Abstract
Yeast xylose metabolism is generally considered to be restricted to respirative conditions because the two-step oxidoreductase reactions from xylose to xylulose impose an anaerobic redox imbalance. We have recently developed, however, a Saccharomyces cerevisiae strain that is at present the only known yeast capable of anaerobic growth on xylose alone. Using transcriptome analysis of aerobic chemostat cultures grown on xylose-glucose mixtures and xylose alone, as well as a combination of global gene expression and metabolic flux analysis of anaerobic chemostat cultures grown on xylose-glucose mixtures, we identified the distinguishing characteristics of this unique phenotype. First, the transcript levels and metabolic fluxes throughout central carbon metabolism were significantly higher than those in the parent strain, and they were most pronounced in the xylose-specific, pentose phosphate, and glycerol pathways. Second, differential expression of many genes involved in redox metabolism indicates that increased cytosolic NADPH formation and NADH consumption enable a higher flux through the two-step oxidoreductase reaction of xylose to xylulose in the mutant. Redox balancing is apparently still a problem in this strain, since anaerobic growth on xylose could be improved further by providing acetoin as an external NADH sink. This improved growth was accompanied by an increased ATP production rate and was not accompanied by higher rates of xylose uptake or cytosolic NADPH production. We concluded that anaerobic growth of the yeast on xylose is ultimately limited by the rate of ATP production and not by the redox balance per se, although the redox imbalance, in turn, limits ATP production.
The yeast Saccharomyces cerevisiae serves as the paradigm model for lower eukaryotes and often also higher eukaryotes. In sharp contrast to almost all other organisms, however, S. cerevisiae has been domesticated for millennia and has been used for uncountable generations in human food processing. This premier status as a workhorse was earned, and continuously selected for, by the ability to anaerobically ferment sugars to ethanol and carbon dioxide at high rates. Perhaps for this reason, it is often forgotten that only some fungi (e.g., fungi in the rumen) (39) and few yeasts grow anaerobically and that S. cerevisiae stands out as a yeast that is capable of rapid anaerobic growth, provided that it is supplemented with membrane sterols (45). This requirement for sterols like ergosterol is related to biosynthetic hydroxylation reactions that require molecular oxygen, so oxygen can be seen as a growth factor (1). However, there must be further limitations that explain the impaired anaerobic growth of yeasts other than S. cerevisiae even on the most common substrate, glucose, when they are supplemented with sterols. One such case is pyrimidine biosynthesis because expression of the URA1 gene from S. cerevisiae allows Pichia stipitis to grow anaerobically on glucose (37). Another, perhaps more general limitation for anaerobic growth is a sufficiently high rate of ATP formation from fermentation of a given substrate (44).
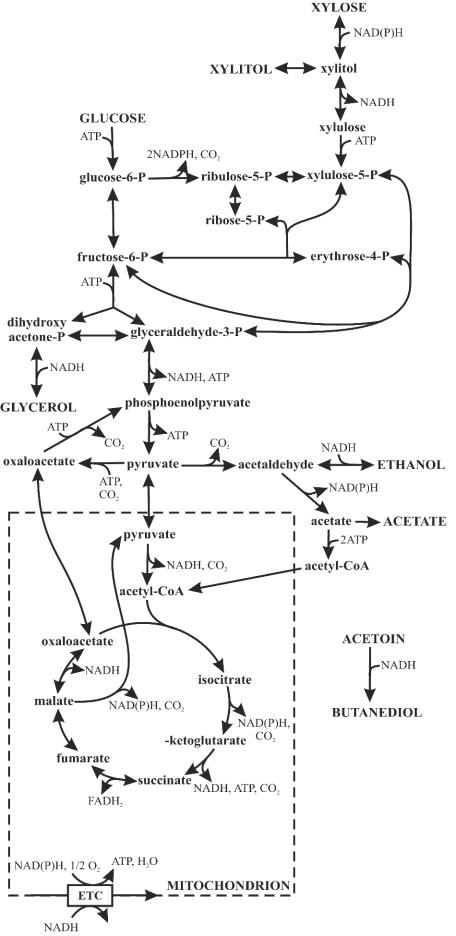
With the commercial interest in fermenting renewable resources to ethanol, significant efforts have been focused on metabolic engineering of efficient anaerobic pentose metabolism in S. cerevisiae (3, 11, 14, 29). A particular focus has been on xylose, which is not normally utilized by S. cerevisiae. Although a few xylose-metabolizing yeasts are found in nature, none can grow anaerobically on xylose (15, 20). In contrast to direct isomerization of xylose to xylulose in bacteria and possibly a few fungi (13, 22) that can grow on xylose in the absence of oxygen, yeast xylose catabolism generally proceeds through the consecutive redox reactions catalyzed by NADPH-dependent xylose reductases and NAD+-dependent xylitol dehydrogenases, with xylitol as the intermediate (Fig. 1). By respiring NADH and supplying NADPH through the pentose phosphate pathway, which is rather active in the xylose-utilizing yeast P. stipitis (8), yeast metabolism can efficiently drive these coupled redox reactions for respirative growth but not for fermentative growth. Functional expression of xylose isomerase in recombinant xylose-utilizing S. cerevisiae, however, does not enable anaerobic growth on xylose (13, 21, 33, 50), which indicates that there must be limitations beyond redox balancing.
FIG. 1.
Bioreaction network of S. cerevisiae central carbon metabolism. Extracellular metabolites are indicated by uppercase letters. ETC, electron transport chain; acetyl-CoA, acetyl coenzyme A; FADH2, reduced flavin adenine dinucleotide.
While the ability to grow anaerobically on pentoses apparently has not evolved in yeasts, we recently isolated a mutant with this ability from a 460-generation continuous culture (38). This evolutionary engineering experiment (34) was initiated with the xylose-utilizing strain S. cerevisiae TMB3001, which overexpresses the NAD(P)H-dependent xylose reductase and the NAD+-dependent xylitol dehydrogenase from P. stipitis and the endogenous xylulokinase (7). With carbon-limited selective pressure for anaerobic growth on xylose, TMB3001 evolved into a population with improved xylose utilization (38). One representative of the major subpopulation in the evolved culture, the C1 mutant, was capable of anaerobic growth on xylose alone. To the best of our knowledge, S. cerevisiae C1 is at present the only yeast that is capable of strictly anaerobic growth on xylose.
The C1 strain provides a unique opportunity to elucidate the molecular mechanisms that are necessary for yeast xylose metabolism under anaerobic conditions. Using DNA microarrays (6) and metabolic flux analysis (35, 42, 47), in this study we identified the key components that enable this phenotype. While the results revealed that balancing redox equivalents is one crucial component, we provide strong evidence that it is ultimately the rate of ATP formation that limits anaerobic growth on xylose.
MATERIALS AND METHODS
Strains, media, and cultivation conditions.
S. cerevisiae strain TMB3001 (CEN.PK 113-7A [MATa his3-Δ1 MAL2-8c SUC2] his3::YIpXR/XDH/XK) (7) and its evolved mutant C1 (=DSM 15519) (38) were used throughout this study. Recombinant strain TMB3001 contains a chromosomally integrated cassette with the NAD(P)H-dependent xylose reductase gene and the NAD+-dependent xylitol dehydrogenase gene from P. stipitis combined with an additional copy of the endogenous xylulokinase gene (7). Expression of xylose reductase is controlled by the ADH1 promoter and terminator, and expression of the other two enzymes is controlled by the PGK1 promoter and terminator. Cultures were stored in aliquots supplemented with 15% glycerol at −80°C and were revived by growth in YPD medium (10 g of yeast extract per liter, 20 g of peptone per liter, 20 g of glucose per liter). All cultures used for physiological and DNA microarray experiments were grown in minimal medium (38, 43). For anaerobic cultivation, the medium was supplemented with ethanol-dissolved ergosterol (Fluka) and Tween 80 (Sigma) at final concentrations of 0.01 and 0.42 g liter−1, respectively.
