Abstract
The cytosolic sulfotransferases (SULTs) are a multigene family of enzymes that catalyze the transfer of a sulfonate group from the physiologic sulfate donor, 3′-phosphoadenosine-5′-phosphosulfate, to a nucleophilic substrate to generate a polar product that is more amenable to elimination from the body. As catalysts of both xenobiotic and endogenous metabolism, the SULTs are major points of contact between the external and physiological environments, and modulation of SULT-catalyzed metabolism can not only affect xenobiotic disposition, but it can also alter endogenous metabolic processes. Therefore, it is not surprising that SULT expression is regulated by numerous members of the nuclear receptor (NR) superfamily that function as sensors of xenobiotics as well as endogenous molecules, such as fatty acids, bile acids, and oxysterols. These NRs include the peroxisome proliferator-activated receptors, pregnane X receptor, constitutive androstane receptor, vitamin D receptor, liver X receptors, farnesoid X receptor, retinoid-related orphan receptors, and estrogen-related receptors. This review summarizes current information about NR regulation of SULT expression. Because species differences in SULT subfamily composition and tissue-, sex-, development-, and inducer-dependent regulation are prominent, these differences will be emphasized throughout the review. In addition, because of the central role of the SULTs in cellular physiology, the effect of NR-mediated SULT regulation on physiological and pathophysiological processes will be discussed. Gaps in current knowledge that require further investigation are also highlighted.
Keywords: SULT, peroxisome proliferator-activated receptor, pregnane X receptor, constitutive androstane receptor, vitamin D receptor, liver X receptor, farnesoid X receptor, retinoid-related orphan receptor, estrogen-related receptor
Introduction
One way that the body responds to its chemical environment is by modulating the expression of xenobioticmetabolizing enzymes. Multiple enzyme systems that catalyze the metabolism of thousands of endogenous and xenobiotic molecules may be affected. Adaptation to dietary components and environmental contaminants may interface with the biotransformation of endogenous compounds to alter physiological processes ranging from hepatic metabolism to neural transmission.
Sulfate conjugation (sulfation or sulfonation) was first recognized as an important metabolic pathway by Bauman in 1876 (Jancova et al., 2010). The cytosolic sulfotransferases (SULTs) are a multigene family of enzymes that catalyze the transfer of a sulfonate group from the physiologic sulfate donor, 3′-phosphoadenosine-5′-phosphosulfate (PAPS), to a nucleophilic O-, N-, or S-substrate to generate a polar endproduct that is more amenable to elimination from the body (Strott, 2002). The SULTs are among a handful of conjugating enzyme systems that are expressed from the earliest periods of gestation onward in the developing human (Strassburg et al., 2002; Duanmu et al., 2006). The SULTs are widely expressed in hepatic tissue and in metabolically active or hormonally responsive extrahepatic tissues (Dooley et al., 2000; Gamage et al., 2006). These enzymes display a broad range of substrate specificities that include pharmaceuticals, procarcinogens, hormones, neurotransmitters, and intermediates of endogenous metabolism (Jancova et al., 2010). It is estimated that up to 80% of all human cancers may be related to lifestyle choices, such as diet, tobacco usage, obesity, and environmental chemical exposures (Gooderham et al., 2001; Sankpal et al., 2012). Environmental “xenoendocrine” agents, dietary constituents, and pollutants can co-opt cellular signaling pathways and reprogram gene expression in favor of carcinogenesis. In addition, unstable sulfate conjugates of procarcinogenic heterocyclic amines that are generated during the cooking of meat at high temperatures can produce reactive intermediates that are mutagenic to DNA (Lewis et al., 1998; Banoglu, 2000; Gooderham et al., 2001). These findings suggest that the combined influence of lifestyle and interindividual differences in SULT gene expression in cancer target tissues may have serious implications for the genesis of cancer in humans (Gooderham et al., 2001).
An active area of investigation is the regulation of SULT expression by the nuclear receptor (NR) superfamily of transcription factors, in particular, those receptors that recognize diverse xenobiotic and endogenous compounds to regulate the transcription of metabolic genes. This review addresses the developing role of the regulation of human and rodent SULTs by members of the NR family with the intention of shedding light on the role of SULT xenobiotic-metabolizing enzymes in normal physiology and in disease mechanisms.
The SULT gene superfamily
There are two principal categories of SULTs: the arylsul-fotransferase (SULT1) and the hydroxysteroid sulfotransferase (SULT2) gene families (Dooley et al., 2000; Glatt et al., 2001; Runge-Morris and Kocarek, 2009). There are five SULT1 subfamilies: SULT1A-E. SULT1A enzymes preferentially metabolize phenolic substrates, including drugs such as acetaminophen and troglitazone (Nagata and Yamazoe, 2000; Strott, 2002; Gamage et al., 2006; Runge-Morris and Kocarek, 2009). Humans express four SULT1A enzymes, including a high-affinity cate-cholamine-conjugating enzyme, SULT1A3/4, that is not present in rodents (Blanchard, 2005). Rodents express a single SULT1A enzyme that is most similar in activity to the human SULT1A1 phenol-sulfating enzyme (Falany et al., 1990; Duffel et al., 1991). Rodent SULT1B catalyzes the sulfation of 3,3′,5-triiodo-L-thyronine (T3), the active form of thyroid hormone (Rutgers et al., 1991; Runge-Morris and Kocarek, 2009), whereas a variety of SULT1 enzymes (SULTs 1A, 1B1, 1C2, 1C4, and 1E1) are capable of cata-lyzing iodothyronine sulfation in humans (Peeters et al., 2005). SULT1C enzymes bioactivate the procarcinogen, N-hydroxy-2-acetylaminofluorene, to DNA-damaging intermediates (Sakakibara et al., 1998; Runge-Morris and Kocarek, 2009). In contrast to humans, the hepatic levels of the SULT1C isoforms in rodents are much greater than they are in humans, resulting in significant species differences in sulfation activities (Nagata et al., 2005). Estrogen sulfotransferase (SULT1E1) displays a high affinity for estrogen (Falany et al., 1995b) and, because sulfonated estrogens do not activate the estrogen receptor, represents a major estrogen-inactivating enzyme in endocrine target tissues, such as the breast (Falany et al., 2002b). Rodent and canine species express an additional SULT1 enzyme termed SULT1D1, which has a role in the sulfation of small phenolic compounds (Tsoi et al., 2001; Nagata et al., 2005). In humans, SULT1D1 is a nonexpressed pseudogene at the same gene locus as SULT1E1 and SULT1B1 (Meinl and Glatt, 2001).
There are only two members of the human SULT2 family that are expressed in tissues. SULT2A1 is highly expressed in the liver and adrenal cortex, with lower levels of expression in the intestinal tract (Falany et al., 1989; Her et al., 1996; Rainey et al., 2002; Runge-Morris and Kocarek, 2009). SULT2B1b is expressed in tissues that do not express SULT2A1, including skin, brain, prostate, breast, and lung (Geese and Raftogianis, 2001). Although both human SULT2 enzymes conjugate 3ß-hydroxysteroids, including pregnenolone and dehydroepian-drosterone (DHEA), SULT2A1 has broader substrate reactivity, possibly associated with its role in xenobiotic and bile-acid sulfation in the liver. SULT2B1b has greater selectivity for 3ß-hydroxysteroid sulfation, although it also sulfates a number of xenobiotics. SULT2B1b does not sulfonate bile acids.
Less well-characterized SULTs include SULTs 3A1, 3A2, 4A1, 5A1, and 6B1, but only SULTs 4A1 and 6B1 have been detected in humans (Runge-Morris and Kocarek, 2009). SULT4A1 is highly conserved throughout vertebrates and is localized to neurons of the central nervous system (CNS) in humans and rats (Liyou et al., 2003). SULT6B1 has been detected primarily in human testes and kidneys; however, the mouse ortholog is more widely expressed and demonstrates sulfation activity with thyroxine (Takahashi et al., 2009). To date, the physiological functions of SULT4A1 and SULT6B1 are not known.
Species differences in SULT expression
Investigation of the regulation of SULT expression by NRs is complicated by both genetic and regulatory differences between the NRs in humans and laboratory species as well as between the SULT genes that are expressed. Significant differences in the SULT isoforms that are expressed exist between humans and the laboratory species that are primarily used to investigate in vivo regulation of SULT expression by NRs. To understand the role of NRs in SULT regulation, an appreciation of the species differences that exist between humans and rodents is required. Several of these differences have been mentioned briefly above. As another major example, in contrast to humans, which have only one SULT2A family member, SULT2A1, rodents express multiple SULT2A transcripts (Blanchard, 2005). Analysis of the mouse genome indicates the presence of seven SULT2A genes clustered on chromosome 7 (Kocarek et al., 2008). However, the kinetic properties and substrate reactivities of the individual enzymes have not been characterized. Studies of SULT2A regulation in mice are complicated by the specificity of the messenger RNA (mRNA) analysis, the lack of antibodies (Abs) to detect and discriminate the proteins, and the lack of information about substrates.
Structural differences, even between orthologous human and rodent SULTs, may have significant effects on regulation and function. The SULT2B1 family in humans generates two transcripts that, when expressed in Escherichia coli, give rise to two active enzymes with overlapping, but different, kinetic properties (Her et al., 1998; Fuda et al., 2002). However, only the longer SULT2B1b isoform has been detected in any human tissue examined. SULT2B1b has a greater affinity for cholesterol sulfation than does the expressed SULT2B1a isoform; this higher affinity is associated with the presence of the unique amino-terminal peptide (Fuda et al., 2002). SULT2B1b is structurally different from other SULT proteins by the presence of both amino- and carboxy-terminal extensions (Fuda et al., 2002). SULT2B1b is the first SULT to be identified as being partially localized in the nuclei of some tissues, including the placenta and breast (He and Falany, 2006; Dumas et al., 2008), and nuclear localization is associated with serine phosphorylation within the unique carboxy-terminal extension (Salman et al., 2011b). Serine phosphorylation has also been reported to stabilize SULT2B1b and increase its activity (Salman et al., 2011b).
Although the basic structure of the SULT2B isoforms is maintained between human and rodents, there are significant differences in the length and structure of both the amino- and carboxy-terminal ends that may affect regulation, localization, and activity. The unique carboxy-terminus of rodent SULT2B is approximately 50% shorter than the corresponding human sequence (Shimizu et al., 2003; Kohjitani et al., 2006), and there have been no reports of the phosphorylation of the carboxy-peptide or of any effects that the shortened peptide may impart to enzyme activity or localization. Human SULT2B1b has been difficult to characterize functionally, because the active native enzyme has not been isolated from tissues or cells (Falany et al., 2006). SULT2B1b activity can be detected in intact cultured breast cancer cells and isolated nuclei from placenta or breast cancer cells; however, SULT activity is lost upon lysis of the cells or nuclei (Falany et al., 2006). Therefore, the kinetic characterization of human SULT2B1b has been performed using expressed tagged enzyme.
Another major complication in the analysis of SULT regulation in rodents is the complex nature of the developmental and sex-related expression of the enzymes. In rodents, adult males have significantly higher levels of phenol SULT activity than do females, whereas females have higher levels of hydroxysteroid SULT activity. A careful analysis of the age- and sex-related expression of the murine SULT2A genes indicates different profiles of hepatic expression (Kocarek et al., 2008). For most of the SULTs, expression of the SULT1 and SULT2A genes in juvenile hepatic tissues is similar between males and females, and the changes in expression occur after sexual maturity. The association of the changes in SULT activity with the changes in SULT transcript expression pattern has not been extensively characterized. In contrast to rodents, sex-related differences in human SULT expression have not been reported in nongonadal tissues. Therefore, there is a significant sex-associated difference in the expression of the hepatic rodent SULTs that needs to be considered in the interpretation of regulatory studies.
Sulfotransferases in endogenous metabolism and regulation by NRs
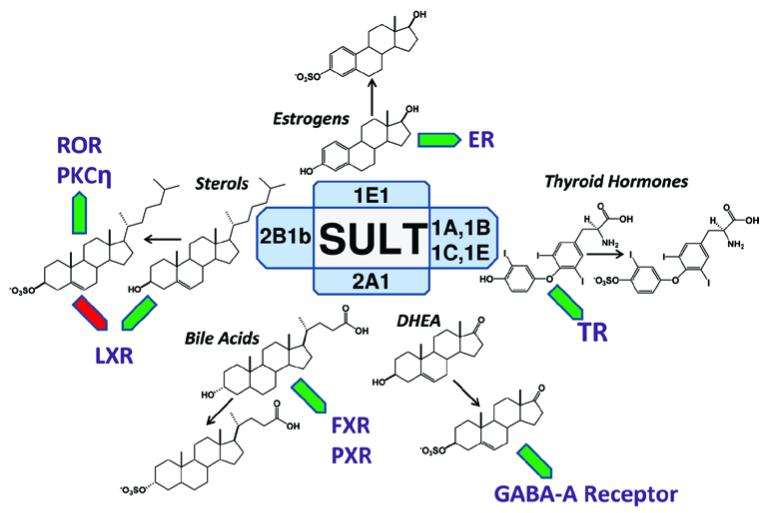
There is an increasing awareness of the role of sulfation in normal physiology and disease susceptibility. Sulfation of hormones and endogenous intermediates of metabolism may actually represent the lion’s share of SULT-catalyzed activity in humans when detoxification demands are under control. DHEA and its sulfonated metabolite, DHEA sulfate (DHEAS), are the most abundant circulating steroids in the human bloodstream (Bertoni et al., 2012). DHEAS, in synergy with nitrous oxide, inhibits platelet activation and aggregation, a result that would predictably affect atherogenic plaque formation and the progression of coronary artery disease in humans (Bertoni et al., 2012). The sulfation of thyroid hormone plays a critical role in energy metabolism. Sulfate conjugation of T3 both reduces its affinity for thyroid hormone receptors and accelerates its rate of degradation by type I deiodinase (Spaulding et al., 1992; Visser, 1996). The ectopic expression of SULT1E1 inhibits estrogen-dependent growth in the estrogen-receptor-positive MCF7 breast cancer cell line and underscores an important role for SULT1E1 in the regulation of in situ estrogen activity within the breast (Falany et al., 2002b). In the skin, estrogen operates to preserve epidermal thickness and hydration and also stimulates proliferation (Kushida et al., 2011). During keratinocyte differentiation, the expression of SULT1E1 is substantially upregulated and the proproliferative effects of estrogen action are held to a minimum (Kushida et al., 2011). Similarly, epidermal cholesterol sulfate produced by SULT2B1b is an amphipathic sulfolipid that is required at high concentrations in the terminally differentiated layers of skin, where SULT2B1b expression is also localized (Higashi et al., 2004). In the brain, neurosteroids DHEA and DHEAS are synthesized and modulate the activity of gamma-aminobutyric acid-A (GABA-A) receptors, suggesting a role for sulfonated neuroactive steroids in the pathogenesis of Parkinson’s disease and other CNS disorders where critical changes in GABAergic neurotransmission have been described (Luchetti et al., 2010; Gartside et al., 2010). Figure 1 summarizes several of the endogenous metabolic reactions where SULT catalysis can affect cell signaling.
