Abstract
The post-translational attachment of one or several ubiquitin molecules to a protein generates a variety of targeting signals that are used in many different ways in the cell. Ubiquitination can alter the activity, localization, protein-protein interactions or stability of the targeted protein. Further, a very large number of proteins are subject to regulation by ubiquitin-dependent processes, meaning that virtually all cellular functions are impacted by these pathways. Nearly a hundred enzymes from five different gene families (the deubiquitinating enzymes or DUBs), reverse this modification by hydrolyzing the (iso)peptide bond tethering ubiquitin to itself or the target protein. Four of these families are thiol proteases and one is a metalloprotease. DUBs of the Ubiquitin C-terminal Hydrolase (UCH) family act on small molecule adducts of ubiquitin, process the ubiquitin proprotein, and trim ubiquitin from the distal end of a polyubiquitin chain. Ubiquitin Specific Proteases (USP) tend to recognize and encounter their substrates by interaction of the variable regions of their sequence with the substrate protein directly, or with scaffolds or substrate adapters in multiprotein complexes. Ovarian Tumor (OTU) domain DUBs show remarkable specificity for different Ub chain linkages and may have evolved to recognize substrates on the basis of those linkages. The Josephin family of DUBs may specialize in distinguishing between polyubiquitin chains of different lengths. Finally, the JAB1/MPN+/MOV34 (JAMM) domain metalloproteases cleave the isopeptide bond near the attachment point of polyubiquitin and substrate, as well as being highly specific for the K63 poly-Ub linkage. These DUBs regulate proteolysis by: directly interacting with and co-regulating E3 ligases; altering the level of substrate ubiquitination; hydrolyzing or remodeling ubiquitinated and poly-ubiquitinated substrates; acting in specific locations in the cell and altering the localization of the target protein; and acting on proteasome bound substrates to facilitate or inhibit proteolysis. Thus, the scope and regulation of the ubiquitin pathway is very similar to that of phosphorylation, with the DUBs serving the same functions as the phosphatase.
Keywords: Deubiquitinating enzyme, Ubiquitin, Poly-Ubiquitin, Proteolysis, Regulation
1. Ubiquitination is a post-translational targeting signal
Ubiquitin (Ub) is a highly conserved 76-residue protein present in all eukaryotic cells. Through a series of enzymatic reactions, the C-terminus of Ub becomes activated and conjugated to the ε-amino group of lysine or the N-terminal α-amino group of another Ub, forming poly-Ub chains, or conjugated to target proteins to form a ubiquitinated protein [1]. The conjugation pathway begins with an E1 activating enzyme that uses ATP to first adenylate Ub’s C-terminal carboxylate and transfer it to an E2 conjugating enzyme (~35 in humans) forming an E2-Ub thioester intermediate (E2~Ub) [2, 3]. E3 Ub ligases (>500 putative E3s in humans) provide substrate specificity within the conjugation pathway by selectively binding both E2~Ub and the target protein to catalyze the transfer of Ub to a lysine or α-amino group of the target protein. E3s fall into two general categories, RING domain E3s (Really Interesting New Gene) which catalyze direct transfer of the E2~Ub to a lysine group within substrate/Ub, and HECT (homologous to E6AP Carboxyl-terminus) domain or RBR (RING-between-RING) E3s which contain active site thiols and form an additional E3~Ub thioester intermediate prior to ligation onto Ub/substrate [1, 4-6].
Ubiquitination can generate many different types of covalent modifications [7]. The conjugation of a single Ub to a protein amino group is called mono-ubiquitination. This modification was first described when the chromosomal protein A24 was identified as histone H2A mono-ubiquitinated at K119 [8]. This histone modification is an epigenetic mark that regulates chromosome structure and transcription [9], while mono-ubiquitination of many cell surface receptors is used as a sorting signal to direct these endocytosed proteins to lysosomal degradation [10]. Poly-Ub chains can be assembled when additional ubiquitins are conjugated to the protein-bound monoubiquitin using any of the seven lysines within Ub or the N-terminal α-amino group (forming linear poly-Ub). Thus, ubiquitination of proteins can result in many structurally unique polymers that direct the modified proteins to different fates. Proteins modified with poly-Ub chains linked through K48 or K11 of Ub are recognized and degraded by the 26S proteasome, while K63 poly-Ub functions in regulating other cellular processes such as signal transduction, lysosome-directed protein sorting and the DNA damage response [10-14]. Linear poly-Ub is assembled during inflammatory signaling [15, 16]. Thus, Ub is a post-translational modification similar to phosphorylation or glycosylation and regulates the stability, localization, or activity of modified proteins. DUBs play a role very similar to that of the phosphatases in kinase/phosphatase pathways.
It is worth noting that this system of modification is so useful to the cell that several other Ub-like proteins have evolved. Thus, Ub-like proteins such as Nedd8, SUMO, and others undergo virtually identical activation and conjugation reactions to modify a large number of proteins [17, 18]. A separate family of DUBs containing the ULP (Ubiquitin-like protease) domain exhibit specificity for SUMOylated proteins [19]. This review will concentrate on Ub-dependent processes but will briefly mention Nedd8 modifications since it is required for optimal activity of one family of E3 ligases.
Like all regulatory post-translational modifications, ubiquitination is reversible. A class of proteases called deubiquitinating enzymes (DUBs) removes Ub from target proteins and disassembles polyubiquitin chains [20, 21]. Deubiquitination is the process of hydrolyzing the (iso)peptide bond linking Ub to a substrate or to itself in a poly-Ub chain. Most often the bond hydrolyzed is an isopeptide linkage between a lysine ε-amino group and the C-terminal carboxylate of Ub. Some DUBs display specificity toward different chain linkages, such as K48 or K63 poly-Ub, while some act less specifically and are capable of cleaving multiple chain types or even Ub-like modifiers [20]. Like many other proteases, DUBs are often inactive or autoinhibited, remaining inactive until they are recruited to their site of activity or bind to the proper substrates. To achieve proper localization and specificity DUBs are modular, requiring domains outside the catalytic core to associate with scaffolds, substrate adapters, or the substrates themselves [20].
This review will discuss several of these deubiquitinating enzymes and highlight a number of ways in which they can regulate proteolysis and other Ub-dependent processes (Figure 1). It is not comprehensive, but only exemplary of the different modes of action observed to date. We will concentrate on those DUBs that have been extensively characterized, where structures are known, and where their mechanisms of action highlight different aspects of cellular regulatory strategies.
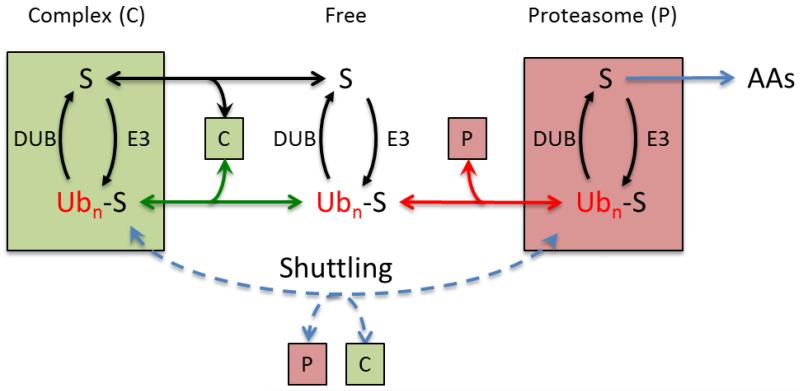
Figure 1.
A given substrate (S) can exist in the free state or bound in a complex (C), each location containing its own distinct E3 and DUB. The strength of binding can be different for the free (black line) or ubiquitinated (green line) substrate. Depending on the value of these binding constants, ubiquitination can either recruit or release S from C. Ubiquitinated protein can be directly bound by the proteasome (red line) or shuttled to the proteasome (dashed blue line) if C is a ubiquitin receptor.
2. The five families of deubiquitinating enzymes
An early bioinformatics approach identified 95 putative DUBs in the human genome [22], yet several lack an active site cysteine or have been shown to act on Ub-like protein conjugates. A more recent estimate puts the number of human ubiquitin-specific DUBs at 86 [23]. DUBs can be grouped into five families based on their conserved catalytic domains. Four of these families are thiol proteases and comprise the bulk of DUBs, while the fifth family is a small group of Ub specific metalloproteases (see below).
2.1 Thiol protease DUBs
Most DUBs are thiol proteases that utilize a catalytic mechanism analogous to that of the plant cysteine protease papain [24, 25]. Thiol-containing DUBs contain a Cys-His-Asp/Asn catalytic triad in which the Asp/Asn functions to polarize and orient the His, while the His serves as a general acid/base by both priming the catalytic Cys for nucleophilic attack on the (iso)peptide carbonyl carbon and by donating a proton to the lysine ε-amino leaving group. The nucleophilic attack of the catalytic Cys on the carbonyl carbon produces a negatively charged transition state that is stabilized by an oxyanion hole composed of hydrogen bond donors. A Cys-carbonyl acyl intermediate ensues and is then hydrolyzed by nucleophilic attack of a water molecule to liberate a protein C-terminal carboxylate and regenerate the enzyme. A striking feature of the thiol protease DUBs is that despite divergent tertiary folds, crystal structures in complex with Ub have revealed the positions of the catalytic dyad/triad discussed above are nearly superimposable [21, 26]. Upon binding Ub, the catalytic domains often undergo structural rearrangements to order regions involved in catalysis. Recently it has been found that many DUBs are inactivated by oxidation of the catalytic cysteine to sulphenic acid (-SOH) [27-29]. The sulphenic acid can be further oxidized to generate sulphinic acid (-SO2H), sulphonic acid (-SO3H), a disulfide, or a sulphenyl amide, which occurs when a sulphenic acid reacts with a nearby backbone amide. Like the disulfide bond, the suphenic acid and sulphenyl amide forms can be reduced with DTT or glutathione.
The thiol proteases are reversibly inhibited by Ub C-terminal aldehyde, forming a thio-hemiacetal between the aldehyde group and the active site thiol. They are irreversibly inactivated by alkylation or oxidation of the catalytic cysteine or reaction of the active site thiol on Ub derivatives containing electrophilic groups near the C-terminus of Ub (i.e., Ub-vinylsulfone, -vinylmethyl ester, -chloroethylamine, and more recently - propargylamine)[30-34].
2.1.1 Ub C-terminal Hydrolase (UCH) domain
DUBs of the UCH family are thiol proteases that contain an N-terminal, 230-residue catalytic domain, sometimes followed by C-terminal extensions that mediate protein-protein interactions. In humans there are four UCH DUBs (UCH-L1, UCH-L3, UCH37/UCH-L5, and BAP1) and these can be sub-grouped based on their substrate specificity. The smaller UCH DUBs (UCH-L1 and UCH-L3) prefer cleaving small leaving groups from the C-terminus of ubiquitin, while the larger UCH DUBs (UCH37 and BAP1) can disassemble poly-Ub chains.
UCH-L1 and UCH-L3 are composed entirely of the UCH domain and are capable of cleaving small molecules and amino acids linked by ester, thioester and peptide bonds to the C-terminus of Ub, yet they are inactive towards di-Ub [35]. In contrast, BAP1 and UCH37 are capable of acting on di-Ub and poly-Ub chains [36-38]. The basis of this specificity stems from a loop that crosses over the UCH catalytic site, forming a pore through which the C-terminus of Ub must be threaded. The length of this crossover loop, and hence the diameter of the pore, varies amongst the enzymes. Engineered UCH-L1 and UCH-L3 are able to cleave di-Ub only when insertions extend these loops [39, 40]. Conversely when the UCH37 loop is shortened by 3-6 amino acids it can no longer cleave di-Ub [39].
In addition to longer crossover loops, UCH37 and BAP1 have C-terminal extensions of ~100 and ~500 residues respectively. In UCH37, the C-terminal extension mediates association with Adrm1/Rpn13 of the proteasomal 19S regulatory subunit and with NFRKB of the INO80 chromatin remodeling complex [41-44]. When associated with the proteasome, UCH37 disassembles poly-Ub chains by hydrolyzing the distal ubiquitin from a chain [38] (see Figure 2A for proximal/distal nomenclature). The extreme C-terminal segment of BAP1 is 38% identical to the C-terminus of UCH37 (defining the UCH37-like domain, ULD) and is necessary for binding the YY1 transcription factor and BRCA1 [45, 46]. The N-terminal portion of the BAP1 extension shares little homology to other proteins, but binds BARD1 and the transcriptional regulator HCF-1 [36, 37, 47].
