Abstract
Relationship loci (rQTL) exist when the correlation between multiple traits varies by genotype. rQTL often occur due to gene-by-gene (G × G) or gene-by-environmental interactions, making them a powerful tool for detecting G × G. Here we present an empirical analysis of apolipoprotein E (APOE) with respect to lipid traits and incident CHD leading to the discovery of loci that interact with APOE to affect these traits. We found that the relationship between total cholesterol (TC) and triglycerides (ln TG) varies by APOE isoform genotype in African-American (AA) and European-American (EA) populations. The e2 allele is associated with strong correlation between ln TG and TC while the e4 allele leads to little or no correlation. This led to a priori hypotheses that APOE genotypes affect the relationship of TC and/or ln TG with incident CHD. We found that APOE*TC was significant (P = 0.016) for AA but not EA while APOE*ln TG was significant for EA (P = 0.027) but not AA. In both cases, e2e2 and e2e3 had strong relationships between TC and ln TG with CHD while e2e4 and e4e4 results in little or no relationship between TC and ln TG with CHD. Using ARIC GWAS data, scans for loci that significantly interact with APOE produced four loci for African Americans (one CHD, one TC, and two HDL). These interactions contribute to the rQTL pattern. rQTL are a powerful tool to identify loci that modify the relationship between risk factors and disease and substantially increase statistical power for detecting G × G.
Keywords: relationship loci (rQTL), gene-by-gene (G × G), pleiotropy, apolipoprotein E (APOE), lipids
CORONARY heart disease [CHD (MIM 608901)] is challenging because it is a complex trait with a complicated genetic architecture. The MIM number is a reference in the Online Mendelian Inheritance in Man (OMIM) database. Recent genome-wide association studies (GWAS) have been successful in identifying regions of the genome with significant marginal effects. However, the combined effect of these loci explains only a small portion of the estimated total heritability. The traditional approach in association studies has been to test one phenotype at a time, even when multiple interrelated phenotypes are available for each individual. Because biological systems are organized in highly interactive pathways, changes at one level are likely to affect multiple traits throughout the system. Pleiotropy occurs when a single gene influences the variation of multiple phenotypes. Pleiotropic loci are common in complex biological systems (Stearns 2010) and tend to interact with other loci affecting traits within the same modular units (Wagner et al. 2007; Kenney-Hunt and Cheverud 2009). Pleiotropy is thought to play a primary role in the evolution of complex structures and systems (Wagner and Zhang 2011).
A conundrum in evolutionary biology is how a complex multitrait system with pleiotropy can evolve when a beneficial mutation for one trait may have detrimental consequences for another. Recent work (Pavlicev et al. 2011a; Pavlicev and Wagner 2012) has shown that relationship loci (rQTL) creates variation in pleiotropy that can be selected upon to further couple or uncouple trait variation and allow joint or separate evolution to occur in complex systems. rQTL occur when the correlation between multiple traits varies by genotype. They are the product of differential G × G or gene-by-environment interactions (see Pavlicev et al. 2011b, Figure 2). For example, differential gene interaction occurs when the pattern of interaction between two loci is different for multiple traits. This creates variation in the pleiotropic effects of a single locus. It also creates a pattern in which the correlation between two traits varies by genotype at the single locus level (rQTL). Empirical work has shown that most rQTL do not exhibit marginal effects., making them invisible to a typical association study. We can take advantage of this single locus pattern to identify rQTL and use them as a priori hypotheses to identify other loci that interact with them. This approach to detecting G × G greatly increases statistical power by reducing the multiple testing burden, and it connects multiple loci to each other and to multiple interrelated traits. A theoretical basis for and methodologies to detect relationship loci has been established in animal QTL work (Ehrich et al. 2003; Cheverud et al. 2004; Pavlicev et al. 2008, 2011b). An early example comes from a French population (Boerwinkle et al. 1987), where the correlation between total cholesterol and triglycerides varied among the most common genotypes of the three isoforms of apolipoprotein E (APOE) (MIM 107741). In another case (Sing et al. 1995), the relationship between the tertiles of cholesterol and coronary artery disease (CAD) changed with respect to APOE genotypes and the order of CAD risk among genotypes changed by cholesterol tertile.
A typical study would use genome-wide single locus tests to identify rQTL for a pair of traits. Each significant rQTL would then be used as a locus in a two-locus interaction model to identify other interacting loci for the two traits. In this article we work with a single rQTL by using the Atherosclerosis Risk in Communities Study (ARIC) to replicate in European-Americans the observation by Boerwinkle et al. (1987) that the correlation between total cholesterol (TC) and triglycerides (TG) varies by APOE genotype. This motivated three subsequent hypotheses. The first hypothesis is that APOE is also an rQTL for TC and TG in African Americans. The second is that APOE influences the relationship between TC and/or TG with incident CHD in both populations. Using genome-wide association data in ARIC, the last hypothesis is that other loci interact with APOE to effect these traits and produce the observed APOE rQTL patterns.
Materials and Methods
Study population
The ARIC study is a very well-phenotyped and ongoing prospective cohort study primarily focused on heart disease (ARIC Investigators 1989). Cohort members, totaling 15,792 persons aged 45–64 years at baseline (1987–89), were randomly chosen from four U.S. communities: Forsyth County, North Carolina; Jackson, Mississippi; suburban Minneapolis, Minnesota; and Washington County, Maryland. While the Jackson sample includes only African Americans, the other field centers samples are representative of the populations in these communities (i.e., mostly non-Hispanic whites in Minneapolis and Washington County, and about 15% African American in Forsyth County). The ARIC study includes large numbers of African Americans (N ∼ 5000) and non-Hispanic whites (N ∼ 11,000).