Figure 1.
Examples of how SULT-catalyzed sulfation of endogenous molecules can affect cell-signaling processes. Green block arrows indicate activation, whereas red block arrows indicate inhibition.
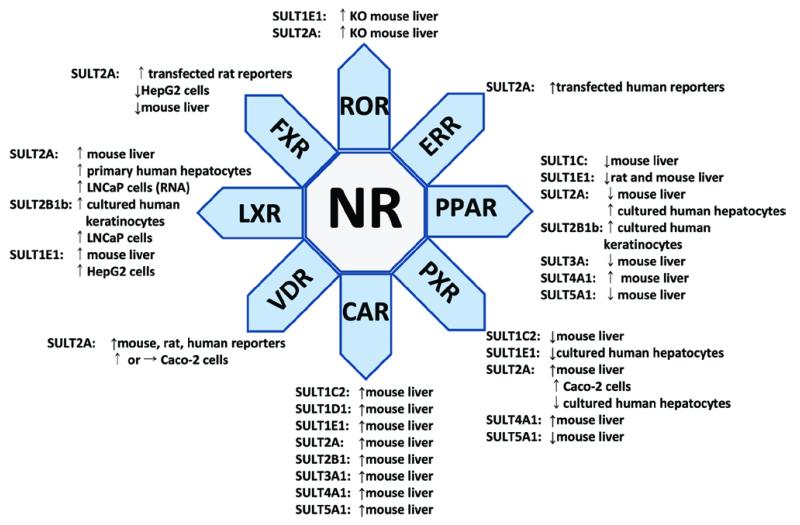
Functionally, the SULTs are emerging as dual integrators of both xenobiotic and endogenous metabolizing pathways. Therefore, it stands to reason that lipid- and xenobiotic-sensing NRs should play a major role in the transcriptional regulation of the cytosolic SULTs. A special class of “orphan” NR has emerged in recent years that, unlike steroid receptors, heterodimerize with the retinoid X receptor (RXR) and are activated either by an RXR ligand, such as 9-cis-retinoic acid, or by xenobiotics or metabolic intermediates that bind and activate the RXR partner receptor. Other orphan NRs, such as hepatocyte nuclear factor 4 alpha (HNF4-α) and retinoid-related orphan receptor (ROR), function as homodimeric or monomeric transcription factors and also figure prominently as regulators of lipid-metabolism networks (Giguere et al., 1994; Bogan et al., 2000; Lau et al., 2008; Yin et al., 2011). Modern clinicians are acutely aware of the blurred line between chronic metabolic disturbances, such as obesity-related diabetes, dyslipidemia, inflammation, and vascular dysfunction, and the pathogenesis of increasingly common clinical conditions, such as liver disease, renal insufficiency, atherosclerotic heart disease, and cancer. Figure 2 summarizes much of the information regarding NR regulation of SULT expression that will be described below.
Figure 2.
. Summary of regulation of SULT expression by NRs. The SULT genes and biological systems are indicated. For the murine SULTs, in many cases, there are sex differences in expression.
SULT regulation by peroxisome proliferator-activated receptors (PPARs)
The PPAR/RXR heterodimer transcription factor family regulates gene-expression networks involved in energy balance, lipid metabolism, and inflammation (Runge-Morris and Kocarek, 2009). PPAR-α is primarily expressed in liver, kidney, and heart tissues and is ligand activated by a major energy source, fatty acids (Runge-Morris and Kocarek, 2009; Robinson and Grieve, 2009), as well as by oxidized fatty-acid intermediates that may serve as signaling molecules (Shiraki et al., 2005; Runge-Morris and Kocarek, 2009). PPAR-γ is highly expressed in adipose tissue and has been demonstrated to regulate adipocyte differentiation (Spiegelman et al., 1997; Runge-Morris and Kocarek, 2009; Robinson and Grieve, 2009), whereas PPAR-δ/β is ubiquitously expressed and regulates diverse gene-expression networks ranging from insulin resistance to embryo implantation (Runge-Morris and Kocarek, 2009; Robinson and Grieve, 2009).
It was previously reported that rats treated with PPAR-α agonists (WY-14,643, gemfibrozil, or di-n-butylphthalate) demonstrated a pronounced suppression of hepatic SULT1E, which is normally robustly expressed in male rat liver (Fan et al., 2004). By contrast, Alnoutis and Klaassen conducted a large study in mice examining the effects of treatment with PPAR-α activators on hepatic SULT expression (Alnouti and Klaassen, 2008). Male and female 8-week-old mice were treated intraperitoneally (i.p.) for 4 days with corn-oil vehicle or a PPAR-α activator, clofibrate, ciprofibrate, or bis(2-ethylhexyl) phthalate (Alnouti and Klaassen, 2008). Overall, PPAR-α agonist treatment had little effect on male mouse hepatic SULT expression, but in female mouse liver, clofibrate treatment suppressed the mRNA levels of several of the SULTs, including the Sult1c, Sult1e, Sult2a, Sult3a, and Sult5a family members, whereas Sult4a1 mRNA content was increased (Alnouti and Klaassen, 2008). In a later study, Aleksunes and Klaassen (2012) examined effects on xenobiotic-metabolizing enzyme and transporter expression in male and female 8-week-old wild-type (WT) mice and in mice with nullified expression of the aryl hydrocarbon receptor (AhR), nuclear factor erythroid 2-related factor 2 (Nrf2), constitutive androstane receptor (CAR), pregnane X receptor (PXR), or PPAR-α. Treatment effects on murine hepatic SULTs and on the sulfate supply enzyme, PAPS synthase 2 (PAPSS2), were quantified using panels of branched DNA-signal amplification assays (Aleksunes and Klaassen, 2012). To examine the regulatory effects of PPAR-α, mice were treated with corn-oil vehicle or 500 mg/kg of clofibrate i.p. for 4 days (Aleksunes and Klaassen, 2012). Clofibrate treatment effects were not striking in PPAR-α-null mice (Aleksunes and Klaassen, 2012). In WT mice, clofibrate treatment increased hepatic PAPSS2 mRNA content by ~1.4-fold in male mice and suppressed SULT5a1 mRNA levels by 42% in female mice.
Our laboratory reported that human, unlike rat, hepatic SULT2A expression is induced by a PPAR-α-mediated mechanism (Fang et al., 2005). Treatment of primary cultured human hepatocytes with ciprofibrate produced a ~2-fold increase in SULT2A1 mRNA, protein, and enzyme activity, whereas ciprofibrate treatment had no effect on SULT2A expression in primary cultured rat hepatocytes (Fang et al., 2005). Computational and deletion analyses performed on SULT2A1 5′-flanking region reporter gene constructs identified two putative PPAR response elements (PPREs) (Fang et al., 2005). Site-directed mutagenesis, electrophoretic mobility shift analysis (EMSA), and chromatin immunoprecipitation (ChIP) revealed that the more-distal PPRE, a direct repeat with one intervening nucleotide (DR1 motif) located at nucleotides −5949 to −5929 relative to the SULT2A1 transcription start site, was a functional PPRE (Fang et al., 2005). Overall, these results suggest a role for the lipid-sensing PPAR-α transcription factor in the transcriptional upregulation of human hepatic SULT2A1 gene transcription, and indicate that there are important species differences that govern SULT2A gene regulation in response to PPAR-α-activating lipid intermediates.
Cholesterol and cholesterol sulfate are important components of dermal lipid homeostasis and skin-barrier function. As keratinocytes move upward through differentiating layers of epithelium, they produce increasing amounts of cholesterol-3-sulfate (Lampe et al., 1983). Cholesterol sulfate is not only a component of the epidermal barrier, but it also functions as a signaling molecule that promotes keratinocyte differentiation through activation of the novel protein kinase C (PKC) isoform, PKCη (Kashiwagi et al., 2002). In the stratum corneum, cholesterol sulfate is hydrolyzed to free cholesterol by steroid sulfatase (Lampe et al., 1983; Epstein et al., 1984). SULT2B1b catalyzes the sulfation of cholesterol with high affinity (Lee et al., 2003) and is expressed in differentiating human keratinocytes where the concentrations of cholesterol-3-sulfate are the highest (Higashi et al., 2004). Given the role of SULT2B1b in the dermal lipid cycle, it stands to reason that SULT2B1b should be a target for transcriptional regulation by lipid-sensing transcription factors in the skin. SULT2B1b mRNA, protein, and enzyme activity increased with calcium-induced differentiation in keratinocytes (Higashi et al., 2004; Jiang et al., 2005). The levels of PPAR-γ, liver X receptor (LXR)-α, and LXR-β mRNA increased with calcium-induced differentiation in cultured human keratinocytes, whereas the amounts of PPAR-α and PPAR-δ/β did not (Jiang et al., 2005). Treatment of cultured human keratinocytes with activators of PPAR-α, PPAR-δ/β, PPAR-γ, and LXR significantly increased the expression of SULT2B1b mRNA and enzyme activity (Jiang et al., 2005). These findings suggest that in the differentiating epidermis, SULT2B1b transcription becomes temporally transactivated by PPAR and LXR transcription factors responding to local changes in intradermal lipid homeostasis.
SULT regulation by PXR
PXR was first described by Kliewer et al. in 1998 (Kliewer et al., 1998). The discovery of PXR originated from an expressed sequence tag database search aimed at identifying complementary DNA (cDNA) sequences that displayed homology to the ligand-binding domains (LBDs) of known NRs (Kliewer et al., 1998). This partial sequence information was used to isolate two cDNA clones (PXR.1 and its splice variant, PXR.2) from a mouse cDNA library (Kliewer et al., 1998). A survey of PXR expression in adult and fetal mouse tissues demonstrated robust expression of PXR in the liver and intestine, with weaker expression in the kidney and stomach (Kliewer et al., 1998). The complete PXR transcription factor is a heterodimer with RXR that binds to a response element consisting of a repeat (direct, inverted, or everted) of AG(G/T)TCA, recruits coactivators, and transactivates target genes, such as cytochrome P450 (CYP)3A family members (Kliewer et al., 1998). To identify potential PXR ligands, PXR LBDs were tested in a chimeric transactivation assay performed in CV-1 cells (Kliewer et al., 1998). PXR is known as a “promiscuous but selective” NR that can be ligand activated by a diverse range of compounds, including steroid hormones such as the C21 steroids, pregnenolone and progesterone, synthetic glucocorticoids, antiglucocorticoids, bile acids, herbal remedies, vitamins, and xenobiotics, including pharmaceuticals and environmental chemicals, such as polychlorinated biphenyls and phthalates (Kliewer et al., 1998; Ngan et al., 2009). The diversity of PXR ligands is conferred by PXR’s large spherical hydrophobic ligand-binding pocket that has five “hot spot” regions of amino-acid residues that interact with ligands (Watkins et al., 2001; Ngan et al., 2009). These hot-spot residues are conserved across species and also facilitate the structural rearrangements that are required for the receptor’s ability to accommodate a diverse array of ligands (Ngan et al., 2009).
In an investigation of PXR-null mice, the basal hepatic expression of the PXR target gene, Cyp3a11, was not changed, but Cyp3a11 induction in response to the prototypical PXR ligands, pregnenolone-16α-carbonitrile (PCN) and dexamethasone (DEX), was abolished (Xie et al., 2000). The rodent and human PXRs differ with respect to their ligand-activation profiles in that rodent, but not human, PXR is activated by PCN and human, but not rodent, PXR is activated by rifampicin (RIF) (Lehmann et al., 1998). Transgenic (Tg) mice that were genetically engineered to express constitutively active human PXR in liver displayed increased hepatic Cyp3a11 expression and were resistant to drug-induced anesthesia as a consequence of increased drug metabolism (Xie et al., 2000).
The livers of mice treated with PCN to activate PXR were surveyed for the presence of novel PXR target genes (Willson et al., 2001). In addition to Cyp3a induction, the expression of murine hepatic Cyp7a, the rate-limiting step in bile-acid synthesis, was downregulated in response to PXR activation and the bile-acid transporter, organic anion-transporting polypeptide 2 (OATP2) was upregulated (Willson et al., 2001). The hepatotoxic secondary bile-acid, lithocholic acid (LCA), and its 3-keto metabolite are responsible for producing intrahepatic cholestasis, and both of these compounds bind and activate PXR (Willson et al., 2001; Staudinger et al., 2001). Therefore, as an inherent defense mechanism against cholestatic liver damage, LCA activates PXR and stimulates its own detoxification and elimination through the induction of CYP3A-mediated hydroxylation, the inhibition of further bile-acid synthesis, and the induction of bile-acid-transporter expression that reduces the hepatic concentration of LCA (Xie et al., 2000). This was demonstrated experimentally when WT or PXR-null mice were treated i.p. with corn-oil vehicle, PCN, or LCA either alone or in combination (Staudinger et al., 2001). In this study, mice with preserved or augmented hepatic PXR expression activated gene-expression programs that were protective against the hepatotoxic detergent effects of LCA (Staudinger et al., 2001). However, in another study, when PXR-null or WT mice were treated with LCA, either through dietary supplementation or i.p. injection, PXR-null mice, compared to WT, were found to be more resistant to LCA-induced hepatotoxicity (Owen et al., 2010). In WT mice, hepatic SULT2A mRNA expression was repressed by LCA feeding, but increased by i.p. injection, a liver-targeted route of administration that bypasses the gut (Owen et al., 2010). In PXR-null mice, hepatic SULT2A expression was not induced by either dietary or i.p. LCA, but both basal and LCA-modulated expression of PAPSS2, one of the major sulfate donor enzymes in the liver, was ~2-fold higher in PXR-null, relative to WT, mice, suggesting that the increased expression of this critical sulfate supply enzyme could account for the enhanced ability of PXR-null mice to detoxify LCA and avert hepatotoxicity (Owen et al., 2010); however, the levels of PAPS within the liver were not reported on. In aggregate, these results suggest that murine hepatic drug-metabolizing enzyme expression, which is orchestrated by bile-acid-sensing orphan NRs, is a protective measure against the hepatotoxic detergent effects of bile acids (Kitada et al., 2003).