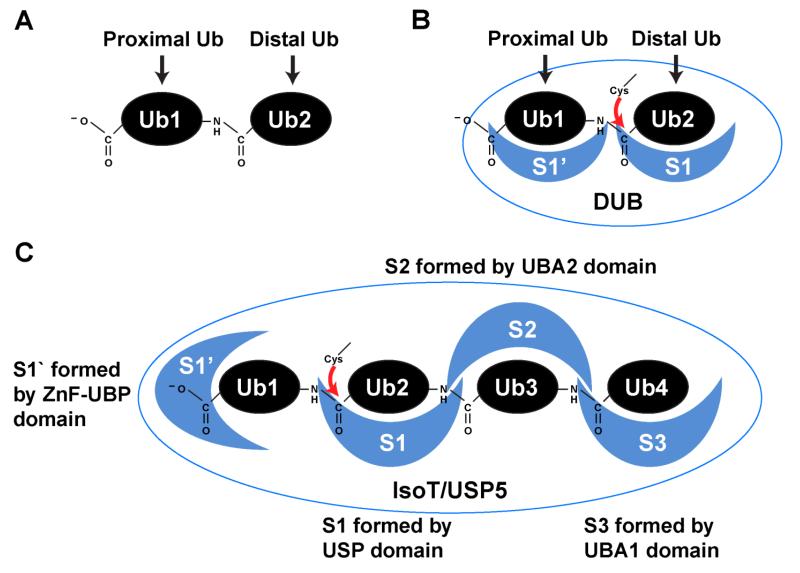
Figure 2.
Ubiquitin binding sites within deubiquitinating enzymes. (A) Di-ubiquitin is depicted to exemplify the proximal ubiquitin (Ub1) and distal ubiquitin (Ub2). The proximal ubiquitin is the initial ubiquitin within a chain, while the distal ubiquitin is final ubiquitin within a chain. The proximal ubiquitin is tethered to the substrate in an anchored poly-Ub chain, or contains a free C-terminal carboxylate in an unanchored chain. (B) A general scenario depicting a thiol DUB catalyzing the hydrolysis of a di-ubiquitin chain. The nomenclature used to describe ubiquitin binding sites within DUBs stems from work on the serine protease papain and its recognition sites for peptide substrates. Like papain, the DUB active site must contact residues flanking the scissile bond, and the S1 and S1’ sites are attributed to the acyl-intermediate side and the leaving group side of the scissile bond respectively. The S1 site binds the ubiquitin containing carboxylate of the hydrolyzed (iso)peptide bond, and the S1’ site binds the ubiquitin containing the amine of the peptide bond. Thus S1 site binds to the distal ubiquitin within a chain, and the S1’ site binds the proximal ubiquitin. It should be noted the S1’ site in DUBs can be formed by residues outside the catalytic domain or S1‘ can reside within the catalytic domain. (C) IsoT/USP5 has four ubiquitin binding sites. IsoT/USP5 is one example of a DUB containing S2 and S3 binding sites which, in this case, are formed by two UBA domains that are inserted within the catalytic USP domain. IsoT/USP5 cannot hydrolyze substrate-anchored poly-Ub as its N-terminal ZnF-UBP domain forms direct interactions with the ubiquitin C-terminal carboxylate.
2.1.2. Ub-Specific Processing Protease (USP) domain
USPs constitute the largest of the DUB families; there are ~56 USP members in humans and 16 in yeast. The USP catalytic domain can vary considerably in size, between 295-850 residues, and consists of six conserved motifs with N- or C-terminal extensions and insertions occurring between the conserved motifs [23]. Two highly conserved regions comprise the catalytic triad, the Cys-box (Cys) and His-box (His and Asp/Asn) [22, 23, 48]. These DUBs tend to recognize and encounter their substrates by interaction of the variable regions of sequence with the substrate protein directly, or with scaffolds or substrate adapters in multiprotein complexes.
The first USP structure described, that of USP7, revealed three subdomains that resemble the thumb, palm and fingers of a right hand [49]. The cleft formed between the palm and the thumb forms the catalytic center, with the thumb containing the Cys-box and the palm the His-box. The finger subdomain forms interactions with Ub to position its C-terminus within the catalytic center. The structure of USP5/IsoT shows how 2 UBL domains inserted within a USP domain provide additional Ub binding sites that allow the enzyme to bind and disassemble poly-Ub chains [50]. The apo structure of USP7 showed a misaligned catalytic triad, yet when complexed with Ub-aldehyde, USP7 undergoes conformational changes within the catalytic cleft, including movement of the catalytic Cys and His residues [49]. In contrast, the structure of USP14, with and without Ub-aldehyde, revealed a well-aligned catalytic triad but two surface loops that occlude the active site in the apo form are displaced upon Ub-aldehyde binding [51]. Could the active site geometry of unbound DUBs reflect a tendency for their oxidation, which requires deprotonation of the catalytic Cys? The USP7 enzyme showed enhanced activity in the presence of DTT, however the USP14 enzyme with its prealigned catalytic triad was inactive, even after addition of DTT, suggesting its catalytic Cys is readily oxidized to the sulphinic/sulphonic acid form [27].
2.1.3 Ovarian Tumor (OTU) domain
Interest in the Drosophila ovarian tumor gene OTU sparked a bioinformatics search that identified several OTU homologs in eukaryotes and viruses, and predicted that the ~180 residue OTU domain encoded a novel family of cysteine protease DUBs [52]. Shortly thereafter OTUB1 and OTUB2 were isolated from HeLa cells and shown to cleave isopeptide linked Ub [53]. In humans there are 15 OTU DUBs that can be evolutionally divided into three classes, the OTUs, the Otubains (OTUBs), and the A20-like OTUs [21].
Members of the OTU DUB family display remarkable specificity for different poly-Ub chain linkages. OTUB1 is highly specific for K48-linked chains, even in mixed chain linkages, whereas OTUB2 can cleave both K63 and K48-linked poly-Ub [54, 55]. The A20 enzyme is specific for K48-linked chains, Cezanne prefers K11-linked chains, and TRABID acts on both K29 and K33-linked poly-Ub [56-58]. Crystal structures of the human OTUB1, OTUB2, TRABID, A20, and yeast Otu1 enzymes have revealed the conserved catalytic OTU domain which contains the S1 site, and N-terminal residues in TRABID and OTUB1 that form the S1’ site [55-57, 59-61] (see Figure 2B S1/S1’ nomenclature). The active site of the OTU domain contains an unusual loop not seen in other thiol-DUBs and can lack an obvious catalytic Asp/Asn [57, 60, 61]. In OTUB1, Ub-aldehyde binding to the S1 active site induces structural rearrangements at the S1’ site, suggesting only K48 poly-Ub linkages productively engage both sites yielding a positioning of the isopeptide bond that permits catalysis [54]. The A20 and OTUB1 enzymes have displayed unusual modes of activity (discussed in later sections) as they directly bind to E2 enzymes [62, 63]. OTU DUBs show remarkable specificity for different Ub chain linkages and may recognize substrates on the basis of those linkages.
2.1.4 Josephin domain
In humans there are four proteins that contain the ~180 residue Josephin domain (Ataxin-3, Ataxin-3L, Josephin-1 and Josephin-2) and all have been shown to possess DUB activity, though to different extents, towards Ub-AMC, Ub-peptide fusions, and K48 poly-Ub or K63 poly-Ub [64, 65]. Ataxin-3 and -3L contain C-terminal extensions composed of two tandem UIMs (Ub-interacting motif), a poly-Gln stretch, and an additional UIM in ataxin-3. The UIMs in Ataxin-3 have been shown to promote Ub-binding, its ubiquitination, and its K63 chains specificity [66-68].
Ataxin-3 is the best studied of the Josephin family members as an expansion of its poly-glutamine stretch gives rise to the neurodegenerative disorder Machado-Joseph disease (also known as spinocerebellar ataxia type 3) [69]. Attempts to gain insights into Ataxin-3 function led to a bioinformatifcs study that predicted Ataxin-3 was a cysteine protease DUB [70]. Shortly thereafter this was confirmed when Ataxin-3 was shown to bind long K48 poly-Ub chains and trim Ub from poly-ubiquitinated lysozyme, an activity inhibited by Ub-aldehyde [71]. Analysis of Ataxin-3 substrate specificity found it can bind longer K63 and K48 poly-Ub (≥5), but its activity is highly specific towards K63 linkages in homogenous and mixed chains [66]. Thus, the Josephin domain DUBs may specialize in distinguishing between polyubiquitin chains of different lengths.
The solution structures of the Ataxin-3 Josephin domain, alone and in complex with Ub, and a crystal structure of the Ataxin-3L Josephin domain covalently bound to Ub-chloroethylamine have been reported [64, 67, 72]. The two Josephin domains are 85% identical, and adopt a similar overall fold, yet the binding site for Ub is quite different between the two [64] (Figure 3). Differences are also observed in the C-terminus of bound Ub; in the crystal structure with covalently bound Ub the catalytic Cys-His-Asn triad is aligned, whereas in the solution structure the C-terminus of Ub splits the catalytic Cys and His yielding a non-productive catalytic conformation. The distorted triad could be a characteristic of a product complex, since the product Ub contains a C-terminal carboxylate not present in a poly-Ub substrate. Finally, the solution structure shows a second free Ub bound to the opposite face of the Josephin domain (S2 site) with its C-terminus positioned towards K48 of Ub bound in the active site. This could represent yet another Ub binding site (in addition to the catalytic site and UIM sites) capable of binding K48-linked polyubiquitin.
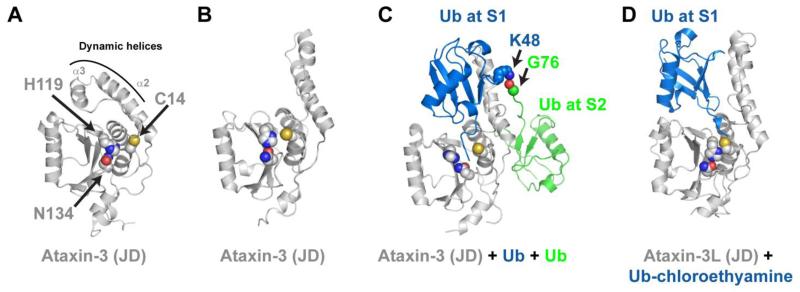
Figure 3.
The Josephin domain (JD) contains multiple ubiquitin binding sites. (A) NMR solution structure of the Ataxin-3 Josephin domain with residues of the catalytic triad shown as spheres (PDB 2AGA). (B) An adopt an alternative conformation (PDB 1YZB). (C) Solution structure of Ataxin-3 JD non-covalently associated with two ubiquitin molecules (PDB 2JRI). One ubiquitin is bound at the S1 site and its C-terminal tail bisects the catalytic Cys-His pair, possibly a reflection of the product complex that follows hydrolysis of the isopeptide bond. A second ubiquitin is bound at the S2 site of the Josephin domain, and its C-terminus is juxtaposed with K48 of ubiquitin bound at the S1 site. (D) Crystal structure of the Ataxin-3L JD covalently bound to Ub-chloroethylamine (PDB 3O65). This structure reveals the Ataxin-3L JD binds ubiquitin at the S1 site using different residues and positions ubiquitin in an alternate conformation.
2.2 Metalloprotease DUBs
Sequence alignments within the JAMM domain identified a Glu-x[N]-His-x-His-x[10]-Asp motif Zn2+ binding sites [73, 74] and soon thereafter the RPN11 subunit of the proteasome was shown to possess DUB activity dependent on these coordinating residues and a bound Zn2+ atom [75, 76]. The JAMM domain is present in bacterial and archaeal proteins as well [73], and crystal structures of the AF2198 JAMM protein from Archaeoglobus fulgidus revealed that the domain adopts a fold most similar to the metallohydrolase cytidine deaminase, while the arrangement of Zn2+ ligands is most similar to the metalloprotease thermolysin [74, 77]. Catalysis requires nucleophilic attack on the carbonyl carbon of the isopeptide bond by an activated water molecule bound to Zn2+ and a conserved glutamate. A negatively charged tetrahedral transition state ensues, and a nearby conserved Ser/Thr in the JAMM domains stabilizes the oxyanion. The tetrahedral intermediate then collapses and the Glu serves as a general base donating a proton to the leaving Lys side chain [77, 78]. Metallo DUBs are insensitive to alkylating agents, Ub aldehyde, or Ub-electrophiles but can be inhibited by removing the catalytic zinc.