During a baseline home interview, persons were invited to participate in the study, and information was collected on health status, selected risk factors, family medical history, employment and educational status, diet, and physical activity. Cohort members completed four clinic examinations, conducted 3 years apart, in 1987–89, 1990–92 (93% overall returned), 1993–95 (86% overall returned), and 1996–98 (80% overall returned; 90% of those who examined in 1993–95 returned). Surveillance of the ARIC cohort for morbidity and mortality has been carried out by annual phone interviews with subsequent abstraction of hospital records to validate cardiovascular events, and completeness of this annual follow-up has been high. For the last complete contact cycle available, 95% of still-living cohort members were contacted and completed a phone interview. The cardiovascular endpoints of interest to ARIC are CHD deaths, nonfatal myocardial infarction, coronary, revascularization, hospitalized congestive heart failure, and stroke.
We focus on incident CHD as an endpoint and measures of plasma levels of TC, TG, low-density lipoprotein (LDL), and high-density lipoprotein (HDL). After excluding individuals on primary cholesterol medications, we used age, sex, body mass index (BMI), and medications with a secondary effect on cholesterol as covariates in all analyses. Most studies that use TG levels use a natural log transformation (ln TG) for analyses; however, the original analysis by Boerwinkle et al. (1987) did not. In the first model described below, we performed both for comparison while using only ln TG in the subsequent interaction analyses. Analyses with TG and ln TG were significant and comparable; in fact, ln TG had slightly smaller P-values. Table 1 and Table 2 give a summary of characteristics of individuals with each APOE genotype included in this study. The measure of incident CHD includes follow-up time and defined events as a definite or probable myocardial infarction (MI), fatal CHD, revascularization procedure, or electrocardiogram (ECG) evidence of MI. Follow-up time is from visit 1 until death, loss to follow-up, or censoring at 2007. Participants included in this study all gave written informed consent for study participation, including genetic research.
Table 1. Summary of characteristics of African-American individuals with APOE genotype data in the ARIC study after adjusting for age, sex, BMI, and secondary cholesterol medication.
| African American | All | e2e2 | e2e3 | e2e4 | e3e3 | e3e4 | e4e4 |
|---|---|---|---|---|---|---|---|
| Count | 3149 | 37 | 427 | 160 | 1411 | 976 | 138 |
| Genotype % | 1 | 1.2 | 13.6 | 5.1 | 44.8 | 31.0 | 4.4 |
| Sex (% male) | 0.38 | 0.41 | 0.37 | 0.41 | 0.36 | 0.4 | 0.36 |
| Age (mean ± SD) | 53.35 ± 5.81 | 53.32 ± 6.12 | 53.26 ± 5.71 | 53.16 ± 5.64 | 53.4 ± 5.8 | 53.32 ± 5.91 | 53.64 ± 5.84 |
| BMI | 29.16 ± 5.81 | 29.01 ± 6.73 | 29.85 ± 6.3 | 29.38 ± 6.17 | 29.23 ± 5.89 | 28.8 ± 5.44 | 28.63 ± 5.12 |
| Triglyceride (mg/dl) | 1.2 ± 0.59 | 1.47 ± 0.75 | 1.19 ± 0.55 | 1.29 ± 0.61 | 1.18 ± 0.59 | 1.22 ± 0.58 | 1.25 ± 0.59 |
| Total chol (mg/dl) | 213.79 ± 43.69 | 193.30 ± 49.48 | 197.55 ± 39.05 | 210.70 ± 39.43 | 214.18 ± 42.53 | 219.59 ± 45.23 | 225.77 ± 45.62 |
| LDL-C (mg/dl) | 136.56 ± 42.15 | 104.99 ± 44.31 | 119.33 ± 36.75 | 132.4 ± 37.42 | 136.66 ± 41.11 | 144.15 ± 42.83 | 148.4 ± 43.43 |
| HDL-C (mg/dl) | 55.79 ± 16.69 | 62.35 ± 21.18 | 57.28 ± 16.81 | 55.71 ± 15.86 | 56.53 ± 17.13 | 53.9 ± 15.62 | 55.26 ± 16.36 |
| inc CHD per 1000 | 135 | 108 | 126 | 151 | 133 | 142 | 138 |
| ln(triglycerides) | 53.35 ± 0.44 | 53.32 ± 0.48 | 53.26 ± 0.42 | 53.16 ± 0.44 | 53.4 ± 0.44 | 53.32 ± 0.43 | 53.64 ± 0.43 |
Table 2. Summary of characteristics of European-American individuals with APOE genotype data in the ARIC study after adjusting for age, sex, BMI, and secondary cholesterol medication.
