Abstract
With the emergence of the mouse as the predominant model system for studying mammalian brain development, in utero imaging methods are urgently required to analyze the dynamics of brain growth and patterning in mouse embryos. To address this need, we combined synthetic focusing with a high-frequency (38-MHz) annular-array ultrasound imaging system for extended depth-of-field, coded excitation for improved penetration, and respiratory-gated transmit-receive. This combination allowed noninvasive in utero acquisition of motion-free, three-dimensional data from individual embryos in approximately 2 minutes, and data from 4 or more embryos in a pregnant mouse in less than 30 minutes. Data were acquired from 148 embryos spanning 5 days of early-to-mid gestational stages of brain development. The results showed that brain anatomy and cerebral vasculature can be imaged with this system, and that quantitative analyses of segmented cerebral ventricles can be used to characterize volumetric changes associated with mouse brain development.
Keywords: annular array, central nervous system, synthetic focusing, coded excitation, respiratory gating, mouse embryo
INTRODUCTION
The central nervous system (CNS) begins during embryogenesis as a simple neural tube and develops into the morphologically complex three-dimensional (3D) brain and spinal cord through cell proliferation, differentiation and migration, spatially and temporally coordinated by a host of signaling molecules. An understanding of the molecular and genetic basis of the morphological changes in the embryonic CNS has been of great interest to developmental neurobiologists. Due to the high degree of homology between the mouse and human genomes and the numerous methods available for manipulating the mouse genome, the mouse has been the genetic organism of choice for studying mammalian brain development. As a result, many genes have been identified that are critical for normal brain development, and large-scale efforts are underway to produce knockout mice for each of the approximately 23,000 mouse genes, many of which are expressed in the CNS (Collins et al., 2007). While these advances have led to critical new insights into the molecular basis of brain development, morphological changes in mouse mutants are usually examined with time-consuming histological methods, generating two-dimensional (2D) sections in standard orientations for qualitative, descriptive analysis by expert anatomists. There is a clear need for high-resolution, high-throughput 3D imaging methods that are capable of acquiring in vivo data for quantitative analysis of the mouse embryonic brain, allowing in utero assessment of dynamic changes in brain morphology during normal brain development and in a wide variety of neurological mouse mutants.
During the critical periods of early embryonic development, the morphology of the mammalian CNS is dominated by the cerebral ventricles, filled with cerebral spinal fluid (CSF) and surrounded by a thin neuroepithelium. Since the first reports of knockout mice with brain defects, it has been recognized that among the earliest detectable phenotypes is the altered size and shape of the cerebral ventricles (Thomas and Capecchi, 1990; McMahon and Bradley, 1990; Wurst et al., 1994). Indeed, there is growing evidence that normal brain development relies on the coordinated growth and patterning of CSF and neuroepithelium during embryogenesis (Gato and Desmond, 2009). Defects in embryonic neurogenesis in mouse models of Down syndrome have been linked to postnatal enlargement of the cerebral ventricles (Ishihara et al., 2010), and changes in ventricle volume have been observed in children with neurological disorders such as Tourette syndrome (Peterson et al., 2001). Clinical ultrasound (5- to 10-MHz) studies have used 3D ventricle segmentation to assess normal (Monteagudo and Timor-Tritsch, 2009; Kim et al., 2008) and abnormal fetal brain development (Timor-Tritsch et al., 2008), starting from late first trimester. In one of the few quantitative reports, a 7.5-MHz transvaginal annular-array probe was used to measure volumes of the human cerebral ventricles between 7–10 weeks of gestation (Blaas et al., 1998). Despite these successes, fetal ultrasound currently has very little applicability in genetic screening for abnormal brain development, due to a basic lack of understanding of how abnormal morphological changes relate to neurodevelopmental diseases. Thus, animal models are essential to increase our understanding of the relationships between altered CNS morphology and specific diseases.
High-frequency (30- to 50-MHz) ultrasound (HFU) imaging has become a standard tool for real-time, in vivo imaging of mouse embryos (Phoon, 2006; Nieman and Turnbull, 2010) at high spatial resolution (< 100-μm). In the first report of HFU for mouse embryo imaging (Turnbull et al., 1995), the potential for imaging the developing CNS was already recognized, including 3D visualization of the cerebral ventricles in normal and Wnt1 null mutant embryos, the first knockout mice with recognized brain defects (Thomas and Capecchi, 1990; McMahon and Bradley, 1990). However, this early work was based on an imaging technology that used fixed-focus transducers with narrow depth-of-field (DOF; 1–2 mm at 40-MHz), which limited the 3D morphometric analysis to images acquired ex vivo after careful positioning of the excised uterus and embryos within the DOF of the 40-MHz transducer (Turnbull et al., 1995). In utero HFU has also been used to quantify (live) mouse embryo growth parameters over a wide range of fetal stages (Mu et al., 2008), although the imaging system employed single-element, fixed-focus transducers with a narrow DOF and the reported in vivo measurements were limited to 2D parameters. Recent improvements in HFU transducer technology have significantly improved the focusing characteristics, making it feasible to perform in vivo 3D analysis of mouse embryo development. Specifically, HFU annular arrays (Brown et al., 2004; Ketterling et al., 2005; Snook et al., 2006) and linear arrays (Cannata et al., 2006; Brown et al., 2007; Foster et al., 2009) have increased the DOF to 6-mm or more at 40 MHz. Both theoretical simulations (Ramachandran and Ketterling, 2007) and phantom experiments (Filoux et al., 2011; Filoux et al., 2012) indicate that the annular arrays produce a more symmetrically-focused beam, resulting in better image quality and increased 3D spatial resolution compared to linear arrays. We previously showed the potential of HFU annular arrays for in utero 3D brain imaging, providing more accurate segmentations of cerebral ventricles compared to an equivalent fixed-focus transducer (Aristizábal el al., 2006). However, due to limitations in the scanner frame rate and a lack of motion-gated acquisition, those previous results were acquired using a semi-invasive, non-in vivo method.
In this report, we expand on our previous results by implementing an improved HFU annular-array system and by acquiring a more extensive set of 3D data that span five days of early-to-mid gestational mouse brain development. The current HFU annular-array system provides improved acoustic penetration and signal-to-noise ratio through the implementation of chirp-coded excitation (Mamou et al., 2009), and respiratory-gated acquisition has been implemented to remove motion artifacts. The combination of these two techniques now permits noninvasive in utero imaging of multiple live embryos in each pregnant mouse, and combined with other system software and hardware modifications, allows full, motion-free 3D data sets to be acquired from each embryo in approximately two minutes. The aim of this study was to first validate our imaging protocol, and then to acquire in utero and in vivo 3D data sets and to establish quantitative 3D parameters to characterize mouse embryonic brain development between gestational days 10 to 14.
