Abstract
The clinical application of siRNA is limited largely by the lack of efficient, cell-specific delivery systems. Antibodies are attractive delivery vehicles for targeted therapy due to their high specificity. In this study we describe the use of a humanized monoclonal antibody (mAb), hu3S193, against Lewis-Y (Ley), as a delivery vehicle for STAT3 siRNA. This mAb is rapidly internalized into Ley expressing cancer cells via antigen recognition, and when coupled to STAT3 siRNA, a potentially powerful molecularly targeted delivery agent is created. Selective silencing of STAT3 is associated with tumor suppression. Two hu3S193 based siRNA delivery systems using STAT3 siRNA as a prototype were developed and tested in Ley-positive cancer cells: (a) a covalent construct based on a reductive disulfide linker that is expected to undergo cleavage within cells and (b) a non-covalent construct based on (D-Arginine)9 (9r) modified hu3S193. Ley-specific binding and internalization of both the covalent and non-covalent constructs were confirmed by flow cytometry and confocal microscopy. Both the covalent and the non-covalent system led to efficient STAT3 silencing in Ley-positive cancer cells (A431), but not in Ley-negative cancer cells (MDA-MB-435). The covalent construct, however, required co-treatment with reagents such as chloroquine or 9r that facilitate the escape of the siRNA from endosomes to achieve significant gene silencing. The 9r modified non-covalent construct, induced ~70% STAT3 knockdown at sub-micromolar siRNA concentrations when used at an optimal vehicle-to-siRNA ratio of 5:1. The STAT3 knockdown also led to ~50% inhibition of cell proliferation of Ley-positive cells. Non-covalent linked STAT3 siRNA-hu3S193 has great promise for targeted knockdown of STAT3 in tumor cells.
INTRODUCTION
Small interfering RNAs (siRNAs) are a class of short double-stranded RNAs that can induce RNA interference (RNAi). After processing and incorporation into the RNA induced silencing complex (RISC), the antisense strand of the siRNA selectively binds to its complementary mRNA and induces its degradation in the cytoplasm (1–4). Synthetic siRNAs of 19–23 bp in length can also induce RNAi (5–6); however, it has been reported that 25 to 27-nt long siRNAs have greater potency than the corresponding 21-nt siRNAs. These longer, so called Dicer substrate siRNAs (D-siRNAs) must be processed by Dicer before they can be incorporated into RISC (7). The increased potency of D-siRNAs has been attributed to a more efficient RISC incorporation by Dicer processing (3, 7).
siRNAs have been considered by many scientists as potential “superdrugs”(8); however, the lack of efficient and specific delivery methodologies have impeded the development of this promising class of therapeutics (2). Initial clinical trials focused on locally administered “naked” siRNA (2). The first targeted siRNA delivery in humans was recently reported and employed human transferrin (Tf) modified cyclodextrin polymer nanoparticles for the specific recognition and treatment of Tf receptor expressing cancers (9). Successful siRNA delivery systems require efficient transport to the target organs, uptake by the target cells and escape of the siRNA from endosome vesicles into the cytoplasm (1, 10). The size and the negative charge of siRNA make the accomplishment of this goal difficult. Viral and nonviral delivery vectors along with physical interventions (e.g. electroporation) have been used to facilitate siRNA delivery (1–2, 11–12). Most of the current research is focused on the development of nonviral vehicles such as liposomes, nanoparticles and peptides due to their lower costs, easier assembly and greater safety compared to viral vectors (2). From a synthetic point of view, there are two kinds of strategies for building nonviral-based delivery vehicles: covalent and non-covalent constructs with siRNA (13). Covalent constructs typically contain a cleavable (disulfide or other sessile bond) or a non-cleavable linker between the siRNA and its vehicle. Most of the non-covalent constructs are based on electrostatic interactions between the positively charged vehicles and negatively charged siRNA. Conjugation of those covalent or non-covalent constructs with specific ligands would generate targeted delivery systems (12).
In principle, ligands that could facilitate targeted siRNA delivery include antibodies, aptamers, small molecules, and other proteins or short peptides (12). Although costly, monoclonal antibodies or antibody fragments are still considered excellent delivery platforms for siRNAs because of their high specificity (2, 14). For example, Lieberman’s group used a protamine modified antibody fragment to deliver non-covalently bound siRNA into target cells (15), Kumar et al. reported a siRNA delivery strategy based on a CD7 single-chain antibody fragment (scFv) conjugated to (D-Arginine)9 (9r) (16). Chang’s group used a cationic liposome mixture modified with an anti-transferrin receptor scFv for the delivery of anti-HER-2 siRNA (17), while Pardridge’s group conjugated a streptavidin modified antibody with biotin modified siRNA to deliver siRNA (18).
Hu3S193, an anti-Lewis-Y (Ley) monoclonal antibody (mAb) (19), was used in our study for the development of a siRNA delivery vehicle for several reasons: First, Ley antigen is a blood group-related antigen expressed in over 70% of epithelial cancers including breast, colon, ovary, prostate and lung cancers. However, its expression on normal cells and normal tissues is limited (19–23). Second, this humanized Ley-specific mAb is well tolerated and selectively targets Ley-expressing tumors, with minimal normal tissue uptake, as demonstrated in a recent first-in-human trial (24). Third, the Ley-hu3S193 complex is a feasible choice for drug delivery since it is internalized after antigen recognition (25). Selective delivery and promising bioactivity have been previously reported when a hu3S193-calicheamicin conjugate was used as specific drug delivery vehicle in a xenograft mouse model (26).
It has been reported that STAT3 is overexpressed and/or persistently activated in a wide diversity of solid tumors and blood malignancies (27–31). Activated STAT3 enhances tumor cell proliferation and survival, stimulates tumor angiogenesis and suppresses antitumor immune responses in the tumor microenvironment (27–31). Thus, the STAT3 pathway contributes to both tumor-cell intrinsic and extrinsic mechanisms of malignancy, making it an important target for cancer therapy. There are several strategies to inhibit STAT3 (or its signaling pathway) including reducing STAT3 levels (knockdown of gene expression), decreasing upstream phosphorylation (tyrosine kinase inhibitors) and blocking protein-protein interactions (inhibition of STAT3 recruitment to activated receptor complex, dimerization or STAT3-DNA promoter binding) (27–31). Initial studies using siRNA/shRNA for the knockdown of STAT3 expression in cancer cells or in the tumor microenvironment have already showed promising results in cancer regression in vitro or in vivo (32–40). For example, an intratumoral injection of Stat3 siRNA with a transfection reagent effectively inhibited tumor growth in a mouse study (39). CpG oligonucleotide-conjugated D-siRNA selectively silenced Stat3 expression in toll-like receptor 9 positive cells, which led to the activation of tumor-associated immune cells and potent antitumor immune responses in vivo (40).
Although both covalent and non-covalent constructs have previously been used for siRNA delivery, these two strategies have not been compared within the same system. Especially for mAb based siRNA delivery, studies on covalent systems are very limited compared with non-covalent systems (13, 18). Herein, we report the construction and efficacy of two hu3S193 based STAT3 D-siRNA delivery vehicles: (a) a covalent construct consisting of mAb and siRNA conjugated by a reductive disulfide linker, which could be cleaved within the cells and (b) a non-covalent construct, which could bind siRNA via electrostatic interactions after the mAb was modified with a (D-Arginine)9 peptide. The tumor cell binding, internalization and STAT3 knockdown efficiencies were evaluated for both constructs.
