Abstract
Quorum sensing via autoinducer-2 (AI-2) has been identified in different strains, including those from Escherichia, Vibrio, Streptococcus, and Bacillus species, and previous studies have suggested the existence of additional quorum-sensing signals working in the stationary phase of Escherichia coli cultures. To investigate the presence and global effect of these possible quorum-sensing signals other than AI-2, DNA microarrays were used to study the effect of stationary-phase signals on the gene expression of early exponential-phase cells of the AI-2-deficient strain E. coli DH5α. For statistically significant differential gene expression (P < 0.05), 14 genes were induced by supernatants from a stationary culture and 6 genes were repressed, suggesting the involvement of indole (induction of tnaA and tnaL) and phosphate (repression of phoA, phoB, and phoU). To study the stability of the signals, the stationary-phase supernatant was autoclaved and was used to study its effect on E. coli gene expression. Three genes were induced by autoclaved stationary-phase supernatant, and 34 genes were repressed. In total, three genes (ompC, ptsA, and btuB) were induced and five genes (nupC, phoB, phoU, argT, and ompF) were repressed by both fresh and autoclaved stationary-phase supernatants. Furthermore, supernatant from E. coli DH5α stationary culture was found to repress E. coli K-12 AI-2 concentrations by 4.8-fold ± 0.4-fold, suggesting that an additional quorum-sensing system in E. coli exists and that gene expression is controlled as a network with different signals working at different growth stages.
Quorum sensing, the regulation of gene expression by producing and responding to secreted autoinducers (AIs) whose concentrations reflect the population density (2), commonly exists in bacteria. Gram-negative bacteria use acylated homoserine lactones as AIs, and gram-positive bacteria use oligopeptides (2, 16). When the cell density is high, the binding of AIs to cell receptors regulates gene expression for a variety of phenotypes, such as production of virulence factors (4), protein production (7), siderophore synthesis (29), bioluminescence (5), biofilm formation (6), and plasmid conjugation (15). Generally, each bacterial species uses its own signal; however, a common AI-2 signal has been discovered for interspecies communication (30, 31, 37). Escherichia coli does not produce acylated homoserine lactone, but it possesses the AI-2 quorum-sensing system (30). Recent studies have found that E. coli O157:H7 uses AI-2 to control the expression of virulence factors, type III secretion, chemotaxis, flagellar synthesis, and motility (24-26) and that E. coli K-12 uses AI-2 to control chemotaxis, motility, and flagellar synthesis (D. Ren, A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood, submitted for publication). In addition, E. coli RP37 uses AI-2 to control cell aggregation (17). In Salmonella enterica serovar Typhimurium, AI-2 concentrations are maximal in mid-exponential-phase growth, and it is degraded in the stationary phase by an unknown mechanism (30). E. coli DH5α does not produce AI-2 due to a 60-amino-acid deletion stemming from a 1-bp deletion that results in early truncation of luxS (formerly ygaG) (31).
Several lines of evidence suggest the existence of additional E. coli quorum-sensing signals besides AI-2. Previously, Withers and Nordström (36) reported that E. coli produces an extracellular factor in late exponential phase to early stationary phase that represses chromosomal replication. By using the bicistronic reporter transposon mini-Tn5 lacZ-tet/1, Baca-DeLancey et al. (1) found some E. coli genes (cysK, astD, tnaB, and gabT) that were activated by extracellular signals from stationary-phase, 0.5× Luria-Bertani (LB) medium cultures that are stable at pH 12 (20 min) and 100°C (10 min). These genes have functions for pyruvate and succinate production (1). Recently, indole, purified from stationary-phase E. coli 0.5× LB medium cultures, was found to induce the expression of gabT, astD, and tnaB (32). In addition, Sperandio et al. (27) found a new AI (AI-3) in E. coli O157:H7 involved in communication between the bacterium and its eukaryotic host.
With the ability to simultaneously quantify the expression of all genes in one organism whose genome has been sequenced (39), DNA microarrays have been successfully used to study bacterial responses to different stimuli such as heat shock and other stresses (12, 35, 40), quorum sensing (8, 26), anaerobic metabolism (38), sporulation (10), and biofilm formation (20, 23, 28, 34). To further study the roles of E. coli stationary-phase extracellular signals, DNA microarrays were used here to study E. coli DH5α (AI-2−) (30) gene expression with and without the addition of E. coli-conditioned medium. The effect of stationary-phase quorum-sensing signals on AI-2 synthesis in the exponential phase was also investigated by using E. coli K-12 (AI-2+). This is the first study to investigate the effect of E. coli stationary-phase quorum-sensing signals on E. coli global gene expression by using DNA microarrays and to show that stationary-phase quorum-sensing signals repress AI-2 concentrations in the exponential phase.
