Significance
Rafflesiaceae produce the world’s largest flowers, but the developmental nature of their floral organs has remained a mystery. Most members of the family have a large floral chamber, which encloses their reproductive organs. We used comparative studies of development and gene-expression patterns to investigate the homology of their floral organs. Our results demonstrate that the similar floral chambers in two Rafflesiaceae subclades are constructed very differently. Thus, the characteristic features that define the floral chamber in these closely related clades are not homologous. Instead, these data indicate that similar floral chambers represent two distinct derivations of this morphology, which may have contributed to the explosive growth in floral diameter that arose secondarily within one subclade, Rafflesia.
Keywords: ABC model, comparative gene expression, evo-devo, gigantism, parasitic plants
Abstract
Rafflesiaceae, which produce the world’s largest flowers, have captivated the attention of biologists for nearly two centuries. Despite their fame, however, the developmental nature of the floral organs in these giants has remained a mystery. Most members of the family have a large floral chamber defined by a diaphragm. The diaphragm encloses the reproductive organs where pollination by carrion flies occurs. In lieu of a functional genetic system to investigate floral development in these highly specialized holoparasites, we used comparative studies of structure, development, and gene-expression patterns to investigate the homology of their floral organs. Our results surprisingly demonstrate that the otherwise similar floral chambers in two Rafflesiaceae subclades, Rafflesia and Sapria, are constructed very differently. In Rafflesia, the diaphragm is derived from the petal whorl. In contrast, in Sapria it is derived from elaboration of a unique ring structure located between the perianth and the stamen whorl, which, although developed to varying degrees among the genera, appears to be a synapomorphy of the Rafflesiaceae. Thus, the characteristic features that define the floral chamber in these closely related genera are not homologous. These differences refute the prevailing hypothesis that similarities between Sapria and Rafflesia are ancestral in the family. Instead, our data indicate that Rafflesia-like and Sapria-like floral chambers represent two distinct derivations of this morphology. The developmental repatterning we identified in Rafflesia, in particular, may have provided architectural reinforcement, which permitted the explosive growth in floral diameter that has arisen secondarily within this subclade.
It has been long recognized that parasitism elicits fundamental changes to an organism’s body plan (1, 2). Similarly, extreme changes in body size can result in dramatic morphological modifications, which in some cases rise to the level of what we term “novelty’” (3–5). Either of these circumstances can pose challenges to understanding structural homology. One lineage that exhibits both complications is the holoparasitic plant family Rafflesiaceae, which produces the world’s largest flowers. Despite their fame, however, the developmental basis of these giants has remained a mystery for nearly two centuries (6, 7). Their floral structure, in particular, is highly modified with respect to most angiosperms, so much so that confusion over their flowers has resulted in Rafflesiaceae-centric terminology to evade statements of homology. This uncertainty has obscured our understanding of their evolutionary origin, which until recently has been unknown (8–10).
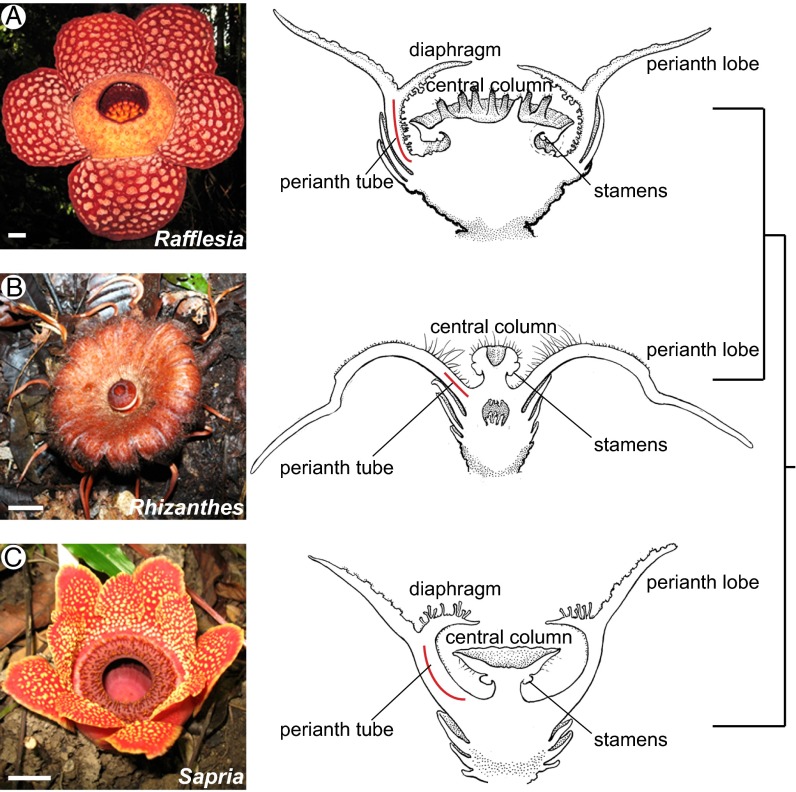
Most members of the family possess a large, bowl-shaped floral chamber [sometimes referred to as a chamber blossom by pollination biologists (11, 12)]. The floor and walls of this chamber are formed by a perianth tube and the roof is defined by an organ called the diaphragm (Fig. 1 and Fig. S1 A and C–E). The opening of the diaphragm serves as the entrance for carrion fly pollinators (13, 14). The chamber is in turn surrounded by a series of attractive sterile organs, termed perianth lobes (Fig. 1 and Fig. S1 A, C–E, and G–K). The central part of the chamber accommodates the central column, which expands distally to form a disk bearing the reproductive organs (Fig. 1 and Fig. S1). Like their closest relatives, Euphorbiaceae, the flowers of Rafflesiaceae are typically unisexual (9). In female flowers, a stigmatic belt forms around the underside of the reproductive disk (13); in male flowers this is where the stamens are borne.
Fig. 1.
