Abstract
The crystal structure of the K1 domain, an adhesin module of the lysine gingipain (Kgp) expressed on the cell surface by the periodontopathic anaerobic bacterium, Porphyromonas gingivalis W83, is compared to the previously determined structures of homologues K2 and K3, all three being representative members of the cleaved adhesin domain family. In the structure of K1, the conformation of the most extensive surface loop is unexpectedly perturbed, perhaps by crystal packing, and is displaced from a previously reported arginine-anchored position observed in K2 and K3. This displacement allows the loop to become free to interact with other proteins; the alternate flipped-out loop conformation is a novel mechanism for interacting with target host proteins, other bacteria, or other gingipain protein domains. Further, the K1 adhesin module, like others, is found to be haemolytic in vitro, and so, functions in erythrocyte recognition thereby contributing to the haemolytic function of Kgp. K1 was also observed to selectively bind to haem-albumin with high affinity, suggesting this domain may be involved in gingipain-mediated haem acquisition from haem-albumin. Therefore, it is most likely that all cleaved adhesin domains of Kgp contribute to the pathogenicity of P. gingivalis in more complex ways than simply mediating bacterial adherence.
Keywords: gingipain adhesins, oral infection, Porphyromonas gingivalis, protein structure, virulence factor
Introduction
Chronic periodontitis is an inflammatory disorder of the supporting structures of the teeth in response to the buildup of dental plaque, which is most commonly a result of poor oral hygiene. Progression to this advanced form of periodontal disease from the preventable gingivitis gives rise to the destruction of the periodontium and subsequent loss of teeth [1–3]. This disease is the primary cause of tooth loss in the human population [4], and there is a growing concern that it may be associated with other chronic disorders such as cardiovascular diseases [5], diabetes mellitus [6, 7], rheumatoid arthritis [8], and some forms of cancer [9]. This suggests that severe periodontal disease not only affects the oral cavity but may influence the overall well-being of an individual.
Selected microorganisms have been implicated in the pathogenesis of periodontal disease, but it is the Gram-negative anaerobic bacterium, Porphyromonas gingivalis, that is most strongly associated with disease severity [1, 10, 11]. P. gingivalis expresses a large array of proteolytic enzymes that bind, lyse, and degrade erythrocytes to release haemoglobin, providing a source of peptides, iron, and haem for this asaccharolytic porphyrin-auxotroph [2, 12–15]. Because of this, P. gingivalis is often abundant in bleeding chronic periodontal lesions that provide haemoglobin released from ruptured erythrocytes [1].
P. gingivalis can either passively survive within oral epithelial cells or it can aggressively disrupt the host immune response through a number of mechanisms, of which a group of cysteine proteases, known as gingipains, are believed to play a key role [1]. Gingipains are expressed by P. gingivalis and significantly contribute to pathogenicity as they play important roles in nutrient acquisition [16], defensin degradation [17–19], complement depletion [20, 21], coagulation cascade manipulation [22], and cytokine signalling network dysregulation [23, 24]. The gingipains effectively allow the bacterium to successfully evade the host immune response and increase nutrient availability. Further, gene knock-out studies found that gingipain-deficient mutants of P. gingivalis were less virulent and unable to acquire haem [25–28]. Together, it can be argued that gingipains are the most important virulence factors of P. gingivalis.
Arginine-specific (Arg-gingipain; RgpA and RgpB) and lysine-specific (Lys-gingipain; Kgp) gingipains are expressed on the cell surface of most strains of P. gingivalis (with the exception of HG66, where they are secreted into the culture medium) and are encoded by individual gene loci (rgpA, rgpB, and kgp, respectively) [29–31]. All gingipains contain a catalytic domain, but only RgpA and Kgp contain a multi-domain haemagglutinin/adhesin (HA) region (Kgp shown in Fig. 1A), which facilitates bacterial adherence to extracellular matrix proteins and host cells [13]. This common function of the HA regions of gingipains is consistent with its markedly high sequence similarity to haemagglutinin A (HagA), which is also expressed by P. gingivalis [32]. Upon secretion and auto-proteolytic maturation, the multiple domains of the HA region of RgpA and Kgp non-covalently associate with each other and their respective catalytic domain to form a stable complex, where solvent-accessible loop regions of each domain are poised to bind potential targets [13, 33].
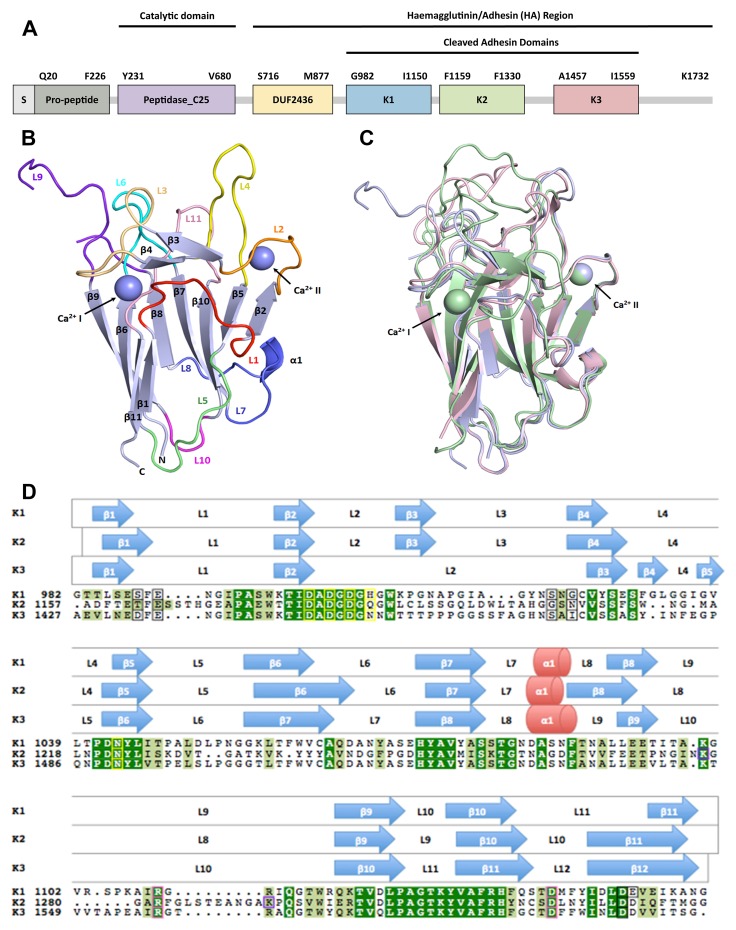
Fig. 1.