MATERIALS AND METHODS
Annular array imaging system
A custom HFU system, which was described previously in detail (Ketterling et al., 2006; Filoux et al., 2011), was utilized for these experiments. The annular arrays were made by bonding a 9-μm poly(vinylidene difluoride-co-trifluoroethylene), P(VDF-TrFE) membrane (Piezotech S.A.S., Hésingue France) to a single-sided, copper-clad-polyimide film (RFlex 1000L810, Rogers Corp., Chandler AZ) after etching the copper with an array pattern. The transducer array was packaged in a brass tube and electrical connections were incorporated with standard coaxial cabling (Fig. 1a). The annular-array transducer was mounted to a high-speed linear actuator (LAS35, SMAC, Carlsbad CA) and scanned over the imaging plane. One element was pulsed and the received radio-frequency (RF) echoes were simultaneously digitized on all five elements. This pulsing scheme was repeated for each of the five array elements, and a complete 2D image required five passes with a total scan time of 0.5 s. Beamforming was accomplished in post processing using a synthetic-focusing algorithm (Ketterling et al., 2006; Aristizábal et al., 2006), where the appropriate time delays were applied to the RF lines for each of the 25 transmit/receive combinations at a series of focal depths and then summed to create the final image. As previously reported (Aristizábal et al., 2006), this imaging scheme increased the DOF from 1-mm to near 6-mm, with the lateral resolution of 80-μm maintained over the entire DOF. The annular-array transducer used for these experiments had five, equal-area elements, with a 38-MHz center frequency, a 12-mm geometric focus and a 6-mm aperture (Ketterling et al., 2005). All ultrasound images were compressed logarithmically and displayed with a dynamic (gray-scale) range of 60-dB.
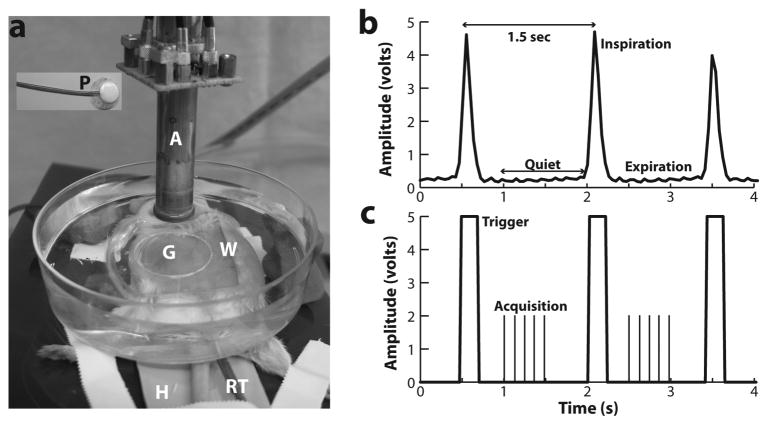
Figure 1. Annular array HFU imaging setup and data acquisition.
(a) The experimental setup is shown, with the array transducer (A) scanning through a water bath (W) and gel membrane (G) mounted over the shaved abdomen of an anesthetized pregnant mouse. Labels: H, heating pad; P, respiratory pillow; RT, rectal temperature probe. (b) A typical measured respiratory signal (inspiration + expiration), with a period close to 1.5s. (c) The digital waveform, derived from the respiratory signal, and used to trigger respiratory-gated acquisition. The vertical lines between triggers represent the acquisition of data, during the “quiet” (expiration) phase of the respiratory cycle, for each of the 5 scans of the transducer.
Chirp-coded Excitation
The annular-array was excited with a linear, chirp-coded excitation pulse engineered for high-frequency imaging (Mamou et al., 2009). A linear chirp was parameterized by the frequency band, B=f2−f1, where f1 and f2 were the starting and ending frequencies, respectively, and the sweep time was T. The set of optimal parameters were determined empirically to be f1=15MHz, f2=65MHz and T=4μs, where the chirp was modulated with a 9%-Tukey tapering window. Before processing the array data with the synthetic-focusing algorithm, a compression filter consisting of a time-reversed excitation chirp, weighted by a Chebyshev window, was applied to each RF line to recover the axial resolution.
Respiratory gating
During in vivo imaging, the mother’s respiratory motion imparted a displacement to the embryo of up to 1-mm in the depth direction. The respiration rate was approximately one breath each 1.5 s with a quiet phase of about 1.1 s and a movement phase of 0.4 s (Fig. 1b). With the improvements in scan rate of our system, we were able to acquire a full set of data for one scan plane (5 passes in 0.5s) during the quiet phase of the respiratory cycle (Fig. 1c). This greatly improved the frame-rate compared to the earlier system, which permitted just one pass for each respiration cycle or roughly 8s to collect all the data for each scan plane.
For respiratory-gated acquisition, a respiratory pillow was placed under the mouse (Fig. 1a) and connected to a differential pressure transducer (TSD160A, BIOPAC Systems, Goleta CA). Using BIOPAC acquisition software (Version 3.73), respiratory waveforms were digitized at 100 samples per second (Fig. 1b), and a threshold was used to generate a transistor-transistor logic (TTL) pulse corresponding to the active phase of the respiration cycle (Fig. 1c). The high-to-low transition of the TTL pulse was detected by our imaging system, and an extra time delay of 300ms was applied before a complete array data set was acquired and stored.
Animals and in utero data acquisition
All animals used in these studies were maintained under protocols approved by the Institutional Animal Care and Use Committee at New York University School of Medicine. Timed pregnant Swiss Webster mice (Taconic, Hudson NY) were imaged between embryonic day (E)10.5 and E14.5, where E0.5 was defined as noon of the day after successful overnight mating. Under isoflurane anesthesia (4% in air for induction, 1–2% in air for maintenance), pregnant mice were placed on a custom stage and heated to maintain the internal body temperature at 37±1°C. To acoustically couple the annular-array transducer to the mouse, the array was lowered into a water bath positioned over the shaved abdomen. The self-contained water bath was made by cutting a 25-mm hole in a plastic Petri dish and creating a water-tight seal by gluing a 40-mm square, 2-mm thick membrane (Aquaflex@ultrasound gel pad; Parker Laboratories, Fairfield NJ) over the hole in the Petri dish.
To locate the desired image plane and to position the embryo for a 3D scan, the central (circular) array element was used in a real-time, fixed-focus scout mode that operated at 10 frames per second. After positioning with the 2D scout mode, a full 3D chirp data set was acquired with 50-μm or 100-μm spacing between adjacent scan planes at a sampling rate of 250MHz. Each B-scan was approximately 7-mm wide and 9-mm deep, and complete 3D data sets (100 slices) were acquired in 2.2 minutes. Data were acquired from a total of 45 pregnant mice (Table 1), where 1–6 embryos were imaged in each mouse (average 3–4 embryos per mouse). High-quality imaging of 4–6 embryos (approximately 50% of the litter) from a single mouse could be achieved in less than 30 minutes (22/45 mice).
Table 1.
