Abstract
Autosomal dominant diseases such as Huntington’s disease (HD) are caused by a gain of function mutant protein and/or RNA. An ideal treatment for these diseases is to selectively suppress expression of the mutant allele while preserving expression of the wild-type variant. RNase H active antisense oligonucleotides (ASOs) or small interfering RNAs can achieve allele selective suppression of gene expression by targeting single nucleotide polymorphisms (SNPs) associated with the repeat expansion. ASOs have been previously shown to discriminate single nucleotide changes in targeted RNAs with ∼5-fold selectivity. Based on RNase H enzymology, we enhanced single nucleotide discrimination by positional incorporation of chemical modifications within the oligonucleotide to limit RNase H cleavage of the non-targeted transcript. The resulting oligonucleotides demonstrate >100-fold discrimination for a single nucleotide change at an SNP site in the disease causing huntingtin mRNA, in patient cells and in a completely humanized mouse model of HD. The modified ASOs were also well tolerated after injection into the central nervous system of wild-type animals, suggesting that their tolerability profile is suitable for advancement as potential allele-selective HD therapeutics. Our findings lay the foundation for efficient allele-selective downregulation of gene expression using ASOs—an outcome with broad application to HD and other dominant genetic disorders.
INTRODUCTION
Autosomal dominant diseases such as Huntington’s disease (HD), retinitis pigmentosa, achondroplasias, cerebellar ataxias, myotonic dystrophy and some forms of familial amyotrophic lateral sclerosis are caused by a gain of function mutant protein and/or RNA (1). An ideal treatment for these diseases would be an allele-selective therapeutic that selectively prevents expression of the disease allele while maintaining expression of the wild-type (wt) variant. Oligonucleotide (ON)-based therapeutics are uniquely suited for targeting autosomal diseases, as they can suppress production of the mutant protein or RNA by targeting the mRNA directly through Watson–Crick interactions (2,3).
HD is an example of autosomal dominant disease caused by an expansion of a CAG repeat in the first exon of the huntingtin (HTT) gene (4). This results in a mutant protein (muHTT) with an expanded polyglutamine tract, which is thought to cause toxicity in neurons. Disease onset typically occurs in middle age and is characterized by neuronal degeneration resulting in profound changes in behavior, cognitive dysfunction, uncontrolled motor movements and dementia (5). Wild-type HTT (wtHTT) is a large protein (350 KDa) that is essential for normal neuronal development in early embryonic stages, but its precise role in the adult brain remain poorly understood (6–9).
Recent reports have demonstrated the utility of ON therapeutics as potential treatments for HD in preclinical animal disease models. Chemically modified RNase H antisense oligonucleotides (ASOs) (10), cholesterol conjugated small interfering RNA (siRNAs) (11) and virally delivered short hairpin RNA (12,13), which partially suppress expression of both, mu and wtHTT, effectively ameliorate the disease phenotype in transgenic mouse models of HD. However, although the suppression of both alleles was therapeutically beneficial in these studies, it would be prudent to preserve the function of wtHTT in the central nervous system (CNS) of adult HD patients if possible.
Allele-selective silencing of muHTT can be achieved by directly targeting the CAG expansion with ONs that act via the steric block or the microRNA pathway (14–16). In animal experiments however, large doses of the CAG targeting ON drug were required to produce ∼50% reduction of muHTT (17). Moreover, expanded CAG repeats occur in numerous other transcripts, and their non-specific downregulation could be deleterious.
An alternative strategy reduces muHTT by targeting SNPs associated with the expanded CAG repeats using either the RNAi mechanism (18–20) or ASOs (21,22). However, the siRNA approach suffers from delivery problems that will require the drug to be injected directly into brain tissue, limiting the efficacy of gene silencing to areas in the immediate vicinity of the site of injection. Given that HD affects most brain regions (23), this approach could have limited utility. Furthermore, ∼93% of validated SNPs in HD are found in intronic regions (22,24). These SNPs appear not to be accessible to siRNAs and short hairpin RNAs, which typically target the mature mRNA transcript in the cytoplasm (25).