| European American | All | e2e2 | e2e3 | e2e4 | e3e3 | e3e4 | e4e4 |
|---|---|---|---|---|---|---|---|
| Count | 9053 | 66 | 1145 | 210 | 5399 | 2048 | 185 |
| Genotype % | 1 | 0.7 | 12.6 | 2.3 | 59.6 | 22.6 | 2.0 |
| Sex (% male) | 0.45 | 0.48 | 0.45 | 0.48 | 0.45 | 0.44 | 0.46 |
| Age (mean ± SD) | 54.15 ± 5.72 | 54.65 ± 6.26 | 54.06 ± 5.8 | 53.88 ± 5.63 | 54.11 ± 5.71 | 54.26 ± 5.72 | 54.67 ± 5.33 |
| BMI | 26.74 ± 4.72 | 26.8 ± 3.91 | 26.96 ± 4.83 | 26.22 ± 4.71 | 26.8 ± 4.73 | 26.54 ± 4.65 | 26.42 ± 4.66 |
| Triglyceride (mg/dl) | 1.43 ± 0.69 | 1.81 ± 0.9 | 1.51 ± 0.77 | 1.48 ± 0.71 | 1.4 ± 0.67 | 1.44 ± 0.66 | 1.64 ± 0.77 |
| Total chol (mg/dl) | 213.02 ± 39.05 | 194.46 ± 52.58 | 201.03 ± 38.66 | 203.74 ± 37.11 | 213.40 ± 38.27 | 219.59 ± 37.89 | 223.07 ± 35.57 |
| LDL-C (mg/dl) | 136.05 ± 36.82 | 113.23 ± 42.78 | 121.61 ± 35.78 | 124.48 ± 34.99 | 136.82 ± 36.24 | 143.19 ± 36.02 | 145.27 ± 34.81 |
| HDL-C (mg/dl) | 51.66 ± 14.21 | 49.24 ± 10.36 | 52.66 ± 13.99 | 53.1 ± 14.61 | 51.75 ± 14.15 | 51.03 ± 14.47 | 48.71 ± 13.08 |
| Inc CHD per 1000 | 155 | 182 | 142 | 124 | 157 | 156 | 185 |
| ln(Triglycerides) | 54.15 ± 0.45 | 54.65 ± 0.49 | 54.06 ± 0.47 | 53.88 ± 0.45 | 54.11 ± 0.44 | 54.26 ± 0.43 | 54.67 ± 0.43 |
Genetic data
APOE genotypes were obtained by using TaqMan assays (Applied Biosystems, Foster City, CA) to genotype the 112 (rs429358) and 158 (rs7412) amino acid variants from exon 4 of APOE (Morrison et al. 2002; Hsu et al. 2005). Genome-wide assocation data for the ARIC participants consist of genotypes for nearly one million SNPs using the Affymetrix 6.0 platform (Ikram et al. 2009). An additional million SNPs were imputed in the Europoean-American sample using MaCH with HapMap as a reference panel (Dehghan et al. 2009). We used only observed genotypes in the African-American population. Quality control criteria for SNPs and individuals matched previous studies with these data (Dehghan et al. 2009; Ikram et al. 2009). All data use and analyses are approved by an institutional IRB HSC-SPH-11-0320. The GWAS data for the ARIC study are available in dbGAP.
Genetic analyses
All analyses were performed using the R statistical software environment (R Development Core Team 2012) with the addition of the Survival package (Therneau 2012). In addition, for the GWAS data, we used the Rserve (Urbanek 2012) package in R to link with the PLINK software package (Purcell et al. 2007; Purcell 2012). After excluding those on primary cholesterol medications, we fit the following linear model separately in both populations,
| (1) |
where APOE is a variable with six levels (one for each genotype). The significance of the interaction term was assessed using full vs. reduced models. Significance of this test rejects the null hypothesis that the correlation between the two traits is equal across genotype classes and that the beta coefficients from each bivariate regression within genotypes do not differ. Because there are three alleles (e2, e3, e4), APOE isoform data are often collapsed to two alleles (e4, non-e4) to make it easier to paramaterize (i.e., additive, dominance). We did not do this because there was no linear additive relationship among the genotypes in Boerwinkle et al. (1987). As a result, we used all five degrees of freedom available from the six possible genotypes. The ARIC study is large enough (see Table 1 and Table 2) to have many individuals for even the rarest genotypic classes (i.e., e2e2 and e4e4).
Because both TC and TG are positively correlated with CHD in the general population, we hypothesized that the APOE genotypes change the relationship of one or both of these lipids with CHD. A Cox proportional hazards model was used to test whether TC and/or ln TG interacts with APOE with respect to CHD separately in each population. A full vs. reduced-model likelihood-ratio test was used to test for significance of the APOE*TC and APOE*ln TG terms in each model. The Cox proportional hazards model is a semiparametric survival method that uses a partial log-likelihood to estimate the effect of independent variables on the hazard function in relation to time to event data. It assumes a parametric form for the effect of the predictors on the hazard function; but unlike parametric models, it does not make any assumptions about the shape of the baseline hazard function (Agresti 2002). Below is the model including ln TG; the other model replaces ln TG with TC:
| (2) |
The null hypothesis posits that TC or ln TG-related risk for incident CHD is equivalent across APOE genotypes.
For each phenotype (TC, ln TG, incident CHD), we performed a genome-wide scan for loci that interact with APOE. GWAS data were not available for many of the individuals with APOE genotypes (795 AA, 1376 EA), leaving 2354 AA and 7677 EA individuals for the APOE*SNP interaction analyses. We used two different traditional genome-wide significance thresholds for the European-American (P < 5 × 10−8) and the African-American (P < 4 × 10−7) scans because of the large difference in the number of tests (∼2.2 million for EA vs. ∼800,000 for AA). Since there is an a priori hypothesis for each phenotype, we need correct for only genome-wide significance within each phenotype. Instead of creating two additive continuous variables (0, 1, 2) for the two SNPs that define the APOE isoforms, we treated it as a variable with six levels (one for each genotype) represented by five (0, 1) indicator variables for n − 1 genotypes and five degrees-of-freedom. The second locus paramaterized as a simple additive locus with one degree of freedom limiting the interaction test to 5 degrees of freedom. Below is the general model where we are interested in the APOE*SNPadd interaction term:
| (3) |
A standard linear model was used for TC and ln TG and an analogous Cox proportional hazards model for incident CHD. Secondarily, we performed analyses for LDL and HDL because they are components of TC and because APOE has known direct effects on both. The null hypothesis for the interaction term is that the relationship of the SNP with the trait is the same among APOE genotypes.