Summary of the embryos imaged and analyzed.
| Embryonic Stage | # Mothers/Litters | # Embryos |
|---|---|---|
|
| ||
| E10.5 | 7 | 24 |
| E11.5 | 10 | 39 |
| E12.5 | 13 | 42 |
| E13.5 | 10 | 29 |
| E14.5 | 5 | 14 |
3D image analysis and quantification
After offline synthetic focusing, 3D stacks of B-mode images were imported into Amira visualization software (v5.2, Mercury Computer Systems, San Diego CA) for volumetric visualization, segmentation and quantitative analysis. Before segmentation, the volumetric data were processed with a 3D Gaussian filter and a contrast-limited adaptive histogram equalization (CLAHE) filter (Zuiderveld, 1994) was applied to optimize contrast between cerebral ventricles and the surrounding tissue. Semi-automatic segmentation of the head and ventricles was performed by choosing voxels within the region of interest and applying a 3D threshold detection algorithm with an empirically-determined window of image gray levels. After the contours were segmented, Amira software was used to reconstruct the head and ventricles in 3D, and volume and surface areas were computed for each data set. In addition to surface area and volume, other morphometric parameters such as the pontine flexure angle were also measured from 3D HFU image data.
RESULTS
Respiratory-gated in utero acquisition
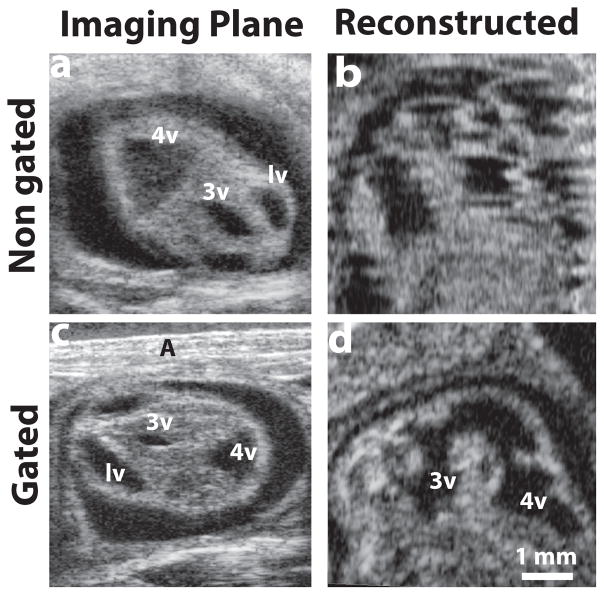
High-quality, 3D ultrasound imaging depends critically on acquiring motion-corrected data. For in utero HFU imaging of early stage mouse embryos, the major source of embryonic motion was the cyclic displacement caused by the mother’s respiration. Other sources of motion included slow contractions of the uterus and the independent motion of the embryo within the uterus, especially at later stages (≥E13.5). With 3D volume acquisition time reduced to ~2 minutes, these uterine/embryonic motions perpendicular to the image plane were generally negligible. Transient out-of-plane motion also resulted from the resistance of the water bath to the mouse abdomen during the active phase of the respiratory cycle, which was most noticeable in the real-time 2D imaging mode. We found empirically that adding a 300-ms time delay, after the gate signal before data acquisition, was sufficient to allow the uterus and embryo to relax back into its resting position after each breathing event. Without gating, respiratory motion resulted in significant distortion in reconstructed image planes (Fig 2a, b). By implementing respiratory gating, motion artifacts were significantly reduced or eliminated, resulting in high-quality reconstructions (Fig. 2c, d). Occasionally, random (non-cyclic) gasping of the mouse generated motion artifacts in both the acquisition and reconstructed planes (not shown), but these were easily detected during data acquisition and were corrected by reacquiring the full 3D dataset during the imaging session. Using the respiratory-gated protocol, high-quality volumetric data were acquired from mouse embryos at each stage between E10.5 and E14.5.
Figure 2. Respiratory-gating to minimize motion artifacts.
(a) A selected plane from the acquired 3D stack without gating, and (b) a reconstructed sagittal plane of the same E12.5 mouse embryo, demonstrating obvious motion artifacts. After implementing respiratory gating, (c) a similar acquired plane of an E12.5 mouse embryo, and (d) the motion-corrected sagittal reconstruction. Labels: A, abdominal wall; lv, lateral ventricle; 3v, third ventricle; 4v, fourth ventricle.
In utero neuroimaging in mouse embryos
In utero 3D images were acquired, covering the head and brain of 148 mouse embryos in 45 mothers, distributed between stages E10.5 to E14.5 (Table 1). For each embryo/3D dataset, the RF line spacing and inter-plane spacing were both set to 50-μm, and 100 image planes were acquired in 2.2 minutes. A dominant feature of the embryonic mouse CNS is the cerebral ventricular system, which are cavities filled with CSF surrounded by the neuroepithelium (presumptive brain) and mesechymal tissue (embryonic head). The ventricles make up a significant volume of the embryonic brain, largely determining the shape at early stages (E10.5–11.5), and undergoing dramatic morphological changes over the developmental stages investigated (Fig. 3). On HFU, the hypo-intense fluid-filled ventricles had high contrast to the surrounding tissues, and could be easily segmented from 3D images for qualitative and quantitative analyses (Fig. 3).
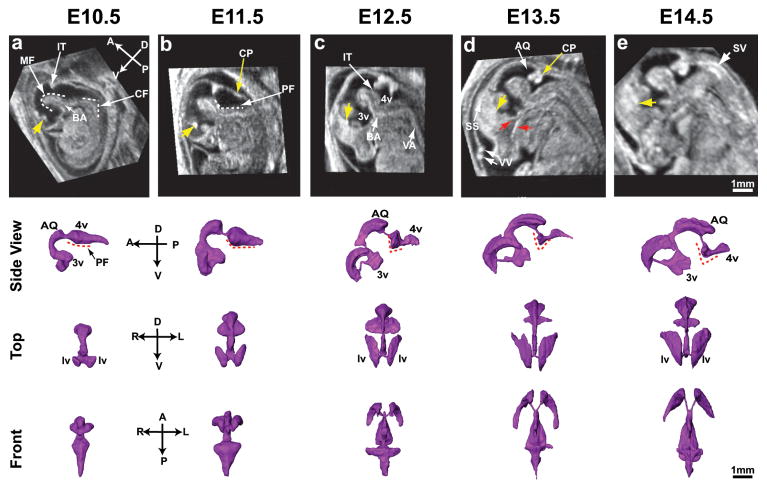
Figure 3. 3D in utero neuroimaging of the embryonic mouse brain.