In contrast, RNase H active ASOs act predominantly in the nucleus and have access to the pre-mRNA (introns, exons and untranslated regions (UTRs)) (25), thus greatly increasing the number of SNP sites available for identifying safe and efficacious ASO therapeutics. RNase H ASOs are also chemically modified for improved drug-like properties (26). ASOs distribute throughout the brain and spinal cord following injection into the cerebrospinal fluid and silence gene expression in most parts of the CNS (10). Furthermore, chemically modified ASOs have successfully completed early stage human clinical trials for the treatment of familial amyotrophic lateral sclerosis (27) and spinal muscular atrophy (28). However, despite significant advances in the design of new ON chemical modifications (26), design strategies that allow for efficient discrimination of a single nucleotide mismatch using ASOs have remained elusive.
In this report, we demonstrate highly allele-selective downregulation of mutant huntingtin mRNA and protein in HD patient-derived fibroblasts and in the CNS of a fully humanized mouse model of HD by using ASOs that target an SNP in the HTT gene. In addition, we also outline some general design principles for the effective targeting of SNPs using RNase H active ASOs—an outcome with broad application for the treatment of dominant genetic disorders.
MATERIALS AND METHODS
ON synthesis
ONs on a 2 µmol scale were made on an ABI 394 DNA/RNA synthesizer using polystyrene-based VIMAD unylinker™ support. Fully protected nucleoside phosphoramidites were incorporated using standard solid-phase oligonucleotide synthesis, i.e. 3% dichloroacetic acid in DCM for deblocking, 1 M 4,5-dicyanoimidazole 0.1 M N-methylimidazole in acetonitrile as activator for amidite couplings, acetic acid in THF and 10% 1-methylimidazole in THF/pyridine for capping and 0.2 M phenylacetyl disulfide in pyridine:acetonitrile 1:1 (v:v) for thiolation. DNA building blocks were dissolved in acetonitrile (0.1 M) and incorporated using 2 × 4 min coupling time, whereas other building blocks were dissolved in acetonitrile:toluene 1:1 (v:v) if necessary and coupled for 2 × 6 min. After conclusion of the synthesis, the 5′ DMT group was removed and cyanoethyl protecting groups removed using triethylamine:acetonitrile 1:1 (v:v). Remaining protecting groups were removed in 25% aq. ammonia at 55°C for 8 h or at room temperature for 48 h when sensitive modifications were used. ONs were purified by ion-exchange high performance liquid chromatography (HPLC) using a linear gradient of buffer A and B. Buffer A: 50 mM NaHCO3 in acetonitrile:water 3:7 (v:v), buffer B: 1.5 M NaBr, 50 mM NaHCO3 in acetonitrile:water 3:7 (v:v). Purified ONs were desalted using C18 reverse-phase cartridges. ONs used in vivo were made on a 40 µmol scale on an AKTA Oligopilot Synthesizer using the same reagents as described for the 2 µmole scale synthesis, except that 15% dichloroacetic acid in toluene was used for deblocking. DNA phosphoramidites were coupled for 3 min, whereas all other building blocks were coupled for 12 min. ONs were purified as described earlier in the text, except that the 5′ DMT group was retained after full-length synthesis and cleaved on the ion-exchange column.
Thermal denaturation studies
ON and RNA was mixed in 1:1 ratio (4 µM duplex) in buffer containing 10 mM phosphate, 100 mM NaCl and 10 mM EDTA at pH 7.0. Duplex was denatured at 85°C and slowly cooled to the starting temperature of the experiment (15°C). Thermal denaturation temperatures (Tm values) were measured in quartz cuvettes (pathlength 1.0 cm) on a Cary 100 ultraviolet (UV)/visible spectrophotometer equipped with a Peltier temperature controller. Absorbance at 260 nm was measured as a function of temperature using a temperature ramp of 0.5°C per min. Tm values were determined using the hyperchromicity method incorporated into the instrument software.