Gross morphology, longitudinal sections, and accepted phylogenetic relationships of Rafflesiaceae. Rafflesia (A) and Sapria (C) exhibit floral chambers, defined by a diaphragm, where the central reproductive column resides. The central column of Rhizanthes (B) is exposed because no floral chamber is formed. (Scale bars, ∼2 cm.) Photo credits: (A) D. Boufford, (B) C.C.D., (C) L.A.N.
Each of the three genera of Rafflesiaceae produces flowers that vary on this general theme. Rafflesia and Sapria have a similar floral architecture, but differ in their perianths: Rafflesia has one whorl of five perianth lobes (Fig. 1A) and Sapria has two whorls (Fig. 1C), each with five similar lobes. Because of the striking similarity in floral morphology of these two genera, which represent the bulk of species diversity in the family, their floral chambers have been assumed to have originated once in the common ancestor of Rafflesiaceae (15) (Fig. 1). The exception is the species-poor clade Rhizanthes, which lacks the floral chamber found in most Rafflesiaceae (16) (Fig. 1B). Rhizanthes has 16 similar perianth lobes and does not form a diaphragm or chamber closure as in Rafflesia and Sapria. The perianth lobes in Rhizanthes also differ considerably in morphology: they are much narrower, have elaborate hairy “pads,” and terminate with distinct tail-like appendages (Fig. 1 and Fig. S1 G–K). Based on its more nested phylogenetic placement within the family, it has been assumed that the morphology of Rhizanthes is uniquely derived (15). The unusual floral organs of Rafflesiaceae pose a serious challenge to understanding their homology and, thus, their evolution.
Like so many branches in the Tree of Life, Rafflesiaceae are not amenable to traditional genetic manipulation, even less so than most plants. These holoparasites grow vegetatively inside their hosts and lack obvious leaves, stems, and roots. The plants have never been successfully propagated, and do not occur outside of their host range in the rainforests of Southeast Asia. Under these circumstances, only multiple independent lines of inquiry can be used to understand the floral structure and evolution of these charismatic plants, including comparative structural, developmental, and gene-expression data. In terms of this last source of data, the ABC model of floral development provides a predictive framework for understanding the identities of the floral organs of Rafflesiaceae (17, 18). The model involves three gene activities: A activity alone specifies sepal identity, A+B define petals, B+C confer stamen identity, and C determines carpel identity. Although it has been recognized that some aspects of the ABC model do not apply to all angiosperms (19), the specification of petal, stamen, and carpel identity by B- and C-class genes is generally conserved in core eudicots (20). In general, great care must be taken when using gene-expression patterns as criteria for establishing homology (21); however, when such correlative data are combined with explicit phylogenetic and developmental data, this approach has been shown to provide critical insights into organ homology (e.g., ref. 22). Along these lines, we cloned homologs of the ABC class genes, which largely belong to the pan-eukaryotic MADS-box (after the four founding members of the family: MCM1, AGAMOUS, DEFICIENS, and SRF) gene family (20), from Rafflesiaceae and studied their expression in different floral organs. We complemented these analyses with an investigation of floral organ initiation and development in all three genera in the family.
Our analyses surprisingly demonstrate that the otherwise similar floral chambers in Rafflesia and Sapria are constructed very differently. In Rafflesia, the diaphragm is derived from the petal whorl, whereas normal petals are absent. In contrast, in Sapria the diaphragm is derived from elaboration of a unique ring structure located between the perianth and the stamen whorl. This structure is analogous to sterile organs that have been independently derived in other angiosperms, such as the trumpet in daffodils (23) and the corona in passionflowers (24). The structure is not present in the closest relatives of Rafflesiaceae, and is elaborated to varying degrees in all three genera of Rafflesiaceae. Thus, the structures that define these similar floral chambers are not homologous. In Rhizanthes, which lacks a diaphragm, a derivative of the ring primordium appears to be adnate to the perianth. The development of this organ is more similar to that of Sapria, and is likely a shared symplesiomorphic feature in these two genera. These differences in construction among the three genera refute the simplistic scenario in which the similarities between Sapria and Rafflesia are interpreted as ancestral in the family.
Results
Cloning of MADS-Box Transcription Factors in Rafflesiaceae.
We cloned 28 MADS intervening keratin-like and C-terminal (MIKC) MADS-box genes from Rafflesiaceae and 62 MIKC MADS-box genes from representatives of their closest relatives: Peraceae, Euphorbiaceae, and Phyllanthaceae (Fig. S2A) (9, 10). We initially confirmed gene orthology using phylogenetic reconstruction from a broad sample of MADS-box genes, followed by more focused taxon-rich analyses within particular MADS-box subfamilies (Fig. S2 B–F; see Supporting Information for details). Single copies of the B-class lineages PISTILLATA (PI) and APETALA3 (AP3), and the C/D-class genes AGAMOUS (AG) and SEEDSTICK (STK), were recovered from Rafflesia (RfPI, RfTM6, RfSTK, and RfAG), Rhizanthes (RhPI, RhTM6, RhSTK, and RhAG), and Sapria (SapPI, SapTM6, and SapAG) (Fig. S2A). Our degenerate cloning approach was successful in recovering members of several other MADS-box subfamilies (e.g., single homologs of SEPALLATA1, FRUITFUL, AGL3, AGL15, and SVP in Rafflesiaceae, one copy of AGL24 in Rafflesia, and duplicate copies of AGL24 in Rhizanthes). As additional verification of gene identity, all homologs possessed the expected C-terminal motifs characteristic of their respective gene lineages (Fig. S3). We did not detect duplicate copies in the floral identity MADS-box repertoire of Rafflesiaceae, even for representatives of the core eudicot hexaploidization event, such as euAP3 or PLENA (25, 26). Thus, these paralogs have either been lost from the genome or are expressed at negligibly low levels. However, species-specific duplications were detected in a number of outgroup taxa. We identified recent duplicates or alleles of PI from Clutia and Pera (Peraceae) (Fig. S2B); euAP3 is duplicated in Garcia (Euphorbiaceae); and TM6 is present in two copies in Clutia (Peraceae) and Jatropha (Euphorbiaceae) (Fig. S2C). AG is duplicated in all outgroup taxa, although most of these duplications appear to be species-specific (Fig. S2D).