Similarly oriented mid-sagittal sections of embryonic heads and surface renderings (viewed from the side, top and front) of the cerebral ventricles at five gestational stages: (a) E10.5; (b) E11.5; (c) E12.5; (d) E13.5; (e) E14.5. Anatomic axes are indicated as anterior-posterior (A-P), dorsal-ventral (D-V) and right-left (R-L). Labels: AQ, aqueduct; BA, basilar artery; cervical flexure, CF; choroid plexus, CP; IT, isthmus; lv, lateral ventricle; MF, mesencephalic flexure; PF, pontine flexure; SS, superior sagittal sinus; SV, spinal vein; VA, vertebral artery; VV, vitelline vessels; 3v, third ventricle; 4v, fourth ventricle. Yellow arrows (a–d) indicate a cross section of the CP in the lateral ventricles; red arrows (d) indicate facial blood vessels. Dashed lines show the MF, PF and CF flexure angles. Scale bars (e) = 1-mm.
A number of anatomical features in the developing mouse brain were readily visualized on 2D and 3D HFU annular-array images (Fig. 3). The anterior-posterior organization of forebrain, midbrain and hindbrain was obvious from the earliest stages. The patterning of the CSF into lateral ventricles, third ventricle, midbrain aqueduct and fourth ventricle was particularly well visualized, as were brain structures such as the mid-hindbrain isthmus that project into the CSF. 3D segmentations of the cerebral ventricles made it much easier to visualize the profound changes in ventricular patterning during embryonic brain development, which was difficult to appreciate from 2D images. From both 3D segmentations and mid-sagittal images, it was apparent that a number of anatomical parameters could be derived as potential quantitative indices of mouse embryonic brain development, including ventricular volume, surface area and angles such as the mesencephalic, pontine and cervical flexures. The pontine flexure angle was particularly interesting as a potential marker of embryonic stage, starting as a nearly flat (wide angle) indentation at E10.5, and becoming more angular with increasing stages.
Interestingly, many hyper-intense cerebral blood vessels were also visualized (Fig. 3), including the choroid plexus in the lateral (at all stages) and fourth (starting at E12.5) ventricles, the basilar, vertebral and other arteries, and the sagittal sinus, nasal vein and other venous vessels in the brain and head. These results suggest that HFU annular-array imaging has potential for future in utero analyses of interrelated brain and vascular patterning during embryogenesis.
Quantitative analysis of cerebral ventricles
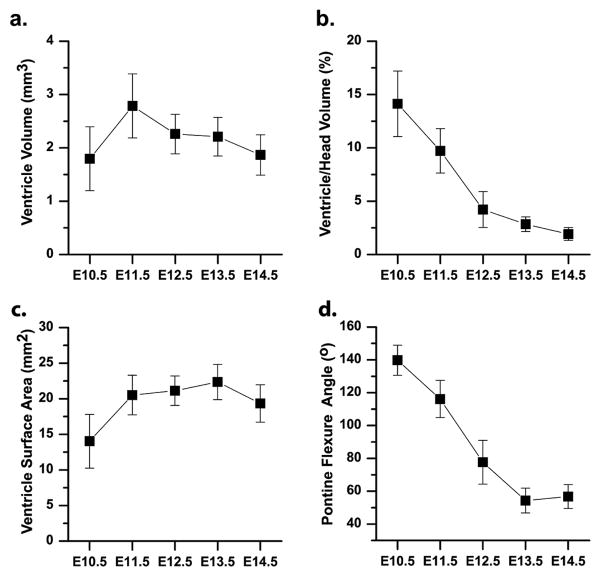
The 3D in utero HFU annular-array data acquired from E10.5–14.5 mouse embryos were analyzed to establish quantitative parameters characteristic of normal brain development. The cerebral ventricles were segmented from each of the 148 data sets (Table 1), and geometrical parameters such as the ventricular volume, surface area, and pontine flexure angle were computed and plotted (mean ± standard deviation) as a function of embryonic stage (Fig. 4). Ventricular volume peaked at E11.5, increasing by close to 75% from E10.5 to E11.5, and then decreasing slowly until E14.5, to a value close to that at E10.5 (Fig. 4a).
Figure 4. Quantitative analysis of developmental morphometric parameters.
(a) The volume of the cerebral ventricles was measured as a function of embryonic stage, from E10.5 to E14.5 (Table 1). (b) In selected embryos (N=6 at each stage), the ventricle volume was normalized to the head volume. (c) The surface area of the brain ventricles and (d) the pontine flexure angle were also measured at each stage (Table 1). In each case, the error bars indicate the standard deviation of the measurement.
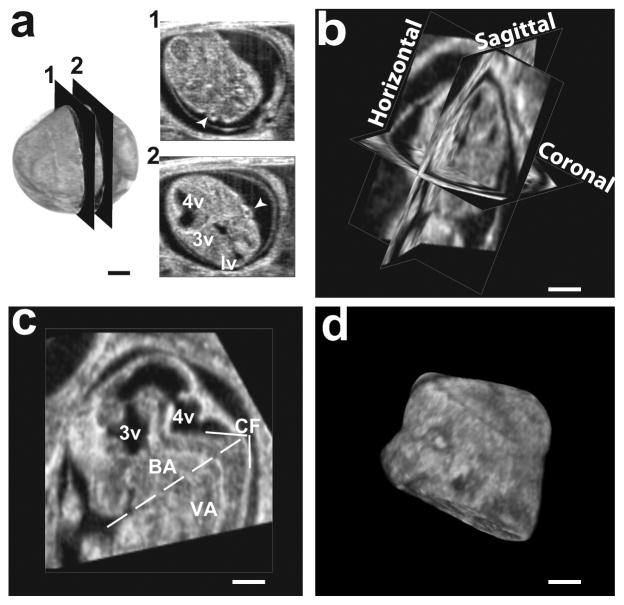
To assess changes in the ventricles relative to the surrounding tissues, we segmented the head and normalized the ventricular volume to the head volume in selected embryos (N=6 embryos, distributed over 6 different mothers, at each embryonic stage; Fig. 4b). Consistent segmentation of the embryonic heads required transformation of each 3D dataset from the acquired oblique sections to a standard orientation in which the horizontal and coronal planes both bisected the embryonic eyes (Fig. 5a, b). Using mid-sagittal sections after this reorientation, each head was digitally separated from the body by a plane that bisected the cephalic flexure and was tangential to the snout (Fig. 5c). The embryonic heads were then manually segmented from the remaining extra-embryonic tissue (Fig. 5d), and the volumes calculated. The ventricular volume was close to 15% of the head volume at E10.5, and decreased over the developmental stages up to E14.5 (Fig. 4b).
Figure 5. Protocol used to orient the embryonic heads for segmentation.
(a) 3D data from an E12.5 head, where the acquisition planes are in non-standard orientations, as demonstrated by the left and right eyes (arrowheads) appearing in separated sections 1 and 2. Labels: lv, lateral ventricle; 3v, third ventricle; 4v, fourth ventricle. (b) After rigid-body transformation, the 3D data was in standard orientation, with the eyes bilaterally symmetric in both coronal and horizontal sections. (c) The bottom of the head was defined by a plane, orthogonal to the sagittal plane and containing the line that bisected the cervical flexure (CF) and was tangential to the base of the snout. Other labels: BA, basilar artery; VA, vertebral artery. (d) Volumetric rendering of the segmented head. Scale bars (a–d) = 1-mm.