Human RNase H1 cleavage pattern using liquid chromatography coupled mass spectrometry
Two hundred nanomolar duplex (A1 and fully complementary or SNP G mismatched RNA) was added to reaction buffer [20 mM Tris–HCl, 50 mM KCl, 5 mM MgCl2 (pH 7.5), 540 µl) and heated to 85°C for 2 min and then slowly cooled to room temperature over 1 h. Human RNase H1 solution (0.4 mg/ml, 4 µl) was added to dilution buffer [50 mM Tris–HCl, 50 mM KCl, 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (pH 7.5) in 30% glycerol, 56 µl], incubated at rt for 60 min and then added to duplex solution. Aliquots were removed at different time points and reaction quenched by mixing with quenching buffer (8 M urea and 50 mM EDTA) and snap-frozen on dry ice. RNA fragments were analyzed by ion-pairing HPLC-electrospray/mass spectrometry using a 1100 HPLC-MS system (Agilent Technologies, Wilmington, DE) containing a quaternary pump, variable wavelength UV detector, a column oven, an autosampler and a single quadrupolar mass spectrometer (Agilent Technologies). RNA fragments were separated on an XBridge C18 reverse phase column (2.1 × 15 mm, 2.5 µmol pore size) maintained at 35°C. Full-length RNA and corresponding fragments were eluted using a 0.1 ml/min flow rate using a gradient of buffer A and B; buffer A: 5 mM tributylammonium acetate in acetonitrile:water 1:5 (v:v), buffer B: 5 mM tributylammonium acetate in acetonitrile:water 9:1 (v:v). The mass spectrometer was set to scan a m/z window of 900–2000. Mass spectra were obtained using a spray voltage of 4 kV, a sheet gas flow of 35 pounds per square inch gauge, a drying gas flow rate of 12 l/min at 350°C and a capillary voltage of −150 V. Chromatograms were analyzed using ChemStation software (Agilent Technologies) and the m/z value compared with a table containing the calculated m/z values of expected RNase H cleavage products (Supplementary Figure S3). RNA sequences used for the experiment were as follows: RNAmu 5′-AGACUUUUUCUGGUGAUGACAAUUUAUUAA; RNAwt 5′-AGACUUUUUCUGGUGAUGGCAAUUUAUUAA where the underlined nucleotides represent the SNP site.
Human RNase H1 cleavage pattern using 32P labeled RNA
RNA was 5′-end labeled with 32P using 20 U of T4 polynucleotide kinase, 120 pmol (7000 Ci/mmol) of [γ-32P]ATP, 40 pmol RNA, 70 mM Tris–HCl, 10 mM MgCl2 and 50 mM dithiothreitol (DTT), at pH 7.6. The labeling reaction was incubated at 37°C for 30 min followed by heating at 90°C for 1 min. Labeled RNA was purified using 12% denaturing polyacrylamide gel. ASO (200 nM), unlabeled 19mer RNA (100 nM) and a small amount of 32P-labeled RNA was mixed in hybridization buffer [20 mM Tris, 20 mM KCl (pH 7.5)] and heated to 90°C for 2 min. To the hybridization mixture was added 0.1 mM TCEP, 1 mM MgCl2 and 40 U of RNaseOUT and incubated at 37°C for 1 h. The human RNase H1 enzyme was incubated in dilution buffer [20 mM Tris, 50 mM KCl and 2 mM TCEP (pH 7.5)] for 1 h at rt. Enzyme solution (1/25 µL duplex solution) was added to duplex solution and incubated at 37°C. At time point’s aliquots were removed and reaction quenched in loading buffer and snap-frozen on dry ice. Cleavage products were separated by 12% denaturing polyacrylamide gel, and products were quantitated on a Phosphor-Imager. RNA sequences used for the experiment were as follows: 19mer RNAmu 5′-CUGGUGAUGACAAUUUAUU; 19-mer RNAwt 5′-CUGGUGAUGGCAAUUUAUU where the underlined nucleotides represent the SNP site. The 19mer RNA was used to facilitate separation of bands by gel electrophoresis.