2.2.1 The JAB1/MPN+/MOV34 (JAMM) domain
The JAMM domain is found in eight human proteins, however PRPF8 is predicted to lack protease activity [21]. Two multisubunit complexes, the proteasome 19S lid complex and the COP9-Signalosome contain JAMM DUBs (POH1/hRpn11 and CSN5/Jab1 respectively). As discussed later, RPN11 is an endopeptidase that cleaves poly-Ub chains en bloc from substrates as they are degraded by the proteasome [75, 76]. CSN5/Jab1 deconjugates the Ub-like modifier Nedd8 to modulate the activity of the SCF E3 ligase [79]. The roles of BRCC36 in the DNA damage response and AMSH in endocytic trafficking are discussed in later sections. An emerging theme of JAMM domains is that they function by cleaving at the base of the chain, between proximal Ub and substrate (RPN11, CSN5/Jab1), and/or they are highly specific for K63 poly-Ub linkages (RPN11, AMSH, AMSH-LP, BRCC36) [75, 79-82]. To date there are 3 crystal structures of human JAMM domains; CSN5/JAB1 [83], AMSH [84], and AMSH-LP in complex with K63 di-Ub, which has yielded insights into K63 poly-Ub specificity [82].
3. How do DUBs regulate Ub-dependent processes?
It is now widely understood that ubiquitination generates a targeting signal that can be used to alter the properties or localization of the ubiquitinated protein. The first discovered, and perhaps still most prominent, role for ubiquitination is in delivering ubiquitinated proteins to the proteasome, a large compartmentalized multi-catalytic protease that is responsible for much of the regulated proteolysis in cells [85, 86]. We can use this system as an analogy for all Ub-dependent processes. Figure 1 represents a general model for regulating Ub dependent processes. A protein can exist in a ubiquitinated or deubiquitinated form interconverted by the action of an E3 and a DUB. In principal, the ubiquitination state can alter the activity of the target protein, its localization (by altering the stability of a protein complex such as Ub-S in complex 1) or its half-life (by delivering it to the proteasome). Furthermore, each location can contain a different set of E3s and DUBs leading to location specific ubiquitination or deubiquitination.
Given this very general model, we can predict several modes of regulation by DUBs. DUBs can act by: directly interacting with and co-regulating E3 ligases; altering the level ubiquitination; hydrolyzing or remodeling ubiquitinated and poly-ubiquitinated substrates; by acting only in certain locations in the cell and altering the localization of the target protein; or by acting at the proteasome itself to facilitate or inhibit proteolysis.
3.1. DUBs affecting the rate of ubiquitination
It has been noted that many DUBs exist in complexes with E3 ligases and regulate the accumulation of ubiquitinated substrates. Well-known DUB/E3 pairs are; Usp2a and Usp7/Mdm2, Usp7/ICP0, Usp8/Ndrp1 and GRAIL, Usp20 and Usp33/VHL, and Ataxin-3/Parkin [87]. In principle, the DUBs could act catalytically to deubiquitinate the E3 or the substrate, and could also have non-catalytic effects by altering the stability or composition of the E3 complex. While there are numerous examples of this type of regulation we have chosen just three, in part because each also has other modes of regulation that we highlight. The examples chosen here emphasize that a given DUB can have more than one mode of action with respect to a single substrate and can participate in the regulation of several different substrates.
3.1.1. A deneddylating DUB activity is required for optimal SCF E3 activity
The catalytic activity of the Skp, cullin, F-box (SCF) family of E3 ligases is highly dependent on a DUB, albeit one acting on the cullin subunit of this ligase conjugated to the Ub-like protein Nedd8. This DUB activity is contributed by the CSN5 subunit (a JAMM domain DUB) of the eight subunit COP9 Signalosome (CSN) [79, 88]. Its activity is required for SCF catalytic activity and the cyclical NEDDylation and deNEDDylation of Cullins is required for optimal SCF activity [89]. CSN is involved in multiple cellular pathways, such as cell cycle control, transcriptional regulation, and the DNA damage response, and the CSN5/Jab1 subunit can function in non-CSN complexes [90]. This pathway of modification has recently been implicated in a variety of cancers and an inhibitor of Nedd8 activating enzyme is in clinical trials [91, 92].
3.1.2. DUBs acting to deubiquitinate E3s
A characteristic hallmark of the E3 mechanism is autoubiquitination. In the absence of substrates many (most?) E3s ubiquitinate themselves and are then subject to degradation by the proteasome. Alternatively, these ligases can be ubiquitinated by other E3s to regulate their degradation. DUBs present in the same protein complexes can reverse these ubiquitination events, sparing the E3 so that it can respond to increases in substrate. For example, USP7 deubiquitinates autoubiquitinated Mdm2, the p53 Ub ligase (see below). USP7 also deubiquitinates autoubiquitinated RING2 ligase of the polycomb complex and RING2 that has been marked for degradation by the E6AP ligase.
3.1.3. E3/DUB co-regulation by reciprocal ubiquitination/deubiquitination of a substrate
A large number of DUBs have been shown to hydrolyze protein bound K48-linked polyubiquitin chains and prevent the degradation of the attached proteins. Two illustrative examples are discussed here.
3.1.3.1. USP7
USP7 is a versatile DUB, with an ever expanding list of substrates that are involved in various cellular pathways (see Table 1) [93]. USP7 is also a key regulator of the p53 tumor suppressor, a sequence specific transcription factor that becomes activated upon various cellular stresses and elicits according cellular responses such as cell cycle arrest, DNA repair, apoptosis and senescence [94]. The cellular level and activity of p53 are tightly regulated, in part by an E3 ligase Mdm2 which binds the p53 transactivation domain inhibiting activation, shuttles nuclear p53 into the cytoplasm where it is inactive, and ubiquitinates p53 promoting its degradation [95]. USP7 is critical component of this pathway as it deubiquitinates and stabilizes both p53 and Mdm2; reduction of USP7 levels destabilizes p53 by promoting the ubiquitinated form, yet ablation of USP7 increases p53 levels by destabilizing Mdm2 [96, 97]. The levels of p53 are also regulated by Mdmx, a structural homolog Mdm2 that lacks E3 activity, but binds p53 and prevent ubiquitination and degradation by Mdm2. Like p53, Mdmx is co-regulated by reciprocal ubiquitination/deubiquitination by Mdm2/USP7 [98].
Table 1.
DUBs discussed. Where available, substrates, functions and additional structural data are noted.
| DUB | Res | Substrate(s) | PDB (notes) | Cellular role(s) | Additional domain(s) |
|---|---|---|---|---|---|
| USP DUBs | |||||
| USP3 | 520 | Histone H2A [137] Histone H2B [137] |
None | Cell cycle progression | ZnF-UBP |
| USP5/IsoT | 858 | Unanchored poly-Ub [115] | 3IHP (+Ub-ethylamine) [50] 2G45 (ZnF-UBP + Ub) [119] |
Ub recycling | Novel ZnF-UBP-like domain ZnF-UBP binds free C-terminus of Ub UBA-1 forms S3 site UBA-2 forms S2 site |
| USP7/HAUSP | 1102 | p53 [97] MDM2 [96] MDMX [98] FOXO4 [196] PTEN [197] Histone H2B [198] Claspin [199] UHRF1 [200] HSV ICP0 [201] EBV EBNA1 [202] BMI1 [153] MEL18 [153] RING2 [171] |
1NB8 (apo USP domain) [49] 1NBF (USP + Ub-aldehyde) [49] |
Cell cycle progression Apoptosis DNA damage response Transcription Immune response Viral replication |
TRAF/Math domain binds p53, MDM2, EBNA1 5 C-terminal UBL domains, additional interactions with p53 and MDM2 |
| USP8/UBPY | 1118 | Membrane receptors/ ESCRT cargo [10, 172] GRAIL [203] STAM2 [179] NRDP1 [181] BRUCE [204] PAR2 [205] |
2GFO (apo USP domain) [206] 3N3K (USP domain + specific inhibitor) [207] 2A9U (MIT/dimerization domain) [206] 2GWF (Rhodanese domain + NRDP1) [206] |
Endocytosis Endosome sorting Cell cycle progression |
MIT/dimerization domain Rhodanese domain binds NRDP1 |
| USP14 | 494 | Proteasome substrates [33] | 2AYN (apo) [51] 2AYO (+ Ub-aldehyde) [51] |
Ub recycling | UBL domain |
| USP16/UBP-M | 823 | Histone H2A [154] | 2I50 [168] | Mitotic progression Transcription |
ZnF-UBP binds C-terminus of Histone H4 |
| USP21 | 565 | Histone H2A [139] RIP1 [208] |
2Y5B (+ linear di-Ub aldehyde) [209] | Transcription Signal transduction Microtubule dynamics |
N-terminus mediates microtubule and centrosome binding |
| USP22 | 525 | Histone H2A [141, 142] Histone H2B [141-143] |
None | Transcription | ZnF-UBP |
| UCH DUBs | |||||
| UCH-L1/ PGP 9.5 |
223 | Proubiquitin? | 2ETL (apo) [210] 3KW5 (+Ub-VME) [211] |
Proubiquitin processing? Ub recycling? |
None |
| UCH-L3 | 230 | Proubiquitin? | 1UCH (apo) [24] 1XD3 (+Ub-VME) [212] |
Proubiquitin processing? Ub recycling? |
None |
| UCH37/ UCH-L5 |
329 | Proteasome substrates [38] | 3A7S (apo UCH domain) [213] 3IHR (apo) [214] |
Chain editing Ub recycling |
ULD binds Rpn13 of 19S regulatory particle ULD binds NRFKB of INO80 |
| BAP1 | 729 | Histone H2A [215] HCF-1 [36, 37] OGT [157] |
None | Cell cycle progression Transcription Hematopoiesis |
ULD binds YY1 HBM binds HCF-1 |
| OTU DUBs | |||||
| OTUB1 | 271 | GRAIL [203] TRAF3 [99] TRAF6 [99] P53 [103] ERa [100] RhoA [101] |
2ZFY (apo) [55] 4DDI (fusion to UbcH5b~Ub + Ub) [104] 4DHI (worm OTUB1 + UBC13) [105] 4DHJ (worm OTUB1 + Ub-aldehyde + Ubc13~Ub) [105] |
Signal transduction T-cell anergy DNA damage response E2 enzyme inhibition |
N-terminal helix binds Ub |
| A20 | 790 | RIP1 [63] TRAF6 [216] MALT1 [217] |
2VFJ (apo OTU domain) [57] 3DKB (apo OTU domain) [61] 3OJ3 (ZnF-4 + Ub) [133] 3OJ4 (ZnF-4 + UbcH5a + Ub) [133] |
Signal transduction Immune response |
7A20-ZnF domains bind poly-Ub and TAXBP1 ZnF-1 binds RIP1, ZnF-4 binds Ub, ZnF-5 and −6 bind UbcH5a, |
| Josephin DUBs | |||||
| Ataxin-3 | 364 | Misfolded ER proteins [69, 218] | 1YZB (NMR, apo Josephin domain) [72] 2AGA (NMR, apo Josephin domain) [67] 2JRI (NMR, Josephin + Ub + Ub) [NP] 2KLZ (NMR, UIM domain + Ub) [NP] |
ERAD Aggresome formation Transcription |
UIM1 UIM2 binds VCP Poly-Q rich UIM3 |
| Ataxin-3L | 355 | Undetermined | 3O65 (Josephin domain + Ub- chloroethylamine) [64] |
Similar to Ataxin-3? | UIM1 UIM2 Poly-Q rich |
| JAMM DUBs | |||||
| 2A-DUB/ MYSM1 |
828 | Histone H2A [140] | 2CU7 (SANT domain) [219] 2DCE (SWIRM domain) [219] |
Transcription Hematopoiesis |
SANT SWIRM |
| RPN11/POH1 | 310 | Proteasome substrates [75, 76] | 4B4T (yeast Rpn11 within 26S proteasome) [220] |
Ub Recycling Chain editing |
C-terminal extension |
| BRCC36 | 316 | Histone H2A/H2A.x [126] NLRP3 [127] |
None | DNA damage response Inflammatory response |
Coiled-coil |
| AMSH | 424 | Membrane receptors/ ESCRT cargo [10, 172] STAM [221] HRS [221] |
None | Endosome sorting | MIT Clathrin binding domain |
| AMSH-LP | 436 | Undetermined | 2ZNR (apo JAMM domain) [82] 2ZNV (JAMM domain + K63 di-Ub) [82] |
Similar to AMSH? | MIT |
| CSN5/JAB1 | 334 | SCF E3 ligases [222] | 4F7O (apo JAMM domain) [83] | Activation of SCF ligases | C-terminal extension |
3.1.3.2. OTUB1
DUBs that deubiquitinate proteasomal substrates should exhibit significant activity on K48-linked chains. OTUB1 has been shown to stabilize substrates by catalytic and non-catalytic mechanisms. It has deubiquitinating activity and exhibits high specificity for K48 isopeptide linkages, even in mixed linkage chains [54, 55]. OTUB1 and its paralog OTUB2, deubiquitinate TRAF3 and TRAF6 to inhibit virus-triggered signaling pathways that ultimately result in IRF3 and NF-κB activation [99]. OTUB1 has also been shown to stabilize the estrogen receptor α [100] and RhoA [101] and in both cases stabilization is dependent on OTUB1’s catalytic Cys91.
