Significance
Chromatin immunoprecipitation (ChIP) is a gold standard technique for genomic protein localization. We have discovered an artifact in ChIP that leads to reproducible but biologically meaningless enrichment of proteins at highly expressed genes, caused by high levels of polymerase II and polymerase III transcription. These findings call into question reports of unexpected localization of transcription factors, repressors, and cytosolic proteins to highly expressed genes. We suggest caution when interpreting ChIP enrichment at highly expressed genes and suggest a heterologous protein control in ChIP experiments to discern biologically meaningful from artifactual enrichment.
Keywords: ChIP-chip, HOT regions, yeast, tRNA
Abstract
Chromatin immunoprecipitation (ChIP) is the gold-standard technique for localizing nuclear proteins in the genome. We used ChIP, in combination with deep sequencing (Seq), to study the genome-wide distribution of the Silent information regulator (Sir) complex in Saccharomyces cerevisiae. We analyzed ChIP-Seq peaks of the Sir2, Sir3, and Sir4 silencing proteins and discovered 238 unexpected euchromatic loci that exhibited enrichment of all three. Surprisingly, published ChIP-Seq datasets for the Ste12 transcription factor and the centromeric Cse4 protein indicated that these proteins were also enriched in the same euchromatic regions with the high Sir protein levels. The 238 loci, termed ”hyper-ChIPable“, were in highly expressed regions with strong polymerase II and polymerase III enrichment signals, and the correlation between transcription level and ChIP enrichment was not limited to these 238 loci but extended genome-wide. The apparent enrichment of various proteins at hyper-ChIPable loci was not a consequence of artifacts associated with deep sequencing methods, as confirmed by ChIP-quantitative PCR. The localization of unrelated proteins, including the entire silencing complex, to the most highly transcribed genes was highly suggestive of a technical issue with the immunoprecipitations. ChIP-Seq on chromatin immunoprecipitated with a nuclear-localized GFP reproduced the above enrichment in an expression-dependent manner: induction of the GAL genes resulted in an increased ChIP signal of the GFP protein at these loci, with presumably no biological relevance. Whereas ChIP is a broadly valuable technique, some published conclusions based upon ChIP procedures may merit reevaluation in light of these findings.
Chromatin immunoprecipitation, followed either by microarrays (ChIP-chip) or deep sequencing (ChIP-Seq) is the standard method for in vivo genome-wide protein localization analysis (reviewed in refs. 1–3). Since the first applications of deep sequencing to ChIP in 2007, the ChIP-Seq technique has quickly become accepted as superior to ChIP-chip hybridization and is now the dominant and most preferred approach for studying DNA- and chromatin-interacting proteins (2, 4, 5). Because of known biases in chromatin preparation and sequencing, nearly all ChIP-Seq studies compare the mapped reads of the immunoprecipiated (IP) sample to an input control with chromatin that is cross-linked but not immunoprecipitated (2, 5, 6). We applied ChIP-Seq to study the distribution of the silencing protein complex, consisting of Sir2, Sir3, and Sir4, in Saccharomyces cerevisiae. Unexpectedly, the well-characterized biology of silencing enabled the resulting data to illuminate a technical artifact introduced by the ChIP technique.
Silencing in S. cerevisiae is established by the Sir2, Sir3, and Sir4 protein complex that binds and deacetylates key positions on nucleosomes, forming a heterochromatic structure that inhibits transcription (reviewed in ref. 7). Prior work raised the possibility that the Sir proteins could occasionally be recruited in error to euchromatic regions (8). Hence, we asked whether there were any signals of euchromatic silencing in Sir2, Sir3, and Sir4 ChIP-Seq data.
In line with a recent report of euchromatic Sir3 localization (9), we identified numerous loci with peaks of Sir2, Sir3, and Sir4 proteins, suggesting the presence of the entire silencing complex. However, the nature of the euchromatic signals and their overlap with highly transcribed genes were suspicious and inconsistent with the expectations from decades of study of silencing proteins and previously published literature (8, 10, 11). Careful investigation of the euchromatic binding sites of the Sir complex led us to discover that strongly expressed loci were reliably hyper-ChIPable; multiple unrelated proteins exhibit increased ChIP levels at these sites. There was no meaningful biology in these ChIP signals. Because of the widespread use of ChIP-based methods and the range of conclusions drawn, this discovery has wide applicability.
Results
Unrelated Proteins Show Overlapping Enrichment in ChIP-Seq Datasets.