To ascertain whether murine and human “anticholestatic” defense mechanisms are similar, patients with primary biliary cirrhosis (PBC) were examined for their metabolic adaptive responses to cholestasis, including their capacity to repress basolateral bile-acid uptake in the liver, block bile-acid synthesis, and induce bile-acid metabolism and transport (Zollner et al., 2007). PBC is a progressive metabolic disease typified by the fibrotic destruction of the intrahepatic bile ducts driven by an immune-mediated activation of hepatic stellate cells by inflammatory cytokines (Tanaka et al., 2012). Elevated levels of hydroxylated, sulfonated, and glucuronidated bile acids have been measured in the urine of patients with PBC (Frohling and Stiehl, 1976; Stiehl et al., 1980; Shoda et al., 1990; Zollner et al., 2007). In patients with PBC, the mRNA expression of hepatic CYP7A1, the rate-limiting step in bile-acid synthesis, was markedly diminished, whereas the expression of alternative bile-acid synthesis pathway enzymes (CYP27A1 and CYP8B1) was not changed (Zollner et al., 2007). The expression of bile-acid-metabolizing enzymes CYP3A4, SULT2A1, UDP glucuronosyltransferase (UGT)2B4, and UGT2B7 was either unchanged or moderately reduced in patients with PBC, but the expression of the basolateral bile-acid export transporter, multidrug-resistance protein 4 (MRP4) protein, was induced in patients with PBC (Zollner et al., 2007). The expression of orphan and liver-enriched NRs involved in the regulation of bile-acid metabolism and transport, such as PXR, farnesoid X receptor (FXR), CAR, RXR, and HNF4-α, was either unchanged or markedly reduced in livers of patients with PBC, suggesting that drug-metabolism responses in certain cases of bile-acid overload may be impaired (Zollner et al., 2007). These results suggest that there are conserved responses to bile-acid overload that occur across species and indicate that NR-mediated regulation of bile-acid synthesis, detoxification, and disposition may represent novel therapeutic targets in the management of patients with cholestatic liver disease.
Chronic and acute inflammation affects the course of metabolic disease progression and also influences NR activation. The acute-phase inflammatory response has been associated with the suppression of a panel of NRs involved in SULT gene regulation, including CAR, FXR, RXR, and PXR (Kim et al., 2004). Patients with chronic inflammatory diseases, such as polymyalgia rheumatica and primary fibromyalgia, display decreased levels of circulating DHEAS, suggesting that this sign may serve as a valuable biomarker of disease (Nilsson et al., 1994; Kim et al., 2004). To determine whether there may be a linkage between DHEAS in the bloodstream and NR-mediated changes in hepatic SULT2A1 expression, mice with inherently high DHEAS levels were treated with lipopolysac-charide (LPS) to induce an acute-phase response (Kim et al., 2004). Eight-week-old female mice were injected i.p. with saline vehicle, 100 μg of LPS, or 40 mg/kg of PCN for 3 days, followed by LPS, and effects on hepatic Sult2a expression and serum DHEA and DHEAS concentrations were determined (Kim et al., 2004). Beginning at 4 hours and extending to 24 hours from the time of LPS administration, hepatic Sult2a1 mRNA and activity levels were suppressed and circulating levels of DHEAS were reduced (Kim et al., 2004). LPS administration also suppressed hepatic PAPSS2 expression and PCN-inducible Sult2a transcription (Kim et al., 2004). The treatment of cultured Hep3B human hepatoma cells with tumor necrosis factor or interleukin-1 inflammatory cytokines also decreased the amount of SULT2A1 mRNA, although the precise mechanism underlying this effect was not characterized. These results attest to the understudied role of “liver immunology” (Tanaka et al., 2012) and inflammatory responses as mediators of NR-regulated pathways that govern the transcription of SULTs and other target drug-metabolizing enzyme genes important to drug-drug interactions and disease susceptibility in humans.
Two surveys conducted by Alnouti and Klaassen were focused on the role of NRs in the transcriptional regulation of xenobiotic-metabolizing enzymes, including the SULTs and the PAPS synthase (PAPSS) sulfate supply enzymes (Alnouti and Klaassen, 2008, 2012). In the first study, 8-week-old male C57BL/6 mice were treated for 4 days with either vehicle control or a panel of PXR activators, including 200 mg/kg of PCN, 200 mg/kg of spirono-lactone, or 75 mg/kg of DEX (Alnouti and Klaassen, 2008). In general, these treatments produced few consistent changes in hepatic SULT expression in male mice. PCN treatment did produce a significant increase in Sult1e1 mRNA content, whereas DEX treatment produced a much larger increase; however, it is possible that this latter effect was not PXR mediated (Alnouti and Klaassen, 2008). In female mice, only the effects of PCN treatment were evaluated. PCN treatment increased the hepatic levels of Sult2a1/2a2 and Sult4a1 mRNA and decreased Sult1c2 and Sult5a1 mRNA levels. PXR activator treatments increased hepatic PAPSS2 mRNA levels in both male and female mice (Alnouti and Klaassen, 2008). In the second study, WT and PXR-null mice were utilized to test the specificity of PXR activation on SULT and PAPSS2 expression (Aleksunes and Klaassen, 2012). In this study, hepatic levels of Sult2a2, Sult3a, and PAPSS2 mRNA were significantly higher in the livers of female PXR-null mice than they were in female WT mice (Aleksunes and Klaassen, 2012). Treatment with PCN increased hepatic Sult2a2 and PAPSS2 expression in female WT mice (Aleksunes and Klaassen, 2012).
In 2004, Echchgadda et al. investigated the role of PXR in the transactivation of murine hepatic gene transcription. Male C57 black mice were injected i.p. with dimethyl sulfoxide or PCN and sacrificed 24 hours later (Echchgadda et al., 2004a). Quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) revealed that Sult2a1 mRNA content was 4-fold higher in PCN-treated mice than in controls (Echchgadda et al., 2004a). Transient transfections of murine Sult2a1 reporter constructs in HepG2 cells, complemented by DNase I footprinting and EMSA analyses, indicated that PXR transactivates murine hepatic Sult2a1 gene transcription at an IR0 (inverted repeat with zero intervening nucleotides) motif located in the proximal 5′-flanking region of the Sult2a1 gene (Echchgadda et al., 2004a). Murine hepatic Sult2a has long been known as an “androgen-repressible” gene that demonstrates increases in mRNA expression with the decline of hepatic androgen receptor levels in the livers of aging male mice (Echchgadda et al., 2004a). In Echchgadda et al.’s study, the expression of neither murine hepatic PXR nor RXR mRNA changed as male mice aged from 8 to 24 months, suggesting that the PXR transcription factor does not play a significant role in the age-related expression of this gene (Echchgadda et al., 2004a).
Unlike rodent SULT2A genes, human SULT2A1 does not contain an IR0 motif that acts as a cis-acting control node for PXR-mediated transcription. Echchgadda et al. treated the human intestinal cell line (Caco-2 cells) with activators of either PXR or CAR and found that SULT2A1 was induced by ~1.7-fold and by 3.5-fold in response to PXR and CAR activation, respectively (Echchgadda et al., 2007). DNase I footprinting and ChIP analyses revealed the presence of a composite cis-acting response element in the 5′-flanking region of the human SULT2A1 gene that contained an IR2 (inverted repeat with 2 intervening bases) and an adjacent DR4 (direct repeat with four intervening bases) motif that bound to both CAR and PXR and also included a proximal HNF4-α-binding site (Echchgadda et al., 2007). Cotransfection of Caco-2 cells with SULT2A1 reporters and expression constructs for PXR, CAR, and HNF4-α produced marked increases in PXR- or CAR-inducible SULT2A1 transcription, suggesting an interaction between the PXR, CAR, and HNF4-α NRs in the regulation of inducible SULT2A1 gene transcription in intestinal cells (Echchgadda et al., 2007).
In primary cultured human hepatocytes, regulation of SULT2A1 gene transcription in response to PXR activation proved to be complex. Primary cultures of human hepatocytes isolated from 23 different liver donors were treated with RIF to activate PXR (Fang et al., 2007). In all cases, CYP3A4, a gene that is known to be transactivated by PXR, demonstrated increased mRNA levels in response to RIF treatment, although the magnitude of induction varied considerably among individual donors (Fang et al., 2007). By contrast, SULT2A1 mRNA expression in response to RIF treatment ranged from modest induction to no change or frank suppression, in comparison to vehicle-treated controls (Fang et al., 2007). Two RIF-responsive regions were identified in the 5′-flanking region of the human SULT2A1 gene, one proximal and one distal, and both of these cis-acting elements bound avidly to HNF4-α, but not to PXR/RXR (Fang et al., 2007). Cotransfection of HepG2 cells with additional PXR increased RIF-inducible CYP3A4 reporter gene expression (positive control), but suppressed SULT2A1 reporter gene transcription (Fang et al., 2007). The results suggest that the HNF4-α NR plays a positive regulatory role in the control of SULT2A1 transcription under basal conditions, but in the presence of RIF activation of PXR, the positive influence of HNF4-α on SULT2A1 transcription is disrupted, either through a physical interaction between the two transcription factors or a repositioning of essential coactivators, and a net suppressive effect on human hepatic SULT2A1 transcription is produced (Fang et al., 2007).
PXR activation by RIF also produces a suppressive effect on human hepatic SULT1E1 transcription through a mechanism that involves HNF4-α (Kodama et al., 2011). SULT1E1 catalyzes the sulfation and inactivation of estrogens and has been shown to play a role in hepatic energy homeostasis (Kodama et al., 2011). In the 1990s, the treatment of female mice carrying the Avy mutation with DEX was observed to produce hyperin-sulinemia, hyperglycemia, induced Sult1e1 expression, and a shift in hepatic sex-hormone balance toward an androgenized liver environment (Gill et al., 1994). Similarly, hepatic Sult1e1 was induced and Sult2a was reciprocally repressed in the livers of diabetes-susceptible C57BL/KsJ mice with a recessive obesity mutation in the diabetes gene (db), a condition that creates an androgenized hepatic microenvironment and obesity-related diabetes (Leiter and Chapman, 1994). More recently, the Tg expression of SULT1E1 in murine adipose tissue has been demonstrated to regulate fat and glucose energy balance (Khor et al., 2010; Kodama et al., 2011). PXR activation by RIF represses human hepatic SULT1E1 transcription through a mechanism that does not involve the direct binding of the PXR transcription factor to the SULT1E1 5′-flankng region (Kodama et al., 2011). Deletion analyses revealed the presence of a 100-nucleotide RIF-responsive region in the SULT1E1 5′-flanking region that is localized to a distal enhancer element (−1000/−901) containing overlapping DR1 and DR2 motifs and two forkhead factor recognition sites (Kodama et al., 2011). As with SULT2A1 (Fang et al., 2007), the RIF-responsive regions in the SULT1E1 gene bind primarily to HNF4-α (Kodama et al., 2011). In the transcriptional regulation of human hepatic SULT1E1, HNF4-α functionally loops the distal 100-nucleotide enhancer region toward the proximal promoter region of SULT1E1 to activate SULT1E1 transcription (Kodama et al., 2011). Ligand binding of RIF to PXR disrupts the looped structure of the SULT1E1 promoter region, histone 3 is deacetylated, and SULT1E1 gene transcription is repressed, thus tipping the hepatocellular androgen/ estrogen balance toward greater estrogenicity (Kodama et al., 2011).
SULT regulation by CAR
In 1994, Baes et al. identified MB67 as a human orphan NR that constitutively transactivated a subset of retinoic acid response elements (RAREs) as a heterodimer with RXR when transfected into cell lines (Baes et al., 1994). A closely related murine receptor was then identified and named mCAR in recognition of its ability to cause constitutive activation of RAREs, and MB67 was renamed hCAR (Choi et al., 1997). After 3α-hydroxy, 5α-reduced androstanes, such as 5α-androstan-3α-ol and 5α-androst-16-ene-3α-ol, were subsequently found to inhibit the constitutive activity of CAR, the acronym was redesignated to stand for constitutive androstane receptor (Forman et al., 1998). CAR’s role as a “xenosensor” and an important regulator of xenobiotic-metabolizing enzyme transcription was first demonstrated by Honkakoski et al., who found that hepatic nuclear extracts from phenobarbital (PB)-treated mice showed enriched binding of RXR and CAR to an affinity probe corresponding to a DR4 motif within the PB-responsive enhancer module of the Cyp2b10 gene (Honkakoski et al., 1998). These investigators subsequently showed that mCAR could mediate the induction of CYP2B6 by PB and PB-type inducers in HepG2 cells if the constitutive activity of CAR was first repressed by androstane treatment (Sueyoshi et al., 1999). The CAR transcription factor transactivates not only target genes involved in xenobiotic detoxification, such as CYP2B, but also genes involved in the detoxification of endogenous metabolites, such as bilirubin and bile acids, and genes involved in apoptosis and endocrine, glucose, and lipid metabolism (Kachaylo et al., 2011). Unlike other NRs where ligand binding prompts the recruitment of coactivators, the CAR LBD’s activation function-2 is in a constitutively active position (Kachaylo et al., 2011). Treatment with either a CAR ligand agonist or a nonligand activator, such as PB, causes cytoplasmic CAR to undergo dephosphorylation, disengage from a complex of chaperone proteins, and translocate into the nucleus, where it heterodimerizes with RXR and binds to cis-acting response elements in CAR target genes (Kachaylo et al., 2011).
In the study by Alnouti and Klaassen that has been described above, to evaluate the effect of CAR activation on mouse hepatic SULT expression, 8-week-old male C57BL/6 mice were treated for 4 days with either vehicle control or a panel of CAR activators, including 300 μg/kg of 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP), 200 mg/kg of diallyl sulfide (DAS), or 100 mg/kg of PB (Alnouti and Klaassen, 2008). Female mice were treated with TCPOBOP only. In general, the CAR activators were found to induce the mRNA expression of several hepatic SULTs and PAPSS2, an effect that was more prominent in female, relative to male, mice (Alnouti and Klaassen, 2008). The specificity of inducer treatments represented a concern in this study, because treatment with DAS induced murine hepatic Sult3a1 expression in male mice, but treatment with the other CAR activators failed to alter Sult3a1 expression (Alnouti and Klaassen, 2008). Treatment of female mice with TCPOBOP increased the expression of several hepatic SULTs, including Sult1c2, Sult1d1, Sult1e1, Sult2a1/2a2, Sult2b1, Sult3a1, and Sult4a1 (Alnouti and Klaassen, 2008). Murine hepatic PAPSS2 mRNA content was increased by TCPOBOP treatment in female, but not in male, mice (Alnouti and Klaassen, 2008), suggesting that gender differences contribute to CAR-mediated regulation of hepatic SULTs and the sulfate supply enzyme, PAPSS2, in mice (Alnouti and Klaassen, 2008). In a second study, WT and CAR-null mice were utilized to test the specificity of CAR activation on SULT and PAPSS2 expression (Aleksunes and Klaassen, 2012). Treatment with TCPOBOP increased the expression of PAPSS2 and all SULTs examined, including Sult1e1, Sult2a2, Sult3a1, and Sult5a1 (Aleksunes and Klaassen, 2012).