Kgp and the structural comparison of its cleaved adhesin modules, K1, K2 and K3. A: The domain structural model of Kgp from P. gingivalis strain W83 proposed by Li & Collyer [32] as the C25 protease domain followed by an undefined region of sequence (designated as DUF2436) and then the cleaved adhesin family modules, K1, K2 and K3. B: A cartoon representation of the fold of K1. The β-sandwich core is coloured light blue and numbered increasing loops/helices are highlighted in a spectrum of colours from red to blue to pink. Ca2+ ion positions are indicated by purple spheres. C: Superimposition of cartoon representations of K1, K2 (PDB code: 3KM5) and K3 (PDB code: 3M1H) shows the structural similarities between the three crystal structures. The K1 structure and its Ca2+ ions are in light blue, the K2 in pale green and K3 in light pink. K1 superimposed onto K2 and K3 with a Cα rmsd of 1.7 Å and 1.3 Å, respectively. The designation of the Ca2+ site I and Ca2+ site II are as shown. D: Structural sequence alignment of the sequences of K1, K2 and K3. Secondary structural elements are grouped above each row, with the aligned sequences on the bottom. β-sheets are represented by blue arrows, α-helices are represented by red cylinders and length of the protein indicated by grey lines. K1 has 173 residues (Gly982-Gly1154), K2 has 178 residues (Ala1157-Gly1334) and K3 has 176 residues (Ala1427-Gly1602). Conserved residues are highlighted in green. The residues which coordinate to Ca2+ ions are identified by a black border (site I) or a yellow border (site II). Kgp auto-proteolysis sites in K2 [34] are enclosed in purple-bordered boxes. Arginine anchors (R) and associated anchoring sites (D) are indicated by a pink border
The crystal structures of the cleaved adhesin domains, K2 and K3, of W83 Kgp were recently reported [33, 34] establishing that the Kgp HA region contains homologous structural domains with a Ca2+-stabilized β-sandwich core, with a structural topology that is typical of MAM domains and carbohydrate-binding modules (CBM). Additionally, these two domains were shown to be haemolytic in vitro and, thus, may be involved in Kgp-mediated erythrocyte recognition, haemolysis, and haem acquisition.
This study reports the crystal structure determination of the third Kgp adhesin module, K1 (see crystallographic data in Appendix A1), and confirms that all three adhesin domains are homologous and have an almost identical β-sandwich core stabilised by two Ca2+ ions. This folded K1 recombinant construct displays similar in vitro binding and haemolytic activities to those previously reported for the K2 and K3 modules.
Materials and method
Plasmid construction, protein expression, and purification
The gene fragment encoding the K1 domain (Gly982–Gly1154) of Kgp from P. gingivalis W83 (gene code AF017059, protein ID O52050) was cloned into pGEX-5X-3 (GE Healthcare) at the cloning sites BamHI/XhoI. Nucleotides encoding three additional residues, Gly–Ile–Pro, were added to the N-terminus of the construct, following the GST tag and the Factor Xa cleavage site. The construct was sequenced to confirm that there was no mutation present.
The constructed plasmid was transformed into Escherichia coli BL21 (DE3) competent cells for protein expression. Cells were grown in Luria–Bertani medium with ampicillin (100 μg/ml) at 37 °C until OD600 = 0.6 and temperature was reduced to 20 °C. Protein overexpression was induced by the addition of 0.1 mM IPTG. The recombinant GST-K1 protein was purified using Glutathione Sepharose 4B affinity chromatography. Protein-bound beads were washed with Factor Xa Cleavage Buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM CaCl2), and K1 was cleaved from the GST tag by Factor Xa protease treatment. The eluted protein was purified by size exclusion chromatography using Superdex 75 HiLoad 16/60 (GE Healthcare) with running buffer (10 mM Tris-HCl pH 7.6, 150 mM NaCl) which yielded a >95% pure 18.5 kDa protein (as estimated by sodium dodecyl sulfate–polyacrylamide gel [SDS–PAGE]). The de-tagged recombinant K1 could be readily concentrated to 10 mg/ml and was monomeric and monodisperse as observed by Dynamic Light Scattering (Appendix A2). The purified protein is very stable, and no proteolysis was detected by SDS–PAGE after storage at −80 °C for 3 months.
Crystallisation, data collection, and determination of K1 structure
Crystallisation of proteins was carried out at room temperature. Crystallisation screens were performed using Mosquito (TTP LabTech) and 96-well screening kits (Qiagen). Bunches of small needle-shaped crystals were observed from several conditions in the Classic and JCSG+ suites (Qiagen) after ~24 h. The conditions were optimised to 40 mM KH2PO4, 20% (v/v) glycerol, 16% (w/v) PEG 8000, and 1 M GuHCl to grow larger crystals using the hanging drop vapour diffusion crystallisation method. Two microlitres of 5 mg/ml protein solution was mixed with 2 μl of reservoir solution. Needle-shaped crystals grew in ~5 days. Crystals were subsequently flash cooled in a N2 stream at 100 K, with no additional cryoprotection. Diffraction data were initially collected using an in-house X‑ray source with a rotating anode X-ray generator (Rigaku RU200H) with an image plate detector (Marresearch scanner 345 mm plate). Data were also measured on the 2BM1 beam line, Australian Synchrotron, Melbourne, using an ADSC Quantum 210r CCD detector. Intensities were integrated and processed using HKL2000 [45]. Solvent-content analysis revealed that each asymmetric unit contained one unit with a Matthews coefficient of 2.05 Å3/Da (39.96% solvent content). The structure was solved by molecular replacement using Phaser [46] with the K3 structure (Protein Data Bank [PDB] entry 3M1H) as the search model. The model was further manually built using COOT [47], and cycles of refinement were performed using REFMAC5 [48]. The data reduction and refinement statistics are summarised in Appendix A1.
Haemolysis
Trizma base, tris-hydrochloride (Tris-HCl), and Tween 20 were purchased from Sigma (St. Louis, MO, USA). Phosphate-buffered saline (PBS) was purchased from Oxoid (Basingstoke, UK). Fresh blood was drawn from human Blood Group 0 donors into 0.1 M citrate anticoagulant. Erythrocytes were suspended from platelet-rich plasma and the buffy coat by differential centrifugation at 350 × g, washed twice with PBS pH 7.4, and resuspended to 1% v/v in PBS. Various concentrations of K1 and heat-treated K1 up to 3 μM were added to the erythrocytes in a total volume of 200 μl and incubated at 25 °C or 37 °C. After periods of incubation, the microtitre plate was centrifuged at 1000 × g for 10 min and the supernatants (100 μl) were transferred into a new microtitre plate. Haemoglobin release was determined spectrophotometrically using a microtitre plate reader (at 405 nm, corresponding to the Soret region of haem absorbance).
Preparation of stoichiometric complexes of HSA and haemin
A complex of rHSA (Prospec, Rehovot, Israel) and haemin (Sigma) was prepared as previously described [49]. A stock solution of haemin at 12 mM was prepared in 100 mM NaOH. A solution of rHSA with a concentration of 0.1 mM was prepared in 0.1 M sodium phosphate buffer, pH 7.0. A solution mixture with a molar ratio of rHSA:haemin = 1:1.2 of the complex had a light brown colour and was stable at room temperature for 20 min. It was centrifuged for 10 min, and a dark precipitate discarded. The resulting supernatant was used to purify the complex by gel filtration using a Superdex 200 HR 10/30 column in a 1× PBS buffer, pH 7.4.
HSA binding experiments
The 96-well ELISA plates were coated with K1 polypeptide (0.4 μg/well in PBS) and incubated overnight at 4 °C. The wells were blocked with 100 μl of 1% skim milk in PBS for 1 h. The binding affinity between the polypeptides and rHSA and rHSA-haem was detected by adding different levels of rHSA in PBS buffer or rHSA-haem in buffer containing 0.1 μM haem to the coated plates. Thereafter, anti-HSA mAb (15C7, AbCam) was added, followed by alkaline phosphatase-conjugated rabbit anti-mouse IgG. The plates were washed with 0.05% Tween 20 in PBS solution except for the wells incubated with rHSA-haem solution. In these experiments, the wells were washed with 0.1 μM haem in Tween 20 solution three times between each step. Colour development was detected at 405 nm for p-nitrophenyl using the phosphatase substrate p-nitrophenyl phosphate. Data were fitted by non-linear regression using GraphPad Prism 4.0 software (GraphPad, La Jolla, CA, USA). Apparent Kd values were determined from hyperbolic binding data using the formula Y = Bmax·X /(Kd + X ).