Flask cultures were grown in 500-ml baffled shake flasks with 50 ml of medium at 30°C and 300 rpm. Carbon-limited chemostat cultures were grown in 1 liter of medium in a 2-liter stirred tank reactor (Bioengineering, Wald, Switzerland) at a dilution (growth) rate of 0.05 h−1. The volume was kept constant by continuous removal of excess culture broth through a sterile needle that was fixed at a predetermined height. A constant pH of 5.0 was maintained by automatic addition of 2 M KOH. Sparging with air at a rate of 1 liter min−1 established aerobic conditions, whereas sparging with pure N2 (O2 concentration, <5 ppm) at a rate of 0.35 liter min−1 established anaerobiosis. Constant gas flow rates were controlled with a mass flow meter (Inceltech, Toulouse, France). To ensure anaerobiosis, the feed medium was also maintained under an N2 atmosphere. The stirrer speed was set at 1,000 and 500 rpm under aerobic and anaerobic conditions, respectively. Culture aliquots for metabolic flux and transcript analysis were withdrawn in a physiological steady state, which was defined as a stable cell density and a stable rate of CO2 evolution for at least three volume changes.
Anaerobic batch experiments for metabolic flux analysis were done in Hungate tubes (Bellco Glass Inc., Vineland, N.J.) that were sealed with butyl rubber septa by using inocula that were washed twice with phosphate-buffered saline (8 g of NaCl per liter, 0.2 g of KCl per liter, 1.44 g of Na2HPO4 per liter, 0.24 g of KH2PO4 per liter; pH 7.0). The basic salt solution was sparged with pure N2 (O2 concentration, <5 ppm) for 15 min, and, after autoclaving, the remaining filter-sterilized, N2-sparged medium components and 10 g of xylose per liter or 10 g of glucose per liter were added. Where indicated below, acetoin was added at a final concentration of 0.5 g liter−1. Culture aliquots were withdrawn with purging with pure N2.
RNA isolation and DNA microarray analysis.
Two 50-ml culture aliquots were harvested in liquid N2-precooled polypropylene tubes (Greiner, Kremsmünster, Austria) and immediately centrifuged at 3,000 × g and 4°C for 3 min. The pellets were washed twice with ice-cold AE buffer (50 mM sodium acetate, 10 mM EDTA; pH 5.2) and rapidly frozen in liquid N2 for storage at −80°C. Total RNA was extracted by the hot-phenol method (36), and the absorbance at 260 and 280 nm was used for quantification and purity control. RNA integrity was assessed in formaldehyde-containing agarose gels (32). mRNA isolation, cDNA synthesis, in vitro transcription (cRNA synthesis), and cRNA fragmentation were performed by using the methods described in the Affymetrix expression analysis technical manual. Hybridization, washing, staining, and scanning of the Gene Chip Yeast Genome S98 arrays (Affymetrix) were performed by using a hybridization oven (Affymetrix), a model 400 fluidics station (Affymetrix), and a GeneArray scanner (Affymetrix).
Gene expression data were analyzed by using the Microarray Suite 5.0 software (Affymetrix). The average fluorescence of each array was normalized to a common value of 100. Of the 9,335 transcripts present on the YG-S98 array, only the 6,383 yeast open reading frames were considered (4). The coefficient of variation (standard deviation divided by the mean) was calculated for all 6,383 transcripts by using the data from duplicate experiments, and the average coefficient of variation over the entire array was used to assess the experimental error for the gene expression analysis in each experiment. Since low transcript levels are inherently difficult to quantify, all expression values that were less than 20 were adjusted to 20 for a fold change analysis. Statistical analysis of differential gene expression was performed with the Significance Analysis of Micro-arrays (SAM) version 1.21 EXCEL add-in software (40), because it scales down better to small numbers of replicates than the standard t test (25). Since a fold change of at least two with an expected median false-positive rate of 1% determined by the SAM software was the minimum requirement for interlaboratory microarray reproducibility (4, 25), we chose these cutoff parameters for the first genome-wide gene expression analysis. To identify significant expression changes below the twofold cutoff for genes of central carbon metabolism, we used the SAM software with a median false-positive discovery rate of 1% but without a fold change cutoff.
Analytical methods.
Cell growth was monitored by determining the optical density at 600 nm (OD600). Cellular dry weight was determined for at least five 10-ml culture aliquots that were centrifuged at 5,000 rpm for 20 min in preweighed glass tubes, washed once with water, and dried at 110°C for 24 h to a constant weight. Commercially available kits were used for enzymatic determination of glucose (Beckman), xylose (Medichem, Steinenbronn, Germany), xylitol (R-Biopharm, Darmstadt, Germany), acetate (R-Biopharm), and glycerol (Sigma) concentrations. Ethanol, acetoin, and butanediol concentrations were determined by gas chromatography as described previously (35). Pyruvate and succinate concentrations were determined by high-performance liquid chromatography (Perkin-Elmer, Shelton, Conn.) with a Supelco H column (Supelco, Bellefonte, Pa.) and 0.15% H3PO4 as the mobile phase. CO2 and ethanol concentrations in the reactor off-gas were determined online with a Prima 600 mass spectrometer (Fisons Instruments, Uxbridge, England).
Determination of physiological parameters and intracellular metabolic fluxes.
Maximum exponential growth rates in batch cultures were determined by log-linear regression of OD600 versus time with growth rate as the regression coefficient. The specific biomass yield was determined by determining the coefficient of linear regression of biomass concentration versus substrate concentration during the exponential growth phase. The biomass concentration was estimated from predetermined OD600-dry weight correlations during the mid-exponential growth phase of aerobic cultures growing on glucose for TMB3001 and C1 (0.530 and 0.479 g unit of OD600−1, respectively). During the exponential growth phase, specific glucose or xylose uptake rates were calculated by determining the ratio of growth rate to specific biomass yield. Ethanol, xylitol, acetate, glycerol, and butanediol yields were calculated by linear regression of by-product concentration versus substrate concentration, and the specific production rates were calculated by determining the product of the specific xylose or glucose uptake rate and the by-product yield.
In chemostat cultures, biomass and by-product yields were determined by determining the ratio of the molar carbon in the product considered to the total molar carbon in the consumed substrates in the steady state, assuming a ratio of 0.476 g of C g of biomass−1 (47). Specific consumption and production rates were calculated by determining the ratios of the molar production rates to the steady-state biomass concentration. The fractions of evaporated ethanol, O2, and CO2 in the bioreactor off-gas were determined by online mass spectrometry analysis.
A previously developed stoichiometric model (47) was used to estimate intracellular carbon fluxes in anaerobic chemostat and batch cultures. The fluxes to ethanol and CO2 were defined as free fluxes, whose computed values were compared with the redundant experimental ethanol and CO2 production rates. The computed free fluxes were always within 15% of the experimental values, thus confirming the reliability of the stoichiometric model used. This model was extended with acetoin reduction to butanediol (48) for cases in which acetoin was added. The macromolecular cell compositions were assumed to be 39% (wt/wt) polysaccharides, 50% (wt/wt) protein, and 6% (wt/wt) RNA in chemostat cultures, 40% (wt/wt) polysaccharides, 52% (wt/wt) protein, and 3% (wt/wt) RNA in batch cultures with xylose, and 31% (wt/wt) polysaccharides, 56% (wt/wt) protein, and 9% (wt/wt) RNA in batch cultures with glucose (47). Since the recombinant xylose reductase in our strains can also catalyze the NADPH-dependent reduction of dihydroxyacetone-P (17), we assessed the effect of changing the cosubstrate specificity of the dihydroxyacetone-P dehydrogenase from NADH to NADPH. With the exception of a higher fraction of NADH-dependent xylose reduction, there was no further effect on the estimated intracellular metabolic flux distribution (data not shown). Since the cofactor use of xylose reductase may change with cultivation conditions or genetic manipulations (7), we were not able to determine the proportions of the NADH- and NADPH-dependent xylose reduction fluxes.
Enzymatic assays.