3.1.4. Modulation of E2 activity
In principle, DUBS could interfere with Ub activation, formation of the E2~Ub intermediate, or reactivity of the intermediate to inhibit ubiquitination. Two examples of the later mechanism are discussed; one catalytic and one non-catalytic.
3.1.4.1. Ataxin-3
One mechanism of interfering with ubiquitination by modulating E2 activity is afforded by the Ataxin-3 mediated inhibition of Parkin autoubiquitination. Parkin, its cognate E2 UbcH7, and Ataxin-3 form a tight complex preventing the autoubiquitination of Parkin and the release of UbcH7 [102]. Interestingly, the inhibition of autoubiquitination and the formation of a tight complex require the active site thiol of the DUB domain. Ataxin-3 is unable to act on pre-ubiquitinated Parkin or on E2~Ub [102].
Parkin is a Parkinson’s disease associated E3 containing a RING-between-RING (RBR) domain. Recently it has been recognized that RBR ligases actually use a mechanism characteristic of the HECT-domain family of ligases, that is they first transfer Ub from E2~Ub to an active site thiol and then pass it on to a protein amino group [4] (editor: please reference RBR review in this volume). UbcH7, the E2 that works with Parkin, is unable to transfer directly to an amino group via the usual RING mechanism. Thus, it is likely that Ataxin-3 inhibits parkin autoubiquitination by intercepting the Ub from E2~Ub with its own active site thiol and the resulting DUB thioester intermediate is protected from hydrolysis by the stable ternary complex.
3.1.4.2. OTUB1
As discussed in section 3.1.3.2, OTUB1’s is highly specific for K48-linked poly-Ub and stabilizes its substrates by disassembling these proteasome-targeting chains. OTUB1 also functions non-catalytically to inhibit K63 ubiquitination of histone H2A by the E3 RNF168 during the DNA damage response [62]. Depletion of OTUB1 led to continuous ubiquitination of histone H2A following ionizing radiation, and overexpression of OTUB1 or the catalytically inactive mutant both suppressed H2A polyubiquitination [62]. This non-canonical mode of regulation was also reported when OTUB1 was shown to stabilize and activate p53 independent of catalytic activity [103]. Insights into this unusual mode of regulation began with the identification of E2 conjugating enzymes that co-purify with OTUB1, including Ubc13 an E2 that generates K63 poly-Ub (in conjunction with the E2 variant UVE1) and functions with RNF168 in the DNA Damage Response (DDR) pathway [62]. OTUB1 was shown to directly bind Ubc13, preferring to bind the Ub thiolester Ubc13 intermediate (Ubc13~Ub), and this interaction was stabilized by OTUB1 N-terminal domain. Similar preferential binding to Ub charged UbcH5b was shown, and activity assays with E3 enzymes concluded that OTUB1 functions as an E2 inhibitor, preventing autoubiquitnation of the E3 TRAF6 [62].
Structures of apo OTUB1 and OTUB1 in complex with the E2s UbcH5b/UBE2D2 and Ubc13 have also been reported (Figure 4A). A UbcH5b(C85S)-OTUB1 fusion protein was generated and reacted with E1 and Ub to generate a stable E2-Ub oxyester bond [104]. In this structure the E2 residues that contact OTUB1 are also known to mediate binding to E3s, thus explaining how binding to the DUB inhibits the E2/E3 interaction. The Ub conjugated to UbcH5b predominately interacts with OTUB1; one of these interactions is mediated by the N-terminus of OTUB1 discussed above, which forms an extended helix (Figure 4B). The OTU domain also contacts the UbcH5-linked Ub (S1’ site) and positions K48 towards the catalytic cleft. Unexpectedly a second, free Ub was bound to OTUB1 (S1 site) and its C-terminal tail was juxtaposed near K48 of UbcH5-conjugated Ub within the catalytic cleft [104]. Thus OTUB1 simultaneously binds to E2-charged Ub and a free Ub, and the arrangement of these two ubiquitins mimics K48 di-Ub. Contemporaneously, two additional OTUB1/Ubc13 structures were reported; human Ubc13 in complex with C.elegans OTUB1, and human Ubc13~Ub analog in complex with C.elegans OTUB1/Ub-aldehyde [105] (Figure 4C). The residues needed for Ubc13 to generate K63 poly-Ub and transfer it to substrates (via binding to UEV1 and RNF168) participate in OTUB1 binding, displaying a mode of competitive inhibition analogous to that of UbcH5b [105]. Another notable finding from this study is that free Ub binding to OTUB1 (at S1) allosterically regulates the enzyme by increasing its affinity for Ubc13~Ub (at S1’) [105].
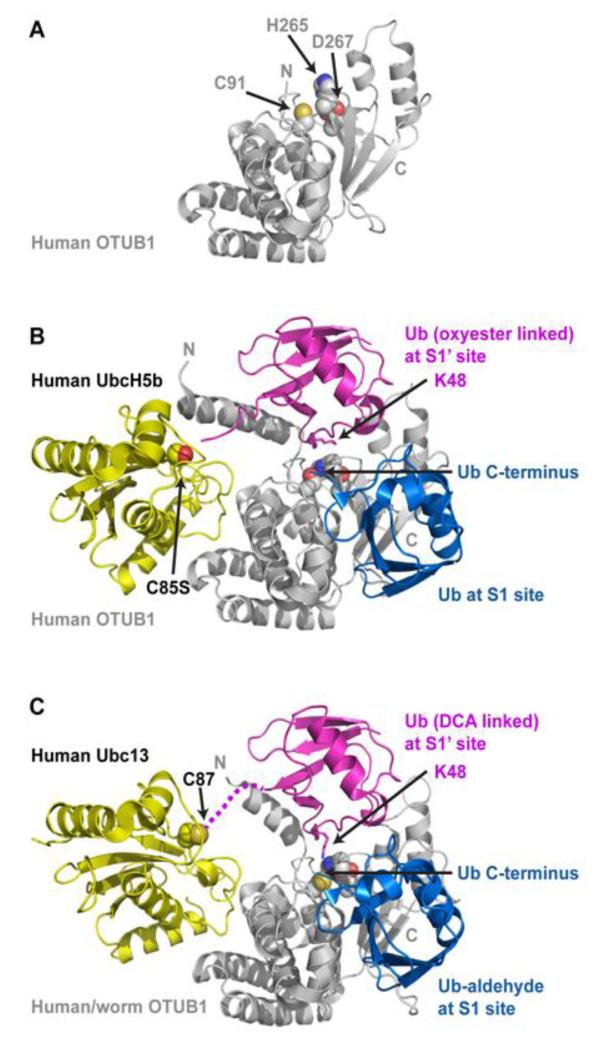
Figure 4.
The OTUB1 deubiquitinase binds charged E2s and inhibits their transfer of ubiquitin to E3. (A) The human OTUB1 structure (PDB 2ZFY) with side chains of the catalytic triad (C91, H265, and D267) depicted as spheres. The structure was solved using an OTUB1 deletion lacking the N-terminal 39 residues, which form the N-terminal helix and the S1‘ site. (B) The structure of human OTUB1 bound to UbcH5bC85S-Ub (covalently linked to UbcH5b S85 by an oxyester bond) and free Ub (PDB 4DDI). The E2 UbcH5b and its linked Ub form contacts with the N-terminal helix, and this E2-conjugated Ub binds the S1’ site with K48 positioned towards the catalytic triad and the C-terminus of ubiquitin at the S1 site. (C) The structure of human/worm OTUB1 bound to Ubc13-UbG75C (covalently linked to Ubc13 C87 with dichloroacetone) and Ub-aldehyde (PDB 4DHJ). The hybrid OTUB1 contains the first 45 residues from human OTUB1 followed by the OTU domain from worm OTUB1. Ub aldehyde is covalently bound at the S1 site. OTUB1 binds both Ub-charged E2s in a similar fashion, with K48 of the E2-linked Ub positioned towards the active site and the C-terminus of Ub bound at the S1 site. The binding and arrangement of two ubiquitin molecules at the S1 and S1’ sites in these two structures mimics OTUB1’s natural substrate K48-linked poly-Ub.
3.2. Processing, recycling, and remodeling polyubiquitin chains
A variety of DUB activities are required to initiate and sustain Ub-dependent processes. These include processing of the primary gene products to yield Ub, disassembling the polyubiquitin chains to down regulate signaling and prevent competitive inhibition of Ub receptors, and recovery of Ub from chains and other inadvertently trapped Ub derivatives.
3.2.1. UCHL1/L3-processing pro-Ub and removal of adventitious Ub derivatives
UCHL1 and UCHL3 are proposed to liberate small molecule nucleophiles that may have inadvertently reacted with Ub C-terminal thiolesters [35]. Because these enzymes can cleave small peptides from the C-terminus of Ub, they could also function in recycling Ub from incomplete proteasomal or lysosomal protein degradation [35]. Another possible role is the co-translational processing of proubiquitin. In most organisms, Ub is expressed as a linear polymer, proubiquitin, consisting of multiple copies of Ub and one or more amino acids appended to the C-terminus of the final Ub. For example, in humans polyubiquitin-C is expressed as 9 Ub monomers followed by a Val, and polyubiquitin-B as 3 monomers followed by a Cys [106]. It is possible that the smaller UCH DUBs function in removing these terminal amino acids from proubiquitin.
While the precise cellular substrate of these enzymes remains unclear, UCH-L1 is cytosolic, highly expressed in the brain, accounting for 1-2% of soluble brain protein, and expressed at low levels in ovaries and testes [107, 108]. UCH-L3 is cytosolic and highly expressed in the heart and in skeletal tissue [109]. UCH-L1 has been linked to neurodegenerative disorders in mice and in humans. In mice, spontaneous deletion of exons 7 and 8 results in a recessive disorder called gracile axonal dystrophy (gad) and the accumulation of β-amyloid protein and ubiquitinated proteins [110]. In humans UCH-L1 is found in neurofibrillary tangles of Alzheimer’s disease patients [111] and is down regulated and oxidatively inactivated in brains of Alzheimer’s and Parkinson’s disease patients [112]. Farnesylation of UCH-L1 promotes ER membrane association and its localization correlates with α-synuclein accumulation and toxicity [113].
3.2.2. USP5/IsoT-recycling polyubiquitin chains
IsoT functions in recycling monomeric Ub by disassembling unanchored poly-Ub chains. These unanchored chains are produced from two sources, proteasomal DUBs that cleave poly-Ub from substrates or from E2/E3 enzymes that synthesize these chains for conjugation to substrates. Deletion of yeast IsoT (UBP14) leads to the accumulation of polyubiquitin and inhibition of proteasomal degradation due to competitive inhibition by the accumulated chains. Knockdown of the mammalian IsoT (USP5) causes a similar accumulation of polyubiquitin as well as an increase in the proteasomal substrate p53 [114].
Mechanistic studies on IsoT found it preferred cleaving longer K48 poly-Ub chains (≥4) over shorter chains and linear poly-Ub, and that it acts as an exopeptidease, cleaving the proximal Ub from unanchored poly-Ub chains [115-117]. IsoT shows little specificity for Ub-chain linkages, as it can hydrolyze tetra-Ub linked through K48, K63, K6 and K29 [118]. Early studies predicted multiple Ub binding sites; Ub-aldehyde was shown to slow the dissociation of free Ub, and high levels of free Ub were capable of inhibiting disassembly of poly-Ub in a chain dependent manner [115, 117]. IsoT contains two Ub-binding UBA domains inserted within its USP domain, an N-terminal domain, and a ZnF-UBP domain. A crystal structure of the isolated ZnF-UBP domain revealed that IsoT binds Ub or unanchored polyubiquitin chains by forming extensive contacts with the free C-terminal Gly-Gly motif [119]. Mutating the C-terminal Gly of Ub to Ala (G76A) or deleting the di-Gly motif abolishes binding to the ZnF-UBP domain [119]. Thus the ZnF-UBP domain binds the proximal Ub of a poly-Ub chain in the S1’ site, and subsequent studies, using UBA mutants and quantitative binding assays, determined UBA-2 forms the S2 site and UBA-1 the S3 site [120] (Figure 2C). The crystal structure of the full length enzyme in complex with Ub-ethylamide was recently reported and confirmed the arrangement of the 4 Ub binding sites [50]. However the structure does not represent a catalytically competent state, as modeling of Ub into the S1’ ZnF-UBP site found K48 to be ~45 Ǻ from the catalytic Cys of the S1 site containing Ub-ethylamide. Conformational flexibility within a disordered loop that tethers the ZnF-UBP domain to the USP domain likely allows rearrangements that both close this gap and permit the indiscriminate hydrolysis of various chain linkages. The N-terminal domain of IsoT was found to adopt a novel ZnF-UBP-like fold, but it cannot bind free Ub and lacks conserved Zn2+ coordinating residues [50].
