Abstract
Mutations in GBA1, the gene for glucocerebrosidase (GCase), are genetic risk factors for Parkinson disease (PD). α-Synuclein (α-syn), a protein implicated in PD, interacts with GCase and efficiently inhibits enzyme activity. GCase deficiency causes the lysosomal storage disorder Gaucher disease (GD). We show that saposin C (Sap C), a protein vital for GCase activity in vivo, protects GCase against α-syn inhibition. Using NMR spectroscopy, site-specific fluorescence, and Förster energy transfer probes, Sap C was observed to displace α-syn from GCase in solution and on lipid vesicles. Our results suggest that Sap C might play a crucial role in GD-related PD.
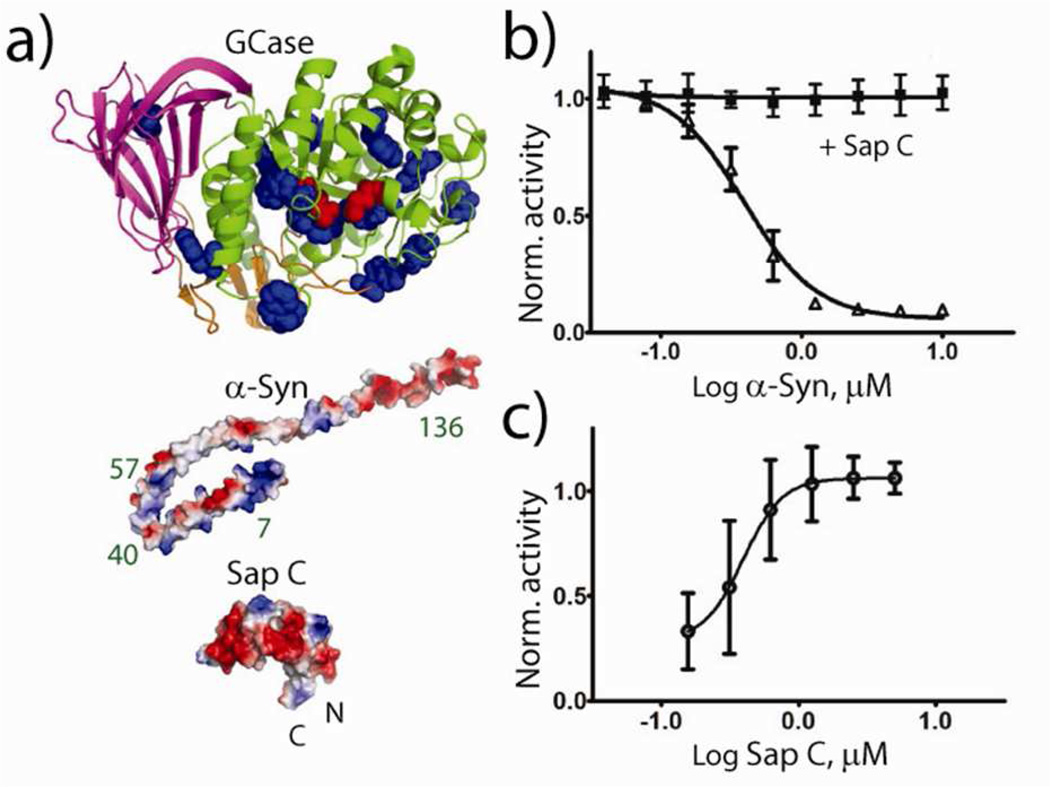
Mutations in the GBA1 gene are the most common of the known risk factors for Parkinson disease (PD).1 Despite a wealth of studies linking GBA1 mutations and PD, the underlying mechanisms have yet to be fully defined.2 GBA1 encodes glucocerebrosidase (GCase, Figure 1a top), a lysosomal enzyme which hydrolyzes glucosylceramide (GluCer) into glucose and ceramide and is deficient in Gaucher disease (GD). Both GCase and α-synuclein (α-syn, Figure 1a middle), a membrane binding protein associated with PD, have been observed in Lewy bodies,3 a classic PD hallmark.
Figure 1.