MATERIALS AND METHODS
Bacterial strains and culture media.
E. coli DH5α [luxS supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (22, 31) was used to investigate the effect of stationary-phase signals on the gene expression of early exponential luxS mutant cells. E. coli K-12 (ATCC 25404) was used to study the effect of stationary-phase signals on AI-2 production. Vibrio harveyi BB170 (AI-1 sensor−, AI-2 sensor+) was used as the reporter to measure AI-2 activity (21, 30). LB medium (22) containing 10 g of tryptone/liter, 5 g of yeast extract/liter, and 10 g of NaCl/liter with pH adjusted to 7.5 (1) was used to grow the E. coli cells. AI bioassay (AB) medium (11) was used to grow V. harveyi BB170, and LM medium (3) was used to determine the number of V. harveyi CFU.
Preparation of conditioned media.
E. coli DH5α was grown in LB medium (pH 7.5) overnight and was diluted 1:100 in the same fresh medium. When the culture grew to an optical density at 600 nm (OD600) (DU 640 spectrophotometer; Beckman, Fullerton, Calif.) of 1.65 (1.2 × 108 cells/ml), the cells were precipitated by centrifuging at 10,000 × g for 10 min at 4°C and the supernatant was collected. Note that an OD of 0.2 indicated exponential growth of the E. coli cultures, an OD of 0.9 indicated transition from late exponential phase to early stationary phase, and an OD of 1.6 indicated stationary-phase growth. The harvested supernatant was split equally into two portions. One portion was supplemented with tryptone and yeast extract to 0.5 times of that of LB medium, and the pH was adjusted to 7.5. The resulting solution was sterilized by filtering through a 0.2-μm-pore-size filter and will be referred to as fresh conditioned medium (FCM). Another portion of the supernatant was autoclaved for 2 h (121°C, 15 lb/in2), tryptone and yeast extract were added, the pH was adjusted to 7.5, and the supernatant was filter sterilized; this medium is termed autoclaved conditioned medium (ACM). Sterile 0.5× LB medium (pH 7.5) was used as the negative control medium to investigate the effect of FCM and ACM on gene expression.
Growth in conditioned media and RNA isolation.
To prepare cell cultures for RNA isolation, 200 μl of an overnight E. coli DH5α culture (grown in LB, pH 7.5) was added to 150 ml of the same fresh medium and was grown to an OD of 0.2 (1.4 × 107 cells/ml). Then the culture was split equally into three portions and was centrifuged at 10,000 × g for 5 min at 4°C. The cell pellets were washed once with 40 ml of ice-cold 0.85% NaCl buffer and were centrifuged at 10,000 × g for 5 min at 4°C. The cell pellets were resuspended in 40 ml each of 0.5× LB (pH 7.5), ACM, or FCM and were incubated with shaking for 30 min at 37°C. All three cultures grew to an OD of around 0.3 after incubation. Then the cells were centrifuged at 10,000 × g for 3 min at −10°C, resuspended in 6 ml of ice-cold 0.85% NaCl buffer, split into four 1.5-ml portions, centrifuged at 10,000 × g for 15 s at room temperature, and flash-frozen in a dry-ice-ethanol bath. The cells were kept at −80°C until RNA isolation.
To lyse the cells, 1.0 ml of RLT buffer (Qiagen, Inc., Valencia, Calif.) and 0.2 ml of 0.1-mm zirconia/silica beads (Biospec, Bartlesville, Okla.) were added to the frozen bead beater tubes containing the cell pellets. The tubes were closed tightly and were beat for 30 s at the maximum speed in a mini-bead beater (catalog no. 3110BX; Biospec). The total RNA was isolated by following the protocol of the RNeasy Mini Kit (Qiagen), including an on-column DNase digestion with RNase-free DNase I (Qiagen). An OD260 reading was used to quantify the RNA yield. OD260/OD280 and 23S/16S rRNA readings were measured to check the purity and integrity of RNA (RNeasy Mini handbook; Qiagen).
DNA microarrays.