Results
K1 structure overview
The crystal structure of the K1 adhesin domain (residues Gly982–Gly1154) is composed of 173 residues with a molecular weight of 18.5 kDa. K1 folds into a β-sandwich composed of two anti-parallel β-sheets, one containing four β-strands, β9–β6–β11–β1, and the other with five β-strands, β8–β7–β10–β5–β2 (Fig. 1B). Loops are found on either end of the structure that connects the β-strands together, with one end containing loops notably longer than the other. A small α-helix (H1) formed by residues Ala1085–Asn1087 is observed between loop 7 (L7) and 8 (L8) on the short-loop end of the β-sandwich. An eight-residue anti-parallel β-bridge, β3–β4, is observed on the long-loop end and is connected to β-strands, β2 and β5, via L2, L3, and L4. Two Ca2+ ions flank the long-loop end of the structure, connecting the β-sandwich core to the loop residues. As Ca2+ ions were not present in the crystallisation buffers, it is most likely that they were incorporated into the structure during protein expression and folding.
At a high resolution of 1.55 Å, the observed electron densities of the K1 structure were generally well-defined, except for a portion of the N-terminal half of L9. Consisting of 21 residues (Ile1097–Arg1117), L9 is the longest loop in the structure. The weak or missing electron densities of residues Thr1098–Ser1104 suggest that this portion of L9 may not be a single conformer in the crystal. The C-terminal half of this loop can, however, be observed in the structure, which is most likely to be a consequence of stabilisation by specific crystal packing arrangements.
A single K1 molecule is tightly packed into the asymmetric unit of the monoclinic space group P21. Three guanidinium ions and two glycerol molecules have been modelled on the surface of each K1 molecule, both of which are present in the crystallisation solution. These ligands appear to play a role in the stabilisation of the crystal packing of the K1 molecule and are most likely not to be biologically relevant.
Common features of the adhesin modules
The cleaved adhesin domains, K1, K2, and K3, share more than 30% sequence identity, with K1 having the highest sequence identity with K3 (71%). Pairwise structural alignment using the program DALI [35] confirms that the previously solved domains are almost identical to K1 as demonstrated by the low root mean square (rmsd) values ~1.5 Å and Z-scores above two (22.9 and 27.8 for K2 and K3, respectively). These values reflect the fact that sequence identity between the adhesin modules is very high, and their folds have significant similarities, especially in terms of core folding. Superimposition of all three modules using PyMOL [35] shows that the β-sandwich core, short-loop end residues, and Ca2+ ion binding sites closely correspond to each other (Fig. 1C). Most of the residues forming the β-strands are conserved, particularly those of β2, β3, β5, β7, β9, and β10 of K1 and K2 and β2, β6, β8, β10, and β11 of K3, respectively (Fig. 1D). As previously reported [33], many of these β-strand residues are not solvent accessible and they form the hydrophobic core of the adhesin domains.
The short-loop end of the β-sandwich of the three structures is highly conserved in sequence and conformation, with L7, L10, and H1 of K1 superimposing to L7, L9, and H1 of K2 and L8, L11, and H1 of K3, respectively. However, the residues of L8 in K1 and L9 in K3, although not formally designated as part of a β-strand (as defined by hydrogen bonding), adopt a conformation that matches the residues of the N-terminal half of β8 in K2.
Less similarity is observed between the sequences and structures of the loops at the long-loop end of the β-sandwich cores of the three adhesin modules, but some portions of the loops correspond closely in conformation. For example, the eight N-terminal residues of L2 in K1 and K3 closely mimic the conformation of L2 in K2. Other areas that closely correspond are loops L4–L4–L4/5, L6–L6–L7, and L11–L10–L12 of K1, K2, and K3, respectively. Further, the six C-terminal residues of L1 of K1 are similar in conformation to β11 of K2 and β12 of K3. Despite significant differences in the loop conformations at the long-loop end of the adhesin modules, it has been suggested that the conserved features are involved in interactions with common binding partners [33].
Interestingly, weak or missing electron densities have been observed in the N-termini of all the longest loops in each of the three adhesin modules; L9 in K1, L8 in K2, and L10 in K3. In addition to Thr1098–Ser1104 of L9 of K1, the residues Gly1273–Lys1276 and Ser1284–Gly1289 in L8 of K2 and Leu1544–Pro1553 of L10 in K3 have poorly defined electron densities. The crystal structures of K2 and K3, however, incorporated a number of additive ligands that partially stabilise the conformations of these loops in the solid state and have quite different crystal packing interactions, which permitted the missing residues to be observed, albeit weakly. This was not the case for K1, and therefore, the missing residues are shown as a break in the structure (Fig. 1B). Weak or missing electron densities in these loops of the high-resolution structures (1.55 Å, 1.4 Å, and 1.56 Å for K1, K2, and K3, respectively) suggest that they might also be highly flexible in solution. According to electrostatic potential analysis (data not shown), these portions of the loops are also highly negatively charged, which may indicate that they could be involved in some binding interactions.
Arginine anchoring site
One notable feature that is conserved between the three adhesin modules is an anchored arginine found in the longest loop of each adhesin domain. Our previous study [33] found that, when the sequences of K2 and K3 are optimally aligned, Arg1280 of L8 in K2 does not match with Arg1557 of L10 in K3 (Fig. 1D). Upon structural superimposition, however, these two arginines lie in the same position in space and in an identical type of interacting environment (Fig. 2B). Despite this, the loops they originate from have markedly different overall conformations (Fig. 1C). Very similar arginine anchoring sites are found in the three structures located near the surface of the long-loop end region and are characterized by two flanking planar aromatic residues, a salt bridge and hydrogen bonding interactions with a conserved internal core aspartic acid residue.
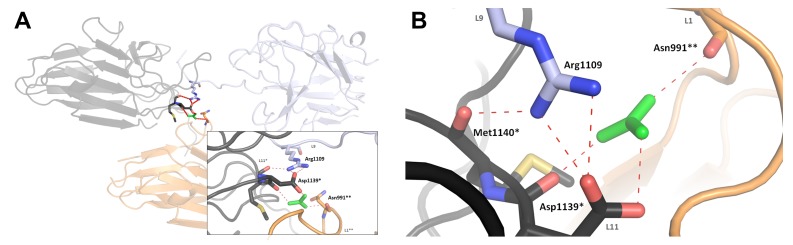
Fig. 2.