For morphologically complex 3D anatomical structures such as the cerebral ventricles, surface area is another traditional parameter that can be measured (Fig. 4c). The surface area increased by close to 45% between E10.5 and E11.5, and then remained constant up to E13.5, slightly decreasing between E13.5 and E14.5. As a quantitative index of embryonic stage, measurements of the pontine flexure angle in the mid-sagittal section showed a steady linear decrease between E10.5 and E13.5, and then remained constant between E13.5 and E14.5 (Fig. 4d).
3D registration decreases segmentation errors
Segmentation errors leading to inaccurate surface morphology of the embryonic cerebral ventricles were mostly due to reduced contrast as a result of the irregular contour of the ventricle-tissue interface caused by the ultrasound speckle pattern. A potential strategy to smooth the speckle pattern and increase the ventricle-tissue contrast is to average multiple 3D HFU images after alignment through an image registration process. To test this, we acquired images of the heads of four E12.5 littermate embryos, which were then aligned (as shown in Fig. 6) and registered in 3D prior to averaging (Amira). Anatomical variability between different embryos resulted in significant blurring after volume averaging with 6-parameter, rigid-body registration (not shown). To correct for this inherent variability between embryos, a nonlinear 12-parameter affine registration algorithm was employed, using normalized mutual information as the metric (Amira). Mid-sagittal sections after 3D registration and averaging showed increased ventricle-tissue contrast compared to images of individual embryos (Fig. 6a, b). Surface renderings of the segmented cerebral ventricles showed reduced local artifacts in the averaged images (Fig. 6c, d). The registration-averaging process also improved the visualization of blood vessels in the mouse embryonic brain.
Figure 6. 3D registration and averaging reduces segmentation errors.
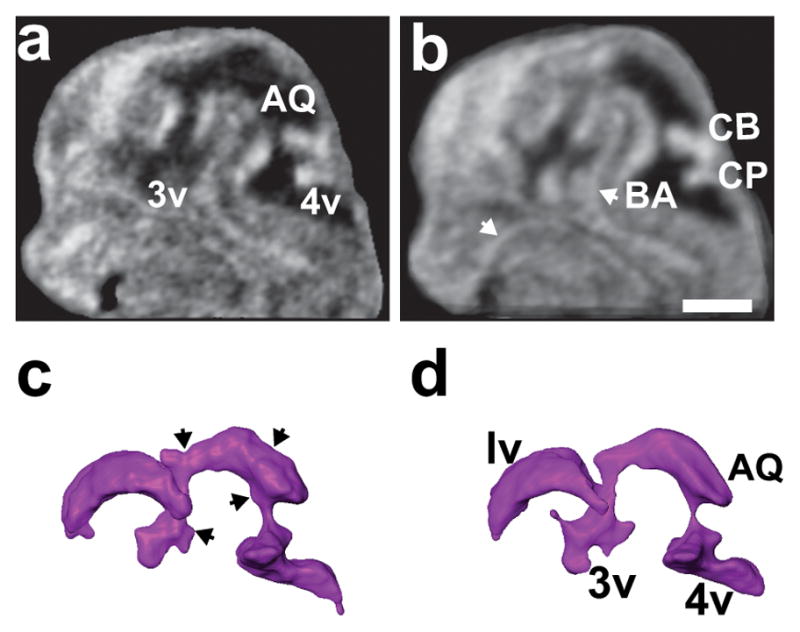
Mid-sagittal sections of (a) an individual E12.5 embryo head, and (b) the average of four littermate embryos after voxel-based intensity registration. The registration-averaging significantly smoothed the borders of the ventricles and improved visualization of cerebral blood vessels (white arrows). (c, d) Reconstructions of the brain ventricles in the individual embryo (c) showed a number of surface irregularities (black arrows) that were eliminated by registration-averaging (d). Labels: AQ, aqueduct; BA, basilar artery; CB, cerebellum; CP, choroid plexus; 3v, third ventricle; 4v, fourth ventricle. Scale bar (b) = 1-mm.
Whole embryo imaging
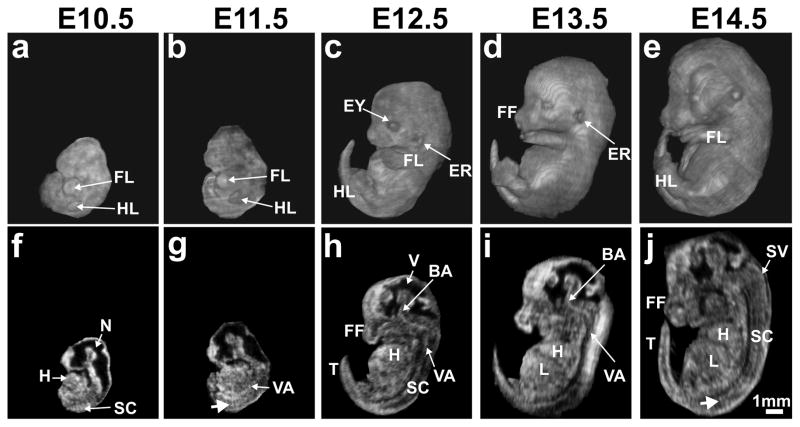
Although the main focus of the current study was to establish the utility of 3D HFU annular array imaging for in utero analysis of mouse brain development, we also acquired whole embryo images at each stage between E10.5 and E14.5 to assess the range of anatomical features and phenotypes that might be examined in the future with this approach. In the acquisition planes, RF line spacing of 50-μm was the same for all stages. Inter-plane spacing was chosen as 50-μm for early (E10.5–11.5) and 100-μm for later (E12.5–14.5) stage embryos, and the number of image planes was kept constant at 100, which required 2.2 minutes per 3D acquisition. For all stages, the volumetric data were transformed to a common coordinate system in Amira, and each embryo was manually segmented from the surrounding tissues. Whole embryos were then visualized using texture-based volume renderings and 2D sections at each stage (Fig. 7).
Figure 7. Whole embryo 3D in utero annular array HFU imaging.
Volume reconstructions (a–e) and the corresponding mid-sagittal sections (f–j) from array-focused data for the following embryonic stages: (a, f) E10.5; (b, g) E11.5; (c, h) E12.5; (d, i) E13.5; (e, j) E14.5. Labels: BA, basilar artery; ER, ear; EY, eye; FF, facial features; FL, forelimb; H, heart; HL, hindlimb; L, liver; N, neural tube; SC, spinal cord; SV, spinal vein; T, tail; V, (brain) ventricle; VA, vertebral artery. White arrows (g, j) indicate intersomitic blood vessels. Scale bar (j) = 1-mm.