Allele selective PCR conditions
GM04022 fibroblasts, obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research, which are heterozygous at SNP rs7685686_A, were used to measure the in vitro potency and selectivity of the modified ASOs. The cells were trypsinized and resuspended at a density of 400 000 cells per ml in growth medium before transfecting varying concentrations of ASOs with electroporation (Harvard Apparatus ECM830, 115 V, 6 msec). Treated cells were maintained at 37°C and 5% CO2 in minimal essential medium containing 15% fetal bovine serum, non-essential amino acids and penicillin/streptomycin. Approximately 24 h post-transfection, the cells were washed with Dulbecco's Phosphate Buffered Saline buffer and lysed. RNA was extracted using the Qiagen RNeasy96 kit, and levels of the human HTT mRNA alleles were determined using the qPCR assay C_2231945_10 at SNP rs362331 from Life Technologies. The mu and wtHTT mRNA levels were measured simultaneously by using two different fluorophores, 6-carboxyfluorescein (FAM) for mutant allele and VIC® for wild-type allele. Quantitative RT-PCR reactions were run on the ABI 7900HT instrument using the Quantitect Probe RT-PCR kit following the manufacturer’s instructions. The HTT mRNA levels were normalized to total RNA content, as measured by Ribogreen. Data are expressed as means ± SD. For the dose-response curves, results were analyzed using non-linear regression with normalized response and variable slope, and IC50 values were calculated with GraphPad Prism Ver.5. Allele selectivity was calculated by dividing the IC50 for inhibiting wtHTT by the IC50 for inhibiting muHTT. If the IC50 for reducing wtHTT was greater than the highest ASO concentration tested, then the allele selectivity was calculated by dividing the highest ASO concentration tested by the IC50 for inhibiting muHTT and expressed as >× fold.
Primary cortical neuron culture
Embryos were dissected from E15.5-E16.5 pregnant Hu97/18 females. Cortical and striatal tissue was isolated from the embryos and cultured as follows. Briefly, cortical and striatal tissue from single embryos were dissected into ice-cold divalent-free Hank’s Balanced Salt Solution (Invitrogen). The tissue from each embryo was minced and digested with 0.05% Trypsin-EDTA (Invitrogen) at 37°C for 8 min. Digestion was halted by addition of complete neurobasal media (Invitrogen) supplemented with 10% HBSS. Cells were resuspended in media and treated with DNAse I (Invitrogen). After trituration through a 200 µl pipette tip, cells were resuspended in neurobasal media with B27 supplement (Invitrogen), counted and seeded at 1.0 × 10E6 cells/well in 6-well plates pre-coated with poly-d-lysine (BD Biosciences). Neurons were incubated in a humidified 37°C incubator in the presence of 5% CO2.
Genotyping
Tissue was isolated from the embryos post-dissection, and DNA was extracted with the DNeasy blood and tissue extraction kit (Qiagen). Purified DNA was used as template for genotyping by PCR with the AccuPrime GC-Rich DNA Polymerase kit (Invitrogen) using Httg010 primers (Httg009_F ATTACAGTCTCACCACGCCC and Httg010_RCACTTGGGTCTTCCCTTGTC). Products were resolved on 1% agarose gel.
Primary neuron screen
Hu97/18 primary neurons were treated with ASO on day 2 in vitro. Each 6-well culture plate consisted of two untreated wells, and four ASO treatments of increasing dose (0–1000 nM). The neurons were collected by scraping into ice-cold phosphate buffered saline (PBS), pelleting cells (500 g, 5 min, 4°C), aspirating the supernatant and snap-freezing the dry cell pellet.
Immunoblotting
Proteins were extracted by lysis with buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 1% Igepal, 40 mM B-glycerophosphate, 10 mM NaF, 1× Roche complete protease inhibitor, 1 mM sodium orthovanadate and 800 mM phenylmethanesulfonylfluoride] containing 0.1% SDS followed by incubation on ice for 30 min with occasional vortexing (19). Debris was removed by centrifugation (15 min, 20 000 g, 4°C) and the supernatant retained. Protein concentrations were determined by detergent compatible assay (BioRad). Samples were prepared for immunoblotting by denaturing the lysates in Lithium Dodecyl Sulfate sample buffer (Invitrogen) with 100 mM DTT and heating to 70°C for 10 min.