Some types of G × G interactions can be classified as spreading or sign epistasis. Spreading epistasis occurs when the effect for locus 1 exists in one genotypic context but not another. Sign epistasis occurs when the allelic effects change direction across genetic backgrounds. Pavlicev et al. (2011b) found that rQTL have a higher proportion of sign epistatic interactions than non-rQTL. Sign epistasis sometimes involve compensatory mutations, which occur when the deleterious effect of a mutation at one locus is alleviated with a context dependency at another locus. In terms of pleiotropy, a mutation at a pleiotropic locus may have a beneficial impact on trait 1 while having a deleterious impact on trait 2. A mutation at another locus may create an interaction that alleviates the deleterious effects of the pleiotropci locus on trait 2 while preserving the beneficial impact on trait 1. Based on their pleiotropy and compensation model, Pavlicev and Wagner (2012) suggest that most adaptive signatures in genome scans could be the result of compensatory changes.
Results
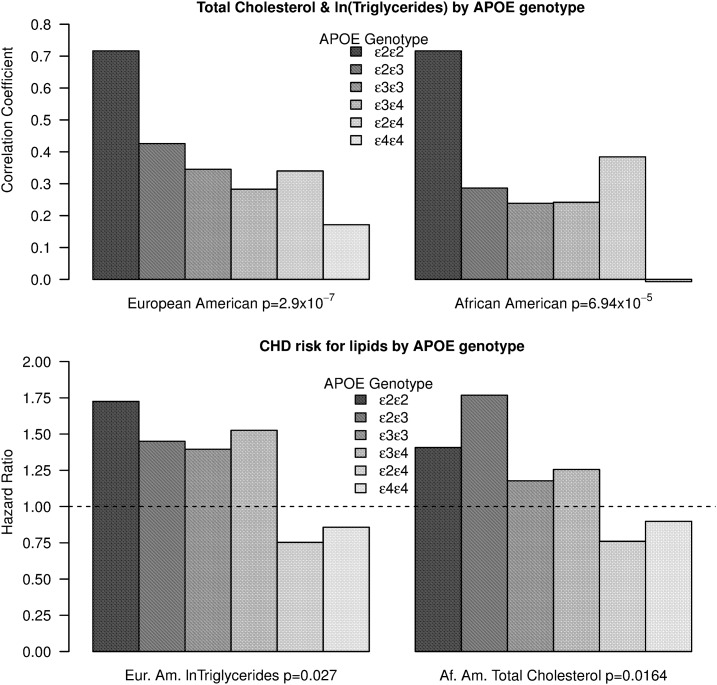
Table 1 and Table 2 give counts and summary characteristics for the ARIC participants with APOE genotype data. The relationship between TC and TG significantly differed by APOE genotypes based on the model in Equation 1 (P = 10–7 EA, 10–5 AA) and the pattern was very similar in the two populations (see Figure 1). Boerwinkle et al. (1987) did not have enough individiuals with e2e2 genotypes to estimate the correlation between TC and TG. Here we see that the correlation is very high (0.716 in both populations). The results using ln TG are virtually the same.
Figure 1.
(Top) Within APOE genotype correlations between total cholesterol and triglycerides after correcting for age, sex, BMI, and secondary cholesterol medication for European-American (EA) and African-American (AA) populations in the ARIC study. The P-values are for the APOE*triglyceride interaction in the full model. (Bottom) Within each APOE genotype, a Cox proportional hazards model for incident CHD was performed with age, sex, BMI, and secondary cholesterol medication as covariates and total cholesterol for African Americans and triglycerides for European Americans. The hazard ratio is plotted where values >1 suggest risk and values <1 suggest a protective effect. The dotted line is at 1. The P-values are for the APOE*triglyceride and APOE*total cholesterol interaction in the full model. Here we report TG but results for ln TG are comparable.
From the models based on Equation 2, APOE*TC was significant for AA (P = 0.016) and not in EA (P = 0.57) and APOE*TG was significant for EA (P = 0.027) but not AA (P = 0.34) (see Figure 1). All results were the same when using a natural log transformation of TG (P = 0.016 EA; P = 0.29 AA). The hazard ratio (exponential of the coefficient) from Cox regression within genotypes (including covariates) is shown in Figure 1. When the hazard ratio is >1 it denotes a positive correlation between CHD and TC or ln TG and values <1 suggest a negative correlation between CHD and TC or ln TG. The pattern of change in TC/ln TG correlation across APOE genotypes loosely resembles the hazard ratio changes in the CHD/ln TG and CHD/TC models with the primary differences related to where the heterozygotes lie with respect to the homozygotes.
In a post-hoc analysis with the components of TC, we found that LDL also has a strong relationship (P = 9.6 × 10−6 EA; P = 1.4 × 10−5 AA) with ln TG as observed before but the relationship of ln TG with HDL was weak or negligible (P = 0.013 EA; P = 0.264 AA). In African-Americans LDL had a stronger interaction (yet similar pattern) with APOE genotype to affect CHD than APOE*TC (P = 0.0066 for APOE*LDL vs. P = 0.016 for APOE*TC) while there was no evidence for an APOE*HDL interaction. There was no evidence for an APOE*LDL or APOE*HDL interaction in the European-American population.