In 2004, Assem et al. reported that CAR is required for the coordinate regulation of hepatic SULT2A and the ABC transporter, MRP4 (Assem et al., 2004). Hepatic SULT2A in rodents has long been known to be “androgen repressible” at the level of transcription (Song et al., 1998), and MRP4 knockout (KO) mice were observed to have reduced SULT2A expression in the liver, presumably resulting from the build-up of androgenic steroids that would normally have been transported out of the liver by MRP4 (Assem et al., 2004). In hepatic failure, intrahepatic cholestasis, or FXR KO mice that display intrahepatic cholestasis resulting from bile duct ligation, toxic bile acids accumulate and expression of hepatic MRP4 is markedly induced (Assem et al., 2004). In support of the hypothesis that mouse hepatic MRP4 and SULT2A are coordinately induced by CAR as a defensive measure to prevent the build-up of toxic bile acids, Assem et al. showed that both MRP4 and Sult2a expression are induced in the livers of mice treated with TCPOBOP, an effect that is ablated in CAR-null mice (Assem et al., 2004). The stable expression of CAR in Hepa1c1c7 mouse hepatoma cells increased the expression of MRP4 and Sult2a (Assem et al., 2004), suggesting that these gene products work together to sulfonate bile acids, making them more polar, and actively export them out of the liver to avoid the detergent effects associated with bile-acid toxicity. In another study, Tg mice expressing a constitutively active CAR (VP-CAR) in the liver were resistant to the hepatotoxic effects of LCA and had increased hepatic Sult2a and PAPSS2 expression (Saini et al., 2004). There are seven murine SULT2A genes (Kocarek et al., 2008). In EMSA analyses, the CAR transcription factor bound to an IR0 motif located in the proximal promoter region of a gene the investigators termed Sult2a9 (Saini et al., 2004); this gene appears to be equivalent to the gene that has been named Sult2a3 by the National Center for Biotechnology Information.
Several xenobiotic CAR modulators have been reported to regulate SULT expression in animal models with downstream effects on hormone metabolism. For example, xanthohumol, a flavonoid from the hop plant, suppressed CAR expression in rat liver and was associated with the repression of CAR target genes involved in thyroid hormone metabolism, including SULT1A1, UGT1A1, and CYP2B15 (Radovic et al., 2010). Sulfation accelerates thyroid hormone deiodination and inactivation (Rutgers et al., 1991), and when liver tumor promoter-susceptible mice were treated with PB, hepatic Sult2a1 mRNA content was increased by 260-fold and serum free thyroxine levels were significantly decreased (Pakharukova et al., 2010). In 2011, Sueyoshi et al. investigated the effects of DAS, a constituent of garlic extract that has been observed to induce xenobiotic-metabolizing enzymes much the same as PB (Sueyoshi et al., 2011). DAS-treated WT mice demonstrated striking increases in hepatic SULT1E1 mRNA, protein, and activity, and these effects were all but eliminated in CAR-null mice (Sueyoshi et al., 2011). In this study, prototypical CAR targets genes, such as CYP2b10 and CYP3a11, were also induced by DAS in WT, but not CAR-null, mice (Sueyoshi et al., 2011). The time course for DAS-inducible SULT1E1 expression was more gradual, spanning over 48 hours, compared to that observed for CYP2b10, which displayed robust induction within 6 hours of DAS treatment (Sueyoshi et al., 2011). Ironically, in this study, serum estrogen levels, as a systemic measure of SULT1E1 estrogen-inactivating effects, were slightly elevated in DAS-treated, relative to control, mice and estrone sulfate levels were unchanged (Sueyoshi et al., 2011). These results suggested that despite a large induction of the estrogen-inactivating enzyme, SULT1E1, in mouse liver after DAS treatment, the effect on circulating estrogen levels was minimal, indicating that SULT1E1 may target alternative endogenous substrates in liver (Sueyoshi et al., 2011). Changes in the circulating levels of estrogens were also not observed in Sult1e1 KO mice (Qian et al., 2001).
The plasticizer, di-n-butyl phthalate (DBP), activates both PXR and CAR in in vitro transcription assays and, in vivo, causes reproductive abnormalities, increased liver weight, and liver lesions in rodent models (Wyde et al., 2005). In pregnant rats treated with high doses of DBP (50-500 mg/kg), hepatic SULT1E1 mRNA content was increased by ~2- to 3-fold in the dams, but not the fetuses (Wyde et al., 2005). CAR has also been implicated in the inducible regulation of murine hepatic Sult1a1 that occurs in response to oral gavage treatment with the toxic halogenated chemical byproduct, octachlorostyrene (OCS), a compound with structural similarities to AhR ligands (Yanagiba et al., 2009). OCS treatment induced murine hepatic SULT1E1 mRNA expression in AhR-null mice and, in EMSA analysis, increased CAR binding to a prototypical PB response element, suggesting that OCS is a xenobiotic CAR activator (Yanagiba et al., 2009). Overall, there is evidence, in rodent systems, that the xenosensor CAR is a modulator of SULT gene expression, but more in-depth analyses are required to define the role of CAR as a regulator of SULT gene transcription in humans.
The role of CAR in cellular energy metabolism has revealed the integrated role of SULTs and other xeno-biotic-metabolizing enzymes in the control of energy balance and signaling pathways leading to obesity. CAR-null mice are resistant to weight loss prompted by calorie restriction (Ding et al., 2006). In mice, fasting increases cyclic adenosine monophosphate and also induces the expression of CAR, NR PPAR-γ coactivator 1 alpha (PCG1-α), and CAR target genes, including Cyp2b10, Ugt1a1, Sult2a1, and the Oatp2 transporter (Ding et al., 2006). HNF4-α directly transactivates CAR transcription, and in CAR reporter gene assays, cotransfection of PGC1-α and HNF4-α produced an augmented effect on CAR transcription (Ding et al., 2006). Fasted mice demonstrated increased Sult2a1 expression in the liver, but this effect was blunted in the livers of HNF4-α-null mice, indicating an essential interaction between CAR and HNF4-α in the induction of Sult2a1 (Ding et al., 2006). By contrast, the mRNA expression of CAR and PXR, but not RXR, was reduced in CD-1 mice fed a high-fat diet (60% kcal fat), relative to low-fat-diet-fed mice (10% kcal fat), and hepatic Sult1a1 expression was reduced in the high-fat-diet-fed mice, which also displayed a general suppression of CAR target genes (Ghose et al., 2011). Overall, these studies on energy metabolism in rodent models suggest that CAR regulates SULT enzymes that catalyze steps in energy homeostasis through their effects on thyroid hormone metabolism and lipid balance.
SULT regulation by the vitamin D receptor (VDR)
The VDR/RXR transcription factor regulates calcium homeostasis in the kidney, bone, and intestine and is also expressed in the liver (Echchgadda et al., 2004b). VDR is activated by its prototypic ligand, 1α,25-dihydroxyvitamin D3 (VitD3), the biologically active form of vitamin D. However, vitamin D analogs (Fan et al., 2009) and the hepatotoxic secondary bile-acid LCA (Makishima et al., 2002; Echchgadda et al., 2004b) have also been described as VDR ligands. Echchgadda et al. reported that VitD3-treated HepG2 cells cotransfected with VDR and RXR had increased SULT2A1 mRNA and protein levels, thus suggesting a role for VDR in the transactivation of human hepatic SULT2A1 (Echchgadda et al., 2004b). To examine the potential for species differences in gene regulation, reporter constructs for human, mouse, and rat SULT2A genes were transfected into HepG2 or Caco-2 cells that were supplemented with exogenous VDR and treated with VitD3 (Echchgadda et al., 2004b). These treatments were associated with the induction of SULT2A reporter gene expression across species lines, and the effect appeared to be dependent upon the presence of cotransfected VDR (Echchgadda et al., 2004b). Further DNaseI footprinting and EMSA analyses revealed that VDR/RXR binds to the same IR0 motif in murine and rat SULT2A that also binds to PXR and FXR (Echchgadda et al., 2004b). Overall, because the VDR, FXR, and PXR NRs can all undergo activation by bile-acid intermediates, these studies suggest an integrated role for bile-acid-sensing NR pathways in the transcriptional regulation of SULT2A. In a later study, Song et al. reported that VitD3-inducible human SULT2A1 transcription is activated by VDR/RXR through a mechanism involving a “composite” transcription factor complex (Song et al., 2006). This mechanism involves the binding of VDR/RXR to an IR2 motif located at nucleotides -184 to -171 of the SULT2A1 5′-flanking sequence and also requires the binding of CAAT/enhancer-binding protein to a site nine nucleotides downstream of the IR2 (Song et al., 2006). These investigators showed that VitD3-inducible SULT2A1 reporter gene transcription could occur in HepG2 cells and also demonstrated, by ChIP, that VDR and RXR were recruited to VitD3-responsive regions in SULT2A1 in Caco-2 cells (Song et al., 2006). The Caco-2 cell line differentiates in cell culture to enterocytes and is used as a model for the study of xenobiotic-metabolizing enzymes and transporters in the intestine (Song et al., 2006). Because the metabolic conversion of the precursor to active VitD3 occurs both in the liver and intestine, the Caco-2 cell line was used by Fan et al. to characterize VitD3 target genes in human intestinal cells (Fan et al., 2009). After treatment of Caco-2 cells with VDR ligands, microarray analysis, in combination with real-time RT-PCR and western blotting, indicated that expression of CYP3A4 and the P-glycoprotein, MRP2, and MRP4 transporters was significantly increased after VitD3 treatment, but that expression of SULT1E1, SULT2A1, and the MRP3 transporter was unchanged (Fan et al., 2009). It is possible that differences in cell-culture conditions affecting the differentiation status of Caco-2 cells may account for the observed differences between the two studies (Song et al., 2006; Fan et al., 2009).
SULT regulation by LXR
Regulation of SULT expression by LXR activation has primarily focused on the SULTs that catalyze hydroxysteroid and estrogen sulfation. These SULTs include human SULT2A1 and SULT2B1b, as well as SULT1E1, which is the high-affinity ß-estradiol-conjugating SULT (Falany et al., 1995b; Zhang et al., 1998). LXR-mediated regulation of the steroid-selective SULTs requires that the tissue-selective expression of the SULTs in humans as well as their substrate selectivities and kinetic properties be understood to appreciate the function of the regulation. Cross-species comparisons of LXR-mediated SULT regulation need to be interpreted with regard to differences in isoform multiplicity, tissue-selective expression, unique isoforms, and basal expression.
LXR-mediated regulation of the SULT2A and SULT2B enzymes is closely associated with the possible role of these SULTs in the metabolism of LXR regulators, primarily oxysterols. Multiple oxysterols are known to activate the LXRs and are considered to be the physiological ligands for the LXRs (Gill et al., 2008). Most of the oxysterols that are LXR ligands are also substrates for the SULT2A and SULT2B enzymes (Fuda et al., 2007; Cook et al., 2009; Bai et al., 2012). There is a better understanding of the properties and activities of the human SULT enzymes than for the rodent SULTs. Detailed studies of oxysterol sulfation by the multiple rodent isoforms have not been reported on.
Human SULT2B1b has been termed the cholesterol SULT because of its greater affinity for cholesterol sulfation, as compared with SULT2B1a (Fuda et al., 2002). However, SULT2B1a protein expression has not been identified in any of the human tissues examined, although SULT2B1a mRNA is detectable in several tissues and active enzyme can be generated by heterologous expression in E. coli (Her et al., 1998; Fuda et al., 2002). Although human SULT2A1 is also capable of sulfating cholesterol, only SULT2B1b is expressed in human skin and keratinocytes (Higashi et al., 2004), which generate high levels of cholesterol sulfate. Cholesterol sulfate has a major role in regulating several keratinocyte functions, including corneocyte desquamation (Strott and Higashi, 2003). In addition, cholesterol sulfate is hydrolyzed by steroid sulfatase in the stratum corneum, where it has a role in the generation of the cutaneous permeability barrier (Higashi et al., 2004; Jiang et al., 2005). SULT2A1 expression is limited to hepatocytes, the reticular layer of the adrenal, and the gastrointestinal tract—tissues that do not express SULT2B1b (Falany et al., 1995a; Tashiro et al., 2000; Falany et al., 2006; Riches et al., 2009).
SULT2B1b expression is regulated by LXR and PPAR activation in human keratinocytes (Jiang et al., 2005). Activation of both cultured differentiated and undifferentiated human keratinocytes with selective LXR and PPAR agonists showed induction of SULT2B1b mRNA and cholesterol SULT activity (Jiang et al., 2005), although cholesterol sulfation activity was lower than expected from the message levels. The lower levels of activity are probably associated with the inability to isolate SULT2B1b from tissues or cells in an active form (Falany et al., 2006; He and Falany, 2006). Selective activation of LXR with 22(R)-hydroxycholesterol and PPAR-γ with ciglitazone had an additive effect on SULT2B1b mRNA expression (Jiang et al., 2005). Although LXR activation plays a role in SULT2B1b expression in keratinocytes, the physiological ligand(s) of LXR in the keratinocytes have not been established. Also, both LXR-α and LXR-β are activated by oxysterols (Gill et al., 2008), and both LXRs are expressed in human keratinocytes in a manner that increases with differentiation, which leaves the question of the relative roles of LXR-α and LXR-β in SULT2B1b induction open for investigation. Although both SULT2A1 and SULT2B1b can sulfate cholesterol, SULT2B1b has a more-limited reactivity with other hydroxysteroids and xenobiotics than SULT2A1. Regulation of SULT2B1b expression by LXR in keratinocytes provides a more specific role in the generation of cholesterol sulfate in a tissue where it is involved with the generation and maintenance of epidermal integrity.
Naturally occurring oxysterols, especially those with an oxygenated side chain, act as LXR ligands and meet the criteria for physiological LXR ligands (Gill et al., 2008). Cholesterol is apparently not an LXR ligand allowing for the generation of a number of oxygenated metabolites that are important in regulating cholesterol levels in different tissues. The “oxysterol hypothesis” proposes that oxysterol regulation of LXR activation is an important factor in coordinating many of the aspects of reverse cholesterol transport (Gill et al., 2008). Several of the oxysterols, including 22(R)- and 25-hydroxycholesterol, have been investigated as regulators of LXR (Janowski et al., 1999; Wong et al., 2008). In addition, intermediates in cholesterol metabolism, such as zymosterol and desmosterol, are reported to be LXR regulators (Yang et al., 2006). This suggests an integrated role for LXR activation in different tissues that may involve the SULTs.