Comparative Organ Identity Gene Expression.
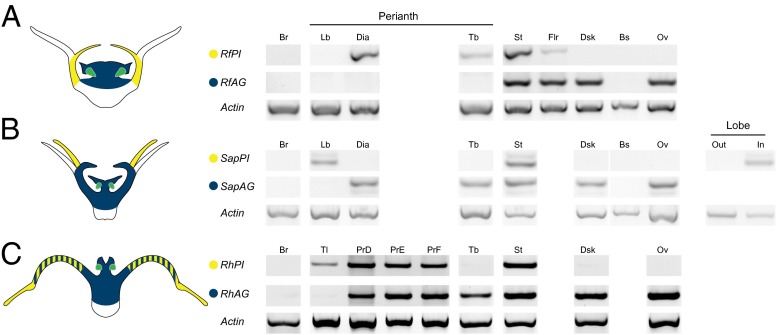
We sampled all major organs and regions of Rafflesiaceae flowers for expression of organ identity genes (Fig. S4). Our initial survey showed that most of the MADS-box homologs we identified are broadly expressed across individual floral organs (Fig. S5 A–C). Two notable exceptions were Rafflesiaceae PI and AG homologs, which are differentially expressed in different floral organs (Fig. 2). We therefore focused our analysis on these loci because the B- and C-class genes are the most conservative elements of the floral organ identity module in core eudicots (17). Thus, they have greater predictive power for interpreting floral organ homology.
Fig. 2.
PISTILLATA (PI, B-class) and AGAMOUS (AG, C-class) gene expression in Rafflesiaceae flowers. (A) Rafflesia PI and AG homologs abbreviated RfPI and RfAG. (B) Sapria, SapPI, and SapAG; In, inner perianth lobes; Out, outer perianth lobes. (C) Rhizanthes, RhPI, and RhAG; PrD, -E, and -F are perianth regions indicated in Fig. S6. Tl, tails. Actin is used as a concentration control. Abbreviations of dissected floral organs are as follows: Br, bracts; Bs, base of the flower; Dia, diaphragm; Dsk, disk; Flr, floor of the floral chamber; Lb, perianth lobes; Ov, ovary; St, stamens; Tb, perianth tube, (see Fig. S4 for the position of these regions). Gene-expression summaries illustrated to the left are shaded as follows: PI expression in yellow, AG expression in dark blue. The overlap of PI and AG in stamens is shown in green and in the proximal lobes of Rhizanthes with blue and yellow hatching.
RfPI is strongly expressed in the diaphragm and stamens with detectable signal seen in the perianth tube (Fig. 2A). RfAG expression is restricted to the central column, including the stamens in male flowers and the inferior ovary of female flowers. The expression of Sapria PI is limited to the inner perianth whorl (Fig. 2B). A signal is also seen in the stamens of male flowers. SapAG exhibits a broad pattern of expression, and is detected in the diaphragm, perianth tube, throughout the central disk, including the stamens of male flowers and the ovary of female flowers. Expression studies in Rhizanthes (Fig. 2C) are complicated by the complex nature of the perianth (see below). RhPI is expressed throughout the perianth lobes, including the tails, and stamens of male flowers. RhAG is broadly expressed throughout the floral organs with the exception of the tails. In addition to the stamens, RhAG expression overlaps with RhPI expression in the proximal portion of the perianth lobes.
We also examined B- and C-class gene expression in the close relative of Rafflesiaceae, Clutia (Peraceae), which bears more typical flowers. Clutia has unisexual flowers with both sepals and petals (Fig. S5D). In female flowers, the carpels are surrounded by stamen-derived nectaries borne opposite the sepals (27). We examined expression in dissected sepals, petals, and carpels from several female flowers. As expected, the B-class gene CluPI is expressed only in the petals. CluAP3 and CluTM6 are expressed throughout the flower, with CluTM6 most strongly expressed in the petals. CluAG is expressed in the carpels. Weak expression of CluAG was also detected in the sepals but this is likely a result of cross-contamination with the nectary, which is closely associated with the base of the sepals. Given that the nectaries are likely stamen-derived (27), they may express C-class genes, but these organs could not be reliably dissected.
Developmental Morphology of Rafflesiaceae.
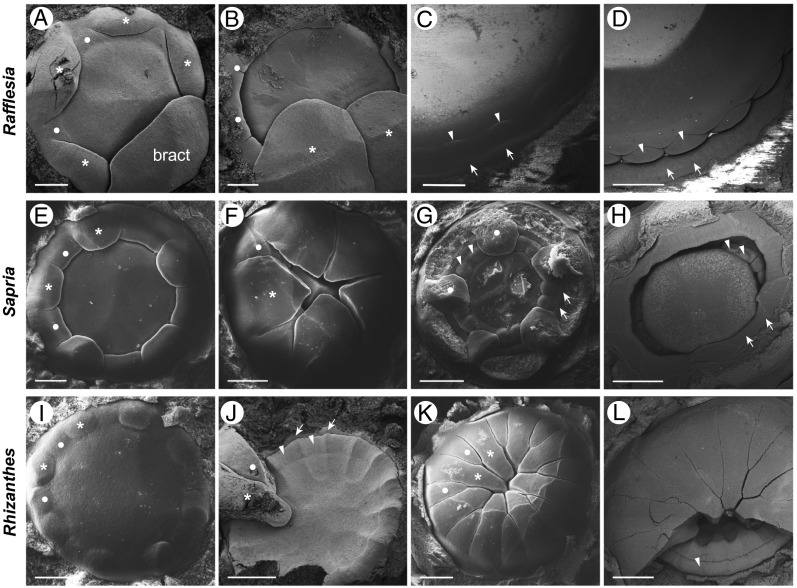
We performed extensive sampling of developmental stages from all three genera of Rafflesiaceae to further understand floral organ identity. In Rafflesia, the five perianth lobes appear sequentially in a spiral (Fig. 3A). Closely following the appearance of the perianth lobes, but preceding the appearance of the stamens, the diaphragm in Rafflesia arises as a ring that extends toward the center of the apex (Fig. 3 A and B). The perianth lobes and the diaphragm are raised above the apex by intercalary growth, which forms the perianth tube. Stamens are initiated on the flanks of the broad, convex floral apex (Fig. 3 C and D). At about the same time, an additional ring structure appears just outside of the stamens, which seems to be derived from the floor of the chamber (Fig. 3 C and D). Although at an early stage both the stamens and this ring are of comparable size (Fig. S6A), the ring does not expand much further and becomes comparatively inconspicuous. The ring is present at the base of the disk in advanced floral buds and in open flowers (Fig. S1 A and B).
