Abstract
Prolonged cultivation (>25 generations) of Saccharomyces cerevisiae in aerobic, maltose-limited chemostat cultures led to profound physiological changes. Maltose hypersensitivity was observed when cells from prolonged cultivations were suddenly exposed to excess maltose. This substrate hypersensitivity was evident from massive cell lysis and loss of viability. During prolonged cultivation at a fixed specific growth rate, the affinity for the growth-limiting nutrient (i.e., maltose) increased, as evident from a decreasing residual maltose concentration. Furthermore, the capacity of maltose-dependent proton uptake increased up to 2.5-fold during prolonged cultivation. Genome-wide transcriptome analysis showed that the increased maltose transport capacity was not primarily due to increased transcript levels of maltose-permease genes upon prolonged cultivation. We propose that selection for improved substrate affinity (ratio of maximum substrate consumption rate and substrate saturation constant) in maltose-limited cultures leads to selection for cells with an increased capacity for maltose uptake. At the same time, the accumulative nature of maltose-proton symport in S. cerevisiae leads to unrestricted uptake when maltose-adapted cells are exposed to a substrate excess. These changes were retained after isolation of individual cell lines from the chemostat cultures and nonselective cultivation, indicating that mutations were involved. The observed trade-off between substrate affinity and substrate tolerance may be relevant for metabolic engineering and strain selection for utilization of substrates that are taken up by proton symport.
Maltose, a disaccharide consisting of two glucose molecules linked via an 1,4-α bond, is the main carbon source for Saccharomyces cerevisiae during beer fermentation and leavening of bread dough (4, 13, 21, 48). In addition to being of significance for applied use, the maltose regulon in S. cerevisiae serves as a paradigm for metabolic regulation in this model eukaryote (19, 24, 38, 39).
The metabolism of maltose differs from that of glucose only in the first two steps, namely, its transport and subsequent hydrolysis into glucose (25). Maltose is taken up via a maltose-one-proton symport mechanism (56). Extrusion of the symported proton via the plasma membrane ATPase costs one ATP molecule per proton (61, 65). As a result, the net ATP yield from the alcoholic fermentation of maltose is lower than that during glucose fermentation (1.5 instead of 2 ATP molecules per glucose unit). In S. cerevisiae, the intracellular hydrolysis of maltose into two glucose molecules is catalyzed by maltase (α-glucosidase [EC 3.2.1.20]).
The genes encoding the maltose permease and the maltase are located on five highly homologous loci (MAL1, MAL2, MAL3, MAL4, and MAL6) (3, 11). The number and identity of MAL loci is strain dependent (37). Every MAL locus consists of three genes. The first gene (MALx1) encodes the maltose-proton symporter (12). Maltase is encoded by the MALx2 gene (14, 23). The third MAL gene (MALx3) encodes a DNA-binding, maltose-dependent transcriptional activator that specifically acts at the MALx1 and MALx2 genes (10, 22).
Maltose metabolism in S. cerevisiae is strongly negatively regulated by glucose, both at the transcriptional level and at the enzyme activity level (5, 26, 27, 33-35, 49, 69). Despite this multilayered regulation of maltose metabolism, several reports indicate that S. cerevisiae cells can have difficulties in coping with sudden changes in the extracellular maltose concentration. Exposure of aerobic, maltose-limited chemostat cultures to excess maltose has even been reported to result in maltose-accelerated death (47). This loss of viability, which was accompanied by the release of glucose in the medium, was interpreted to be a result of nonrestricted maltose uptake and hydrolysis, with the resulting accumulation of glucose and protons in the cells leading to cell death and lysis (47). Release of glucose upon exposure to excess maltose was also observed in S. cerevisiae mutants that were defective in glucose catabolite repression (15, 16).
During prolonged cultivation in nutrient-limited chemostats, natural genetic selection often leads to the accumulation of mutants with an improved affinity for the growth-limiting nutrient (6, 64). As a rule of thumb, it has been proposed that chemostat cultivations that last for fewer than 20 generations allow the detailed investigation of physiological responses to defined, specific conditions. Conversely, the genetic adaptation that occurs in more prolonged chemostat runs can provide insight in evolutionary processes (17, 29). On one hand, the selection pressure in chemostat cultures may be used to enhance the selection of desirable genetic and physiological properties. On the other hand, prolonged cultivation (also batch cultivation) may have detrimental effects, such as the gradual loss of productivity (53). This is relevant for the fermentation industry, in which prolonged cultivation (e.g., repeated batch cultivation) is often applied.
In a recent study (25) we analyzed glucose efflux upon exposure of S. cerevisiae to excess maltose, with yeast cells originating from “young” chemostat cultures (<20 generations). In these experiments no cell lysis was observed upon exposure to excess maltose. However, in further work on this subject, we observed an apparent effect of chemostat culture age on transport capacity. The aim of the present study was to further investigate the effect of prolonged maltose-limited chemostat cultivation on the physiology of S. cerevisiae. To this end we monitored the affinity for maltose, genome-wide transcript levels, activities of key enzymes, and physiological responses to maltose excess during long-term cultivation in maltose-limited chemostat cultures.
MATERIALS AND METHODS
Strains and maintenance.
The haploid, prototrophic S. cerevisiae strain CEN.PK113-7D (MATa MAL2-8c SUC2) was obtained from P. Kötter, Frankfurt, Germany. Precultures were grown to stationary phase in shake flask cultures on synthetic medium (63) adjusted to pH 6.0 and containing 2% (wt/vol) glucose. After addition of sterile glycerol (30%, vol/vol), 2-ml aliquots were stored in sterile vials at −80°C. These frozen stock cultures were used to inoculate precultures for chemostat cultivation.
Media.
Synthetic medium containing mineral salts and vitamins was prepared and sterilized as described previously (63). For chemostat cultivation, the maltose concentration in reservoir media was 7.5 g · liter−1 (0.25 mol of C · liter−1).
Chemostat cultivation.
Aerobic chemostat cultivation was performed at a dilution rate of 0.10 h−1 at 30°C in 2.0-liter laboratory fermentors (Applikon, Schiedam, The Netherlands) with stirring at 800 rpm. The working volume of the cultures was kept at 1.0 liter by a peristaltic effluent pump coupled to an electrical level sensor. This setup ensured that under all growth conditions, biomass concentrations in samples taken directly from the culture differed by <1% from biomass concentrations in samples taken from the effluent line (41). The exact working volume was measured after each experiment. The pH was kept at 5.0 ± 0.1 by an ADI 1030 biocontroller via the automatic addition of 2 mol of KOH · liter−1. The fermentor was flushed with air at a flow rate of 0.5 liter · min−1 by using a Brooks 5876 mass flow controller. The dissolved oxygen concentration was continuously monitored with an oxygen electrode (model 34 100 3002; Ingold) and remained above 60% of air saturation. A steady state was defined as the situation in which at least five volume changes had passed after the last change in growth conditions and in which the biomass concentration and the specific rates of carbon dioxide production and oxygen consumption had remained constant (<2% variation) over two volume exchanges. In case of spontaneous oscillatory behavior, a pulse of ethanol (50 mM) was added to the culture to dampen out the oscillations. Chemostat cultures were routinely checked for purity by phase-contrast microscopy.