Several oxysterols have been implicated in the regulation of human SULT2 expression in different tissues. Both human SULT2A1 and SULT2B1b are also capable of sulfating many of the oxysterols (Fuda et al., 2007; Cook et al., 2009), as well as cholesterol (Aksoy et al., 1993a; Fuda et al., 2002). In most tissues, the kinetics for oxysterol sulfation by both SULT enzymes are considerably more favorable than for cholesterol sulfation. Although sulfation of oxysterols has been regarded as an inactivation mechanism, similar to estrogen sulfates not being able to bind to estrogen receptors, there are currently several reports that oxysterol sulfates are physiologically active molecules. Cook et al. (Cook et al., 2009) demonstrated that 24(S)-hydroxycholesterol-sulfate was a higher affinity inhibitor of LXR-α in a coactivator recruitment assay than the unconjugated oxysterol was as an LXR agonist. Bai et al. (2012) reported that 5-cholesten-3β-25-diol-3-sulfate suppresses LXR/sterol regulatory element-binding protein (SREBP)-1c signaling with properties opposite to those demonstrated by the unsulfated oxysterol. In cultured human aortic endothelial cells, 25-hydroxycholesterol and 25-hydroxycholesterol-sulfate regulated aspects of lipid metabolism in opposing directions, suggesting that regulation of SULT2B1b expression by LXR activation may be involved (Xu et al., 2010). Studies of interaction of oxysterols and oxysterol sulfates in the regulation of LXR activity is an area in need of investigation; however, the initial data are suggestive of oxysterols and oxysterol sulfates having opposing roles in the activation of LXRα. Combined with the ability of LXR to induce SULT1E1 and SULT2B1b in different tissues and cell types, the possible development of an interactive pattern of SULT and substrate regulation is developing.
Synthesis of 24(S)-hydroxycholesterol is the only known mechanism for the removal of cholesterol from human brain, and LXR activation has a role in increasing 24(S)-hydroxycholesterol transport. Activation of LXR in primary rat oligodendrocytes with T0901317 resulted in the induction of several LXR-target genes, including ABCA1, ABCG1, and apolipoprotein E (Nelissen et al., 2012); however, the expression of SULT2B1b was not reported on. Recently, SULT2B1b has been detected in both fetal and adult human brain, suggesting a possible role for 24(S)-hydroxycholesterol sulfation in cholesterol metabolism in the brain (Salman et al., 2011a). Therefore, LXR regulation of SULT2B1b activity in brain tissues could have a significant role in metabolism and excretion of cholesterol from the brain as well as the modulation of neurosteroids, such as dehydroepiandrosterone and pregnenolone and their sulfates (Gibbs et al., 2006; Zheng, 2009).
The SULT2A family in rodents is sexually dimorphic with significantly higher expression of the SULT2A isoforms in females, as compared to males (Kocarek et al., 2008; Waxman and Holloway, 2009). This sexual dimorphism presents at puberty and occurs in concert with an increase in SULT1A and SULT1C expression in males. Lee et al. (2008) have reported that LXR activation inhibits androgen-dependent prostate regeneration in male mice in a SULT2A-dependent manner. However, the expression of SULT2A in the prostate is not well established. Expression of murine Sult2a9 was not detected in mouse prostate tissues, nor did LXR activation induce its expression. LXR activation did induce Sult2a9 expression in the liver, suggesting that hepatic sulfation of androgen may be responsible for the decreased androgenic activity (Lee et al., 2008). In general, testosterone sulfation is associated with SULT2A isoforms; however, the ability to conjugate the 17-position of the D-ring is not as efficient as conjugation at the 3α- and 3ß-hydroxyl positions (Falany et al., 1994). In contrast, expressed SULT2B1b has little or no activity toward androgens (Geese and Raftogianis, 2001; Meloche and Falany, 2001). LXR activation also resulted in the repression of steroid sulfatase (sulfatase C) expression in mouse prostate, suggesting an additional mechanism for androgen inactivation in conjunction with increased androgen sulfation (Lee et al., 2008).
Human SULT2A1 is the only SULT that is capable of sulfating testosterone on the D-ring hydroxyl, allowing for a role in androgen sulfation. LXR activation is associated with SULT2A1 induction in primary human hepatocytes (Uppal et al., 2007). Lee et al. (Lee et al., 2008) reported that LXR activation increased SULT2A1 mRNA content in human LNCaP prostate cancer cells. However, SULT2A1 protein or activity is not detectable in LNCaP cells (He and Falany, 2007). In contrast, SULT2B1b is readily detectable in healthy prostate epithelium, benign prostatic hyperplasia, and prostatic adenocarcinoma, as well as LNCaP cells (He and Falany, 2007). LXR activation resulted in a modest increase in SULT2B1b message levels in LNCaP cells, as compared to strong induction in keratinocytes (Jiang et al., 2005). The role of LXR in the regulation of SULT expression is still not well understood. Human SULT1E1 is also present in normal and prostate cancer tissues, allowing for the possibility of its regulation by LXR activation (Nakamura et al., 2006; Suzuki et al., 2011). LXR may have multiple roles in the regulation of androgenic activity in the prostate, so a more-careful analysis of SULT expression and regulation in human prostate needs to be considered.
Although several human SULTs can sulfate estrogens, SULT1E1 is responsible for the sulfation and inactivation of E2 at physiological concentrations (Falany et al., 1995b; Zhang et al., 1998). SULT1E1 has a Km for E2 sulfation of 4 nM and regulates estrogen receptor alpha (ER-α) activation by E2 in endometrial cells down to low picomolar concentrations (Kotov et al., 1999). Induction of SULT1E1 activity provides a high-affinity mechanism for the intracellular inactivation of E2 (Zhang et al., 1998). SULT1E1 is highly and specifically overexpressed in the liver of strains of obese and diabetic mice (Leiter and Chapman, 1994). In cystic fibrosis, transmembrane conductance regulator KO (CFTR−/−) mice, SULT1E1 activity is significantly increased in the livers of female CFTR−/− mice and half of male CFTR−/− mice (Falany et al., 2002a). Hepatic SULT1E1 activity may be induced up to 100-fold in mice; however, the levels and mechanism of induction in humans are not well described.
Gong et al. (2007) reported that LXR activation in mice resulted in the liver-specific transcriptional induction of SULT1E1 that inhibited estrogen-dependent uterine epithelial proliferation. In addition, these investigators demonstrated that LXR activation inhibited the growth of human MCF-7 breast cancer xenografts in nude mice (Gong et al., 2007). These responses were not observed in SULT1E1 KO mice after treatment with LXR agonists. SULT1E1 expression in human breast cancer cell lines and breast cancers is very low or absent, compared to the levels in healthy breast epithelial cells and tissues (Suzuki et al., 2011). SULT1E1 expression is not detectable in human MCF-7 cells (Falany and Falany, 1996; Falany et al., 2002b); LXR activation inhibited MCF-7 proliferation, although it was not reported whether SULT1E1 expression was increased (Gong et al., 2007). MCF-7 cells do express significant levels of SULT2B1b, which is inducible by LXR activation in other tissues (Higashi et al., 2004; Dumas et al., 2008).
LXR activation has been implicated in the down regulation of growth-hormone stimulation of insulin-like growth factor 1 (IGF-1) expression in human HepG2 hepatocytes by the induction of SULT1E1 (Falany et al., 2009). E2 is required for optimal growth-hormone stimulation of signal transducer and activator of transcription (STAT)5b phosphorylation and IGF-1 transcription (Li et al., 2009). LXR activation of the HepG2 cells induced SULT1E1, but not SULT1A1, expression, resulting in a decreased level of growth-hormone-stimulated STAT5b phosphorylation (He et al., 2008). In Buffalo rat liver (BRL) cells, LXR activation also downregulated growth-hormone-stimulated STAT5b activation (Zadjali et al., 2011). In BRL cells, LXR activation decreased STAT5b protein levels through a SREBP1-mediated mechanism; however, in human HepG2 cells, a decrease in STAT5b levels was not detected after LXR activation (Li et al., 2009).
SULT regulation by the FXR
The involvement of FXR in the regulation of SULT expression has been investigated because of its linkage to the sulfation of bile acids. FXR is one of the major bile-acid sensors involved in the regulation of bile-acid levels in the intrahepatic circulation (Cui et al., 2012; Matsubara et al., 2012). Conjugation of bile acids with a sulfonate group increases the water solubility of the bile acids and decreases their toxicity that is closely associated with their hydrophobicity (Alnouti, 2009). Initial studies identified high levels of sulfated bile acids in the serum of patients with obstructive jaundice and acute hepatitis, but significantly lower levels in patients with chronic hepatitis and cirrhosis (Makino et al., 1974, 1975). Bile-acid sulfates are generally low in patients with a normal intrahepatic circulation of bile acids. In healthy individuals administered primary and secondary bile acids, the highest rate of sulfation was observed with lithocholate, with the subsequent transport of lithocholate 3-sulfate into the bile (Alnouti, 2009). However, in hepatic conditions that disrupt the intrahepatic circulation, sulfation is viewed as one mechanism for increasing bile-acid solubility, and secretion into the circulation across the sinusoidal surface of the hepatocyte is enhanced so that bile-acid sulfates can be excreted by the kidneys (Kitada et al., 2003).
Bile-acid sulfation was initially associated with what is currently considered the SULT2 family in rats (Barnes et al., 1989). Utilizing pure human liver SULT2A1 or “DHEA-sulfotransferase,” Radominska et al. (1990) demonstrated that DHEA-sulfotransferase was responsible for bile-acid sulfation in the human liver. SULT2A1 was capable of sulfating all of the bile acids as well as the taurine- and glycine-conjugated bile acids that were tested. Lithocholate and its amidated metabolites were the most rapidly sulfated bile acids. Lithocholate is a monohydroxylated derivative of CDCA and the most hydrophobic and toxic member of the bile acids. In the human liver, the basal levels of SULT2A1 expression are relatively high (Aksoy et al., 1993b; Falany et al., 1995a) and it is the only SULT enzyme with a high affinity for bile acid and bile-amidate sulfation (Radominska et al., 1990).
Initial characterization of SULT2A regulation indicated that CDCA was a potent transcriptional inducer, mediated through the binding of FXR/RXR heterodimers (Song et al., 2001). Promoter-reporter studies in HepG2 and Caco-2 cells indicated that FXR stimulated rat SULT2A transcription by binding to the same IR0 motif that has been described above under PXR (Song et al., 2001). Induction was detected utilizing insertion of sequential promoter fragments as well as multiple copies of the IR0 motif in reporter transfection assays in HepG2 and Caco-2 cells. In contrast to the reporter-assay-based induction of rat SULT2A, Miyata et al. (2006b) demonstrated that treatment of HepG2 cells with FXR agonists, CDCA, or GW4064 resulted in a repression of human SULT2A1 expression.
The in vivo repression of bile-acid sulfation and SULT2A1 expression associated with FXR activation was also observed in mice. Kitada et al. (2003) reported that in mice fed 1% lithocholate in the diet, Sult2a protein and bile-acid sulfation activity were higher in livers of FXR-null mice than WT mice. Markers of LCA toxicity, including transaminase levels, were also inversely correlated to SULT2A expression, indicating that increased LCA sulfation had a protective role. From the FXR-null mouse studies, FXR expression apparently had a repressive role on basal SULT2A expression. Subsequent studies by Miyata et al. (2006a) using WT and FXR-null mice supported that CDCA activation of FXR resulted in the repression of SULT2A expression. Because several of the NRs reported to regulate SULT2A expression may bind to the same IR0 motif in rodent SULT2A promoters, treatment of intact cells and animals with pharmacologic doses of FXR agonists may be inhibiting the binding of other NRs that are involved with the induction or regulation of SULT2A expression.
It is increasingly recognized that a complicated inter-play of NRs is involved in the regulation of bile-acid levels in normal and pathological conditions. The response to rising bile-acid levels involves numerous metabolic and transport systems, including SULTs. In patients with stage III or IV PBC, a prototypic chronic cholestatic liver disease, the high rate of bile-acid sulfate excretion was not associated with changes in SULT2A1 expression (Zollner et al., 2007). The lack of altered SULT2A1 expression may be the result of a loss of responsiveness in late-stage disease to FXR or other NR regulation. In addition, most of the patients in this study received standard ursodeoxy-cholate treatment to maintain biliary function without any observed effects on FXR activation (Zollner et al., 2007).
SULT regulation by RORs
The ROR transcription factors bind as monomers to AT-rich ROR response elements (ROREs) in the 5′-flanking regions of target genes (Giguere et al., 1994). There is evidence that these NRs are ligand activated by intermediates in cholesterol and retinoid metabolism, such as cholesterol, cholesterol sulfate, 7-oxygenated sterols, and retinoids (Kallen et al., 2002; Stehlin-Gaon et al., 2003; Kallen et al., 2004; Wang et al., 2010). Differential splicing and alternative promoter usage enables a diversity of expression of ROR-α, ROR-β, and ROR-γ isoforms in tissues (Jetten et al., 2001). Because ROR-α and ROR-γ are frequently coexpressed in metabolically active tissues, such as the liver, kidney, and brown fat, their functional roles were examined in single- and double-ROR-α/ROR-γ KO mouse models (Jetten et al., 2001; Kang et al., 2007). Both ROR-α and ROR-γ were abundantly expressed in the livers of WT AKR/J control mice and displayed fluctuations in expression that were consistent with the periodicity of circadian rhythm (Kang et al., 2007). ROR-α/ROR-γ double-KO mice displayed significant reductions in blood triglyceride and cholesterol levels, relative to WT mice (Kang et al., 2007). Microarray analysis revealed striking increases in the expression of Sult1e1 (16.9-fold) and Sult2a1 (43.5-fold) in the livers of ROR-α/ROR-γ double-KO mice, relative to WT controls (Kang et al., 2007). Double-ROR-α/ROR-γ KO mice also displayed disturbances in the expression of a range of hepatic target genes involved in lipid, bile-acid, steroid, and xenobiotic metabolism, including Cyp7a1, Cyp8b1,Cyp27a1, Cyp7b1 (bile-acid metabolism), Cyp4a10/14 (fatty-acid metabolism), and Cyp2b10 (xenobiotic metabolism) (Kang et al., 2007). These results suggest that ROR-α and ROR-γ play an important role in regulating metabolism and that they suppress murine hepatic Sult1e1 and Sult2a1 transcription under basal conditions.