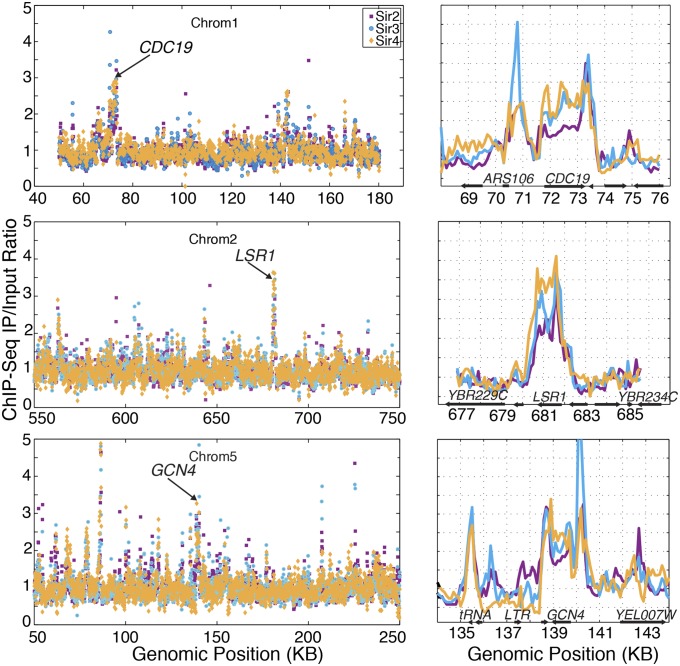
The well-described Sir protein silencing complex in yeast is important for silencing the cryptic mating-type loci and some subtelomeric regions. However, we were interested in the potential for euchromatic Sir-protein enrichment because the silencer-binding proteins that recruit the Sir proteins to silencers also bind at many positions in euchromatin. To investigate this possibility, we performed ChIP-Seq experiments on myc-tagged Sir2, Sir3, and Sir4 and analyzed the genome-wide IP/input ratios in euchromatin, operationally defined as sequences 50 kilobases (kb) or farther from chromosome ends. To identify euchromatic loci with Sir complex occupancy, we imposed a stringent standard for enrichment requiring Sir2, Sir3, and Sir4 to each be enriched twofold or greater, with many of these loci exhibiting substantially higher enrichment of individual Sir proteins. Using this strict standard we found a total of 238 distinct loci (Fig. 1 and Table S1). Given that no euchromatic function has been ascribed to the Sir complex, this was a surprisingly high number of distinct euchromatic loci to be enriched for all Sir proteins.
Fig. 1.
Euchromatic enrichment of the Sir complex in ChIP-Seq datasets. Examples of the 238 euchromatic loci with twofold or higher ChIP-Seq enrichment of the Sir2, Sir3, and Sir4 proteins. (Left) Chromosome-level views. (Right) Zoomed in tracks for the CDC19, LSR1, and GCN4 loci. For each protein, the IP/input ratios are normalized to the genome-wide median.
These peaks reflected high read coverage in the immunoprecipitated DNA, rather than low representation in the input sample (Fig. S1). We also used the popular peak-calling software model-based analysis of ChIP-Seq (MACS) on the three Sir datasets, and 76 of the 238 peaks were confirmed by the program as statistically significant at the default P value of 10−5 (Table S1). Additionally, this euchromatic enrichment was not due to an artifact resulting from next-generation sequencing methods as ChIP–quantitative PCR (qPCR) of myc-tagged Sir4 reproduced the ChIP-Seq enrichment at the highly-expressed LSR1, CDC19, and GCN4 genes, which were prominent among the enriched genes in the ChIP-Seq datasets; Sir4-myc was not enriched at MET3, a gene not identified as enriched in the ChIP-Seq samples (Fig. S2). To check for statistical significance, we asked how many loci had the inverse – input/IP ratio greater than 2 for all three proteins. In contrast to the 238 loci with high IP/input, only a single locus passed the inverse threshold. The discovery of a significant fraction of euchromatin being occupied by all three members of the silencing complex suggested either unanticipated roles of Sir proteins or the existence of a systematic bias in the data.
The sites of the Sir complex enrichment across the yeast genome were inconsistent with prior knowledge of Sir-mediated silencing. The Sir proteins play roles in silencing telomeric and subtelomeric loci (10, 12–15), and in control of rDNA recombination, but no reproducible derepression of euchromatic genes is observed in sir mutants (11). Moreover, predictions for possible euchromatic Sir targets in a prior study (8) did not overlap with the 238 Sir-enriched loci reported here. Hence, we considered the possibility of a ChIPability issue that could result in strong but misleading peaks, despite normalization to the “input-Seq” sample, a commonly used method of normalization (2, 5, 6).
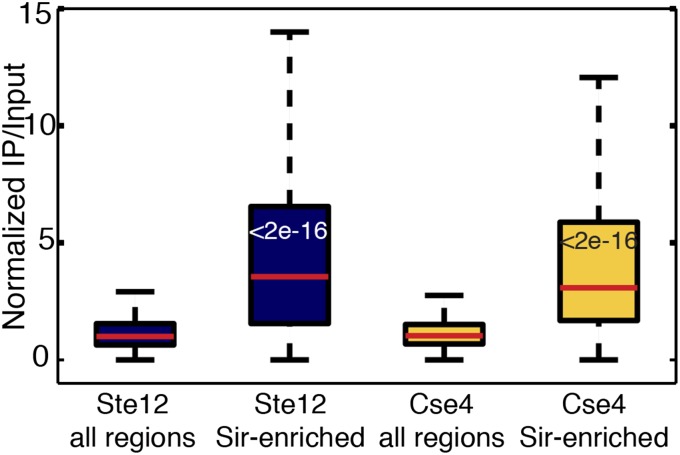
We reasoned that if certain genomic regions were more susceptible to immunoprecipitation per se, they would show increased ChIP signals for unrelated DNA-binding and chromatin-binding proteins. To test this hypothesis, we analyzed published ChIP-Seq datasets for the Ste12 transcription factor and the Cse4 centromere protein specifically at the 238 euchromatic loci already identified as being enriched for Sir2, Sir3, and Sir4 (16). These 238 euchromatic loci also exhibited increased Ste12 and Cse4 signals (Fig. 2, Fig. S3, and Table S1). Therefore, these regions were not specifically bound by Sir proteins and did not reveal new Sir-protein biology. Rather they revealed loci that were commonly enriched in ChIP datasets. We called these regions of overlapping peaks of unrelated proteins “hyper-ChIPable.”
Fig. 2.
Sir-enriched euchromatic loci are hyper-ChIPable. Boxplots of ChIP-Seq levels for the transcription factor Ste12 and the centromeric Cse4 protein, comparing the genome-wide distribution versus the 238 Sir-enriched loci. The top of each box indicates the 75th percentile, the bottom the 25th percentile, and the thick bar inside the box is the median. The whiskers extend out to the most extreme data point that is at most 1.5 times the interquartile range from the box. Wilcoxon–Mann–Whitney P values comparing the genome-wide and Sir-enriched distributions are shown in the Sir-enriched boxes.