The antisense E. coli DNA microarrays were prepared as described previously (33). In detail, each gene probe was synthesized by PCR and has a size of the full open reading frame of the corresponding gene (200 to 2,000 nt). The double-strand PCR products were denatured in 50% dimethyl sulfoxide and were spotted onto aminosilane slides (Full Moon Biosystems, Sunnyvale, Calif.) as probes to hybridize with the mRNA-derived cDNA samples. It has been shown that each array can detect 4,228 of the 4,290 E. coli open reading frames (33). Each gene has two spots per slide. Eight amplified Klebsiella pneumoniae genes and 12 cDNA clones from plants were spotted on each slide as negative controls, while there were 76 E. coli PCR products spotted on each slide as positive controls (33).
Synthesis of Cy3- or Cy5-labeled cDNA.
To convert the total RNA into labeled cDNA from the E. coli DH5α samples treated with FCM, ACM, or 0.5× LB, reverse transcription was performed in 1.5-ml microcentrifuge tubes (Fisher) to which 6 μg of total RNA and 6 μg of random hexamer primers (Invitrogen Corp., Carlsbad, Calif.) were added. The volume was adjusted to 24 μl with RNase-free water (Invitrogen). The mixture was incubated for 10 min at 70°C, followed by 10 min at room temperature for annealing; then the reaction components were added, consisting of 8 μl of 5× SuperScript II reaction buffer (Invitrogen), 4 μl of 0.1 M dithiothreitol (Invitrogen), 1 μl of a deoxynucleoside triphosphate mix (a 2 mM concentration each of dATP, dGTP, and dTTP and 1 mM dCTP), 1 μl of 0.5 mM Cy3- or Cy5-labeled dCTP (Amersham Biosciences, Piscataway, N.J.), and 2 μl of SuperScript II reverse transcriptase (10 U/μl, Invitrogen). cDNA synthesis was conducted at 42°C for 2 h and was stopped by heating at 94°C for 5 min. After cDNA synthesis, the RNA template was removed with 2 μl of 2.5 M NaOH. The pH was neutralized with 10 μl of 2 M HEPES buffer, and the cDNA was purified with a Qiaquick PCR Mini Kit (Qiagen). The efficiency of labeling was checked via absorbance at 260 nm for the cDNA concentration, at 550 nm for Cy3 incorporation, and at 650 nm for Cy5 incorporation.
Hybridization and washing.
The cDNA samples of E. coli DH5α treated with FCM or 0.5× LB (6 μg of each) were each labeled with both Cy3 and Cy5 dyes to remove artifacts related to different labeling efficiencies; hence, each experiment required at least two slides. The Cy3-labeled FCM sample and Cy5-labeled 0.5× LB sample were hybridized on the first slide. Similarly, the Cy5-labeled FCM sample and Cy3-labeled 0.5× LB sample were hybridized on the second slide. Since each gene has two spots on a slide, the two hybridizations generated eight data points for each gene (four points for the FCM sample and four points for the 0.5× LB sample). DNA microarrays for the E. coli DH5α treated with ACM or 0.5× LB were performed in an analogous manner.
The DNA microarrays were incubated in prehybridization solution (3.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] [Invitrogen], 0.1% sodium dodecyl sulfate [SDS] [Invitrogen], and 0.1% bovine serum albumin [Invitrogen]) at 45°C for 20 min. The arrays were rinsed with double-distilled water and were spun dry by centrifugation. Labeled cDNA (6 μg) was concentrated to 10 μl of total volume and was mixed with 10 μl of 4× cDNA hybridization solution (Full Moon Biosystems) and 20 μl of formamide (EM Science, Gibbstown, N.J.). The hybridization mix was heated to 95°C for 2 min and was added to the DNA microarrays; each array was covered with a coverslip (Corning, Big Flats, N.Y.) and was incubated overnight at 37°C for hybridization. When the hybridization was finished, the coverslips were removed in 1× SSC-0.1% SDS at room temperature, and the arrays were washed once for 5 min in 1× SSC-0.1% SDS at 40°C, twice for 10 min in 0.1× SSC-0.1% SDS at 40°C, and twice for 1 min in 0.1× SSC at 40°C. The arrays were quickly rinsed by dipping in room-temperature double-distilled water and were then spun dry by centrifugation.
Image and data analysis.
