Significance
This study demonstrates mismatch repair (MMR) reactions reconstituted in vitro with purified Saccharomyces cerevisiae proteins. Biochemical analysis of MMR in vitro showed that MMR required mispair binding by the MutS homolog 2–MutS homolog 6 complex and corresponded to the Exonuclease 1-dependent subpathway of MMR. The reactions observed involved the formation of long excision tracts whose length was consistent with the length of MMR-dependent gene conversion tracts in vivo. The availability of this reconstituted MMR reaction now allows the wealth of mutations affecting MMR generated from the genetic analysis of S. cerevisiae MMR mechanisms in vivo to be used in biochemical reconstitution studies whose ultimate goal is to reconstitute MMR linked to both DNA replication and recombination.
Keywords: DNA replication fidelity, genome instability, mutator phenotype, cancer, mutagenesis
Abstract
A problem in understanding eukaryotic DNA mismatch repair (MMR) mechanisms is linking insights into MMR mechanisms from genetics and cell-biology studies with those from biochemical studies of MMR proteins and reconstituted MMR reactions. This type of analysis has proven difficult because reconstitution approaches have been most successful for human MMR whereas analysis of MMR in vivo has been most advanced in the yeast Saccharomyces cerevisiae. Here, we describe the reconstitution of MMR reactions using purified S. cerevisiae proteins and mispair-containing DNA substrates. A mixture of MutS homolog 2 (Msh2)–MutS homolog 6, Exonuclease 1, replication protein A, replication factor C-Δ1N, proliferating cell nuclear antigen and DNA polymerase δ was found to repair substrates containing TG, CC, +1 (+T), +2 (+GC), and +4 (+ACGA) mispairs and either a 5′ or 3′ strand interruption with different efficiencies. The Msh2–MutS homolog 3 mispair recognition protein could substitute for the Msh2–Msh6 mispair recognition protein and showed a different specificity of repair of the different mispairs whereas addition of MutL homolog 1–postmeiotic segregation 1 had no affect on MMR. Repair was catalytic, with as many as 11 substrates repaired per molecule of Exo1. Repair of the substrates containing either a 5′ or 3′ strand interruption occurred by mispair binding-dependent 5′ excision and subsequent resynthesis with excision tracts of up to ∼2.9 kb occurring during the repair of the substrate with a 3′ strand interruption. The availability of this reconstituted MMR reaction now makes possible detailed biochemical studies of the wealth of mutations identified that affect S. cerevisiae MMR.
DNA mismatch repair (MMR) is a critical DNA repair pathway that is coupled to DNA replication in eukaryotes where it corrects misincorporation errors made during DNA replication (1–9). This pathway prevents mutations and acts to prevent the development of cancer (10, 11). MMR also contributes to gene conversion by repairing mispaired bases that occur during the formation of recombination intermediates (3, 4, 12). Finally, MMR acts to suppress recombination between divergent but homologous DNA sequences, thereby preventing the formation of genome rearrangements that can result from nonallelic homologous recombination (4, 13–15).
Our knowledge of the mechanism of eukaryotic MMR comes from several general lines of investigation (3–9). Studies of bacterial MMR have provided a basic mechanistic framework for comparative studies (5). Genetic and cell-biology studies, primarily in Saccharomyces cerevisiae, have identified eukaryotic MMR genes, provided models for how their gene products define MMR pathways, and elucidated some of the details of how MMR pathways interact with replication (1–4). Reconstitution studies, primarily in human systems, have identified some of the catalytic features of eukaryotic MMR (7–9, 16, 17). Biochemical and structural studies of S. cerevisiae and human MMR proteins have provided information about the function of individual MMR proteins (6–9).
In eukaryotic MMR, mispairs are bound by MutS homolog 2 (Msh2)–MutS homolog 6 (Msh6) and Msh2–MutS homolog 3 (Msh3), two partially redundant complexes of MutS-related proteins (3, 4, 18, 19). These complexes recruit a MutL-related complex, called MutL homoloh 1 (Mlh1)–postmeiotic segregation 1 (Pms1) in S. cerevisiae and Mlh1–postmeiotic segregation 2 (Pms2) in human and mouse (3, 4, 20–23). The Mlh1–Pms1/Pms2 complex has an endonuclease activity suggested to play a role in the initiation of the excision step of MMR (24, 25). Downstream of mismatch recognition is a mispair excision step that can be catalyzed by Exonuclease 1 (Exo1) (26–28); however, defects in both S. cerevisiae and mouse Exo1 result in only a partial MMR deficiency, suggesting the existence of additional excision mechanisms (26, 27, 29). DNA polymerase δ, the single-strand DNA binding protein replication protein A (RPA), the sliding clamp proliferating cell nuclear antigen (PCNA), and the clamp loader replication factor C (RFC) are also required for MMR at different steps, including activation of Mlh1–Pms1/Pms2, stimulation of Exo1, potentially in Exo1-independent mispair excision, and in the gap-filling resynthesis steps of MMR (3, 16, 17, 24, 27, 30–36). Although much is known about these core MMR proteins, it is not well understood how eukaryotic MMR is coupled to DNA replication (1, 2), how excision is targeted to the newly replicated strand (1, 25, 37–39), or how different MMR mechanisms such as Exo1-dependent and -independent subpathways are selected or how many such subpathways exist (1, 24, 27, 29).
S. cerevisiae has provided a number of tools for studying MMR, including forward genetic screens for mutations affecting MMR, including dominant and separation-of-function mutations, the ability to evaluate structure-based mutations in vivo, cell biological tools for visualizing and analyzing MMR proteins in vivo, and overproduction of individual MMR proteins for biochemical analysis. However, linking these tools with biochemical systems that catalyze MMR reactions in vitro for mechanistic studies has not yet been possible. Here, we describe the development of MMR reactions reconstituted using purified proteins for the analysis of MMR mechanisms.
Results
MMR Is Catalyzed in Vitro by Purified S. cerevisiae Proteins.