Arginine anchoring sites for arginines of loops K1 L9 (R1109), K2 L8 (R1280) and K3 L10 (R1557). Salt bridge hydrogen bonding interactions with a conserved aspartic acid residue (D1139, D1319 and K1588 in K1, K2 and K3, respectively) are shown as dashed lines. A: Guanidinium ion (green) mediating an interaction between one K1 molecule (light blue) and two other crystallographic symmetrically related K1 molecules, coloured black(*) and orange(**), respectively (inset: detailed view of guanidinium ion-mediated reaction). B: Superimposition of arginine anchors and their anchoring sites in K1 (light blue), K2 (light green) and K3 (pink). Met1140, Asn991 and guanidinium ion of K1 are excluded. The positions of the K2 and K3 arginine atoms overlay with K1 R1109 with an rmsd of 1.0 Å and 1.3 Å, respectively
Knowing that Arg1109 of L9 of K1 aligns with Arg1557 of L10 of K3, it would be expected that the arginine anchor of K1 would behave in a similar manner as observed in the other two adhesin structures (i.e., bind to a conserved internal aspartic acid). Instead, the arginine anchor interacts with an aspartic acid of another crystallographic symmetrically related molecule (designated * in Fig. 2) via a bound guanidinium ion, which is present in the crystallisation conditions (Fig. 2A). In the crystal, this salt bridge interaction is repeated, forming continuous links throughout the lattice. This guanidinium ion mediates an interaction between three symmetrically related molecules – Arg1109 interacts with an anchoring site composed of Asp1139* and Met1140* in one molecule (designated * in Fig. 2A) and Asn991** of another molecule (designated as ** in Fig. 2A). The observation of a continuous network of salt-bridge links is possibly an artifact of crystallisation and, therefore, not biologically relevant. It is most probable that, in the solution state, the K1 arginine anchor is also stable in a configuration similar to its counterparts in K2 and K3, as we observe that the K1 module is present as a monomer in solution and not an oligomer (see Appendix A2). If we assume that in the monomeric solution state Arg1109 is indeed anchored to Asp1139 (in the same K1 molecule), then we might expect that the anchoring might be equivalent to that observed in the K2 and K3 structures. This hypothesis is supported by a structural alignment of the three observed anchoring sites, demonstrating that the equivalent residues involved appear to be in almost the same position in space in all of the three adhesin domain structures (Fig. 2B). However, in gingipain complexes on the surface of the bacterium, a labile salt bridge could be an important structural feature for adhesin function.
Calcium binding sites and loop conformations
When the structures of the adhesin domains are superimposed, the Ca2+ ions occupy similar positions (Fig. 1D). Two sites in the adhesin domains are responsible for ligating to the Ca2+ ions, with the responsible residues generally conserved within the Kgp cleaved adhesin domains, but with minor differences (Appendix A3).
In Ca2+ binding site I, six residues coordinate the binding of the Ca2+ ion in K1, rather than the five residues plus one water molecule observed in K2 and K3. Glu1447 of K1 (present as an aspartic acid in K2 and K3, but not bound directly to the Ca2+ ion) replaces the water molecule. Overall, three out of seven metal-ligating residues are conserved; the ligating main chain carbonyls of Ser988 and Gly1146 in K1 are replaced with Thr1162 and Asn1205 in K2 and Asp1433 and Ile1472 in K3. The conformations of the loops, L1–L1–L1 and L3–L3–L2 of K1, K2, and K3, respectively, and β-strands, β2 of K2 and β3 of K1 and K2 (Fig. 1D), that surround this binding site do not correspond well in either sequence or in the structure alignments. Despite these apparent sequence differences, the Ca2+ is located in K1 in almost the same position within the molecule indicating a probable structural role for this bound ion.
Six of the seven metal-ligating residues are conserved in the Ca2+ binding site II of the adhesin domains. The seventh ligand, the main chain carbonyl of His1007 (K1) is substituted with Glu1185 (K2) or Asn1452 (K3). β2, β5, and the N-terminus of L3 in K1 are almost identical in sequence and in conformation to β2, β5, and L2 of K2 and β2, β6, and the N-terminus of L3 in K3. All of the metal-ligating residues of site II are located in these conserved areas, demonstrating a probable role for the bound cation in stabilizing loop conformations. The aspartic acids found in this binding site are conserved over all sequences of the adhesin domains, including K3* and A1, which may suggest that all of the adhesin domains are structurally dependent on bound Ca2+. A previous study of the thermal stability of the Kgp adhesin modules demonstrated that the thermal denaturation of K1 was specifically Ca2+-dependent [33]. Significantly, structural roles for Ca2+ are observed in many of the other domains found in the galactose-binding domain-like (GBD) superfamily.
Haemolytic activities
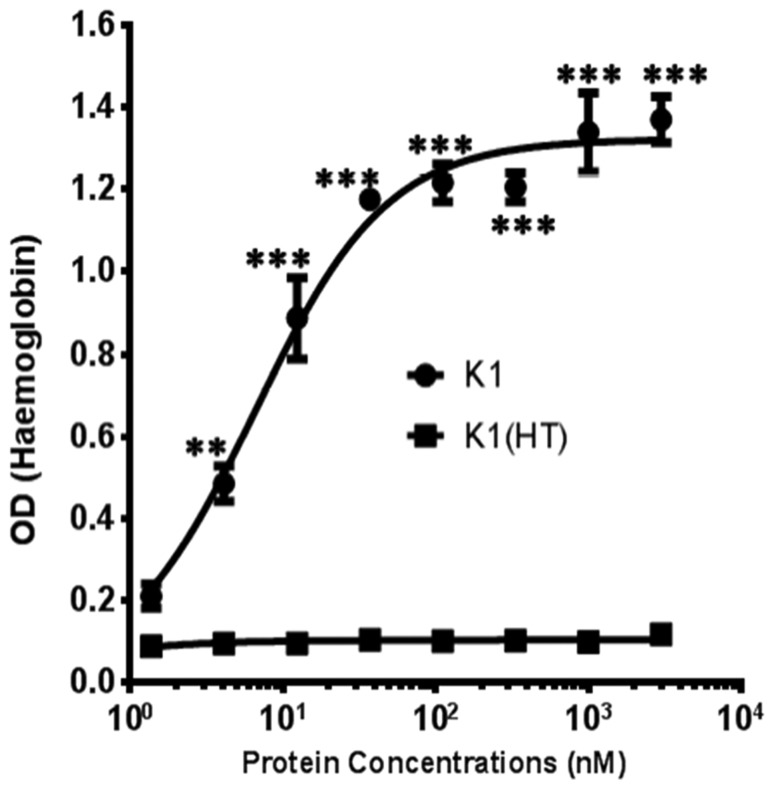
The current study demonstrates that the K1 protein construct used here induces concentration- and time-dependent haemolysis (K1, Fig. 3). K1 is effective in a range of 5 to 5000 nM with 50% haemolysis observed at 6.6 nM. The Kgp catalytic domain was absent in the K1 polypeptide, indicating that haemolysis occurred independently of proteolytic activity. K1 was able to perform haemolysis as compared to the negative control, heat-treated K1, which has lost haemolytic ability.
Fig. 3.