The covariate adjusted correlation between TC and ln TG in the general population (0.24 for AA and 0.33 EA) is reflective of the e3e3 genotype, which is by far the most common genotype. This is also true for hazard ratios for CHD/TC (in AA) and CHD/ln TG (in EA). The risk relationship seen in the general population is a weighted average with e3e3 providing the largest influence while other less common genotypes pull in opposite directions (Figure 1). TC in AA and ln TG in AA have much a stronger positive CHD risk for individuals with the e2e2 and e2e3 genotypes while there is no risk (or negative) for those with e2e3 or e4e4.
Initially, 10 loci that significantly interact with APOE in African Americans were found, one for incident CHD, six for TC, one for LDL, and two for HDL. Six loci were found for European Americans and they were all for TC. However, from QQ plots we found the scans for TC and LDL showed P-value inflation while CHD and HDL did not (see Supporting Information, Figure S3). The inflation does not appear to be due to stratificaiton for two reasons: because the use of principle compenents as covariates did not the affect the analyses (see below) and because traditional marginal single SNP tests with those traits did not show QQ plot inflation. G × G interaction tests sometimes exhibit type I error inflation due to underestimates of the covariance matrix (Voorman et al. 2011). Work done by Bůžková et al. (2011) and Voorman et al. (2011) showed that sandwich estimators and the parametric bootstrap can be used to obtain valid P-values, with the parametric bootstrap as the gold standard.
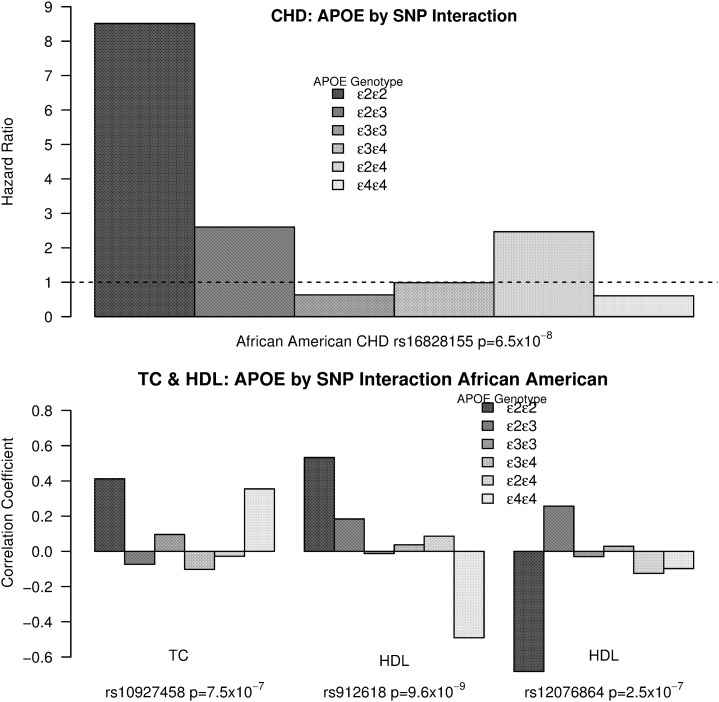
To obtain an empirical estimate of the P-values for the TC (six EA, six AA) and LDL (one AA) loci, for each locus we simulated 100 million parametric bootstraps to compare with the original statistic. Only one TC locus remained significant, leaving four significant loci that interact with APOE in African Americans (see Figure 2 and Table 3). Plots and descriptions of the other loci just under significance can be seen in Table S1, Figure S1, and Figure S2.
Figure 2.
(Top) Interaction of rs16828155 with APOE genotypes in African Americans using a Cox proportional hazards model and incident CHD. The hazard ratio was plotted for the rs16828155 additive variable (0, 1, 2) from within the APOE genotype models. The dotted line is at 1. The P-value is for the APOE*rs16828155 interaction term in the full model with the APOE genotypes as factors. (Bottom) Three SNPs referenced in the discussion with a genome-wide significant interaction in African Americans with APOE to affect total cholesterol (rs5758267) and HDL (rs912618 and rs12076864). Similar to the top, however, the traits are quantitative and we plot within genotype correlation coefficients after adjusting for the covariates. The P-values refer the to APOE*SNP interaction term in the full model where the APOE genotypes are treated as factors and the SNP is treated as additive (0, 1, 2).
Table 3. The most significant SNP from each of the four genome-wide significant locations that interact with APOE in African Americans.
| Trait | CHD | TC | HDL | HDL |
|---|---|---|---|---|
| SNP | rs16828155 | rs5758267 | rs12076864 | rs912618 |
| Chromosome | 1 | 22 | 1 | 14 |
| Location (build 36.3) | 167051924 | 39949296 | 110968915 | 61003344 |
| Maj/Min (MAF) | T/C (0.475) | T/A (0.271) | C/T (0.216) | A/G (0.378) |
| P-value | 6.51E-08 | 1.6–E–07a | 2.54E–07 | 9.62E–09 |
| Gene | L3MBTL2 (intron) | PRKCH (intron) | ||
| Left gene | DPT | C22:RP1-85F18.2 | KCNA2 | TMEM30B |
| Right gene | SUMO1P2 | CHADL | KCNA3 | LOC729637 |
P-value estimated with 100 million parametric bootstrap replicates and also. Significant in EA (P = 0.03).
Stratification could be a source of confounding for analyses in the African-American population. Principal components were available for a subset of the individuals with APOE and GWAS data (1986 AA). Because of the loss of individuals (368), we decided to do the analyses with all of the available data (without the principal components) and for each genome-wide significant locus we used the smaller data set and the first two principal components as covariates to test for consistency. Using the smaller data set and including the principal components, each of the loci found in African Americans showed the same effects and remained highly significant. The original rQTL models with APOE in African Americans were also tested using the smaller data set and each retained significance.