RESULTS AND DISCUSSION
Synthesis and characterization of hu3S193 siRNA constructs: (a) covalent construct (hu3S193-siRNA)
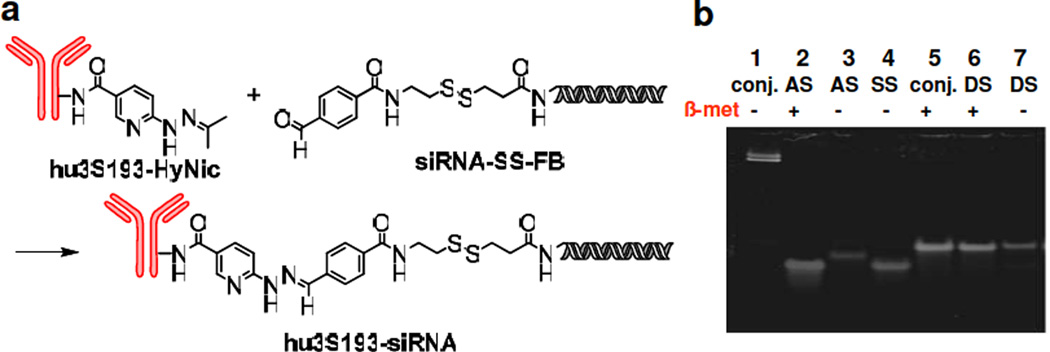
The linker in the covalent construct contains a disulfide bond, which is expected to be reduced within the cytoplasm of the cell, and a stable bis-arylhydrazone bond with a specific UV wavelength at 350 nm, which facilitated detection and purification (Fig. 1a). To synthesize this construct, hu3S193 and siRNA were first modified with commercially available linkers hydrazinonicotinate acetone hydrazone (HyNic) and disulfide formyl benzoate (SS-FB), respectively. This afforded the corresponding products hu3S193-HyNic and siRNA-SS-FB. Hu3S193-HyNic batches with different linker numbers (N = 1 – 5) per mAb were prepared by modulating the concentration of the reactants. Individual hu3S193-HyNic batches with different linker ratios were allowed to react with siRNA-SS-FB which produced the covalent hu3S193-siRNA constructs (see Table 1 for description of antibody siRNA constructs). Regardless of the number of HyNic per mAb, the primary products after reaction with excess siRNA were mono hu3S193-siRNA conjugates. Lowering the HyNic-to-mAb ratio resulted in significantly reduced coupling yields with the siRNA (Fig. S1). Considering the significant reduction in chemical coupling yields with the siRNA when a low number of HyNic (N<5) was attached to hu3S193, hu3S193-HyNic (N = 5) was used for all further syntheses. PAGE analysis of hu3S193-siRNA after disulfide reduction showed a signal corresponding to double stranded siRNA, which suggests the feasibility of a similar release mechanism of siRNA within cells (Fig. 1b). For flow cytometry and confocal microscopy studies we also synthesized a fluorescein-labeled hu3S193-siRNA (hu3S193-siRNA(FL)) covalent conjugate using a fluorescein-labeled amino modified RNA strand.
Figure 1. Synthesis and characterization of the covalent construct hu3S193-siRNA.
a) Synthetic scheme for the preparation of the covalent construct b) PAGE analysis of mAb-siRNA conjugate. Lane 1 and 5: hu3S193-siRNA conjugates; lane 2 and 3: SS-FB-antisense strand (AS); lane 4: sense strand (SS); lane 6 and 7: double strand siRNA-SSFB (DS). Lanes 2, 5 and 6 treatment with β-mercaptoethanol (5%).
Table 1.
Description of antibody siRNA constructs.
| Construct | Description |
|---|---|
| hu3S193 | parent antibody |
| hu3S193-siRNA | covalent antibody siRNA construct |
| hu3S193-HyNic | HyNic linker modified antibody |
| hu3S193-siRNA(FL) | fluorescein-labeled hu3S193-siRNA |
| 9r | (D-arginine)9 peptide |
| 15r | (D-arginine)15 peptide |
| hu3S193-9r(1) | 9r modified antibody with antibody:9r ratio = 1:1 |
| hu3S193-9r(4) | 9r modified antibody with antibody:9r ratio = 1:4 |
| siRNA(FL) | fluorescein-labeled siRNA |
| hu3S193-9r (1):siRNA | Non-covalent construct based on hu3S193-9r (1) |
| hu3S193-15r(1) | 15r modified antibody with antibody:15r ratio = 1:1 |
| siSTAT3 | STAT3 siRNA |
| siLuci | luciferase siRNA |
(b) Non-covalent construct (hu3S193-9r:siRNA)
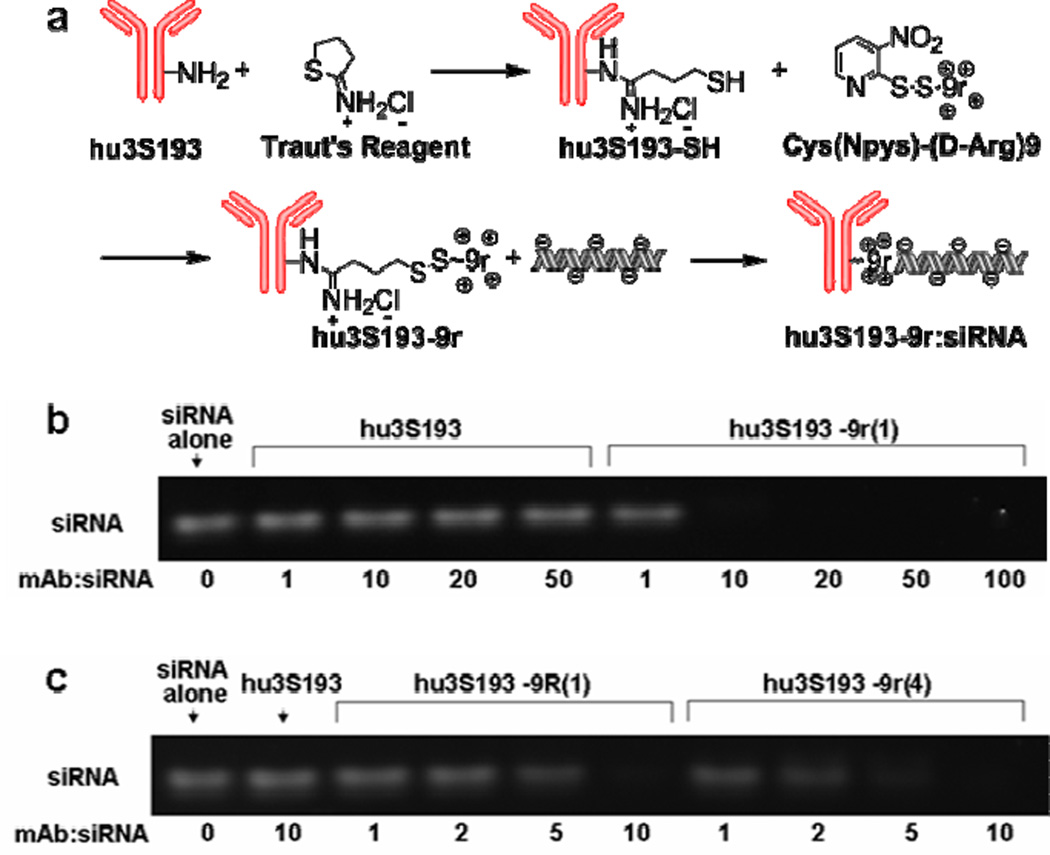
In addition to the covalent system a (D-Arginine)9 based non-covalent siRNA delivery strategy was developed. Arginine-rich peptides have been used for siRNA delivery as free peptide (41) or conjugated with target ligands (16, 42). The delivery mechanisms are not fully understood. It has been suggested that arginine-rich peptides improve siRNA delivery by facilitating the escape of siRNAs from endosomes (43–44) or by protecting siRNAs from degradation (42). d-Peptides were favored due to their increased stability toward proteolysis (45). To synthesize the non-covalent construct, hu3S193 was first thiolated using Traut’s reagent, then allowed to react with activated (D-Arginine)9 (9r) to produce hu3S193-9r (Fig. 2a). Two batches of 9r modified hu3S193 with 9r-to-mAb ratios of 1:1 (hu3S193-9r(1)) and 4:1 (hu3S193-9r(4)) were synthesized. Upon mixing with siRNA, hu3S193-9r associates with siRNA via electrostatic interactions. Electrophoretic gel mobility-shift assays (EMSA) confirmed that siRNA efficiently binds to hu3S193-9r(1), but not to the unmodified hu3S193 (Fig. 2b). Due to the increased number of 9r per mAb, hu3S193-9r(4) displayed stronger binding to siRNA (Fig. 2c). A 15r peptide modified hu3S193, hu3S193-15r(1), was also synthesized using the same methodology. EMSA results indicated that hu3S193-15r(1) binds siRNA similarly to hu3S193-9r(1) (data not shown). For flow cytometry or confocal imaging studies, fluorescein-labeled siRNA (siRNA(FL)) was used instead of regular siRNA in the preparation of the non-covalent construct.