Fig. 3.
Floral development in Rafflesia (A–D), Sapria (E–H), and Rhizanthes (I–L). (A) Spiral initiation sequence of the perianth lobes (asterisks) of Rafflesia, with all but one bract removed; the diaphragm appears as a ring (dots). (B) The diaphragm (dots) extends horizontally toward the center of the flower; all but the last two perianth lobes (asterisks) removed. (C) Stamen (arrowheads) and (inner) ring (arrows) initiation. (D) Later stage of stamen (arrowheads) and ring (arrows) development; the diaphragm has been removed but is apparent as the darker band around the lighter center. (E) Two whorls of perianth lobes are initiated at alternate positions in Sapria (asterisks and dots). (F) The outer whorl perianth lobes (pointed tips, asterisk) cover the inner whorl (dot). (G) Five clawed inner perianth lobes (dots) and the initiation of the stamen whorl (arrowheads) and the (inner) ring (arrows); five outer perianth lobes removed. (H) The diaphragm (arrows) is lifted above the stamens (arrowheads) by intercalary growth. (I) Appearance of perianth lobes in Rhizanthes at the flanks of a broad floral apex; asterisks and dots indicate alternating perianth organs. (J) Initiation of stamens (arrowheads) and a ring outside of the stamens (arrows); all but two perianth lobes removed (asterisk and dot). (K) Two whorls of 8+8 alternating lobes (asterisks and dots). (L) A more advanced stage of stamen development (arrowhead) and the formation of a concave floral apex. (Scale bars: 500 μm in A–C, E, I, J, L; 1 mm in D, F–H, K.)
In Sapria, the perianth lobes appear nearly simultaneously as two alternating whorls (Fig. 3E). The perianth lobes of the outer whorl are acute at the apex and broad at the base near the level of attachment (Fig. 3F). In contrast, lobes of the inner whorl are rounded at the apex and narrower at the base (Fig. 3G). Although the two whorls differ conspicuously at this early stage, these differences diminish as development proceeds to anthesis (Fig. 1C). In contrast to Rafflesia, the diaphragm in Sapria appears later in development, at the same time or shortly after the stamens appear, from a ring primordium outside of the stamen whorl (Fig. 3G). Intercalary growth below the diaphragm lifts the diaphragm together with the two whorls of perianth lobes forming the perianth tube (Fig. 3H).
The 16 perianth lobes of Rhizanthes appear in two whorls of eight lobes each (Fig. 3I). The lobes form nearly simultaneously in a circle on the flanks of a broad floral apex and then grow in horizontal direction toward each other (Figs. 3 J and K). Their round tips touch, bend downward, and continue elongating toward the floral apex, which is slightly concave at this stage (Fig. 3K). The lobes’ descending portions, which correspond to the tails, are tightly appressed to each other in bud (Fig. 4C and Fig. S1J). At this later stage, the stamen whorl also appears and, concomitantly, a ring structure arises outside the stamen whorl (Fig. 3J). The ring becomes congenitally fused to, and elongates with, the perianth lobes as they grow (Fig. S6 B–F), ultimately forming the pads at the base of each perianth lobe (Fig. S1 H–K). The adnation between the ring and the perianth is discernible histologically (Fig. S6 B–F).
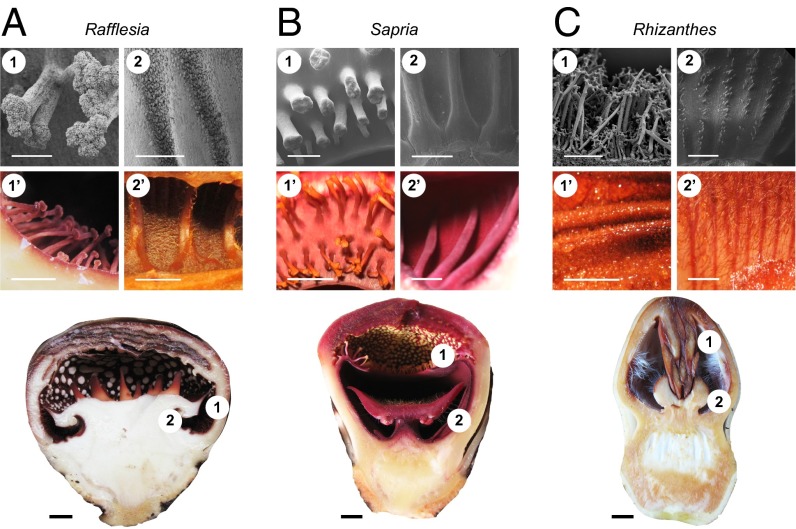
Fig. 4.
Gross floral morphological landmarks in Rafflesia (A), Sapria (B), and Rhizanthes (C). The positions of the ramenta (in Rafflesia and Sapria) or trichomes (in Rhizanthes) (1 and 1′) and ridges/grooves (2 and 2′) across the three genera are compared using SEM (1 and 2) and light photography (1′ and 2′). [Scale bars: SEM micrographs, 1 mm (except C1, which equals 500 μm); photographs, 5 mm.] Circles numbered 1 and 2 in the bottom photographs indicate the position of these key morphological landmarks in advanced floral buds. (Scale bars, 1 cm.)