Exhaust gas analysis.
The exhaust gas was cooled in a condenser (2°C) and dried with a Perma Pure dryer (type PD-625-12P). O2 and CO2 concentrations were determined with a Rosemount NGA2000 analyzer. Determination of the exhaust gas flow rate and calculations of specific rates of CO2 production and O2 consumption were performed as described previously (62, 66).
Determination of culture dry weight.
Culture samples (10 ml) were filtered over preweighed nitrocellulose filters (pore size, 0.45 μm; Gelman Sciences). After removal of medium, the filters were washed with demineralized water, dried in a Whirlpool Easytronic M591 microwave oven for 20 min at a 360-W output, and weighed. Duplicate determinations varied by <1%.
Extracellular metabolite analysis.
Glucose, maltose, ethanol, glycerol, acetate, and pyruvate present in the supernatants of chemostat cultures were determined by high-pressure liquid chromatography (HPLC) analysis with an HPX-87H Aminex ion-exchange column (300 by 7.8 mm; Bio-Rad) at 60°C. The column was eluted with 5 mM sulfuric acid at a flow rate of 0.6 ml · min−1. Pyruvate and acetate were detected with a Waters 441 UV meter, coupled to a Waters 741 data module, at 214 nm. Glucose, maltose, ethanol, and glycerol were detected with an ERMA type ERC-7515A refractive-index detector coupled to a Hewlett-Packard type 3390A integrator. Maltose concentrations in reservoir media were also analyzed by HPLC.
Off-line anaerobic fermentation assays.
Samples containing exactly 200 mg (dry weight) of biomass were harvested from a steady-state chemostat culture by centrifugation (5,000 × g, 3 min) and resuspended in 10 ml of fivefold-concentrated synthetic medium (pH 5.6). Subsequently, these cell suspensions were introduced into a thermostatted (30°C) vessel. The volume was adjusted to 40 ml with demineralized water. After 10 min of incubation, 10 ml of a maltose solution (20 g · liter−1 [final concentration]) was added, and two 1-ml samples were taken at appropriate time intervals for 2 h. The 10-ml headspace was continuously flushed with water-saturated carbon dioxide at a flow rate of approximately 30 ml · min−1. Sugar concentrations and metabolite levels in the supernatants were determined by HPLC analysis. At the end of the experiments (after 2 h), some growth had taken place (data not shown). Consequently, carbon recoveries were only ca. 90% when growth of biomass was not taken into account.
Preparation of cell extracts.
For preparation of cell extracts, culture samples were harvested by centrifugation, washed twice with 10 mM potassium phosphate buffer (pH 7.5) containing 2 mM EDTA, concentrated fourfold, and stored at −20°C. Before being assayed, the samples were thawed, washed, and resuspended in 100 mM potassium phosphate buffer (pH 7.5) containing 2 mM MgCl2 and 1 mM dithiothreitol. Extracts were prepared by sonication with 0.7-mm-diameter glass beads at 0°C in a MSE Soniprep 150 sonicator (150-W output, 8-μm peak-to-peak amplitude) for 4 min at 0.5-min intervals. Unbroken cells were removed by centrifugation (20 min at 36,000 × g) at 4°C. The supernatant was used as the cell extract.
Maltase activity assays.
Maltase (EC 3.2.1.20) activity was measured with a discontinuous assay. The reaction mixture contained a McIlvain buffer (a combination of 100 mM citric acid and 200 mM Na2HPO4) at pH 6.6 and 120 mM maltose (43). The reaction was carried out in a total volume of 2 ml at 30°C and was started by the addition of cell extract. At different time intervals, a sample (100 μl) was withdrawn from the reaction mixture and the reaction was stopped by the addition of 10 μl of 1 M Tris. The samples were analyzed enzymatically for glucose by using the UV method (Boehringer kit no. 716251). Activities were expressed as micromoles of maltose converted per minute and per milligram of protein (units · milligram of protein−1).
Protein determination.
Protein concentrations in the supernatants of anaerobic fermentation assay mixtures and in cell extracts were determined by the Lowry method (32). Dried bovine serum albumin (fatty-acid free; obtained from Sigma, Zwijndrecht, The Netherlands) was used as a standard.
Fluorescent staining for yeast viability.
A commercial live-dead yeast viability kit (L-7009; Molecular Probes, Leiden, The Netherlands) was used to estimate the fraction of dead cells in samples from anaerobic fermentation assays (36). FUN-1 and Calcofluor White M2R cell stain were added to yeast cell suspensions (106 to 107 cells · ml−1) at final concentrations of 5 to 20 μM and 25 μM, respectively. After staining, the suspensions were mixed thoroughly and incubated in the dark at 30°C for 30 min. Five microliters of the stained yeast suspension was trapped between a coverslip and an object slide and analyzed with a fluorescence microscope (Axioplan 2 Imaging; Zeiss, Weesp, The Netherlands) with appropriate filter sets (fluorescein isothiocyanate [Zeiss 09 450-490 FT510 LP515] and DAPI [4′,6′-diamidino-2-phenylindole] [Zeiss 02 G365 FT395 LP420]).
Determination of viable counts.
Viable counts were determined on 2% (wt/vol) YPD agar plates. This complex medium contained, per liter, 10 g of yeast extract (Difco, Detroit, Mich.), 20 g of peptone from casein (Merck, Darmstadt, Germany), 20 g of d-glucose, and 20 g of agar (Difco). Viable counts of S. cerevisiae strain CEN.PK.113-7D were obtained by counting the number of colonies on YPD agar prepared after proper dilution of the culture (50 to 400 colonies per plate). The colonies were counted after 48 h of incubation at 30°C. In all experiments, the average number on three plates was used to calculate the viable counts.
Proton transport measurements.