Figure 2. Synthesis and characterization of the non-covalent construct hu3S193-9r:siRNA.
a) Synthetic scheme for the preparation of the non-covalent construct. b) siRNA binding to hu3S193-9r(1). Hu3S193 or hu3S193-9r(1) were mixed with siRNA at the indicated molar ratios and analyzed by electrophoresis. c) Comparison of siRNA binding to hu3S193-9r(1) or hu3S193-9r(4). The constructs hu3S193-9r(1) and hu3S193-9r(4) (modified with one or four 9r peptides per antibody, respectively) were incubated with siRNA at the indicated molar ratios and analyzed by electrophoresis.
Specific delivery of siRNA into Ley-expressing cells by covalent and non-covalent constructs
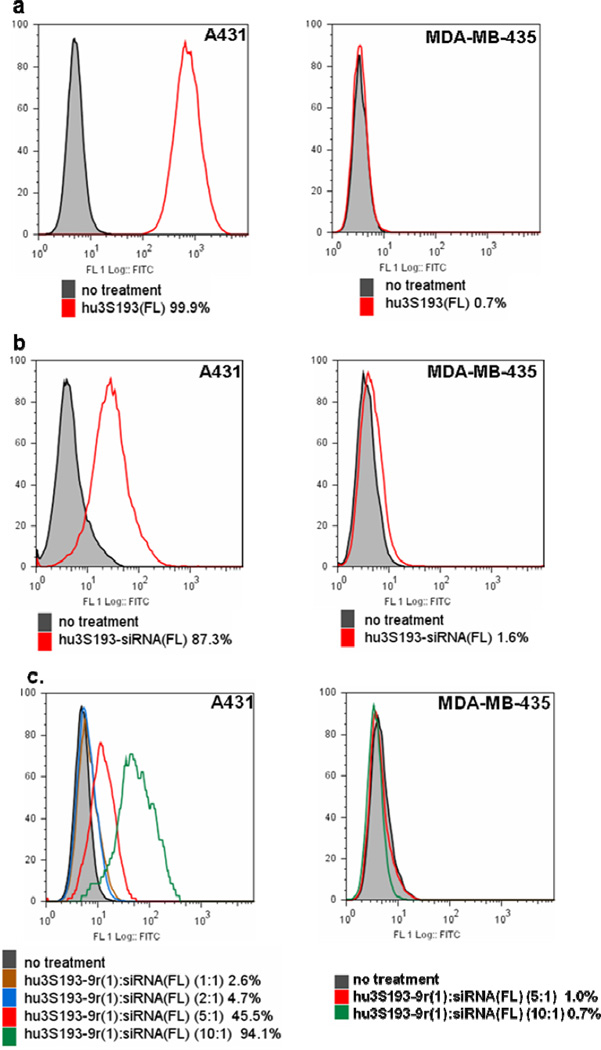
Using fluorescein-labeled hu3S193 (hu3S193(FL)), we first confirmed by flow cytometry analysis previous reports (46–47) that A431 cells (human squamous cell carcinoma) expressed high levels of Ley while MDA-MB-435 cells (breast cancer) were Ley-negative (Fig. 3a). To determine whether the constructs could efficiently and specifically bind to Ley-positive cells, MDA-MB-435 and A431 cells were incubated with hu3S193-siRNA(FL) or hu3S193-9r:siRNA(FL) and analyzed by flow cytometry. As shown in Fig. 3b, the covalent conjugate could specifically bind to Ley-expressing A431 cells, but not to Ley-negative MDA-MB-435 cells. Flow cytometry analysis of the two non-covalent constructs, hu3S193-9r(1) and hu3S193-9r(4), modified with either one or four 9r per mAb respectively showed dramatically different results: When A431 and MDA-MB-435 cells were treated with various ratios of hu3S193-9r(1)-to-siRNA(FL) ranging from 1:1 to 1:10, only the percentage of fluorescein-positive A431 cells increased with higher hu3S193-9r(1)-to-siRNA(FL) ratios (Fig. 3c). No significant siRNA binding was observed after treatment of A431 cells with siRNA(FL) alone or siRNA(FL) mixed with hu3S193 and/or 9r (Fig. S2a). In addition, pretreatment of A431 cells with the unmodified hu3S193 antibody partially blocked cell binding of hu3S193-9r(1):siSTAT3(FL)(5:1). The percentage of positive cells was reduced by ~70 % by the pretreatment (Fig.S2b).
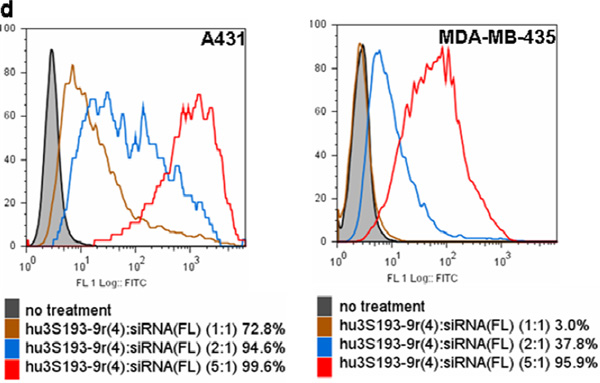
Figure 3. Flow cytometry analysis of cell binding.
a) Ley expression in A431 and MDA-MB-435. The cells were stained with hu3S193(FL); b) Cell binding of hu3S193-siRNA(FL); c) Cell binding of hu3S193-9r(1):siRNA(FL) and d) hu3S193-9r(4):siRNA(FL). The constructs hu3S193-9r(1) or hu3S193-9r(4) were incubated with siRNA(FL) at various molar ratios. The mixtures were then used to stain the cells. The siRNA concentration was 300 nM for all binding studies. The percentage of fluorescein positive cells is indicated for each treatment.
On the other hand, when the cells were treated with hu3S193-9r(4): siRNA(FL) the percentage of both fluorescein-positive A431 and MDA-MB-435 cells increased with higher hu3S193-9r(4)-to-siRNA(FL) ratios (Fig. 3d). These results suggest that increasing the amount of 9r per mAb led to nonspecific binding to Ley-negative cells. Thus, hu3S193-9r(1) modified with one 9r peptide per mAb was used for all further experiments with the non-covalent construct.