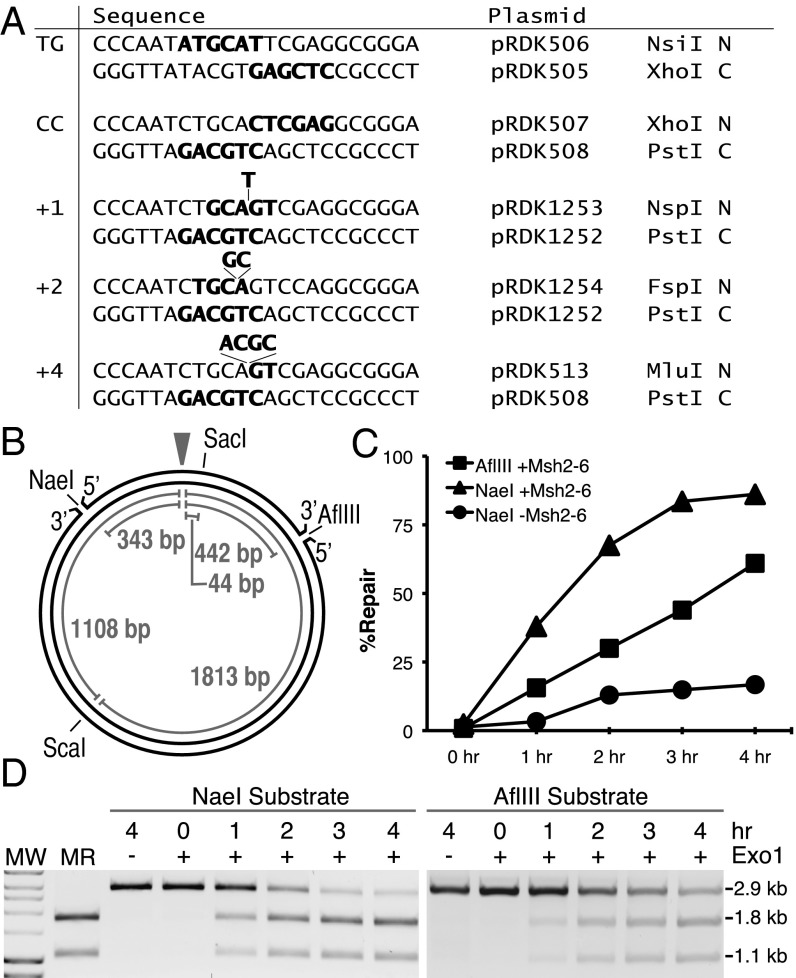
In the studies described here, we used a series of circular phagemid-based mispair-containing substrates that allow detection of MMR directed by a strand interruption called a nick. These substrates contained TG, CC, +1 (+T), +2 (+GC), or +4 (+ACGA) mispairs that disrupted a restriction endonuclease cleavage site on the continuous strand, which was restored upon MMR-mediated excision and resynthesis of the nicked strand (Fig. 1A). A nick was present either at an NaeI site 343 bp 5′ to the mispair or at an AflIII site 442 bp 3′ to the mispair (Fig. 1B); these nicks are further from the mispair than those used in human MMR reactions that were 128 bp 5′ and 141 bp 3′ from the mispair, respectively. Repair of the nicked strand was detected by cleavage with the restriction endonuclease whose recognition sequence in the continuous strand was restored at the mispair site and ScaI to produce a diagnostic pair of 1.1-kb and 1.8-kb fragments. We also constructed a homoduplex control substrate with the sequence of pRDK1252 (Fig. 1A) and a 5′ nick at the NaeI site.
Fig. 1.
Repair of pBluescript-based mispair-containing plasmids in a reconstituted in vitro MMR system. (A) Sequence of the polylinker region between the ApaI and BamHI sites of different substrates indicating the mispair, the restriction sites in each strand, and the plasmid from which each strand was derived. C, continuous strand N, nicked strand. (B) Map of the pBluescript plasmid showing the positions of the various features used in the assays presented and the relevant distances between key sites. The mispair is indicated by the arrowhead. (C and D) Repair of the +1 (+T) substrates containing either a 5′ nick at the NaeI site or a 3′ nick at the AflIII site in reactions for the indicated times containing Msh2–Msh6, Mlh1–Pms1, Exo1, PCNA, RFC-Δ1N, RPA, and DNA polymerase δ with presence/absence of Msh2–Msh6 or Exo1 as indicated. Repair was detected by digestion with PstI and ScaI, and the repair products were visualized after agarose gel electrophoresis (D) and the DNA species seen on the gels were quantified (C). MR, markers for repair products; MW, molecular weight markers. Note: 100% repair is repair of 200 ng or 105.5 fmol of substrate.
It was possible to observe a repair reaction with substrates containing either a strand interruption at the NaeI site (5′ nick) or a strand interruption at the AflIII site (3′ nick) with a +1 insertion mispair (+T) in the nicked strand catalyzed by a combination of Msh2–Msh6, Mlh1–Pms1, Exo1, RPA, RFC-Δ1N (a form of RFC containing an N-terminal truncation of subunit 1), PCNA, and DNA polymerase δ (Fig. 1 C and D). Robust repair of the AflIII substrate required fourfold greater amounts of Exo1 than repair of the NaeI substrate. The reaction was linear for up to 3 h (NaeI substrate) or 4 h (AflIII substrate), resulting in repair of as much as 85% of the substrate. Omission of Msh2–Msh6 markedly reduced the fraction of substrate repaired (Fig. 1C). The residual repair in the absence of Msh2–Msh6 was likely due to Exo1 as omission of Exo1 completely eliminated repair (Fig. 1D).
