52; Esrrb/Nr5a2, nuclear receptors linked to the ESC state 8, 53; and Tcf3, the most abundant of the Lef/Tcf family of canonical Wnt transcriptional effectors in mESCs 7.
Through pair-wise co-binding analyses, we were able to classify binding patterns of these regulatory factors into several clusters; notably Tcf3, Nanog, Sox2, Smad1, and Oct4 interactions most closely resembled those observed through β-catenin ChIP-seq (Figure 2A). Given that β-catenin regulates gene expression through Tcf transcription factors, of which Tcf3 is most abundantly expressed in the mESCs, we first did two-way intersection of β-catenin and Tcf3 binding peaks taking only the β-catenin::Tcf3 regions to increase the credibility of binding events. A further intersection with NOS peaks, produced Group-A (β-catenin::Tcf3) and Group-B (β-catenin::Tcf3::NOS) (Figure 2B): comparison of these two categories provides an insight into whether canonical Wnt signaling action differs in the presence of NOS. We explored motif enrichment, chromatin state, and functional properties of predicted target genes adjacent to these binding regions. A clear consensus Lef/Tcf motif was the most over-represented motif in the Group-A (p-value < 1e-336, two-proportion z test) (Figure 2C), while the most enriched motif in Group-B closely resembled the published Oct-Sox motif (p-value < 1e-561, two-proportion z test) (Figure 2D). Stem cell- and ectoderm-related terms were enriched in Group-B targets, while axis specification and mesoderm terms were over-represented in Group-A targets (Figure 2E and 2F; Table S2). In terms of chromatin state, both groups displayed a strong H3K4me2 signature, an indicator of poised or active enhancer regions 55, but the signature was more prominent amongst Group B regions, suggestive of a more active state in ESCs (Figure 2G). Consistent with this view, Group-B displayed a stronger H3K4me1 and H3K27ac active enhancer signature than Group-A (Figure 2G) 56–58. We also intersected Group-A and Group-B associated genes with Group-1 and Group-2 targets as defined by Yi et al 33. Group-1 consists of genes where the response to Wnt3a addition to wild-type ESCs is similar to that observed on Tcf3 ablation (Tcf3 KO): i.e., genes predicted to be regulated by Wnt3a antagonism of Tcf3 repression of target activity. Group-2 consists of genes that show a similar response to Wnt3a independent of Tcf3 activity: i.e., Wnt3-dependent and Tcf3-independent targets. We observed a strong enrichment (p-value < 1e-14.4, two proportion z-test) of Group-A and Group-B regulated genes with the genes showing a transcriptional response to Wnt3a in a Tcf3-dependent or Tcf3-independent manner lending additional evidence to support the conclusion that β-catenin ChIP identifies likely regulatory regions mediating transcriptional regulation by Wnt3a signaling and Tcf3 binding. However, we did not observe a clear pairwise segregation between Group-A/B genes and Group-1/2 genes.
Figure 2. Characterization of β-catenin and ESC pluripotency factors binding.
(A) Heat map depicting the correlation of β-catenin and ESC factors bindings. Red: positive correlation; blue: negative correlation.
(B) Venn diagram of β-catenin, Tcf3, and intersection of Nanog/Oct4/Sox2 peak regions. Two groups of peaks are highlighted: Group-A: β-catenin::Tcf3, and Group-B: β-catenin::Tcf3::NOS.
(C) (D) Enriched motifs in Group-A and Group-B. Red: motif occurrence in β-catenin peaks; grey: motif occurrence in matched control regions with the same coverage. P-value was calculated according to two proportion z-test.
(E) (F) Functional annotation of Group-A and Group-B regions using GREAT. The –log10 of the raw binomial p-value is shown.
(G) Aggregation plots of H3K4me1, H3K4me2, and H3K27ac signals ±3 kb around the peak summit for binding regions in Group-A (red) and Group-B (blue) as well as corresponding matched control regions with standard error bars (black and grey). The analysis is done using HOMER 75. Bin size 100 bp.
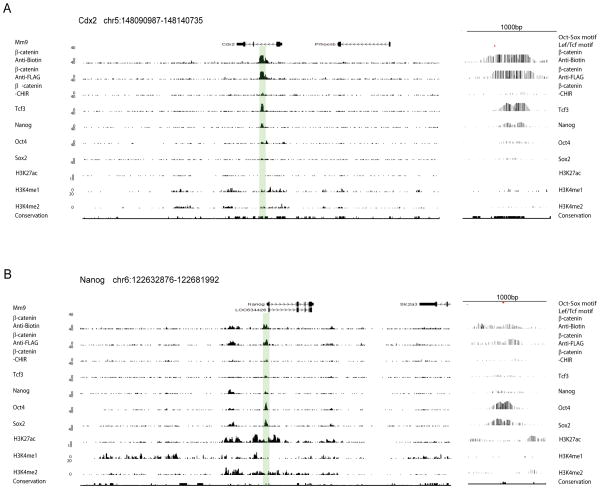
In summary, the data suggests that two groups of β-catenin binding regions are involved in transcription of two distinct categories of target genes through two distinct mechanisms, one through a Lef/Tcf-mediated DNA interaction of β-catenin with poised enhancers around differentiation-related genes (Group A) and one through cooperative interactions of NOS and β-catenin/Lef/Tcf with active enhancers around stem cell- and ectoderm-related genes (Group B). A CisGenome browser screenshot of representative genes of Group-A (Cdx2, Figure 3A; Axin2, Figure S4), and Group-B (Nanog, Figure 3B) show the strong relative signal intensity of β-catenin over Nanog/Oct4/Sox2 for Group-A versus Group-B associated genes.
Figure 3. CisGenome browser screenshots showing combinatorial binding pattern of β-catenin and core pluripotency factors in CM.
β-catenin binding to known Wnt target genes related to differentiation (Cdx2, A), and pluripotency (Nanog, B). Endogenous association of β-catenin is also displayed in the absence of CHIR stimulation (CHIR-). Tcf3, Nanog, Oct4, Sox2 and histone modification profiles displayed here are from published datasets (see Results). A zoom in on highlighted regions with motif annotation is displayed to the right side.