(a) Molecular structures of GCase, α-syn, and Sap C. GCase (PDB code 2NSX) with its 12 Trp residues (used as Förster energy transfer donors) shown in blue and active site residues (E235 and E340) in red. α-Syn (PDB code 1XQ8) and Sap C (PDB code 1SN6) with positive (blue) and negative (red) electrostatic potentials shown. Cys-mutation sites of α-syn used for Dns labeling are noted. (b) GCase activity (50 nM GCase, 1 mM 4-methylumbelliferyl β-D-glucopyranoside, 350 µM POPC:POPS vesicles, pH 5.5) with increasing α-syn concentration in the absence (triangles) and presence of 5 µM Sap C (squares). (c) GCase activity titrated by increasing concentrations of Sap C in the presence of 10 µM α-syn. Activity levels are normalized to GCase alone and error bars indicate standard deviations from two independent measurements.
A growing number of studies show a correlation between GCase deficiency and increased α-syn levels,4 leading some to speculate that GluCer accumulation affects normal α-syn turnover.4b Intriguingly, we discovered that α-syn physically interacts with GCase under acidic conditions found in lysosomes,5 a site of α-syn degradation.6 In further substantiating this relationship, we found that α-syn inhibits GCase activity on the membrane;5b although, it is currently unresolved whether reduced GCase activity alone leads to increased α-syn levels.7 Since only a minority of GD patients and carriers develop PD, other factors are also expected to play a role in promoting pathogenesis. Obvious molecules of interest include those that modulate GCase activity and α-syn-GCase interaction.
In vivo degradation of GluCer by GCase is facilitated by the co-factor saposin C (Sap C),8 a 9 kDa membrane-interacting protein (Figure 1a bottom).9 Sap C has been proposed to function by altering lipid bilayer properties or through direct association with GCase.10 Although rare, Sap C deficiency alone can result in GD symptoms in patients,11 demonstrating its essential role in GluCer metabolism. Sap C deficiency was shown to cause severe GD phenotypes and enhanced storage of GluCer in a GD-mouse model.12
Here, we investigated whether Sap C, a vital co-factor in vivo, can affect GCase activity inhibition by α-syn in vitro.5b Consistent with our previous study,5b an IC50 ~ 0.4 µM is obtained for α-syn inhibition using a fluorogenic substrate and vesicle assay (Figure 1b, equal molar phosphatidylcholine and phosphatidylserine, POPC:POPS, pH 5.5). Remarkably, inhibition is completely abolished in the presence of Sap C (5 µM) at all measured concentrations of α-syn (Figure 1b). At the highest α-syn concentration (10 µM), Sap C protects GCase in a concentration dependent manner, yielding an AC50 of 0.4 µM (Figure 1c). Under the high POPS condition used here, activity enhancement by Sap C is negligible, making this rescue even more remarkable.
Since Sap C had been proposed to bind to GCase directly, we questioned whether Sap C displaces α-syn from GCase, thus affording rescue of its activity. In prior work, using nuclear magnetic resonance (NMR) spectroscopy with isotopically labeled α-syn, a C-terminal region, residues 118–137, rich in acidic (3 Asp and 5 Glu) residues was identified as the site of interaction with GCase.5a Association with GCase (~60 kD) slows the molecular tumbling of these C-terminal residues, reducing their NMR signal intensities while N-terminal residues remain in the highly disordered unbound state with intensities unchanged (Figure 2a, top panel). Upon addition of Sap C, partial recovery of intensity is seen for the C-terminal α-syn residues, indicating an increase in the fraction of unbound α-syn. Since Sap C and α-syn do not interact in solution,5a the displacement of α-syn is due to Sap C binding to GCase.
Figure 2.