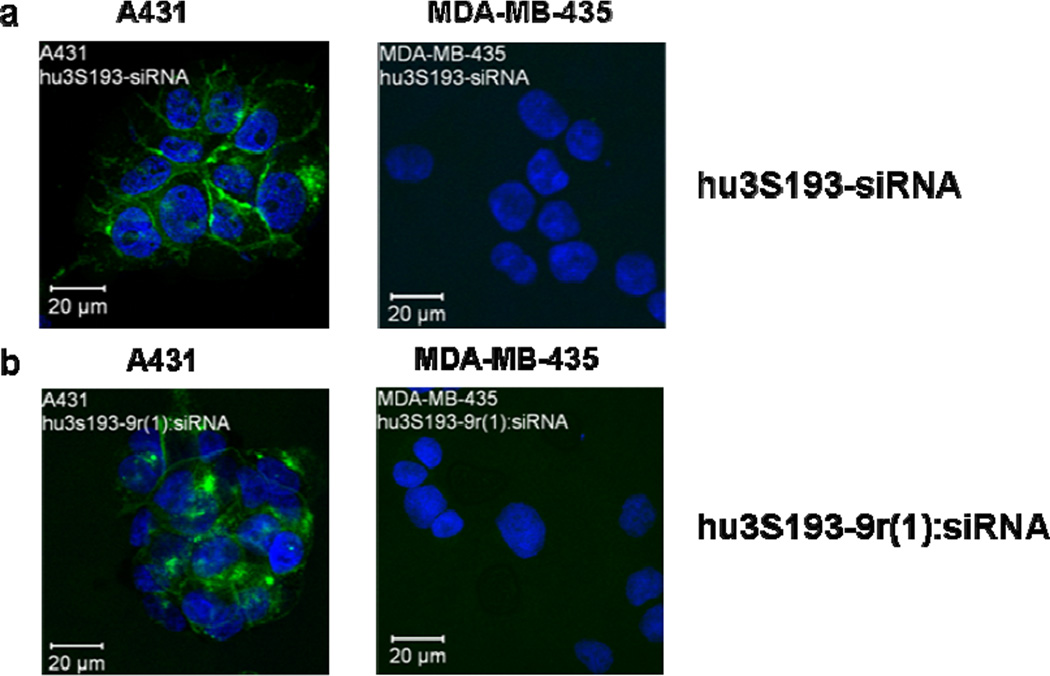
Cell internalization of the covalent and non-covalent constructs was confirmed by confocal imaging. Fluorescence signals were observed on the surface and within Ley-expressing A431 cells, but not within Ley-negative MDA-MB-435 cells for both hu3S193-siRNA(FL) (Fig. 4a) and hu3S193-9r(1):siRNA(FL) (Fig. 4b). No internalization was observed in Ley-expressing cells with siRNA(FL) alone or siRNA(FL) mixed with hu3S193 and/or 9r (data not shown).
Figure 4. Confocal microscopy.
Ley expressing A431 and Ley-negative MDA-MB-435 cells were treated for 5 h with a) hu3S193-siRNA(FL) or b) hu3S193-9r(1):siRNA(FL) (5:1). The siRNA concentration was 600 nM for all experiments.
Silencing efficiency of the covalent and non-covalent constructs
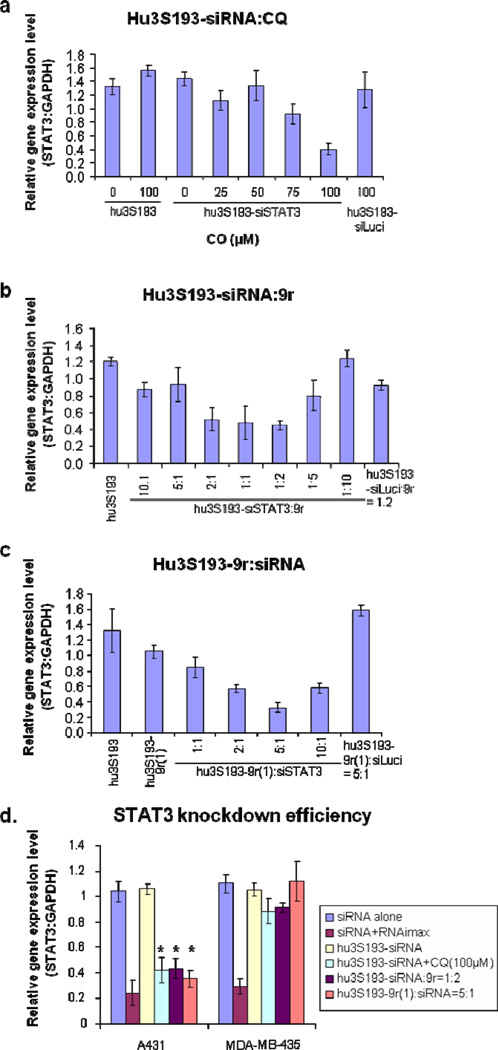
To test STAT3 silencing efficiency Ley-expressing cell lines were treated with both covalent and non-covalent constructs and STAT3 expression levels were determined by quantitative PCR using GAPDH as reference gene. Surprisingly, the covalent construct hu3S193-siRNA did not show any significant gene silencing effects, even when used at a concentration of 1500 nM (data not shown). Since significant binding and internalization was observed by flow cytometry and confocal imaging, we speculated that the siRNA was stuck in the endosomes and inefficient endosome release is the reason for the lack of gene silencing. To test this hypothesis, we treated A431 and MDA-MB-435 cells with hu3S193-siRNA in the presence of chloroquine (48–50), a known endosome disrupting reagent. At a concentration of 100 µM chloroquine STAT3 expression levels were decreased by ~60 % in the Ley-positive cell line A431, but not in the Ley-negative cell line MDA-MB-435 (Fig. 5a and 5d). STAT3 levels did not decrease after adding chloroquine to A431 cells treated with the negative control siRNA (siLuci) (Fig.5a).
Figure 5. Knockdown efficiency.
Cells were treated with the covalent or non-covalent construct for 24 h. The effect of the treatment on STAT3 expression levels was measured by quantitative PCR. The data were normalized to the GAPDH expression. a) Influence of chloroquine (CQ) on the knockdown efficiency of the covalent construct. A431 cells were treated with hu3S193-siSTAT3 and CQ at the indicated concentrations. b) Influence of 9r peptide on the knockdown efficiency of the covalent construct. A431 cells were treated with hu3S193-siSTAT3 and 9r peptide at the indicated molar ratios. Cells treated with non-covalent constructs containing luciferase siRNA (hu3S193-siLuci:9r (molar ratio 1:2)) were used as a negative control. c) Knockdown efficiency of the non-covalent construct. A431 cells were treated with hu3S193-9r(1):siSTAT3 at the indicated molar ratios. Cells treated with hu3S193-9r(1):siLuci at a molar ratio of 5:1 were used as a negative control. d) Comparison of the knockdown efficiency in Ley-positive and Ley-negative cells. A431 (Ley-positive) and MDA-MB-435 (Ley-negative) cells were treated with siSTAT3, hu3S193-siSTAT3 alone, hu3S193-siSTAT3+CQ (100 µM), hu3S193-siSTAT3:9r(1:2), hu3S193-9r(1):siSTAT3 (5:1) or transfected with 50 nM siRNA using RNAiMAX as transfection reagent. The siRNA concentration was 300 nM for all experiments (except for the transfection reaction).*P-values compared with siRNA alone treatment on A431 cell line were < 0.0006.
Unfortunately, the cytotoxicity of chloroquine prohibits its broad clinical application for siRNA delivery (51). So instead, our attention turned towards arginine-rich peptides, which also have been suggested to promote siRNA delivery by facilitating the escape of siRNAs from endosomes (43–44). Moreover, no significant toxicity has been reported with arginine peptide administration in vivo (16, 41–42, 44). When the covalent construct hu3S193-siRNA was pre-incubated with 9r peptide, a significant improvement in gene knockdown was observed. Optimal gene silencing was achieved when hu3S193-siRNA was mixed with 9r at a ratio of 1:2 (Fig. 5b). The decrease in knockdown efficiency at higher hu3S193-siRNA:9r ratios could be attributed to an inefficient release of the siRNA from the peptide complex (44). STAT3 expression in the Ley-negative MDA-MB-435 cell line was not influenced by the same treatment (Fig. 5d).