Activation of canonical Wnt signaling directs early mesoderm differentiation
To connect DNA association profiles of these factors with canonical Wnt signaling-mediated gene expression, we intersected β-catenin ChIP peaks with neighboring genes, with a focus on those genes that displayed differential expression between mESCs cultured in CM+CHIR and CM+XAV939 (XAV) (Table S3). XAV is a tankyrase inhibitor antagonizing Wnt signaling 59. Among the intersected gene set, canonical Wnt signaling target genes, early mesoderm-, and TE-related genes ranked top on the most up-regulated genes. However pluripotency-related genes showed no strong differential expression (Table S3).
The integrity of our microarray data set was supported by two analyses, prediction of canonical Wnt signaling target genes and correlation with published expression data sets. First, we applied an approach based upon an empirical finding that the potential of a gene being a direct target of a given TF decreases monotonically as a function of the distance of the binding site to that gene’s TSS60. Second, using a rank product based ranking method, we made a probabilistic prediction of β-catenin target gene list (Figure 4A Table S4) 61. Using a false discovery rate (FDR) of 10%, we obtained 376 and 362 putative direct target genes that were positively and negatively regulated by CHIR, respectively, in CM. This method accurately predicted known target genes of canonical Wnt signaling, such as Axin2, T, Sp5, Lef1, Cdx2, and Tcfcp2l1. Interestingly, the identification of Porcn, which encodes a key factor in the palmitoylation and secretion of Wnt ligands62, suggests a hitherto unrecognized positive feedback loop in canonical Wnt signaling.
Figure 4. Integration of β-catenin ChIP-seq and expression profiling in mESCs treated with an activator or inhibitor of canonical Wnt signaling.
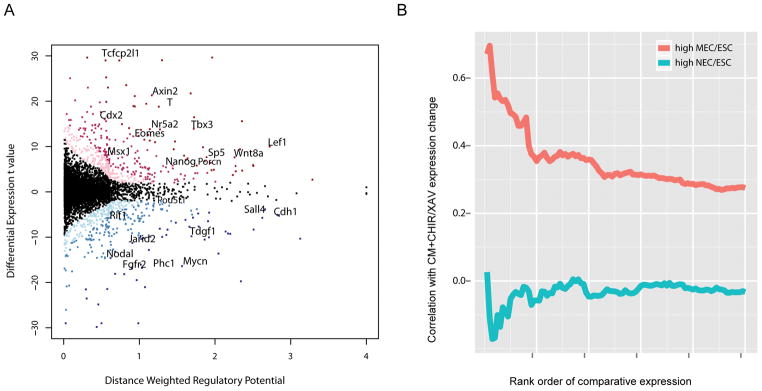
(A) Scatter plot of β-catenin direct target gene prediction based on distance weighted regulatory potential score from ChIP-seq and t-value of differential CHIR/XAV expression in CM. Red dots: up-regulated genes with FDR < 0.10; blue dots: down-regulated genes with FDR < 0.10. The darker red/blue represents the higher likelihood for a gene being β-catenin direct target. The horizontal and vertical histograms reflect the distribution of the index for distance weighted regulatory potential and differential expression t value, respectively. Representative genes are labeled.
(B) Correlation of top 1000 genes of high MEC/ESC or NEC/ESC expression ratio (from microarray data in Shen et al.63) (x-axis) with their differential expression fold changes in CM+CHIR/CM+XAV (y-axis). Genes were ranked by their expression ratio in MEC versus ESC or NEC versus ESC from high to low. For the top 1000 genes in the two ranks, their expression ratio in CM+CHIR versus CM+XAV were checked. Bins represent the top 20 genes, then the top 40 genes, etc., as determined by the MEC/ESC ratio or NEC/ESC ratio. The correlation of MEC/ESC or NEC/ESC ratio with CM+CHIR/CM+XAV ratio for each bin were calculated and plotted.
We calculated the correlation of the up-regulated genes in our data with the published microarray data comparing gene expression between ESCs, mesendoderm cells (MECs), and neural ectoderm cells (NECs) 63. The top 100 genes displaying a high MEC/ESC expression ratio showed some correlation (> 0.5) with genes exhibiting a high CHIR/XAV expression ratio. No correlation was observed with genes associated with neural ectoderm development (high NEC/ESC) (Figure 4B), consistent with the known role of canonical Wnt signaling in inducing mesendoderm and suppressing neural ectoderm development 43, 64, 65.
Similarity of β-catenin chromatin binding between CM+CHIR and 2i
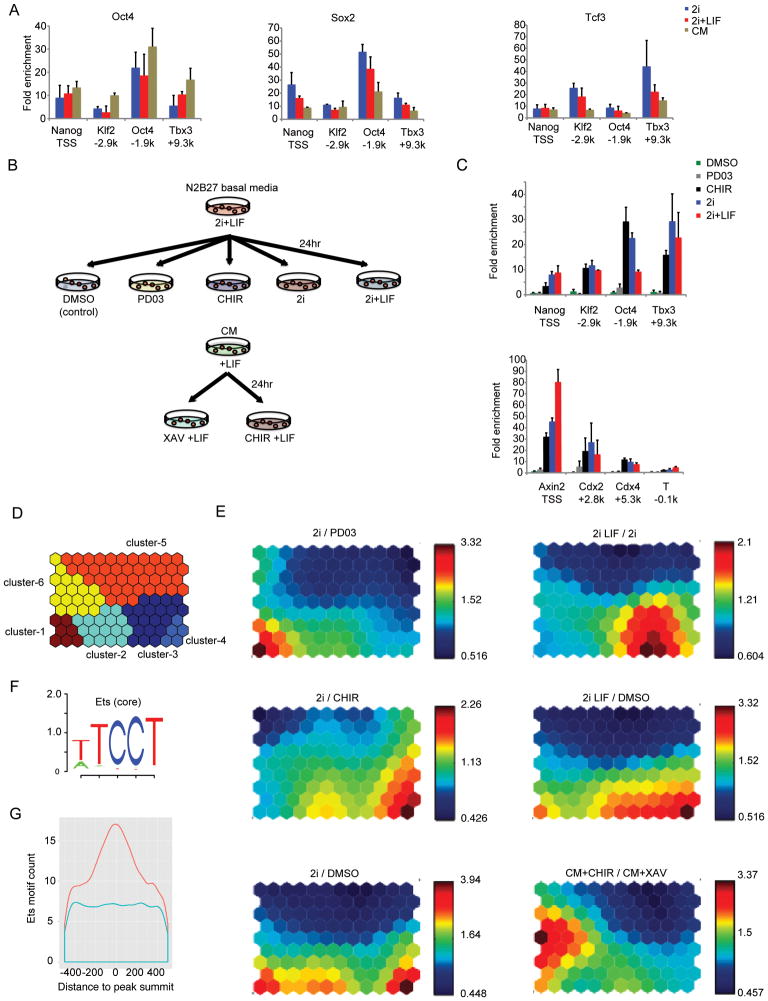
To examine the interactions of β-catenin and pluripotency network components under the 2i conditions, Ctnnb1-BioFLneo; BirA ESCs were cultured under 2i supplemented with LIF to enhance colonogenicity 13. ChIP-qPCR for Sox2, Oct4, and Tcf3 binding were conducted at cis-elements near pluripotency genes in 2i, 2i+LIF and CM conditions. The bindings of the three factors were comparable in all three conditions (Figure 5A, p-value > 0.05 from two sample t-test).