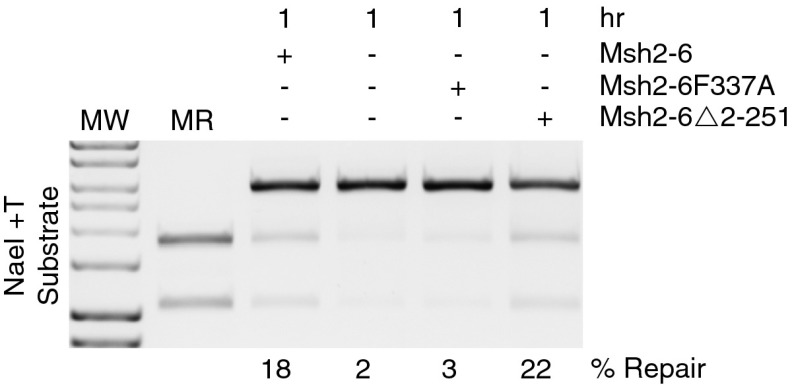
To further evaluate the requirement for Msh2–Msh6, the ability of two different mutant Msh2–Msh6 proteins to substitute for the wild-type complex in the repair of the NaeI +T mispair substrate was evaluated (Fig. 2). The mutant Msh2–Msh6 complex containing the Msh6-F337A substitution that eliminates mispair recognition (40) did not support repair, demonstrating that mispair recognition was required for repair. In contrast, the mutant Msh2–Msh6 complex containing a deletion of Msh6 residues 2 through 251 that eliminates the Msh6 PCNA interacting site but is proficient for mispair binding (41) fully supported repair; this lack of a defect is consistent with studies showing that mutant human Msh2–Msh6 complexes lacking the ability of Msh6 to interact with PCNA could fully (42) or partially (43) complement the in vitro MMR defect of extracts of Msh6-defective HCT15 cells. This lack of a defect indicates that the Msh2–Msh6–PCNA interaction is not required for MMR in vitro and that the requirement for PCNA demonstrated below likely reflects a role for PCNA at the gap-filling step of MMR.
Fig. 2.
Reconstituted MMR reactions in vitro require the ability of Msh2–Msh6 to bind mispairs but not PCNA. Reconstituted mismatch repair of the +1 (+T) substrate containing a 5′ nick at the NaeI site was performed for 1 h as described in Fig. 1. The presence or absence of Msh2–Msh6, the mispair binding defective Msh2–Msh6–F337A protein, and the PCNA binding defective Msh2–Msh6–Δ2–251 protein and the % repair are as indicated. MR, markers for repair products; MW, molecular weight markers.
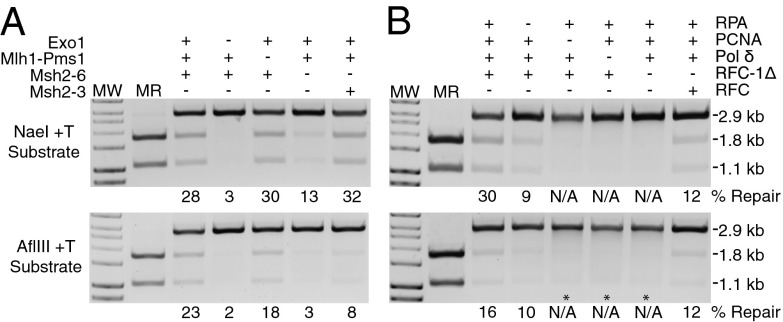
By omitting individual proteins, it was found that repair of both the NaeI and AflIII +1 (+T) mispair substrates required Msh2–Msh6, Exo1, RFC-Δ1N, PCNA, and DNA polymerase δ, (Fig. 3 A and B) whereas omission of RPA caused a partial repair defect (Fig. 3B). Msh2–Msh3 was able to substitute for Msh2–Msh6 (Fig. 3A), consistent with genetic results indicating that both Msh2–Msh6 and Msh2–Msh3 can function in the repair of single-base insertion/deletion mispairs in vivo (18, 44). Native RFC could substitute for RFC-Δ1N (Fig. 3B). Mlh1-Pms1 was not required for repair of either substrate (Fig. 3A), even though our Mlh1–Pms1 preparations have RFC-PCNA–stimulated endonuclease activity (45). The lack of an Mlh1–Pms1 requirement is consistent with the fact that both substrates are repaired by a 5′ excision reaction (see The NaeI and AflIII Substrates Are Repaired by Short and Long Patch 5′ to 3′ Excision Repair, Respectively). In both sets of experiments in which RFC-Δ1N, PCNA, or DNA polymerase δ were omitted, there was a diffuse region of what appeared to be degraded DNA migrating faster than the 2.9-kb ScaI linearized substrate. This affect was more evident in the reactions with the AflIII substrates where also visible was a small amount of a DNA species that migrated at the position of single-stranded circular pRDK1252 DNA (* in Fig. 3B, Lower). These excision products will be discussed under The NaeI and AflIII Substrates Are Repaired by Short and Long Patch 5′ to 3′ Excision Repair, Respectively.
Fig. 3.
Repair of the +T substrate containing a 5′ nick at the NaeI site or a 3′ nick at the AflIII site in vitro requires Msh2–Msh6 or Msh2–Msh3, and Exo1, PCNA, RFC, RPA, and DNA polymerase δ but not Mlh1–Pms1. Repair of the indicated substrate in 3-h reactions was assayed by digestion with PstI and ScaI as indicated in Fig. 1. The effect of omission of Exo1, Mlh1–Pms1, and Msh2–Msh6 or substitution of Msh2–Msh3 for Msh2–Msh6 is shown in A, and the effect of omission of RPA, PCNA, DNA polymerase δ, and RFC-Δ1N or substitution of RFC for RFC-Δ1N is shown in B. The * in B shows the position of a DNA species formed in the −PCNA, −DNA polymerase δ, and −RFC-Δ1N reactions with the AflIII substrate that has the same mobility as single-stranded pBluescript circular DNA. MR, markers for repair products; MW, molecular weight markers; N/A, no visible repair.