Additional Floral Morphological Landmarks in Rafflesiaceae.
Finally, we made a detailed study of gross morphological landmarks in all three genera of Rafflesiaceae. In Rafflesia, the reproductive column is notable for the presence of alternating longitudinal grooves and ridges that begin at the ring and extend to the base of the disk (Fig. 4A). The grooves accommodate individual anthers in male flowers but are also present in female flowers, which have a highly reduced stamen whorl. Similar grooves and ridges are found on the inner surface of the perianth tube in Sapria, and here they extend from the undersurface of the diaphragm to the base of the central column (Fig. 4B and Fig. S1 D and E). Finally, the perianth tube of Rhizanthes exhibits shallow grooves and ridges demarcated by dark striations, from the attachment of the central column to the beginning of the perianth lobes, which similarly alternate with the stamens in male flowers and staminodia in female flowers (Fig. 4C).
Another peculiar morphological feature in Rafflesia and Sapria is the presence of a series of multicellular, vascularized, branched structures called ramenta. In Rafflesia, the ramenta line the inner surface of the perianth tube and extend from outside the ring at the base of the central column to the undersurface of the diaphragm (Fig. 4A and Fig. S1A). In contrast, the ramenta in Sapria are restricted to the upper surface of the diaphragm and are absent from the inside of the chamber (Fig. 4B and Fig. S1 C–F). Ramenta, similar to those observed in Rafflesia and Sapria, are absent in Rhizanthes, which instead exhibits a variety of unicellular trichomes covering the adaxial side of the perianth (Fig. 4C and Fig. S1 G–K).
Discussion
Integrating Molecular, Developmental, and Morphological Data to Elucidate Floral Organ Homologies in Rafflesiaceae.
In all three genera of Rafflesiaceae, the reproductive column expresses AG homologs with PI homologs restricted to the stamens. This similarity in expression is consistent with the conserved structure of the column, which is likely homologous across the family. The perianth organs of Rafflesia, Sapria, and Rhizanthes, however, exhibit very distinct patterns of organ identity gene expression, despite their positional similarity. In Rafflesia, the diaphragm and perianth tube express the B-class gene RfPI, whereas in Sapria SapPI is restricted to the inner perianth lobes (Fig. 2). The broad expression found in other Rafflesiaceae B-class genes, namely the TM6 homologs, is not significant because PI and TM6 proteins typically function as obligate heterodimers (28). Restricted expression of PI, therefore, limits the breadth of B-class gene function to the diaphragm and perianth tube in Rafflesia and to the inner perianth lobes in Sapria. B-class function specifies petals in model core eudicots (17) and, as expected, CluPI expression is also specific to the petals in the close relative of Rafflesiaceae, Clutia. These observations suggest that the diaphragm in Rafflesia and the inner perianth lobes in Sapria possess petal identity. Gene expression in Rhizanthes more closely resembles Sapria than Rafflesia in that RhAG expression is expanded to include the perianth tube and even portions of the perianth lobes, but not the distal-most region of the lobes that form the tails. Although RhAG expression appears to overlap with that of RhPI in the more proximal pads of the perianth lobes, we hypothesize that this overlap likely reflects RhAG expression in the ring-derived adaxial pads, whereas RhPI is expressed in the abaxial layers of the perianth proper (Fig. 2 and Fig. S6D). This interpretation indicates a likely homology between the ring-derived diaphragm of Sapria and the ring-derived series of pads of Rhizanthes.
Differential gene expression among the genera of Rafflesiaceae, particularly between Sapria and Rafflesia, which look superficially similar, can be explained in two ways. First, they could reflect dramatic shifts in gene expression that are independent of the homology of the structures themselves (21). This is the case for the sepal-derived and highly modified perianth of Aristolochia, which appears to express B-class genes (29). Second, they could represent the actual identity, and thus homology, of the organs (30, 31). To determine which of these two scenarios is more plausible, we examined the initiation and elaboration of the floral organs in Rafflesiaceae. In all three genera, organ initiation is organized into two waves. The perianth organs appear first, including the perianth lobes and diaphragm in Rafflesia and the two whorls of perianth lobes in Sapria and Rhizanthes. Second, we see initiation of the stamens along with an adjacent ring primordium between the perianth and the stamen whorl that does not correspond to any canonical floral whorl. This ring remains a relatively minor feature in Rafflesia. In contrast, in Sapria the ring structure becomes prominent in giving rise to the diaphragm; in Rhizanthes it forms the series of pads on the perianth lobes. The distinct fates and positions of the ring derivatives in Rafflesia versus Sapria/Rhizanthes reflect a combination of differential elaboration of the structures themselves (less in Rafflesia, more in Sapria and Rhizanthes). In Rafflesia, intercalary expansion occurs between the ring and the perianth whorl, whereas in Sapria it occurs between the ring and the base of the column. In Rhizanthes, some intercalary growth also occurs between the ring and the base of the column, but the deep lobing of the perianth makes identification of intercalary growth in this case more difficult to interpret.
These developmental patterns correlate perfectly with our gene-expression data: early arising inner perianth organs (i.e., the diaphragm in Rafflesia and the inner lobes of Sapria) express PI homologs alone but ring-derived structures express AG homologs (the chamber floor in Rafflesia, diaphragm in Sapria, and series of pads of Rhizanthes). Further evidence for this correspondence comes from our assessment of gross morphology: grooves and ridges are always present between the ring-derived structures and the base of the reproductive column; in Rafflesia and Sapria ramenta line the region between the ring and the inner perianth organs. The positions of these key floral landmarks—ridges/grooves and ramenta—are most easily explained by the hypothesis that intercalary growth in different regions gave rise to the similar floral chambers in Rafflesia and Sapria. In terms of our identification of the Rafflesia diaphragm as the petal whorl, it is interesting to note that considerable variation in diaphragm morphology is observed in many species of Rafflesia. Some have distinctly lobed diaphragms, and others have reduced diaphragms that form a small shelf at the chamber opening (32). Elucidating the developmental basis of this variation may shed light on the modification of this floral organ.