Figure 5. Roles of small molecules CHIR and PD03 in 2i.
(A) ChIP-qPCR for Oct4, Sox2 and Tcf3 interaction at defined regulatory regions surrounding pluripotency target genes in mESCs cultured in 2i, 2i+LIF, and CM. Data represent the mean of biological replicates.
(B) Experimental scheme for studying the role of CHIR and PD03 in CM and 2i-adapted mESCs. ChIP-qPCR and microarray analysis were performed in 2i-adapted mESCs cultured for 24 hours with DMSO (control), PD03, CHIR, 2i, and 2i+LIF. Cells at passage 20 under the 2i+LIF condition were subjected to each assay. Cells that had been maintained in CM on feeder cells were cultured for 24 hours in CM with CHIR or XAV prior to microarray analysis.
(C) ChIP-qPCR for β-catenin at selected loci near pluripotency-related genes (upper), differentiation-related and Wnt target genes (lower) on 2i-adapted mESCs. ChIP using anti-FLAG antibodies was performed according to the experimental scheme described in (B). Data show the mean and standard error of the mean (s.e.m.) for three biological replicates.
(D) K-means clustering was used to classify genes with expression fold change > 2 in at least one comparison group of 2i/PD03, 2i/CHIR, 2i/DMSO, 2iLIF/2i, 2iLIF/DMSO, and CM+CHIR/CM+XAV. A total of 388 genes were clustered into six clusters.
(E) Individual component maps are shown for each pair-wise comparison. Top left: 2i/PD03; middle left: 2i/CHIR; bottom left: 2i/DMSO; top right: 2iLIF/2i; middle right: 2iLIF/DMSO; bottom right: CM+CHIR/CM+XAV. In general, red indicates up-regulation and blue down-regulation. The number by each color bar is the actual number of fold change.
(F) Five bp core Ets motif logo TCCTW from TRANSFAC motif M00339.
(G) Enrichment of Ets core motif ± 500bp around β-catenin peak summit (orange) compared with matched control regions (blue).
To understand which gene category was regulated by each 2i component, and how β-catenin may participate in this regulation, we performed ChIP-qPCR for β-catenin in 2i-adapted Ctnnb1-BioFLneo; BirA ESCs cultured with DMSO, PD03, CHIR, 2i, or 2i+LIF for 24 hours (Figure 5B). A strong enrichment of β-catenin was observed in CHIR-, 2i-, and 2i+LIF-treated cells at the same pluripotency-related gene regions as those bound by β-catenin in CM+CHIR (Figure 5C, upper panel); the binding was dependent on the activation of canonical Wnt signaling, since it was lost within 24-hour of the removal of CHIR (Figure 5C; DMSO and PD03 in upper panel). Thus, stabilization of β-catenin leads to similar interactions at the DNA level in quite different culture regimens. Thus, β-catenin likely contributes to expression of pluripotency-related genes under 2i conditions by the direct association with cis-regulatory modules governing expression of the pluripotency network. Interestingly, the MEK inhibitor PD03 did not affect β-catenin association with differentiation-related gene regions (Figure 5C, lower panel).
Differentiation genes fail to be up-regulated by CHIR in 2i as in CM
Predominant activation of a mesendoderm lineage differentiation program by CHIR in CM (Figure 4B), and the association of β-catenin with differentiation-related gene regions in 2i (Figure 5C) raised a question about how differentiation is inhibited in 2i. We performed microarray analysis in 2i-adapted Ctnnb1-BioFLneo; BirA ESCs cultured with DMSO, PD03, CHIR, 2i, and 2i+LIF according to the same experimental scheme as in Figure 5B (Table S5). In this, the effect of CHIR is distinguished by comparing the relative gene expression level in the ESCs cultured in 2i to those cultured in PD03 where CHIR is absent (2i / PD03). Comparing CM+CHIR over CM+XAV (CM+CHIR / CM+XAV) provides another metric of CHIR activity, while a comparison of 2i+LIF over 2i (2iLIF / 2i) sheds light on any direct effect of LIF on gene expression. Using Kohonen Self-Organizing Maps (SOM) 66, we identified groups of genes that shared similar patterns of expression changes between such comparisons (Figure 5D), and visualized target gene expression change in a series of heat map (Figure 5E). Genes that showed a greater than 2-fold differential expression in at least one comparison were clustered (Figure 5D): the six cluster associated gene names are presented in Figure S5. In Figure 5D, each hexagonal map unit represents a SOM node, which has an underlying vector of a pair-wise fold change under different conditions. In Figure 5E, the gradient of each pair-wise fold change is displayed separately while maintaining the same topological structure as in Figure 5D. On comparing Figure 5D and 5E, clusters-1,-3, and -4 represented genes strongly up-regulated by CHIR (2i / PD03), LIF (2i LIF / 2i), and PD03 (2i / CHIR), respectively, in 2i.. Cluster-2 represented genes moderately up-regulated by CHIR (2i / PD03), LIF (2i LIF / 2i) and PD03 (2i / CHIR). Genes in cluster-6 were up-regulated by CHIR in serum conditions (CM+CHIR / CM+XAV), and those in cluster-5 were down-regulated in all comparison groups.