We also tested the knockdown efficiency of the non-covalent system, hu3S193-9r(1):siRNA. This construct effectively decreases STAT3 expression under optimized conditions. The molar ratio of vehicle-to-siRNA is critical for achieving efficient knockdowns. The best gene silencing (>70%) was observed at a molar ratio of hu3S193-9r(1) to siRNA of 5:1 (Fig. 5c). The decrease in knockdown efficiency that was observed at higher vehicle-to-siRNA ratios might be due to the inefficient release of siRNA from the 9r peptide (44). We also tested the influence of the peptide length on the gene knockdown efficiency by comparing a 15r peptide modified hu3S193 (hu3S193-15r(1)) with hu3S193-9r(1). Similar knockdown efficiencies were observed, suggesting that increasing the peptide length from 9 to 15 amino acids does not significantly alter the siRNA delivery efficiency for the non-covalent system (Fig. S3a).
To determine the siRNA concentration necessary to achieve optimal silencing, A431 cells were treated with various siRNA concentrations. The ratio of hu3S193-9r(1) to siRNA was kept at 5:1. Maximal silencing was observed at a siRNA concentration of 300 nM. Increasing this siRNA concentration did not lead to improved gene silencing. (Fig. S3b).
To confirm the specificity of siRNA delivery into Ley-positive cells we treated MDA-MB-468 cells, a Ley-positive cell line (with reduced Ley expression levels compared to A431) and Ley-negative MDA-MB-435 cells using the same conditions. STAT3 expression levels were reduced in MDA-MB-468 by ~50 % (data not shown). No significant gene knockdown was observed in MDA-MB-435 cells (Fig. 5d). siRNA alone or the covalent conjugate hu3S193-siRNA alone also did not induce gene knockdown regardless of the cell type (Fig. 5d). On the other hand, using RNAiMAX as transfection reagent (siRNA 50 nM) led to a 70–80% reduction in STAT3 expression levels in all the cell lines similarly (Fig. 5d).
Taken together, our data indicate that the covalent construct hu3S193-siRNA requires chloroquine or 9r peptide for efficient gene silencing. The non-covalent construct hu3S193-9r(1):siRNA shows promising, target cell-specific gene knockdown. Both the covalent and the non-covalent system are capable of delivering siRNA into Ley-positive cells. The 9r peptide in the non-covalent system not only mediates siRNA binding and delivery, but also helps the siRNA escape from the endosomes and/or protects the siRNA from degradation (42–44). Since the hu3S193-9r(1):siRNA non-covalent delivery system has general applicability for delivering different siRNAs or multiple siRNAs without modification of the delivery vehicle, the following experiments were focused on this 9r modified non-covalent system.
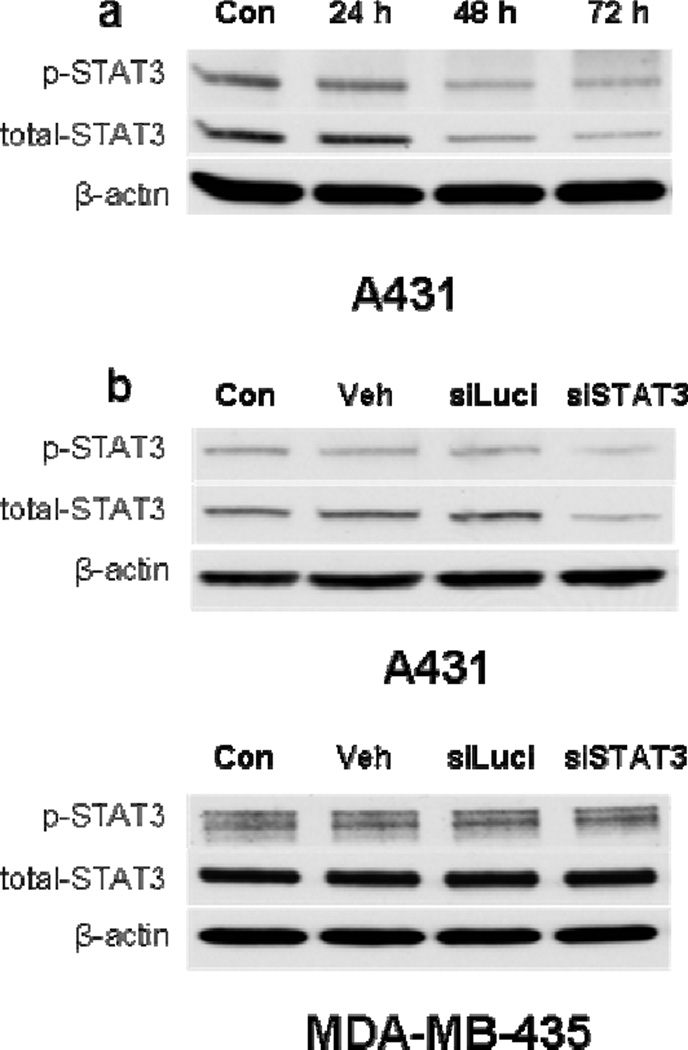
To confirm the knockdown efficiency and specificity at the protein level, we treated cells with hu3s193-9r(1):siRNA (5:1) and determined the STAT3 levels by Western blot analysis after 24, 48 and 72 h. A significant decrease in STAT3 protein levels was observed at 48 and 72 h (Fig. 6a). No decrease in STAT3 was observed upon treatment with hu3S193-9r(1) alone or with hu3S193-9r(1):luciferase siRNA (siLuci) (Fig. 6b). The same treatment did not change the STAT3 expression in MDA-MB-435 cells (Fig. 6b).
Figure 6. Western blot analysis of STAT3 expression.
a) Time course of STAT3 knockdown: A431 cells were treated with hu3S193-9r(1):siSTAT3(5:1) for 24, 48 and 72 h. b) Specificity of STAT3 knockdown: Ley-positive A431 and Ley-negative MDA-MB-435 cells were treated with hu3S193-9r(1) alone (Veh), hu3S193-9r(1):siLuci(5:1) (siLuci) or hu3S193-9r(1):siSTAT3(5:1) (siSTAT3) for 72 h. Untreated cells were used as a control (Con). The siRNA concentration was 300 nM for all experiments.