Samples were resolved on 10% low-BIS acrylamide gels (200:1 acrylamide:BIS) with tris-glycine running buffer (25 mM Tris, 190 mM Glycine, 0.1% SDS) containing 10.7 mM beta-mercaptoethanol added fresh. Gels were run at 100 V for 304 min through the stack, then 190 V for 2.5 h or until the 75 kDa molecular weight marker band was at the bottom of the gel. Proteins were transferred to nitrocellulose at 24 V for 2 h with NuPage transfer buffer [Invitrogen: 25 mM Bicine, 25 mM Bis–Tris, 1.025 mM EDTA, 5% MeOH (pH 7.2)]. Membranes were blocked with 5% milk in PBS, and then blotted with for HTT with the Miilipore anti-HTT (mAb 2166). Anti-calnexin (Sigma C4731) immunoblotting was used as loading control. Proteins were detected with IR dye 800CW goat anti-mouse (Rockland 610-131-007) and AlexaFluor 680 goat anti-rabbit (Molecular Probes A21076)-labeled secondary antibodies, and the LiCor Odyssey Infrared Imaging system. Data are expressed as means ± SEM. Where appropriate, results were analyzed using non-linear regression with normalized response and variable slope and IC50 values were calculated with GraphPad Prism Ver.5.
Evaluation of ASOs in humanized mouse brain
For intracerebroventricular (ICV) injection of ASO or PBS vehicle, Hu97/18 mice (N = 4/group) were anesthetized with isofluorane and placed into a stereotaxic frame. A Hamilton syringe with sharpened 26 Ga needle was used to punch through the skull at 0.3 mm anterior and 1.0 mm lateral to Bregma and lowered to a depth of 3 mm. ASO (25, 75, 150, 300 or 500 µg) in a total volume of 10 µl of sterile PBS was injected into the right lateral ventricle at a rate of 1 µl/s. Mice were sacrificed 28 days later. Brains were removed, briefly incubated on ice and placed into a 2 mm coronal brain matrix (ASI instruments). Slab 2, which included portions of anterior cortex and striatum, was then removed, divided into right and left hemisphere portions and snap frozen in liquid nitrogen. Frozen tissue was then lysed and evaluated for levels of mu and wtHTT protein by allelic separation immunoblotting as previously described19. The remaining posterior portion of the brain was post-fixed for 24 h in 4% paraformaldehyde, cryoprotected in 30% sucrose and cut by cryostat into 25 µm free-floating coronal sections. Sections were then processed for ASO immunohistochemistry as previously described (19).
Evaluation of ASOs in C57B16 mouse and Sprague–Dawley rat brain
All procedures were performed under isofluorane anesthesia and in accordance with IACUC regulations. For mouse tolerability studies, ASO was injected into the right lateral ventrical of C57Bl6 female mice (n = 4 per treatment). Briefly, through the use of steriotaxic guides, a needle attached to a Hamilton syringe was inserted at bregma coordinates + 1.0 ML, +0.3 AP and −3.0 DV. Ten microliters of solution containing 300 µg of ASO in PBS was injected over ∼10 s. For repeated dosing in rats, male Sprague–Dawley rats were instrumented with subcutaneous ports (Instech) to deliver dosing solutions to the lumbar intrathecal space (n = 6 per treatment). Briefly, 8 cm long PE-10 catheters inserted 2 cm into the spine from the lumbar-sacrum junction were secured to dosing ports placed subcutaneously and sutured to underlying muscle. Animals were allowed to recover for 1 week post-surgery before beginning injections. Intrathecal (IT) bolus injections were delivered in 30 µL volumes and administered to the subcutaneous port, through the skin of awake animals.
Behavioral analysis
All behavioral analyses were performed during the light-cycle.
Body weight and adverse events
For 8 weeks after the initial dose, each animal was weighed and evaluated weekly by a trained observer for adverse events. Adverse events were defined as any behavior not typical in a na ïve matched control animal, including, but not limited to: limb clasping, abnormal limb splay, abnormal gait, tremors, abnormal respiration, paralysis, spasticity, impaired righting reflex, hyperactivity and lethargy.
Open field
Animals were placed in a 90 × 90 gray chamber in a lit behavior room. Fifteen-minute overhead videos were recorded, and open field activity was assessed by subsequent analysis of the videos with AnyMaze software (Stoelting Inc.).