There are three key insights from this analysis. First, CHIR, PD03, and LIF up-regulated different sets of genes in 2i, as shown in cluster-1, cluster-3, and cluster-4, respectively. Second, CHIR up-regulated different sets of genes in 2i from those in CM (cluster-1 vs. cluster-6). Several differentiation-related genes, such as T, Cdx2, Cdx1 appear in cluster-6 that were down-regulated in 2i compared to CHIR (see 2i / CHIR) (Figure S5). We hypothesize that the lineage promoting action of canonical Wnt signaling as observed in CM may be suppressed through the deprivation of potential collaborating factors for β-catenin in 2i, or through secondary modification of transcriptional complexes without the down-regulation of β-catenin binding to cis-regulatory control regions for differentiation associated targets (Figure 5C). In support of the former, an Ets motif was significantly enriched in β-catenin binding regions – Ets factors provide the transcriptional output to MEK/ERK signaling (Figure 5F and 5G; see Discussion). Thirdly, the effect of 2i over DMSO (2i / DMSO, left bottom) appears to be an additive result of the effect of CHIR (2i / PD03, left top) and PD03 (2i / CHIR, left middle). Similarly, the effect of 2i+LIF over DMSO (2i LIF / DMSO, right middle) appears to be an additive result of CHIR (2i / PD03, left top), PD03 (2i / CHIR, left middle), and LIF (2i LIF / 2i, right top).
β-catenin complexes with Oct4 and Tcf3 at Oct-Sox motifs in 2i cultured mESCs
Binding of Oct4 and Sox2 to pluripotency-related gene regions was unaltered under 2i condition in contrast to the effect of CHIR on cells grown in CM (see earlier). To investigate the physical interaction of β-catenin and Tcf3 with these two pluripotency determinants we performed co-immunoprecipitation (Co-IP) analysis with nuclear extracts from mESCs cultured in 2i+LIF and 2i with antibodies specific to β-catenin, Tcf3, Sox2 and Oct4. Oct4 and β-catenin were co-immunoprecipitated with anti-Oct4 antibodies (Figure 6A); the failure of a reciprocal IP likely reflects epitope masking of the epitope recognized by the β-catenin antibody in an Oct4 complex 34. Tcf3 was associated with β-catenin and Oct4, but not Sox2 (Figure 6A). Consistently, Sox2 only pulled down Oct4, not β-catenin or Tcf3. These results suggest that two regulatory complexes exist under 2i conditions: one containing β-catenin, Tcf3, and Oct4 and another with Sox2 and Oct4.
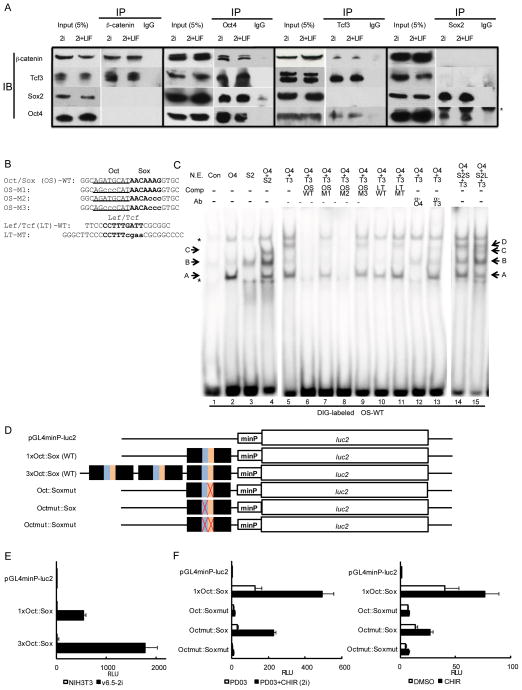
Figure 6. Physical association of β-catenin, Oct4, Sox2, and Tcf3 and in vitro binding properties of Oct4, Sox2, and Tcf3 to the Oct-Sox composite motif.
(A) Co-immunoprecipitation analysis of β-catenin, Tcf3, Oct4, and Sox2 complexes in mESCs. IP, immunoprecipitation; IB, immunoblotting. Asterisk indicates heavy chains of antibody used in IP.
(B) Sequence of oligonucleotide probes used in EMSA. Oct motif is underlined; Sox and Lef/Tcf motifs are bolded. Mutations are shown in lowercases.
(C) Cooperative bindings of Oct4, Sox2, and Tcf3 to the Oct-Sox (OS) composite motif, determined by EMSA. The indicated combinations of nuclear extracts isolated from 293T cells overexpressing Pou5f1 (Oct4), Sox2, or Tcf7l1 (Tcf3) were analyzed by EMSA using DIG-labeled OS probes. Bands A, B, C, and D denoted with arrows indicate Oct4-binary, Sox2-binary, Oct4-Sox2-ternary, and Oct4-Tcf3-ternary complexes with the Oct/Sox probe, respectively. Asterisks indicate non-specific bands. NE, nuclear extracts; Con, extracts from mock-transfected cells; O4, extracts from Oct4-overexpressing cells; S2, extracts from Sox2-overexpressing cells; S2S, smaller amount of extracts (0.2 μg) from Sox2-overexpressing cells; S2L, larger amount of extracts (2 μg) from Sox2-overexpressing cells. T3, extracts from Tcf3-overexpressing cells; Comp, unlabeled competitors; Ab, antibodies; α-O4, anti-Oct4 antibody; α-T3, anti-Tcf3 antibody. Data here is extracted from Figure S7B which provides more extensive competitor experiments and antibody supershift experiments to identity each shifted band.
(D) Schematic of luciferase reporter constructs used in (E) and (F). Pou5f1 distal enhancer region containing the Oct-Sox composite motif drives the luciferase gene with a minimal TATA-box promoter element under pGL4 vector backbone. Each mutation corresponds to mutant motifs in EMSA analysis.
(E) Luciferase reporter assay using Pou5f1 distal enhancer region in NIH3T3 cells and 2i-cultured mESCs (v6.5). Forty-eight hours after transfection with reporter constructs, cells were subjected to the assay. mESCs were maintained under 2i+LIF condition for 11 passages prior to the assay.
(F) Luciferase reporter assay in mESCs (v6.5) cultured under 2i condition (left) or CM (right). mESCs were maintained under 2i+LIF condition for 11 passages prior to the assay. Upon transfection with reporter constructs, cells were switched into basal media of 2i culture (mixture of neurobasal media, DMEM/F12, N2, and B27 supplements) with PD03 or 2i (left), or CM in the presence or absence of CHIR (right). The assay was performed 48 hours after transfection. RLU, relative light unit.