Cell extracts were prepared from mid-exponential-phase cultures in minimal medium with glucose. Cell pellets were harvested by centrifugation, washed with deionized water, and resuspended in 0.1 M triethanolamine buffer (pH 7.0) containing 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, and 0.5 mM EDTA. The suspension was vortexed with glass beads (diameter, 0.5 mm) at 4°C for 5 min, incubated on ice for 5 min, and vortexed again for 5 min. The cell debris and glass beads were separated by centrifugation at 20,000 × g and 4°C for 5 min. In vitro activities of xylose reductase (with NADPH and NADH), xylitol dehydrogenase (with NADH), and xylulokinase in the supernatant were determined by using a previously described method (7), which was adapted for measurement in 96-well plates with a total volume of 200 μl. The sole difference was the use of triethanolamine buffer at pH 7.0 instead of glycine buffer at pH 9.0 for the xylitol dehydrogenase assay. The total protein content of the supernatant was determined with a commercially available Pyrogallol red-based kit (Beckman). Specific activities were expressed in units per milligram of protein; 1 U was defined as reduction or oxidation of 1 μmol of NAD(P)H per min.
RESULTS
Chemostat cultivation.
For reliable and meaningful identification of specific differences between mutants, particularly when genome-wide methodologies are used, it is important to ensure that environmental conditions are identical and to minimize physiological differences that cause nonspecific responses (6, 25). To avoid nonspecific, growth rate-related effects, we grew S. cerevisiae TMB3001 (7) and the evolved C1 strain (38) in carbon-limited chemostat cultures at the same growth rate for genome-wide transcript level and metabolic flux analyses. The dilution rate used, 0.05 h−1, allowed reliable establishment of a physiological steady state for both strains under the conditions used. Anaerobic growth solely on xylose was not investigated directly because the control strain could not grow at all under these conditions and C1 can grow only at a maximum growth rate of about 0.012 h−1 (38). In the steady state, the C1 strain consumed xylose at a higher specific rate and left significantly lower residual xylose concentrations in the medium (Table 1). This higher coutilization of xylose enabled much greater accumulation of biomass in the aerobic C1 culture. Compared to anaerobically grown TMB3001, C1 had a 20% lower biomass concentration and generated significantly more metabolic by-products, particularly glycerol and xylitol, from the xylose consumed. In contrast to C1 (Table 1), TMB3001 was incapable of maintaining an aerobic steady state at a dilution rate of 0.05 h−1 on 20 g of xylose per liter because its maximum growth rate was only 0.016 h−1 under these conditions (38). Since even this low growth rate on xylose was achieved by TMB3001 only after a lengthy adaptation process that probably introduced additional mutations (38) and since we focused on anaerobic growth, we did not directly compare aerobic growth rates on xylose alone for the two strains. Moreover, growth rates that are so low typically do not lead to stable steady states in chemostat cultures and also reduce the quality of the transcript data.
TABLE 1.
Metabolite concentrations and physiological parameters of S. cerevisiae TMB3001 and C1 in steady-state chemostat cultures at a dilution rate of 0.05 h−1
| Conditions | Strain | Feed concn (g liter−1)
|
Steady-state concn (g liter−1)a
|
Specific consumption or production rates (g g of biomass−1 h−1)
|
Carbon balance (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Xylose | Glucose | Xylose | Biomass | Ethanol | Xylitol | Glycerol | Xylose | Ethanol | CO2 | |||
| Aerobic | TMB3001 | 9.6 ± 0.1b | 9.7 ± 0.5 | 0 | 5.5 ± 0.2 | 6.5 ± 0.3 | 0 | 0 | 0 | −0.03 ± 0.00 | 0 | 0.09 ± 0.00 | 114 ± 0 |
| C1 | 9.6 ± 0.1 | 9.7 ± 0.1 | 0 | 0.1 ± 0.0 | 10.0 ± 0.2 | 0 | 0 | 0 | −0.05 ± 0.00 | 0 | 0.07 ± 0.00 | 115 ± 1 | |
| 0 | 20.9 ± 0.0 | 0 | 0.8 ± 0.2 | 8.7 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | −0.11 ± 0.00 | <0.01 | 0.08 ± 0.00 | 100 ± 0 | ||
| Anaerobic | TMB3001 | 10.0 ± 0.0 | 10.2 ± 0.1 | 0.1 ± 0.0 | 7.4 ± 0.0 | 1.0 ± 0.0 | 4.0 ± 0.2 | 1.0 ± 0.0 | 1.2 ± 0.0 | −0.14 ± 0.01 | 0.24 ± 0.00c | 0.28 ± 0.00 | 107 ± 3 |
| C1 | 10.0 ± 0.0 | 10.2 ± 0.1 | 0.1 ± 0.0 | 4.7 ± 0.0 | 0.8 ± 0.0 | 4.1 ± 0.0 | 1.9 ± 0.3 | 2.6 ± 0.1 | −0.31 ± 0.00 | 0.29 ± 0.00c | 0.34 ± 0.00 | 104 ± 2 | |
The acetate concentration was below the detection level (0.05 g liter−1) except for the anaerobic C1 culture, in which the acetate concentration was 0.1 ± 0.0 g liter−1.
Average ± standard deviation for two independent cultures.
The values include an approximately 20% fraction of evaporated ethanol that was determined by online mass spectrometry of the reactor off-gas.
Global gene expression analysis of chemostat culture cells.
DNA microarray analysis was performed with RNA isolated from cells in the chemostat cultures described above to indirectly elucidate molecular changes that underlie the ability to grow on xylose as the sole carbon source, because a direct comparison on xylose alone was not feasible. For each strain and condition, transcript levels were quantified by using duplicate experiments with average coefficients of variation between 0.13 and 0.34 (Table 2). A total of 577 genes exhibited a statistically significant greater twofold differential expression pattern in C1 than in TMB3001 (as determined with SAM, version 1.21, with a maximal expected median false-positive rate of 1%) (4, 25) under at least one cultivation condition, and 119 of these genes were differentially expressed under all three conditions (data not shown). For growth on xylose alone, differential gene expression was determined by comparison with TMB3001 under the same conditions but with an additional carbon source, glucose, since TMB3001 cannot grow at this rate on xylose (12, 38). By comparing the differential expression for TMB3001 and C1 on the mixture, we obtained indirect information concerning genes that are specifically affected in C1 during growth on xylose alone.
TABLE 2.
Average coefficients of variation for the microarray experiments
| Cultivation conditions | Strain | Average coefficient of variationa |
|---|---|---|
| Aerobic cultures | ||
| Glucose-xylose | TMB3001 | 0.16 |
| C1 | 0.13 | |
| Xylose | C1 | 0.34 |
| Anaerobic cultures | ||
| Glucose-xylose | TMB3001 | 0.17 |
| C1 | 0.17 |
The values are the average coefficients of variation (standard deviation divided by the mean for two independent cultures) for all genes.
A significant portion of the differentially expressed genes encoded metabolic functions, and the strongest up-regulation by far was evident for the galactose metabolism genes GAL1, GAL2, GAL7, and GAL10 under all conditions (Table 3). Expression of the central carbon metabolism genes SOL3, MAE1, GPD2, ADH5, and particularly PYK2 was generally significantly increased, whereas PDC6, ADH4, PGM2, SDH1, ZWF1, YAT1, ACH1, and RHR2 were significantly overexpressed under at least one condition (Table 3). Although reported to transport xylose (12), the hexose transporter HXT2 was down-regulated under all conditions, and HXT4, HXT5, and STL1 were down-regulated under aerobic conditions (data not shown). The HXT16 gene, in contrast, was strongly up-regulated under aerobic conditions (Table 3). Generally, we observed lower aerobic expression of several genes encoding minor or putative isoenzymes involved in central carbon metabolism (TKL2, SOL4, YGR043C, and ALD3) and lower anaerobic expression of the peroxisomal genes ICL1 and IDP3, as well as the glycerol kinase gene GUT1 and the mitochondrial glycerol-3-phosphate dehydrogenase gene GUT2 in C1 (data not shown). This down-regulation of genes that are responsible for glycerol consumption matches the greater expression of the glycerol-producing genes GPD2 and RHR2 and the significantly increased glycerol production in the anaerobic C1 culture (Table 1), although glycerol 3-phosphatases have been reported previously to not be rate limiting for glycerol production (28). Furthermore, increased expression of the high-capacity low-affinity ammonium transporter MEP3 and concomitant decreased expression of the low-capacity high-affinity ammonium transporter MEP2 demonstrated that C1 had adapted to the nitrogen-excess, carbon-limited conditions in the evolution chemostat (38). Among the most consistently and strongly down-regulated genes were the mating type-specific genes MFA1 and STE2. Since decreased expression of mating type-specific genes is characteristic of diploid or polyploid strains (10), we confirmed the haploidy and unaltered mating type of the C1 strain by PCR (data not shown).