SULT regulation by the Estrogen-Related Receptors (ERRs)
The orphan NRs known as ERRs (ERR-α, ERR-β, and ERR-γ) share sequence similarities with the estrogen receptor and can bind to extended NR half-sites as monomers (Johnston et al., 1997) or homodimers (Vanacker et al., 1999). The role of ERR transcription factors in the regulation of xenobiotic-metabolizing enzymes is just emerging. The existing data suggest that under basal conditions, ERRs assume the conformation of a transcriptionally active NR (Greschik et al., 2002). Natural estrogens do not activate ERRs, and only synthetic ligands have been identified to date (Coward et al., 2001; Tremblay et al., 2001; Willy et al., 2004). ERR-α is expressed in both fetal and adult adrenal tissue and its expression in the adrenal cortex colocalizes with that of SULT2A1, a major steroidogenic enzyme in the adrenal gland (Seely et al., 2005). Transient cotransfections of SULT2A1 reporter constructs with ERR-α expression plasmid were performed in CV-1 cells (Seely et al., 2005). These studies demonstrated a marked induction of SULT2A1 reporter gene transcription in response to ERR-α stimulation (Seely et al., 2005). Serial deletions revealed the presence of three ERR-α-responsive sites in the 5′-flanking region of SULT2A1 located at nucleotides −1191, −85, and −65, relative to the core promoter region (Seely et al., 2005). Site-directed mutagenesis of each site impaired ERR-α-inducible SULT2A1 transcription, but the sites located at −85 and −65 had the greatest effect (Seely et al., 2005). These sites were demonstrated by EMSA to bind proteins in adult adrenal nuclear extracts, and complex formation was retarded by the addition of ERR-α Ab (Seely et al., 2005). Because steroidogenic transcription factor 1 (SF1) was previously shown to regulate SULT2A1 transcription in the adrenal gland, the effects of ERR-α and SF1 cotransfection were also examined in a series of cotransfection studies conducted in CV-1 cells (Seely et al., 2005). The results indicated that both SF1 and ERR-α induced SULT2A1 reporter gene expression to a comparable extent and that cotransfection neither augmented nor impaired SULT2A1 inducibility, suggesting that ERR-α and SF1 likely regulate SULT2A1 through independent mechanisms, but may share the same cis-acting response elements (Seely et al., 2005). In HepG2 cells, the cotransfection of increasing amounts of ERR-α expression plasmid has been reported to produce dosedependent decreases in SULT2A1 reporter expression, and cis-acting regions in the SULT2A1 5′-flanking region mediating this effect have been identified (Huang et al., 2011). However, the biological significance of ERR-α in the liver and other tissues where ERR-α levels are measurably lower than those demonstrated in adrenal and heart issues, for example, is not clear and will require further investigation.
Conclusion
SULTs have important functions in the metabolism of many drugs and xenobiotics, primarily in the inactivation of their pharmacological effects by conjugation with the charged sulfonate group. Many of the SULT enzymes also have important roles in the metabolism of endogenous compounds, including steroids, oxysterols, and bile acids. The initial characterization of the “orphan” NRs established their important functions as xenosensors to modulate or adapt metabolism to the exposure of environmental compounds. Subsequent characterization of these NRs described equally important roles in the regulation of myriad endogenous processes. The NR regulation of the SULTs is primarily associated with endogenous physiological activities of the SULTs, in contrast to their xenobiotic/drug-conjugation activities.
NR regulation of the SULTs focuses mainly on three enzymes (SULT1E1, SULT2A1, and SULT2B1b), which are responsible for estrogen, hydroxysteroid, oxysterol, and cholesterol sulfation. The SULT1 enzymes (SULTs 1A, 1B, 1C, and 1D) associated with phenolic compound conjugation are not generally responsive to regulation by the NRs. The role of SULT2A1 in decreasing bile-acid/ amidate toxicity in situations of disrupted bile synthesis and/or enterohepatic circulation has been extensively studied. Several NRs regulate SULT2A1 expression and, in several instances, apparently by the same promoter elements. The complex regulation of SULT2A1 expression in the human liver may act to dampen the overall regulation of its expression in vivo, an interesting, important direction for future research. The interpretation of NR regulation of the SULT2A family in rodents has been significantly hampered by the multiplicity of isoforms, sex-dependent expression, and lack of understanding of the activity, substrate selectivity, and regulation of the individual isoforms.
Evolving areas of nuclear regulation of SULT expression involve extrahepatic tissues, where the involvement of different NRs is becoming apparent. ERR-α appears to have a major role in the regulation of SULT2A1 expression in human adrenal tissue, where it is involved with the synthesis of high levels of DHEAS. SULT2B1b regulation, by PPAR-α and/or LXR, in human keratinocytes is linked to the regulation of cholesterol sulfate formation in the skin. Oxysterol regulation of SULT2B1b expression and function in tissues, such as prostate and aortic endothelial cells, is in need of investigation.
SULT1E1 is responsible for high-affinity sulfation of estradiol at physiological concentrations. Sult1e1 expression has been observed to be highly induced (>100-fold) in mouse liver in genetic conditions, such as ob/ob or db/db mice or CFTR−/− mice. PXR, CAR, LXR, and ROR have been implicated in the regulation of Sult1e1 expression in mice; however, the large variability of SULT1E1 expression has not been reported on in humans. LXR induction of SULT1E1 expression was implicated in the regulation of growth-hormone signaling in human HepG2 cells.
To date, NR regulation of the SULTs appears to focus on the steroid/sterol-conjugating enzymes. The effects of NR/SULT regulation and function in rodents may provide insights into regulation in humans; however, direct comparisons should be made cautiously. Investigation of the role of the individual NRs in the expression and regulation of the SULTs in extrahepatic tissues is in need of investigation.
Footnotes
Declaration of interest
The authors report no declarations of interest. The authors alone are responsible for the content and writing of the paper.
References
- Aksoy IA, Otterness DM, Weinshilboum RM. Cholesterol sulfation in human liver. Catalysis by dehydroepiandrosterone sulfotransferase. Drug Metab Dispos. 1993a;21:268–276. [PubMed] [Google Scholar]
- Aksoy IA, Sochorova V, Weinshilboum RM. Human liver dehydroepiandrosterone sulfotransferase: nature and extent of individual variation. Clin Pharmacol Ther. 1993b;54:498–506. doi: 10.1038/clpt.1993.181. [DOI] [PubMed] [Google Scholar]
- Aleksunes LM, Klaassen CD. Coordinated regulation of hepatic phase-I and -II drug metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARalpha-, and Nrf2-null mice. Drug Metab Dispos. 2012;40:1366–1379. doi: 10.1124/dmd.112.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y. Bile acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol Sci. 2009;108:225–246. doi: 10.1093/toxsci/kfn268. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J Pharmacol Exp Ther. 2008;324:612–621. doi: 10.1124/jpet.107.129650. [DOI] [PubMed] [Google Scholar]
- Assem M, Schuetz EG, Leggas M, Sun D, Yasuda K, Reid G, et al. Interactions between hepatic Mrp4 and Sult2a as revealed by the constitutive androstane receptor and Mrp4 knockout mice. J Biol Chem. 2004;279:22250–22257. doi: 10.1074/jbc.M314111200. [DOI] [PubMed] [Google Scholar]
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1552. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Zhang X, Xu L, Kakiyama G, Heuman D, Sanyal A, et al. Oxysterol sulfation by cytosolic sulfotransferase suppresses liver X receptor/sterol regulatory element binding protein-1c signaling pathway and reduces serum and hepatic lipids in mouse models of nonalcoholic fatty liver disease. Metabolism. 2012;61:836–845. doi: 10.1016/j.metabol.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banoglu E. Current status of the cytosolic sulfotransferases in the metabolic activation of promutagens and procarcinogens. Curr Drug Metab. 2000;1:1–30. doi: 10.2174/1389200003339234. [DOI] [PubMed] [Google Scholar]
- Barnes S, Buchina ES, King RJ, McBurnett T, Taylor KB. Bile acid sulfotransferase I from rat liver sulfates bile acids and 3-hydroxy steroids: purification, N-terminal amino acid sequence, and kinetic properties. J Lipid Res. 1989;30:529–540. [PubMed] [Google Scholar]
- Bertoni A, Rastoldo A, Sarasso C, Di VC, Sampietro S, Nalin M, et al. Dehydroepiandrosterone-sulfate inhibits thrombin-induced platelet aggregation. Steroids. 2012;77:260–268. doi: 10.1016/j.steroids.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Blanchard RL. Nomenclature and molecular biology of the human sulfotransferase family. In: Pacifici GM, Coughtrie MWH, editors. Human cytosolic sulfotransferases. Taylor & Francis; Boca Raton, Florida, USA: 2005. pp. 1–26. [Google Scholar]
- Bogan AA, Dallas-Yang Q, Ruse MD, Jr., Maeda Y, Jiang G, Nepomuceno L, et al. Analysis of protein dimerization and ligand binding of orphan receptor HNF4alpha. J Mol Biol. 2000;302:831–851. doi: 10.1006/jmbi.2000.4099. [DOI] [PubMed] [Google Scholar]
- Choi HS, Chung M, Tzameli I, Simha D, Lee YK, Seol W, et al. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J Biol Chem. 1997;272:23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- Cook IT, Duniec-Dmuchowski Z, Kocarek TA, Runge-Morris M, Falany CN. 24-hydroxycholesterol sulfation by human cytosolic sulfotransferases: formation of monosulfates and disulfates, molecular modeling, sulfatase sensitivity, and inhibition of liver x receptor activation. Drug Metab Dispos. 2009;37:2069–2078. doi: 10.1124/dmd.108.025759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward P, Lee D, Hull MV, Lehmann JM. 4-Hydroxytamoxifen binds to and deactivates the estrogen-related receptor gamma. Proc Natl Acad Sci U S A. 2001;98:8880–8884. doi: 10.1073/pnas.151244398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Aleksunes LM, Tanaka Y, Fu ZD, Guo Y, Guo GL, et al. Bile acids via FXR initiate the expression of major transporters involved in the enterohepatic circulation of bile acids in newborn mice. Am J Physiol Gastrointest Liver Physiol. 2012;302:G979–G996. doi: 10.1152/ajpgi.00370.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Lichti K, Kim I, Gonzalez FJ, Staudinger JL. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Biol Chem. 2006;281:26540–26551. doi: 10.1074/jbc.M600931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley TP, Haldeman-Cahill R, Joiner J, Wilborn TW. Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem Biophys Res Commun. 2000;277:236–245. doi: 10.1006/bbrc.2000.3643. [DOI] [PubMed] [Google Scholar]
- Duanmu Z, Weckle A, Koukouritaki SB, Hines RN, Falany JL, Falany CN, et al. Developmental expression of aryl, estrogen, and hydroxysteroid sulfotransferases in pre- and postnatal human liver. J Pharmacol Exp Ther. 2006;316:1310–1317. doi: 10.1124/jpet.105.093633. [DOI] [PubMed] [Google Scholar]
- Duffel MW, Binder TP, Hosie L, Baden HA, Sanders JA, Knapp SA, et al. Purification, immunochemical characterization, and immunohistochemical localization of rat hepatic aryl sulfotransferase IV. Mol Pharmacol. 1991;40:36–44. [PubMed] [Google Scholar]
- Dumas NA, He D, Frost AR, Falany CN. Sulfotransferase 2B1b in human breast: differences in subcellular localization in African American and Caucasian women. J Steroid Biochem Mol Biol. 2008;111:171–177. doi: 10.1016/j.jsbmb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B. The xenobiotic-sensing nuclear receptors pregnane X receptor, constitutive androstane receptor, and orphan nuclear receptor hepatocyte nuclear factor 4alpha in the regulation of human steroid-/bile acid-sulfotransferase. Mol Endocrinol. 2007;21:2099–2111. doi: 10.1210/me.2007-0002. [DOI] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Oh TS, Cho SH, Rivera OJ, Chatterjee B. Gene regulation for the senescence marker protein DHEA-sulfotransferase by the xenobiotic-activated nuclear pregnane X receptor (PXR) Mech Ageing Dev. 2004a;125:733–745. doi: 10.1016/j.mad.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Roy AK, Chatterjee B. Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin D receptor. Mol Pharmacol. 2004b;65:720–729. doi: 10.1124/mol.65.3.720. [DOI] [PubMed] [Google Scholar]
- Epstein EH, Williams ML, Elias PM. The epidermal cholesterol sulfate cycle. J Am Acad Dermatol. 1984;10:866–868. doi: 10.1016/s0190-9622(84)80144-9. [DOI] [PubMed] [Google Scholar]
- Falany CN, Comer KA, Dooley TP, Glatt H. Human dehydroepiandrosterone sulfotransferase. Purification, molecular cloning, and characterization. Ann N Y Acad Sci. 1995a;774:59–72. doi: 10.1111/j.1749-6632.1995.tb17372.x. [DOI] [PubMed] [Google Scholar]
- Falany CN, He D, Dumas N, Frost AR, Falany JL. Human cytosolic sulfotransferase 2B1: isoform expression, tissue specificity and subcellular localization. J Steroid Biochem Mol Biol. 2006;102:214–221. doi: 10.1016/j.jsbmb.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falany CN, He D, Li L, Falany JL, Wilborn TW, Kocarek TA, et al. Regulation of hepatic sulfotransferase (SULT) 1E1 expression and effects on estrogenic activity in cystic fibrosis (CF) J Steroid Biochem Mol Biol. 2009;114:113–119. doi: 10.1016/j.jsbmb.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol. 1995b;52:529–539. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]
- Falany CN, Vazquez ME, Heroux JA, Roth JA. Purification and characterization of human liver phenol-sulfating phenol sulfotransferase. Arch Biochem Biophys. 1990;278:312–318. doi: 10.1016/0003-9861(90)90265-z. [DOI] [PubMed] [Google Scholar]
- Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulphotransferase. Biochem J. 1989;260:641–646. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falany CN, Wheeler J, Oh TS, Falany JL. Steroid sulfation by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 1994;48:369–375. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res. 1996;56:1551–1555. [PubMed] [Google Scholar]