All three sources of evidence—gene expression, development, and morphological landmarks—indicate that the diaphragm of Rafflesia is derived from the petal whorl, but the diaphragm of Sapria is derived from the unique ring structure. This conclusion suggests that diaphragm formation is not homologous in Rafflesia and Sapria. It appears that in Rafflesia, the chamber is primarily formed via expansion of the congenitally fused sepal and petal bases, which constitute the perianth tube. In contrast, in Sapria, the sepal and petal whorls develop as free lobes that do not contribute to the formation of the floral chamber, which instead forms by elaboration of the ring structure. The modifications seen in Rhizanthes are more complex and involve fusion of the ring to the perianth as well as deep dissection of this compound structure. Regardless, in all three genera the structures derived from the ring primordium express AG homologs, which is not surprising because they originate in close proximity to the base of the reproductive column. In this sense, the ring of Rafflesia, the diaphragm of Sapria, and the series of pads of Rhizanthes bear parallels to peculiar outgrowths, called coronas, seen in divergent angiosperm clades, including the radial filaments (commonly termed “crown of thorns”) in passionflowers (24) and the trumpet of daffodils (23). In these cases, the coronas are also initiated late in development, after significant elaboration of the other floral organs, and they express AG homologs, despite the fact that they are sterile organs.
Evolution of the Floral Chamber in Rafflesiaceae.
Our results refute the simplistic scenario of floral evolution proposed for Rafflesiaceae, in which the floral chamber in Rafflesia and Sapria is thought to represent the ancestral condition within the family, which was then lost in Rhizanthes (15). Instead, our data suggest that the floral chambers in Rafflesia and Sapria are constructed differently and the involvement of the petal whorl is a Rafflesia-specific invention. Given the evolutionary and phenotypic distance between Rafflesiaceae and its common ancestor with Euphorbiaceae (∼95 My) (15) and the widely accepted fact that parasitism can lead to drastic changes that confound assessment of homology (2, 8, 33), outgroup comparisons with Euphorbiaceae are problematic for understanding floral evolution in Rafflesiaceae.
If we instead focus on Rafflesiaceae, one model is that the closed floral chambers characteristic of the family have arisen independently in Rafflesia and Sapria, perhaps because of similar selective pressures imposed by fly pollinators (Fig. S7A). In this case, the common ancestor of Rafflesiaceae lacked an organized floral chamber and this ancestral condition was retained in Rhizanthes. A similar example has been documented in two related groups, Aristolochia and Hydnora, which have evolved floral chambers with different construction. In Hydnora, the roof of the chamber is formed by the stamens; in Aristolochia it is formed by the perianth (29, 34). A second model would be that the common ancestor of Rafflesiaceae possessed a floral chamber similar to that in Sapria [i.e., derived from expansion between the ring and the column (Fig. S7B)]. This floral chamber was then lost along the branch leading to Rhizanthes and remodeled along the branch leading to Rafflesia, such that the diaphragm was derived from the petal whorl. Under the second model there would have been relatively little change in the gross morphology of these floral chambers. Despite this superficial similarity, however, the underlying development of the Rafflesia and Sapria-like floral chambers would have been completely repatterned. Variation in the timing of these changes can also produce a third model (e.g., Fig. S7C).
This second model might seem unlikely, especially because the fly pollinators that visit these plants appear to be shared between the three Rafflesiaceae genera (13–16). However, such changes are not without precedent and have been explained by developmental system drift (35). This is a process by which characters that are thought to be homologous have diverged in their morphogenetic or gene regulatory underpinnings. For example, developmental system drift underlies the conservation of the segmented body plan in insects, which nonetheless exhibits variability in the underlying mechanisms of segmentation (36, 37). Detailed investigation of this phenomenon requires functional comparison between two species with the same body plan to understand changes in the underlying molecular machinery (38), and as such it is not feasible in Rafflesiaceae. Moreover, although such studies may illuminate important mechanistic features of the developmental system, they cannot explain the underlying reasons for such a striking change in development.
So why then has Rafflesia, which contains the largest of all flowers (Rafflesia arnoldii R.Br., 1 m in diameter), undergone such profound developmental repatterning? The developmental genetic mechanisms that permit flower size to increase rapidly remain unknown for this or any other lineage (12). Interestingly, although the early ancestors of Rafflesiaceae exhibited a dramatic increase in floral size (9), it appears that within the family, rates of floral size evolution were further elevated in Rafflesia (39). One potential explanation for this pattern is that the change we have posited for Rafflesia floral development could have further reduced constraints on floral diameter and permitted its additional burst in gigantism. Several factors, including an earlier shift in the developmental timing of diaphragm initiation, the effective reduction in the perianth lobes, and the architectural remodeling of the chamber itself, could have contributed to the stabilization of these enormous flowers. Support for this hypothesis comes from the occurrence of lobed (imperfectly formed) diaphragms in Rafflesia, which are restricted to species of small size [e.g., Rafflesia lobata Barcelona & Pelser (32)]. Further investigation within Rafflesia will help to better address this question, but overall, our study highlights the surprising and dynamic nature of morphological evolution among the “greatest prodigy of the vegetable kingdom” (6). In particular, the study underscores the degree to which seemingly similar morphologies can be fundamentally remodeled among closely related species.
Materials and Methods
Plant Material.