Grip-strength
Hindlimb grip-strength was assessed by holding the animal horizontal to a 2 mm diameter metal bar attached to a force meter (World Precision Instruments). Once the animals grasped the bar with both hindpaws, they were pulled away until they released their grip. The maximum force achieved was calculated with LabScribe2 software (iWorx Systems Inc.). This was repeated six times consecutively for each animal, and the mean score of all six trials was reported.
RNA analysis for damage markers
Eight weeks after the initial dose, animals in tolerability studies were sacrificed and cortex, striatum and spinal cord tissues were harvested by blunt dissection. Subsequent quantitative real-time PCR analysis was performed as described previously (9). The additional primer probe sets were used: Rat Aif1 5′-AGGAGAAAAACAAAGAACACCAGAA-3′; 5′-CAATTAGGGCAACTCAGAAATAGCT-3′; 5′-Fam-CCAACTGGTCCCCCAGCCAAGA-Tamra-3′; Rat Mpeg 5′-TTCCTCAGAAGGAGAGCAACCT-3′; 5′-TTGCCATTGACTTTGGAGAAAA-3′ 5′-Fam-AAGTACCACCTCATTTTCCATCAACACGGA-IOWA-3′; Rat Gfap 5′-GAAACCAGCCTGGACACCAA-3′ 5′-TCCACAGTCTTTACCACGATGTTC-3′; 5′-Fam-TCCGTGTCAGAAGGCCACCTCAAGA-Tamra-3′; Rat Cyclophilin A 5′-CCCACCGTGTTCTTCGACA-3′; 5′-AAACAGCTCGAAGCAGACGC-3′; 5′-Fam-CACGGCTGATGGCGAGCCC-Tamra-3′; Mouse Aif1 5′-TGGTCCCCCAGCCAAGA-3′; 5′-CCCACCGTGTGACATCCA-3′; 5′-Fam-AGCTATCTCCGAGCTGCCCTGATTGG-Tamra-3′.
RESULTS
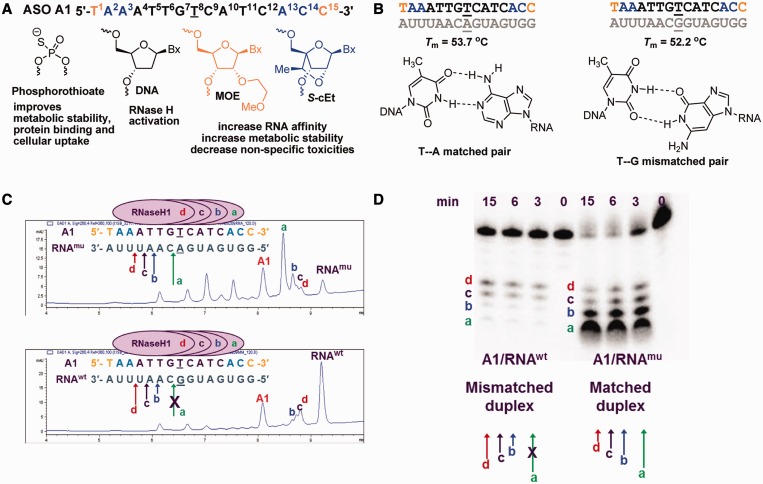
We recently reported ∼5-fold selectivity for reducing muHTT compared with wtHTT in HD patient-derived fibroblasts, cultured primary neurons and in murine BACHD brain using gapmer ASOs targeting SNPs associated with expanded CAG repeats (22). One of the ASOs (A1) identified in this study targeted SNP rs7685686_A located within intron 42 of the HTT gene. ASO A1 is a fully phosphorothioate (PS) modified 15mer oligonucleotide with a 9-base deoxynucleotide gap flanked on each end with 2′-O-methoxyethyl (MOE) (29) and 2′,4′-constrained-[S]-2′-O-ethyl (S-cEt) (30) modified nucleotides (Figure 1A). The central gap region supports RNase H-mediated degradation of target mRNA, whereas the modified nucleotides in the flanks improve affinity of the ASO for target RNA and stabilize the ASO from exo-nuclease mediated degradation. The PS backbone modification also stabilizes the ASO from nuclease digestion and facilitates internalization of the ASO into cells by binding to cell-surface proteins (31). ASO A1 is perfectly matched to muHTT while it has a single G:T mismatch to wtHTT positioned at the center of the oligonucleotide. Thus, the 5-fold selectivity observed with ASO A1 in cell culture was surprising considering that the GT wobble base pair is only slightly less stabilizing as compared with the AT cognate base pair (Figure 1B) (32).