Given the overlap in ChIP peaks between Oct4, Sox2, and Tcf3, the Oct-Sox composite motif recovered in β-catenin ChIP-seq data, sequence similarity within this consensus motif between Sox and Tcf DNA binding sites and their structural similarities sharing an HMG DNA binding domain, we tested whether Tcf3 directly bound to the Oct-Sox composite motif in vitro by electrophoretic mobility shift assay (EMSA). Nuclear extracts (NE) were prepared from 293T cells over-expressing Pou5f1 (Oct4), Sox2, or Tcf7l1 (Tcf3) (Figure S6) and tested for their ability to bind a double-stranded DNA probe incorporating an Oct-Sox composite motif located within a distal Pou5f1 enhancer (Figure 6B) The complete data for all EMSA is shown in Figure S7A and B and key lanes extracted from this data set to Figure 6C.
Oct4 and Sox2 alone complex with the Oct/Sox probe (Figure S7A, lanes 2–7) whereas no binding was observed for Tcf3 (Figure S7A, lanes 8–10). In contrast, Tcf3 complexed with a Lef/Tcf binding motif (LT probe) under the same conditions (Figure S7A, lanes 12–20; band E). Next, we examined co-binding for cooperative interactions. Oct4 and Sox2 co-binding led to additional band (C) not seen when Oct4 (A) or Sox2 (B) bound alone (Figure 6C, lane 2–4; Figure S7B, lane 2–8 68). As above, no Tcf3 interaction was observed with the Oct/Sox motif and Tcf3 failed to compete with Sox2 at the Sox2 binding site (Figure S7B, lanes 9–13). However, in the presence of Oct4 and Tcf3, additional bands were observed that likely reflect ternary complexes of Oct4 and Tcf3, with Tcf3 bound at the Sox2 site (band D in Figure 6C, lanes 5–13).
Several lines of evidence support this view. First, the additional band was eliminated by unlabeled LT probe or anti-Tcf3 antibodies (Figure 6C, lanes 10 and 13), but not by unlabeled mutated LT probe (Figure 6C, lane 11). Second, the additional band was competed with the unlabeled WT Oct/Sox probe, and by one in which the Oct-motif was mutated, but not by a probe containing mutations in both Oct and Sox motifs (Figure 6C, lanes 6, 7, and 9). Finally, when Oct4 binding was competed by unlabeled Sox-mutated probe, or blocked with anti-Oct4 antibodies, the additional band disappeared (Figure 6C, lane 8 and lane 12, respectively). Together, these results suggest that Tcf3 binds to the Sox site in the Oct/Sox composite motif in an Oct4-dependent manner whereas Oct and Sox factors can independently associate with their target sites.
To clarify possible patterns of complex formation when all of the three proteins were present we incubated the Oct-Sox composite motif with Oct4, Sox2, and Tcf3 (Figure 6C, lanes 14–15). When low amounts of Sox2 were present with the other two proteins, we observed band shifts indicative of Oct4-DNA, Sox2-DNA, and Oct4-Tcf3-DNA complexes (Figure 6C, lanes 14; bands A, B, and D, respectively). With higher concentrations of Sox2 we observed the formation of an additional Oct4-Sox2-DNA complex (Figure 6C, lane 15, band C). A competition between Sox2 and Tcf3 has been computationally predicted by Mason et al69. Together these data suggest a mutually exclusive competitive interaction for Tcf3 and Sox2 at Oct-Sox motifs where the association of Tcf3 requires a cooperative interaction with Oct factors to overcome the less favored consensus of a Sox versus a Lef/Tcf binding motif.
Functional relevance of canonical Wnt signaling with the Oct-Sox motif was supported by in vitro luciferase reporter assay using the Pou5f1 distal enhancer region (Figure 6D). The region belongs to Group B in Figure 2B–G, and sequence of the Oct/Sox probe used in the above EMSA was derived from the region. We confirmed that the region had enhancer activity in 2i-cultured mESCs (v6.5), but not in NIH3T3 cells, in a copy number-dependent manner (Figure 6E). 2i-cultured mESCs showed up-regulation of the enhancer activity compared to PD03-treated one, suggesting the positive effect of CHIR stimulus, i.e., canonical Wnt input, on the enhancer activity; the up-regulation was diminished upon mutation of the Sox motif to the same extent as mutation of both Oct and Sox motifs. In contrast, when the Oct motif was mutated, we still observed elevated enhancer activity when 2i-culture was compared to PD03-treated alone (Figure 6F, left). The similar trend was recapitulated in mESCs cultured CM (Figure 6F, right). Together, these data support the conjecture that canonical Wnt signaling contributes to the transcription of pluripotency genes via the Sox site within an Oct-Sox composite motif.
Discussion
Our transcriptional analysis of Wnt pathway action in ESCs has generated several new insights into pluripotency and differentiation networks. First, ChIP-seq analysis enabled the identification of genomic targets of β-catenin activity in mESCs. Second, a strong association is observed between β-catenin bound regions and those occupied by core pluripotency factors (NOS) and Tcf3. Third, there are marked differences in bound regions and candidate target genes between regions where only β-catenin::Tcf3 overlap, and those where β-catenin::Tcf3 also intersect with the core pluripotency networks (NOS). This is observed in motif enrichment (Lef/Tcf motif vs. Oct-Sox motif) suggesting distinct binding modes, the function of associated genes (axis specification-and mesoderm-related genes vs. stem cell- and ectoderm-related genes) suggestive of different biological outcomes, and the activity status of likely enhancer regions in mESCs (high activity for β-catenin::Tcf3::NOS and low activity for β-catenin::Tcf3). Fourth, under standard culture condition, the activation of canonical Wnt signaling elevated expression of differentiation-related genes, while activity of pluripotency-related genes was maintained. Fifth, under 2i condition, β-catenin also engaged at likely enhancers for TE lineage- and axis specification-related genes but under 2i conditions, these targets are not activated. Inhibition of MEK/ERK signaling by PD03 is critical in blocking these differentiation pathways and the enrichment of Ets motifs within differentiation related enhancers suggest a cooperative interplay of Ets and canonical Wnt complexes for gene activity. Sixth, β-catenin, Tcf3, and Oct4 interact under 2i conditions. Finally, canonical Wnt signaling up-regulated transcription in vitro via Oct-Sox motifs that could be engaged directly by Oct4-Tcf3 or Oct4-Sox2. Considering all data, we propose that under the 2i condition, canonical Wnt signaling participates in the pluripotency network via Oct4/β-catenin/Tcf3 complex formation and although Sox2 is absent, engagement of β-catenin still favors activity of pluripotency-associated genes (Figure 7, upper).