The Hyper-ChIPable Regions Coincided with Highly Expressed Genes.
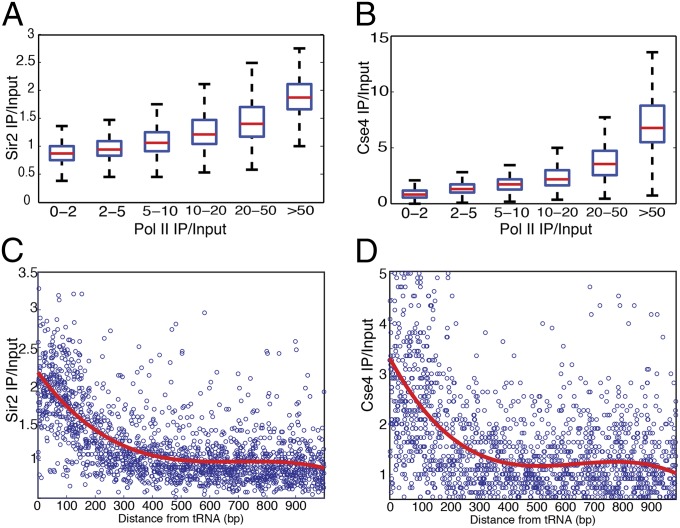
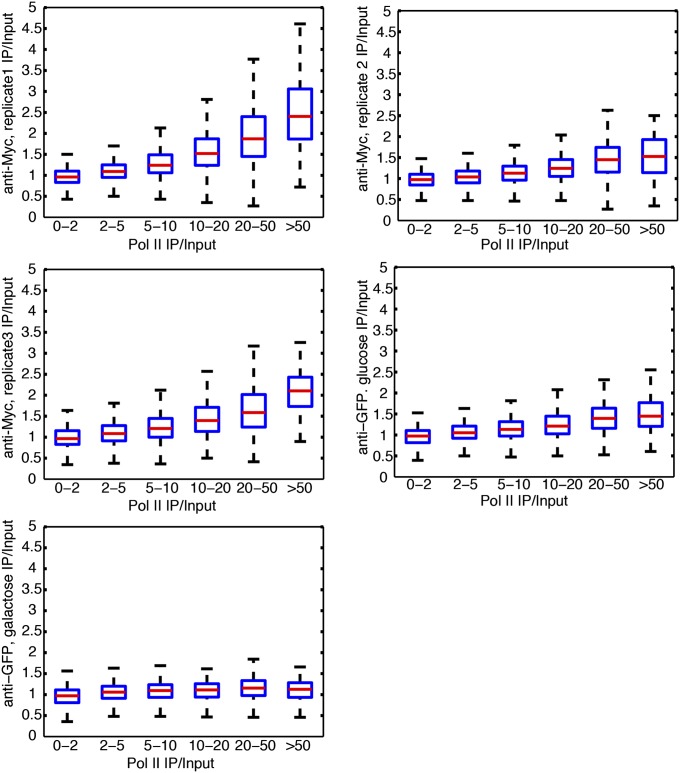
Investigations of common characteristics about these hyper-ChIPable loci where many unrelated factors were enriched revealed that these loci were frequently coincident with highly expressed Pol II-transcribed genes and tRNA genes, also known to be heavily transcribed by Pol III (17–19). Of the 238 regions, 145 (61%) overlapped tRNA genes, and another 47 (20%) had more than 10-fold enrichment of Pol II (Table S1). Additionally, this relationship was also evident with qPCR analyses, in which three genes with high expression showed Sir4 signal, but MET3, which is not expressed under these conditions, did not show enrichment of Sir4 (Fig. S2). This concordance of the 238 Sir-enriched euchromatic loci being coincident with highly expressed genes led us to investigate the genome-wide ChIP-to-expression-level correlation by comparing the ChIP levels of the Sir2, Sir3, and Sir4 and Cse4 proteins to RNA Pol II occupancy or tRNA gene proximity genome-wide. There was a striking positive relationship between ChIP enrichment and the level of RNA Pol II (Fig. 3 A and B and Fig. S4), extending the correlation between increased ChIP capacity with increased transcription beyond the originally identified 238 loci. Similarly, the ChIP levels of Cse4, Ste12, Sir2, Sir3, and Sir4 correlated with proximity to tRNAs (Fig. 3 C and D and Fig. S5).
Fig. 3.
Highly expressed loci are hyper-ChIPable. Boxplots of the Sir2 (A) and Cse4 (B) genome-wide ChIP-Seq levels, as a function of Pol II enrichment. Sir2 (C) and Cse4 (D) genome-wide ChIP-Seq levels, as a function of the distance from all S. cerevisiae annotated tRNAs. The red curve is a polynomial fit to the scatterplot.
This positive genome-wide relationship between ChIP enrichment and RNA Pol II occupancy suggested that the hyper-ChIPability of loci was due to increased transcription. If so, the ChIP-specific enrichment signal would be expected to extend across the entire gene body. Representative loci showed consistent enrichment of Sir proteins over a continuous stretch of 1,000–2,000 base pairs (bp) (Fig. 1, Right). To test the generality of this trend, we analyzed the ChIP levels of the Sir2, Sir3, and Sir4 proteins as a function of distance from the 5′ end of the 200 most highly transcribed genes in the S. cerevisiae genome. As predicted, a metagene analysis of these loci showed unambiguous enrichment of all three silencing proteins deep inside the ORFs (Fig. S6). Therefore, the 238 initially identified hyper-ChIPable loci illuminated the genome-wide phenomenon that more highly expressed genes showed increased enrichment independent of the transcription factor or chromatin protein assayed. This presence of the Sir protein silencing complex over the most heavily transcribed genes was exactly the opposite of the expected distribution for Sir-based gene silencing. Thus, combined with the ChIP signals for the distribution of the unrelated Ste12 and Cse4 proteins over the most expressed genes, the data were strongly suggestive of a technical issue, rather than interesting new biology.