Cell proliferation
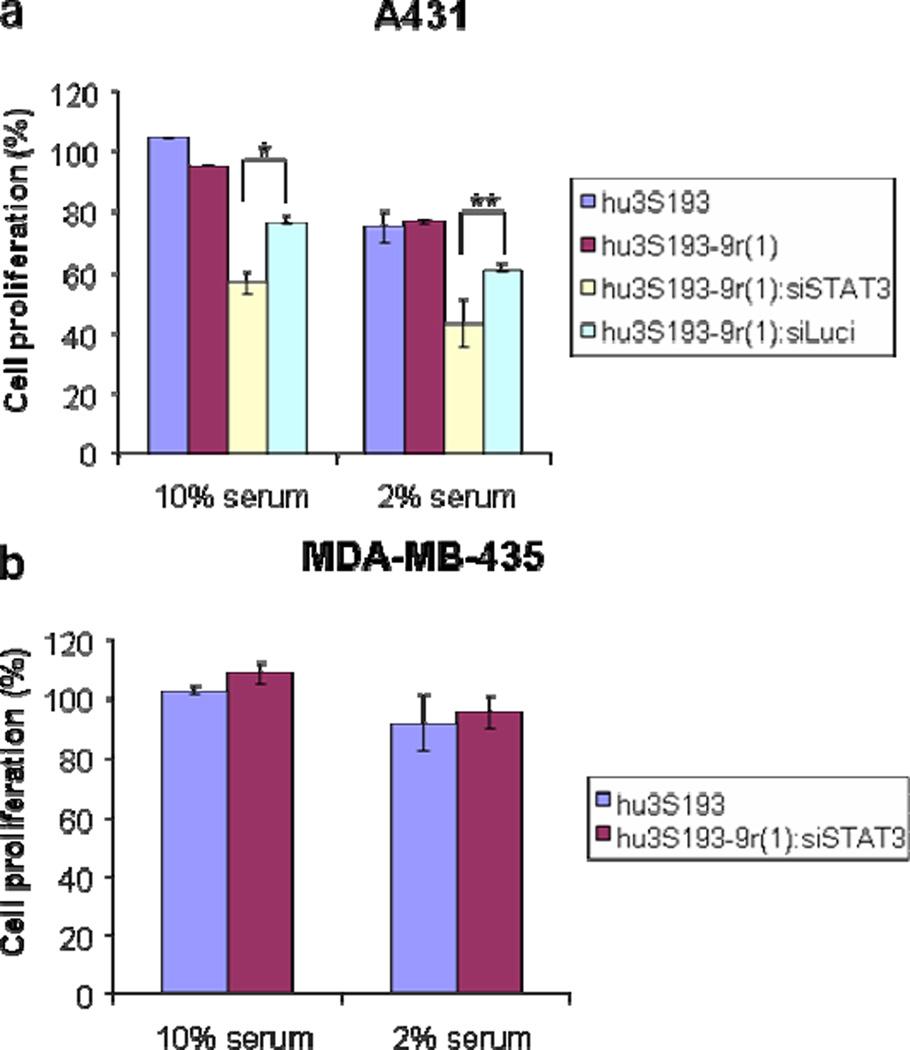
Since a strong correlation exists between the overexpression of STAT3 and cell proliferation (27–31), we further examined the effects of the treatment with hu3S193-9r(1):siSTAT3 on A431 cells in vitro. After 48 h, there is a slight, but consistent reduction of cell proliferation with hu3S193-9r(1):siSTAT3 treatment compared to vehicle alone or hu3S193-9r(1):siLuci (Fig. 7a). Reducing the serum level from 10% to 2% resulted in greater levels of inhibition. These results are consistent with data previously reported by Bonner et al. who used STAT3 shRNA for the treatment of A431 cells (52). The proliferation of MDA-MB-435 cells was not influenced by the treatment with hu3S193-9r(1):siSTAT3 (Fig. 7b). These results are consistent with our earlier observations on the cell-specific binding and gene knockdown in Ley-positive cells.
Figure 7. Cell proliferation assay.
a) A431 cells treated with hu3S193, hu3S193-9r(1), hu3S193-9r(1):siSTAT3(5:1) and hu3S193-9r(1):siLuci(5:1) and b) MDA-MB-435 cells were treated with hu3S193 and hu3S193-9r(1):siSTAT3(5:1) for 48 h and then analyzed using the MTS assay. The siRNA concentration was 300 nM.
*, **P-value between hu3S193-9r(1):siSTAT3(5:1) and hu3S193-9r(1):siLuci(5:1) treatment on A431 cell line with 10% serum is 0.0006*, with 2% serum is 0.008**.
STAT3 has been suggested to not only influence the cell viability and proliferation, but more importantly, the tumor cell microenvironment and/or angiogenesis in vivo (28). In vivo experiments including biodistribution and anti-tumor activity studies are currently underway. In conclusion, our results suggest that hu3S193-9r(1) is a promising vehicle for efficient cell-specific delivery of siRNAs (e.g. STAT3 or other tumor-related targets) into Ley-expressing tumor cells.
METHODS
Preparation of hu3S193-siRNA and hu3S193-9r:siRNA
D-siRNAs were designed according to published guidelines (7, 53–54). An initial screening was performed using RNAiMAX (invitrogen) as transfection reagent. The siRNAs (50 nM, final concentration) were transfected according to the manufacturer’s protocol for reverse transfections. The siRNA with best STAT3 knockdown efficiency was used in the study (data not shown). Sequence of STAT3 D-siRNA: 5’-GGAAGCUGCAGAAAGAUACGACUdGdA-3’ (sense strand); 5’-UCAGUGGUAUCUUUCUGCAGCUUCCGU-3’ (antisense strand). Sequence of luciferase siRNA: 5’-GGUUCCUGGAACAAUUGCUUUUAdCdA-3’ (sense strand); 5’-UGUAAAAGCAAUUGUUCCAGGAACCAG-3’ (antisense strand).
For the synthesis of the covalent construct, hu3S193 was allowed to react with S-HyNic (Solulink) and the average HyNic linker number per antibody was determined by reacting the resulting product with 2-sulfobenzaldehyde sodium salt according to manufacturer’s protocol (Solulink). Hu3S193-HyNic batches with different linker numbers (N = 1 – 5) per mAb were obtained after purification via Zeba™ Desalt Spin Columns (Thermo Scientific). The antisense strand was 5’-amino modified during solid phase synthesis and further reacted with linker S-SS-FB (Solulink). The linker modified antisense strand was purified by HPLC, annealed with the sense strand and conjugated with hu3S193-HyNic. Products were purified by FPLC. The stoichiometry of hu3S193-siRNA was calculated based on the BCA protein assay (Thermo Scientific) and A280, A260 measurements The RNA component of hu3S193-siRNA conjugate was confirmed by disulfide reduction (β-mercaptoethanol, Thermo Scientific) and analyzed on 15% PAGE in 1× TBE. For the fluorescein modified covalent conjugate, 5’-fluorescein-labeled, amino modified antisense RNA was prepared during solid phase synthesis using 6-fluorescein phosphoramidite (Glen Research, Cat#10-1094-XX). The fluorescein-labeled hu3S193-siRNA (hu3S193-siRNA(FL)) conjugate was similarly synthesized according the aforementioned methods.
For the non-covalent construct, hu3S193 was thiolated using Traut’s reagent (Thermo Scientific) and then modified with Cys(Npys)-(D-Arg)9 (9r, AnaSpec). The stoichiometry of thiol modification on hu3S193 was calculated based on signals resulting from reaction with Ellman’s reagent (Thermo Scientific). The hu3S193-9r:siRNA complex was obtained by mixing hu3S193-9r and siRNA in PBS at the indicated molar ratios and incubating the mixtures at room temperature for 20 min. Fluorescein-labeled siRNA (siRNA(FL) from IDT) was used instead of regular siRNA in the preparation of the non-covalent construct for flow cytometry and confocal imaging studies.
SYBR® Gold (Invitrogen) was used for siRNA staining and SimplyBlue™ SafeStain (Invitrogen) was used for protein staining in all gel studies.
Gel shift assay
Hu3S193, hu3S193-9r(1) or hu3S193-9r(4) were incubated with siRNA at different ratios for 20 min at room temperature. The binding of siRNA to vehicle was analyzed on 6% PAGE in 0.5 × TBE.
Cell lines
A431 and MDA-MB-435 cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% heat-inactivated FBS.
Flow cytometry
For cell binding analysis, 5×105 cells suspended in PBS were incubated with hu3S193(FL), hu3S193-siRNA(FL) or hu3S193-9r:siRNA(FL) at 4 °C for 30 min, washed 3 times with PBS and then fixed with 2% paraformaldehyde for 10 min. Fluorescence data were collected on a FACScalibur (Beckton Dickinson) and analyzed using FlowJo software (Tree Star).