As the ratio of maltose-proton symport is 1:1 (56, 61), it was possible to calculate the maltose transport capacity, by measuring the rapid alkalinization of the medium (56). After harvesting of cells from steady-state cultures by centrifugation, the cells were resuspended in 1 ml of 1.25 mM phthalate buffer (pH 5.0) and transferred to a small magnetically stirred and thermostatted (30°C) vessel. This suspension was adjusted to a total volume of 4 ml with the same phthalate buffer. The final concentration of biomass was 3.5 to 4.5 g (dry weight) · liter−1. At time zero, a maltose pulse (100 μl, 1 M) was added. The rapid alkalinization of the extracellular environment was monitored with an Applikon XD5533501C sensitive pH electrode which was connected to a Philips PW 9421 pH meter. The signal was recorded with a Kipp BD40 recorder. The pH meter was used in the millivolt mode. To calculate initial proton uptake rates from the recordings, the system was calibrated with defined pulses of NaOH (20 and 40 μl, 10 mM NaOH). Maltose transport capacity was calculated from the initial slopes of the curves.
Measurements of residual maltose in continuous cultures.
Five-milliliter samples of cells were rapidly (within 3 s) transferred from the chemostat culture into a syringe containing 62.0 g of cold steel beads (diameter of 4 mm and temperature of −20°C). After withdrawal from the fermentor, the sample was directly filtered (0.25-μm-pore-size filter). The supernatant was analyzed for maltose by HPLC with a HPX-87K column (300 by 7.8 mm; Bio-Rad) at 85°C. The column was eluted with demineralized water at a flow rate of 0.5 ml · min−1. Maltose concentrations were determined with a Dionex ED50 detector in IPAD mode.
Total RNA isolation.
Cells were rapidly (within 3 s) transferred from the chemostat culture into liquid nitrogen to immediately quench the metabolism. The frozen cell suspension (about 40 g of cell broth) was thawed gently on ice. After complete thawing, the cell suspension was centrifuged at 0°C for 5 min. Total RNA extraction from the pellets was performed by the hot-phenol method (54).
Probe preparation and hybridization to arrays.
Further handling of RNA samples for genome-wide transcriptome analysis with the Affymetrix GeneChip was performed as described previously (44). Acquisition and quantification of array images as well as primary data analysis were performed with the Affymetrix Microarray Suite, version 4.01. Before comparison, all arrays were globally scaled to a target value of 1,000 by using the average signal of all gene features. From the 9,335 transcript features on the YG-S98 arrays, a filter was applied to extract 6,383 yeast open reading frames. Statistical analysis of genome-wide data sets was performed with Microsoft Excel running the Significance Analysis of Microarrays (version 1.12) add-in (59). Statistical analysis of small subsets of maltose-related transcripts was done with a regular Student t test (P < 0.05).
Restart of chemostat cultivation.
Stored glycerol stocks (−80°C) of a prolonged maltose-limited chemostat cultivation culture (containing 30% [vol/vol] sterile glycerol) were streaked out once for purity on a synthetic medium plate containing 0.8% (wt/vol) glucose. Single colonies were used for inoculation of a shake flask containing 0.8% (wt/vol) glucose, which, after growth to stationary phase, was used as the inoculum for the chemostat culture.
RESULTS
Physiology of S. cerevisiae in prolonged maltose-limited chemostat cultivations.
Prolonged (>25 generations) chemostat cultivation of S. cerevisiae CEN.PK113-7D under maltose limitation at a dilution rate of 0.10 h−1 revealed no detectable changes in cell yield, exhaust gas rates, or, consequently, respiration quotient. The morphology of the cells also remained constant during the cultivation time as monitored microscopically (data not shown). This was in contrast to results of selection experiments under glucose limitation, in which cells become smaller and/or elongated, leading to an increased surface-to-volume ratio (2, 7).
Prolonged cultivation under maltose limitation leads to an increased substrate affinity.
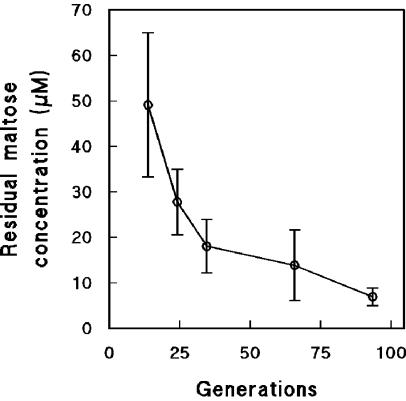
The affinity of microorganisms for a growth-limiting nutrient is determined by the ratio of their maximum substrate consumption rate and their substrate saturation constant (μmax · Ks−1). Nutrient-limited cultivation of microorganisms in chemostat cultures leads to a strong selective pressure for cells with an improved affinity for the growth-limiting nutrient (67). A typical consequence of selection for cells with an improved substrate affinity for the growth-limiting nutrient is a decrease of its residual concentration in the chemostat (6, 17). Indeed, during prolonged cultivation of S. cerevisiae in maltose-limited chemostat cultures, the residual maltose concentrations gradually decreased from 50 μM at 10 generations to 10 μM after 90 generations of chemostat cultivation (Fig. 1).
FIG. 1.
Residual maltose concentrations in aerobic, maltose-limited chemostat cultures of S. cerevisiae CEN.PK113-7D as a function of culture age. Data are represented as averages ± mean deviations from two experiments with cells from independent chemostat cultures.
Maltose transport capacity increases during prolonged cultivation.
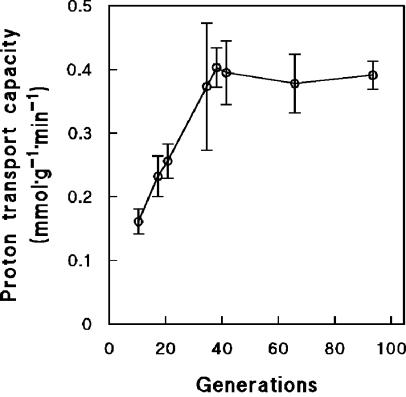
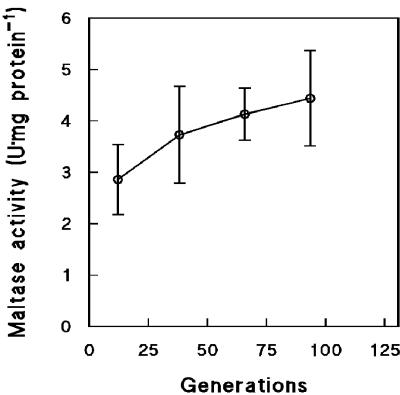
To investigate a possible role of maltose transport in the increasing affinity for maltose during long-term maltose-limited cultivation, we analyzed the capacity of the maltose-proton symporter. Transport of maltose was measured indirectly by measuring the uptake of protons, based on the known 1:1 stoichiometry of maltose/H+ symport in yeast (61, 65). After maltose-limited cultivation for 35 generations, the maltose transport capacity of the cells had increased by 2.5-fold (Fig. 2). This correlated well with a similar decrease of the residual maltose concentration over the same time interval (Fig. 1). Prolongation of the chemostat runs beyond 40 generations did not result in a higher maltose transport capacity (Fig. 2), even though the residual maltose concentration continued to decrease (Fig. 1). Maltase activity was also monitored during prolonged chemostat cultivation under maltose limitation. Maltase activities increased only slightly, from 2.9 ± 0.7 U · mg of protein−1 after 10 generations to 4.4 ± 0.9 U · mg of protein−1 after 90 generations of cultivation (Fig. 3).