Green Fluorescent Protein Was Enriched at Highly Expressed Loci by ChIP-Seq Analyses in an Expression-Dependent Manner.
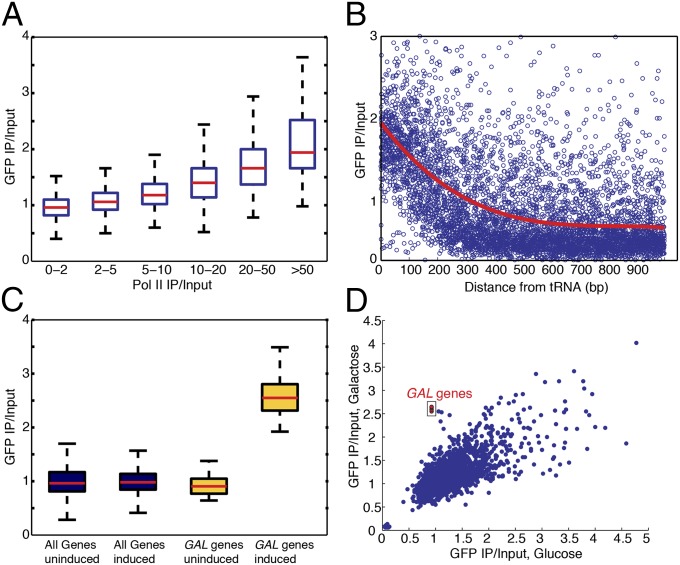
The analyses described above linked high expression level with increased IP/input ChIP signals for unrelated chromatin- and DNA-interacting proteins. To challenge the possibility that this correlation might reflect an unappreciated dimension of chromatin protein biology, we tested whether immunoprecipitation of the heterologous green fluorescent protein (GFP) with a nuclear localization signal (NLS) and lacking any histone or DNA-binding domain, would reproduce ChIP peaks over the highly expressed genes. The GFP ChIP-Seq data recapitulated the Cse4-, Ste12-, and Sir-protein results: GFP enrichment signals were higher in the 238 Sir-enriched loci than the rest of the genome (Fig. S7A). Additionally, the GFP enrichment correlated with Pol II levels and increased with proximity to tRNA genes (Fig. 4 A and B). Again, we confirmed these enrichments of GFP by ChIP-qPCR: GFP enrichments were higher at the LSR1, CDC19, and GCN4 genes, compared with the nonexpressed MET3 locus (Fig. S7B). The GFP antibody was the third antibody displaying hyper-ChIPability, illustrating the pervasiveness of this enrichment at highly expressed loci (anti-Myc for Sir2, Sir3, Sir4, and Cse4; and anti-GFP and anti-HA for Ste12). Therefore, by performing ChIP-Seq of a protein that we did not expect to be associating with DNA, we demonstrated that hyper-ChIPable loci were not regions of unexpected binding of chromatin proteins, but rather an artifact of the ChIP method.
Fig. 4.
ChIP-Seq of nuclear-localized GFP shows hyper-ChIPability. (A) GFP ChIP-Seq levels, from YPD (2% glucose) media, as a function of Pol II enrichment. (B) GFP ChIP-Seq levels, from YPD (2% glucose) media, as a function of increasing distance from tRNAs. (C) Boxplots of GFP ChIP-Seq levels, showing increase for the GAL1, GAL2, GAL7, and GAL10 genes, specifically under the induced galactose condition. For the induction, cells were shifted from overnight growth in raffinose to galactose media for 4 h. (D) Average GFP ChIP-Seq levels for all genes, comparing dextrose versus galactose-shifted media. The GAL1, GAL2, GAL7, and GAL10 genes are highlighted in the red rectangle; they show normal levels in dextrose but are enriched after the shift to galactose.
The correlation between ChIP levels and highly expressed loci suggested that high-level expression per se could contribute to hyper-ChIPability. Moreover, Fan and Struhl reported artifactual ChIP signal under inducing conditions of TBP and Hsf1 over the GAL1 coding region and of TBP and Gal4 over a heat shock gene (20). However, it was possible that some other feature of genes with the capacity for high-level expression led to their enrichment during chromatin immunoprecipitations. To directly test the effect of expression on the enrichment of these loci in the ChIP immunoprecipitates, we performed ChIP-Seq of chromatin from cells expressing the NLS-GFP shifted from medium with glucose to medium with galactose as the sole carbon source. There was a clear spike in the GFP ChIP levels at the galactose-inducible GAL genes (GAL1, GAL2, GAL7, and GAL10), upon induction of these genes (Fig. 4C). The GFP results were highly reproducible genome-wide, with the exception of the activated GAL loci, which were not enriched in glucose media (where the genes are tightly repressed), but enriched in chromatin from galactose-grown cells (Fig. 4D). Similarly, as measured by ChIP-qPCR, the GFP signal also increased after induction of the GAL1 gene by shifting from glucose to galactose-containing medium (Fig. S7C). This result established that increased transcription was sufficient for hyper-ChIPability.
No-Tag Controls Did Not Eliminate Hyper-ChIPability.