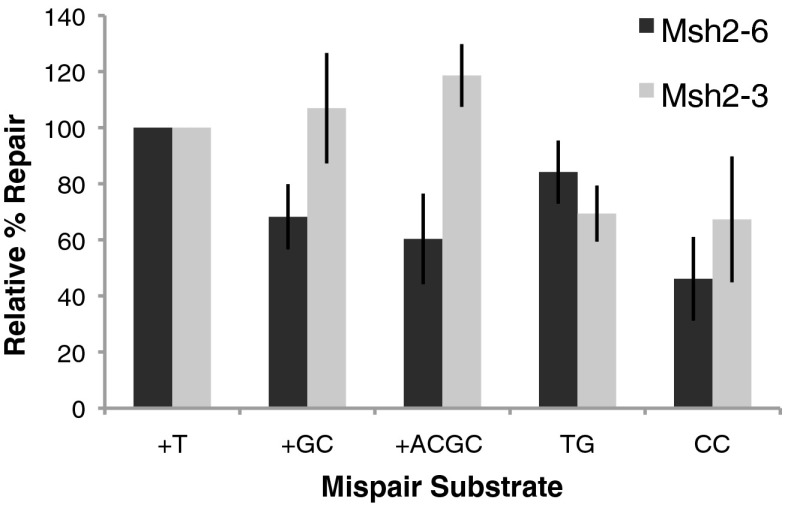
Substrates containing +1 (+T), +2 (+GC), or +4 (+ACGA) insertion mispairs or TG or CC mispairs and a nick at the NaeI site were tested in repair reactions containing either Msh2–Msh6 or Msh2–Msh3 (Fig. 4). All five substrates were efficiently repaired in both the Msh2–Msh6- and Msh2–Msh3-containing reactions although there were small but measurable differences in the efficiency of repair. In the Msh2–Msh6 reaction, the +1 and TG substrates were repaired at close to the same levels and to a modestly greater extent than repair of the +2, +4, and CC substrates. In contrast, in the Msh2–Msh3 reactions, the +2 and +4 substrates were repaired to a modestly greater extent than the +1, TG, and CC substrates. These results are consistent with the results of analysis of MMR in vivo indicating that Msh2–Msh6 repairs insertion mispairs with a preference for +1 versus +2 and +4 mispairs and repairs base:base mispairs with CC mispairs being less well repaired whereas Msh2–Msh3 repairs insertion/deletion mispairs with a preference for +2 and +4 versus +1 mispairs and inefficiently repairs base:base mispairs with a preference for CC mispairs versus TG mispairs (18, 44, 46).
Fig. 4.
Mispair specificity of Msh2–Msh6 and Msh2–Msh3 dependent repair. Reactions containing either Msh2–Msh6 or Msh2–Msh3 were performed for 3 h with substrates containing a 5′ nick at the NaeI site and containing +1 (+T), +2 (+GC), +4 (+ACGC), TG, and CC mispairs as indicated (See Figs. 1 and 3). The extent of repair indicated was that relative to the repair of the +1 (+T) substrate present in each set of reactions. The average extent of repair of the +1 (+T) substrate in the Msh2–Msh6 reactions was 49% and in the Msh2–Msh3 reactions was to 67%.
The NaeI and AflIII Substrates Are Repaired by Short and Long Patch 5′ to 3′ Excision Repair, Respectively.
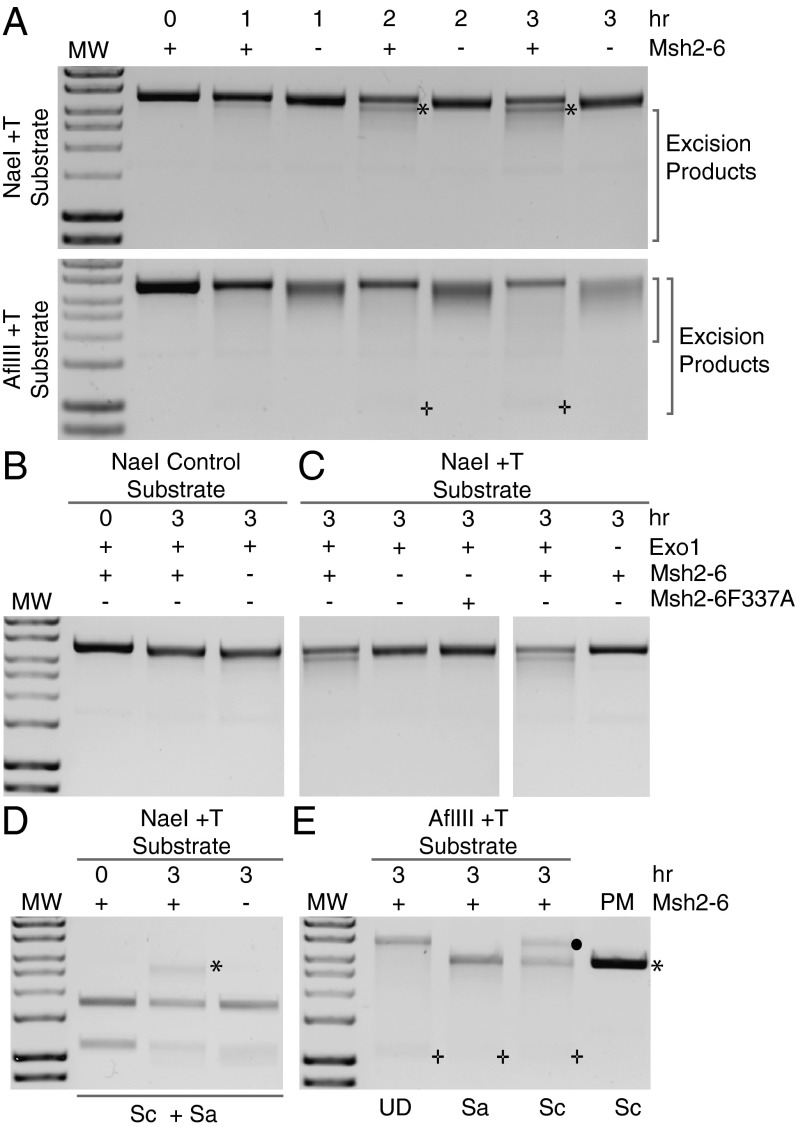
In the experiments with the +1 (+T) mispair substrates in which individual proteins were omitted (Fig. 3), in the absence of Msh2–Msh6 or Exo1, the ScaI-digested DNA was present as the full-length linear DNA species whereas, in the absence of RPA, and to an even greater extent in the absence of RFC, PCNA, or DNA polymerase δ, less of the DNA was present as the full-length linear species and there was a smear of more rapidly migrating DNA species suggestive of excision products formed in the absence of DNA repair synthesis. To further analyze this excision, reactions with the +1 (+T) mispair substrates lacking DNA polymerase δ with or without Msh2–Msh6 were performed for different times and analyzed by agarose gel electrophoresis without digestion of the DNA with a restriction endonuclease (Fig. 5A). For both the NaeI substrate and the AflIII substrate, there was a time-dependent conversion of the substrate to species that migrated more rapidly than the substrate DNA. With each substrate, the presence of Msh2–Msh6 resulted in conversion of the substrate to more of the rapidly migrating species than in the absence of Msh2–Msh6. In the case of the NaeI substrate, a discrete excision species that ran slightly ahead of the substrate DNA was evident in the reactions with Msh2–Msh6 whereas, in the AflIII reactions, a small amount of a discrete species migrating at or near the position of a single-stranded circular DNA marker was evident in the reactions with Msh2–Msh6. Excision reactions performed with an NaeI substrate lacking a mispair showed no stimulation of excision by Msh2–Msh6, indicating that a mispair was required for Msh2–Msh6-stimulated excision (Fig. 5B). Other control experiments performed with the NaeI substrate showed that the excision reaction was eliminated by substituting the mispair binding defective Msh2–Msh6–F337A complex (40) for wild-type Msh2–Msh6 or by omitting Exo1 (Fig. 5C) and that the extent of excision was not affected by the presence or absence of Mlh1-Pms1 (Fig. S1).