TABLE 3.
Metabolism-related genes with significantly increased levels of expression in C1 relative to the levels of expression in TMB3001 under at least one chemostat conditiona
| Fold change in gene expression in chemostat culture
|
Gene description | ||||
|---|---|---|---|---|---|
| Aerobic conditions
|
Anaerobic conditions with glucose-xylose | ||||
| Gene name | Gene no. | Xyloseb | Glucose-xylose | ||
| Significant in C1 under all conditions | |||||
| ADH5 | YBR145W | 3 | 2.6 | 2.1 | Alcohol dehydrogenase isoenzyme V |
| ASN1 | YPR145W | 5.1 | 4 | 3.8 | Asparagine synthetase |
| CDC21 | YOR074C | 3.3 | 2.6 | 2.2 | Thymidylate synthase |
| DFR1 | YOR236W | 3.5 | 2.3 | 2 | Dihydrofolate reductase |
| GAL1 | YBR020W | 288 | 388 | 140 | Galactokinase |
| GAL10 | YBR019C | 87 | 119 | 67 | UDP-glucose 4-epimerase |
| GAL2 | YLR081W | 285 | 211 | 339 | Galactose permease |
| GAL7 | YBR018C | 248 | 226 | 242 | Galactose-1-phosphate uridyl transferase |
| GPD2 | YOL059W | 2.3 | 2.8 | 2.5 | Glycerol-3-phosphate dehydrogenase |
| HIS3 | YOR202W | 2.5 | 2.4 | 2.4 | Imidazoleglycerol-phosphate dehydratase |
| ILV6 | YCL009C | 3.2 | 2.7 | 2.6 | Acetolactate synthase, small subunit |
| MAE1 | YKL029C | 3.8 | 5.5 | 2.4 | Mitochondrial malic enzyme |
| MCT1 | YOR221C | 3.6 | 3.4 | 2.5 | Malonyl-coenzyme A carrier protein transferase |
| MEP3 | YPR138C | 2.9 | 2 | 2.5 | Ammonia transport (high capacity, low affinity) |
| PUT4 | YOR348C | 6.6 | 5 | 3.4 | Proline permease |
| PYK2 | YOR347C | 12 | 9.1 | 3.1 | Pyruvate kinase, glucose-repressed isoform |
| SOL3 | YHR163W | 2.6 | 2.2 | 2.6 | Possible 6-phosphogluconolactonase |
| SUL2 | YLR092W | 2.2 | 2 | 2.1 | Sulfate permease |
| YEL041W | 4 | 3.5 | 26 | NAD kinase | |
| YER053C | 2.7 | 2.6 | 2.5 | Similar to Caenorhabditis elegans mitochondrial phosphate carrier | |
| Significant in C1 only under aerobic conditions | |||||
| ACH1 | YBL015W | 2.2 | 2.7 | 1.5 | Acetyl-coenzyme A hydrolase |
| ADH4 | YGL256W | 4.3 | 2.6 | 2.5 | Alcohol dehydrogenase isoenzyme IV |
| BNA2 | YJR078W | 2.7 | 3 | 1.5 | Tryptophan 2,3-dioxygenase |
| CAR2 | YLR438W | 3.1 | 2.6 | 1.3 | Ornithine transaminase |
| GDH3 | YAL062W | 2.2 | 2.5 | 0.9 | NADP-linked glutamate dehydrogenase |
| HEM1 | YDR232W | 2.1 | 2 | 1 | 5-Aminolevulinate synthase |
| HXT16 | YJR158W | 4.6 | 8 | 1.4 | Hexose permease |
| MSE1 | YOL033W | 2.1 | 2.2 | 1.7 | Mitochondrial glutamyl-tRNA synthetase |
| MSK1 | YNL073W | 3.7 | 3.1 | 1.7 | Mitochondrial lysyl-tRNA synthetase |
| NCP1 | YHR042W | 2.9 | 2.9 | 0.9 | NADPH-ferrihemoprotein reductase |
| PDC6 | YGR087C | 3 | 2.8 | 1.3 | Pyruvate decarboxylase |
| PGM2 | YMR105C | 4.8 | 3.9 | 1.6 | Phosphoglucomutase |
| PMT3 | YOR321W | 2.1 | 2.3 | 0.9 | Dolichyl-phosphate-mannose-protein mannosyltransferase |
| SDH1 | YKL148C | 2.6 | 2.4 | 0.6 | Succinate dehydrogenase cytochrome b |
| YAT1 | YAR035W | 4.9 | 3.7 | 1.8 | Carnitine o-acetyltransferase |
| ZWF1 | YNL241C | 3.3 | 2.7 | 1.6 | Glucose-6-phosphate dehydrogenase |
| Significant in C1 only under aerobic conditions on the glucose-xylose mixture | |||||
| COR1 | YBL045C | 1.7 | 2.4 | 1.3 | Ubiquinol-cytochrome c reductase core subunit 1 |
| CYT1 | YOR065W | 1.6 | 2.1 | 2 | Ubiquinol-cytochrome c reductase cytochrome c1 subunit |
| HEM13 | YDR044W | 1.6 | 2.6 | 1.1 | Coproporphyrinogen oxidase, aerobic |
| RHR2 | YIL053W | 1.8 | 2.1 | 1.7 | dl-Glycerol-3-phosphatase |
| THR1 | YHR025W | 1.4 | 2.2 | 1.3 | Homoserine kinase |
| Significant in C1 only under anaerobic conditions | |||||
| ARG5 | YER069W | 1.9 | 1.6 | 2.4 | N-Acetyl-γ-glutamyl-phosphate reductase and acetylglutamate kinase |
| ECM40 | YMR062C | 1.1 | 1.2 | 2.3 | Amino acid N-acetyltransferase |
| MET22 | YOL064C | 1.5 | 1.6 | 2.1 | 3′,5′ Bisphosphate nucleotidase |
Gene expression levels were considered significant when there was at least a twofold SAM-determined change at an expected median false-positive rate of 1%.
Compared to TMB3001 cultivated under aerobic conditions on xylose and glucose.
Upon detailed inspection of metabolic functions specified by differentially expressed genes, we noted a general pattern: the central carbon metabolism genes directly involved in cytosolic NADPH formation (ZWF1) or NADH consumption (ADH4, ADH5, and GPD2) were up-regulated and the genes involved in NADH formation (ALD3 and GUT2) were down-regulated. Consistently, increased aerobic expression of the respiratory genes COR1 and CYT1 indicated that there was increased NADH oxidation in C1. In addition, we observed higher levels of expression of the NADPH-producing mitochondrial malic enzyme, which has previously been hypothesized to constitute a transhydrogenation system that reoxidizes cytosolic NADH by reduction of mitochondrial NADP+ (2). Similarly, up-regulation of the NADPH-dependent glutamate dehydrogenase gene GDH3 indicates that that there is a transhydrogenation cycle catalyzed by the GDH1, GDH2, and GDH3 products (5), which may reoxidize NADH by reducing NADP+. The putative transhydrogenation mechanisms, however, would be irrelevant under fermentative conditions because of the very low tricarboxylic acid cycle flux (9, 23, 24).
Comparison of transcript levels and intracellular carbon fluxes.