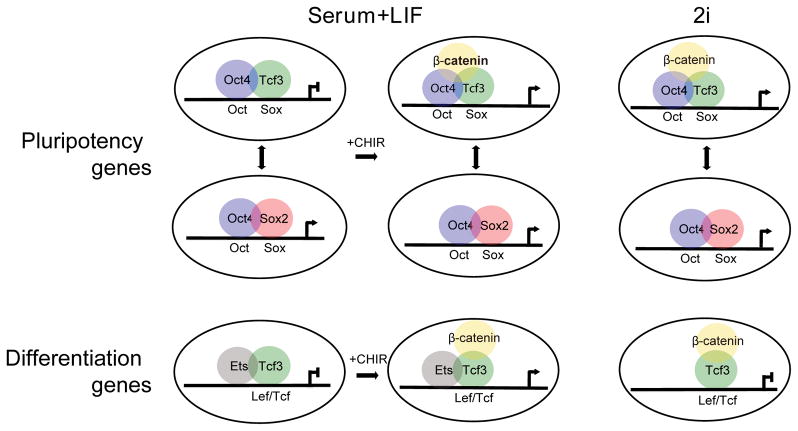
Figure 7. Schematic model of β-catenin-dependent regulation of pluripotency network.
Oct4-Sox2 binding to Oct-Sox composite motifs maintains activity of key regulators of pluripotency. Tcf3 interaction with Oct factors at the same motif is predicted to destabilize this circuit. CHIR-mediated stabilization of β-catenin has opposing actions. Entry of β-catenin into Oct4/Tcf3 complexes abrogates Tcf3 actions thereby promoting pluripotency. However, the production of active canonical Wnt transcriptional complexes engages differentiation targets destabilizing pluripotency. PD03-mediated inhibition of MEK/ERK signaling restores a pluripotency balance blocking the activation of Wnt dependent differentiation genes enabling culture under 2i conditions. Given the role of MEK/ERK signaling downstream of receptor tyrosine kinases in the regulation of the Ets-family of transcriptional regulators, and the enrichment of Ets motifs in predicted cis-regulatory, we propose the combined action of Wnt and RTK signaling in the differentiation of ES cells.
The analysis of canonical Wnt pathway mutants sheds additional light on this process. First, β-catenin activity is essential under 2i conditions for the derivation and maintenance of mESCs 32. Second, our work, and that of others indicate that Tcf proteins are required for occupancy at Oct4-dependent promoter 32–34 and engagement of β-catenin can enhance Oct4 promoter activity 32–34. Tcf3 is a key component in the transcriptional complex; Tcf3 actions are linked to groucho-mediated gene silencing 70. Tcf3 inhibits pluripotency and the removal of Tcf3 can functionally substitute for the actions of β-catenin under 2i conditions 71, 72. Thus, the main action of β-catenin appears to be to neutralize the destabilizing activity of Tcf3 in the pluripotency network. Interestingly, mutant forms of β-catenin lacking the transcriptional activating domain are effective in maintaining pluripotency 32 suggesting that β-catenin acts by abrogating Tcf3 silencing rather than forming a β-catenin-dependent activation complex. The conclusion that Tcf3 and β-catenin do not form an active transcriptional complex is supported by recent studies of Tcf3 mutant mouse embryos 73.
Wnt actions in maintaining a state of pluripotency have also been linked to the control of telomerase activity through direct regulation of Tert promoter activity in mESCs. β-catenin binding was reported to be enriched around the Tert gene in ChIP analysis of ESCs and binding further enhanced by Wnt3a treatment, or expression of a stabilized form of β-catenin 74. In contrast, we see no enrichment at the Tert locus in our whole genome analysis (data not shown).
Summary
ESCs are in an inherently unstable state wherein endogenous Tcf3 activity antagonizes the core pluripotency network through competition at Oct/Sox motifs for Sox2 binding. Surprisingly, analysis of Sox2, Nanog, and Oct4 binding suggest that Tcf3 is not a transcriptional target of this network (data not shown). Under standard culture conditions of serum and LIF, β-catenin plays no significant role in maintaining pluripotency: its activity is not essential 30,32 and stimulation of the pathway favors differentiation through engagement at “classic” Lef/Tcf motifs around differentiation associated target genes. Presumably, LIF actions are dominant and the levels of activation of target genes are not sufficient to trigger widespread differentiation. In contrast, in 2i medium Tcf3 actions are critical and β-catenin is essential to overcome Tcf3’s inhibitory effects within the pluripotency network. What remains to be determined is how the inhibition of MEK/ERK signaling blocks the differentiation promoting arm of Lef/Tcf::β-catenin -directed gene regulation. Our data on motif recovery and β-catenin engagement suggest a cooperative role for MEK/ERK directed Ets factors independent of Lef/Tcf::β-catenin binding to activate target differentiation promoting genes consistent with the critical actions of Fgf and Wnt signaling in promoting lineage commitment in early mammalian development (Figure 7, lower).
Supplementary Material
Table S1: Annotation of b-Catenin ChIP-seq binding regions from anti-FLAG and anti-Biotin replicates.
Table S2a: GREAT analysis of cis-regulatory elements classified by 3-way intersection of β-catenin, Nanog, Oct4, and Sox2 (Marson et al.)
Table S2b: GREAT analysis of cis-regulatory elements classified by 3-way intersection of β-catenin, Nanog, Oct4, and Sox2 (Marson et al.)
Table S3a: RMA normalized expression index of CHIR99021 and XAV939-treated mESC.
Table S3b: Up-regulated genes between CHIR versus XAV-treated mESC analyzed by LIMMA (adj.P.Val < 0.005)
Table S3c: Down-regulated genes between CHIR versus XAV-treated mESC analyzed by LIMMA (adj.P.Val < 0.005)
Table S3d: Gene Ontology of up-regulated genes between CHIR versus XAV-treated mESC analyzed by DAVID.