The hybridized slides were scanned with the Generation III Array Scanner (Molecular Dynamics Corp.). Readings at 570 and 670 nm was used to quantify the probes labeled with Cy3 and Cy5 separately. The signal was quantified with Array Vision 4.0 or 6.0 software (Imaging Research, St. Catherines, Ontario, Canada). Genes were identified as differentially expressed if the expression ratio was greater than 2.4-fold (for the data set of FCM versus 0.5× LB) or 1.9-fold (for the data set of ACM versus 0.5× LB) and if P (found by t test) was less than 0.05. P values were calculated on log-transformed, normalized intensities. Including the P criterion ensures the reliability of the induced- and/or repressed-gene list. Normalization was relative to the median total fluorescent intensity per slide per channel. The gene functions were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/).
RNA dot blotting.
Digoxigenin-labeled DNA probes of six genes, phoB, ompC, ptsA, btuB, nupC, and b1498, were synthesized by using the PCR DIG Probe Synthesis Kit (Roche Applied Science, Mannheim, Germany). The PCR was performed in 30 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 40 s. The final extension was at 72°C for 7 min. The probes have lengths of 172 to 400 bp (see Table 5 for specific primers used). Total RNA (1.25, 2.5, or 5 μg) from independent cell cultures (different experiments from those used for the DNA microarrays but identical culture conditions) was blotted on positively charged nylon membranes (Boehringer Ingelheim, Ridgefield, Conn.) by using a Bio-Dot Microfiltration Apparatus (Bio-Rad, Richmond, Calif.). Total RNA was fixed by baking for 2 h at 80°C. DNA probes (about 400 ng; a serial dilution of RNA samples was tested to ensure excess of the DNA probes) were denatured in boiling water for 5 min before hybridizing to RNA. Hybridization (50°C, 16 h) and washing were according to the protocol for digoxigenin labeling and detection (Roche Applied Science). To detect the signal, disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2-[5-chloro]tricyclo[3.3.1.1,7]decan}-4-yl) phenyl phosphate (Roche Applied Science) was used as a substrate to give chemiluminescence, and the light was recorded by Biomax X-ray film (Kodak, Rochester, N.Y.).
TABLE 5.
RNA dot blotting confirmation of E. coli DH5α gene expression as a result of the addition of FCM and ACM supernatants containing stationary-phase signalsa
| Gene | DNA primers used for probe synthesis | DNA microarray expression ratio | RNA dot blotting expression ratio |
|---|---|---|---|
| phoB | 5′-TATTCTGGTCGTAGAAGATGAAGCT-3′ | −5 by FCM | −5 by FCM |
| 5′-TAATCCCTGCATCTCAATCACCTCT-3′ | −36 by ACM | −1.5 by ACM | |
| ompC | 5′-TAAAGTACTGTCCCTCCTGGTCCCA-3′ | +2.5 by ACM | +2 by ACM |
| 5′-GTAGGTGTCACCACCGAATTCTGGC-3′ | |||
| ptsA | 5′-GATTCTGGAAGCTCACCGATCCCTG-3′ | +4 by ACM | +1.5 by ACM |
| 5′-ACCAACCAGCGTTGGAATGTTGAAC-3′ | |||
| btuB | 5′-GCACAGGATACCAGCCCGGATACTC-3′ | +3 by ACM | +2 by ACM |
| 5′-GCGTCGTGATGATATTCACCACCCC-3′ | |||
| nupC | 5′-ACTGCTTGTTATCGAAGTGTTACTG-3′ | −5 by FCM | −2.5 by FCM |
| 5′-GGTGTACATACGATTACGGGAGATT-3′ | |||
| b1498 | 5′-GAAGAGGAACATTTATGAAGTCTGC-3′ | +4 by FCM | +2 by FCM |
| 5′-TGTGGCACATAGCCGTTAGTAAAAC-3′ |
RNA samples were independent of those used in the DNA microarray experiments. Relative expression ratios are shown.
Effect of stationary-phase signals on AI-2 concentrations in E. coli K-12 cultures.
To study if the unknown signals from the stationary phase of E. coli culture may cause the degradation of AI-2, FCM was supplemented with 0.5% glucose and was used to grow E. coli K-12 from an OD of 0.02 to one of 0.9 in 2.25 h. As the control sample, sterile 0.5× LB medium was supplemented with 0.5% glucose and was used to grow E. coli K-12 to an OD of 0.9 under identical conditions. Then both samples were centrifuged at 10,000 × g for 10 min at 4°C to remove the cells, and the supernatants were filter sterilized and were kept at −20°C until the AI-2 assay.
AI-2 AI bioassay.