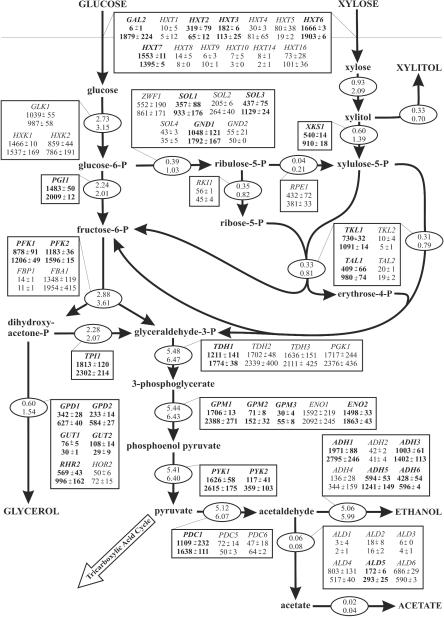
The large number of differentially expressed genes involved in central carbon and redox metabolism suggested that there are significant metabolic differences between TMB3001 and C1. Hence, we estimated the intracellular flux distribution from the anaerobic chemostat data for both strains grown on the glucose-xylose mixture (Table 1) by using a previously described stoichiometric model (47). Enhanced xylose catabolism in the C1 culture resulted in about 20% increased fluxes throughout the entire network, but the fluxes through the pentose phosphate pathway and into glycerol formation were more than doubled (Fig. 2). Notably, the flux of glucose 6-phosphate into the pentose phosphate pathway was strongly increased in C1, which is consistent with the notion that more cytosolic NADPH is required in the mutant to drive xylose reduction.
FIG. 2.
Comparison of molar carbon fluxes (ovals; values are expressed in millimoles per gram of biomass per hour) and transcript levels of the related enzymes (boxes; values are expressed in arbitrary units) in S. cerevisiae TMB3001 (upper values) and C1 (lower values) during anaerobic chemostat cultivation on 10 g of glucose per liter and 10 g of xylose per liter. The averages ± standard deviations from a DNA microarray analysis of two independent cultures are shown. Independent of the fold change, changes in gene expression were considered significant (indicated by boldface type) upon SAM analysis with an expected median false-positive rate of 1%. The standard deviations for all carbon fluxes were less than 10% of the averages from two independent cultures. Extracellular metabolites are indicated by uppercase letters. Fluxes from the following intracellular metabolites are required for biomass formation but are not shown explicitly: glucose 6-phosphate, ribose 5-phosphate, erythrose 4-phosphate, 3-phosphoglycerate, phosphoenolpyruvate, pyruvate, and acetate (24, 47).
Generally, increased carbon fluxes in the C1 strain were in excellent agreement with increased transcript levels of the corresponding genes from the same chemostat culture (Fig. 2). Albeit in many cases the differences were below the twofold significance level, it is remarkable that statistically significant and consistent increases in expression levels were discernible for most relevant central metabolic genes. The scale of these increases in expression was about the same as the scale of the flux increase; i.e., the increases involved SOL1, SOL3, GND1, XKS1, TKL1, and TAL1 in the pentose phosphate pathway, PFK1, PFK2, TDH1, GPM1, GPM2, GPM3, ENO2, PYK1, and PYK2 in the glycolysis pathway, and GPD1, GPD2, RHR2, PDC1, ADH1, ADH3, and ADH5 in the by-product pathways. One of the generally overexpressed ADH genes, ADH2, was clearly unchanged. This is fully consistent with the function of ADH2 in metabolizing ethanol (27, 41) that is not required for anaerobic ethanol formation. Among the ALD genes involved in acetate formation, in contrast, only ALD5 expression was increased significantly, as would be expected from the very low increase in flux through this pathway.
Since the recombinant xylose reductase and xylitol dehydrogenase genes in the TMB3001 genome (7) were not present on the YG-S98 array, we determined the in vitro enzymatic activities of all xylose catabolism enzymes in batch cultures of TMB3001 and C1 (Table 4). The C1 strain exhibited fivefold-higher activity of the ADH1 promoter-controlled xylose reductase. Concomitantly, the in vitro activities of the PGK1-controlled xylitol dehydrogenase and xylulokinase were more than twofold higher. This increased expression of the xylose genes was probably related to the high levels of expression of the ADH1 and PGK1 genes in the C1 mutant. Increased expression of xylose reductase has previously been reported to increase the rate of xylose uptake (17). The redox cofactor specificity of xylose reductase remained unaltered, with an approximately 100% higher activity with NADPH. While uncontrolled xylulokinase overexpression is detrimental to S. cerevisiae (18, 19), activity that is more than double the activity of TMB3001 does not have any apparent negative effect in C1, presumably because during evolution all three enzyme activities increased simultaneously.
TABLE 4.
Specific enzyme activities in the catabolic xylose pathways of S. cerevisiae TMB3001 and C1 during aerobic batch growth on glucose
| Strain | Enzyme activities (U mg of protein−1)
|
|||
|---|---|---|---|---|
| Xylose reductase
|
Xylitol dehydrogenase | Xylulokinase | ||
| NADPH | NADH | |||
| TMB3001 | 0.08 ± 0.00a | 0.04 ± 0.00 | 0.8 ± 0.0 | 0.5 ± 0.0 |
| C1 | 0.42 ± 0.03 | 0.24 ± 0.01 | 1.9 ± 0.0 | 1.1 ± 0.0 |
Average ± standard deviation for two independent experiments.
The large number of differentially expressed genes involved in central carbon and xylose metabolism and their consistent levels of expression suggest that there are a few regulatory mutations rather than many specific mutations in the C1 strain. Supporting this view, we found statistically significant increased expression of the positive regulators of glycolysis and ethanol formation, GCR1, REB1, RAP1, and PDC2 (Table 5). The extraordinary up-regulation of GAL3 appears to override the regulation exerted by the moderately increased expression of the galactose repressors GAL80 and GAL6 (Table 5) and thus is responsible for the strong induction of the GAL genes (Table 3). Increased expression of many components of the high-osmolarity glycerol regulatory pathway (51) was probably responsible, at least in part, for the up-regulation of genes involved in glycerol formation. Lastly, increased expression of the negative regulator of hexose transport, MTH1, may have been the common cause for the down-regulation of HXT2, HXT5, HXT4, and STL1.
TABLE 5.
Regulatory genes of sugar transport, osmotic stress response, and central carbon and galactose metabolism with significantly different levels of expression in C1 than in TMB3001 under at least one chemostat conditiona
| Fold change in gene expression in chemostat culture
|
Function | ||||
|---|---|---|---|---|---|
| Aerobic conditions
|
Anaerobic conditions with glucose-xylose | ||||
| Gene name | Gene no. | Xyloseb | Glucose-xylose | ||
| Sugar transport | |||||
| MTH1 | YDR277C | 2.1 | 2.6 | 1.0 | Negative regulator of HXT gene expression |
| Central carbon metabolism | |||||
| ADR1 | YDR216W | 1.0 | 0.8 | 0.1 | Positive regulator of ADH2 and peroxisomal genes |
| CYC8 | YBR112C | 2.2 | 2.0 | 1.5 | Mediates glucose repression |
| GCR1c | YPL075W | 1.9 | 1.6 | 1.6 | Positive regulator of glycolytic genes |
| HAP1 | YLR256W | 1.7 | 2.0 | 1.4 | Activator of CYC1 and CYP3 transcription |
| HAP5 | YOR358W | 2.5 | 2.8 | 2.1 | Regulates respiratory functions |
| PDC2c | YDR081C | 1.9 | 1.7 | 1.2 | Regulates transcription of PDC1 and PDC5 |
| RAP1 | YNL216W | 2.0 | 1.9 | 1.0 | Involved in glycolytic gene regulation |
| REB1 | YBR049C | 1.6 | 2.1 | 0.8 | Involved in glycolytic gene regulation |
| SNF1 | YDR477W | 2.0 | 1.8 | 1.0 | Required for release from glucose repression |
| TUP1 | YCR084C | 1.8 | 2.6 | 1.0 | Mediates glucose repression |
| Galactose metabolism | |||||
| GAL3 | YDR009W | 14.6 | 21.1 | 7.5 | Involved in galactose induction of GAL genes |
| GAL4 | YPL248C | 2.1 | 2.1 | 0.4 | Positive regulator of GAL genes |
| GAL6 | YNL239W | 2.7 | 2.6 | 2.7 | Negative regulator of the GAL genes |
| GAL80 | YML051W | 3.5 | 4.1 | 2.1 | Inhibits activation by Gal4p in the absence of galactose |
| High-osmolarity glycerol pathway (osmotic stress response) | |||||
| ASK10 | YGR097W | 2.2 | 1.7 | 2.5 | Activator of the SKN7 regulatory system |
| MSN1 | YOL116W | 2.6 | 2.3 | 1.3 | Expressed during hyperosmotic response |
| MSN4 | YKL062W | 2.6 | 2.4 | 2.1 | Key regulator of stress-responsive gene expression |
| PBS2 | YJL128C | 1.4 | 1.3 | 2.9 | Involved in osmolarity sensing |
| PTP2 | YOR208W | 2.9 | 2.0 | 2.1 | Involved in osmolarity sensing |
| RIM15 | YFL033C | 2.2 | 1.7 | 1.1 | Regulation of meiosis and response to stress |
| SKN7 | YHR206W | 1.8 | 2.0 | 0.8 | Expressed during oxidative and osmotic stress |
| SSK22 | YCR073C | 2.9 | 2.1 | 1.8 | Involved in osmolarity sensing |
Gene expression levels were considered significant when there was at least a twofold SAM-determined change at an expected median false-positive rate of 1%.