Haemolytic activity of the K1 module. Titrations of K1 up to 3 μM were added to a 0.2% erythrocyte suspension in a total volume of 200 μL. Incubation with erythrocytes with K1 was done in PBS at 25 °C for 48 h. After incubation the microtitre plate was centrifuged then at 1000 × g for 10 min and the supernatants (100 μL) transferred into a new microtitre plate. Haemoglobin release was measured by absorbance at 405 nm in a microtitre plate reader. Results are representative of three separate experiments. Error bars indicate the means and SE. **, P < 0.01, ***, P < 0.001 compared with heat-treated (HT) K1 polypeptide. The results from a fitted curve with an R2 value of 0.99 show that the half-maximum haemolysis induced by the K1 adhesin was at a concentration of 6.6 nM
Human serum albumin (HSA) binding experiments
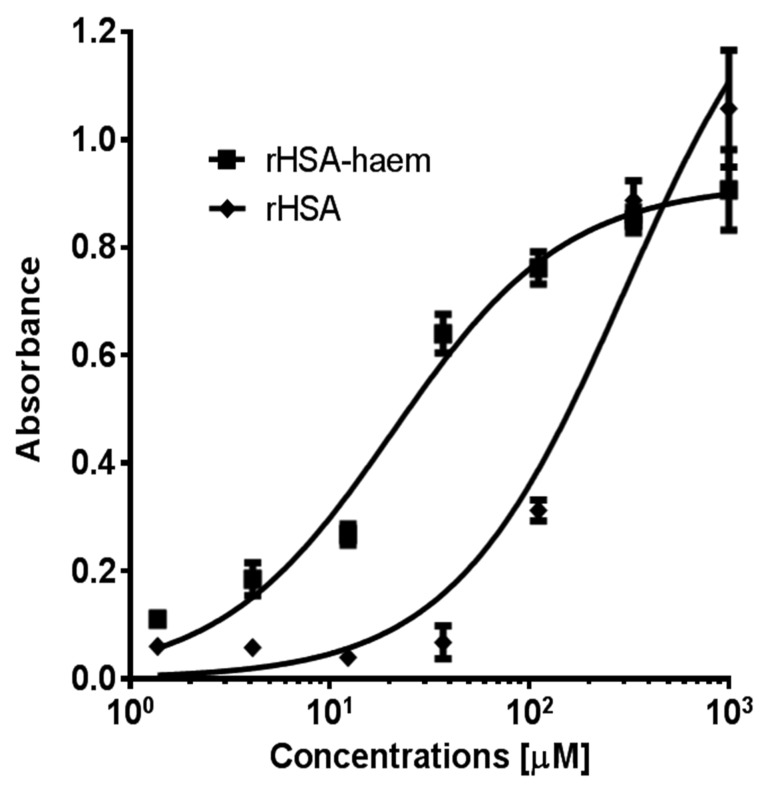
Haemoglobin is the most preferred source of iron for P. gingivalis [1]. When unavailable, this siderophore-lacking bacterium is able to sequester iron via other means, such as the degradation of albumin, the most abundant protein in plasma (~640 μM). Albumin is a major protein component of gingival crevicular fluid and the periodontal pocket [36]. When bound to haem (Kd ~ 10−8 M), haem-albumin can serve as a carbon, nitrogen, porphyrin, and iron source for P. gingivalis [37–39]. The exact mechanism of haem acquisition from haem-albumin is unclear, but it is known that the recombinant cleaved adhesin domains, K2, K3, and K1K2 have the capacity to bind to haem-loaded recombinant human serum albumin (rHSA-haem) [33]. This study reports that K1 too can bind to rHSA-haem (Fig. 4). Binding affinity for rHSA-haem was fifteen-fold higher than to apo-HSA for K1.
Fig. 4.
Interaction of rHSA and rHSA-haem with K1 polypeptide. 96-well ELISA plates were coated with K1 protein (0.4 μg/well in PBS) and incubated overnight at 4 °C. The wells were blocked with 1% skim milk in PBS for 1 h. Purified rHSA or rHSA-haem was added to the plate in various concentrations. Thereafter, anti-HSA mAb (15C7, AbCam) was added, followed by alkaline phosphatase-conjugated rabbit anti-mouse IgG. Colour development was detected with phosphatase substrate. Data were fitted by non-linear regression using GraphPad Prism 4.0 software (GraphPad, La Jolla, CA, USA). Apparent Kd value determined for binding of K1 to rHSA was 300 μM and for rHSA-haem, 20 μM. The fitted curves for rHSA and rHSA-haem with R2 values of 0.97 and 0.98, respectively
Discussion
K1 is the third Kgp cleaved adhesin module family member to be structurally characterized. The crystal structure has been solved and refined to resolution 1.55 Å. K1 conforms to the β-sandwich fold of the cleaved adhesin family and contains the same Ca2+ ion binding sites in homologous locations. Structural comparison among the adhesin domains confirms that the most variable area of the domain structures is on the long-loop end of the β-sandwich core. This solvent-accessible area may be indicative of unique binding functions of individual domains. Other adhesin modules in P. gingivalis (such as K3* of Kgp HG66, R1 and R2 of Rgp, and A2-10 of HagA) have high sequence identity with the Kgp cleaved adhesin modules [33], especially with the residues in Ca2+ binding site II and those that form the structural core. It can be postulated that these modules will have a similar structure, that is, a Ca2+ ion-stabilised β-sandwich motif.
It is now known that all three Kgp cleaved adhesin modules can induce haemolysis and can selectively bind to haem-albumin, but it is unclear which regions of the proteins are responsible for these activities. Li et al. [33] reported that K2-induced haemolysis was dependent on the integrity of the loop L8 bound to an arginine binding site near the surface of the protein. When cleaved, the L8 arginine anchor is lost, and so, can no longer induce haemolysis. It has not yet been tested if the arginine anchors in L9 of K1 and L10 of K3 are also required for haemolytic activity. Interestingly, recombinant K1 was comparable in haemolytic activity to a K1K2 construct and was found to be more potent than K2 or K3 alone [32]. rHSA-haem binds to all three recombinant modules with the K1 interaction being the weakest [33]. In comparison, all the interactions with apo-rHSA (i.e. in the absence of haem) were observed to be weaker, with a K1K2K3 multi-domain construct showing the least affinity. This then calls into question whether interactions between the adhesin modules in gingipains have a synergistic and/or an antagonistic effect on each other.
The outward facing conformation of the longest loop of K1, L9, is atypical of those equivalents previously observed in other crystals of the cleaved adhesin domain family. It was expected that the arginine anchor (Arg1109) originating from this loop would bind to an internal anchoring site (Asp 1139), but we found that in our crystals this was not the case. Rather, the arginine anchor binds to an arginine anchoring site of a neighbouring symmetrically related molecule within the crystal structure. It seems that the high concentration of guanidine hydrochloride present in the crystallisation buffer contributed to this event, as in each unit cell, a guanidinium ion displaces the guanidinium group of the arginine anchor of L9, causing it to bind to another available arginine anchoring site. It should be noted, however, that crystals of the K1 adhesin module were observed in other conditions without guanidinium hydrochloride, but only with its addition could we obtain diffracting crystals. As other aspartic acids are present on the surface of K1, it seems that the characteristic properties of the arginine anchoring site of the adhesin modules are essential for the binding of the arginine anchor: being two flanking planar aromatic residues, a salt bridge/hydrogen-bonding interaction with a conserved aspartic acid residue. This K1-L9 conformer was not previously detected in K1 by the method of small angle X-ray solution scattering applied to samples of the K1K2 or K1K2K3 multi-domain constructs [33], possibly reflecting limitations of this technique or more likely, that this conformer is a minor contributor to the overall solution structure under these particular experimental conditions. Despite the likelihood of the observation of this conformer being crystallographic artifact, it may also be possible that this interaction is representative of the manner in which individual domains of the RgpA/Kgp gingipain complex non-covalently associate. This can be implied from the sequence of RgpA (in the homologous R1 and R2 domains) which also contains the conserved arginine anchors and their corresponding binding sites [32, 40]. Further structural information on the cleaved adhesin domains of RgpA is required to confirm this. Another possibility is that the observed open conformation of the loop corresponds to conformers relevant to functional complexes of gingipains with host target proteins or those of other associated bacteria. When combined with the structure of the (crystallographic) symmetrically related molecule, a model can be constructed that demonstrates the conformation of the K1 adhesin module if the arginine anchor interacted with an internal anchoring site, assuming it would superimpose to the same space as the equivalent sites found in the other modules (data not shown). It is unknown which predominant conformation L9 would take in the domain’s solution state nor if a conformational change is induced upon binding. Nevertheless, it appears that K1 is capable of haemolysis while the entire loop is intact.