(a) NMR 15N HSQC spectra backbone amide intensities plotted by residue of α-syn (top) and Sap C (bottom). (Top) Data for α-syn with GCase (Taliglucerase alfa) alone and in the presence of Sap C are shown in blue and pink, respectively. Data for α-syn with GCase (Imiglucerase) are shown in green. The portion of the acidic C-terminal region of α-syn (residues 101–140) that experiences the greatest intensity quenching is expanded. The α-syn intensity quenching due to interaction with both FDA approved versions of recombinant GCase is the same within error. They differ in the composition of glycans and amino acids at the N- and C-termini.15 Protein concentrations were 50 µM for GCase, α-syn and Sap C. (Bottom) Data for Sap C with GCase alone and in the presence of α-syn are shown in blue and pink, respectively. Protein concentrations for GCase, α-syn, and Sap C were 33, 76, and 38 µM, respectively. (b) Spectral changes of Dns136-α-syn–GCase as a function of increasing Sap C. Mean wavelength, <λ>, is plotted with left and right axes for proteins in solution (squares, 50 mM MES pH 5.5, and 25 mM NaCl) and vesicles (circles, 0.9 mM POPC:POPS), respectively. Dns136-α-syn (2 µM) and GCase (5 µM) were used. Data for α-syn alone in vesicles are also shown (triangles, left axis). In the absence of vesicles, <λ> = 523 nm. Error bars indicate standard deviations from two independent measurements. (Inset) Dns136-α-syn–GCase titration with Sap C. (c) Förster energy transfer from GCase (2 µM) to Dns136-α-syn (5 µM, red curve) in the absence of vesicles followed by the addition of Sap C. Fluorescence spectra of GCase and Dns136-α-syn alone are shown in purple and blue, respectively. Intensity scale for Dns wavelengths have been expanded for clarity.
The association between Sap C and GCase was detected by NMR spectroscopy using isotopically labeled Sap C (Figure 2A, bottom panel). Intensities for all residues of Sap C were reduced in the presence of GCase. Since Sap C is a folded protein, all of its residues undergo slower tumbling upon interaction. Adding α-syn results in partial recovery for all residues, indicating that α-syn also can cause Sap C to dissociate from GCase. Given that Sap C has regions that are rich in acidic residues (Figure 1a), some of which have been previously proposed to interact with GCase,13 Sap C and α-syn could have overlapping GCase binding sites (Figure S1), though displacement via an allosteric effect cannot be ruled out.
To study the α-syn-GCase interaction both in solution and in the presence of vesicles, we turned to site-specific fluorescence spectroscopy. In the GCase-bound form, fluorescence of dansyl (Dns)-labeled α-syn at residue 136 exhibits an intensity increase and a spectral blue-shift upon enzyme interaction both in solution and on lipid vesicles reflecting reduced mobility and increased hydrophobic environment.5 Adding Sap C reverses these trends with a concomitant decrease in intensity and spectral red shift (Figure 2b inset). We confirmed that Sap C has negligible effect on Dns136 fluorescence in the absence of GCase in solution5a and on the membrane (Figure 2b, triangles). By comparison, the concentration of Sap C required to dissociate α-syn from GCase on the membrane is higher than that in solution. This observation may result from differences in membrane binding affinities between α-syn and Sap C.
Several GCase:α-syn and GCase:Sap C binding models were tested, and our data are best described by a 1:1 and 2:1 GCase to α-syn and Sap C model, respectively (Figure S2). Using the previously determined α-syn dissociation constants (KD = 1 and 4 µM in the absence and presence of vesicles, respectively), the changes in Dns136 mean wavelength (<λ>) and intensity can be fit to yield apparent KD values for Sap C binding to GCase: 0.26 ± 0.13 and 2.4 ± 0.5 µM in solution and in the presence of vesicles, respectively.
To further characterize the dissociation of α-syn from GCase induced by Sap C, we performed Förster energy transfer experiments using intrinsic Trp residues in GCase as donors and Dns-α-syn as the acceptor. Sap C does not contain any native Trp residues. Unlike in solution, sites in both N- and C-terminal regions of α-syn interact with and are in close proximity (~11–33 Å) to GCase on the lipid membrane.5b Using Dns-sites (7, 40, 57, and 136), the extent of energy transfer between Trp and Dns was monitored with added Sap C (Figure 2c and Figure S3). As expected the addition of Sap C (10 µM) reduced the efficiency of energy transfer as a result of α-syn displacement, with de-quenching and quenching of Trp and Dns fluorescence, respectively. We note that Trp intensity from GCase in the presence of vesicles is slightly changed upon the addition of Sap C, indicating that Sap C interaction may influence the environment of one or more Trp residues in GCase (Figure S3 and S4). Nevertheless, energy transfer is lost as measured from Dns fluorescence, confirming that Sap C impedes the interaction of GCase both on and off the membrane.