E. coli supernatants were assayed using the method of Surette and Bassler (21, 30). The reporter strain V. harveyi BB170 was grown in AB medium overnight and was diluted 1:5,000 into the fresh AB medium; then the cell-free supernatants from the E. coli samples were added at a concentration of 10% (vol/vol). The time course of bioluminescence was measured with a 20/20 luminometer (Turner Design, Sunnyvale, Calif.) and was reported in relative light units. The cell density of the V. harveyi reporter strain was measured by spreading the cells on LM plates and by counting CFU after 24 h. The experiment was conducted in duplicate.
RESULTS
Comparison of gene expression profiles affected by FCM and ACM.
To investigate the effect of stationary-phase signals on a genetic basis and to study the stability of the signals, DNA microarrays were used to obtain the gene expression profiles of E. coli treated with FCM, ACM, or 0.5× LB. Since the unknown signal studied by Baca-DeLancey et al. (1) was stable at 100°C for 10 min, the ACM was prepared to investigate the heat sensitivity of the signals in the present study. In addition, nutrients (tryptone and yeast extract) were added in FCM and ACM to ensure that the differential gene expression was not caused by starvation.
According to the microarray results, FCM significantly induced 14 genes (Table 1) and significantly repressed 6 genes (Table 2). Similarly, ACM induced 3 genes (Table 3) and repressed 34 genes (Table 4). The gene expression patterns under these two sets of conditions agree well with each other. All three genes (ompC, ptsA, and btuB) induced by ACM were also induced by FCM. Similarly, five out of the six genes (nupC, phoB, phoU, argT, and ompF) repressed by FCM were also repressed by ACM. However, FCM induced more genes than ACM, while ACM repressed more genes than FCM. Hence, the signals may have been partially degraded by autoclaving.
TABLE 1.
Genes induced by the fresh E. coli stationary-phase supernatanta
| Gene | b no. | Expression ratio | Description |
|---|---|---|---|
| Known functions | |||
| ptsA | b3947 | 21.4 | PEP-protein phosphotransferase system enzyme I |
| btuB | b3966 | 14.2 | Vitamin B12 receptor precursor |
| tnaL | b3707 | 10.6 | Tryptophanase leader peptide, degradation of small molecules |
| ompC | b2215 | 4.1 | Outer membrane protein c precursor |
| tnaA | b3708 | 3 | Tryptophanase |
| cysA | b2422 | 2.6 | Transport of small anionic molecules |
| fliK | b1943 | 2.6 | Flagellar hook-length control protein |
| aldA | b1415 | 2.5 | Carbon energy metabolism |
| Unknown functions | |||
| b1498 | b1498 | 3.9 | Putative enzyme, not classified |
| b1285 | b1285 | 3.8 | ORF, unknown |
| evgA | b2369 | 3 | Putative positive transcription regulator (sensor EvgS) |
| yqgD | b2941 | 2.8 | ORF, unknown |
| b2845 | b2845 | 2.6 | Putative transporter protein |
| ybaJ | b0461 | 2.4 | ORF, unknown |
ORF, open reading frame. Boldfaced designations indicate those genes induced by both FCM and ACM.
TABLE 2.
Genes repressed by the fresh E. coli stationary-phase supernatanta
| Gene | b no. | Expression ratio | Description |
|---|---|---|---|
| nupC | b2393 | −5.3 | Transport of small molecules: nucleosides, purines, pyrimidines |
| phoB | b0399 | −4.5 | Positive response regulator for pho regulon; sensor is PhoR (or CreC) |
| phoU | b3724 | −4.4 | Negative regulator for pho regulon and putative enzyme in phosphate metabolism |
| argT | b2310 | −4.2 | Transport of small molecules: amino acids and amines |
| ompF | b0929 | −3.8 | Membrane and outer membrane constituents |
| phoA | b0383 | −2.7 | Alkaline phosphatase |
Boldfaced designations indicate genes repressed by both FCM and ACM.
TABLE 3.
Genes induced by the autoclaved E. coli stationary-phase supernatanta
| Gene | b no. | Expression ratio | Description |
|---|---|---|---|
| ptsA | b3947 | 3.9 | PEP-protein phosphotransferase system enzyme I |
| btuB | b3966 | 3.0 | Vitamin B12 receptor precursor |
| ompC | b2215 | 2.5 | Outer membrane protein c precursor |
Boldfaced designations indicate genes induced by both FCM and ACM.
TABLE 4.