For some protein domains, flexible or disordered states in solution tend to be represented in high-resolution crystal structures as weak or missing electron densities. Flexible regions are illustrative of an active/binding site, allowing for conformational change to occur upon substrate binding. Interestingly, residues of the highest apparent motion in K1, K2, and K3 are located at the same structural position (in a different fashion for K1) and in similar positions during sequence alignment. These residues are located in the N-termini of the most extensive loop of each domain (L9 of K1, L8 of K2, and L10 of K3), coincidentally, the same loops that contain anchored arginines.
Ca2+ ion binding is a common feature of the galactose-binding domain-like (GBD) superfamily. As a member, the cleaved adhesins also contain bound calcium which contributes to structural stability. Structurally homologous members, such as the MAM domain of human receptor-type tyrosine-protein phosphatases, ephrin receptors, and various carbohydrate binding modules (CBMs), also bind Ca2+ ions [34]. A conserved motif is shared among these, which corresponds to Ca2+ ion binding site I in the adhesin domains. A conserved aspartic acid in this motif is equivalent to Asp1146, Asp1326, and Asp1295 in K1, K2, and K3, respectively. Additionally, a glutamic acid, corresponding to Glu990, Glu1164, and Glu1435, is common to most CBMs. The conserved nature of both Ca2+ ion binding sites and the full occupancy of these sites by Ca2+ ions in the crystals indicate that these cations principally play a structural role. A previous study of the thermal stability of the Kgp adhesin modules indicated that thermal denaturation of K1 was specifically Ca2+-dependent [33].
Further identification of host proteins that interact with K1 and the cleaved adhesin modules will be critical in elucidating the biological role of the HA region in the pathogenesis of periodontal disease. A previous study [33] reported that the adhesin modules can bind haemoglobin, HSA, and fibrinogen, proteins that are present in high levels in blood and most likely targeted by P. gingivalis during early colonisation. A recent study reported that K1 may be essential for the binding of a K1K2 polypeptide to IFN-γ, and it is hypothesized that the adhesin domains can directly regulate the biological activities of cytokines, contributing to the bacterium’s dysregulation of the host immune response [43]. A study on another major periodontal pathogen, Treponema denticola, discovered that co-incubation with P. gingivalis was followed by the overexpression of HA1 (sequence encompassing the K1 domain [32]) from Kgp, RgpA, and HagA [44]. Further, HA1 was identified as a key adhesion factor for co-aggregation with T. denticola and was associated with increased haemagglutination activity. The same study also found that HA1 could bind to fibronectin, fibrinogen, and collagen type V. In contrast, Li et al. [33] reported that no adhesin module had any haemagglutinating activity, but it was observed that K1 could bind to fibrinogen. It is not yet confirmed if K1 can also bind to collagen type V.
Now that the structures of all three Kgp adhesin modules are available, detailed comparisons between the adhesin domains and other structural homologues can be made, thereby providing the opportunity to infer putative functions for each domain and the entire HA region. Further, this knowledge may provide insight regarding the capacity of gingipain biovars (that vary in Kgp HA region sequences [50]) to contribute to virulence of a particular strain of P. gingivalis. Further studies of the cleaved adhesin domains and the full-length HA region will yield a greater understanding of how the gingipains contribute to the pathogenesis of P. gingivalis. This could facilitate novel drug and vaccine development in the treatment of periodontitis.
Acknowledgments
This study was supported by Institute of Dental Research Trust Funds. Some of the data for this research were recorded on beamlines at the Australian Synchrotron, Victoria, Australia. Thanks to Dr Ana Silva for the recording of the data at the synchrotron. PDB deposition: Coordinates of K1 have been deposited in the Protein Data Bank (PDB ID: 4ITC).
Appendix 1
Appendix A1
Table A1.
Data processing and refinement statistics
| Data processing | |
| Wavelength (Å) | 0.95370 |
| Space group | P21 |
| Unit-cell parameters (Å) | a = 34.0, b = 33.3, c = 67.2 |
| Resolution (Å) | 1.55 |
| Total reflections | 104,423 |
| Unique reflections | 42,331 |
| Completeness (%) | 99.4 (98.8) |
| (I/σ(I)) | 15.7 (2.9) |
| Rmerge | 0.055 (0.310) |
| Refinement | |
| No. of molecules in asymmetric unit | 1 |
| No. of atoms | 1450 |
| No. of water molecules | 123 |
| Resolution (Å) | 1.55 |
| Unique reflections | 20,886 |
| rmsd bonds (Å) | 0.013 |
| rmsd angles (°) | 1.59 |
| Isotropic B factor (Å2) | 14.4 |
| Ramachandran plot | |
| Favoured (%) | 95.7 |
| Allowed (%) | 3.7 |
| Rwork | 0.1533 |
| Rfree | 0.1814 |
| PDB code | 4ITC |
| Values in parentheses are for the highest resolution shell | |
Appendix 2
Appendix A2
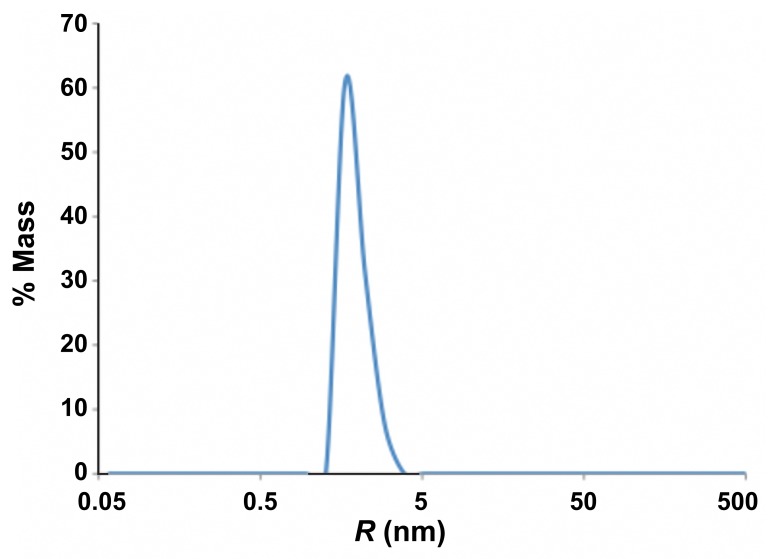
Dynamic Light Scattering (DLS) of K1 protein
Fig. A2.