FIG. 2.
Maltose transport capacities of cells from aerobic, maltose-limited chemostat cultures of S. cerevisiae CEN.PK113-7D as a function of culture age. Maltose transport capacity was measured indirectly by monitoring off-line uptake of protons after a pulse of maltose (25 mM). Since the ratio of the maltose-H+ symport system is 1:1 (55, 59), proton transport equals maltose transport capacity. Data are represented as averages ± mean deviations from three experiments with cells from independent chemostat cultures.
FIG. 3.
Maltase activities of cells harvested from aerobic, maltose-limited chemostat cultures of S. cerevisiae CEN.PK113-7D as a function of culture age. Activities were measured by a discontinuous assay measuring the formation of glucose over time. Data are represented as averages ± mean deviations from two experiments with cells from independent chemostat cultures.
Prolonged chemostat cultivation leads to maltose hypersensitivity.
It has been previously proposed that a high capacity for maltose-proton symport may lead to maltose-accelerated death when cells are exposed to excess maltose (47). We therefore investigated the effect of prolonged cultivation in maltose-limited cultures on physiological responses after exposure to excess maltose.
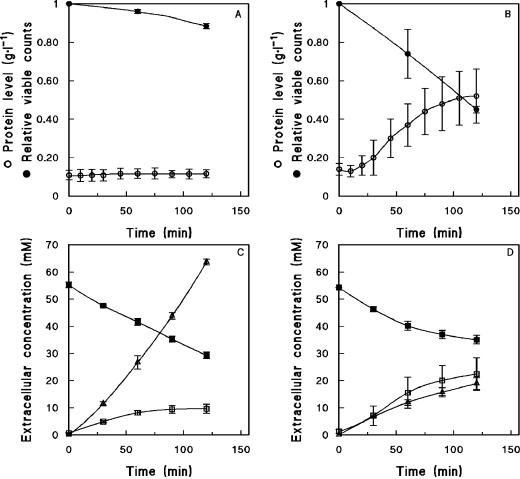
When samples from young (<20 generations) maltose-limited chemostat cultures of S. cerevisiae CEN.PK113-7D were exposed to excess maltose, an efflux of glucose was observed, but cell viability was hardly affected (Fig. 4A and C). We recently investigated this glucose efflux phenomenon and demonstrated that it is caused by efflux of glucose via one or more HXT hexose transporters, rather than by cell lysis or extracellular maltase activity (25). When samples from older (>25 generations) maltose-limited chemostat cultures were exposed to excess maltose, a significant decrease of cell viability was observed. Plating on complex glucose media revealed a dramatic decrease in the number of CFU after exposure of culture samples to 20 g of maltose · liter−1 maltose. Viable counts were reduced by 55% ± 2% after 35 generations (Fig. 4B) and even by 62% after 60 generations of cultivation (data not shown). This massive loss of cell viability was confirmed with a fluorescent live-dead staining technique (data not shown).
FIG. 4.
Extracellular metabolite concentrations and cell integrity (viability) during off-line anaerobic maltose fermentation assays with cells harvested from aerobic, maltose-limited chemostat cultures of S. cerevisiae CEN.PK113-7D. (A and C) Cells sampled after 10 generations of chemostat cultivation. (B and D) Cells sampled after 35 generations of chemostat cultivation. Symbols: ○, extracellular protein concentrations; •, fraction of viable cells as determined from plate counts; □, ▪, and ▵, concentrations of the extracellular metabolites glucose, maltose, and ethanol, respectively. Relative viable counts are represented as averages ± mean deviations from two experiments with cells from independent chemostat cultures. Extracellular protein and metabolite concentrations are represented as averages ± mean deviations from three experiments with cells from independent chemostat cultures.
As another indicator of cell lysis, extracellular protein concentrations were analyzed after exposure of culture samples to excess maltose. In cells from young cultures, the extracellular protein concentration did not change after exposure to maltose (Fig. 4A). However, in cells from a 35-generation-old culture, the extracellular protein concentration increased by ca. fivefold during exposure to excess maltose (Fig. 4B).
Further confirmation of cell lysis was found in the changes in metabolite production and maltose consumption during exposure to excess maltose. A 10-generation-old culture consumed 26.0 ± 1.2 mM maltose during a 2-h pulse experiment, while a 35-generation-old culture consumed only 19.2 ± 1.5 mM maltose. Ethanol production decreased even more drastically, from 63.6 ± 1.6 to 18.9 ± 2.1 mM after 10 and 35 generations of cultivation, respectively. The release of glucose was twofold higher in cells from the long-term culture (Fig. 4C and D).
Transcriptome analysis of long-term maltose-limited cultures.
To check whether the observed physiological adaptations to long-term maltose-limited cultivation were caused by transcriptional (up-)regulation of MAL genes, a genome-wide transcript analysis was performed on young (10 generations) and old (45 generations) maltose-limited chemostat cultures. Several of the structural genes encoding maltose permeases (MALx1) and maltases (MALx2) exhibited significantly increased transcript levels in the old cultures (Table 1). The pooled transcript levels of these genes also were significantly higher (Table 1). In the interpretation of these data, it should be taken into account that the complement of MALx1 and MALx2 genes in the CEN.PK113-7D strain is likely to differ from that on the DNA microarrays. Moreover, the signal for some of these transcripts in the old cultures may have been underestimated due to saturation. A further transcriptome-wide and nonbiased analysis using the Significance Analysis of Microarrays algorithm (minimum fold change of 2 and false discovery rate of 2.5%) did not yield additional transcripts that were significantly up- or down-regulated during prolonged cultivation.
TABLE 1.