3.2.3. BRCC36 downregulates DSB signaling by removing K63-linked polyubiquitin
The DNA Damage Response (DDR) to double strand breaks (DSB) leads to the phosphorylation of histone H2A.x at Ser139 by the ATM and DNA-PKcs kinases [121]. This phosphorylation event results in the recruitment of MDC1 and the E3 ligases RNF8 and RNF168 which assemble K63 poly-Ub chains on H2A.x [122]. This modification on H2A.x serves to both relax chromatin and to create a binding site for the Rap80 complex, which binds K63 poly-Ub using tandem UIMs and assembles repair complexes containing BRCA1 [122]. BRCC36 is a K63 specific metallo-DUB and core component of the five subunit Rap80 complex [80, 123-125]. BRCC36 functions in the disassembly of K63 poly-Ub on H2A/H2A.x and termination of RNF8/RNF168 ubiquitination events [126]. Depletion of BRCC36 led to the accumulation of ubiquitinated H2A.x following IR, and overexpression of BRCC36 decreases Ub-H2A at DSBs, an effect dependent on Zn2+ coordinating residues [126]. BRCC36 also functions in a four subunit cytoplasmic complex, BRISC, that shares similar components of the RAP80 complex [80]. BRCC36 within BRISC functions in disassembling poly-Ub chains on NLRP3 (but not the proximal ubiquitin) in response to viral and bacterial infection, promoting the assembly of the NLRP3 inflammasome complex [127].
3.2.4. A20-remodels K63-linked chains to form K48-linked chains and terminate NFκB signaling
A20 is an OTU DUB that contains a C-terminal extension harboring 7 ZnF domains that endow A20 with E3 Ub ligase activity. A20 is a key regulator of the immune and inflammatory response pathways that trigger transcriptional activation of NFκB family of transcription factors. It deubiquitinates components (RIP1, TRAF6, MALT1) in multiple immune signaling cascades including TNFR1, IL-1R, and TLR4 to down regulate the NFκB response [128]. In humans mutations within the A20 gene have been linked to a host of inflammatory and malignant diseases [128]. In response to TNF signaling, K63 poly-ubiquitination of RIP1 promotes the assembly of a complex that phosphorylates the NFκB inhibitor IκBα. Phosphorylation of the cytoplasmic NFκB/IκBα complex results in the proteasomal degradation of IκBα and release of NFκB to allow its entry into the nucleus and transcriptional activities. A20 acts directly on RIP1 to disassemble K63 poly-Ub, a prerequisite for its E3 activity that subsequently polyubiquitinates RIP with K48 chains to target it to the proteasome for degradation [63]. Precise details of this mechanism are still poorly understood, as A20 also binds TAXBP1 and the E3 ligase ITCH, an E3 necessary for RIP1 ubiquitination and degradation [129]. The E3 activity of A20 also functions in dampening NFκB signaling by targeting the E2 enzymes UbcH5a and UBC13 for degradation [130]. These E2s function during different stages of the TLR4 and IL-R1 signaling cascades to promote NFκB activation by ubiquitination and activation of TRAF6 (using UBC13) and IKK (using UbcH5a) [131, 132]. The E3 ITCH is not required for UBC13 degradation [130], suggesting A20 has intrinsic E3 activity as well as a second E3 activity mediated by the TAXBP1/ITCH complex.
In vitro A20 shows low DUB activity and prefers K48 poly-Ub as a substrate over K63 poly-Ub, yet it deubiquitinates K63 poly-ubiquitinated TRAF6 by clipping at the base of the chain, removing it en bloc [61]. Crystal structures of the A20 OTU domain revealed a minimal catalytic site that rationalizes its generally weak DUB activity [57, 61]. In place of the conserved catalytic Asp/Asn found in other thiol DUBs, the A20-like OTU DUBs utilize a nearby Asp/Glu to bind a water molecule which fulfills the role of His-polarization [56, 57]. A thorough analysis of the A20 ZnF domains further defined their roles in binding to Ub, E2s, and substrates; ZnF-1 promotes RIP1 binding, ZnF4 binds Ub, and ZnF-5 and -6 bind UbcH5a [133].
3.3. DUBs acting at the level of localization
As suggested by Figure 1, the regulation of ubiquitination and deubiquitination is often very dependent on localization. To illustrate this point we have chosen to discuss the regulation of a single ubiquitination event, the modification of Histone H2A, in a variety of contexts involved in the structure of chromatin and transcriptional regulation. Histone H2A was the first protein shown to be modified by Ub when in 1977 it was found to contain an unusual structure with two N-termini and a single C-terminus [8]. We now know that in humans ~10% of histone H2A is ubiquitinated at K119, and ~1% of H2B is ubiquitinated at K120 [134]. H2A ubiquitination at K119 was understood to be the only site of modification, but very recently two groups have reported a second site, K13/K15, as the site of ubiquitination by RNF168 during DDR [135, 136]. Regulation of H2A and H2B ubiquitination status plays a role in several nuclear processes in addition to DDR including transcriptional activation, gene silencing, cell cycle progression, and mitosis. While the precise functions of H2A/H2B ubiquitination in transcription remain largely ambiguous, ubiquitination of H2B is generally associated with actively transcribed genes and thought to function in transcriptional initiation, while ubiquitination of H2A is usually associated with silenced genes, including X-inactivated genes and developmentally regulated genes [20, 134]. Ubiquitination of chromatin is one of several post-translational modifications to occur on histones, and the cross-talk between these epigenetic marks collectively orchestrates the aforementioned processes.
3.3.1 USP7, USP16, and BAP1 are Chromatin-Associated DUBs regulating HOX genes
There are nine DUBs in humans that have been shown to act upon ubiquitinated H2A or H2B – USP3, USP7, USP16, USP21, USP22, USP44, 2A-DUB, BRCC36 and BAP1 (see Table 1). USP3 was identified in HeLa chromatin extracts and its depletion elevates the levels of ubiquitinated H2A and H2B, delays S-phase progression and induces the DNA damage response [137]. USP21 deubiquitinates H2A during hepatocyte regeneration to activate gene transcription, and it localizes to centrosomes ensuring proper microtubule dynamics [138, 139]. 2A-DUB, a JAMM family DUB, was found to deubiquitinate H2A and positively regulate transcription of androgen receptor regulated genes in concert with the histone acetylase p/CAF complex [140]. USP22 is a component of the SAGA transcriptional coactivator complex and can deubiquitinate H2A and H2B [141-143]. USP44 negatively regulates H2B ubiquitination during embryonic stem cell development [144]. Histone deubiquitination has been the subject of recent reviews [20, 134, 145], and here we highlight three DUBs, USP7, USP16, and BAP1, that function in polycomb group (PcG) complexes and modulate transcription of PcG target genes.
The ubiquitination of H2A-K119 by the E3 ligase RING2 (Ring1b) and its coactivator BMI1 has an established role in transcriptional repression of homeotic genes and in X chromosome inactivation [146-148]. Repression of these genes is accomplished by a group of polycomb group proteins (PcG) that were identified in Drosophila genetic screens as necessary to silence the expression of HOX genes and prevent homeotic transformations. PcG proteins assemble to form three distinct complexes in Drosophila, PhoRC, PRC1 and PRC2 [149-151]. PhoRC directly binds to polycomb response elements (PREs) within DNA and recruits PRC2 which contains H3-K27 trimethylase activity, and PRC1, which contains the H2A-K119 Ub E3 ligase complex Sce/Psc (RING2 and BMI1 in humans). An expansion of the PcG proteins in humans has led to multiple orthologs of their fly counterparts; for example, the PRC1 E3 ligase proteins Sce has two human paralogs (RING1 and RING2) and Psc has three (BMI1, MEL18, and NSPC1) [150]. Deubiquitination of H2A-K119 at PcG-regulated genes in flies has been attributed to a UCH DUB called Calypso, the homolog of human BAP1, which associates with the PRC2 complex by binding to the Asx protein [152]. In humans USP7 and USP11 co-purify with PRC1 proteins and indirectly regulate expression of PcG target genes [153]. Another DUB, USP16, has been shown to regulate the expression of human HOXD10 [154], but its recruitment to PcG complexes is less understood.
3.3.1.1. BAP1
In flies, chromatin-IP (ChIP) studies found the Calypso/Asx complex co-localized with PcG proteins Pho (of PhoRC) and Ph (of PRC1) at the PREs of several PcG protein targets including HOX genes [152]. Examination of the HOX Ubx gene in cells where expression is either active or inactive found that Calypso/Asx bound to the Ubx PRE in both cases [152]. Loss of Calypso in larval imaginal discs, where Ubx is normally repressed, led to activation of Ubx expression and this was rescued by transgene expression of wild type Calypso but not the active site Cys mutant. Thus the localization of Calypso/Asx alone does not dictate whether Ubx is activated or repressed, but loss of Calypso leads to transcriptional activation. Loss of Asx in flies led to an increase in Ub-H2A levels without influencing other chromatin marks (H3K4 me3, H3K27me3), and assays using purified proteins found Asx stimulates Calypso activity towards Ub-AMC, and that Asx/Calypso and the human orthologs BAP1/ASXL1 deubiquitinate H2A but not H2B in reconstituted nucleosomes [152]. The influence of BAP1 and ASXL1 on HOX gene expression has also been examined by ChIP in human hematopoietic cells. In these studies, depletion of BAP1 does not influence expression from the HoxA gene cluster, however depletion of ASXL1 reduces H3K27me3 levels and the presence of PRC2 components while enhancing H3K4me3 levels, Ub-H2A levels, and transcription of HoxA genes [155]. Taken together, these results show that the BAP1/ASXL1 complex in both humans and flies functions in repressing Hox gene expression, although the precise temporal epigenetic modifications differ between organisms.
BAP1 is believed to have gained additional functions in eukaryotes because, unlike Calypso, it contains an HCF-1 binding motif (HBM) known to mediate BAP1 binding to HCF-1 in mice and humans [36, 37]. HCF-1 is a transcriptional regulator that can bind a host of transcription factors as well as activating and repressing chromatin-modifying complexes [156]. ChIP studies in mice have found that BAP1 and HCF-1 co-localize to 3800 gene promoters, though it is not known whether ASXL1 is also present in these complexes [157]. The large number of genes thought to be regulated by BAP1 suggests it plays critical role in the cell, and this is proving to be true as mutations within the BAP1 gene have been linked to a number of cancers, including lung adenocarcinoma, uveal melanoma, clear cell renal cell carcinomas, malignant mesothelioma, and novel melanocytic tumors [46, 158-161]. Germline mutations to BAP1 predisposes to some of the aforementioned cancers [162-165]. BAP1 knockout mice are embryonic-lethal, and inducible knockout of BAP1 led to myeloid transformations characteristic of human chronic myelocytic leukemia, a disease recently linked to ASXL1 mutations in humans [155, 157].