Table S3e: Gene Ontology of down-regulated genes between CHIR versus XAV-treated mESC analyzed by DAVID.
Table S4a: bCatenin direct target prediction: positively regulated direct target genes in serum+LIF+CHIR vs serum+LIF+XAV.
Table S4b: bCatenin direct target prediction: negatively regulated direct target genes in serum+LIF+CHIR vs serum+LIF+XAV.
Table S5a: RMA normalized expression index of 2i-adapted mESCs cultured in DMSO, PD03, CHIR, 2i and 2i+LIF for 24 hours.
Table S5b:Differential gene expression analysis of 2i-adapted mESCs cultured in 2iLIF versus 2i medium using LIMMA (adj.P.Val < 0.005).
Table S5c:Differential gene expression analysis of 2i-adapted mESCs cultured in 2i versus CHIR medium using LIMMA (adj.P.Val < 0.005).
Table S5d:Differential gene expression analysis of 2i-adapted mESCs cultured in 2i versus PD03 medium using LIMMA (adj.P.Val < 0.005).
Table S5e:Differential gene expression analysis of 2i-adapted mESCs cultured in 2i versus DMSO medium using LIMMA (adj.P.Val < 0.005).
Table S6: Sequences for ChIP-qPCR primers and genomic coordinates for peaks tested.
Acknowledgments
We thank Drs. Philippe Soriano, Alan B. Cantor, and Taku Saito for provision of experimental materials (specified in Supplementary Methods); Laurie Chen, Joe Vaughan, Jill McMahon, Christian Daly, Jennifer Couget, Qianzi Tang, and Genome Modification Facility, Harvard Stem Cell Institute for providing technical assistance. We are also grateful to Dr. Ung-il Chung for critical comments and helpful discussions. Work in A.P.M.’s laboratory was supported by a grant from the NIH (DK056246). X.S.L was supported by a grant from the NIH (2R01HG4069). S.O. was supported by Grants-in-Aid for young scientist (#23689079) from the Japan Society for the Promotion of Science (JSPS), Daiwa Securities Health Foundation Grant, and Nakatomi Foundation Research Grant.
Footnotes
Disclosure of potential conflicts of interest
The authors indicate no potential conflicts of interest.
Author contributions: X.Z. and S.O.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; K.A.P.: collection and/or assembly of data, data analysis and interpretation; X.S.L.: data analysis and interpretation; APM: conception and design, data analysis and interpretation, financial support, manuscript writing, final approval of manuscript.
References
- 1.Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the3mouse. Proceedings of the National Academy of Sciences. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J. An Extended Transcriptional Network for Pluripotency of Embryonic Stem Cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loh Y-H, Wu Q, Chew J-L, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 4.Jiang J, Chan Y-S, Loh Y-H, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez F, Boue S, Belmonte JCI. Methods for making induced pluripotent stem cells: reprogramming a la carte. Nature Reviews Genetics. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 9.Ying QL, Nichols J, Chambers I, et al. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 10.Williams RL, Hilton DJ, Pease S, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 11.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 12.Niwa H, Burdon T, Chambers I, et al. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Gene Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P, Tong C, Mehrian-Shai R, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bain J, Plater L, Elliott M, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 17.Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- 18.Aberle H, Bauer A, Stappert J, et al. beta-catenin is a target for the ubiquitin-proteasome pathway. Faseb Journal. 1997;11:A1409–A1409. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubinfeld B, Albert I, Porfiri E, et al. Binding of GSK3 beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 20.Murray JT, Campbell DG, Morrice N, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taelman VF, Dobrowolski R, Plouhinec JL, et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa K, Nishinakamura R, Iwamatsu Y, et al. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Bioph Res Co. 2006;343:159–166. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- 24.Ten Berge D, Kurek D, Blauwkamp T, et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singla DK, Schneider DJ, LeWinter MM, et al. wnt3a but not wnt11 supports self-renewal of embryonic stem cells. Biochem Bioph Res Co. 2006;345:789–795. doi: 10.1016/j.bbrc.2006.04.125. [DOI] [PubMed] [Google Scholar]
- 26.Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 27.Lluis F, Pedone E, Pepe S, et al. Periodic Activation of Wnt/beta-Catenin Signaling Enhances Somatic Cell Reprogramming Mediated by Cell Fusion. Cell Stem Cell. 2008;3:493–507. doi: 10.1016/j.stem.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Marson A, Foreman R, Chevalier B, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anton R, Kestler HA, Kuhl M. beta-Catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. Febs Lett. 2007;581:5247–5254. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Lyashenko N, Winter M, Migliorini D, et al. Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole MF, Johnstone SE, Newman JJ, et al. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Gene Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wray J, Kalkan T, Gomez-Lopez S, et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi F, Pereira L, Hoffman JA, et al. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat Cell Biol. 2011;13:762–770. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly KF, Ng DY, Jayakumaran G, et al. beta-Catenin Enhances Oct-4 Activity and Reinforces Pluripotency through a TCF-Independent Mechanism. Cell Stem Cell. 2011;8:214–227. doi: 10.1016/j.stem.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakre MM, Hoi A, Mong JCY, et al. Generation of multipotential mesendodermal progenitors from mouse embryonic stem cells via sustained Wnt pathway activation. J Biol Chem. 2007;282:31703–31712. doi: 10.1074/jbc.M704287200. [DOI] [PubMed] [Google Scholar]
- 36.Lindsley RC, Gill JG, Kyba M, et al. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 37.He SY, Pant D, Schiffmacher A, et al. Lymphoid enhancer factor 1-mediated wnt signaling promotes the initiation of trophoblast lineage differentiation in mouse embryonic stem cells. Stem Cells. 2008;26:842–849. doi: 10.1634/stemcells.2007-0356. [DOI] [PubMed] [Google Scholar]
- 38.Morkel M, Huelsken J, Wakamiya M, et al. beta-Catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development. 2003;130:6283–6294. doi: 10.1242/dev.00859. [DOI] [PubMed] [Google Scholar]