In addition to the obvious increase in size with increasing embryonic stage, volume renderings from in utero HFU annular-array images showed clearly the marked morphological changes in areas such as the head and limbs (Fig. 7a–e), consistent with the known developmental time-courses (Kaufman 1992). Specifically, the embryonic head and facial features underwent distinct changes in size and shape, and the emergence of the eyes and ears was apparent at these developmental stages. Similarly, the limbs continually grew in length between E10.5 and E14.5, with the forelimb approximately one day ahead of the hindlimb. The distal appendages were well visualized, beginning as paddle-like limb-buds at E10.5, developing into webbed structures by E12.5, and emerging as recognizable paws with separated digits by E14.5. Internal anatomical features were best appreciated in sections extracted from the 3D image data (Fig. 7f–j), which clearly demonstrated the developing brain, spinal cord, heart, liver and a number of blood vessels, including the aorta, cerebral vessels, vertebral arteries, and even the intersomitic vessels in the developing spinal cord. Although sagittal sections are shown in Fig. 3, the 3D HFU data could be sectioned in other standard orientations (coronal, horizontal) or in oblique sections to study a variety of developing organs and anatomical structures in mouse embryos, which should be useful for phenotype analysis in a wide variety of mouse mutants in the future.
DISCUSSION AND CONCLUSION
Previously, we demonstrated the feasibility of in utero 3D neuroimaging in mouse embryos using HFU (Turnbull et al., 1995; Turnbull, 1999), and showed the clear advantages of annular-array imaging (Aristizábal el al., 2006) and coded excitation (Mamou et al., 2009) for this application. This report extends our previous work, establishing a fully noninvasive, high-throughput acquisition system, based on HFU annular-array imaging with coded excitation. By incorporating respiratory gating and optimizing RF data acquisition within the respiratory cycle, the time to acquire volumetric data from each mouse embryo was reduced by a factor of five to just over 2 minutes. With our current annular array system, it is now possible to acquire high-resolution (80 μm lateral), high-quality 3D image data of the brain in 50% or more of the embryos in each mouse in less than half an hour. This is a significant advance for studies of normal mouse embryonic development (Phoon, 2006; Mu et al, 2008), and makes HFU annular-array imaging a feasible approach for future 3D in utero mutant phenotype analysis (Nieman and Turnbull, 2010).
As a tool for analyzing morphological phenotypes in mouse embryos, the HFU annular-array system has several advantages over other methods. First and foremost, HFU provides in vivo data, allowing the possibility of longitudinal imaging over several days of development in individual embryos. Although longitudinal imaging was not part of the current study, we did verify that specific embryos could be re-imaged during an acquisition session. In addition, previous reports have shown that longitudinal studies are feasible by careful mapping of embryo position relative to adjacent anatomical landmarks such as the bladder and other internal organs (Ji and Phoon, 2005). The most common approach for morphological analysis of mouse embryos is histology, which provides higher resolution data than HFU but is limited to ex vivo analysis at one stage per embryo. Histology also has inherent problems of tissue shrinkage and distortion that are avoided using in vivo ultrasound. Comparing data acquisition times, HFU requires just over 2 minutes to acquire 3D data from an embryo, while histological analysis requires several days to fix, dehydrate, embed, section and stain an embryo plus additional time to image/photograph the sections.
It is also worth comparing HFU annular-array imaging to MRI, which has been used for studying fixed brains of normal and mutant mice (Zhang et al., 2003; Chuang et al., 2011), and which is the only other imaging modality currently capable of noninvasive in utero acquisition of high-resolution (100–125 μm isotropic) whole-brain 3D data in live mouse embryos (Deans et al, 2008; Nieman et al., 2009; Berrios-Otero et al., 2012; Parasoglou et al., 2013). MRI has been demonstrated recently for in utero imaging over the same early stages of brain development as the current report (E10.5–14.5), providing sufficient resolution and endogenous contrast to segment the cerebral ventricles and to visualize cerebral vasculature (Parasoglou et al., 2013). However, imaging time is 2–2.5 h per embryo, which limits the utility of MRI since it is only practical currently to image a single embryo within each pregnant mouse. In the same amount of time required to obtain one 3D MRI dataset (2h), the HFU annular array system could potentially acquire 3D data from four pregnant mice, and from four or more embryos per mouse. Thus, HFU annular array imaging now provides a feasible approach for in vivo mutant-brain-phenotype analysis, similar to previous demonstrations of 2D HFU and Doppler ultrasound for in vivo analysis of cardiovascular phenotypes (Srinivasan et al., 1998; Phoon et al., 2004; Ji and Phoon, 2005). The ability to collect 3D data over short time frames also makes it practical to assess normal developmental variability in a statistically significant fashion. Interestingly, clinical studies have shown that 3D MRI and ultrasound provide similar results in assessing infant ventricle volume (Gilmore et al., 2001). These results suggest that there is no inherent disadvantage for ultrasound-based 3D analyses compared to MRI, at least in the cerebral ventricles, but obvious advantages in terms of acquisition time and throughput.
Our results provide important quantitative data for future studies of normal mouse brain development and for analysis of mutant phenotypes. Specifically, we characterized the volume and surface area of the cerebral ventricles, as well as changes in the pontine flexure angle, between the critical early-to-mid gestational stages E10.5–14.5 (Fig. 5). Surprisingly, despite marked changes in the morphology of the ventricles through these developmental stages, both the volume and surface area were relatively constant. Ventricle volume and surface area reported in adult mice are also only about 2x larger than these results in embryos (Ma et al, 2008), suggesting that the amount of CSF is highly regulated from very early in development and remains remarkably constant despite the enormous expansion of the brain and head. As biomarkers of gestational stage more reliable than the nominal “embryonic day” (E), we found that the normalized ventricle (ventricle/head) volume and the pontine flexure angle were promising indices that can be easily measured from in utero HFU images. These data should provide an important baseline for characterizing brain defects in a wide range of mouse mutant embryos. Our results also provide very useful landmarks for in utero manipulations of mouse embryos using HFU-guided injections. Indeed, future implementation of in utero injections on the 3D HFU annular-array system, together with a streamlined and rapid set of 3D visualization tools, should significantly improve the targeting accuracy of these procedures, which have traditionally used 2D HFU with single-element, fixed-focus transducers (Olsson et al., 1997; Liu et al., 1998).
In addition to the clear visualization and 3D quantification of the developing mouse cerebral ventricles, the synthetically-focused HFU annular-array data provided excellent visualization of a number of cerebral blood vessels such as the basilar artery, as well as the highly vascularized choroid plexus in the lateral and fourth ventricles. Although choroid plexus has been reported to be present in the lateral ventricles starting at E11.5 (Sturrock, 1979), we saw clear evidence that it begins to form as early as E10.5 (Fig. 4). Relevant to the visualization of blood vessels on ultrasound images is the fact that embryonic red blood cells are nucleated up to E14.5 (Henery and Kaufman, 1992), which results in an increased ultrasound backscatter signal at high frequencies that peaks at E13.5 (Le Floc’h et al., 2004). With fixed-focus HFU transducers, backscatter from blood shows similar grey scale level as tissue, but because blood is circulating the blood vessels can sometimes be differentiated from tissue on real-time images due to the changing speckle pattern. With the annular-array acquisition and synthetic-focusing approach, each image plane represents an average of 25 transmit-to-receive RF echoes and the speckle size in the vasculature is reduced, resulting in a slightly higher amplitude and more homogenous signal from the blood compared to the surrounding tissue, which provides an improved ability to image the embryonic vasculature based on the endogenous contrast in blood. It should also be noted that our recent generation of reporter mice for vascular imaging, based on biotinylation of vascular endothelial cells, provides new opportunities to analyze gene expression changes in the embryonic cerebral vasculature based on contrast enhancement from biotin-targeted microbubbles (Bartelle et al., 2012).