Transcript levels of young (10 generations) and old (45 generations) aerobic, maltose-limited chemostat cultures of S. cerevisiae CEN.PK113-7D
| Enzyme | Locus
|
Transcript levela
|
Fold change (old vs young culture) | P valueb | ||
|---|---|---|---|---|---|---|
| Systematic name | Standard name | Young culture | Old culture | |||
| Maltose permease | YGR289C | MAL11 | 20,015 ± 2,127 | 24,930 ± 248 | 1.2 | 0.19 |
| YBR298C | MAL31 | 12,721 ± 813 | 15,587 ± 1,056 | 1.4 | 0.04 | |
| YDL247W | 258 ± 152 | 430 ± 455 | 1.7 | 0.69 | ||
| YJR160C | 229 ± 85 | 363 ± 187 | 1.6 | 0.49 | ||
| Total | 33,223 ± 2284 | 41,310 ± 1,191 | 1.2 | 0.04 | ||
| Maltase | YBR299W | MAL32 | 17,626 ± 310 | 20,011 ± 404 | 1.1 | 0.03 |
| YJL221C | FSP2 | 3,018 ± 545 | 8,232 ± 2,208 | 2.7 | 0.17 | |
| YGR287C | 1,698 ± 384 | 5,059 ± 863 | 3.0 | 0.08 | ||
| YJL216C | 813 ± 207 | 1,084 ± 37 | 1.3 | 0.31 | ||
| Total | 23,155 ± 764 | 34,386 ± 2,405 | 1.5 | 0.00 | ||
| Maltose activator | YGR288W | MAL13 | 282 ± 2 | 337 ± 76 | 1.2 | 0.49 |
| MAL23 | Ac | A | ||||
| YBR297W | MAL33 | 865 ± 287 | 834 ± 38 | −1.0 | 0.90 | |
| YPR196W | 2,863 ± 106 | 3,670 ± 378 | 1.3 | 0.18 | ||
| YFL052W | A | A | ||||
| Total | 3,728 ± 306 | 4,504 ± 380 | 1.2 | 0.15 | ||
Average transcript levels and their mean deviations were calculated from two independent chemostat cultures.
Determined by Student's t test (a P value of ≤0.05 is considered significant).
A, transcript defined as absent by the algorithm implemented in version 4.01 of the Affymetrix Microarray Suite software.
The maltose-sensitive phenotype of long-term maltose-limited cultures involves stable mutations.
To test whether the changes observed after long-term maltose-limited cultivation were the result of physiological adaptation or of the selection of mutants, a new chemostat was started with a single-cell culture isolated from a 42-generation chemostat culture. After 10 generations of maltose-limited chemostat cultivation of this strain, samples were taken and analyzed for maltose sensitivity. Off-line exposure to excess maltose led to a 3.5-fold increase of the extracellular protein concentration (from 0.14 ± 0.01 to 0.48 ± 0.10 g · liter−1), which is comparable to the protein release by cells from of 35-generation-old cultures of the reference strain (Fig. 4B). After 2 h of exposure of the selected strain to excess maltose, 27.9 ± 1.9 mM maltose was consumed and 36.3 ± 8.8 mM glucose and 21.9 ± 8.8 mM ethanol were produced. These concentrations are in good agreement with those observed for prolonged cultivations of the reference strain (Fig. 4D).
DISCUSSION
Affinity for maltose during prolonged maltose-limited growth.
During prolonged maltose-limited chemostat cultivation of S. cerevisiae, the residual substrate (maltose) concentration (s) decreased from an initial value of ca. 18 mg · liter−1 (0.05 mM) to a value of ca. 3 mg · liter−1 (0.01 mM). This was indicative of a gradually increasing affinity (μmax · Ks−1) for maltose (8).
In chemostat cultures at constant specific growth rates, the rate of maltose transport via the MALx1-encoded maltose-proton symporters equals the specific in situ rate of maltose consumption (qmaltose [millimoles · gram−1 · hour−1) (65). In an analysis of growth kinetics, we assume that, at the low residual maltose concentration in the chemostat culture, the specific rate of maltose-proton symport is controlled solely by the extracellular maltose concentration. This is equivalent to the assumption that maltose transport over the plasma membrane controls the specific growth rate. If, furthermore, maltose transport obeys Michaelis-Menten kinetics for extracellular maltose, then
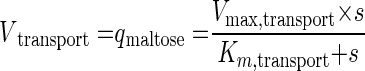 |
(1) |
The maltose uptake assays were performed at a maltose concentration of 25 mM, which is an order of magnitude above the substrate saturation constant (Km,transport) in wild-type S. cerevisiae (47). Consequently, the observed maltose transport activities should give a reliable estimate of the maximum transport capacity (Vmax,transport). Hence, the Km,transport can be calculated from
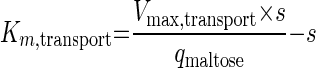 |
(2) |
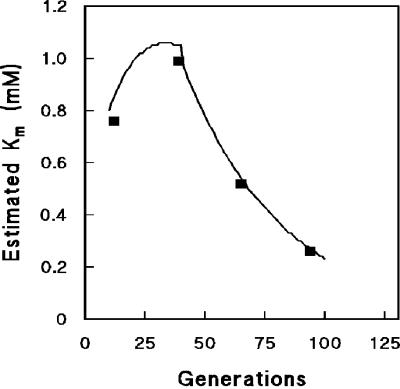
During the initial phase of the long-term cultivation experiments, Vmax,transport increased, while Km,transport remained at ca. 1 mM (Fig. 2 and 5). This estimated Km,transport corresponds well with the values of 1 to 5 mM reported in the literature (47, 61). After about 35 generations, Vmax,transport did not increase further. The observed further increase in maltose affinity was consistent with a gradual decrease of Km,transport (Fig. 5). Thus, the development of maltose consumption kinetics indicates that long-term adaptation of S. cerevisiae to maltose-limited growth involves two distinct phases. In the first phase, affinity is increased by increasing the capacity of maltose transport and hydrolysis (Fig. 2 and 3). The resulting maximum transport capacity may, for example, be determined by the maximum available membrane space (30). In a second phase, the increase of maltose uptake affinity is consistent with a decrease of the Km of the maltose transporter(s). Alternatively, the decrease of the calculated Km indicated that, after 35 generations, control of the specific growth rate was no longer controlled by maltose-proton symport.
FIG. 5.
Estimated Km of transport during prolonged maltose-limited chemostat cultivation. Km values were estimated by substituting the regression formula of residual maltose concentrations (s) and maltose transport capacities (Vmax) versus generations of chemostat cultivation into equation 2. Substrate saturation constants from the data points were also calculated by using equation 2.
Between culture ages of 10 and 45 generations, the capacities of maltose transport and maltase increased by ca. 2.5- and 1.5-fold, respectively (Fig. 2 and 3). This correlated with changes in the transcript levels of several of the structural genes encoding maltose transporters and maltases (Table 1), suggesting that the rapid increase in enzyme activity was at least to some extent achieved by increasing the number of maltose carriers in the plasma membrane rather than exclusively by increasing the turnover number of the individual carriers via point mutations or changes in their membrane environment. A transcriptome analysis of long-term glucose-grown chemostat cultures also revealed important changes in the expression of sugar transporters (17). We have not investigated the molecular basis for the further increase of the affinity for maltose that occurred after 45 generations. Mechanisms that might contribute to this further increase include a different relative expression level of maltose transporter genes, mutations in one or more of these genes leading to a reduced Km,transport, and changes in the lipid composition of the plasma membrane (60).