3.3.1.2. USP16 (Ubp-M)
In a search for DUBs that could deubiquitinate H2A, fractionation of HeLa cell H2A DUB activity led to the isolation of USP16 [154]. USP16 is specific for Ub-H2A, as it deubiquitinates nucleosomal Ub-H2A in vitro and its depletion in cells elevates Ub-H2A levels without influencing Ub-H2B [154]. Examination of the HOXD10 gene expression found depletion of USP16 led to an increase in its expression, and this defect was rescued by re-expression of the wild type enzyme, but not the active site Cys mutant. ChIP studies on HOXD10 binding of USP16 and the BMI1 subunit of PRC1 found both proteins are localized to the HOXD10 promoter, yet H2A was not ubiquitinated unless USP16 was depleted. Because BMI1 promoter occupancy was unaffected in USP16-depleted cells, these finding suggest DUB activity counteracts PRC1-mediated ubiquitination to maintain a repressed state of transcription [154]. USP16 was also identified in a mitotic phosphoprotein screen where it was shown to be phosphorylated in prometaphase and metaphase, to bind mitotic chromosomes and to deubiquitinate isolated chromatin [166]. USP16 regulates chromatin condensation during mitosis by deubiquitinating H2A, an activity that precedes H3-S10 phosphorylation by the Aurora B kinase [154], a hallmark of condensed metaphase chromosomes [167]. Intriguingly, USP16 contains an N-terminal ZnF-UBP domain known to recognize the C-terminal residues of unanchored Ub (-RLRGG) [119, 168]. This is an unexpected feature for an enzyme that doesn’t involve acting on a free Ub chain. However, a recent study has found that ZnF-UBP domains can bind C-terminal diglycine sequences present in other proteins with similar affinity to Ub, and that USP16 binds favorably to such a motif present in histone H4 (-YGFGG) [169]. USP16 was shown to pull-down recombinant H3/H4 tetramer, suggesting it is recruited to its target H2A by the Znf-UBP-histone H4 interaction. In support of this finding, a USP3 ZnF-UBP domain mutation in a conserved histidine that coordinates Zn2+ abolished its ability to IP histones H2A and H2B [137].
3.3.1.3 USP7/HAUSP
Purification of the Psc orthologs BMI1 and MEL18 identified several PRC1 components along with two DUBs, USP7 and USP11. Pull-downs with recombinant proteins found both DUBs are capable of directly associating with other PRC1 members and each other suggesting they bind multiple proteins within the PRC1 complex. Examination of the PRC1-regulated INK4a locus found depletion of both USP7 and USP11 resulted in expression of p16INK4a at the transcript and protein level, and decreased binding of PRC1 members at the INK4a locus as assessed by ChIP. Though recombinant USP7 was capable of deubiquitinating H2A in nucleosomes, its depletion had little effect on cellular Ub-H2A or Ub-H2B levels, but did destabilize BMI1 and MEL18 protein levels [153]. Thus these DUBs influence expression from PcG-regulated promoters by stabilizing PRC1 components rather than directly acting on Ub-H2A. Although overexpression or depletion of USP7 had no effects on Ub-H2A or Ub-H2B levels in this study, USP7 has been shown to shown to form a complex with the Epstein-Barr virus (EBV) protein EBNA1and human GMP synthase that deubiquitinates histone H2B leading to expression of EBV genes [170]. USP7 was also found to associate with and deubiquitinate the PRC1 E3 ligase RING2, and this activity functions to stabilize RING2. USP7 was indiscriminate towards chain types, cleaving proteasome-targeting K48 chains catalyzed by the E3 E6AP, and branched K6-, K27-, and K48 chains catalyzed by auto-ubiquitination [171].
3.4. Vectoral Processes
Because of the spatial distribution of E3s and DUBs, and the existence of numerous ubiquitin receptors, this modification offers an ideal system for regulating vectoral processes that result in transport of a protein from one part of a cell to another. A classic example is in the endocytic pathway where transport and degradation of cargo proteins depends on ubiquitination at the cell surface, ubiquitin receptor binding in early endosomes, and deubiquitination at the late endosome [10, 172]. A variation of this pathway is also important in viral budding [173], autophagy [174] and cytokinesis [175].
3.4.1. Sorting of proteins to the vacuole/lysosome
A variety of cell surface receptors, especially the receptor tyrosine kinases such as EGFR, are ubiquitinated by E3 ligases such as the oncogene c-Cbl in response to receptor engagement, and this Ub is used as a sorting tag to direct the protein through the endocytic pathway to the lysosome for degradation [10, 176]. Monoubiquitination and K63-linked polyubiquitination are most often observed. A number of endosomal sorting complexes required for transport (ESCRTs) containing Ub-binding domains are thought to ferry the ubiquitinated cargo to the multivesicular body (MVB) where it is internalized before the MVB fuses with the lysosome [176]. This Ub must be removed from the cargo for efficient internalization by the MVB. The timing of deubiquitination is critical; if it occurs early then the receptor can be recycled to the cell surface, while failure to remove it can consume Ub and slow lysosomal degradation [10, 176].
3.4.1.1. USP8 and AMSH regulate endocytosis and lysosomal degradation of endocytic cargo
Two DUBs, USP8 and AMSH, have been implicated in this pathway based on genetic and biochemical evidence. Both bind to the STAM subunit of ESCRT-0 at the sorting endosome and to CHMPS components of ESCRT-III during formation of the MVB [10, 172]. AMSH exhibits specificity for K63-linked chains while USP8 can cleave most types of poly-Ub [81, 177]. A precise definition of the roles of these two DUBs is complicated by the fact that their effects on endocytosis are dependent on the identity of the substrate and ubiquitination can occur at several points in the cargo’s journey. Nevertheless, we can generalize that AMSH probably counteracts the activity of membrane localized E3 ligases and enhances recycling of the receptor, as well as inhibiting binding of Vps4 to ESCRT-III, resulting in failure to dissociate ESCRT-III complex necessary for sorting [10]. Endocytic defects observed upon loss of USP8 are believed to primarily impact the ESCRT-0 complex, however misregulated receptor internalization has also been observed. USP8 depletion results in enlarged and aberrant endosomes that contain elevated levels of ubiquitinated proteins, including the sorting protein Eps15, and decreased levels of STAM2 and Hrs [10, 178-180]. USP8 deubiquitinates STAM, preventing its degradation by the proteasome [179], and Nrdp1, an E3 required for the lysosomal degradation of EGFR family members ErbB3 and ErbB4 [181].
3.4.1.2. Ataxin3-Crosstalk between proteasomal and lysosomal autophagy pathways
In addition to endocytosis, substrates can be targeted to the lysosome by formation of autophagic vesicles. Autophagy proceeds by formation of a double-membrane vesicle, often around a cellular organelle or deposit, and then fusion with the lysosome. For many years it was assumed that proteasomal and lysosomal degradation were distinct unrelated pathways. However, there is now significant evidence that the two interact and that ubiquitin-dependent events are important in each [182]. Impairment of each upregulates the other, both utilize polyubiquitin signals (K63 for autophagy and K48 for proteasomal degradation), and many substrates seem to be degraded by both pathways. Further, the p62/sequestosome polyubiquitin binding protein plays a role in delivering substrates to each process [183].
The best understood connection between these pathways is seen when misfolded proteins accumulate in the cell, especially disease-causing polyglutamine repeat proteins that aggregate in Amyotrophic Lateral Sclerosis, Alzheimer, Parkinson, and Huntington diseases [184]. Aggregated proteins can be refolded by chaperones, cleared by the proteasome or autophagy or accumulated at the microtubule organizing center in a large inclusion body called the aggresome. Formation of the aggresome is thought to sequester the aggregates in a non-lethal form [185] and the balance between these pathways probably depends on DUBs that can remodel, remove or edit polyubiquitin chains. The Ataxin-3 DUB associates with parkin, HDAC6 and other aggresome components and its activity enhances aggresome formation by misfolded superoxide dismutase [186] and the cystic fibrosis transmembrane regulator [187]. It is hypothesized that Ataxin-3 trims K63-linked chains from the misfolded ubiquitinated proteins and enhances the rate of aggresome formation [187].
3.5. Proteasome bound DUBs
The 26S proteasome is an ATP-dependent, multi-subunit protease that primarily functions to degrade poly-ubiquitinated proteins. It can be subdivided into two complexes, the 20S core particle (CP) and the 19S regulatory particle (RP). The 28 subunit 20S CP is formed by four heptameric rings that stack to form a barrel-like structure enclosing three protease sites within its interior lumen. Access to the 20S lumen is regulated by the ATP-driven 19S RP which opens a translocation channel, unfolds and directs substrates into the CP interior. The 19S regulatory particle (19 subunits in yeast) also functions in the recognition and deubiquitination of proteasome substrates. In humans three DUBs from different families, UCH37/UCH-L5 (UCH), USP14 (USP), and POH1/RPN11 (JAMM/MPN+), associate with the proteasomal 19S RP. These enzymes are well conserved in eukaryotes with the exception of S.cerevisiae which lacks a UCH37 homolog [188]. They differ in several aspects with regard to their necessity, role, and catalytic mechanism. Of the three, only RPN11 is an essential, stoichiometric component, while UCH37 and USP14 transiently associate and co-purify with proteasomes to different extents in different organisms [41, 189]. A separate review in this issue covers this topic in much more detail (Finley, this volume).
3.5.1. RPN11 removes poly-Ub to facilitate coupled translocation and proteolysis
One function of the proteasome-associated DUBs is to remove the poly-Ub chain from substrates prior to completing degradation. This activity serves to prevent undesired degradation of Ub, but also facilitates unfolding and translocation of the substrate through the small pore at the end of the 20S protease. In the absence of these DUB activities, the proteasome must unfold both Ub and the substrate, translocating both polypeptides into the CP lumen [188]. This significantly slows degradation of the substrate and results in the proteolytic loss of Ub. Conversely, if the Ub tag is removed before substrate is engaged by the protease, degradation may be incomplete or fail entirely due to dissociation of the substrate.
RPN11 is the DUB largely responsible for removing poly-Ub from substrate, although USP14 may also contribute since Ub levels drop in its absence [189-191]. The metalloprotease activity of RPN11 was first noticed when treatment of proteasomes with Ub-aldehyde and Ub-VS failed to inhibit degradation of model substrates [75, 76]. RPN11 acts as an endopeptidase, cleaving poly-Ub chains en bloc from substrates, and its activity within proteasomes is dependent on ATP-hydrolysis and intact proteasomes [75, 76]. Thus RPN11 functions after the proteasome has engaged the substrate and is committed to proteolysis, a mechanism that prevents dissociation of the deubiquitinated substrate and averted degradation. RPN11 is essential for viability in yeast [192], and its depletion in HeLa markedly elevates cellular poly-Ub levels, impairs proteasome assembly, and inhibits cell growth [189].
3.5.2. All three proteasomal DUBs play a role in chain editing to assure fidelity of proteolysis
The K63-linked polyubiquitin chain is not an efficient degradation signal, in spite of the fact that it is efficiently bound by the proteasome, RPN11 displays highly specific K63 poly-Ub endopeptidase activity, as purified 19S particles treated with NEM are capable of cleaving K63 isopeptide bonds in di-Ub and in a mixed K48/K63 tetra-Ub chain [80]. As substrates bearing K63 poly-Ub most often are not destined for proteasomal degradation, RPN11 could contribute a “proof reading” function by disassembling K63-linkages and preventing degradation of substrates with this tag.
UCH37 (and possibly USP14) can act as ATP-independent exopeptidases, trimming distal Ub progressively from a substrate-anchored poly-Ub chain [38]. If the polyubiquitin chain is long enough, it can remain bound until the substrate is productively engaged and then removed by RPN11 during normal proteolysis the proteasome. If translocation and proteolysis stalls, the abortive degradation intermediate needs to be cleared and this trimming will continue to shorten the chain. Substrates that have short poly-Ub chains have a weaker affinity for the proteasome [193] and are more likely to be released from the proteasome rather than degraded. UCH37 associates with the 19S regulatory particle via interaction with ADRM1/hRPN13, and that this interaction requires a KEKE motif within the UCH37 C-terminal extension [42-44]. The C-terminal extension holds UCH37 in an inactive state, and its deletion or engagement with hRPN13 stimulates Ub-AMC hydrolysis [42, 43]. UCH37 is also a component of the INO80 chromatin remodeling complex, where its C-terminal extension mediates binding to the INO80 subunit NFRKB [41]. When bound to INO80, or NFRKB alone, UCH37 is inactive towards Ub-AMC; this inhibition is relieved by co-associating with hRPN13 or purified proteasomes [41]. UCH37 is more abundant in proteasomes from bovine blood compared to HeLa cells, and its high prevalence in HeLa INO80 complexes has suggested it recruits UCH37-less proteasomes to INO80 to degrade yet-to-be identified chromatin targets [41].
USP14, and its yeast ortholog UBP6, require an N-terminal Ub-like (Ubl) domain for association with the 19S particle (to the RPN1 subunit) and their activity towards Ub-AMC is stimulated 300-800-fold when associated with proteasomes [191, 194]. Deletion of yeast UBP6 results in a Ub-depletion phenotype, probably from a failure to remove short polyubiquitin chains from bound substrates and their subsequent degradation by the proteasome. In yeast, UBP6 delays proteasomal degradation of cyclin B, and this delay requires an intact Ubl domain and proteasomal association. Intriguingly, the degradation delay is also observed in the absence of a catalytic cysteine, attributed to a non-catalytic mechanism of RPN11 inhibition [195].