Finally, our results demonstrate that 3D image registration-averaging can be used to improve edge detection, signal-to-noise and segmentation in HFU images (Fig. 7). This is an exciting new development that may lead to advances in 3D ultrasound image processing and in the future creation of atlases of mouse brain development based on in vivo HFU data. It is likely that registration-averaging of 3D images acquired from an individual embryo during each acquisition session could provide improvements in image quality for in utero neuroimaging, similar to approaches currently employed for in utero MRI (Nieman et al, 2009; Berrios-Otero et al., 2012; Parasoglou et al., 2013). A full 3D acquisition on our HFU annular array system currently takes about 2 minutes with a five-pass scanning approach. By implementing a five-channel pulser and a single-pass scanning approach, we should be able to reduce this time by a factor of five. Averaging of 5–10 registered data sets for each embryo could then be accomplished in under 5 minutes (compared to the 2–2.5 h currently required for MRI), and would result in significant gains in signal-to-noise-ratio, contrast and the quality of segmentations from HFU data.
Acknowledgments
This research was supported in part by grants from the National Institutes of Health (EB008606 to JAK; NS038461 and HL078665 to DHT).
Abbreviations
- CNS
central nervous system
- CSF
cerebral spinal fluid
- DOF
depth-of-field
- HFU
high frequency ultrasound
- RF
radio-frequency
- 2D
two-dimensional
- 3D
three-dimensional
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aristizábal O, Ketterling JA, Turnbull DH. 40-MHz annular array imaging of mouse embryos. Ultrasound Med Biol. 2006;32:1631–37. doi: 10.1016/j.ultrasmedbio.2006.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelle BB, Berrios-Otero CA, Rodriguez JJ, Friedland AE, Aristizábal O, Turnbull DH. Novel genetic approach for in vivo vascular imaging in mice. Circ Res. 2012;110:938–47. doi: 10.1161/CIRCRESAHA.111.254375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrios-Otero CA, Nieman BJ, Parasoglou P, Turnbull DH. In utero phenotyping of mouse embryonic vasculature with MRI. Magn Reson Med. 2012;67:251–57. doi: 10.1002/mrm.22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaas HG, Eik-Nes SH, Berg S, Torp H. In vivo three-dimensional ultrasound reconstructions of embryos and early fetuses. Lancet. 1998;352:1182–86. doi: 10.1016/S0140-6736(98)03227-9. [DOI] [PubMed] [Google Scholar]
- Brown JA, Demore CE, Lockwood GR. Design and fabrication of annular arrays for high-frequency ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control. 2004;51:1010–17. doi: 10.1109/tuffc.2004.1324405. [DOI] [PubMed] [Google Scholar]
- Brown JA, Foster FS, Needles A, Cherin E, Lockwood GR. Fabrication and performance of a 40-MHz linear array based on a 1–3 composite with geometric elevation focusing. IEEE Trans Ultrason Ferroelectr Freq Control. 2007;54:1888–94. doi: 10.1109/tuffc.2007.473. [DOI] [PubMed] [Google Scholar]
- Cannata JM, Williams JA, Zhou Q, Ritter TA, Shung KK. Development of a 35-MHz piezo-composite ultrasound array for medical imaging. IEEE Trans Ultrason Ferroelectr Freq Control. 2006;53:224–36. doi: 10.1109/tuffc.2006.1588408. [DOI] [PubMed] [Google Scholar]
- Chuang N, Mori S, Yamamoto A, Jiang H, Ye X, Xu X, Richards LJ, Nathans J, Miller MI, Toga AW, Sidman RL, Zhang J. An MRI-based atlas and database of the developing mouse brain. Neuroimage. 2011;54:80–89. doi: 10.1016/j.neuroimage.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W International Mouse Knockout Consortium. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Deans AE, Wadghiri YZ, Berrios-Otero CA, Turnbull DH. Mn enhancement and respiratory gating for in utero MRI of the embryonic mouse central nervous system. Magn Reson Med. 2008;59:1320–28. doi: 10.1002/mrm.21609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filoux E, Mamou J, Aristizábal O, Ketterling JA. Characterization of the spatial resolution of different high-frequency imaging systems using a novel anechoic-sphere phantom. IEEE Trans Ultrason Ferroelectr Freq Control. 2011;58:994–1005. doi: 10.1109/TUFFC.2011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filoux E, Mamou J, Moran CM, Pye SD, Ketterling JA. Correspondence - Characterization of the effective performance of a high-frequency annular-array-based imaging system using anechoic-pipe phantoms. IEEE Trans Ultrason Ferroelectr Freq Control. 2012;59:2825–30. doi: 10.1109/TUFFC.2012/2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster FS, Mehi J, Lukacs M, Hirson D, White C, Chaggares C, Needles A. A new 15–50 MHz array-based micro-ultrasound scanner for preclinical imaging. Ultrasound Med Biol. 2009;35:1700–08. doi: 10.1016/j.ultrasmedbio.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Gato A, Desmond ME. Why the embryo still matters: CSF and the neuroepithelium as interdependent regulators of embryonic brain growth, morphogenesis and histiogenesis. Dev Biol. 2009;327:263–72. doi: 10.1016/j.ydbio.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Gilmore JH, Gerig G, Specter B, Charles HC, Wilber JS, Hertzberg BS, Kliewer MA. Infant cerebral ventricle volume: a comparison of 3-D ultrasound and magnetic resonance imaging. Ultrasound Med Biol. 2001;27:1143–46. doi: 10.1016/s0301-5629(01)00400-8. [DOI] [PubMed] [Google Scholar]
- Henery CC, Kaufman MH. Relationship between cell size and nuclear volume in nucleated red blood cell of developmentally matched diploid and tetraploid mouse embryos. J Exp Zool. 1992;261:472–78. doi: 10.1002/jez.1402610414. [DOI] [PubMed] [Google Scholar]
- Ishihara K, Amano K, Takaki E, Shimohata A, Sago H, Epstein CJ, Yamakawa K. Enlarged brain ventricles and impaired neurogenesis in the Ts1Cje and Ts2Cje mouse models of Down syndrome. Cereb Cortex. 2010;20:1131–43. doi: 10.1093/cercor/bhp176. [DOI] [PubMed] [Google Scholar]
- Ji RP, Phoon CK. Noninvasive localization of nuclear factor of activated T cells c1−/− mouse embryos by ultrasound biomicroscopy-Doppler allows genotype-phenotype correlation. J Am Soc Echocardiogr. 2005;18:1415–21. doi: 10.1016/j.echo.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. The atlas of mouse development. Academic Press; New York: 1992. [Google Scholar]