There is no obvious functional information about the region around rs16828155 that connect to CHD risk but a study (Lunetta et al. 2007) using the Framingham Heart Study found a GWAS hit (rs1412337) 166 kb upstream for morbidity-free survival at age 65. rs16828155 has a strong additive relationship with CHD risk among African-American individuals that carry at least one APOE e2 allele and none for those without an e2 allele (see Figure 2).
The HDL hit, rs912618, is found in protein kinase C, eta (PRKCH, 605437). A nonsynonymous variant in exon 9 (rs2230500) of PRKCH was shown to be associated with cerebral infarction in a Japanese case/control study, specifically lacunar infarction (Kubo et al. 2007). It is also associated with ischemic stroke (Li et al. 2012), LDL, and coronary heart disease (Zhu et al. 2012). According to the HapMap database, this SNP is of appreciable frequency only in Asian populations. Kubo et al. (2007) found that it was expressed in vascular endothelial cells and foamy macrophages in human atherosclerotic lesions and PKC-eta expression increased as the lesion type progressed. Using an analogous Cox proportional hazards model with incident CHD, the APOE*rs912618 interaction term was not significant (P = 0.11); however, the direction of effects was as expected. In particular, individuals with e4e4 genotypes had the strongest negative relationship (correlation = −0.49, P = 2.47 × 10−7; see Figure 2) between rs912618 and HDL and the expected positive association with CHD (hazard ratio = 2.19, P = 0.09) within the e4e4 genotype. rs912618 was not associated with ischemic stroke directly or through interaction with APOE by use of a Cox proportional hazards model.
The TC hit, rs5758267, resides in an intron of L3MBTL2 (611865) and according to the ENCODE project using the RegulomeDB tool (Boyle et al. 2012), it has a score of 1f, which means it is an eQTL for the PHF5A and a transcription factor binding site/DNase peak. None of these are obvious clues to the biological nature of the interaction. However, as stated above rs5758267 also shows mild evidence for an interaction with APOE for TC in the European-American population. Unsurprisingly, it also shows strong evidence for an interaction with APOE for LDL (P = 1 × 10−5) in AA.
For comparison and possible replication of the interacting loci in African Americans, only rs5758267 had an appreciable minor allele frequency (MAF) in European Americans (AA = 0.27, EA = 0.28) and it also shows evidence for an interaction with APOE in EA (P = 0.035). For the other three, rs912618, the MAF in EA is 0.01 and the other two are <0.01, giving little or no power for replication. However, the remaining three interacting SNPs all had at least one nominally significant SNP interacting with APOE for their respective trait within a 40-kb region.
Differential epistasis can cause the pattern observed in an rQTL (Pavlicev et al. 2011b). The CHD and TC loci found to interact with APOE exhibit differential epistasis by interacting differently among the pairs of traits (TC/ln TG and CHD/TC), which in turn create different relationships between the traits across APOE genotypes (i.e., the APOE rQTL pattern). Within APOE genotypes where the trait relationships are positively correlated, the interacting locus has the same direction of effects on the two traits, leading to a stronger correlation between the two traits within that genotype. Within APOE genotypes where the relationship between the traits is nonexistent (i.e., e2e4 and e4e4), the interacting loci have either opposing effects on the two traits or only an effect on one trait, which breaks up the correlation between the traits.
While the other 12 loci did not meet significance (Table S1, Figure S1, and Figure S2), they had patterns similar to that of the significant loci. Lumping all 16 loci together, for those loci with sufficient counts for e2e4, we found opposing effects for CHD and ln TG in EA (5 of 6) and CHD and TC in AA (4 of 7). All but one (12 of 13) found opposing direction of effects for TC and ln TG in e4e4 while most showed opposing direction of effects for TC and CHD in EA (5 of 6) and ln TG and CHD in AA (5 of 7). It is not expected that all loci that interact with APOE will contribute to this specific rQTL, but these loci appear to contribute to the bivariate relationship differences among APOE genotypes (see Figure 1).
In addition to APOE itself, none of the other interacting loci have even a nominal association directly with the trait itself such that none of these loci would be found by a standard single-locus GWAS. This is consistent with other rQTL studies (Pavlicev et al. 2011b). APOE is the only locus to have a direct association with any of the traits (TC, LDL, ln TG, and HDL), which is already well established in the literature (Templeton et al. 2005). All of the observed interactions involve sign epistasis where the allelic effect changes directions across genetic backgrounds. (See Figure 2, Figure S1 and, Figure S2.)
Discussion
We were able to replicate the work of Boerwinkle et al. (1987) and establish that APOE acts as an rQTL between TC and ln TG in both EA and AA populations. This led to significant a priori tests establishing that APOE modulates the relationship between CHD and ln TG in European Americans and CHD and TC in African Americans. The a priori tests allow for multiple testing to be done only within each genome scan analogous to work on controlling false positives for epistatic QTL done by Wei et al. (2010).
The rQTL approach, as demonstrated here, is a powerful way to identify loci that effect relationships between important biological risk factors and the relationship between these factors and disease. In our case, the rQTL (APOE) was already known; however, the same model used to validate APOE as an rQTL can be used in a genome-wide scan to identify other previously unknown rQTL for a given pair of traits. These loci would typically be undetected in a normal GWAS analysis and even if “seen,” their role in pleiotropic variation and gene-by-gene interactions would not be evident. It is also a unique, efficient, and powerful approach to identifying gene-by-gene interactions. It enhances statistical power by defining a priori loci (rQTL) that are likely to be involved in an interaction and reducing the number of tests to the order of a GWAS instead of all pairwise tests.