- Falany JL, Greer H, Kovacs T, Sorscher EJ, Falany CN. Elevation of hepatic sulphotransferase activities in mice with resistance to cystic fibrosis. Biochem J. 2002a;364:115–120. doi: 10.1042/bj3640115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falany JL, Macrina N, Falany CN. Regulation of MCF-7 breast cancer cell growth by beta-estradiol sulfation. Breast Cancer ResTreat. 2002b;74:167–176. doi: 10.1023/a:1016147004188. [DOI] [PubMed] [Google Scholar]
- Fan J, Liu S, Du Y, Morrison J, Shipman R, Pang KS. Up-regulation of transporters and enzymes by the vitamin D receptor ligands, 1alpha,25-dihydroxyvitamin D3 and vitamin D analogs, in the Caco-2 cell monolayer. J Pharmacol Exp Ther. 2009;330:389–402. doi: 10.1124/jpet.108.149815. [DOI] [PubMed] [Google Scholar]
- Fan LQ, You L, Brown-Borg H, Brown S, Edwards RJ, Corton JC. Regulation of phase I and phase II steroid metabolism enzymes by PPAR alpha activators. Toxicology. 2004;204:109–121. doi: 10.1016/j.tox.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M. Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor alpha transcription factor. Mol Pharmacol. 2005;67:1257–1267. doi: 10.1124/mol.104.005389. [DOI] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Ellis E, Duanmu Z, Fu J, Duniec-Dmuchowski Z, et al. Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4alpha and pregnane X receptor. J Pharmacol Exp Ther. 2007;323:586–598. doi: 10.1124/jpet.107.124610. [DOI] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, et al. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Frohling W, Stiehl A. Bile salt glucuronides: identification and quantitative analysis in the urine of patients with cholestasis. Eur J Clin Invest. 1976;6:67–74. doi: 10.1111/j.1365-2362.1976.tb00495.x. [DOI] [PubMed] [Google Scholar]
- Fuda H, Javitt NB, Mitamura K, Ikegawa S, Strott CA. Oxysterols are substrates for cholesterol sulfotransferase. J Lipid Res. 2007;48:1343–1352. doi: 10.1194/jlr.M700018-JLR200. [DOI] [PubMed] [Google Scholar]
- Fuda H, Lee YC, Shimizu C, Javitt NB, Strott CA. Mutational analysis of human hydroxysteroid sulfotransferase SULT2B1 isoforms reveals that exon 1B of the SULT2B1 gene produces cholesterol sulfotransferase, whereas exon 1A yields pregnenolone sulfotransferase. J Biol Chem. 2002;277:36161–36166. doi: 10.1074/jbc.M207165200. [DOI] [PubMed] [Google Scholar]
- Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- Gartside SE, Griffith NC, Kaura V, Ingram CD. The neurosteroid dehydroepiandrosterone (DHEA) and its metabolites alter 5-HT neuronal activity via modulation of GABAA receptors. J Psychopharmacol. 2010;24:1717–1724. doi: 10.1177/0269881109105836. [DOI] [PubMed] [Google Scholar]
- Geese WJ, Raftogianis RB. Biochemical characterization and tissue distribution of human SULT2B1. Biochem Biophys Res Commun. 2001;288:280–289. doi: 10.1006/bbrc.2001.5746. [DOI] [PubMed] [Google Scholar]
- Ghose R, Omoluabi O, Gandhi A, Shah P, Strohacker K, Carpenter KC, et al. Role of high-fat diet in regulation of gene expression of drug metabolizing enzymes and transporters. Life Sci. 2011;89:57–64. doi: 10.1016/j.lfs.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs TT, Russek SJ, Farb DH. Sulfated steroids as endogenous neuromodulators. Pharmacol Biochem Behav. 2006;84:555–567. doi: 10.1016/j.pbb.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- Gill AM, Leiter EH, Powell JG, Chapman HD, Yen TT. Dexamethasone-induced hyperglycemia in obese Avy/a (viable yellow) female mice entails preferential induction of a hepatic estrogen sulfotransferase. Diabetes. 1994;43:999–1004. doi: 10.2337/diab.43.8.999. [DOI] [PubMed] [Google Scholar]
- Gill S, Chow R, Brown AJ. Sterol regulators of cholesterol homeostasis and beyond: the oxysterol hypothesis revisited and revised. Prog Lipid Res. 2008;47:391–404. doi: 10.1016/j.plipres.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Glatt H, Boeing H, Engelke CE, Ma L, Kuhlow A, Pabel U, et al. Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutat Res. 2001;482:27–40. doi: 10.1016/s0027-5107(01)00207-x. [DOI] [PubMed] [Google Scholar]
- Gong H, Guo P, Zhai Y, Zhou J, Uppal H, Jarzynka MJ, et al. Estrogen deprivation and inhibition of breast cancer growth in vivo through activation of the orphan nuclear receptor liver X receptor. Mol Endocrinol. 2007;21:1781–1790. doi: 10.1210/me.2007-0187. [DOI] [PubMed] [Google Scholar]
- Gooderham NJ, Murray S, Lynch AM, Yadollahi-Farsani M, Zhao K, Boobis AR, et al. Food-derived heterocyclic amine mutagens: variable metabolism and significance to humans. Drug Metab Dispos. 2001;29:529–534. [PubMed] [Google Scholar]
- Greschik H, Wurtz JM, Sanglier S, Bourguet W, van Dorsselaer A, Moras D, et al. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol Cell. 2002;9:303–313. doi: 10.1016/s1097-2765(02)00444-6. [DOI] [PubMed] [Google Scholar]
- He D, Falany CN. Characterization of proline-serine-rich carboxyl terminus in human sulfotransferase 2B1b: immunogenicity, subcellular localization, kinetic properties, and phosphorylation. Drug Metab Dispos. 2006;34:1749–1755. doi: 10.1124/dmd.106.011114. [DOI] [PubMed] [Google Scholar]
- He D, Falany CN. Inhibition of SULT2B1b expression alters effects of 3beta-hydroxysteroids on cell proliferation and steroid hormone receptor expression in human LNCaP prostate cancer cells. Prostate. 2007;67:1318–1329. doi: 10.1002/pros.20615. [DOI] [PubMed] [Google Scholar]
- He D, Wilborn TW, Falany JL, Li L, Falany CN. Repression of CFTR activity in human MMNK-1 cholangiocytes induces sulfotransferase 1E1 expression in co-cultured HepG2 hepatocytes. Biochim Biophys Acta. 2008;1783:2391–2397. doi: 10.1016/j.bbamcr.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her C, Szumlanski C, Aksoy IA, Weinshilboum RM. Human jejunal estrogen sulfotransferase and dehydroepiandrosterone sulfotransferase: immunochemical characterization of individual variation. Drug Metab Dispos. 1996;24:1328–1335. [PubMed] [Google Scholar]
- Her C, Wood TC, Eichler EE, Mohrenweiser HW, Ramagli LS, Siciliano MJ, et al. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53:284–295. doi: 10.1006/geno.1998.5518. [DOI] [PubMed] [Google Scholar]
- Higashi Y, Fuda H, Yanai H, Lee Y, Fukushige T, Kanzaki T, et al. Expression of cholesterol sulfotransferase (SULT2B1b) in human skin and primary cultures of human epidermal keratinocytes. J Invest Dermatol. 2004;122:1207–1213. doi: 10.1111/j.0022-202X.2004.22416.x. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhou T, Chen Y, Sun T, Zhang S, Chen G. Estrogen-related receptor ERRalpha-mediated downregulation of human hydroxysteroid sulfotransferase (SULT2A1) in Hep G2 cells. Chem Biol Interact. 2011;192:264–271. doi: 10.1016/j.cbi.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancova P, Anzenbacher P, Anzenbacherova E. Phase II drug metabolizing enzymes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2010;154:103–116. doi: 10.5507/bp.2010.017. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, et al. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci U S A. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM, Kurebayashi S, Ueda E. The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol. 2001;69:205–247. doi: 10.1016/s0079-6603(01)69048-2. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Kim P, Elias PM, Feingold KR. LXR and PPAR activators stimulate cholesterol sulfotransferase type 2 isoform 1b in human keratinocytes. J Lipid Res. 2005;46:2657–2666. doi: 10.1194/jlr.M500235-JLR200. [DOI] [PubMed] [Google Scholar]
- Johnston SD, Liu X, Zuo F, Eisenbraun TL, Wiley SR, Kraus RJ, et al. Estrogen-related receptor alpha 1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Mol Endocrinol. 1997;11:342–352. doi: 10.1210/mend.11.3.9897. [DOI] [PubMed] [Google Scholar]
- Kachaylo EM, Pustylnyak VO, Lyakhovich VV, Gulyaeva LF. Constitutive androstane receptor (CAR) is a xenosensor and target for therapy. Biochemistry (Mosc.) 2011;76:1087–1097. doi: 10.1134/S0006297911100026. [DOI] [PubMed] [Google Scholar]
- Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, et al. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, et al. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- Kashiwagi M, Ohba M, Chida K, Kuroki T. Protein kinase C eta (PKC eta): its involvement in keratinocyte differentiation. J Biochem. 2002;132:853–857. doi: 10.1093/oxfordjournals.jbchem.a003297. [DOI] [PubMed] [Google Scholar]
- Khor VK, Dhir R, Yin X, Ahima RS, Song WC. Estrogen sulfotransferase regulates body fat and glucose homeostasis in female mice. Am J Physiol Endocrinol Metab. 2010;299:E657–E664. doi: 10.1152/ajpendo.00707.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Shigenaga J, Moser A, Grunfeld C, Feingold KR. Suppression of DHEA sulfotransferase (Sult2A1) during the acute-phase response. Am J Physiol Endocrinol Metab. 2004;287:E731–E738. doi: 10.1152/ajpendo.00130.2004. [DOI] [PubMed] [Google Scholar]
- Kitada H, Miyata M, Nakamura T, Tozawa A, Honma W, Shimada M, et al. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J Biol Chem. 2003;278:17838–17844. doi: 10.1074/jbc.M210634200. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Duanmu Z, Fang HL, Runge-Morris M. Age- and sex-dependent expression of multiple murine hepatic hydroxysteroid sulfotransferase (SULT2A) genes. Biochem Pharmacol. 2008;76:1036–1046. doi: 10.1016/j.bcp.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama S, Hosseinpour F, Goldstein JA, Negishi M. Liganded pregnane X receptor represses the human sulfotransferase SULT1E1 promoter through disrupting its chromatin structure. Nucleic Acids Res. 2011;39:8392–8403. doi: 10.1093/nar/gkr458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohjitani A, Fuda H, Hanyu O, Strott CA. Cloning, characterization, and tissue expression of rat SULT2B1a and SULT2B1b steroid/sterol sulfotransferase isoforms: divergence of the rat SULT2B1 gene structure from orthologous human and mouse genes. Gene. 2006;367:66–73. doi: 10.1016/j.gene.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Kotov A, Falany JL, Wang J, Falany CN. Regulation of estrogen activity by sulfation in human Ishikawa endometrial adenocarcinoma cells. J Steroid Biochem Mol Biol. 1999;68:137–144. doi: 10.1016/s0960-0760(99)00022-9. [DOI] [PubMed] [Google Scholar]
- Kushida A, Hattori K, Yamaguchi N, Kobayashi T, Date A, Tamura H. Sulfation of estradiol in human epidermal keratinocyte. Biol Pharm Bull. 2011;34:1147–1151. doi: 10.1248/bpb.34.1147. [DOI] [PubMed] [Google Scholar]
- Lampe MA, Williams ML, Elias PM. Human epidermal lipids: characterization and modulations during differentiation. J Lipid Res. 1983;24:131–140. [PubMed] [Google Scholar]
- Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Gong H, Khadem S, Lu Y, Gao X, Li S, et al. Androgen deprivation by activating the liver X receptor. Endocrinology. 2008;149:3778–3788. doi: 10.1210/en.2007-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Fuda H, Lee YC, Negishi M, Strott CA, Pedersen LC. Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone and 3′-phosphoadenosine 5′-phosphate. Rationale for specificity differences between prototypical SULT2A1 and the SULT2BG1 isoforms. J Biol Chem. 2003;278:44593–44599. doi: 10.1074/jbc.M308312200. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter EH, Chapman HD. Obesity-induced diabetes (diabesity) in C57BL/KsJ mice produces aberrant trans-regulation of sex steroid sulfotransferase genes. J Clin Invest. 1994;93:2007–2013. doi: 10.1172/JCI117194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AJ, Walle UK, King RS, Kadlubar FF, Falany CN, Walle T. Bioactivation of the cooked food mutagen N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by estrogen sulfotransferase in cultured human mammary epithelial cells. Carcinogenesis. 1998;19:2049–2053. doi: 10.1093/carcin/19.11.2049. [DOI] [PubMed] [Google Scholar]
- Li L, He D, Wilborn TW, Falany JL, Falany CN. Increased SULT1E1 activity in HepG2 hepatocytes decreases growth hormone stimulation of STAT5b phosphorylation. Steroids. 2009;74:20–29. doi: 10.1016/j.steroids.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyou NE, Buller KM, Tresillian MJ, Elvin CM, Scott HL, Dodd PR, et al. Localization of a brain sulfotransferase, SULT4A1, in the human and rat brain: an immunohistochemical study. J Histochem Cytochem. 2003;51:1655–1664. doi: 10.1177/002215540305101209. [DOI] [PubMed] [Google Scholar]
- Luchetti S, Bossers K, Frajese GV, Swaab DF. Neurosteroid biosynthetic pathway changes in substantia nigra and caudate nucleus in Parkinson’s disease. Brain Pathol. 2010;20:945–951. doi: 10.1111/j.1750-3639.2010.00396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino I, Hashimoto H, Shinozaki K, Yoshino K, Nakagawa S. Sulfated and nonsulfated bile acids in urine, serum, and bile of patients with hepatobiliary diseases. Gastroenterology. 1975;68:545–553. [PubMed] [Google Scholar]
- Makino I, Shinozaki K, Nakagawa S, Mashimo K. Measurement of sulfated and nonsulfated bile acids in human serum and urine. J Lipid Res. 1974;15:132–138. [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Li F, Gonzalez FJ. FXR signaling in the enterohepatic system. Mol Cell Endocrinol. 2012 May 17; doi: 10.1016/j.mce.2012.05.004. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinl W, Glatt H. Structure and localization of the human SULT1B1 gene: neighborhood to SULT1E1 and a SULT1D pseudogene. Biochem Biophys Res Commun. 2001;288:855–862. doi: 10.1006/bbrc.2001.5829. [DOI] [PubMed] [Google Scholar]
- Meloche CA, Falany CN. Expression and characterization of the human 3 beta-hydroxysteroid sulfotransferases (SULT2B1a and SULT2B1b) J Steroid Biochem Mol Biol. 2001;77:261–269. doi: 10.1016/s0960-0760(01)00064-4. [DOI] [PubMed] [Google Scholar]
- Miyata M, Matsuda Y, Tsuchiya H, Kitada H, Akase T, Shimada M, et al. Chenodeoxycholic acid-mediated activation of the farnesoid X receptor negatively regulates hydroxysteroid sulfotransferase. Drug Metab Pharmacokinet. 2006a;21:315–323. doi: 10.2133/dmpk.21.315. [DOI] [PubMed] [Google Scholar]
- Miyata M, Watase H, Hori W, Shimada M, Nagata K, Gonzalez FJ, et al. Role for enhanced faecal excretion of bile acid in hydroxysteroid sulfotransferase-mediated protection against lithocholic acid-induced liver toxicity. Xenobiotica. 2006b;36:631–644. doi: 10.1080/00498250600776827. [DOI] [PubMed] [Google Scholar]
- Nagata K, Shimada M, Yamazoe Y. Species differences in cytosolic sulfotransferases. In: Pacifici GM, Coughtrie MW, editors. Human cytosolic sUlfotransferases. Taylor & Francis; Boca Raton, Florida, USA: 2005. pp. 253–278. [Google Scholar]