Compared to TMB3001 cultivated under aerobic conditions on xylose and glucose.
The expression change does not meet the twofold cutoff but was statistically significant with an expected median false-positive rate of 1%.
Increased catabolic rates improve anaerobic growth on xylose.
Directed evolution of the C1 strain apparently relieved the metabolic limitations for anaerobic growth on xylose (38), and the molecular differences in C1 described above suggest that the coordinated changes in carbon and redox metabolism are key components of this phenotype. However, these analyses did not reveal the pivotal limitation of yeast pentose metabolism for anaerobic growth, but the evolved strain provides a unique opportunity to address this question experimentally. Generally, the stoichiometry of anaerobic glucose metabolism was not radically different from that of anaerobic xylose metabolism, although less biomass was generated per gram of substrate consumed in the latter case (Table 6). The maximum specific rate of growth, in contrast, was dramatically lower on xylose (Table 6). This indicates that there is a kinetic problem, which may involve uptake and/or catabolism of xylose or an insufficient rate of ATP production to support growth, as hypothesized previously (30).
TABLE 6.
Physiological parameters of TMB3001 on glucose and of C1 on xylose in anaerobic batch cultures with a carbon source concentration of 10 g liter−1
| Strain | Substrate | Maximum growth rate (h−1)a | Specific substrate uptake rate (g g of biomass−1 h−1)b | Yields (g g−1) of:
|
||||
|---|---|---|---|---|---|---|---|---|
| Biomassa | Ethanolb | Xylitolb | Glycerola | Acetatea | ||||
| TMB3001 | Glucose | 0.373 | 4.51 | 0.083 | 0.397 | 0 | 0.085 | 0.007 |
| C1 | Xylose | 0.014 | 0.60 | 0.024 | 0.277 | 0.321 | 0.041 | 0.007 |
| C1 + acetoinc | Xylose | 0.019 | 0.62 | 0.031 | 0.373 | 0.180 | 0.028 | 0.011 |
The rates were determined over at least three generations of growth, and the average values for duplicate experiments are shown. The standard deviations were less than 10%.
The values are averages for duplicate experiments. The standard deviations were less than 5%.
Acetoin was added at a concentration of 0.5 g liter−1.
To differentiate between these possibilities, we grew C1 anaerobically on xylose in the presence of acetoin, which can be reduced to butanediol by consumption of NADH. This reaction reduces the extensive cytosolic formation of NADH in the xylitol dehydrogenase reaction (Fig. 1) and thus increases the rate of xylose catabolism during anaerobic xylose fermentation (48). Acetoin was quantitatively converted to butanediol in the C1 culture (data not shown), and less xylitol accumulated in the presence of acetoin, presumably as a consequence of NADH oxidation (Table 6). While the specific rate of xylose uptake remained virtually the same, the rate of xylose catabolism increased significantly because ethanol was produced instead of xylitol. Notably, cometabolism of acetoin increased the rate of anaerobic growth and the biomass yield on xylose by about one-third (Table 6), clearly demonstrating that xylose uptake is not the limiting factor for anaerobic growth of yeast on xylose alone.
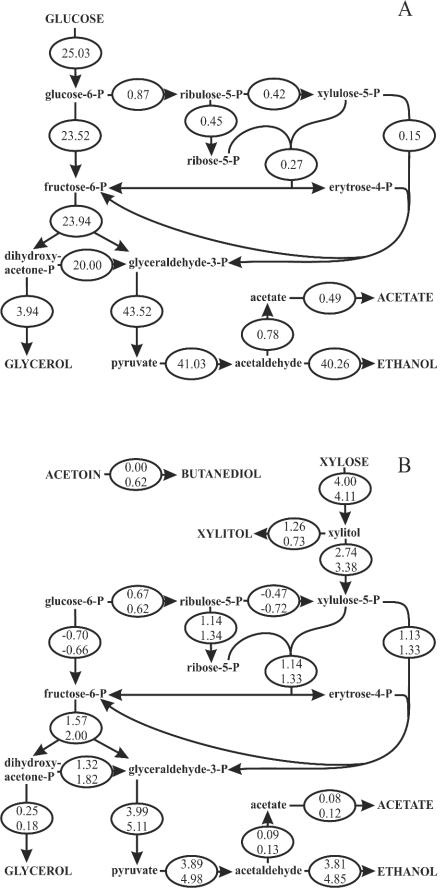
Metabolic flux analysis was then used to distinguish whether increased xylose catabolism per se or an increased rate of ATP formation caused the more rapid anaerobic growth of C1 on xylose plus acetoin. As expected (23, 24), less than 4% of the glucose consumed entered the pentose phosphate pathway in the anaerobic TMB3001 culture, primarily to supply pathway intermediates for biomass formation (Fig. 3A). In the C1 culture, in contrast, about 20% of the consumed xylose reentered the oxidative pentose phosphate pathway to supply NADPH for the xylose reductase reaction (Fig. 3B). From the estimated intracellular fluxes through the known ATP-consuming and -producing biochemical reactions catalyzed by xylulokinase, hexokinase, phosphofructokinase, phosphoglycerate kinase, and pyruvate kinase, we then calculated the specific ATP production rates of the three cultures. The ATP production rate calculated in this way was the maximum rate that was available for biomass formation. The rate of production on xylose for C1 (3.7 mmol of ATP g of biomass−1 h−1) was extraordinarily low compared to the rate of production for TMB3001 during anaerobic growth on glucose (38.7 mmol g of biomass−1 h−1). The latter rate resulted in a yield of about 10 g of biomass mol of ATP−1, which was very similar to the value determined previously for a glucose-limited anaerobic chemostat culture (11 g of biomass mol of ATP−1) (44). Finally, in the presence of acetoin, the production rate increased to 4.9 mmol g of biomass−1 h−1 during anaerobic growth on xylose, and this 32% increase correlates almost perfectly with the 35% increase in the growth rate in the presence of acetoin.
FIG. 3.
Molar carbon fluxes (expressed in millimoles per gram of biomass per hour) during anaerobic exponential batch growth of TMB3001 on 10 g of glucose per liter (A) and during anaerobic exponential batch growth of C1 on 10 g of xylose per liter (B, upper values) or 10 g of xylose per liter plus 0.5 g of acetoin per liter (B, lower values). The averages for duplicate experiments are shown, and the standard deviations were less than 10% in all cases.