Fig. 5.
Excision on the +1 (+T) substrate containing either a 5′ nick at the NaeI site or a 3′ nick at the AflIII site is in the 5′-to-3′ direction and is stimulated by Msh2–Msh6. Excision reactions were performed with +T substrates containing a 5′ nick at the NaeI site or a 3′ nick at the AflIII site in reactions containing Mlh1–Pms1, Exo1, PCNA, RFC-Δ1N, RPA without DNA polymerase δ with or without Msh2–Msh6 or with Msh2–Msh6–F337A as indicated for the indicated times. The reaction products were analyzed by agarose gel electrophoresis without prior digestion with PstI. MW indicates size standards. (A) Excision reactions were performed for 0–3 h. The * indicates a prominent excision product seen with the NaeI substrate in reactions containing Msh2–Msh6. The + shows the position of a DNA species that migrates at the position of single-stranded pBluescript circular DNA seen with the AflIII substrate in reactions containing Msh2–Msh6. (B) Excision reactions performed with a substrate containing a 5′ nick at the NaeI site but lacking a mispair showing the absence of apparent excision products in reactions containing Msh2–Msh6. (C) Excision reactions performed with a +1 (+T) substrate containing a 5′ nick at the NaeI site where Msh2–Msh6 or Exo1 were omitted or the mispair binding defective Msh2–Msh6–F337A protein was substituted for Msh2–Msh6 showing that Msh2–Msh6, mispair binding, and Exo1 were required for the formation of excision products. (D) Analysis of excision products formed with the +T substrate with a 5′ nick at the NaeI site by digestion with SacI (Sa, 387 bp 5′ from the NaeI site) + ScaI (Sc, 2,156 bp 5′ from the NaeI site). The* indicates the full-length linear double-stranded DNA species seen in the presence of Msh2–Msh6. (E) Analysis of excision products formed with the +1 (+T) mispair 3′ nick AflIII substrate by digestion with SacI (Sa, 2,523 bp 5′ from the NaeI site) or ScaI (Sc, 1,371 bp 5′ from the AflIII site) or without digestion (UD). The • indicates the ScaI-resistant nicked circular double-stranded DNA species formed. The * indicates the full-length linear double-stranded DNA species. The + indicates a DNA species that migrates at the position of single-stranded pBluescript circular DNA. PM indicates 100 ng of unincubated substrate DNA digested with ScaI as a control for complete digestion.
To demonstrate that the direction of excision was 5′ to 3′ from the nick, we took advantage of the inability of restriction endonucleases to cleave sites in single-stranded DNA (47). Excision in the 5′-to-3′ direction past the +1 (+T) mispair in the NaeI substrate was monitored by loss of cleavage of an SacI site located 44 bp 3′ to the mispair (Figs. 1A and 5D). In the absence of excision, double digestion with SacI and ScaI produced 1.1- and 1.8-kb species. In excision reactions containing Msh2–Msh6, full-length linear DNA was also generated due to resistance to cleavage by SacI. In the absence of Msh2–Msh6, this full-length linear DNA product was absent, indicating that excision did not reach the SacI site. Identical experiments were used to characterize the excision products formed with the AflIII substrate where the +1 (+T) mispair was located 2.48 kb 5′ of the nick at the AflIII site and 0.44 kb 3′ of the nick. Excision in the 5′-to-3′ direction from the AflIII site was following by monitoring digestion at the ScaI site 1.4 kb 3′ of the nick whereas 3′-to-5′ excision was monitored by digestion at the SacI site 0.4 kb 5′ from the nick (Figs. 1A and 5E). These experiments showed that some of the excision products formed in the reactions with Msh2–Msh6 had extended in the 5′-to-3′ direction past the ScaI site, rendering it resistant to digestion, resulting in the formation of ScaI-resistant circular DNA. In contrast, there appeared to be no excision in the 3′-to-5′ direction leading to the formation of SacI-resistant circular DNA.