Biologically, it provides a framework to link multiple traits together and with the rQTL and other interacting loci. In the context of human medicine, these loci can lead to further insights about conditions where the magnitude of risk for a known risk factor changes. In the case of triglycerides in European Americans and total cholesterol (and LDL) in African Americans, their importance to CHD risk depends on what APOE genotype an individual has. This raises issues of importance for treatments targeting risk factor levels. If treatments for TC and ln TG are based on their association with CHD, then lipid-lowering drugs may be necessary or useful only for individuals with APOE genotypes where the CHD risk is strong for TC or ln TG. In particular, the level of triglycerides matters, in terms of CHD risk, for an EA carrying the e2e2 genotype while it does not for those carrying an e2e4 or e4e4 genotype. The level of cholesterol matters for an AA carrying an e2e3 or e2e2 genotype while it does not for those carrying an e2e4 or e4e4 genotype.
It is well established that both TC and ln TG have positive CHD risk in the general population. The three APOE alleles (e2, e3, e4) are associated with low, medium, and high TC and ln TG levels. These same alleles are associated with low, medium, and high CHD risk. It is tempting to think that the relationship between APOE and CHD is strictly through its linear influence on lipid levels. Our results suggest that this explanation is too simple and masks important relationships between APOE and CHD independent of or at least not linearly related to lipid levels. Here we suggest that APOE genotypes influence the relationship between lipid levels and CHD, not just the actual lipid level itself.
Various studies have shown that APOE alleles respond differently to different types of LDL-lowering treatments. Response (reduction in LDL) to exercise is greater for those with the e3 allele than the e4 allele. Statins produce a similar pattern while Probucol, which has a different target and mechanism for lowering LDL than statins, has the opposite effect with a greater response for those carrying the e4 allele (Hagberg et al. 2000). Gustavsson et al. (2012) found that APOE genotypes interact with both smoking and physical inactivity with respect to CHD. They determined that these interactions were independent of LDL levels and concluded that something other than a direct effect on lipid levels is responsible for this relationship with CHD.
While APOE has a similar effect on the correlation between TC and ln TG in both populations, it is surprising to see that APOE affects the relationship between only TC and CHD in African Americans and only ln TG and CHD in European Americans. This difference between African Americans and European Americans may be another example of observed yet not understood differences in the behavior of lipids and CHD between the two populations (Haffner et al. 1999). In another study (T. J. Maxwell and C. M. Ballantyne, unpublished results) the authors have observed that CETP promoter variants associated with CETP protein concentration in European-Americans are strongly associated with HDL levels, yet those same variants in African-Americans are associated with only CETP protein concentration and not HDL.
Relationship loci present a powerful approach to uncovering the complex genetic architecture of common diseases. They establish a foothold into the world of pleiotropy and interactions, which are the basis for modularity and the inherent organization of biological systems in pathways of interacting factors. Because rQTL create variation in pleiotropy, selection can act upon them to couple and uncouple traits enabling evolution to change multiple traits jointly or separately (Pavlicev et al. 2011a).
Supplementary Material
Acknowledgments
The authors thank the staff and participants of the ARIC study for their important contributions. This work was supported by the National Institutes of Health (NIH) grant HL105502 to TJM. CMB is a member of scientific advisory boards for Pfizer and Merck. All authors declare that there are no conflicts of interest. All research is approved by an institutional IRB HSC-SPH-11-0320. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; and NIH contract HHSN268200625226C. Infrastructure was partly supported by grant UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Footnotes
Communicating editor: L. B. Jorde
Literature Cited
- Agresti A., 2002. Categorical Data Analysis, Ed. 2 Wiley, New York. [Google Scholar]
- ARIC Investigators , 1989. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am. J. Epidemiol. 129: 687–702. [PubMed] [Google Scholar]
- Boerwinkle E., Visvikis S., Welsh D., Steinmetz J., Hanash S. M., et al. , 1987. The use of measured genotype information in the analysis of quantitative phenotypes in man. II. The role of the apolipoprotein E polymorphism in determining levels, variability, and covariability of cholesterol, betalipoprotein, and triglycerides in a sample of unrelated individuals. Am. J. Med. Genet. 27: 567–582. [DOI] [PubMed] [Google Scholar]
- Boyle A. P., Hong E. L., Hariharan M., Cheng Y., Schaub M. A., et al. , 2012. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bůžková P., Lumley T., Rice K., 2011. Permutation and parametric bootstrap tests for gene-gene and gene-environment interactions. Ann. Hum. Genet. 75: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., Ehrich T. H., Vaughn T. T., Koreishi S. F., Linsey R. B., et al. , 2004. Pleiotropic effects on mandibular morphology. II. Differential epistasis and genetic variation in morphological integration. J. Exp. Zool. B Mol. Dev. Evol 302: 424–435. [DOI] [PubMed] [Google Scholar]