Maltose hypersensitivity.
Extreme sensitivity to growth substrates, often leading to “substrate-accelerated death,” has been observed for many combinations of microorganisms and substrates upon a transition from nutrient limitation to nutrient excess. In general, substrate-accelerated death appears to be due to an unrestricted influx of substrate, which subsequently may lead to several problems, including intracellular accumulation of substrate and metabolic intermediates, intracellular acidification, and futile cycling (9, 45, 46, 57).
In earlier studies in which maltose-accelerated death was observed in wild-type strains of S. cerevisiae or in mutants affected in glucose catabolite repression, osmotic problems due to rapid maltose uptake and hydrolysis were proposed as causes for this phenomenon (15, 16). Furthermore, it has been proposed that the unrestricted uptake of maltose might lead to intracellular acidification, contributing to the cell lysis that is observed in maltose-stressed cultures (47).
Effects of prolonged cultivation under maltose-limited conditions on the occurrence of maltose-accelerated death have not been reported previously. This does not necessarily imply that such effects were overlooked in the earlier chemostat studies by Postma et al. (47). Instead, the different response of young chemostat cultures observed in their work may be attributed to the use of different S. cerevisiae strains and/or different experimental conditions. Our observations are consistent with a key role of the maltose-proton symporter (and possibly maltase) in maltose-accelerated death: a clear correlation was observed between an increased capacity for maltose transport and an increased maltose sensitivity. We attempted to further investigate the role of the maltose transporter by constitutively overexpressing the MAL61 gene (encoding an S. cerevisiae maltose-proton symporter) from the PMA1 promoter on a multicopy vector. However, although the maltose transport activity of young chemostat cultures of the transformed strain was significantly higher than that of the reference strain, it remained lower than the value at which maltose sensitivity became detectable in long-term cultures of the reference strain. Consistently, no maltose-accelerated death was observed when young chemostat cultures of the transformed strain were exposed to excess maltose (data not shown).
Selection in steady-state chemostat cultures.
Single-cell cultures isolated from a prolonged maltose-limited culture exhibited a much lower residual substrate concentration than young cultures of the original strain. Selection of mutants with an improved affinity for the growth-limiting nutrient during prolonged chemostat cultivation has been reported for several combinations of microorganisms and growth-limiting nutrients (see, e.g., references 2, 17, 28, 52, 58, 67, and 68). Our observations underline that when parameters other than biomass and product yields are taken into account, “steady states” in chemostat cultures may not really exist. Already within 20 to 25 generations (the rule of thumb proposed in early publications [17]), significant changes in maltose affinity were observed (Fig. 1). This issue is especially relevant when high-information-density analytical tools (e.g., DNA microarrays) are used for quantitative analysis of microbial growth in chemostats. In such experiments, culture age has to be standardized to minimize experimental noise (29, 44).
The observed correlation between improved maltose affinity and increased maltose sensitivity can be explained by a central role of the capacity of maltose-proton symport in these two parameters. This correlation is likely to occur in other cases in which the capacity of proton symporters is increased via metabolic engineering and/or strain selection, e.g., to broaden the substrate range of industrial organisms. For example, problems that have been encountered in attempts to engineer S. cerevisiae for the utilization of lactose via the expression of a heterologous lactose-proton symporter and β-galactosidase may well be related to this phenomenon (1, 50, 51).
The term evolutionary engineering has recently been coined to describe the selection of microbial strains with specific characteristics via selection under rationally designed culture conditions (18, 20, 31, 40, 53, 55). The use of chemostat cultures as “evolutionary devices” for selection of mutant microorganisms was already described in the 1950s and has since been successfully applied to improve numerous physiological traits (42, 64). The results obtained in this study illustrate how, at least in some cases, selection under steady-state chemostat conditions may proceed at the expense of microbial performance under dynamic conditions. In an ecological sense, this underlines how important physiological characteristics such as substrate affinity and substrate tolerance can be interpreted only by taking into account the dynamic environmental conditions to which microorganisms have been exposed during evolution.
Acknowledgments
We thank Hans van Dijken for many stimulating discussions and Serge Donkers for constructing the MAL61-overexpressing strain.
This work was financially supported by the Dutch Ministry of Economic Affairs via the EET program. The research group of J.T.P. is part of the Kluyver Centre for Genomics of Industrial Fermentation, which is supported by The Netherlands Genomics Initiative.