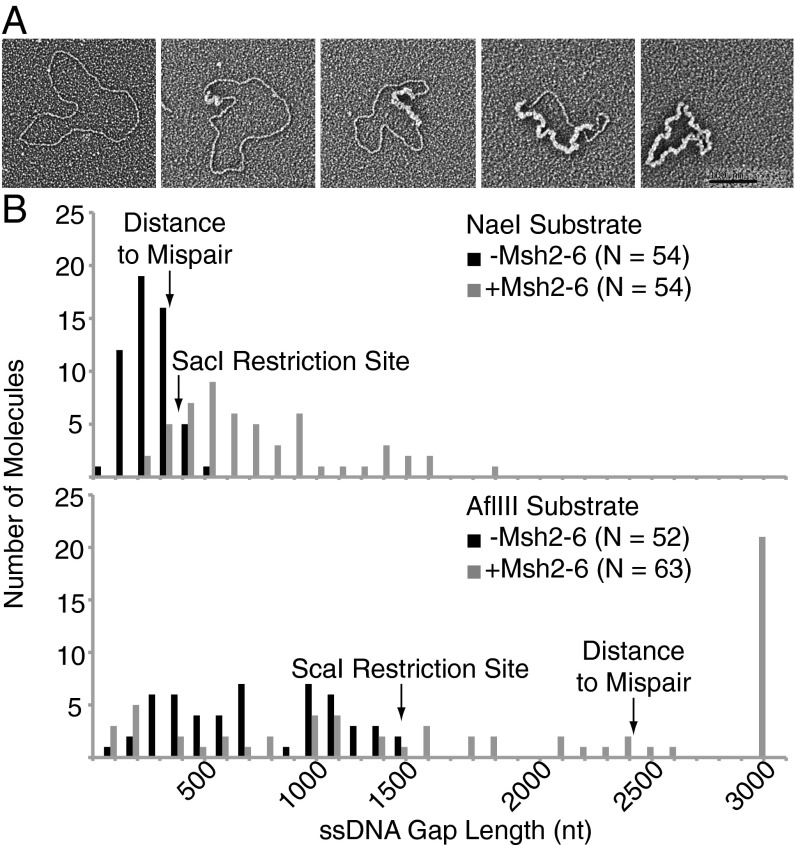
The excision products formed in 3-h reactions with substrates containing a +1 (+T) mispair and nicks at either the NaeI or AflIII sites with or without Msh2–Msh6 were then characterized by electron microscopy using Escherichia coli single-stranded DNA binding protein (SSB) to stain the single-stranded regions of DNA. This analysis revealed a variety of molecules ranging from those lacking apparent single-stranded regions (no excision) to those lacking apparent double-stranded regions (complete excision) (Fig. 6A); however, note that double-stranded regions of fewer than ∼150 bp and single-stranded regions of fewer than ∼100 bp are likely not visible using this technique. The distribution of the lengths of the single-stranded regions formed in these reactions is plotted in Fig. 6B along with the position of the mispair in each substrate in the 5′ direction from the nick in the substrate. With both substrates, there were more molecules with excision tracts extending past the site of the mispair, as well as extending past the SacI site in the substrate containing the nick at the NaeI site or past the ScaI site in the substrate containing the nick at the AflIII site, in reactions containing Msh2–Msh6 compared with reactions lacking Msh2-Msh6. This distribution of excision products is consistent with the Msh2–Msh6 dependence of MMR observed (Figs. 1 and 3) and the electrophoretic and restriction digestion based analysis of the excision products (Fig. 5).
Fig. 6.
Electron microscopy shows that excision results in single-stranded gaps whose length is stimulated by Msh2–Msh6. Excision reactions were performed for 3 h with +1 (+T) substrates containing either a 5′ nick at the NaeI site or a 3′ nick at the AflIII site in reactions containing Exo1, PCNA, RFC-Δ1N, and RPA without DNA polymerase δ with or without Msh2–Msh6 as indicated. In addition, the reaction with the substrate with the strand interruption at the NaeI site also contained Mlh1–Pms1. (A) A series of representative DNA molecules obtained, with single-stranded DNA stained with E. coli SSB and thus appearing thicker. (Left) Double-stranded circular DNA followed by, from left to right, double-stranded circular DNAs with increasing sizes of single-stranded gaps. (Right) Single-stranded circular DNA. (Scale bar: 100 nm.) (B) Plot of the distribution of single-stranded gap sizes observed. The positions of the mispair, SacI, and ScaI sites are indicated relative to the position of the nick assuming excision is in the 5′ to 3′ direction.
Discussion
A critical problem in understanding the mechanisms of MMR is linking insights into MMR mechanisms from genetics and cell-biology studies with biochemical studies of individual MMR proteins and reconstituted MMR reactions. In particular, this type of analysis would allow evaluating how well in vitro MMR reactions correspond to in vivo MMR reactions and facilitate reconstituting features of MMR that have thus far been observed only in vivo. Thus far, reconstitution approaches have been most successful for human MMR whereas analysis of MMR in vivo has been most advanced in S. cerevisiae. Consequently, because many human MMR proteins must be expressed using baculovirus or purified directly from mammalian cells (17, 22, 43, 47–49) and because human and S. cerevisiae MMR proteins are not completely homologous, it can potentially be difficult to evaluate insights from S. cerevisiae genetics in human MMR reconstitution studies. To overcome these barriers, in the present study, we have reconstituted nick-directed MMR reactions using S. cerevisiae proteins. Critical to these efforts has been the optimization of protein overproduction and purification to allow purification of proteins using plasmid-based overproduction in small volumes of cells to facilitate the investigation of mutant MMR proteins.
We observed that a mixture of Msh2–Msh6, Exo1, RPA, PCNA, RFC, and DNA polymerase δ would repair a mispaired phagemid substrate containing a +1 (+T) mispair and either 5′ strand interruption located 343 bp from the mispair or a 3′ strand interruption located 442 bp from the mispair. Msh2–Msh6, Exo1, PCNA, RFC, and DNA polymerase δ were absolutely required for repair whereas RPA was partially required and Msh2–Msh3 could substitute for Msh2–Msh6. The reactions were approximately linear for up to 3–4 h and could approach repair of 100% of the substrate DNA. The slow kinetics most likely reflects the rate of excision because multiple repair reactions occur per Exo1 molecule during the time course (∼11 NaeI substrates and ∼3 AflIII substrates repaired per Exo1) and higher levels of Exo1 are required to observe repair of the AflIII substrate that requires a much longer excision tract than repair of the NaeI substrate. The Msh6–F337A substitution that eliminates mispair recognition by Msh2–Msh6 (40) eliminated repair, demonstrating that the observed reaction was a true mispair-driven repair reaction rather than a nonspecific excision and resynthesis reaction. Eliminating the ability of Msh2–Msh6 to interact with PCNA had no affect on MMR in vitro even though PCNA was required for the complete repair reaction in vitro. This lack of a defect could reflect the fact that Exo1-dependent MMR does not require the Msh2–Msh6–PCNA interaction in vivo (1) and that the requirement for PCNA in the in vitro MMR reactions is most likely at the gap-filling step (17, 32, 34). The ability to perform this type of genetic analysis in vitro demonstrates a key feature of the in vitro MMR system reported here and supports the view that the reconstituted reactions observed reflect some of the properties on MMR in vivo.