- Dehghan A., Yang Q., Peters A., Basu S., Bis J. C., et al. , 2009. Association of novel genetic loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ. Cardiovasc. Genet. 2: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich T. H., Vaughn T. T., Koreishi S. F., Linsey R. B., Pletscher L. S., et al. , 2003. Pleiotropic effects on mandibular morphology. I. Developmental morphological integration and differential dominance. J. Exp. Zool. B Mol. Dev. Evol 296: 58–79. [DOI] [PubMed] [Google Scholar]
- Gustavsson J., Mehlig K., Leander K., Strandhagen E., Björck L., et al. , 2012. Interaction of apolipoprotein E genotype with smoking and physical inactivity on coronary heart disease risk in men and women. Atherosclerosis 220: 486–492. [DOI] [PubMed] [Google Scholar]
- Haffner S. M., D’Agostino R., Jr, Goff D., Howard B., Festa A., et al. , 1999. LDL size in African Americans, Hispanics, and non-Hispanic whites : the insulin resistance atherosclerosis study. Arterioscler. Thromb. Vasc. Biol. 19: 2234–2240. [DOI] [PubMed] [Google Scholar]
- Hagberg J. M., Wilund K. R., Ferrell R. E., 2000. APO E gene and gene-environment effects on plasma lipoprotein-lipid levels. Physiol. Genomics 4: 101–108. [DOI] [PubMed] [Google Scholar]
- Hsu C. C., Kao W. H. L., Coresh J., Pankow J. S., Marsh-Manzi J., et al. , 2005. Apolipoprotein E and progression of chronic kidney disease. J. Am. Med. Assoc. 293: 2892–2899. [DOI] [PubMed] [Google Scholar]
- Ikram M. A., Seshadri S., Bis J. C., Fornage M., DeStefano A. L., et al. , 2009. Genomewide association studies of stroke. N. Engl. J. Med. 360: 1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney-Hunt J. P., Cheverud J. M., 2009. Differential dominance of pleiotropic loci for mouse skeletal traits. Evolution 63: 1845–1851. [DOI] [PubMed] [Google Scholar]
- Kubo M., Hata J., Ninomiya T., Matsuda K., Yonemoto K., et al. , 2007. A nonsynonymous SNP in PRKCH (protein kinase C eta) increases the risk of cerebral infarction. Nat. Genet. 39: 212–217. [DOI] [PubMed] [Google Scholar]
- Li J., Luo M., Xu X., Sheng W., 2012. Association between 1425G/A SNP in PRKCH and ischemic stroke among Chinese and Japanese populations: a meta-analysis including 3686 cases and 4589 controls. Neurosci. Lett. 506: 55–58. [DOI] [PubMed] [Google Scholar]
- Lunetta K. L., D’Agostino R. B., Sr, Karasik D., Benjamin E. J., Guo C.-Y., et al. , 2007. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Med. Genet. 8(Suppl. 1): S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A. C., Ballantyne C. M., Bray M., Chambless L. E., Sharrett A. R., et al. , 2002. LPL polymorphism predicts stroke risk in men. Genet. Epidemiol. 22: 233–242. [DOI] [PubMed] [Google Scholar]
- Pavlicev M., Wagner G. P., 2012. A model of developmental evolution: selection, pleiotropy and compensation. Trends Ecol. Evol. 27: 316–322. [DOI] [PubMed] [Google Scholar]
- Pavlicev M., Kenney-Hunt J. P., Norgard E. A., Roseman C. C., Wolf J. B., et al. , 2008. Genetic variation in pleiotropy: differential epistasis as a source of variation in the allometric relationship between long bone lengths and body weight. Evolution 62: 199–213. [DOI] [PubMed] [Google Scholar]
- Pavlicev M., Cheverud J. M., Wagner G. P., 2011a Evolution of adaptive phenotypic variation patterns by direct selection for evolvability. Proc. Biol. Sci. 278: 1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicev M., Norgard E. A., Fawcett G. L., Cheverud J. M., 2011b Evolution of pleiotropy: epistatic interaction pattern supports a mechanistic model underlying variation in genotype-phenotype map. J. Exp. Zool. B Mol. Dev. Evol. 316: 371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S., 2012 PLINK software package, version 1.07. http://pngu.mgh.harvard.edu/purcell/plink/.
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2012 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
- Sing C. F., Haviland M. B., Templeton A. R., Reilly S. L., 1995. Alternative genetic strategies for predicting risk of atherosclerosis, pp. 638–644 in Atherosclerosis X. Exerpta Medica International Congress Series, edited by Woodford F. P., Davignon J., Sniderman A. D. Elsevier, Amsterdam. [Google Scholar]
- Stearns F. W., 2010. One hundred years of pleiotropy: a retrospective. Genetics 186: 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A. R., Maxwell T., Posada D., Stengård J. H., Boerwinkle E., et al. , 2005. Tree scanning: a method for using haplotype trees in phenotype/genotype association studies. Genetics 169: 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T., 2012 A Package for Survival Analysis in S. R package, version 2.36–14 Available at: http://cran.r-project.org/web/packages/survival/index.html. Accessed: July 20, 2013.
- Urbanek S., 2012 Rserve: Binary R server. R package, version 0.6–8 Available at: http://cran.r-project.org/web/packages/Rserve/index.html. Accessed: July 20, 2013.
- Voorman A., Lumley T., McKnight B., Rice K., 2011. Behavior of QQ-Plots and Genomic Control in Studies of Gene-Environment Interaction. PLoS ONE 6: e19416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P., Pavlicev M., Cheverud J. M., 2007. The road to modularity. Nat. Rev. Genet. 8: 921–931. [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Zhang J., 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12: 204–213. [DOI] [PubMed] [Google Scholar]
- Wei W.-H., Knott S., Haley C. S., de Koning D.-J., 2010. Controlling false positives in the mapping of epistatic QTL. Heredity 104: 401–409. [DOI] [PubMed] [Google Scholar]
- Zhu J., Yan J.-J., Kuai Z.-P., Gao W., Tang J.-J., et al. , 2012. The role of PRKCH gene variants in coronary artery disease in a Chinese population. Mol. Biol. Rep. 39: 1777–1782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.