Genes repressed by the autoclaved E. coli stationary-phase supernatanta
| Gene | b no. | Expression ratio | Description |
|---|---|---|---|
| Known functions | |||
| phoB | b0399 | −35.7 | Phosphate regulon regulatory protein PhoB |
| nupC | b2393 | −8.8 | Nucleoside permease (nucleoside-transport system protein) |
| phoU | b3724 | −6.5 | Phosphate transport system regulatory protein |
| phoR | b0400 | −5.6 | Phosphate regulon sensor protein PhoR |
| argT | b2310 | −5 | Lysine-arginine-ornithine-binding periplasmic protein precursor |
| nlpC | b1708 | −4.5 | Probable lipoprotein N1pC precursor |
| dsdX | b2365 | −4.4 | DsdX permease |
| gltB | b3212 | −3.4 | Glutamate synthetase, large subunit |
| ompF | b0929 | −3.3 | Outer membrane protein f precursor |
| guaC | b0104 | −3.2 | GMP reductase |
| phoE | b0241 | −3.2 | Outer membrane pore protein E |
| bcr | b2182 | −3.1 | Bicyclomycin resistance protein (sulfonamide resistance) |
| nupG | b2964 | −3 | Nucleoside-transporting protein NupG |
| carA | b0032 | −2.7 | Carbamoyl-phosphate synthetase glutamine chain |
| gltD | b3213 | −2.6 | Glutamate synthetase beta subunit |
| carB | b0033 | −2.5 | Glutamine-hydrolyzing |
| ldcC | b0186 | −2.4 | Lysine decarboxylase, constitutive |
| rfbX | b2037 | −2.3 | O-antigen transporter |
| sdiA | b1916 | −2.3 | SdiA regulatory protein |
| grxA | b0849 | −2.1 | Glutaredoxin 1 |
| phoH | b1020 | −2.1 | PhoH protein (phosphate starvation-inducible protein PsiH) |
| aroP | b0112 | −2 | Aromatic amino acid transport protein AroP |
| flgJ | b1081 | −2 | Flagellar basal body protein FlgJ |
| pyrE | b3642 | −2 | Orotate phosphoribosyltransferase |
| Unknown functions | |||
| yciH | b1282 | −5.1 | Hypothetical protein |
| b2063 | b2063 | −4.9 | Putative transport, not classified |
| b2639 | b2639 | −3.9 | Unknown |
| b1017 | b1017 | −3.5 | Unknown |
| b1018 | b1018 | −3.1 | Unknown |
| b1520 | b1520 | −3 | Open reading frame, unknown |
| ycdB | b1019 | −2.9 | Hypothetical protein |
| yffG | b2468 | −2.6 | Putative enzyme, not classified |
| yafL | b0227 | −2.5 | Hypothetical protein |
| ybgC | b0736 | −2.2 | Hypothetical protein |
Boldfaced designations indicate genes repressed by both FCM and ACM.
Genes induced by stationary-phase signals.
The genes induced by FCM have functions for energy metabolism (aldA), anion and carbohydrate transport (cysA, ptsA), flagellar synthesis (fliK), membrane protein (ompC), indole production (tnaA, tnaL), and unknown functions (b1285, b1498, b2845, ybaJ, and yqgD) (Table 1). The induction of tnaA agrees with the recent report that E. coli MG1655 produces indole at 340 μM in the stationary phase (32).
The induction of ompC by the stationary-phase signals is consistent with the fact that the two important outer membrane porin proteins, OmpC and OmpF, are subject to many environmental factors (18). ompC and ompF are controlled by OmpR-phosphate (OmpR-P) in an opposite manner. OmpR-P activates ompF at low osmolarity, while it represses ompF and activates ompC at high osmolarity (18). In addition, ompF was found to be repressed when cells enter the stationary phase (18), and the expression of ompC was found to increase in E. coli biofilms which contain a high density of cells (19). Therefore, the induction of ompC by stationary-phase signals suggests that the E. coli cells are encountering high osmolarity in the stationary phase.
Genes repressed by stationary-phase signals.
As expected, ompF was repressed by stationary-phase signals. In addition, the pho operon was also repressed by stationary-phase signals (Table 2). While five genes were consistently repressed by FCM and ACM, more genes (34 genes) were repressed by ACM. The increase in number of repressed genes may be caused by the heat treatment of the conditioned medium, which may degrade chemicals and produce toxic products. The high temperature may lyse chemicals such as peptides and amino acids. In addition, the high temperature and pressure may facilitate a variety of reactions between reactive groups (such as amines), which may lead to the production of unfavorable compounds for the cells.
Validation of the DNA microarray results with RNA dot blotting.