DISCUSSION
Based on global gene expression and metabolic flux analysis, we identified two distinguishing characteristics that are seemingly necessary for anaerobic growth of S. cerevisiae on xylose alone. First, we showed that enhanced xylose consumption was accompanied by moderately increased carbon fluxes throughout the entire metabolic network of C1, which were most pronounced in the pentose phosphate and glycerol pathways. The calculated changes in flux between intracellular metabolites were in excellent agreement with the 1.5- to 2-fold-higher levels of expression of most central metabolic genes in C1 than in the parent strain TMB3001. Second, the pattern of differentially expressed metabolic genes suggested that increased cytosolic NADPH formation and increased cytosolic NADH consumption are important to drive xylose catabolism under anaerobic conditions. From these results we concluded that organisms, such as yeasts, that rely on two consecutive redox reactions for pentose catabolism are capable of anaerobic growth on this compound only when redox metabolism enables sufficiently high catabolic fluxes. Since we could not detect any mutations in the open reading frames and promoter regions of the most prominently up-regulated GAL2 and PYK2 genes and the ADH1 and PGK1 promoter-controlled xylose reductase, xylitol dehydrogenase, and xylulokinase genes (data not shown), it appears that for the most part altered activity of key regulators is responsible for this phenotype.
A strain that originated from a polyploid parent strain, the randomly generated S. cerevisae mutant TMB3400 with improved coutilization of xylose (49), was recently subjected to global expression analysis under conditions almost identical to those used here (46). Consistent with the data for C1, several pentose phosphate pathway and galactose metabolism genes were up-regulated. Since Gal2p can also transport xylose (12), the consistent observation that GAL2 was among the most strongly up-regulated genes in C1 and in TMB3400 strongly suggests that xylose uptake is, at least in part, mediated by Gal2p. The strong up-regulation of galactose metabolism-related genes was probably induced by the higher level of expression of the dominant positive regulator GAL3 (Table 5) (46), particularly since we were unable to detect any mutation in the upstream region of GAL2 in our mutants. In contrast to the expression in C1, the levels of expression of genes involved in glycolysis, redox metabolism, and ethanol or glycerol production were not altered in TMB3400, which may explain why this mutant cannot grow anaerobically on xylose alone (49).
Our conclusion that increased cytosolic NADPH formation and NADH consumption are important for anaerobic growth of yeast on xylose is consistent with flux and proteome data obtained for recombinant, xylose-fermenting S. cerevisiae during a shift from glucose-containing medium to glucose-xylose-containing medium (26, 31). In particular, it was shown that strains overexpressing the preferentially NADPH-dependent xylose reductase from P. stipitis generate the required NADPH primarily through the oxidative pentose phosphate pathway (16). This causes lower glycolytic fluxes, and the concomitantly reduced NADH reoxidation by reactions downstream of fructose 6-phosphate can be compensated for in respiring yeast but not in fermenting yeast (2); this explains the capacity of such recombinant strains to grow aerobically but not anaerobically on xylose alone (7, 14).
While directed evolution of C1 apparently relieved all metabolic bottlenecks for anaerobic growth on xylose (38), the low rate of growth (doubling time, about 50 h) shows that the biosynthetic components are barely generated at an appropriate rate. Adding the NADH-oxidizing compound acetoin to C1 cultures increased the anaerobic growth rate on xylose by about one-third, which demonstrates that the balance of redox cofactors was still growth limiting. Xylose uptake was clearly not limiting, since the rate of xylose uptake remained constant and xylose catabolism increased only because less of the side product xylitol was produced. Calculating the cofactor balances from a more detailed flux analysis then revealed that the specific cytosolic NADPH production rate remained constant but that the ATP production rate increased by the same factor as the growth rate in the acetoin cofeed culture. This result strongly suggests that the rate of ATP formation is the primary limiting factor for anaerobic growth on xylose alone, although we cannot exclude the possibility that the pentose phosphate pathway operates at its maximum level and thus cannot supply more NADPH.
This raises the general question, what rate of ATP formation is actually necessary to support anaerobic growth solely on xylose? While the specific ATP production rate 3.7 mmol of ATP g of biomass−1 h−1 was sufficient in C1, a rate of 1.8 mmol of ATP g of biomass−1 h−1 in TMB3001 when acetoin was added was apparently insufficient (48). Since the amount of ATP that is necessary to just sustain the viability of the existing cell mass, the so-called maintenance energy, is about 1.2 mmol of ATP g of biomass−1 h−1 in oxygen-limited P. stipitis cultures grown on xylose alone (30), it appears that at least 2 mmol of ATP g of biomass−1 h−1 must be generated from the catabolism of xylose to enable anaerobic growth. Within 460 generations under selective pressure for improved xylose metabolism (38), the C1 strain apparently evolved such that altered redox cofactor metabolism in a number of reactions and higher levels of expression of almost all catabolic genes permit sufficiently high catabolic fluxes of xylose to ethanol.
Acknowledgments
We thank Jörg Hauf for determining the mating type of C1.
This work was funded by the Swiss Bundesamt für Bildung und Wissenschaft within European Commission Framework V project BIO-HUG and by the Swedish Energy Administration.
REFERENCES
- 1.Andreasen, A. A., and T. J. B. Stier. 1953. Anaerobic nutrition of Saccharomyces cerevisiae. 1. Ergosterol requirement for growth in a defined medium. J. Cell. Comp. Physiol. 41:23-36. [DOI] [PubMed] [Google Scholar]
- 2.Bakker, B. M., K. M. Overkamp, A. J. van Maris, P. Kötter, M. A. Luttik, J. P. van Dijken, and J. T. Pronk. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:15-37. [DOI] [PubMed] [Google Scholar]
- 3.Becker, J., and E. Boles. 2003. A modified Saccharomyces cerevisiae strain that consumes l-arabinose and produces ethanol. Appl. Environ. Microbiol. 69:4144-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 5.Boles, E., W. Lehnert, and F. K. Zimmermann. 1993. The role of the NAD-dependent glutamate dehydrogenase in restoring growth on glucose of a Saccharomyces cerevisiae phosphoglucose isomerase mutant. Eur. J. Biochem. 217:469-477. [DOI] [PubMed] [Google Scholar]
- 6.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 7.Eliasson, A., C. Christensson, C. F. Wahlbom, and B. Hahn-Hägerdal. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66:3381-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiaux, J., Z. P. Çakar, M. Sonderegger, K. Wüthrich, T. Szyperski, and U. Sauer. 2003. Metabolic-flux profiling of the yeasts Saccharomyces cerevisiae and Pichia stipitis. Eukaryot. Cell 2:170-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gombert, A. K., M. M. dos Santos, B. Christensen, and J. Nielsen. 2001. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 183:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 11.Hahn-Hägerdal, B., C. F. Wahlbom, M. Gardonyi, W. H. van Zyl, R. R. Cordero Otero, and L. J. Jönsson. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73:53-84. [DOI] [PubMed] [Google Scholar]
- 12.Hamacher, T., J. Becker, M. Gardonyi, B. Hahn-Hägerdal, and E. Boles. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783-2788. [DOI] [PubMed] [Google Scholar]
- 13.Harhangi, H. R., A. S. Akhmanova, R. Emmens, C. Van Der Drift, W. T. De Laat, J. P. Van Dijken, M. S. Jetten, J. T. Pronk, and H. J. Op Den Camp. 2003. Xylose metabolism in the anaerobic fungus Piromyces sp. strain E2 follows the bacterial pathway. Arch. Microbiol. 180:134-141. [DOI] [PubMed] [Google Scholar]