- Nagata K, Yamazoe Y. Pharmacogenetics of sulfotransferase. Annu Rev Pharmacol Toxicol. 2000;40:159–176. doi: 10.1146/annurev.pharmtox.40.1.159. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Suzuki T, Fukuda T, Ito A, Endo M, Moriya T, et al. Steroid sulfatase and estrogen sulfotransferase in human prostate cancer. Prostate. 2006;66:1005–1012. doi: 10.1002/pros.20426. [DOI] [PubMed] [Google Scholar]
- Nelissen K, Mulder M, Smets I, Timmermans S, Smeets K, Ameloot M, et al. Liver X receptors regulate cholesterol homeostasis in oligodendrocytes. J Neurosci Res. 2012;90:60–71. doi: 10.1002/jnr.22743. [DOI] [PubMed] [Google Scholar]
- Ngan CH, Beglov D, Rudnitskaya AN, Kozakov D, Waxman DJ, Vajda S. The structural basis of pregnane X receptor binding promiscuity. Biochemistry. 2009;48:11572–11581. doi: 10.1021/bi901578n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, de la Torre B, Hedman M, Goobar J, Thorner A. Blood dehydroepiandrosterone sulphate (DHEAS) levels in polymyalgia rheumatica/giant cell arteritis and primary fibromyalgia. Clin Exp Rheumatol. 1994;12:415–417. [PubMed] [Google Scholar]
- Owen BM, Milona A, van Mil S, Clements P, Holder J, Boudjelal M, et al. Intestinal detoxification limits the activation of hepatic pregnane X receptor by lithocholic acid. Drug Metab Dispos. 2010;38:143–149. doi: 10.1124/dmd.109.029306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakharukova M, Smetanina M, Kaledin V, Obut T, Merkulova T. The increased CAR-dependent metabolism of thyroid hormones in mice with high cancer susceptibility. Life Sci. 2010;87:439–444. doi: 10.1016/j.lfs.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Peeters RP, Kester MH, Wouters PJ, Kaptein E, van Toor H, Visser TJ, et al. Increased thyroxine sulfate levels in critically ill patients as a result of a decreased hepatic type I deiodinase activity. J Clin Endocrinol Metab. 2005;90:6460–6465. doi: 10.1210/jc.2005-0866. [DOI] [PubMed] [Google Scholar]
- Qian YM, Sun XJ, Tong MH, Li XP, Richa J, Song WC. Targeted disruption of the mouse estrogen sulfotransferase gene reveals a role of estrogen metabolism in intracrine and paracrine estrogen regulation. Endocrinolog. 2001;142:5342–5350. doi: 10.1210/endo.142.12.8540. [DOI] [PubMed] [Google Scholar]
- Radominska A, Comer KA, Zimniak P, Falany J, Iscan M, Falany CN. Human liver steroid sulphotransferase sulphates bile acids. Biochem J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovic B, Hussong R, Gerhauser C, Meinl W, Frank N, Becker H, et al. Xanthohumol, a prenylated chalcone from hops, modulates hepatic expression of genes involved in thyroid hormone distribution and metabolism. Mol Nutr Food Res. 2010;54(Suppl 2):S225–S235. doi: 10.1002/mnfr.200900489. [DOI] [PubMed] [Google Scholar]
- Rainey WE, Carr BR, Sasano H, Suzuki T, Mason JI. Dissecting human adrenal androgen production. Trends Endocrinol Metab. 2002;13:234–239. doi: 10.1016/s1043-2760(02)00609-4. [DOI] [PubMed] [Google Scholar]
- Riches Z, Stanley EL, Bloomer JC, Coughtrie MW. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab Dispos. 2009;37:2255–2261. doi: 10.1124/dmd.109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson E, Grieve DJ. Significance of peroxisome proliferator-activated receptors in the cardiovascular system in health and disease. Pharmacol Ther. 2009;122:246–263. doi: 10.1016/j.pharmthera.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA. Regulation of sulfotransferase and UDP-glucuronosyltransferase gene expression by the PPARs. PPAR Res. 2009;728941 doi: 10.1155/2009/728941. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutgers M, Heusdens FA, Visser TJ. Deiodination of iodothyronine sulfamates by type I iodothyronine deiodinase of rat liver. Endocrinology. 1991;129:1375–1381. doi: 10.1210/endo-129-3-1375. [DOI] [PubMed] [Google Scholar]
- Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, et al. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol. 2004;65:292–300. doi: 10.1124/mol.65.2.292. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y, Yanagisawa K, Katafuchi J, Ringer DP, Takami Y, Nakayama T, et al. Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J Biol Chem. 1998;273:33929–33935. doi: 10.1074/jbc.273.51.33929. [DOI] [PubMed] [Google Scholar]
- Salman ED, Faye-Peterson O, Falany CN. Hydroxysteroid sulfotransferase 2B1b expression and localization in normal human brain. Hormone Mol Biol Clin Invest. 2011a;8:445–454. doi: 10.1515/HMBCI.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman ED, He D, Runge-Morris M, Kocarek TA, Falany CN. Site-directed mutagenesis of human cytosolic sulfotransferase (SULT) 2B1b to phospho-mimetic Ser348Asp results in an isoform with increased catalytic activity. J Steroid Biochem Mol Biol. 2011b;127:315–323. doi: 10.1016/j.jsbmb.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankpal UT, Pius H, Khan M, Shukoor MI, Maliakal P, Lee CM, et al. Environmental factors in causing human cancers: emphasis on tumorigenesis. Tumour Biol. 2012;33:1265–1274. doi: 10.1007/s13277-012-0413-4. [DOI] [PubMed] [Google Scholar]
- Seely J, Amigh KS, Suzuki T, Mayhew B, Sasano H, Giguere V, et al. Transcriptional regulation of dehydroepiandrosterone sulfotransferase (SULT2A1) by estrogen-related receptor alpha. Endocrinology. 2005;146:3605–3613. doi: 10.1210/en.2004-1619. [DOI] [PubMed] [Google Scholar]
- Shimizu C, Fuda H, Yanai H, Strott CA. Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal expression, kinetic analysis of isoforms, and comparison with prototypical SULT2A1. Endocrinology. 2003;144:1186–1193. doi: 10.1210/en.2002-221011. [DOI] [PubMed] [Google Scholar]
- Shiraki T, Kamiya N, Shiki S, Kodama TS, Kakizuka A, Jingami H. Alpha,beta-unsaturated ketone is a core moiety of natural ligands for covalent binding to peroxisome proliferator-activated receptor gamma. J Biol Chem. 2005;280:14145–14153. doi: 10.1074/jbc.M500901200. [DOI] [PubMed] [Google Scholar]
- Shoda J, Tanaka N, Osuga T, Matsuura K, Miyazaki H. Altered bile acid metabolism in liver disease: concurrent occurrence of C-1 and C-6 hydroxylated bile acid metabolites and their preferential excretion into urine. J Lipid Res. 1990;31:249–259. [PubMed] [Google Scholar]
- Song CS, Echchgadda I, Baek BS, Ahn SC, Oh T, Roy AK, et al. Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid X receptor. J Biol Chem. 2001;276:42549–42556. doi: 10.1074/jbc.M107557200. [DOI] [PubMed] [Google Scholar]
- Song CS, Echchgadda I, Seo YK, Oh T, Kim S, Kim SA, et al. An essential role of the CAAT/enhancer binding protein-alpha in the vitamin D-induced expression of the human steroid/ bile acid-sulfotransferase (SULT2A1) Mol Endocrinol. 2006;20:795–808. doi: 10.1210/me.2005-0428. [DOI] [PubMed] [Google Scholar]
- Song CS, Jung MH, Kim SC, Hassan T, Roy AK, Chatterjee B. Tissue-specific and androgen-repressible regulation of the rat dehydroepiandrosterone sulfotransferase gene promoter. J Biol Chem. 1998;273:21856–21866. doi: 10.1074/jbc.273.34.21856. [DOI] [PubMed] [Google Scholar]
- Spaulding SW, Smith TJ, Hinkle PM, Davis FB, Kung MP, Roth JA. Studies on the biological activity of triiodothyronine sulfate. J Clin Endocrinol Metab. 1992;74:1062–1067. doi: 10.1210/jcem.74.5.1533227. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Hu E, Kim JB, Brun R. PPAR gamma and the control of adipogenesis. Biochimie. 1997;79:111–112. doi: 10.1016/s0300-9084(97)81500-3. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorrselaer A, Renaud JP, et al. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nat Struct Biol. 2003;10:820–825. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- Stiehl A, Becker M, Czygan P, Frohling W, Kommerell B, Rotthauwe HW, et al. Bile acids and their sulphated and glucuronidated derivatives in bile, plasma, and urine of children with intrahepatic cholestasis: effects of phenobarbital treatment. Eur J Clin Invest. 1980;10:307–316. doi: 10.1111/j.1365-2362.1980.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Strassburg CP, Strassburg A, Kneip S, Barut A, Tukey RH, Rodeck B, et al. Developmental aspects of human hepatic drug glucuronidation in young children and adults. Gut. 2002;50:259–265. doi: 10.1136/gut.50.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strott CA. Sulfonation and molecular action. Endocr Rev. 2002;23:703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- Strott CA, Higashi Y. Cholesterol sulfate in human physiology: what’s it all about? J Lipid Res. 2003;44:1268–1278. doi: 10.1194/jlr.R300005-JLR200. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Green WD, Vinal K, Woodrum TS, Moore R, Negishi M. Garlic extract diallyl sulfide (DAS) activates nuclear receptor CAR to induce the Sult1e1 gene in mouse liver. PLoS One. 2011;6:e21229. doi: 10.1371/journal.pone.0021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miki Y, Nakamura Y, Ito K, Sasano H. Steroid sulfatase and estrogen sulfotransferase in human carcinomas. Mol Cell Endocrinol. 2011;340:148–153. doi: 10.1016/j.mce.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Sakakibara Y, Mishiro E, Kouriki H, Nobe R, Kurogi K, et al. Molecular cloning, expression and characterization of a novel mouse SULT6 cytosolic sulfotransferase. J Biochem. 2009;146:399–405. doi: 10.1093/jb/mvp087. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Leung PS, Kenny TP, Gershwin ME, Bowlus CL. Immunological orchestration of liver fibrosis. Clin Rev Allergy Immunol. 2012;43:220–229. doi: 10.1007/s12016-012-8323-1. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Sasano H, Nishikawa T, Yabuki N, Muramatsu Y, Coughtrie MW, et al. Expression and activity of dehydroepiandrosterone sulfotransferase in human gastric mucosa. J Steroid Biochem Mol Biol. 2000;72:149–154. doi: 10.1016/s0960-0760(00)00020-0. [DOI] [PubMed] [Google Scholar]
- Tremblay GB, Kunath T, Bergeron D, Lapointe L, Champigny C, Bader JA, et al. Diethylstilbestrol regulates trophoblast stem cell differentiation as a ligand of orphan nuclear receptor ERR beta. Genes Dev. 2001;15:833–838. doi: 10.1101/gad.873401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi C, Falany CN, Morgenstern R, Swedmark S. Identification of a new subfamily of sulphotransferases: cloning and characterization of canine SULT1D1. Biochem J. 2001;356:891–897. doi: 10.1042/0264-6021:3560891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal H, Saini SP, Moschetta A, Mu Y, Zhou J, Gong H, et al. Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology. 2007;45:422–432. doi: 10.1002/hep.21494. [DOI] [PubMed] [Google Scholar]
- Vanacker JM, Bonnelye E, Chopin-Delannoy S, Delmarre C, Cavailles V, Laudet V. Transcriptional activities of the orphan nuclear receptor ERR alpha (estrogen receptor-related receptor-alpha) Mol Endocrinol. 1999;13:764–773. doi: 10.1210/mend.13.5.0281. [DOI] [PubMed] [Google Scholar]
- Visser TJ. Pathways of thyroid hormone metabolism. Acta Med Austriaca. 1996;23:10–16. [PubMed] [Google Scholar]
- Wang Y, Kumar N, Solt LA, Richardson TI, Helvering LM, Crumbley C, et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. J Biol Chem. 2010;285:5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Holloway MG. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol. 2009;76:215–228. doi: 10.1124/mol.109.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson TM, Jones SA, Moore JT, Kliewer SA. Chemical genomics: functional analysis of orphan nuclear receptors in the regulation of bile acid metabolism. Med Res Rev. 2001;21:513–522. doi: 10.1002/med.1023. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Murray IR, Qian J, Busch BB, Stevens WC, Jr., Martin R, et al. Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc Natl Acad Sci U S A. 2004;101:8912–8917. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Quinn CM, Gelissen IC, Brown AJ. Endogenous 24(S),25-epoxycholesterol fine-tunes acute control of cellular cholesterol homeostasis. J Biol Chem. 2008;283:700–707. doi: 10.1074/jbc.M706416200. [DOI] [PubMed] [Google Scholar]
- Wyde ME, Kirwan SE, Zhang F, Laughter A, Hoffman HB, Bartolucci-Page E, et al. Di-n-butyl phthalate activates constitutive androstane receptor and pregnane X receptor and enhances the expression of steroid-metabolizing enzymes in the liver of rat fetuses. Toxicol Sci. 2005;86:281–290. doi: 10.1093/toxsci/kfi204. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- Xu L, Bai Q, Rodriguez-Agudo D, Hylemon PB, Heuman DM, Pandak WM, et al. Regulation of hepatocyte lipid metabolism and inflammatory response by 25-hydroxycholesterol and 25-hydroxycholesterol-3-sulfate. Lipids. 2010;45:821–832. doi: 10.1007/s11745-010-3451-y. [DOI] [PubMed] [Google Scholar]
- Yanagiba Y, Ito Y, Kamijima M, Gonzalez FJ, Nakajima T. Octachlorostyrene induces cytochrome P450, UDP-glucuronosyltransferase, and sulfotransferase via the aryl hydrocarbon receptor and constitutive androstane receptor. Toxicol Sci. 2009;111:19–26. doi: 10.1093/toxsci/kfp130. [DOI] [PubMed] [Google Scholar]
- Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, et al. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J Biol Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- Yin L, Ma H, Ge X, Edwards PA, Zhang Y. Hepatic hepatocyte nuclear factor 4alpha is essential for maintaining triglyceride and cholesterol homeostasis. Arterioscler Thromb Vasc Biol. 2011;31:328–336. doi: 10.1161/ATVBAHA.110.217828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadjali F, Santana-Farre R, Mirecki-Garrido M, Ellis E, Norstedt G, Fernandez-Perez L, et al. Liver X receptor agonist downregulates growth hormone signaling in the liver. Hormone Mol Biol Clin Invest. 2011;8:471–478. doi: 10.1515/HMBCI.2011.125. [DOI] [PubMed] [Google Scholar]
- Zhang H, Varmalova O, Vargas FM, Falany CN, Leyh TS. Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J Biol Chem. 1998;273:10888–10892. doi: 10.1074/jbc.273.18.10888. [DOI] [PubMed] [Google Scholar]
- Zheng P. Neuroactive steroid regulation of neurotransmitter release in the CNS: action, mechanism, and possible significance. Prog Neurobiol. 2009;89:134–152. doi: 10.1016/j.pneurobio.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Zollner G, Wagner M, Fickert P, Silbert D, Gumhold J, Zatloukal K, et al. Expression of bile acid synthesis and detoxification enzymes and the alternative bile acid efflux pump MRP4 in patients with primary biliary cirrhosis. Liver Int. 2007;27:920–929. doi: 10.1111/j.1478-3231.2007.01506.x. [DOI] [PubMed] [Google Scholar]