Buds of Rafflesia tuan-mudae Becc. and Rafflesia cantleyi Solms-Laubach in different developmental stages were collected in Gunung Puey, Sarawak, and in Ulu Geroh, Peninsular Malaysia, respectively. Sapria himalayana Griffith was collected at Queen Sirikit Botanic Garden, Chiang Mai, Thailand. Rhizanthes lowii (Becc.) Harms was collected near Kampung Giam, Sarawak. Vouchers are deposited at the Harvard University Herbaria (A). The historical collection of Rafflesia patma Blume by A. Ernst is housed at the Botanical Institutes of the University of Zurich (Z). Clutia sp. and Pera bumeliifolia Griseb. were collected from the University of California Davis Botanical Conservatory and Fairchild Tropical Botanic Garden, respectively, and vouchers are deposited at the University of California at Davis herbarium (DAV) and Fairchild Tropical Botanic Garden herbarium (FTG). Dalechampia sp., Breynia sp., Acalypha sp., Garcia sp., Monadenium sp., and Jatropha sp. were obtained from living material at the University of Connecticut greenhouse and vouchers are deposited at the University of Connecticut herbarium (CONN).
RNA Isolation, cDNA Synthesis, and Cloning of MADS-Box Genes.
Floral material from both male and female buds of different sizes (diameter of Rafflesia floral buds ranging from 5 to 25 cm; Sapria from 3 to 15 cm; and Rhizanthes from 2 to 13 cm) was dissected and flash-frozen in the field, then kept at −80 °C until RNA isolation. Total RNA was extracted from 0.1- to 0.5-g tissue using Plant RNA Reagent (Invitrogen) following the manufacturer’s recommendations. DNA contamination was eliminated after incubation with TURBO-DNase (Ambion) for 2 h and total DNased RNA were used for cDNA synthesis using polyT primer (40) with SuperScript RT II (Invitrogen). Degenerate primer approach was used to clone MADS-box genes from Rafflesiaceae and outgroups as previously described (for A-class products, see ref. 40; for B- and C-class products, see ref. 41). All sequences were recovered independently from a minimum of three clones, but typically from more than 50 clones (GenBank accession nos. KF730013–KF730100). Sequences were examined and edited manually in Geneious (Geneious).
RT-PCRs.
The floral buds were dissected into several nonoverlapping regions (Fig. S4). One to 5 μg of DNased, organ-specific RNA from the same regions of at least three floral buds (biological replicates) were used separately for cDNA synthesis as described above. Gene-specific primers (Table S2) spanning an intron were used in a PCR with one-tenth dilution of cDNA with 55 °C annealing, 15-s elongation time, and 26 cycles. Products were separated on 1.5–2% agarose gels. Actin amplification was used as an internal control.
Supplementary Material
Acknowledgments
We thank K. Bomblies, D. Boufford, C. Extavour, W. Friedman, D. Haig, A. Knoll, S. Mathews, B. Tomlinson, and members of the C.C.D., E.M.K., and W. Friedman laboratories for valuable comments; S.-Y. Wong and members of the S.-Y.Wong laboratory, M. Wong, R. Pongsattayapipat, and the staff at Queen Sirikit Botanic Garden, S. Sari, and B. Insan for field support; the University of California, Davis conservatory and the University of Connecticut greenhouse for living plant material; M. Matthews for help with microtome sectioning; R. Hellmiss for help with the graphics; B. Angell for the illustrations; and C.-C. Wu for providing the initial MADS box alignments and related information. Imaging support was provided by the Center for Microscopy and Image Analysis at the University of Zurich and the Center for Nanoscale Systems at Harvard University. This research was supported by National Science Foundation Assembling the Tree of Life Grant DEB-0622764 (to C.C.D.), and National Science Foundation Grant DEB-1120243 (to C.C.D. and E.M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database, http://www.ncbi.nlm.nih.gov/genbank/ (accession nos. KF730013–KF730100).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310356110/-/DCSupplemental.
References
- 1.Bush AO, Fernandez JC, Esch GW, Seed JR. Parasitism: The Diversity and Ecology of Animal Parasites. Cambridge, UK: Cambridge Univ Press; 2001. [Google Scholar]
- 2.Kuijt J. The Biology of Parasitic Flowering Plants. Berkeley, CA: Univ of California Press; 1969. [Google Scholar]
- 3.Emlen DJ, Nijhout HF. The development and evolution of exaggerated morphologies in insects. Annu Rev Entomol. 2000;45:661–708. doi: 10.1146/annurev.ento.45.1.661. [DOI] [PubMed] [Google Scholar]
- 4.Sander PM, et al. Biology of the sauropod dinosaurs: The evolution of gigantism. Biol Rev Camb Philos Soc. 2011;86(1):117–155. doi: 10.1111/j.1469-185X.2010.00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanken J, Wake DB. Miniaturization of body size: Organismal consequences and evolutionary significance. Annu Rev Ecol Syst. 1993;24:501–519. [Google Scholar]
- 6.Brown R. An account of a new genus of plants, named Rafflesia. Trans Linn Soc (Lond) 1822;13:201–236. [Google Scholar]
- 7.Brown R. Description of the female flower and fruit of Rafflesia arnoldi with remarks on its affinities; and an illustration of the structure of Hydnora africana. Trans Linn Soc (Lond) 1845;19:221–239. [Google Scholar]
- 8.Barkman TJ, Lim SH, Salleh KM, Nais J. Mitochondrial DNA sequences reveal the photosynthetic relatives of Rafflesia, the world’s largest flower. Proc Natl Acad Sci USA. 2004;101(3):787–792. doi: 10.1073/pnas.0305562101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis CC, Latvis M, Nickrent DL, Wurdack KJ, Baum DA. Floral gigantism in Rafflesiaceae. Science. 2007;315(5820):1812. doi: 10.1126/science.1135260. [DOI] [PubMed] [Google Scholar]
- 10.Wurdack KJ, Davis CC. Malpighiales phylogenetics: Gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot. 2009;96(8):1551–1570. doi: 10.3732/ajb.0800207. [DOI] [PubMed] [Google Scholar]