The discovery of expression-dependent GFP ChIP enrichment led us to ask whether the antibody target itself contributed to the signal or whether this enrichment depended only on the immunoprecipitation with an antibody. We performed ChIP-Seq on chromatin immunoprecipitated with anti-Myc in a strain lacking a Myc tag on any protein. Likewise, we tested the GFP antibody in a strain without any GFP. The resulting hyper-ChIPability was variable in these no-tag or no-GFP experiments. In contrast to the strong enrichment over highly expressed loci observed in seven of seven ChIP-Seq datasets above (Sir2, Sir3, Sir4, Cse4, Ste12, GFP-glucose, and GFP-galactose), only two out of the five no-tag/no-GFP datasets showed an enrichment quantitatively similar to the tagged datasets; another two showed enrichments in the immunoprecipitated chromatin with a weak correlation with published Pol II levels, and one set showed no hyper-ChIPability whatsoever (Fig. 5).
Fig. 5.
Hyper-ChIPability is present but variable in no-tag and no-GFP ChIP-Seq. Boxplots of ChIP-Seq levels for immunoprecipitations in a strain with no protein tags and no GFP, as a function of Pol II enrichment. Shown are three biological replicates using anti-Myc antibody and an IP performed using the anti-GFP antibody for glucose- and galactose-grown cells.
The variable extent of the enrichment at highly expressed loci in these “negative control samples” implied that no-tag controls could not be reliably used to remove spurious cases of hyper-ChIPability. Despite the variability, the presence of hyper-ChIPability in some of the extracts from cells lacking the target of the antibody was striking. It indicated that the enrichment was due, at least in part, to something that happened in vitro from the IP step itself interacting with this more “open” chromatin, rather than due to increased interaction between a nuclear protein and highly transcribed regions in vivo.
Using GFP ChIP-Seq to Remove False Positive Targets.
Our detection of the hyper-ChIPable loci, despite normalization by input-Seq, and the variability in no-tag experiments above, implied that neither no-tag nor input samples are sufficient controls to remove false positive ChIP peaks. Hence, we considered whether the GFP ChIP-Seq data could be used to discriminate between real and artifactual localization signals.
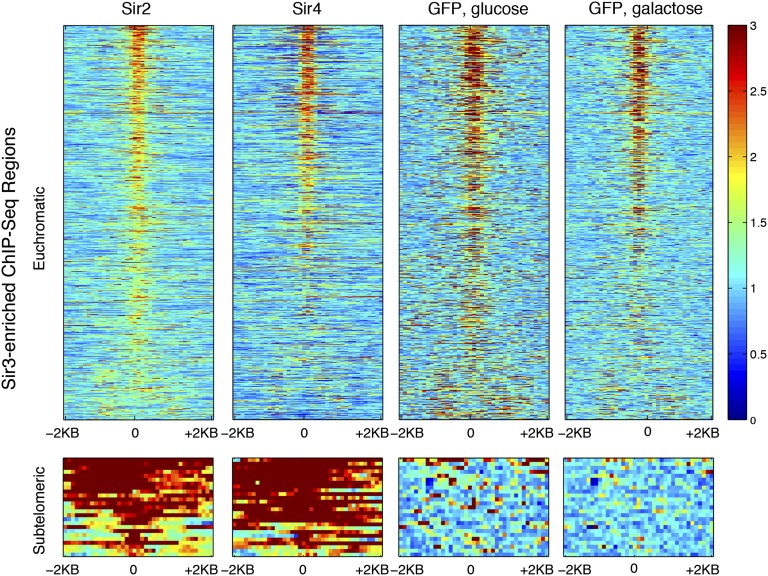
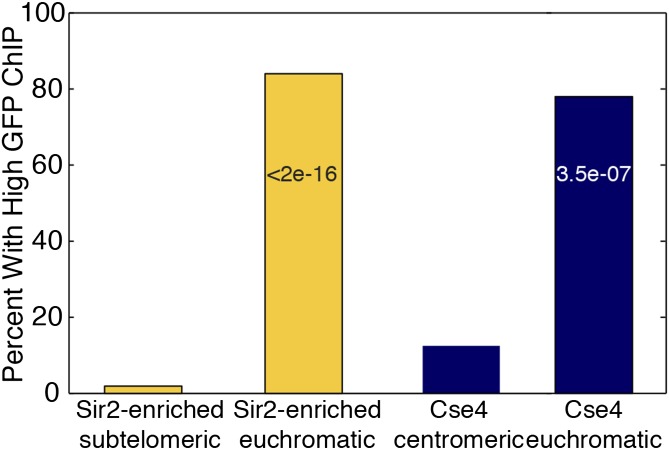
In principle, known silenced chromatin should be specifically enriched for the Sir2, Sir3, and Sir4 proteins, without the corresponding GFP increase, in contrast to the hyper-ChIPable euchromatic loci where Sir proteins and GFP seemed to colocalize. As predicted, subtelomeric sequences, within 10 kb of chromosome ends, had enrichment of all three Sir proteins, without a high GFP signal (Fig. 6). Therefore, we applied a simple GFP-enrichment threshold for all putative Sir-enriched loci, requiring the average IP/input GFP enrichment to be below 1.5-fold before a signal was considered tentatively meaningful. Among subtelomeric Sir targets, only 1 of 53 regions (2%) had a high GFP level; however, in the euchromatic Sir-enriched regions, 201 of 238 (84%) were filtered out because of high GFP enrichment. Similarly, for Cse4, only 2 of the 16 centromeric targets (12.5%) had GFP >1.5×, but 61 of the 78 noncentromeric Cse4 targets (78%) had high GFP enrichment (Fig. 7). These results suggested that it was useful to include a GFP-like ChIP sample as a method for improving the specificity of the ChIP experiments.