To confirm the gene expression profiles obtained from the DNA microarray analysis, total RNA was isolated from independent samples and was prepared in the same manner as the DNA microarray experiments, and the expression levels of six representative genes (phoB, ompC, ptsA, btuB, nupC, and b1498) were confirmed with RNA dot blotting (Table 5). For example, phoB was repressed fivefold by FCM in the DNA microarray results and was also repressed fivefold by FCM in the RNA dot blotting experiment. Hence, the DNA microarray results provided valid information about the differential gene expression profiles due to the addition of FCM or ACM.
Stationary-phase signals repress the AI-2 concentration in E. coli K-12 exponential cultures.
Since AI-2 is degraded when S. enterica serovar Typhimurium cells enter the stationary phase (30), it was hypothesized that the addition of stationary-phase signals to an E. coli culture would repress the AI-2 concentration. To test this hypothesis, FCM and 0.5× LB were each supplemented with 0.5% glucose and were used to grow E. coli K-12 from an OD of 0.02 to an OD of 0.9; glucose was added to produce AI-2 (30). The supernatants were then used for the AI-2 bioassay.
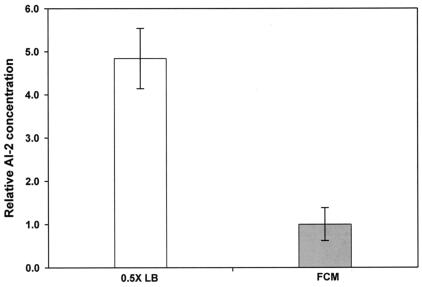
It was found that the stationary-phase signals repressed by 4.8-fold ± 0.4-fold the AI-2 concentration in FCM (79% repressed), compared to the concentration of AI-2 found in 0.5× LB (Fig. 1). This agrees well with a previous study that indicated that 75% of AI-2 was degraded when S. enterica serovar Typhimurium LT2 was grown for 20 h (14 h after the AI-2 reached the maximum) (30).
FIG. 1.
Repression of AI-2 concentrations in E. coli K-12 exponential-phase cultures by stationary-phase signals. The value of 1.0 was set at 2.5 × 10−5 relative light units/CFU.
DISCUSSION
Although the actual compounds need to be identified, this study indicates that E. coli has stationary-phase signals that specifically induce or repress certain genes to adjust to high cell density. Since the effects of FCM and ACM were compared with results for 0.5× LB (both FCM and ACM contain 0.5× LB), the induction or repression of genes was not due to the absence of needed nutrient components. It is well accepted that bacteria use quorum sensing to discern their population and adjust gene expression to take advantage of the resources and attack eukaryotic hosts (2). Given that E. coli AI-2 controls different phenotypes such as flagellar synthesis, motility, and virulence expression (25, 26), an understanding of the mechanism of AI-2 degradation is necessary. In this study, signals from stationary-phase cultures were found to decrease AI-2 concentrations significantly in E. coli K-12 cultures. Hence, there may be different quorum-sensing signals working at different growth stages enabling bacteria to discern their growth state. The genes induced by these stationary-phase signals are candidates for further studies, since it is possible that some of them encode factors that degrade AI-2 or induce enzymes that degrade AI-2. It is also possible that the stationary-phase signals repress AI-2 synthesis (via luxS) and allow basal AI-2 degradation to reduce AI-2 concentrations in the stationary phase. However, the luxS gene of E. coli DH5α was not affected by stationary-phase signals in this study (an average transcription level of luxS mRNA was detected for both the FCM and ACM samples, but luxS was not differentially expressed, since the expression ratios were about one in both the FCM and ACM microarray data sets); hence, the mode of AI-2 regulation appears to be degradation in the stationary phase. Interestingly, AI-1 of Erwinia and Agrobacterium sp., which are similar in structure to furanone, are subject to degradation by lactonase (AiiA) produced by Bacillus cereus (9).