REFERENCES
- 1.Adam, A. C., J. A. Prieto, M. Rubio-Texeira, and J. Polaina. 1999. Construction of a lactose-assimilating strain of baker's yeast. Yeast 15:1299-1305. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J., C. Paquin, P. W. Oeller, and L. W. Lee. 1985. Physiological characterization of adaptive clones in evolving populations of the yeast, Saccharomyces cerevisiae. Genetics 110:173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, J. A. 1981. The utilization of disaccharides and some other sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 39:347-404. [DOI] [PubMed] [Google Scholar]
- 4.Bell, P., V. J. Higgins, and P. V. Attfield. 2001. Comparison of fermentative capacities of industrial baking and wild-type yeasts of the species Saccharomyces cerevisiae in different sugar media. Lett. Appl. Microbiol. 32:224-229. [DOI] [PubMed] [Google Scholar]
- 5.Brondijk, T. H. C., M. E. van der Rest, D. Pluim, Y. de Vries, K. Stingl, B. Poolman, and W. N. Konings. 1998. Catabolite inactivation of wild-type and mutant maltose transport proteins in Saccharomyces cerevisiae. J. Biol. Chem. 273:15352-15357. [DOI] [PubMed] [Google Scholar]
- 6.Brown, C. J., K. M. Todd, and R. F. Rosenzweig. 1998. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 15:931-942. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. M., and J. S. Hough. 1965. Elongation of yeast cells in continuous culture. Nature 206:676-678. [DOI] [PubMed] [Google Scholar]
- 8.Button, D. K. 1991. Biochemical basis for whole-cell uptake kinetics: specific affinity, oligotrophic capacity, and the meaning of the Michaelis constant. Appl. Environ. Microbiol. 57:2033-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calcott, P. H., and J. R. Postgate. 1972. On substrate-accelerated death in Klebsiella aerogenes. J. Gen. Microbiol. 70:115-122. [DOI] [PubMed] [Google Scholar]
- 10.Chang, Y. S., R. A. Dubin, E. Perkins, D. Forrest, C. A. Michels, and R. B. Needleman. 1988. MAL63 codes for a positive regulator of maltose fermentation in Saccharomyces cerevisiae. Curr. Genet. 14:201-209. [DOI] [PubMed] [Google Scholar]
- 11.Charron, M. J., E. Read, S. R. Haut, and C. A. Michels. 1989. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics 122:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, Q., and C. A. Michels. 1991. MAL11 and MAL61 encode the inducible high-affinity maltose transporter of Saccharomyces cerevisiae. J. Bacteriol. 173:1817-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dequin, S. 2001. The potential of genetic engineering for improving brewing, wine-making and baking yeasts. Appl. Microbiol. Biotechnol. 56:577-588. [DOI] [PubMed] [Google Scholar]
- 14.Dubin, R. A., R. B. Needleman, D. Gossett, and C. A. Michels. 1985. Identification of the structural gene encoding maltase within the MAL6 locus of Saccharomyces carlsbergenesis. J. Bacteriol. 164:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Entian, K. D. 1980. A defect in carbon catabolite repression associated with uncontrollable and excessive maltose uptake. Mol. Gen. Genet. 179:169-175. [DOI] [PubMed] [Google Scholar]
- 16.Entian, K. D., and M. C. Loureiro-Dias. 1990. Misregulation of maltose uptake in a glucose repression defective mutant of Saccharomyces cerevisiae leads to glucose poisoning. J. Gen. Microbiol. 136:855-860. [DOI] [PubMed] [Google Scholar]
- 17.Ferea, T. L., D. Botstein, P. O. Brown, and R. F. Rosenzweig. 1999. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl. Acad. Sci. USA 96:9721-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores, N., J. Xiao, A. Berry, F. Bolivar, and F. Valle. 1996. Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat. Biotechnol. 14:620-623. [DOI] [PubMed] [Google Scholar]
- 19.Goldenthal, M. J., M. Vanoni, B. Buchferer, and J. Marmur. 1987. Regulation of MAL gene expression in yeast: gene dosage effects. Mol. Gen. Genet. 209:508-517. [DOI] [PubMed] [Google Scholar]
- 20.Hall, B. G., and B. Hauer. 1993. Acquisition of new metabolic activities by microbial populations. Methods Enzymol. 224:603-613. [DOI] [PubMed] [Google Scholar]
- 21.Hammond, J. R. M. 1995. Genetically-modified brewing yeasts for the 21st century. Progress to date. Yeast 11:1613-1627. [DOI] [PubMed] [Google Scholar]
- 22.Higgins, V. J., M. Braidwood, P. Bell, P. Bissinger, I. W. Dawes, and P. V. Attfield. 1999. Genetic evidence that high noninduced maltase and maltose permease activities, governed by MALx3-encoded transcriptional regulators, determine efficiency of gas production by baker's yeast in unsugared dough. Appl. Environ. Microbiol. 65:680-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong, S. H., and J. Marmur. 1986. Primary structure of the maltase gene of the MAL6 locus of Saccharomyces carlsbergenesis. Gene 41:75-84. [DOI] [PubMed] [Google Scholar]
- 24.Hu, Z., A. W. Gibson, J. H. Kim, L. A. Wojciechowicz, B. Zhang, and C. A. Michels. 1999. Functional domain analysis of the Saccharomyces MAL-activator. Curr. Genet. 36:1-12. [DOI] [PubMed] [Google Scholar]
- 25.Jansen, M. L. A., J. H. de Winde, and J. T. Pronk. 2002. Hxt-carrier-mediated glucose efflux upon exposure of Saccharomyces cerevisiae to excess maltose. Appl. Environ. Microbiol. 68:4259-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein, C. J. L., L. Olsson, B. Ronnow, J. D. Mikkelsen, and J. Nielsen. 1996. Alleviation of glucose repression of maltose metabolism by MIG1 disruption in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 62:4441-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein, C. J. L., J. L. Rasmussen, B. Ronnow, L. Olsson, and J. Nielsen. 1999. Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J. Biotechnol. 68:197-212. [DOI] [PubMed] [Google Scholar]
- 28.Kovarova-Kovar, K., and T. Egli. 1998. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol. Mol. Biol. Rev. 62:646-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubitschek, H. E. 1970. Introduction to research with continuous cultures. Prentice-Hall, Englewood Cliffs, N.J.
- 30.Lenaz, G. 1987. Lipid fluidity and membrane protein dynamics. Biosci. Rep. 7:823-837. [DOI] [PubMed] [Google Scholar]
- 31.Long-McGie, J., A. D. Liu, and V. Schellenberger. 2000. Rapid in vivo evolution of a beta-lactamase using phagemids. Biotechnol. Bioeng. 68:121-125. [DOI] [PubMed] [Google Scholar]
- 32.Lowry, O. H., H. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 33.Medintz, I., H. Jiang, E. Han, and W. Cui. 1996. Characterization of the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. J. Bacteriol. 178:2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medintz, I., H. Jiang, and C. A. Michels. 1998. The role of ubiquitin conjugation in glucose-induced proteolysis of Saccharomyces maltose permease. J. Biol. Chem. 273:34454-34462. [DOI] [PubMed] [Google Scholar]
- 35.Medintz, I., X. Wang, T. Hradek, and C. A. Michels. 2000. A pest-like sequence in the N-terminal cytoplasmic domain of Saccharomyces maltose permease is required for glucose-induced proteolysis and rapid inactivation of transport activity. Biochemistry 39:4518-4526. [DOI] [PubMed] [Google Scholar]