Fig. 6.
GFP and Sir ChIP-Seq levels are correlated only in euchromatin. Heat maps, plotting IP/input ChIP-Seq levels for euchromatic (Upper) and subtelomeric (Lower) Sir3-enriched regions. On the y axis, the regions are sorted in descending order of average Sir3 ChIP ratios. On the x axis, the regions are centered at the Sir3 peak, showing 2 kb to the left and right of the Sir3 peak center. (Upper) A total of 739 euchromatic regions, 50 kb or more from chromosome ends. (Lower) A total of 29 subtelomeric regions within 10 kb of chromosome ends.
Fig. 7.
Using GFP ChIP-Seq to improve ChIP localization analysis. Percent of Sir- and Cse4-enriched loci with average GFP ChIP ratio >1.5. For Sir proteins, the comparison is between the subtelomeric regions where the Sir proteins are known to localize and the euchromatic loci where the ChIP signal is artifactual. For the Cse4 centromere-binding protein, the comparison is between centromeric and euchromatic/noncentromeric regions. χ2 P values are shown within the euchromatic bars.
Discussion
Through interrogation of the genome-wide distribution of the yeast Sir-protein silencing complex, Ste12 and Cse4 we found regions that exhibited ChIP enrichment nonspecifically. Characterization of these loci revealed that they correlated with regions of high expression. To test whether this enrichment was due to unforeseen biology, we assayed the enrichment of a completely heterologous protein in yeast, GFP, and found that the GFP enrichment paralleled the nonspecific signal identified for the five endogenous proteins above. Thus, this was a pervasive problem not indicative of new biology, but rather a systematic enrichment across datasets. We term these loci “hyper-ChIPable.”
Our results agree with the work of Fan and Struhl, questioning reports of pervasive Mediator complex binding across the yeast genome (20). The recapitulated hyper-ChIPability of all unrelated proteins that we analyzed, including silencing factors and GFP, make it clear that this spurious signal was not limited to the Mediator, but is a general and underappreciated problem with the ChIP procedure per se.
There are many possible causes of the hyper-ChIPability. The detection of increased enrichment signal in no-tag and no-GFP controls suggests that something is happening during immunoprecipitation specifically, causing chromatin from highly expressed genes to be selectively recovered. The input nonimmunoprecipitated samples would of course lack that enrichment, leading to the genesis of biologically artifactual enrichments. It may be that DNA from actively transcribed regions, because of nucleosome depletion, is more exposed and likely to interact with beads or antibodies during the IP through something as simple as electrostatic interactions. Other possibilities include nonspecific interactions between some antibodies and RNA polymerase II and RNA polymerase III.
Regardless of the cause, our findings clearly argue for caution in interpreting ChIP signals in regions that are highly expressed. One way of ascertaining whether an enrichment is biologically relevant is to evaluate the underlying expression state of a locus. Genes expressed at the highest levels would be most vulnerable to the artifact described here. This sort of enrichment has already been reported in the literature for Sir proteins. For example, Sir2 was reported to be enriched at CDC19 by Li et al., a locus we find to be hyper-ChIPpable, but no biological consequence for this enrichment was found (21). Additionally, Sir3 was reported to associate with the GAL1-10 locus specifically upon induction by galactose, which is consistent with our findings that this nonspecific enrichment occurs only when genes are highly expressed (9). By inference, interpreting the enrichment profiles of proteins that cause high-level expression may be especially problematic because the consequence of their function is to set the stage for this artifact. One discriminator of the real signal is that enrichment distributed over a gene body, as shown in the examples here, is indicative of this artifact.
Because input samples do not show the same enrichment as IP, and because no-tag experiments were so variable, obtaining genome-wide ChIP samples for multiple different proteins of different functions would appear to be the safest way to avoid errors of interpretation. A GFP or an endogenous nuclear-localized protein that does not bind to DNA or chromatin, or a protein such as Cse4 known to have just a handful of targets, can serve as a good control to identify biologically meaningless ChIP peaks and to estimate the intrinsic level of hyper-ChIPability of a given locus. More work is necessary to identify the biophysical basis of hyper-ChIPability, but the fundamental biological cause of the artifact is high-level transcription as reported by RNA polymerase ChIP signals, RNA-seq read counts, and directly by the induction-specific appearance of the artifact at the GAL genes.
Clearly at highly expressed genes, ChIP can be used successfully to identify proteins critical for the high expression. However, very likely, published reports of ChIP signals at highly expressed genes should be reanalyzed, given the above results. Moreover, given the pervasiveness of this artifact as established here for yeast, it is reasonable to expect that hyper-ChIPability is a general problem of ChIP applications in all organisms. Particularly, the highly expressed HOT (Highly Occupied Target) regions, where unrelated transcription factors seem to bind without any underlying sequence specificity, may be a consequence of the same hyper-ChIPability. Although there may still be protein- or antibody-specific nuances, a workable normalization standard is needed for each organism and possibly for each tissue type. It would seem that an adequate number of biological repeats of ChIP-Seq analyses of a heterologous protein, such as GFP, to establish the noise level of the hyper-ChIPable peaks would provide a foundation for normalizing ChIP-Seq datasets. The work described here provides such a foundation for yeast studies.
Materials and Methods
Yeast Strains and Plasmids.
Strains are listed in Table S2. All yeast strains were generated in the W303 background. Deletions and tags were constructed through one-step integration of knockout cassettes (22). C-terminal tagging of Sir genes was also conducted using one-step gene replacement with the 13xMyc tag. The NLS-GFP plasmid was acquired from Addgene (24038).