In summary, Sap C protects GCase from α-syn inhibition and competes with α-syn binding. Taken together with the reported inverse correlation between GCase and α-syn levels derived from tissue culture, animal models, and patients,4 Sap C could act as a modifier in the homeostasis of α-syn. Our results might suggest two possible disease implications. First, the high levels of α-syn in neurons could lead to greater attenuation of neuronal GCase activity in patients deficient in Sap C, and this association could potentially explain why GBA1 mutations cause neuronopathic GD in some patients, but not in others. Second, if α-syn-GCase interaction promotes PD pathology via activity inhibition,5b then Sap C could play a protective role by removing α-syn from GCase. In this scenario, Sap C deficiency would be a risk factor for PD. Alternatively, if interaction of α-syn with GCase is involved in its normal lysosomal degradation as previously hypothesized,5a then increased Sap C levels displacing α-syn could potentially be harmful. In fact, high levels of Sap C have been observed in the spleen and blood of GD patients,14 though this has not been evaluated in the brain. Further investigation is clearly needed to determine if and to what extent Sap C and/or the interplay between Sap C, α-syn, and GCase is involved in PD. Resolution of these different viewpoints will require quantification of the physiological concentrations of α-syn, Sap C, and GCase in lysosomes from brain samples of patients with GBA1 mutations as well as PD, GD, and healthy individuals.
Supplementary Material
ACKNOWLEDGMENT
Recombinant GCase was a gift from Protalix Biotherapeutics, Carmiel, Israel. The Sap C plasmid was provided by Gilbert Privé (University of Toronto, Canada). We thank Nico Tjandra (NHLBI) for the use of NMR spectrometer, Duck-Yeon Lee (NHLBI Biochemistry Core Facility) for technical assistance with mass spectrometry and Zhiping Jiang (NHLBI) for the expression of isotopically labeled Sap C.
Funding Sources
Supported by the Intramural Research Program at the NIH, NHLBI and NHGRI.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental details and Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
T.L.Y. and J.M.G. contributed equally. The authors declare no competing financial interest.
REFERENCES
- 1.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, et al. New Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westbroek W, Gustafson AM, Sidransky E. Trends Mol. Med. 2011;17:485–493. doi: 10.1016/j.molmed.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goker-Alpan O, Stubblefield BK, Giasson BI, Sidransky E. Acta Neuropathol. 2010;120:641–649. doi: 10.1007/s00401-010-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Cullen V, Sardi SP, Ng J, Xu YH, Sun Y, Tomlinson JJ, Kolodziej P, Kahn I, Saftig P, et al. Ann. Neurol. 2011;69:940–953. doi: 10.1002/ana.22400. [DOI] [PubMed] [Google Scholar]; (b) Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, Krainc D. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sardi SP, Clarke J, Kinnecom C, Tamsett TJ, Li L, Stanek LM, Passini MA, Grabowski GA, Schlossmacher MG, et al. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12101–12106. doi: 10.1073/pnas.1108197108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gegg ME, Burke D, Heales SJR, Cooper JM, Hardy J, Wood NW, Schapira AHV. Ann. Neurol. 2012;72:455–463. doi: 10.1002/ana.23614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Yap TL, Gruschus JM, Velayati A, Westbroek W, Goldin E, Moaven N, Sidransky E, Lee JC. J. Biol. Chem. 2011;286:28080–28088. doi: 10.1074/jbc.M111.237859. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yap TL, Velayati A, Sidransky E, Lee JC. Mol. Genet. Metab. 2013;108:56–64. doi: 10.1016/j.ymgme.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, Di Monte DA. J. Biol. Chem. 2010;285:13621–13629. doi: 10.1074/jbc.M109.