The Msh2–Msh6 and Msh2–Msh3 mispair recognition complexes were essentially equally active in promoting MMR in vitro. Both complexes promoted the repair of two different base–base mispairs and three different insertion mispairs. Msh2–Msh6 promoted the repair of the +1 base insertion and TG mispairs to a greater extent than the +2 and +4 base insertion and CC mispairs whereas Msh2–Msh3 promoted the repair of the +2 and +4 base insertion mispairs to a greater extent than the +1 base insertion mispair and to an even greater extent than the TG and CC mispairs. The observed repair of the insertion mispairs parallels the results of genetic studies in S. cerevisiae indicating that Msh2–Msh6 and Msh2–Msh3 can repair +1 base insertion mispairs with relatively equal efficiency whereas repair of larger insertion mispairs is more dependent on Msh2–Msh3 than on Msh2–Msh6 (18, 44, 46). The relatively efficient repair of the +2 and +4 base insertion mispairs by Msh2–Msh6 was greater than predicted from genetic studies but was similar to the efficient repair of large insertion mispairs promoted by human cell extracts complemented by either Msh2–Msh6 or Msh2–Msh3 (50). The observation that Msh2–Msh3 could promote repair of TG and CC mispairs and that Msh2–Msh6 could promote repair of TG and to a lesser extent CC mispairs is consistent with genetic studies that have indicated that Msh2–Msh3 can repair some base–base mispairs in vivo (46); note that inefficient repair of a TG mispair mediated by human Msh2–Msh3 has been observed in a reconstituted MMR system (16). Interestingly, the relative differences between the efficiency of repair of the different mispairs was less than might be predicted from the results of genetic studies of MMR in S. cerevisiae, suggesting that mispair affinity may not be rate-limiting in MMR reactions in vitro and that other features of MMR in vivo, such as sequence context affects (51, 52), may determine the efficiency of repair of different mispairs in vivo.
In the absence of DNA polymerase δ as well as other proteins required for DNA synthesis (RFC or PCNA), an excision reaction was observed. This excision reaction required Exo1 and was stimulated by both a mispair and Msh2–Msh6, where mispair binding by Msh2–Msh6 was required for stimulating excision. Strikingly, the excision tracts observed were very long, as long as 1,750 bp in reactions with the substrate containing a nick at the NaeI site and 2,900 bp with the substrate containing a nick at the AflIII site, which approximates the length of MMR excision tracts predicted from genetic studies of gene conversion (12, 53). The electron microscopy experiments indicated that the length of the longest excision tract was threefold longer in the presence of Msh2–Msh6. These excision tract lengths were much longer than seen in human excision reactions in vitro (16, 47) even though the reactions described here used less Exo1 (20% and 80% of the amount of Exo1 used in human MMR reactions for the substrate containing a nick at the NaeI site and at the AflIII site, respectively) than used in human excision reactions (16, 17). Although these differences could reflect the longer incubations used here, it is also possible that human Exo1 is less active than S. cerevisiae Exo1. In contrast to the results of one study of excision by human MMR proteins that Mlh1–Pms2 promotes termination of excision (16), Mlh1–Pms1 did not appear to have any effect on excision, and, even in the presence of Mlh1–Pms1, mispair-stimulated excision could extend well past the mispair. These results raise the possibility that, in the S. cerevisiae system, termination of excision may involve some type of competition between excision and the DNA synthesis machinery or coupling of excision to the DNA synthesis machinery.
The lack of an Mlh1–Pms1 requirement in the S. cerevisiae system, even though our Mlh1–Pms1 preparations are fully active for RFC–PCNA-dependent endonuclease activity (45), conflicted with the results of some human MMR studies where an Mlh1–Pms2 endonuclease-dependent MMR reaction can occur (17). The most likely explanations for this lack of an Mlh1-Pms1 requirement are that the Mlh1—Pms1 endonuclease is not active under the reaction conditions used here or that the potent 5′ excision-based MMR reaction seen with the AflIII substrate containing a 3′ nick outcompetes any Mlh1–Pms1 endonuclease-dependent reaction. However, recent studies have indicated that S. cerevisiae Exo1-dependent MMR is not nearly as dependent on the Mlh1–Pms1 endonuclease as Exo1-independent MMR is (1, 39), suggesting that additional components or different reaction conditions may be required for an Mlh1–Pms1-dependent reaction in vitro. Establishing an Mlh1–Pms1-dependent reaction in vitro remains a key challenge for the S. cerevisiae system.
Materials and Methods
Protein Purification.
The proteins used in the described studies were expressed in either E. coli or S. cerevisiae as described in SI Materials and Methods. Yields ranged from 100 μg to 500 μg per L of expressing cells. All of the protein preparations were confirmed to be greater than 95% pure by SDS/PAGE followed by staining the gels with Coomassie Blue.
DNA Substrates.
DNA substrates were constructed by annealing the combinations of mutant single-stranded circular pBluescript SK+ DNAs and linearized, denatured mutant double-stranded pBluescript SK+ DNAs indicated in Fig. 1. The methods used for this purpose are described in SI Materials and Methods.
Repair Assays.