Size distribution of K1. 12 μl of K1 protein (8 mg/ml) in 10 mM Tris-HCl, 150 mM NaCl, pH 7.6, was measured at 20 °C and an average of 20 1s acquisitions are as shown
Table A2.
Average measurements from DLS analysis of K1
| R (nm) | MW-R (kDa) | % Intensity | % Mass |
|---|---|---|---|
| 1.9 | 16 | 100 | 100 |
Appendix 3
Appendix A3
Table A3.
Comparison of the Ca2+ ion binding sites observed in the Kgp adhesin module crystal structures
| K1 |
K2 |
K3 |
||||
|---|---|---|---|---|---|---|
| Ca2+ binding site | Ligands | Distance (Å)* | Ligands | Distance (Å)** | Ligands | Distance (Å)*** |
| Site I | Ser988 O | 2.35 | Thr1162 O | 2.42 | Asp1433 O | 2.40 |
| Glu990 Oε2 | 2.26 | Glu1164 Oε2 | 2.36 | Glu1435 Oε2 | 2.36 | |
| Ser1022 Oγ | 2.44 | Gly1202 O | 2.46 | Ser1470 Oγ | 2.54 | |
| Gly1024 O | 2.31 | Asn1205 O | 2.36 | Ile1472 O | 2.31 | |
| Asp1146 Oδ1 | 2.43 | Asp1326 Oδ1 | 2.48 | Asp1595 Oδ1 | 2.48 | |
| Asp1146 Oδ2 | 2.48 | Asp1326 Oδ2 | 2.61 | Asp1595 Oδ2 | 2.43 | |
| Glu1147 Oε1 | 2.40 | H2O | 2.36 | H2O | 2.39 | |
| Site II | Asp1001 Oδ1 | 2.31 | Asp1179 Oδ1 | 2.28 | Asp1446 Oδ1 | 2.31 |
| Asp1003 Oδ1 | 2.38 | Asp1181 Oδ1 | 2.42 | Asp1448 Oδ1 | 2.37 | |
| Asp1003 Oδ2 | 2.74 | Asp1181 Oδ2 | 2.81 | Asp1448 Oδ2 | 2.82 | |
| Asp1005 Oδ1 | 2.40 | Asp1183 Oδ1 | 2.40 | Asp1450 Oδ1 | 2.39 | |
| His1007 O | 2.30 | Gln1185 O | 2.27 | Asn1452 O | 2.33 | |
| Asn1043 Oδ1 | 2.37 | Asn1221 Oδ1 | 2.42 | Asn1490 Oδ1 | 2.28 | |
| H2O | 2.42 | H2O | 2.44 | H2O | 2.41 | |
|
*Distance between Ca2+ and protein/solvent ligands in the
K1 structure **Distance between Ca2+ and protein/solvent ligands from Chain A as observed in the K2 structure [34] ***Distance between Ca2+ and protein/solvent ligands from Chains A, B, C and D as observed in the K3 structure [33] | ||||||
Contributor Information
L. A. Ganuelas, 1School of Molecular Bioscience, The University of Sydney, Sydney, Australia.
N. Li, 1School of Molecular Bioscience, The University of Sydney, Sydney, Australia.
P. Yun, 2Institute of Dental Research, Westmead Millennium Institute and Centre for Oral Health, Westmead Hospital, Sydney, NSW, Australia.
N. Hunter, 2Institute of Dental Research, Westmead Millennium Institute and Centre for Oral Health, Westmead Hospital, Sydney, NSW, Australia; 3Faculty of Dentistry, The University of Sydney, Sydney, Australia.
C. A. Collyer, 1School of Molecular Bioscience, The University of Sydney, Sydney, Australia.
References
- 1.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998 Dec;62(4):1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000 Oct;24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 3.Armitage GC. Periodontal diagnoses and classification of periodontal diseases. Periodontol 2000. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown LJ, Johns BA, Wall TP. The economics of periodontal diseases. Periodontol 2000. 2002;29:223–234. doi: 10.1034/j.1600-0757.2002.290111.x. [DOI] [PubMed] [Google Scholar]
- 5.Bahekar AA, Singh S, Saha S, Molnar J, Arora R. The prevalence and incidence of coronary heart disease is significantly increased in periodontitis: a meta-analysis. Am Heart J. 2007 Nov;154(5):830–837. doi: 10.1016/j.ahj.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 6.Saremi A, Nelson RG, Tulloch-Reid M, Hanson RL, Sievers ML, Taylor GW, Shlossman M, Bennett PH, Genco R, Knowler WC. Periodontal disease and mortality in type 2 diabetes. Diabetes Care. 2005 Jan;28(1):27–32. doi: 10.2337/diacare.28.1.27. [DOI] [PubMed] [Google Scholar]
- 7.Khader YS, Dauod AS, El-Qaderi SS, Alkafajei A, Batayha WQ. Periodontal status of diabetics compared with nondiabetics: a meta-analysis. J Diabetes Complications. 2006 Jan-Feb;20(1):59–68. doi: 10.1016/j.jdiacomp.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Mercado FB, Marshall RI, Bartold PM. Inter-relationships between rheumatoid arthritis and periodontal disease. A review. J Clin Periodontol. 2003 Sep;30(9):761–772. doi: 10.1034/j.1600-051x.2003.00371.x. [DOI] [PubMed] [Google Scholar]
- 9.Meyer MS, Joshipura K, Giovannucci E, Michaud DS. A review of the relationship between tooth loss, periodontal disease, and cancer. Cancer Causes Control. 2008 Nov;19(9):895–907. doi: 10.1111/j.1399-302X.2009.00532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slots J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in periodontal disease: introduction. Periodontol 2000. 1999 Jun;20:7–13. doi: 10.1111/j.1600-0757.1999.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci. 2003 Dec;4(6):409–426. doi: 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- 12.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999 Jun;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 13.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003 Dec;4(6):397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 14.Kusaba A, Ansai T, Akifusa S, Nakahigashi K, Taketani S, Inokuchi H, Takehara T. Cloning and expression of a Porphyromonas gingivalis gene for protoporphyrinogen oxidase by complementation of a hemG mutant of Escherichia coli. Oral Microbiol Immunol. 2002 Oct;17(5):290–295. doi: 10.1034/j.1399-302x.2002.170505.x. [DOI] [PubMed] [Google Scholar]
- 15.Roper JM, Raux E, Brindley AA, Schubert HL, Gharbia SE, Shah HN, Warren MJ. The enigma of cobalamin (Vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J Biol Chem. 2000 Dec 22;275(51):40316–4023. doi: 10.1074/jbc.M007146200. [DOI] [PubMed] [Google Scholar]
- 16.Sroka A, Sztukowska M, Potempa J, Travis J, Genco CA. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J Bacteriol. 2001 Oct;183(19):5609–5616. doi: 10.1128/JB.183.19.5609-5616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzik K, Bzowska M, Smagur J, Krupa O, Sieprawska M, Travis J, Potempa J. A new insight into phagocytosis of apoptotic cells: proteolytic enzymes divert the recognition and clearance of polymorphonuclear leukocytes by macrophages. Cell Death Differ. 2007 Jan;14(1):171–182. doi: 10.1038/sj.cdd.4401927. [DOI] [PubMed] [Google Scholar]
- 18.Kuula H, Salo T, Pirilä E, Hagström J, Luomanen M, Gutierrez-Fernandez A, Romanos GE, Sorsa T. Human beta-defensin-1 and -2 and matrix metalloproteinase-25 and -26 expression in chronic and aggressive periodontitis and in peri-implantitis. Arch Oral Biol. 2008 Feb;53(2):175–186. doi: 10.1016/j.archoralbio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Odell EW, Wu PJ. Susceptibility of Porphyromonas gingivalis and P. asaccharolytica to the non-oxidative killing mechanisms of human neutrophils. Arch Oral Biol. 1992 Aug;37(8):597–601. doi: 10.1016/0003-9969(92)90121-n. [DOI] [PubMed] [Google Scholar]