- 36.Millard, P. J., B. L. Roth, H. P. Thi, S. T. Yue, and R. P. Haugland. 1997. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 63:2897-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naumov, G. I., E. S. Naumova, and C. A. Michels. 1994. Genetic variation of the repeated MAL loci in natural populations of Saccharomyces cerevisiae and Saccharomyces paradoxus. Genetics 136:803-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Needleman, R. B. 1991. Control of maltase synthesis in yeast. Mol. Microbiol. 5:2079-2084. [DOI] [PubMed] [Google Scholar]
- 39.Needleman, R. B., D. B. Kaback, R. A. Dubin, E. L. Perkins, N. G. Rosenberg, K. A. Sutherland, D. B. Forrest, and C. A. Michels. 1984. MAL6 of Saccharomyces: a complex genetic locus containing three genes required for maltose fermentation. Proc. Natl. Acad. Sci. USA 81:2811-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noack, D., R. Geuther, M. Tonew, R. Breitling, and D. Behnke. 1988. Expression and secretion of interferon-alpha 1 by Streptomyces lividans: use of staphylokinase signals and amplification of a neo gene. Gene 68:53-62. [DOI] [PubMed] [Google Scholar]
- 41.Noorman, H. J., J. Baksteen, J. J. Heijnen, and K. C. A. M. Luyben. 1991. The bioreactor overflow device: an undesired selective separator in continuous cultures? J. Gen. Microbiol. 13:2171-2177. [Google Scholar]
- 42.Novick, A., and L. Szilard. 1950. Experiments with the chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. USA 36:708-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okuyama, M., A. Okuno, N. Shimizu, H. Mori, A. Kimura, and S. Chiba. 2001. Carboxyl group of residue Asp647 as possible proton donor in catalytic reaction of alpha-glucosidase from Schizosaccharomyces pombe. Eur. J. Biochem. 268:2270-2280. [DOI] [PubMed] [Google Scholar]
- 44.Piper, M. D., P. Daran-Lapujade, C. Bro, B. Regenberg, S. Knudsen, J. Nielsen, and J. T. Pronk. 2002. Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 277:37001-37008. [DOI] [PubMed] [Google Scholar]
- 45.Postgate, J. R., and J. R. Hunter. 1963. Acceleration of bacterial death by growth substrates. Nature 198:273. [DOI] [PubMed] [Google Scholar]
- 46.Postgate, J. R., and J. R. Hunter. 1964. Accelerated death of Aerobacter aerogenes starved in the presence of growth-limiting substrates. J. Gen. Microbiol. 34:459-473. [DOI] [PubMed] [Google Scholar]
- 47.Postma, E., C. Verduyn, A. Kuiper, W. A. Scheffers, and J. P. van Dijken. 1990. Substrate-accelerated death of Saccharomyces cerevisiae CBS 8066 under maltose stress. Yeast 6:149-158. [DOI] [PubMed] [Google Scholar]
- 48.Randez-Gil, F., P. Sanz, and J. A. Prieto. 1999. Engineering baker's yeast: room for improvement. TIBTECH 17:237-244. [DOI] [PubMed] [Google Scholar]
- 49.Riballo, E., M. Herweijer, D. H. Wolf, and R. Lagunas. 1995. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J. Bacteriol. 177:5622-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubio-Texeira, M., M. Arevalo-Rodriguez, J. L. Lequerica, and J. Polaina. 2000. Lactose utilization by Saccharomyces cerevisiae strains expressing Kluyveromyces lactis LAC genes. J. Biotechnol. 84:97-106. [DOI] [PubMed] [Google Scholar]
- 51.Rubio-Texeira, M., J. A. Castrillo, A. C. Adam, U. O. Ugalde, and J. Polaina. 1998. Highly efficient assimilation of lactose by a metabolically engineered strain of Saccharomyces cerevisiae. Yeast 14:827-837. [DOI] [PubMed] [Google Scholar]
- 52.Rutgers, M., M. J. Teixeira de Mattos, P. W. Postma, and K. van Dam. 1987. Establishment of the steady state in glucose-limited chemostat cultures of Klebsiella pneumoniae. J. Gen. Microbiol. 133:445-451. [DOI] [PubMed] [Google Scholar]
- 53.Sauer, U. 2001. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 73:129-169. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt, M. E., T. A. Brown, and T. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider, K. H., G. Jakel, R. Hoffmann, and F. Giffhorn. 1995. Enzyme evolution in Rhodobacter sphaeroides: selection of a mutant expressing a new galactitol dehydrogenase and biochemical characterization of the enzyme. Microbiology 141:1865-1873. [DOI] [PubMed] [Google Scholar]
- 56.Serrano, R. 1977. Energy requirements for maltose transport in yeast. Eur. J. Biochem. 80:97-102. [DOI] [PubMed] [Google Scholar]
- 57.Strange, R. E., and F. A. Dark. 1965. “Substrate-accelerated death” of Aerobacter aerogenes. J. Gen. Microbiol. 39:215-228. [DOI] [PubMed] [Google Scholar]
- 58.Tsen, S., S. Lai, C. Pang, J. Lee, and T. H. Wilson. 1996. Chemostat selection of an Escherichia coli mutant containing permease with enhanced lactose affinity. Biochem. Biophys. Res. Commun. 224:351-357. [DOI] [PubMed] [Google Scholar]
- 59.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Rest, M. E., A. H. Kamminga, A. Nakano, Y. Anraku, B. Poolman, and W. N. Konings. 1995. The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol. Rev. 59:304-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Leeuwen, C. C. M., R. A. Weusthuis, E. Postma, P. J. A. van den Broek, and J. P. van Dijken. 1992. Maltose/proton co-transport in Saccharomyces cerevisiae. Biochem. J. 284:441-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Urk, H., P. R. Mak, W. A. Scheffers, and J. P. van Dijken. 1988. Metabolic responses of Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621 upon transition from glucose limitation to glucose excess. Yeast 4:283-291. [DOI] [PubMed] [Google Scholar]
- 63.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous study on regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 64.Weikert, C., U. Sauer, and J. E. Bailey. 1997. Use of a glycerol-limited, long-term chemostat for isolation of Escherichia coli mutants with improved physiological properties. Microbiology 143:1567-1574. [DOI] [PubMed] [Google Scholar]
- 65.Weusthuis, R. A., H. Adams, W. A. Scheffers, and J. P. van Dijken. 1993. Energetics and kinetics of maltose transport in Saccharomyces cerevisiae: a continuous culture study. Appl. Environ. Microbiol. 59:3102-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weusthuis, R. A., M. A. H. Luttik, W. A. Scheffers, J. P. van Dijken, and J. T. Pronk. 1994. Is the Kluyver effect in yeast caused by product inhibition? Microbiology 140:1723-1729. [DOI] [PubMed] [Google Scholar]
- 67.Wick, L. M., H. Weilenmann, and T. Egli. 2002. The apparent clock-like evolution of Escherichia coli in glucose-limited chemostats is reproducible at large but not at small population sizes and can be explained with Monod kinetics. Microbiology 148:2889-2902. [DOI] [PubMed] [Google Scholar]
- 68.Wiebe, M. G., G. D. Robson, B. Cunliffe, A. P. J. Trinci, and S. G. Oliver. 1992. Nutrient-dependent selection of morphological mutants of Fusarium graminearum A3/5 isolated from long-term continuous flow cultures. Biotechnol. Bioeng. 40:1181-1189. [DOI] [PubMed] [Google Scholar]
- 69.Yao, B., P. Sollitti, X. Zhang, and J. Marmur. 1994. Shared control of maltose induction and catabolite repression of the MAL structural genes in Saccharomyces. Mol. Gen. Genet. 243:622-630. [DOI] [PubMed] [Google Scholar]