Confocal microscopy
Cells were grown on slides, incubated with hu3S193-siRNA(FL) or hu3S193-9r(1):siRNA(FL) at 37 °C for 5–24 h, washed with PBS and fixed with 2% paraformaldehyde. The slides were mounted after staining the nuclei with DAPI (Vector) and analyzed by confocal microscopy (Zeiss Upright LSM510 2-Photon Microscope).
Quantitative PCR
Hu3S193-siRNA:9r or hu3S193-9r(1):siRNA were premixed at room temperature for 20 min at the indicated molar ratios. 3×104 A431 or MDA-MB435 cells were seeded on a 48-well plate and treated with hu3S193-siRNA, hu3S193-siRNA with chloroquine, hu3S193-siRNA:9r or hu3S193-9r(1):siRNA for 24 h. Total RNA was extracted from cells using the RNeasy kit (Qiagen). The Express One-Step SuperScript qRT-PCR kit (Invitrogen) was used to reverse-transcribe and amplify 25 ng of total RNA per reaction according to the manufacturer’s protocol. The ProbeFinder software (Roche Applied Science) was used to design the primer sets for STAT3 and GAPDH and to select the respective probes from the Universal ProbeLibrary (Roche). Probe # 17 and the following primers were used for the STAT3 assay: 5’ cgatggagtacgtgcagaaa and 5’ tgagattctgctaatgacgttatcc. Probe #60 and the following primers were used for the GAPDH assay: 5’ agccacatcgctcagacac and 5’ gcccaatacgaccaaatcc. All samples were run in triplicates. Amplifications were performed on a Bio-Rad iCycler iQ5 Multiple-Color Real-time PCR Detection System. The data were normalized to the GAPDH expression and the relative expression levels were calculated using the 2−ΔΔCt method.
Western blot analysis
Cells were lysed with M-per buffer (Thermo Scientific) containing protease and phosphatase inhibitors (Thermo Scientific). 20 µg of protein was resolved in 4%–15% SDS-PAGE (Bio-Rad) and transferred to NuPage nitrocellulose membranes (Invitrogen). The membranes were washed with PBS-0.1% Tween 20, blocked for 1 hr at room temperature with 5% milk in PBS-0.1% Tween 20 and then incubated overnight at 4 °C with the indicated primary antibodies (phospho-STAT3 (Cell Signaling, Cat#9131), total STAT3 (Cell Signaling, Cat#9132), β-actin (Sigma-Aldrich, Cat#A5441)). The membrane was then washed with PBS-0.1% Tween 20, incubated for 1 h at room temperature with HRP-conjugated anti-rabbit (GE, NA9340) or anti-mouse (GE, NA931) secondary antibodies. The blots were developed using an enhanced chemiluminescence detection system (Thermo Scientific). For detection of total STAT3, the corresponding phospho blots were incubated with stripping buffer (Thermo Scientific) and reblotted.
MTS assays
Cell proliferation assays were performed using the CellTiter 96 Aqueous One Solution Cell proliferation Assay (Promega). Each well of a 96-well plate was seeded with 5,000 cells in culture medium. After 16 h culture, the cells were treated with hu3S193-9r(1):siRNA. After 48 h of treatment, MTS was added to the cells according to the supplier's protocol and the absorbance was measured at 490 nm using a 96-well plate reader (Synergy 4, Biotek).
Supplementary Material
ACKNOWLEDGMENT
Ludwig Institute for Cancer Research/Atlantic Philanthropies are gratefully acknowledged for support of this work. We thank the Irell &Manella Graduate School of Biological Sciences at City of Hope for a merit fellowship to Y.M. We also acknowledge the Synthetic and Biopolymer Chemistry core, Light Microscopy Digital Imaging core and Analytical Cytometry core facilities at City of Hope.
Footnotes
ASSOCIATED CONTENT
Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Reference
- 1.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med. 2009;1:142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 4.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci U S A. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amarzguioui M, Lundberg P, Cantin E, Hagstrom J, Behlke MA, Rossi JJ. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat Protoc. 2006;1:508–517. doi: 10.1038/nprot.2006.72. [DOI] [PubMed] [Google Scholar]
- 8.Perkel JM. RNAI Therapeutics: A Two-Year Update. Science. 2009;326:454. [Google Scholar]
- 9.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 10.White PJ. Barriers to successful delivery of short interfering RNA after systemic administration. Clin Exp Pharmacol Physiol. 2008;35:1371–1376. doi: 10.1111/j.1440-1681.2008.04992.x. [DOI] [PubMed] [Google Scholar]
- 11.Tseng YC, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev. 2009;61:721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda Y, Taira K. Ligand-targeted delivery of therapeutic siRNA. Pharm Res. 2006;23:1631–1640. doi: 10.1007/s11095-006-9001-x. [DOI] [PubMed] [Google Scholar]
- 13.Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjug Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- 14.Liu B. Exploring cell type-specific internalizing antibodies for targeted delivery of siRNA. Brief Funct Genomic Proteomic. 2007;6:112–119. doi: 10.1093/bfgp/elm015. [DOI] [PubMed] [Google Scholar]
- 15.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 16.Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, Shankar P. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pirollo KF, Rait A, Zhou Q, Hwang SH, Dagata JA, Zon G, Hogrefe RI, Palchik G, Chang EH. Materializing the potential of small interfering RNA via a tumor-targeting nanodelivery system. Cancer Res. 2007;67:2938–2943. doi: 10.1158/0008-5472.CAN-06-4535. [DOI] [PubMed] [Google Scholar]
- 18.Xia CF, Boado RJ, Pardridge WM. Antibody-mediated targeting of siRNA via the human insulin receptor using avidin-biotin technology. Mol Pharm. 2009;6:747–751. doi: 10.1021/mp800194y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott AM, Geleick D, Rubira M, Clarke K, Nice EC, Smyth FE, Stockert E, Richards EC, Carr FJ, Harris WJ, Armour KL, Rood J, Kypridis A, Kronina V, Murphy R, Lee FT, Liu Z, Kitamura K, Ritter G, Laughton K, Hoffman E, Burgess AW, Old LJ. Construction, production, and characterization of humanized anti-Lewis Y monoclonal antibody 3S193 for targeted immunotherapy of solid tumors. Cancer Res. 2000;60:3254–3261. [PubMed] [Google Scholar]
- 20.Sakamoto J, Furukawa K, Cordon-Cardo C, Yin BW, Rettig WJ, Oettgen HF, Old LJ, Lloyd KO. Expression of Lewisa, Lewisb, X, Y blood group antigens in human colonic tumors and normal tissue and in human tumor-derived cell lines. Cancer Res. 1986;46:1553–1561. [PubMed] [Google Scholar]
- 21.Kim YS, Yuan M, Itzkowitz SH, Sun QB, Kaizu T, Palekar A, Trump BF, Hakomori S. Expression of LeY and extended LeY blood group-related antigens in human malignant, premalignant, and nonmalignant colonic tissues. Cancer Res. 1986;46:5985–5992. [PubMed] [Google Scholar]
- 22.Dettke M, Palfi G, Loibner H. Activation-dependent expression of the blood group-related lewis Y antigen on peripheral blood granulocytes. J Leukoc Biol. 2000;68:511–514. [PubMed] [Google Scholar]