Finally, it should be noted that these observations suggest a complex coupling and interplay between and among the catalytic particle, the 19S regulatory complex, and these three DUBs. These interactions are much more completely discussed elsewhere in this issue (Finley, this volume).
4. Perspective
Ubiquitin-dependent processes are crucial to all cellular functions. The assembly of a Ub or poly-Ub tag is a targeting signal that regulates activity, localization, protein-protein interactions and half-life. Several hundred ubiquitin ligases and nearly a hundred deubiquitinating enzymes control these modifications. These enzymes are temporally and spatially controlled and most often act as part of multi-protein complexes. Thus, there has been much interest in these pathways as drug targets. This survey of DUB action in the proteolysis pathway highlights important problems that must be overcome to achieve the necessary specificity of drug action. A major challenge is designing drugs that will interfere with nearly a thousand enzymes that all act by a handful of chemical mechanisms. Another is the fact that a single DUB can have many substrates and a single substrate can be the target of several DUBs. Nonetheless, very similar challenges exist is manipulating the kinase/phosphatase regulated pathways and those enzymes have proven to be amenable targets in treating important pathologies.
Highlights.
Nearly a hundred deubiquitinating enzymes from five gene families regulate proteolysis by:
directly interacting with and co-regulating E3 ligases;
altering the level of substrate ubiquitination;
hydrolyzing or remodeling ubiquitinated and poly-ubiquitinated substrates;
acting in specific locations in the cell and altering the localization of the target protein;
acting on proteasome bound substrates to facilitate or inhibit proteolysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- [2].Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2011;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun. 2007;354:329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- [7].Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- [8].Goldknopf IL, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci U S A. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wright DE, Wang CY, Kao CF. Histone ubiquitylation and chromatin dynamics. Frontiers in bioscience: a journal and virtual library. 2012;17:1051–1078. doi: 10.2741/3973. [DOI] [PubMed] [Google Scholar]
- [10].J M, Clague S. Urbe, Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- [11].Harhaj EW, Dixit VM. Regulation of NF-kappaB by deubiquitinases. Immunol Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 2009;8:436–443. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
- [13].Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fushman D, Wilkinson KD. Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 biology reports. 2011;3:26. doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Emmerich CH, Schmukle AC, Walczak H. The emerging role of linear ubiquitination in cell signaling. Science signaling. 2011;4:re5. doi: 10.1126/scisignal.2002187. [DOI] [PubMed] [Google Scholar]
- [16].Iwai K. Diverse ubiquitin signaling in NF-kappaB activation. Trends in cell biology. 2012;22:355–364. doi: 10.1016/j.tcb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- [17].van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- [18].Schulman BA. Twists and turns in ubiquitin-like protein conjugation cascades. Protein Sci. 2011;20:1941–1954. doi: 10.1002/pro.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- [22].Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- [23].Ye Y, Scheel H, Hofmann K, Komander D. Dissection of USP catalytic domains reveals five common insertion points. Mol Biosyst. 2009;5:1797–1808. doi: 10.1039/b907669g. [DOI] [PubMed] [Google Scholar]
- [24].Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. Embo J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Storer AC, Menard R. Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 1994;244:486–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- [26].Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- [27].Lee JG, Baek K, Soetandyo N, Ye Y. Reversible inactivation of deubiquitinases by reactive oxygen species in vitro and in cells. Nat Commun. 2013;4:1568. doi: 10.1038/ncomms2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kulathu Y, Garcia FJ, Mevissen TE, Busch M, Arnaudo N, Carroll KS, Barford D, Komander D. Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nat Commun. 2013;4:1569. doi: 10.1038/ncomms2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cotto-Rios XM, Bekes M, Chapman J, Ueberheide B, Huang TT. Deubiquitinases as a signaling target of oxidative stress. Cell Rep. 2012;2:1475–1484. doi: 10.1016/j.celrep.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- [31].Sommer S, Weikart ND, Linne U, Mootz HD. Covalent inhibition of SUMO and ubiquitin-specific cysteine proteases by an in situ thiol-alkyne addition. Bioorg Med Chem. 2013;21:2511–2517. doi: 10.1016/j.bmc.2013.02.039. [DOI] [PubMed] [Google Scholar]
- [32].Ekkebus R, van Kasteren SI, Kulathu Y, Scholten A, Berlin I, Geurink PP, de Jong A, Goerdayal S, Neefjes J, Heck AJ, Komander D, Ovaa H. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J Am Chem Soc. 2013;135:2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. Embo J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wilkinson KD, Gan-Erdene T, Kolli N. Derivitization of the C-terminus of ubiquitin and ubiquitin-like proteins using intein chemistry: methods and uses. Methods Enzymol. 2005;399:37–51. doi: 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
- [35].Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- [36].Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, Liu J, O'Rourke K, Dixit VM, Wilson AC. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Molecular and cellular biology. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. The Journal of biological chemistry. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- [39].Zhou ZR, Zhang YH, Liu S, Song AX, Hu HY. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem J. 2012;441:143–149. doi: 10.1042/BJ20110699. [DOI] [PubMed] [Google Scholar]
- [40].Popp MW, Artavanis-Tsakonas K, Ploegh HL. Substrate filtering by the active site crossover loop in UCHL3 revealed by sortagging and gain-of-function mutations. J Biol Chem. 2009;284:3593–3602. doi: 10.1074/jbc.M807172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- [43].Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. Embo J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. Embo J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, J F, 3rd, Drobetsky E, Milot E, Shi Y, Affar B. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Molecular and cellular biology. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N, Berger SL, Prendergast GC, J F., 3rd BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- [47].Nishikawa H, Wu W, Koike A, Kojima R, Gomi H, Fukuda M, Ohta T. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 2009;69:111–119. doi: 10.1158/0008-5472.CAN-08-3355. [DOI] [PubMed] [Google Scholar]
- [48].Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- [50].Avvakumov GV, Walker JR, Xue S, Allali-Hassani A, Asinas A, Nair UB, Fang X, Zuo X, Wang YX, Wilkinson KD, Dhe-Paganon S. Two ZnF-UBP domains in isopeptidase T (USP5) Biochemistry. 2012;51:1188–1198. doi: 10.1021/bi200854q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. Embo J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Makarova KS, Aravind L, Koonin EV. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci. 2000;25:50–52. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- [53].Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Edelmann MJ, Iphofer A, Akutsu M, Altun M, di Gleria K, Kramer HB, Fiebiger E, Dhe-Paganon S, Kessler BM. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- [56].Licchesi JD, Mieszczanek J, Mevissen TE, Rutherford TJ, Akutsu M, Virdee S, El Oualid F, Chin JW, Ovaa H, Bienz M, Komander D. An ankyrin-repeat ubiquitin-binding domain determines TRABID’s specificity for atypical ubiquitin chains. Nat Struct Mol Biol. 2012;19:62–71. doi: 10.1038/nsmb.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- [58].Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nanao MH, Tcherniuk SO, Chroboczek J, Dideberg O, Dessen A, Balakirev MY. Crystal structure of human otubain 2. EMBO Rep. 2004;5:783–788. doi: 10.1038/sj.embor.7400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, Shanks JR, Reyes-Turcu FE, Wilkinson KD, Marmorstein R. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–11049. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lin SC, Chung JY, Lamothe B, Rajashankar K, Lu M, Lo YC, Lam AY, Darnay BG, Wu H. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol. 2008;376:526–540. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O’Donnell L, Kumakubo A, Munro M, Sicheri F, Gingras AC, Natsume T, Suda T, Durocher D. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- [63].Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- [64].Weeks SD, Grasty KC, Hernandez-Cuebas L, Loll PJ. Crystal structure of a Josephin-ubiquitin complex: evolutionary restraints on ataxin-3 deubiquitinating activity. J Biol Chem. 2011;286:4555–4565. doi: 10.1074/jbc.M110.177360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tzvetkov N, Breuer P. Josephin domain-containing proteins from a variety of species are active de-ubiquitination enzymes. Biol Chem. 2007;388:973–978. doi: 10.1515/BC.2007.107. [DOI] [PubMed] [Google Scholar]
- [66].Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mao Y, Senic-Matuglia F, Di Fiore PP, Polo S, Hodsdon ME, De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc Natl Acad Sci U S A. 2005;102:12700–12705. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Berke SJ, Chai Y, Marrs GL, Wen H, Paulson HL. Defining the role of ubiquitin-interacting motifs in the polyglutamine disease protein, ataxin-3. J Biol Chem. 2005;280:32026–32034. doi: 10.1074/jbc.M506084200. [DOI] [PubMed] [Google Scholar]
- [69].Matos CA, de Macedo-Ribeiro S, Carvalho AL. Polyglutamine diseases: the special case of ataxin-3 and Machado-Joseph disease. Prog Neurobiol. 2011;95:26–48. doi: 10.1016/j.pneurobio.2011.06.007. [DOI] [PubMed] [Google Scholar]
- [70].Scheel H, Tomiuk S, Hofmann K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum Mol Genet. 2003;12:2845–2852. doi: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- [71].Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- [72].Nicastro G, Menon RP, Masino L, Knowles PP, McDonald NQ, Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc Natl Acad Sci U S A. 2005;102:10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tran HJ, Allen MD, Lowe J, Bycroft M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry. 2003;42:11460–11465. doi: 10.1021/bi035033g. [DOI] [PubMed] [Google Scholar]
- [75].Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- [76].Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- [77].Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lipscomb WN, Strater N. Recent Advances in Zinc Enzymology. Chem Rev. 1996;96:2375–2434. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- [79].Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- [80].Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. Embo J. 2009;28:621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- [83].Echalier A, Pan Y, Birol M, Tavernier N, Pintard L, Hoh F, Ebel C, Galophe N, Claret FX, Dumas C. Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1. Proc Natl Acad Sci U S A. 2013;110:1273–1278. doi: 10.1073/pnas.1209345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Davies CW, Paul LN, Kim MI, Das C. Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability. J Mol Biol. 2011;413:416–429. doi: 10.1016/j.jmb.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Tanaka K, Mizushima T, Saeki Y. The proteasome: molecular machinery and pathophysiological roles. Biological chemistry. 2012;393:217–234. doi: 10.1515/hsz-2011-0285. [DOI] [PubMed] [Google Scholar]
- [86].Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem J. 2008;414:161–175. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- [89].Enchev RI, Scott DC, da Fonseca PC, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP. Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2012;2:616–627. doi: 10.1016/j.celrep.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- [91].Lee MH, Zhao R, Phan L, Yeung SC. Roles of COP9 signalosome in cancer. Cell Cycle. 2011;10:3057–3066. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- [93].Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys. 2011;60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- [94].Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- [95].Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- [96].Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- [97].Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- [98].Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- [99].Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, Gao Y, Ran Y, Tien P, Shu HB. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–4297. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Stanisic V, Malovannaya A, Qin J, Lonard DM, O’Malley BW. OTU Domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) alpha and affects ERalpha transcriptional activity. J Biol Chem. 2009;284:16135–16145. doi: 10.1074/jbc.M109.007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Edelmann MJ, Kramer HB, Altun M, Kessler BM. Post-translational modification of the deubiquitinating enzyme otubain 1 modulates active RhoA levels and susceptibility to Yersinia invasion. FEBS J. 2010;277:2515–2530. doi: 10.1111/j.1742-4658.2010.07665.x. [DOI] [PubMed] [Google Scholar]
- [102].Durcan TM, Kontogiannea M, Bedard N, Wing SS, Fon EA. Ataxin-3 deubiquitination is coupled to Parkin ubiquitination via E2 ubiquitin-conjugating enzyme. J Biol Chem. 2012;287:531–541. doi: 10.1074/jbc.M111.288449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. Embo J. 2012;31:576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Juang YC, Landry MC, Sanches M, Vittal V, Leung CC, Ceccarelli DF, Mateo AR, Pruneda JN, Mao DY, Szilard RK, Orlicky S, Munro M, Brzovic PS, Klevit RE, Sicheri F, Durocher D. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol Cell. 2012;45:384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Wiborg O, Pedersen MS, Wind A, Berglund LE, Marcker KA, Vuust J. The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. Embo J. 1985;4:755–759. doi: 10.1002/j.1460-2075.1985.tb03693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Doran JF, Jackson P, Kynoch PA, Thompson RJ. Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis. J Neurochem. 1983;40:1542–1547. doi: 10.1111/j.1471-4159.1983.tb08124.x. [DOI] [PubMed] [Google Scholar]