- 20.Mahtout H, Chandad F, Rojo JM, Grenier D. Porphyromonas gingivalis mediates the shedding and proteolysis of complement regulatory protein CD46 expressed by oral epithelial cells. Oral Microbiol Immunol. 2009 Oct;24(5):396–400. doi: 10.1111/j.1399-302X.2009.00532.x. [DOI] [PubMed] [Google Scholar]
- 21.Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007 Jun 1;178(11):7242–7250. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- 22.Fitzpatrick RE, Wijeyewickrema LC, Pike RN. The gingipains: scissors and glue of the periodontal pathogen, Porphyromonas gingivalis. Future Microbiol. 2009 May;4(4):471–487. doi: 10.2217/fmb.09.18. [DOI] [PubMed] [Google Scholar]
- 23.Yun PL, DeCarlo AA, Hunter N. Modulation of major histocompatibility complex protein expression by human gamma interferon mediated by cysteine proteinase-adhesin polyproteins of Porphyromonas gingivalis. Infect Immun. 1999 Jun;67(6):2986–2995. doi: 10.1128/iai.67.6.2986-2995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharp L, Poole S, Reddi K, Fletcher J, Nair S, Wilson M, Curtis M, Henderson B, Tabona P. A lipid A-associated protein of Porphyromonas gingivalis, derived from the haemagglutinating domain of the RI protease gene family, is a potent stimulator of interleukin 6 synthesis. Microbiology. 1998 Nov;144(Pt 11):3019–3026. doi: 10.1099/00221287-144-11-3019. [DOI] [PubMed] [Google Scholar]
- 25.Gibson 3rd FC, Genco CA. Prevention of Porphyromonas gingivalis-induced oral bone loss following immunization with gingipain R1. Infect Immun. 2001 Dec;69(12):7959–7963. doi: 10.1128/IAI.69.12.7959-7963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grenier D, Roy S, Chandad F, Plamondon P, Yoshioka M, Nakayama K, Mayrand D. Effect of inactivation of the Arg- and/or Lys-gingipain gene on selected virulence and physiological properties of Porphyromonas gingivalis. Infect Immun. 2003 Aug;71(8):4742–4748. doi: 10.1128/IAI.71.8.4742-4748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009 May-Jun;11(6-7):637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999 Jun 18;274(25):17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 29.Curtis MA, Kuramitsu HK, Lantz M, Macrina FL, Nakayama K, Potempa J, Reynolds EC, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 1999 Nov;34(8):464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake DB, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998 Aug 14;273(33):21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 31.Pavloff N, Potempa J, Pike RN, Prochazka V, Kiefer MC, Travis J, Barr PJ. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem. 1995 Jan 20;270(3):1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 32.Li N, Collyer CA. Gingipains from Porphyromonas gingivalis – complex domain structures confer diverse functions. Eur J Microbiol Immunol. 2011;1(1):41–58. doi: 10.1556/EuJMI.1.2011.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N, Yun P, Jeffries CM, Langley D, Gamsjaeger R, Church WB, Hunter N, Collyer CA. The modular structure of haemagglutinin/adhesin regions in gingipains of Porphyromonas gingivalis. Mol Microbiol. 2011 Sep;81(5):1358–1373. doi: 10.1111/j.1365-2958.2011.07768.x. [DOI] [PubMed] [Google Scholar]
- 34.Li N, Yun P, Nadkarni MA, Ghadikolaee NB, Nguyen KA, Lee M, Hunter N, Collyer CA. Structure determination and analysis of a haemolytic gingipain adhesin domain from Porphyromonas gingivalis. Mol Microbiol. 2010 May;76(4):861–873. doi: 10.1111/j.1365-2958.2010.07123.x. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanioka T, Matsuse R, Shigemoto Y, Ojima M, Shizukuishi S. Relationship between periodontal disease status and combination of biochemical assays of gingival crevicular fluid. J Periodontal Res. 2005 Aug;40(4):331–338. doi: 10.1111/j.1600-0765.2005.00807.x. [DOI] [PubMed] [Google Scholar]
- 37.Curry S, Devine DV, Décary F, editors. Abstracts of the 27th Congress of the International-Society-of-Blood-Transfusion Vancouver. Canada: Vox Sanguinis; 2002. Beyond expansion: structural studies on the transport roles of human serum albumin; p. S5.9. [DOI] [PubMed] [Google Scholar]
- 38.Milner P, Batten JE, Curtis MA. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for alpha-ketoglutarate. FEMS Microbiol Lett. 1996 Jul 1;140(2-3):125–130. doi: 10.1016/0378-1097(96)00159-0. [DOI] [PubMed] [Google Scholar]
- 39.Bramanti TE, Holt SC. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalis W50. J Bacteriol. 1991 Nov;173(22):7330–7339. doi: 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo Y, Nguyen KA, Potempa J. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon's knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 2010 Oct;54(1):15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sotomayor M, Schulten K. The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys J. 2008 Jun;94(12):4621–4633. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mishra NK, Kulhánek P, Snajdrová L, Petrek M, Imberty A, Koca J. Molecular dynamics study of Pseudomonas aeruginosa lectin-II complexed with monosaccharides. Proteins. 2008 Jul;72(1):382–392. doi: 10.1002/prot.21935. [DOI] [PubMed] [Google Scholar]
- 43.Alam M, Höglund C, Thorstrand C, Philip A. Atrioventricular plane displacement in severe congestive heart failure following dilated cardiomyopathy or myocardial infarction. J Intern Med. 1990 Dec;228(6):569–575. doi: 10.1128/IAI.00528-12. [DOI] [PubMed] [Google Scholar]
- 44.Orth RK, O'Brien-Simpson NM, Dashper SG, Reynolds EC. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol Oral Microbiol. 2011 Aug;26(4):229–240. doi: 10.1111/j.2041-1014.2011.00612.x. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowsky Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol Macromolecular Crystallography Pt A. 1997;276:307–326. doi: 10.1016/s0076-6879(97)76066-x. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr, Sect D: Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, et al. Features and development of Coot. Acta Crystallogr Sect D: Biol Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D: Biol Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Fanali G, et al. Modulation of heme and myristate binding to human serum albumin by anti-HIV drugs. FEBS J. 2007;274(17):4491–4502. doi: 10.1111/j.1742-4658.2007.05978.x. [DOI] [PubMed] [Google Scholar]
- 50.Nadkarni MA, et al. Distribution of Porphyromonas gingivalis biotypes defined by alleles of the kgp (Lys-gingipain) gene. J Clin Microbiol. 2004;42:3873–3876. doi: 10.1128/JCM.42.8.3873-3876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]