- Ketterling JA, Aristizábal O, Turnbull DH, Lizzi FL. Design and fabrication of a 40-MHz annular array transducer. IEEE Trans Ultrason Ferroelectr Freq Control. 2005;52:672–81. doi: 10.1109/tuffc.2005.1428050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterling JA, Ramachandran S, Aristizábal O. Operational verification of a 40-MHz annular array transducer. IEEE Trans Ultrason Ferroelectr Freq Control. 2006;53:623–30. doi: 10.1109/tuffc.2006.1610571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Jeanty P, Turner C, Benoit B. Three-dimensional sonographic evaluations of embryonic brain development. J Ultrasound Med. 2008;27:119–24. doi: 10.7863/jum.2008.27.1.119. [DOI] [PubMed] [Google Scholar]
- Liu A, Joyner AL, Turnbull DH. Alteration of limb and brain patterning in early mouse embryos by ultrasound-guided injection of Shh-expressing cells. Mech Dev. 1998;75:107–15. doi: 10.1016/s0925-4773(98)00090-2. [DOI] [PubMed] [Google Scholar]
- Ma Y, Smith D, Hof PR, Foerster B, Hamilton S, Blackband SJ, Yu M, Benveniste H. In vivo 3D digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Front Neuroanat. 2008;2:1. doi: 10.3389/neuro.05.001.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamou J, Aristizábal O, Silverman RH, Ketterling JA, Turnbull DH. High-frequency chirp ultrasound imaging with an annular array for ophthalmologic and small-animal imaging. Ultrasound Med Biol. 2009;35:1198–1208. doi: 10.1016/j.ultrasmedbio.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–85. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Monteagudo A, Timor-Tritsch IE. Normal sonographic development of the central nervous system from the second trimester onwards using 2D, 3D and transvaginal sonography. Prenat Diagn. 2009;29:326–39. doi: 10.1002/pd.2146. [DOI] [PubMed] [Google Scholar]
- Mu J, Slevin JC, Qu D, McCormick S, Adamson SL. In vivo quantification of embryonic and placental growth during gestation in mice using micro-ultrasound. Reprod Biol Endocrinol. 2008;6:34. doi: 10.1186/1477-7827-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman BJ, Szulc KU, Turnbull DH. Three-dimensional, in vivo MRI with self-gating and image coregistration in the mouse. Magn Reson Med. 2009;61:1148–57. doi: 10.1002/mrm.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman BJ, Turnbull DH. Ultrasound and magnetic resonance microimaging of mouse development. Meth Enzymol. 2010;476:379–400. doi: 10.1016/S0076-6879(10)76021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Campbell K, Turnbull DH. Specification of mouse telencephalic and mid-hindbrain progenitors following heterotopic ultrasound-guided embryonic transplantation. Neuron. 1997;19:761–72. doi: 10.1016/s0896-6273(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Parasoglou P, Berrios-Otero CA, Nieman BJ, Turnbull DH. High resolution MRI of early-stage mouse embryos. NMR Biomed. 2013;26:224–31. doi: 10.1002/nbm.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BS, Staib L, Scahill L, Zhang H, Anderson C, Leckman JF, Cohen DJ, Gore JC, Albert J, Webster R. Regional brain and ventricular volumes in Tourette syndrome. Arch Gen Psychiatry. 2001;58:427–40. doi: 10.1001/archpsyc.58.5.427. [DOI] [PubMed] [Google Scholar]
- Phoon CK. Imaging tools for the developmental biologist: ultrasound biomicroscopy of mouse embryonic development. Pediatr Res. 2006;60:14–21. doi: 10.1203/01.pdr.0000219441.28206.79. [DOI] [PubMed] [Google Scholar]
- Phoon CK, Ji RP, Aristizábal O, Worrad DM, Zhou B, Baldwin HS, Turnbull DH. Embryonic heart failure in NFATc1−/− mice: Novel mechanistic insights from in utero ultrasound biomicroscopy. Circ Res. 2004;95:92–99. doi: 10.1161/01.RES.0000133681.99617.28. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Ketterling J. A comparison of acoustic beam properties of a high-frequency annular and linear array. Ultrasonics Symposium, 2007. IEEE; Piscataway, NJ, USA. pp. 1689–92. [Google Scholar]
- Snook KA, Hu CH, Shrout TR, Shung KK. High-frequency ultrasound annular-array imaging. Part I: array design and fabrication. IEEE Trans Ultrason Ferroelectr Freq Control. 2006;53:300–08. doi: 10.1109/tuffc.2006.1593368. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Baldwin HS, Kwee L, Labow M, Aristizábal O, Artman M, Turnbull DH. Noninvasive, in utero imaging of mouse embryonic heart development with 40-MHz echocardiography. Circulation. 1998;98:912–18. doi: 10.1161/01.cir.98.9.912. [DOI] [PubMed] [Google Scholar]
- Sturrock RR. A morphological study of the development of the mouse choroid plexus. J Anat. 1979;129:777–93. [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990;346:847–50. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- Timor-Tritsch IE, Monteagudo A, Santos R. Three-dimensional inversion rendering in the first- and early second-trimester fetal brain: its use in holoprosencephaly. Ultrasound Obstet Gynecol. 2008;32:744–50. doi: 10.1002/uog.6245. [DOI] [PubMed] [Google Scholar]
- Turnbull DH, Bloomfield TS, Baldwin HS, Foster FS, Joyner AL. Ultrasound backscatter microscope analysis of early mouse embryonic brain development. Proc Natl Acad Sci USA. 1995;92:2239–43. doi: 10.1073/pnas.92.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull DH. In utero ultrasound backscatter microscopy of early stage mouse embryos. Comput Med Imaging Graph. 1999;23:25–31. doi: 10.1016/s0895-6111(98)00060-3. [DOI] [PubMed] [Google Scholar]
- Wurst W, Auerbach AB, Joyner AL. Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development. 1994;120:2065–75. doi: 10.1242/dev.120.7.2065. [DOI] [PubMed] [Google Scholar]
- Zhang J, Richards LJ, Yarowsky P, Huang H, van Zijl PC, Mori S. Three-dimensional anatomical characterization of the developing mouse brain by diffusion tensor microimaging. Neuroimage. 2003;20:1639–48. doi: 10.1016/s1053-8119(03)00410-5. [DOI] [PubMed] [Google Scholar]
- Zuiderveld K. Graphic gems IV. Academic Press; San Diego CA: 1994. pp. 474–85. [Google Scholar]