- [108].Sekiguchi S, Kwon J, Yoshida E, Hamasaki H, Ichinose S, Hideshima M, Kuraoka M, Takahashi A, Ishii Y, Kyuwa S, Wada K, Yoshikawa Y. Localization of ubiquitin C-terminal hydrolase L1 in mouse ova and its function in the plasma membrane to block polyspermy. Am J Pathol. 2006;169:1722–1729. doi: 10.2352/ajpath.2006.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun. 1998;251:688–692. doi: 10.1006/bbrc.1998.9532. [DOI] [PubMed] [Google Scholar]
- [110].Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, Wada K. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- [111].Lowe J, McDermott H, Landon M, Mayer RJ, Wilkinson KD. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J Pathol. 1990;161:153–160. doi: 10.1002/path.1711610210. [DOI] [PubMed] [Google Scholar]
- [112].Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- [113].Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, Cookson MR, Liu Y, Logan T, Lansbury PT., Jr. Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proc Natl Acad Sci U S A. 2009;106:4635–4640. doi: 10.1073/pnas.0806474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030–5041. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Wilkinson KD, Tashayev VL, O’Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- [116].Falquet L, Paquet N, Frutiger S, Hughes GJ, Hoang-Van K, Jaton JC. A human de-ubiquitinating enzyme with both isopeptidase and peptidase activities in vitro. FEBS Lett. 1995;359:73–77. doi: 10.1016/0014-5793(94)01451-6. [DOI] [PubMed] [Google Scholar]
- [117].Hadari T, Warms JV, Rose IA, Hershko A. A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J Biol Chem. 1992;267:719–727. [PubMed] [Google Scholar]
- [118].Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- [119].Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- [120].Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- [122].Stewart GS. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 2009;8:1532–1538. doi: 10.4161/cc.8.10.8351. [DOI] [PubMed] [Google Scholar]
- [123].Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–728. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci U S A. 2009;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 Critically Regulates Inflammasome Activity. Mol Cell. 2013;49:331–338. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- [128].Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Shembade N, Harhaj NS, Parvatiyar K, Copeland NG, Jenkins NA, Matesic LE, Harhaj EW. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- [130].Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, Kirkpatrick DS, Dixit VM, Hymowitz SG. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- [134].Atanassov BS, Koutelou E, Dent SY. The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett. 2011;585:2016–2023. doi: 10.1016/j.febslet.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Gatti M, Pinato S, Maspero E, Soffientini P, Polo S, Penengo L. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 2012;11:2538–2544. doi: 10.4161/cc.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- [137].Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- [138].Urbe S, Liu H, Hayes SD, Heride C, Rigden DJ, Clague MJ. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol Biol Cell. 2012;23:1095–1103. doi: 10.1091/mbc.E11-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, Ito T. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008;7:1522–1524. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- [143].Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, Feldmesser E, Brik A, Yu X, Hanna J, Aberdam D, Domany E, Oren M. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Vissers JH, Nicassio F, van Lohuizen M, Di Fiore PP, Citterio E. The many faces of ubiquitinated histone H2A: insights from the DUBs. Cell Div. 2008;3:8. doi: 10.1186/1747-1028-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- [147].Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- [148].Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- [149].Majewski IJ, Ritchie ME, Phipson B, Corbin J, Pakusch M, Ebert A, Busslinger M, Koseki H, Hu Y, Smyth GK, Alexander WS, Hilton DJ, Blewitt ME. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood. 2010;116:731–739. doi: 10.1182/blood-2009-12-260760. [DOI] [PubMed] [Google Scholar]
- [150].Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- [151].Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- [152].Scheuermann JC, Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. Embo J. 2010;29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- [155].Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, Pandey S, Patel JP, Chung YR, Koche R, Perna F, Zhao X, Taylor JE, Park CY, Carroll M, Melnick A, Nimer SD, Jaffe JD, Aifantis I, Bernstein BE, Levine RL. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- [157].Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, Wu J, Phu L, Katavolos P, LaFave LM, Abdel-Wahab O, Modrusan Z, Seshagiri S, Dong K, Lin Z, Balazs M, Suriben R, Newton K, Hymowitz S, Garcia-Manero G, Martin F, Levine RL, Dixit VM. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science (New York, N.Y.) 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, Leng N, Pavía-Jiménez A, Wang S, Yamasaki T, Zhrebker L, Sivanand S, Spence P, Kinch L, Hambuch T, Jain S, Lotan Y, Margulis V, Sagalowsky AI, Summerour PB, Kabbani W, Wong SW, Grishin N, Laurent M, Xie X, Haudenschild CD, Ross MT, Bentley DR, Kapur P, Brugarolas J. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44:1–12. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science (New York, N.Y.) 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Wiesner T, Murali R, Fried I, Cerroni L, Busam K, Kutzner H, Bastian BC. A distinct subset of atypical Spitz tumors is characterized by BRAF mutation and loss of BAP1 expression. Am J Surg Pathol. 2012;36:818–830. doi: 10.1097/PAS.0b013e3182498be5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, Creaney J, Lake RA, Zakowski MF, Reva B, Sander C, Delsite R, Powell S, Zhou Q, Shen R, Olshen A, Rusch V, Ladanyi M. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, Windpassinger C, Wackernagel W, Loy S, Wolf I, Viale A, Lash AE, Pirun M, Socci ND, Rütten A, Palmedo G, Abramson D, Offit K, Ott A, Becker JC, Cerroni L, Kutzner H, Bastian BC, Speicher MR. Germline mutations in BAP1 predispose to melanocytic tumors. Nature genetics. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, Cox NJ, Dogan AU, Pass HI, Trusa S, Hesdorffer M, Nasu M, Powers A, Rivera Z, Comertpay S, Tanji M, Gaudino G, Yang H, Carbone M. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Goldstein AM. Germline BAP1 mutations and tumor susceptibility. Nature genetics. 2011;43:925–926. doi: 10.1038/ng.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB, Christopher BN, Boru G, Hovland P, Davidorf FH. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856–859. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Cai SY, Babbitt RW, Marchesi VT. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc Natl Acad Sci U S A. 1999;96:2828–2833. doi: 10.1073/pnas.96.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Johansen KM, Johansen J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 2006;14:393–404. doi: 10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- [168].Pai MT, Tzeng SR, Kovacs JJ, Keaton MA, Li SS, Yao TP, Zhou P. Solution structure of the Ubp-M BUZ domain, a highly specific protein module that recognizes the C-terminal tail of free ubiquitin. J Mol Biol. 2007;370:290–302. doi: 10.1016/j.jmb.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Hard RL, Liu J, Shen J, Zhou P, Pei D. HDAC6 and Ubp-M BUZ domains recognize specific C-terminal sequences of proteins. Biochemistry. 2010;49:10737–10746. doi: 10.1021/bi101014s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog. 2009;5:e1000624. doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].de Bie P, Zaaroor-Regev D, Ciechanover A. Regulation of the Polycomb protein RING1B ubiquitination by USP7. Biochem Biophys Res Commun. 2010;400:389–395. doi: 10.1016/j.bbrc.2010.08.082. [DOI] [PubMed] [Google Scholar]
- [172].Wright MH, Berlin I, Nash PD. Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination. Cell Biochem Biophys. 2011;60:39–46. doi: 10.1007/s12013-011-9181-9. [DOI] [PubMed] [Google Scholar]
- [173].Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Rusten TE, Filimonenko M, Rodahl LM, Stenmark H, Simonsen A. ESCRTing autophagic clearance of aggregating proteins. Autophagy. 2008;4 [Google Scholar]
- [175].Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. Embo J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Niendorf S, Oksche A, Kisser A, Lohler J, Prinz M, Schorle H, Feller S, Lewitzky M, Horak I, Knobeloch KP. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27:5029–5039. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- [180].Mizuno E, Kobayashi K, Yamamoto A, Kitamura N, Komada M. A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes. Traffic. 2006;7:1017–1031. doi: 10.1111/j.1600-0854.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- [181].Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol Cell Biol. 2004;24:7748–7757. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- [183].Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- [184].Olzmann JA, Li L, Chin LS. Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem. 2008;15:47–60. doi: 10.2174/092986708783330692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Swinnen E, Buttner S, Outeiro TF, Galas MC, Madeo F, Winderickx J, Franssens V. Aggresome formation and segregation of inclusions influence toxicity of alpha-synuclein and synphilin-1 in yeast. Biochem Soc Trans. 2011;39:1476–1481. doi: 10.1042/BST0391476. [DOI] [PubMed] [Google Scholar]
- [186].Wang H, Ying Z, Wang G. Ataxin-3 regulates aggresome formation of copper-zinc superoxide dismutase (SOD1) by editing K63-linked polyubiquitin chains. The Journal of biological chemistry. 2012;287:28576–28585. doi: 10.1074/jbc.M111.299990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Burnett BG, Pittman RN. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc Natl Acad Sci U S A. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.R110.003871. R110 003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Chernova TA, Allen KD, Wesoloski LM, Shanks JR, Chernoff YO, Wilkinson KD. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. The Journal of biological chemistry. 2003;278:52102–52115. doi: 10.1074/jbc.M310283200. [DOI] [PubMed] [Google Scholar]
- [191].Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Rinaldi T, Ricci C, Porro D, Bolotin-Fukuhara M, Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol Biol Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Piotrowski J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. The Journal of biological chemistry. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- [194].Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- [195].Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- [196].van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- [197].Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- [199].Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J Cell Biol. 2009;184:13–19. doi: 10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L, Hu S, Zeng P, Guo R, Diao J, Lan F, Harper JW, Shi YG, Xu Y, Shi Y. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci U S A. 2012;109:4828–4833. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. Embo J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, Frappier L. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278:29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- [203].Soares L, Seroogy C, Skrenta H, Anandasabapathy N, Lovelace P, Chung CD, Engleman E, Fathman CG. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat Immunol. 2004;5:45–54. doi: 10.1038/ni1017. [DOI] [PubMed] [Google Scholar]
- [204].Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- [205].Hasdemir B, Murphy JE, Cottrell GS, Bunnett NW. Endosomal deubiquitinating enzymes control ubiquitination and down-regulation of protease-activated receptor 2. J Biol Chem. 2009;284:28453–28466. doi: 10.1074/jbc.M109.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr., Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J Biol Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- [207].Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, Persaud A, Walker JR, Neculai AM, Neculai D, Vorobyov A, Garg P, Beatty L, Chan PK, Juang YC, Landry MC, Yeh C, Zeqiraj E, Karamboulas K, Allali-Hassani A, Vedadi M, Tyers M, Moffat J, Sicheri F, Pelletier L, Durocher D, Raught B, Rotin D, Yang J, Moran MF, Dhe-Paganon S, Sidhu SS. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339:590–595. doi: 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Xu G, Tan X, Wang H, Sun W, Shi Y, Burlingame S, Gu X, Cao G, Zhang T, Qin J, Yang J. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J Biol Chem. 2010;285:969–978. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Ye Y, Akutsu M, Reyes-Turcu F, Enchev RI, Wilkinson KD, Komander D. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson’s disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006;103:4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Boudreaux DA, Maiti TK, Davies CW, Das C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc Natl Acad Sci U S A. 2010;107:9117–9122. doi: 10.1073/pnas.0910870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- [213].Nishio K, Kim SW, Kawai K, Mizushima T, Yamane T, Hamazaki J, Murata S, Tanaka K, Morimoto Y. Crystal structure of the de-ubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain. Biochem Biophys Res Commun. 2009;390:855–860. doi: 10.1016/j.bbrc.2009.10.062. [DOI] [PubMed] [Google Scholar]
- [214].Burgie SE, Bingman CA, Soni AB, Phillips GN., Jr. Structural characterization of human Uch37. Proteins. 2011 doi: 10.1002/prot.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Landreville S, Agapova OA, Matatall KA, Kneass ZT, Onken MD, Lee RS, Bowcock AM, Harbour JW. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin Cancer Res. 2012;18:408–416. doi: 10.1158/1078-0432.CCR-11-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- [217].Duwel M, Welteke V, Oeckinghaus A, Baens M, Kloo B, Ferch U, Darnay BG, Ruland J, Marynen P, Krappmann D. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- [218].Wang Q, Li L, Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Yoneyama M, Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Matsuda T, Watanabe S, Tomo Y, Nishimura Y, Harada T, Terada T, Shirouzu M, Hayashizaki Y, Ohara O, Tanaka A, Kigawa T, Yokoyama S. Structural and functional differences of SWIRM domain subtypes. J Mol Biol. 2007;369:222–238. doi: 10.1016/j.jmb.2007.03.027. [DOI] [PubMed] [Google Scholar]
- [220].Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, Forster F. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Sierra MI, Wright MH, Nash PD. AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR4. J Biol Chem. 2010;285:13990–14004. doi: 10.1074/jbc.M109.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]