Sir-Myc Chromatin Immunoprecipitation.
Seventy OD at 600 nm units of logarithmically growing cells were cross-linked with 1% formaldehyde for 1 h at room temperature. Chromatin was prepared as previously described (23) with minor modifications. Cells were lysed with 0.5-mm zirconia beads in 1 mL FA lysis buffer. Cell lysates were collected by centrifugation at 74,000 × g for 40 min then washed for 1 h at 4 °C in FA lysis buffer. Samples were then centrifuged at 74,000 × g for 22 min and then sonicated as described to an average size of 300–400 bp. Chromatin was immunoprecipitated with 120 μL of anti–c-Myc agarose (Sigma; 7470) overnight at 4 °C. Washes, elution, and isolation of DNA were performed as previously described (23).
GFP Chromatin Immunoprecipitation.
Cells were grown overnight at 30 °C in Complete Synthetic Media lacking leucine (CSM-leucine) with raffinose [2% (wt/vol)] as the sole sugar source. These cultures were then seeded at an OD of 0.2 into 100 mL of YP dextrose (2%) or YP galactose (2%) and grown at 30 °C for two doublings (about 4 h). Cells were fixed for 1 h at room temperature in 1% formaldehyde and chromatin, isolated as described above. Before immunoprecipitation, chromatin was precleared for 1 h at 4 °C with 40 μL Protein A Sepharose bead slurry (17-5280-01; GE Healthcare). Precleared chromatin was immunoprecipitated with 5 μL anti-GFP antibody (Abcam; ab290) overnight at 4 °C. Then a 120-μL aliquot of Protein A Sepharose bead slurry was added and samples were incubated for 4 h at 4 °C.
Sequencing and Mapping.
Sir-Myc libraries were prepared with modifications to the Illumina paired-end library protocol as in ref. 24. Libraries were loaded into one lane each and sequenced using the Illumina Genome Analyzer II as 45-bp paired-end reads.
GFP libraries were constructed using the Illumina Tru-Seq library preparation kit with the following modifications. Upon end repair, samples were cleaned up using a Qiagen MinElute column. Adapters were used at a 10-fold diluted concentration as to that provided. Samples were indexed and sequenced in a single lane on the Illumina HiSeq 2000 as 100-bp paired-end reads.
Sir-myc reads were mapped to the S288C genome using MAQ (25). GFP libraries were mapped with BWA and Samtools (26, 27). The number of reads mapped for each sample is outlined in Table S3. All sequences have been deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive under accession no. SRP030670.
Quantitative PCR.
Quantitative PCR on isolated chromatin from ChIP preparations was conducted with Dynamo HS SYBR green kit (Thermo Scientific) on a MX3000P qPCR machine (Agilent). The following primers were used for amplification: SEN1 (forward, ACCAAAGGTGGTAATGTTGATGTC and reverse, GGGAGGCGATGGTTTAGCCTGTAG), MET3 (forward, TTCTTGCAATTCGGTGGTGG and reverse, GACTCTAGCTGAATATCGGC), GCN4 (forward, CATCAAGACTGAAGAGGATCCAAT and reverse, AGTGGTAACTGGAATGTCATTGTC), and LSR1 (forward, GCTTTCTGTTTCTCCCTTAGTTTG and reverse, GCGAAGAAATCAACAATAAGAGCG).
Data Analysis.
IP/Input ratios.
Every base of the genome was assigned the total number of sequence reads overlapping it, separately for the input and IP sequence reads. Subsequent normalization and analysis was performed on median read coverage across 100-bp windows, sliding along each chromosome in 50-bp steps. The median IP coverage of each 100-bp interval was divided by the median input coverage for the same window. The IP/input ratios of each interval were normalized, dividing by the median genome-wide IP/input. Positions with fewer than 20 input reads were excluded from all subsequent analysis because of unreliable enrichment associated with division by low numbers.
Sir-enriched euchromatic loci.
We searched for all 100-bp euchromatic intervals, 50 kb or more from chromosome ends, with the normalized IP/input ratio of the Sir2, Sir3, and Sir4 greater than 2. All adjacent locations within 2,000 bp were merged into single contiguous interval, giving the 238 Sir-enriched euchromatic loci. We also used the MACS peak-calling software on the Sir ChIP-Seq datasets using the following parameters: a gsize of 1.2 × 107, mfold value of 2, and tag size (tsize) of 45 (28). All other parameters were used with the default settings.
Ste12, Cse4, and Pol II datasets.
We used the published ChIP-Seq datasets for the Ste12, Cse4, and Pol II proteins (16). Mapped reads were downloaded from the NCBI Gene Expression Omnibus, accession no. GSE13322. For each IP, we summed the read counts across the three replicate experiments at each base pair of the S. cerevisiae genome; the same was done for the input samples of each protein.
ChIP levels as a function of expression.
All genomic positions were split into those within 1,000 bp of an annotated tRNA gene start or stop (tRNA-proximal dataset) or more than 1,000 bp of tRNA gene start or stop (tRNA-distal dataset). The tRNA-proximal dataset was used to make the scatterplot of ChIP level versus increasing distance from the tRNA genes, and the tRNA-distal set was used to plot the ChIP levels as a function of Pol II ChIP signal.
Glucose versus galactose GFP ChIP-Seq.
Average IP/input ratios from the GFP-glucose and GFP-galactose ChIP-Seq experiments were calculated for each annotated S. cerevisiae gene. Boxplots were plotted for all genes, compared with the galactose-induced GAL1, GAL2, GAL7, and GAL10. The scatterplot comparing GFP ChIP-Seq of chromatin from cells grown in glucose and galactose media was also plotted on gene-level averages.