- 14.Ho, N. W. Y., Z. Chen, and A. P. Brainard. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffries, T. W. 1983. Utilization of xylose by bacteria, yeasts, and fungi. Adv. Biochem. Eng. Biotechnol. 27:1-32. [DOI] [PubMed] [Google Scholar]
- 16.Jeppsson, M., B. Johansson, B. Hahn-Hägerdal, and M. F. Gorwa-Grauslund. 2002. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl. Environ. Microbiol. 68:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeppsson, M., K. Träff, B. Johansson, B. Hahn-Hägerdal, and M. F. Gorwa-Grauslund. 2003. Effect of enhanced xylose reductase activity on xylose consumption and product distribution in xylose-fermenting recombinant Saccharomyces cerevisiae. FEMS Yeast Res. 3:167-175. [DOI] [PubMed] [Google Scholar]
- 18.Jin, Y. S., H. Ni, J. M. Laplaza, and T. W. Jeffries. 2003. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate d-xylulokinase activity. Appl. Environ. Microbiol. 69:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson, B., C. Christensson, T. Hobley, and B. Hahn-Hägerdal. 2001. Xylulokinase overexpression in two strains of Saccharomyces cerevisiae also expressing xylose reductase and xylitol dehydrogenase and its effect on fermentation of xylose and lignocellulosic hydrolysate. Appl. Environ. Microbiol. 67:4249-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtzman, C. P. 1994. Molecular taxonomy of the yeasts. Yeast 10:1727-1740. [DOI] [PubMed] [Google Scholar]
- 21.Lonn, A., K. L. Traff-Bjerre, R. R. Codero Otero, W. H. van Zyl, and B. Hahn-Hägerdal. 2003. Xylose isomerase activity influences xylose fermentation with recombinant Saccharomyces cerevisiae strains expressing mutated xylA from Thermus thermophylus. Enzyme Microb. Technol. 32:567-573. [Google Scholar]
- 22.Lowe, S. E., M. K. Theodorou, and A. P. Trinci. 1987. Growth and fermentation of an anaerobic rumen fungus on various carbon sources and effect of temperature on development. Appl. Environ. Microbiol. 53:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maaheimo, H., J. Fiaux, Z. P. Çakar, J. E. Bailey, U. Sauer, and T. Szyperski. 2001. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional 13C labeling of common amino acids. Eur. J. Biochem. 268:2464-2479. [DOI] [PubMed] [Google Scholar]
- 24.Nissen, T., U. Schulze, J. Nielsen, and J. Villadsen. 1997. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology 143:203-218. [DOI] [PubMed] [Google Scholar]
- 25.Piper, M. D., P. Daran-Lapujade, C. Bro, B. Regenberg, S. Knudsen, J. Nielsen, and J. T. Pronk. 2002. Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 277:37001-37008. [DOI] [PubMed] [Google Scholar]
- 26.Pitkänen, J.-P., A. Aristidou, L. Salusjärvi, L. Ruohonen, and M. Penttilä. 2003. Metabolic flux analysis of xylose metabolism in recombinant Saccharomyces cerevisiae using continuous culture. Metab. Eng. 5:16-31. [DOI] [PubMed] [Google Scholar]
- 27.Plapp, B. V., A. J. Ganzhorn, R. M. Gould, D. W. Green, and A. D. Hershey. 1987. Structure and function in yeast alcohol dehydrogenases. Prog. Clin. Biol. Res. 232:227-236. [PubMed] [Google Scholar]
- 28.Remize, F., L. Barnavon, and S. Dequin. 2001. Glycerol export and glycerol-3-phosphate dehydrogenase, but not glycerol phosphatase, are rate limiting for glycerol production in Saccharomyces cerevisiae. Metab. Eng. 3:301-312. [DOI] [PubMed] [Google Scholar]
- 29.Richard, P., R. Verho, M. Putkonen, J. Londesborough, and M. Penttilä. 2003. Production of ethanol from l-arabinose by Saccharomyces cerevisiae containing a fungal l-arabinose pathway. FEMS Yeast Res. 3:185-189. [DOI] [PubMed] [Google Scholar]
- 30.Rizzi, M., C. Klein, C. Schultze, N. A. Bui-Thanah, and H. Dellweg. 1989. Xylose fermentation by yeasts. 5. Use of ATP balances for modeling oxygen-limited growth and fermentation of yeast Pichia stipitis with xylose as carbon source. Biotechnol. Bioeng. 34:509-514. [DOI] [PubMed] [Google Scholar]
- 31.Salusjärvi, L., M. Poutanen, J. P. Pitkänen, H. Koivistoinen, A. Aristidou, N. Kalkkinen, L. Ruohonen, and M. Penttilä. 2003. Proteome analysis of recombinant xylose-fermenting Saccharomyces cerevisiae. Yeast 20:295-314. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sarthy, A. V., B. L. McConaughy, Z. Lobo, J. A. Sundstrom, C. E. Furlong, and B. D. Hall. 1987. Expression of the Escherichia coli xylose isomerase gene in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 53:1996-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauer, U. 2001. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 73:129-170. [DOI] [PubMed] [Google Scholar]
- 35.Sauer, U., V. Hatzimanikatis, H.-P. Hohmann, M. Manneberg, A. P. G. M. van Loon, and J. E. Bailey. 1996. Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Appl. Environ. Microbiol. 62:3687-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, N. Q., and T. W. Jeffries. 1998. Anaerobic growth and improved fermentation of Pichia stipitis bearing a URA1 gene from Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 50:339-345. [DOI] [PubMed] [Google Scholar]
- 38.Sonderegger, M., and U. Sauer. 2003. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl. Environ. Microbiol. 69:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trinci, A. P. J., D. R. Davies, K. Gull, M. I. Lawrence, B. B. Nielsen, A. Rickers, and M. K. Theodorou. 1994. Anaerobic fungi in herbivorous animals. Mycol. Res. 98:129-152. [Google Scholar]
- 40.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallari, R. C., W. J. Cook, D. C. Audino, M. J. Morgan, D. E. Jensen, A. P. Laudano, and C. L. Denis. 1992. Glucose repression of the yeast ADH2 gene occurs through multiple mechanisms, including control of the protein synthesis of its transcriptional activator, ADR1. Mol. Cell. Biol. 12:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varma, A., and B. O. Palsson. 1994. Metabolic flux balancing: basic concepts, scientific, and practical use. Bio/Technology 12:994-998. [Google Scholar]
- 43.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 44.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1990. Energetics of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136:405-412. [DOI] [PubMed] [Google Scholar]
- 45.Visser, W., W. A. Scheffers, W. H. Batenburg-van der Vegte, and J. P. van Dijken. 1990. Oxygen requirements of yeasts. Appl. Environ. Microbiol. 56:3785-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahlbom, C. F., R. R. Cordero Otero, W. H. van Zyl, B. Hahn-Hägerdal, and L. J. Jonsson. 2003. Molecular analysis of a Saccharomyces cerevisiae mutant with improved ability to utilize xylose shows enhanced expression of proteins involved in transport, initial xylose metabolism, and the pentose phosphate pathway. Appl. Environ. Microbiol. 69:740-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahlbom, C. F., A. Eliasson, and B. Hahn-Hägerdal. 2001. Intracellular fluxes in a recombinant xylose-utilizing Saccharomyces cerevisiae cultivated anaerobically at different dilution rates and feed concentrations. Biotechnol. Bioeng. 72:289-296. [DOI] [PubMed] [Google Scholar]
- 48.Wahlbom, C. F., and B. Hahn-Hägerdal. 2002. Furfural, 5-hydroxymethyl furfural and acetoin act as external electron acceptors during anaerobic fermentation of xylose by recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 78:172-178. [DOI] [PubMed] [Google Scholar]
- 49.Wahlbom, C. F., W. H. van Zyl, L. J. Jonsson, B. Hahn-Hägerdal, and R. R. Otero. 2003. Generation of the improved recombinant xylose-utilizing Saccharomyces cerevisiae TMB 3400 by random mutagenesis and physiological comparison with Pichia stipitis CBS 6054. FEMS Yeast Res. 3:319-326. [DOI] [PubMed] [Google Scholar]
- 50.Walfridsson, M., X. Bao, M. Anderlund, G. Lilius, L. Bülow, and B. Hahn-Hägerdal. 1996. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl. Environ. Microbiol. 62:4648-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wojda, I., R. Alonso-Monge, J. P. Bebelman, W. H. Mager, and M. Siderius. 2003. Response to high osmotic conditions and elevated temperature in Saccharomyces cerevisiae is controlled by intracellular glycerol and involves coordinate activity of MAP kinase pathways. Microbiology 149:1193-1204. [DOI] [PubMed] [Google Scholar]