- 23.Zhang S, Zhang HS, Cordon-Cardo C, Reuter VE, Singhal AK, Lloyd KO, Livingston PO. Selection of tumor antigens as targets for immune attack using immunohistochemistry: II. Blood group-related antigens. International Journal of Cancer. 1997;73:50–56. doi: 10.1002/(sici)1097-0215(19970926)73:1<50::aid-ijc9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Scott AM, Tebbutt N, Lee FT, Cavicchiolo T, Liu Z, Gill S, Poon AM, Hopkins W, Smyth FE, Murone C, MacGregor D, Papenfuss AT, Chappell B, Saunder TH, Brechbiel MW, Davis ID, Murphy R, Chong G, Hoffman EW, Old LJ. A phase I biodistribution and pharmacokinetic trial of humanized monoclonal antibody Hu3s193 in patients with advanced epithelial cancers that express the Lewis-Y antigen. Clin Cancer Res. 2007;13:3286–3292. doi: 10.1158/1078-0432.CCR-07-0284. [DOI] [PubMed] [Google Scholar]
- 25.Kelly MP, Lee FT, Tahtis K, Smyth FE, Brechbiel MW, Scott AM. Radioimmunotherapy with alpha-particle emitting 213Bi-C-functionalized trans-cyclohexyl-diethylenetriaminepentaacetic acid-humanized 3S193 is enhanced by combination with paclitaxel chemotherapy. Clin Cancer Res. 2007;13:5604s–5612s. doi: 10.1158/1078-0432.CCR-07-1071. [DOI] [PubMed] [Google Scholar]
- 26.Boghaert ER, Sridharan L, Armellino DC, Khandke KM, DiJoseph JF, Kunz A, Dougher MM, Jiang F, Kalyandrug LB, Hamann PR, Frost P, Damle NK. Antibody-targeted chemotherapy with the calicheamicin conjugate hu3S193-N-acetyl gamma calicheamicin dimethyl hydrazide targets Lewisy and eliminates Lewisy-positive human carcinoma cells and xenografts. Clin Cancer Res. 2004;10:4538–4549. doi: 10.1158/1078-0432.CCR-04-0037. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 28.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anticancer Drugs. 2005;16:601–607. doi: 10.1097/00001813-200507000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18:254–267. doi: 10.1038/cr.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao L, Zhang L, Hu J, Li F, Shao Y, Zhao D, Kalvakolanu DV, Kopecko DJ, Zhao X, Xu DQ. Down-regulation of signal transducer and activator of transcription 3 expression using vector-based small interfering RNAs suppresses growth of human prostate tumor in vivo. Clin Cancer Res. 2005;11:6333–6341. doi: 10.1158/1078-0432.CCR-05-0148. [DOI] [PubMed] [Google Scholar]
- 33.Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling X, Arlinghaus RB. Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res. 2005;65:2532–2536. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- 35.Sumita N, Bito T, Nakajima K, Nishigori C. Stat3 activation is required for cell proliferation and tumorigenesis but not for cell viability in cutaneous squamous cell carcinoma cell lines. Exp Dermatol. 2006;15:291–299. doi: 10.1111/j.0906-6705.2006.00407.x. [DOI] [PubMed] [Google Scholar]
- 36.Lee SO, Lou W, Qureshi KM, Mehraein-Ghomi F, Trump DL, Gao AC. RNA interference targeting Stat3 inhibits growth and induces apoptosis of human prostate cancer cells. Prostate. 2004;60:303–309. doi: 10.1002/pros.20072. [DOI] [PubMed] [Google Scholar]
- 37.Gao LF, Wen LJ, Yu H, Zhang L, Meng Y, Shao YT, Xu DQ, Zhao XJ. Knockdown of Stat3 expression using RNAi inhibits growth of laryngeal tumors in vivo. Acta Pharmacol Sin. 2006;27:347–352. doi: 10.1111/j.1745-7254.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- 38.Qiu Z, Huang C, Sun J, Qiu W, Zhang J, Li H, Jiang T, Huang K, Cao J. RNA interference-mediated signal transducers and activators of transcription 3 gene silencing inhibits invasion and metastasis of human pancreatic cancer cells. Cancer Sci. 2007;98:1099–1106. doi: 10.1111/j.1349-7006.2007.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Alizadeh D, Van Handel M, Kortylewski M, Yu H, Badie B. Stat3 inhibition activates tumor macrophages and abrogates glioma growth in mice. Glia. 2009;57:1458–1467. doi: 10.1002/glia.20863. [DOI] [PubMed] [Google Scholar]
- 40.Kortylewski M, Swiderski P, Herrmann A, Wang L, Kowolik C, Kujawski M, Lee H, Scuto A, Liu Y, Yang C, Deng J, Soifer HS, Raubitschek A, Forman S, Rossi JJ, Pardoll DM, Jove R, Yu H. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat Biotechnol. 2009;27:925–932. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang YH, Hou YW, Lee HJ. An intracellular delivery method for siRNA by an arginine-rich peptide. J Biochem Biophys Methods. 2007;70:579–586. doi: 10.1016/j.jbbm.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 42.Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 43.El-Sayed A, Khalil IA, Kogure K, Futaki S, Harashima H. Octaarginine- and octalysine-modified nanoparticles have different modes of endosomal escape. J Biol Chem. 2008;283:23450–23461. doi: 10.1074/jbc.M709387200. [DOI] [PubMed] [Google Scholar]
- 44.Kim SW, Kim NY, Choi YB, Park SH, Yang JM, Shin S. RNA interference in vitro and in vivo using an arginine peptide/siRNA complex system. J Control Release. 2010;143:335–343. doi: 10.1016/j.jconrel.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc Natl Acad Sci U S A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westwood JA, Smyth MJ, Teng MW, Moeller M, Trapani JA, Scott AM, Smyth FE, Cartwright GA, Power BE, Honemann D, Prince HM, Darcy PK, Kershaw MH. Adoptive transfer of T cells modified with a humanized chimeric receptor gene inhibits growth of Lewis-Y-expressing tumors in mice. Proc Natl Acad Sci U S A. 2005;102:19051–19056. doi: 10.1073/pnas.0504312102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee FT, Mountain AJ, Kelly MP, Hall C, Rigopoulos A, Johns TG, Smyth FE, Brechbiel MW, Nice EC, Burgess AW, Scott AM. Enhanced efficacy of radioimmunotherapy with 90Y-CHX-A"-DTPA-hu3S193 by inhibition of epidermal growth factor receptor (EGFR) signaling with EGFR tyrosine kinase inhibitor AG1478. Clin Cancer Res. 2005;11:7080s–7086s. doi: 10.1158/1078-0432.CCR-1004-0019. [DOI] [PubMed] [Google Scholar]
- 48.de Duve C, de Barsy T, Poole B, Trouet A, Tulkens P, Van Hoof F. Commentary. Lysosomotropic agents. Biochem Pharmacol. 1974;23:2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]
- 49.Erbacher P, Roche AC, Monsigny M, Midoux P. Putative role of chloroquine in gene transfer into a human hepatoma cell line by DNA/lactosylated polylysine complexes. Exp Cell Res. 1996;225:186–194. doi: 10.1006/excr.1996.0169. [DOI] [PubMed] [Google Scholar]
- 50.Alimoghaddam K. Effect of chloroquine on transduction of cell lines and baboon CD34 cells by a GALV pseudotyped retrovirus. Arch. Iran. Med. FIELD Full Journal Title:Archives of Iranian Medicine. 2002;5:244–250. [Google Scholar]
- 51.Zhang H, Solomon VR, Hu C, Ulibarri G, Lee H. Synthesis and in vitro cytotoxicity evaluation of 4-aminoquinoline derivatives. Biomed Pharmacother. 2008;62:65–69. doi: 10.1016/j.biopha.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonner JA, Trummell HQ, Willey CD, Plants BA, Raisch KP. Inhibition of STAT-3 results in radiosensitization of human squamous cell carcinoma. Radiother Oncol. 2009;92:339–344. doi: 10.1016/j.radonc.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 54.Amarzguioui M, Rossi JJ. Principles of Dicer substrate (D-siRNA) design and function. Methods Mol Biol. 2008;442:3–10. doi: 10.1007/978-1-59745-191-8_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.