Heat maps.
The y axis is a list of Sir3-enriched regions. The regions were selected as follows. (i) All 100 mers with Sir3 IP/input >2 were merged into a contiguous block if separated by less than 2 kb. (ii) The regions were ordered by descending level of average Sir3 IP/input ratio. (iii) The heat maps were constructed for 2,000 bp to the left and right of the center of each Sir3-enriched region. (iv) The heat maps were plotted separately for euchromatic regions, greater than 50 kb from chromosome ends and subtelomeric regions that were within 10 kb of chromosome ends.
Using GFP to enrich for Sir and Cse4 true binding events.
Sir-enriched regions were defined as above, where there were contiguous blocks of >2× IP/input ratios of Sir2, Sir3, and Sir4. For each such subtelomeric or euchromatic region, we calculated the average GFP IP/input ratio across the entire region. We used the GFP in the galactose ChIP-Seq dataset because it had higher sequence coverage. Regions with average GFP IP/input ratio >1.5 were deemed “high GFP” and those with an IP/input ratio of ≤1.5 were deemed “low GFP.”
Cse4 targets were from the supplementary information of the published ChIP-Seq study (16). Cse4 targets were partitioned into “centromeric” and “euchromatic/noncentromeric” according to the designation in the published supplementary table from the above study. High-GFP and low-GFP thresholding was the same as for the Sir-protein analysis above.
Statistical analyses.
All statistical tests and fits were performed using Matlab.
Supplementary Material
Acknowledgments
We thank Dylan Mooijman, Michael Eisen, Ravi Sachidanandam, Leonid Mirny, Kevin Struhl, Oliver Rando, Michael Snyder, Ting Wu, Audrey Gasch, Fred Winston, and members of the Fred Winston, A.v.O., and J.R. laboratories for helpful comments and discussions and Minyong Chung and the Vincent J. Coates Genomics Sequencing Laboratory at the University of California Berkeley for all high throughput sequencing. This work was supported by the National Institutes of Health (NIH)/National Cancer Institute Physical Sciences Oncology Center at Massachusetts Institute of Technology (U54CA143874), an NIH Pioneer Award (DP1 CA174420), NIH Grant R01 GM 068957, and a Netherlands Organization for Scientific Research Vici award (to A.v.O.), a National Science Foundation predoctoral fellowship (to D.M.T.), University of California Berkeley’s Cellular, Biochemical, and Molecular Sciences training grant from the NIH, and NIH Grant GM31105 (to J.R.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information Short Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. SRP030670).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316064110/-/DCSupplemental.
References
- 1.Hanlon SE, Lieb JD. Progress and challenges in profiling the dynamics of chromatin and transcription factor binding with DNA microarrays. Curr Opin Genet Dev. 2004;14(6):697–705. doi: 10.1016/j.gde.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet. 2012;13(12):840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ho JW, et al. (2011) ChIP-chip versus ChIP-seq: Lessons for experimental design and data analysis. BMC Genomics 12:134. [DOI] [PMC free article] [PubMed]
- 5.Chen Y, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods. 2012;9(6):609–614. doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kidder BL, Hu G, Zhao K. ChIP-Seq: Technical considerations for obtaining high-quality data. Nat Immunol. 2011;12(10):918–922. doi: 10.1038/ni.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 8. Teytelman L, Nishimura EA, Ozaydin B, Eisen MB, Rine J (2012) The enigmatic conservation of a Rap1 binding site in the Saccharomyces cerevisiae HMR-E silencer. G3 (Bethesda) 2(12):1555-1562. [DOI] [PMC free article] [PubMed]
- 9.Radman-Livaja M, et al. Dynamics of Sir3 spreading in budding yeast: Secondary recruitment sites and euchromatic localization. EMBO J. 2011;30(6):1012–1026. doi: 10.1038/emboj.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28(4):327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 11.Marchfelder U, Rateitschak K, Ehrenhofer-Murray AE. SIR-dependent repression of non-telomeric genes in Saccharomyces cerevisiae? Yeast. 2003;20(9):797–801. doi: 10.1002/yea.1001. [DOI] [PubMed] [Google Scholar]
- 12.Barton AB, Kaback DB. Telomeric silencing of an open reading frame in Saccharomyces cerevisiae. Genetics. 2006;173(2):1169–1173. doi: 10.1534/genetics.106.058420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 14.Vega-Palas MA, Martín-Figueroa E, Florencio FJ. Telomeric silencing of a natural subtelomeric gene. Mol Gen Genet. 2000;263(2):287–291. doi: 10.1007/s004380051170. [DOI] [PubMed] [Google Scholar]
- 15.Wyrick JJ, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402(6760):418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 16.Lefrançois P, et al. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84(2):245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 18.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA. 2003;100(25):14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol. 2004;24(10):4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan X, Struhl K. Where does mediator bind in vivo? PLoS ONE. 2009;4(4):e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Valsakumar V, Poorey K, Bekiranov S, Smith JS. Genome-wide analysis of functional sirtuin chromatin targets in yeast. Genome Biol. 2013;14(5):R48. doi: 10.1186/gb-2013-14-5-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23. 23. Aparicio O, et al. (2005) Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol Chap 21:Unit 21.3. [DOI] [PubMed]
- 24.Zill OA, Scannell D, Teytelman L, Rine J. Co-evolution of transcriptional silencing proteins and the DNA elements specifying their assembly. PLoS Biol. 2010;8(11):e1000550. doi: 10.1371/journal.pbio.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, et al. (2008) Model-based analysis of ChIP-seq (MACS). Genome Biol 9(9):R137. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.