MMR assays were based on modification of previously published procedures (16, 17, 35, 54). Proteins were diluted if necessary with 7.5 mM Hepes, pH 7.5, 10% (vol/vol) glycerol, 200 mM KCl, 1 mM DTT, and 0.5 mg/mL BSA. Then, 8.4 fmol of Exo1, 390 fmol of Mlh1–Pms1, 390 fmol of Msh2–Msh6, 290 fmol of PCNA, 80 fmol of DNA Polymerase δ, 220 fmol of RFC-Δ1N, and 1,800 fmol of RPA were combined into 8 μL and mixed with 2 μL of 100 ng/μL NaeI substrate and 10 μL of 33 mM Tris, pH 7.6, 75 mM KCl, 2.5 mM ATP, 1.66 mM Glutathione, 8.3 mM MgCl2, 80 μg/mL BSA, and 200 μM each of the dNTPs. The reactions, containing 118 mM KCL, were then incubated at 30 °C. After 1–4 h, 500 mM EDTA was added to a concentration of 20 mM followed by the addition of 40 μL of 360 μg/mL proteinase K and 0.4 mg/mL glycogen. Reactions were then incubated at 55 °C for 30 min. The DNA products were then purified by phenol extraction and ethanol precipitation and were digested with 5 units each of PstI and ScaI for 1 h at 37 °C. The DNA products were then separated by electrophoresis on a 0.8% agarose gel run in Tris-Acetate-EDTA buffer (BioRad) containing 0.6 μg/mL ethidium bromide, and the gels were photographed using a BioRad ChemiDoc XP imaging system and Image Lab software, version 4.1. The MMR assays containing the AflIII substrate were modified by increasing the amount of Exo1 in each assay to 33.6 fmol. The excision assays used the same protocol as the MMR assays except that the DNA Polymerase δ was omitted and DNA products were analyzed by agarose gel electrophoresis either without or after digestion with ScaI, SacI, or both ScaI and SacI, as indicated. For the undigested excision products generated with the AflIII substrate, Sybr Green I (Invitrogen) was used to stain the DNA in the agarose gels. The method for visualizing the excision products by electron microscopy is described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sandra Martinez for the Exo1 plasmids, Dr. Peter Burgers for strains and plasmids, and Drs. Richard Fishel, Paul Modrich, and Christopher D. Putnam for helpful discussions. This work was supported by National Institutes of Health Grants GM50006 (to R.D.K.) and GM31819 (to J.D.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318971110/-/DCSupplemental.
References
- 1.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147(5):1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011;334(6063):1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9(1):89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 4.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 5.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 6.Martín-López JV, Fishel R. The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome. Fam Cancer. 2013;12(2):159–168. doi: 10.1007/s10689-013-9635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 8.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5(4):a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281(41):30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4(10):769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 11.Peltomäki P, Vasen HF. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer Mutations predisposing to hereditary nonpolyposis colorectal cancer: Database and results of a collaborative study. Gastroenterology. 1997;113(4):1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 12.Detloff P, Petes TD. Measurements of excision repair tracts formed during meiotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12(4):1805–1814. doi: 10.1128/mcb.12.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta A, Adjiri A, New L, Crouse GF, Jinks Robertson S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccaromyces cerevisiae. Mol Cell Biol. 1996;16(3):1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: The role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80(3):507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 15.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460(7258):984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122(5):693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280(48):39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10(4):407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 19.Acharya S, et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93(24):13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prolla TA, Pang Q, Alani E, Kolodner RD, Liskay RM. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265(5175):1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 21.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem. 2005;280(23):22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 22.Blackwell LJ, Wang S, Modrich P. DNA chain length dependence of formation and dynamics of hMutSalpha.hMutLalpha.heteroduplex complexes. J Biol Chem. 2001;276(35):33233–33240. doi: 10.1074/jbc.M105076200. [DOI] [PubMed] [Google Scholar]
- 23.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95(21):12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadyrov FA, et al. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci USA. 2009;106(21):8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pluciennik A, et al. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc Natl Acad Sci USA. 2010;107(37):16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tishkoff DX, et al. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94(14):7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: Model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21(15):5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orans J, et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145(2):212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei K, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17(5):603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet. 2000;26(3):375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 31.Lin YL, et al. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem. 1998;273(3):1453–1461. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- 32.Umar A, et al. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87(1):65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 33.Xie Y, Counter C, Alani E. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics. 1999;151(2):499–509. doi: 10.1093/genetics/151.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem. 1997;272(16):10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 35.Dzantiev L, et al. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15(1):31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J Biol Chem. 2000;275(47):36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 37.Langston LD, O’Donnell M. DNA replication: Keep moving and don’t mind the gap. Mol Cell. 2006;23(2):155–160. doi: 10.1016/j.molcel.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 38.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 39.Liberti SE, Larrea AA, Kunkel TA. Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase α. DNA Repair (Amst) 2013;12(2):92–96. doi: 10.1016/j.dnarep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J Biol Chem. 1999;274(23):16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 41.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol Cell. 2007;26(4):565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyer RR, et al. The MutSalpha-proliferating cell nuclear antigen interaction in human DNA mismatch repair. J Biol Chem. 2008;283(19):13310–13319. doi: 10.1074/jbc.M800606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15(6):724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: Dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17(5):2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith CE, et al. Dominant mutations in S. cerevisiae PMS1 identify the Mlh1-Pms1 endonuclease active site and an Exonuclease 1-independent mismatch repair pathway. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003869. 9(10):e1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27(18):6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12(5):1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 48.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91(7):995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 49.Wilson T, Guerrette S, Fishel R. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J Biol Chem. 1999;274(31):21659–21664. doi: 10.1074/jbc.274.31.21659. [DOI] [PubMed] [Google Scholar]
- 50.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273(31):19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 51.Mazurek A, Johnson CN, Germann MW, Fishel R. Sequence context effect for hMSH2-hMSH6 mismatch-dependent activation. Proc Natl Acad Sci USA. 2009;106(11):4177–4182. doi: 10.1073/pnas.0808572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274(38):26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 53.St Charles J, Petes TD. High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 Mb arm of yeast chromosome IV. PLoS Genet. 2013;9(4):e1003434. doi: 10.1371/journal.pgen.1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muster-Nassal C, Kolodner R. Mismatch correction catalyzed by cell-free extracts of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83(20):7618–7622. doi: 10.1073/pnas.83.20.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.