Although indole has been reported to be a possible signal that upregulates the expression of gabT, astD, and tnaB of E. coli (32), these genes were not induced by FAM or ACM in the present study (but tnaA and tnaL were induced). Hence, it appears that there are signals other than indole that are responsible for the induction and repression of the genes found in the present study (i.e., if indole had been the signal, these genes should have been induced), and these signals induced indole synthesis via tnaA (Table 1). It is also striking that many phosphate-related genes are induced (ptsA) and repressed by the stationary-phase signals (phoA, phoB, phoU [Table 2]; and phoR, phoE, phoH, pyrE [Table 4]). Phosphate is important for bacteria, as evidenced by the fact that a Pseudomonas aeruginosa mutant deficient in polyphosphate synthesis (ppk1 mutant, lacking polyphosphate kinase) has defects in swimming, swarming, twitching motilities, virulence factor production, and biofilm formation (H. Zhang, K. Ishige, M. H. Rashid, and A. Kornberg, Abstr. Pseudomonas 2003, p. XXXVI, 2003 [http://pseudomonas2003.rsvs.ulaval.ca/Abstract%20book.pdf]). Hence, the regulation of these genes by the stationary-phase signals may be critical for cells to adjust their metabolism to survive with high cell density and attack eukaryotic hosts.
It is expected that stationary-phase signals will be discovered that may improve our understanding of E. coli quorum sensing and may promote the discovery of new antagonists against bacterial pathogens. The existence of additional signals is also supported by the study of biofilms. Clearly, E. coli DH5α, which is luxS deficient, makes a normal biofilm (14), and if signaling is involved in biofilms (6, 13), then there are other quorum signals present in E. coli. As for further studies to identify the signals, reporter strains for the most-induced genes found in the present study, ompC, ptsA, and btuB, should be constructed and used to study the compounds isolated from the stationary-phase cultures. In addition, FCM may be prepared from chemically defined medium (if it also supports the production of signals) rather than from the rich medium (like LB medium) to facilitate signal identification. To separate the compounds, FCM may be extracted with different organic solvents, such as ethyl acetate (32) and dichloromethane. The organic phase and water phase could be further purified with chromatography. Then each component may be tested for its effect on expression of the above genes.
REFERENCES
- 1.Baca-DeLancey, R. R., M. M. T. South, X. Ding, and P. N. Rather. 1999. Escherichia coli genes regulated by cell-to-cell signaling. Proc. Natl. Acad. Sci. USA 96:4610-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 4.Beck von Bodman, S., D. R. Majerczak, and D. L. Coplin. 1998. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. USA 95:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, J.-G., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264:21670-21676. [PubMed] [Google Scholar]
- 6.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 7.DeLisa, M. P., J. J. Valdes, and W. E. Bentley. 2001. Quorum signaling via AI-2 communicates the “Metabolic Burden” associated with heterologous protein production in Escherichia coli. Biotechnol. Bioeng. 75:439-450. [DOI] [PubMed] [Google Scholar]
- 8.DeLisa, M. P., C.-F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, Y.-H., J.-L. Xu, X.-Z. Li, and L.-H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenberg, E. P., J. W. Hastings, and S. Ulitzur. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 120:87-91. [Google Scholar]
- 12.Helmann, J. D., M. F. W. Wu, P. A. Kobel, F.-J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 14.Jayaraman, A., A. K. Sun, and T. K. Wood. 1998. Characterization of axenic P. fragi and E. coli biofilms for corrosion inhibition of SAE 1018 steel. J. Appl. Microbiol. 84:485-492. [DOI] [PubMed] [Google Scholar]
- 15.Lithgow, J. K., V. E. Danino, J. Jones, and J. A. Downie. 2001. Analysis of N-acyl homoserine-lactone quorum-sensing molecules made by different strains and biovars of Rhizobium leguminosarum containing different symbiotic plasmids. Plant Soil 232:3-12. [Google Scholar]
- 16.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 17.Park, S., P. M. Wolanin, E. A. Yuzbashyan, P. Silberzan, J. B. Stock, and R. H. Austin. 2003. Motion to form a quorum. Science 301:188. [DOI] [PubMed] [Google Scholar]
- 18.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: Multiple factors influence SYnthesis of the OmpF and OmpC Porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 19.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 21.Ren, D., J. J. Sims, and T. K. Wood. 2001. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 3:731-736. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Schembri, M. A., K. Kjærgaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 24.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stintzi, A., K. Evans, J.-M. Meyer, and K. Poole. 1998. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol. Lett. 166:341-345. [DOI] [PubMed] [Google Scholar]
- 30.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei, Y., J.-M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. Larossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teltzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, M., J. DeRisi, H.-H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Withers, H. L., and K. Nordström. 1998. Quorum-sensing acts at initiation of chromosomal replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:15694-15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 38.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye, R. W., T. Wang, L. Bedzyk, and K. M. Croker. 2001. Application of DNA microarrays in microbial systems. J. Microbiol. Methods 47:257-272. [DOI] [PubMed] [Google Scholar]
- 40.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]