- 39.Merrill BJ, Pasolli HA, Polak L, et al. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 40.Behringer RR, Liu PT, Wakamiya M, et al. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 41.Birchmeier W, Huelsken J, Vogel R, et al. Requirement for beta-catenin in anterior-posterior axis formation in mice. Journal of Cell Biology. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skarnes WC, Kelly OG, Pinson KI. The Wnt co-receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development. 2004;131:2803–2815. doi: 10.1242/dev.01137. [DOI] [PubMed] [Google Scholar]
- 43.Barrow JR, Howell WD, Rule M, et al. Wnt3 signaling in the epiblast is required for proper orientation of the anteroposterior axis. Dev Biol. 2007;312:312–320. doi: 10.1016/j.ydbio.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 44.Dignam JD, Lebovitz RM, Roeder RG. Accurate Transcription Initiation by Rna Polymerase-Ii in a Soluble Extract from Isolated Mammalian Nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Rao S, Chu J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 46.de Boer E, Rodriguez P, Bonte E, et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hernan R, Heuermann K, Brizzard B. Multiple epitope tagging of expressed proteins for enhanced detection. Biotechniques. 2000;28:789–793. doi: 10.2144/00284pf01. [DOI] [PubMed] [Google Scholar]
- 48.Schatz PJ. Use of Peptide Libraries to Map the Substrate-Specificity of a Peptide-Modifying Enzyme - a 13 Residue Consensus Peptide Specifies Biotinylation in Escherichia-Coli. Bio-Technol. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 49.Howard PK, Shaw J, Otsuka AJ. Nucleotide-Sequence of the Bira-Gene Encoding the Biotin Operon Repressor and Biotin Holoenzyme Synthetase Functions of Escherichia-Coli. Gene. 1985;35:321–331. doi: 10.1016/0378-1119(85)90011-3. [DOI] [PubMed] [Google Scholar]
- 50.McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han JY, Yuan P, Yang H, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ku M, Koche RP, Rheinbay E, et al. Genomewide Analysis of PRC1 and PRC2 Occupancy Identifies Two Classes of Bivalent Domains. Plos Genetics. 2008;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heng JC, Feng B, Han J, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Tam WL, Lim CY, Han JY, et al. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–2031. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He HH, Meyer CA, Shin H, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rada-Iglesias A, Bajpai R, Swigut T, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. P Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang SMA, Mishina YM, Liu SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 60.Tang Q, Chen Y, Meyer C, et al. A Comprehensive View of Nuclear Receptor Cancer Cistromes. Cancer Res. 2011;71:6940. doi: 10.1158/0008-5472.CAN-11-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Breitling R, Armengaud P, Amtmann A, et al. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. Febs Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 62.Takada R, Satomi Y, Kurata T, et al. Monounsaturated fatty acid modification of Wnt protein: Its role in Wnt secretion. Developmental Cell. 2006;11:791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Shen X, Liu Y, Hsu Y-J, et al. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takada S, Mcmahon J, Stark K, et al. Wnt-Genes in the Regulation of Mesodermal Development. J Cell Biochem. 1994:465–465. [Google Scholar]
- 65.Yamaguchi TP, Takada S, Yoshikawa Y, et al. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Gene Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohonen T. Essentials of the self-organizing map. Neural Netw. 2012;37:52–65. doi: 10.1016/j.neunet.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 67.Kunath T, Saba-El-Leil MK, Almousailleakh M, et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 68.Chew JL, Loh YH, Zhang WS, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mason MJ, Plath K, Zhou Q. Identification of context-dependent motifs by contrasting ChIP binding data. Bioinformatics. 2010;26:2826–2832. doi: 10.1093/bioinformatics/btq546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brantjes H, Roose J, van de Wetering M, et al. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo G, Huang Y, Humphreys P, et al. A PiggyBac-Based Recessive Screening Method to Identify Pluripotency Regulators. Plos One. 2011;6(4):e18189. doi: 10.1371/journal.pone.0018189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schaniel C, Ang Y-S, Ratnakumar K, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu CI, Hoffman JA, Shy BR, et al. Function of Wnt/beta-catenin in counteracting Tcf3 repression through the Tcf3-beta-catenin interaction. Development. 2012;139:2118–2129. doi: 10.1242/dev.076067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoffmeyer K, Raggioli A, Rudloff S, et al. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336:1549–1554. doi: 10.1126/science.1218370. [DOI] [PubMed] [Google Scholar]
- 75.Heinz S, Benner C, Spann N, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Annotation of b-Catenin ChIP-seq binding regions from anti-FLAG and anti-Biotin replicates.
Table S2a: GREAT analysis of cis-regulatory elements classified by 3-way intersection of β-catenin, Nanog, Oct4, and Sox2 (Marson et al.)
Table S2b: GREAT analysis of cis-regulatory elements classified by 3-way intersection of β-catenin, Nanog, Oct4, and Sox2 (Marson et al.)
Table S3a: RMA normalized expression index of CHIR99021 and XAV939-treated mESC.
Table S3b: Up-regulated genes between CHIR versus XAV-treated mESC analyzed by LIMMA (adj.P.Val < 0.005)
Table S3c: Down-regulated genes between CHIR versus XAV-treated mESC analyzed by LIMMA (adj.P.Val < 0.005)
Table S3d: Gene Ontology of up-regulated genes between CHIR versus XAV-treated mESC analyzed by DAVID.
Table S3e: Gene Ontology of down-regulated genes between CHIR versus XAV-treated mESC analyzed by DAVID.
Table S4a: bCatenin direct target prediction: positively regulated direct target genes in serum+LIF+CHIR vs serum+LIF+XAV.
Table S4b: bCatenin direct target prediction: negatively regulated direct target genes in serum+LIF+CHIR vs serum+LIF+XAV.
Table S5a: RMA normalized expression index of 2i-adapted mESCs cultured in DMSO, PD03, CHIR, 2i and 2i+LIF for 24 hours.
Table S5b:Differential gene expression analysis of 2i-adapted mESCs cultured in 2iLIF versus 2i medium using LIMMA (adj.P.Val < 0.005).
Table S5c:Differential gene expression analysis of 2i-adapted mESCs cultured in 2i versus CHIR medium using LIMMA (adj.P.Val < 0.005).
Table S5d:Differential gene expression analysis of 2i-adapted mESCs cultured in 2i versus PD03 medium using LIMMA (adj.P.Val < 0.005).
Table S5e:Differential gene expression analysis of 2i-adapted mESCs cultured in 2i versus DMSO medium using LIMMA (adj.P.Val < 0.005).
Table S6: Sequences for ChIP-qPCR primers and genomic coordinates for peaks tested.