- 11.Proctor M, Yeo P, Lack A. The Natural History of Pollination. Portland, OR: Timber Press; 2003. [Google Scholar]
- 12.Davis CC, Endress PK, Baum DA. The evolution of floral gigantism. Curr Opin Plant Biol. 2008;11(1):49–57. doi: 10.1016/j.pbi.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Beaman R, Decker P, Beaman J. Pollination of Rafflesia (Rafflesiaceae) Am J Bot. 1988;75(8):1148–1162. [Google Scholar]
- 14.Bänziger H, Hansen B. Unmasking the real identity of Sapria poilanei Gagnepain emend., and description of Sapria ram sp. n. (Rafflesiaceae) Nat Hist Bull Siam Soc. 1997;45:149–170. [Google Scholar]
- 15.Bendiksby M, et al. Elucidating the evolutionary history of the Southeast Asian, holoparasitic, giant-flowered Rafflesiaceae: Pliocene vicariance, morphological convergence and character displacement. Mol Phylogenet Evol. 2010;57(2):620–633. doi: 10.1016/j.ympev.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Bänziger H, Hansen B. A new taxonomic revision of the deceptive flower, Rhizanthes Dumortier (Rafflesiaceae) Nat Hist Bull Siam Soc. 2000;48(1):117–144. [Google Scholar]
- 17.Litt A, Kramer EM. The ABC model and the diversification of floral organ identity. Semin Cell Dev Biol. 2010;21(1):129–137. doi: 10.1016/j.semcdb.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Smaczniak C, Immink RG, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development. 2012;139(17):3081–3098. doi: 10.1242/dev.074674. [DOI] [PubMed] [Google Scholar]
- 19.Irish VF, Litt A. Flower development and evolution: Gene duplication, diversification and redeployment. Curr Opin Genet Dev. 2005;15(4):454–460. doi: 10.1016/j.gde.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Kramer EM, Irish VF. Evolution of the petal and stamen developmental programs: Evidence from comparative studies of the lower eudicots and basal angiosperms. Int J Plant Sci. 2000;161(6 Suppl.):S29–S40. [Google Scholar]
- 21.Jaramillo MA, Kramer EM. The role of developmental genetics in understanding homology and morphological evolution in plants. Int J Plant Sci. 2007;168(1):61–72. [Google Scholar]
- 22.Brockington SF, et al. ‘Living stones’ reveal alternative petal identity programs within the core eudicots. Plant J. 2012;69(2):193–203. doi: 10.1111/j.1365-313X.2011.04797.x. [DOI] [PubMed] [Google Scholar]
- 23.Waters MT, et al. The corona of the daffodil Narcissus bulbocodium shares stamen-like identity and is distinct from the orthodox floral whorls. Plant J. 2013;74(4):615–625. doi: 10.1111/tpj.12150. [DOI] [PubMed] [Google Scholar]
- 24.Hemingway CA, Christensen AR, Malcomber ST. B- and C-class gene expression during corona development of the blue passionflower (Passiflora caerulea, Passifloraceae) Am J Bot. 2011;98(6):923–934. doi: 10.3732/ajb.1100026. [DOI] [PubMed] [Google Scholar]
- 25.Kramer EM, Hall JC. Evolutionary dynamics of genes controlling floral development. Curr Opin Plant Biol. 2005;8(1):13–18. doi: 10.1016/j.pbi.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 26.Jiao Y, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13(1):R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endress PK, Matthews ML. Elaborate petals and staminodes in eudicots: Diversity, function, and evolution. Org Divers Evol. 2006;6(4):257–293. [Google Scholar]
- 28.Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347(2):183–198. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Jaramillo MA, Kramer EM. APETALA3 and PISTILLATA homologs exhibit novel expression patterns in the unique perianth of Aristolochia (Aristolochiaceae) Evol Dev. 2004;6(6):449–458. doi: 10.1111/j.1525-142X.2004.04053.x. [DOI] [PubMed] [Google Scholar]
- 30.Ambrose BA, et al. Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell. 2000;5(3):569–579. doi: 10.1016/s1097-2765(00)80450-5. [DOI] [PubMed] [Google Scholar]
- 31.Álvarez-Buylla ER, et al. B-function expression in the flower center underlies the homeotic phenotype of Lacandonia schismatica (Triuridaceae) Plant Cell. 2010;22(11):3543–3559. doi: 10.1105/tpc.109.069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barcelona J, Pelser P, Balete D, Co L. Taxonomy, ecology, and conservation status of Philippine Rafflesia (Rafflesiaceae) Blumea. 2009;54(1-3):77–93. [Google Scholar]
- 33.Griffith W. On the root-parasites referred by authors to Rhizantheae; And on various plants related to them. Trans Linn Soc (Lond) 1845;19:303–348. [Google Scholar]
- 34.Bolin J, Maass E, Musselman L. Pollination biology of Hydnora africana Thunb. (Hydnoraceae) in Namibia: Brood site mimicry with insect imprisonment. Int J Plant Sci. 2009;170(2):157–163. [Google Scholar]
- 35.True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3(2):109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- 36.Liu PZ, Kaufman TC. Short and long germ segmentation: Unanswered questions in the evolution of a developmental mode. Evol Dev. 2005;7(6):629–646. doi: 10.1111/j.1525-142X.2005.05066.x. [DOI] [PubMed] [Google Scholar]
- 37.Bentley D, Keshishian H, Shankland M, Toroian-Raymond A. Quantitative staging of embryonic development of the grasshopper, Schistocerca nitens. J Embryol Exp Morphol. 1979;54:47–74. [PubMed] [Google Scholar]
- 38.Sommer RJ. In: Evolutionary Systems Biology. Soyer O, editor. Heidelberg: Springer; 2012. pp. 79–92. [Google Scholar]
- 39.Barkman TJ, et al. Accelerated rates of floral evolution at the upper size limit for flowers. Curr Biol. 2008;18(19):1508–1513. doi: 10.1016/j.cub.2008.08.046. [DOI] [PubMed] [Google Scholar]
- 40.Litt A, Irish VF. Duplication and divergence in the APETALA1/FRUITFULL gene lineage: Implications for the evolution of floral development programs. Genetics. 2003;165(2):821–833. doi: 10.1093/genetics/165.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stellari GM, Jaramillo MA, Kramer EM. Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Mol Biol Evol. 2004;21(3):506–519. doi: 10.1093/molbev/msh044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.