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a)Dermentzaki G, Dimitriou E, Xilouri M, Michelakakis H, Stefanis L.Plos One 20138 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Manning-Bog AB, Schule B, Langston JW. Neurotoxicology. 2009;30:1127–1132. doi: 10.1016/j.neuro.2009.06.009. [DOI] [PubMed] [Google Scholar]; (c) Cleeter MWJ, Chau K-Y, Gluck C, Mehta A, Hughes DA, duchen M, Wood NW, Hardy J, Cooper JM, Schapira AH. Neurochem. Int. 2013;62:1–7. doi: 10.1016/j.neuint.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a)Ho MW, O'Brien JS.Proc. Natl. Acad. Sci. U. S. A 1971682810–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kolter T, Sandhoff K. Annu. Rev. Cell Dev. Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- 9.(a) de Alba E, Weiler S, Tjandra N. Biochemistry. 2003;42:14729–14740. doi: 10.1021/bi0301338. [DOI] [PubMed] [Google Scholar]; (b) Rossmann M, Schultz-Heienbrok R, Behlke J, Remmel N, Alings C, Sandhoff K, Saenger W, Maier T. Structure. 2008;16:809–817. doi: 10.1016/j.str.2008.02.016. [DOI] [PubMed] [Google Scholar]; (c) Vaccaro AM, Ciaffoni F, Tatti M, Salvioli R, Barca A, Tognozzi D, Scerch C. J. Biol. Chem. 1995;270:30576–30580. doi: 10.1074/jbc.270.51.30576. [DOI] [PubMed] [Google Scholar]
- 10.(a) Alattia JR, Shaw JE, Yip CM, Prive GG. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17394–17399. doi: 10.1073/pnas.0704998104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vaccaro AM, Tatti M, Ciaffoni F, Salvioli R, Barca A, Scerch C. J. Biol. Chem. 1997;272:16862–16867. doi: 10.1074/jbc.272.27.16862. [DOI] [PubMed] [Google Scholar]; (c) Vaccaro AM, Tatti M, Ciaffoni F, Salvioli R, Maras B, Barca A. FEBS Lett. 1993;336:159–162. doi: 10.1016/0014-5793(93)81631-9. [DOI] [PubMed] [Google Scholar]
- 11.(a) Schnabel D, Schroder M, Sandhoff K. FEBS Lett. 1991;284:57–59. doi: 10.1016/0014-5793(91)80760-z. [DOI] [PubMed] [Google Scholar]; (b) Tamargo RJ, Velayati A, Goldin E, Sidransky E. Mol. Genet. Metab. 2012;106:257–263. doi: 10.1016/j.ymgme.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tylki-Szymanska A, Czartoryska B, Vanier MT, Poorthuis BJMH, Groener JAE, Lugowska A, Millat G, Vaccaro AM, Jurkiewicz E. Clin. Genet. 2007;72:538–542. doi: 10.1111/j.1399-0004.2007.00899.x. [DOI] [PubMed] [Google Scholar]; (d) Tylki-Szymanska A, Groener JEM, Kaminski ML, Lugowska A, Jurkiewicz E, Czartoryska B. Mol. Genet. Metab. 2011;104:627–630. doi: 10.1016/j.ymgme.2011.09.010. [DOI] [PubMed] [Google Scholar]; (e) Vaccaro AM, Motta M, Tatti M, Scarpa S, Masuelli L, Bhat M, Vanier MT, Tylki-Szymanska A, Salvioli R. Hum. Mol. Genet. 2010;19:2987–2997. doi: 10.1093/hmg/ddq204. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, Liou B, Ran H, Skelton MR, Williams MT, Vorhees CV, Kitatani K, Hannun YA, Witte DP, et al. Hum. Mol. Genet. 2010;19:1088–1097. doi: 10.1093/hmg/ddp580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Atrian S, Lopez-Vinas E, Gomez-Puertas P, Chabas A, Vilageliu L, Grinberg D. Proteins: Struct. Funct. Bioinform. 2008;70:882–891. doi: 10.1002/prot.21554. [DOI] [PubMed] [Google Scholar]; (b) Weiler S, Kishimoto Y, Obrien JS, Barranger JA, Tomich JM. Protein Sci. 1995;4:756–764. doi: 10.1002/pro.5560040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Chang MHY, Bindloss CA, Grabowski GA, Qi XY, Winchester B, Hopwood JJ, Meikle PJ. Clin. Chem. 2000;46:167–174. [PubMed] [Google Scholar]; (b) Morimoto S, Yamamoto Y, O'Brien JS, Kishimoto Y. Proc. Natl. Acad. Sci. U. S. A. 1990;87:3493–3497. doi: 10.1073/pnas.87.9.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tekoah Y, Tzaban S, Kizhner T, Hainrichson M, Gantman A, Golembo M, Aviezer D, Shaaltiel Y. Biosci. Rep. 2013;33:771–781. doi: 10.1042/BSR20130081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.