Figure 1.
Chemical design and analysis of human RNase H1 cleavage patterns for ASO A1. (A) Chemical design of starting ASO A1, black letters indicate DNA, orange letters indicate MOE and blue letters indicate S-cEt nucleotides; underlined letter is the nucleotide across from the SNP site; all internucleosidic linkages are PS. (B) Tm of A1 with matched and mismatched RNA complements representing muHTT and wtHTT mRNA and structures of the AT cognate and GT wobble base pairs. (C and D) Analysis of human RNase H1 cleavage patterns of the matched duplex (A1/RNAmu) versus the mismatched duplex (A1/RNAwt) containing a GT wobble base pair positioned in the center of the PS DNA gap. Peaks labeled a, b, c and d correspond to the RNA fragments arising from human RNase H1 cleavage and are depicted using colored arrows. The cleavage products were analysed using (C) LCMS or (D) gel electrophoresis using 5′-radiolabeled RNA.
Chemical modifications that improve binding discrimination for the GT wobble base pair do not improve allele selectivity
We measured the duplex thermal stability of ASO A1 versus a matched RNA complement representing muHTT and a single base mismatched RNA representing wtHTT in UV monitored thermal denaturation experiments (Tm). As expected, there was a small (1.5°C) difference in the Tm of the matched and the mismatched ASO/RNA duplexes (Figure 1B).
To examine whether the allele selectivity of ASO A1 could be increased using chemical modifications known to either improve binding discrimination for the GT wobble base pair or modulate RNase H activity in the vicinity of the mismatch, we prepared ASOs where the dT nucleotide in the ASO across from the SNP site was replaced with chemical modifications (ASOs A2–A14, Supplementary Figure S1A). The gap-modified ASOs were evaluated in Tm (Supplementary Figure S1C) and in cell culture experiments (Supplementary Figure S1B). We found that selected modifications such as C5-propyne (33), 2S-dT (34) and 3′-fluoro hexitol nucleic acid (FHNA) (35) were able to improve G:T discrimination modestly, but this did not result in improved allele selectivity in cell culture (Supplementary Figure S2).
GT wobble base pair alters the preferred cleavage site of human RNase H1
To understand the origins of the selectivity exhibited by ASO A1, we examined the cleavage of the duplexes between ASO A1 with the matched and mismatched RNA complements RNAmu and RNAwt, respectively, using recombinant human RNase H1 in a biochemical assay. Human RNase H1, the key enzyme implicated in the pharmacology of DNA containing ASOs (36), cuts the RNA strand of RNA/DNA heteroduplex 6–7 bp from the 3′-DNA/5′-RNA end of the heteroduplex (37,38). The enzyme binds to different regions of a DNA/RNA heteroduplex and produces distinct cleavage sites based on the initial site of binding (39). Introducing chemical modifications into the DNA strand, which bias the conformation of the sugar furanose ring into a RNA-like C3′-endo sugar pucker typically, do not support RNase H activity (40).
In our experiments, ASO A1/RNAmu and ASO A1/RNAwt duplexes were incubated with recombinant human RNase H1 and analyzed for cleavage using liquid chromatography coupled mass spectrometry (LCMS, Figure 1C and Supplementary Figure S3). We found that, for the matched duplex A1/RNAmu, the preferred cleavage site (a) was adjacent to the centrally positioned AT base pair corresponding to the SNP site. In contrast, for the mismatched duplex A1/RNAwt representing the ASO/wtHTT mRNA heteroduplex, no cleavage was observed adjacent to the SNP site. Instead, cleavage was shifted to less efficient minor cleavage sites (b, c and d) across from the 5′-end of the DNA gap. Results from the LCMS RNase H assay were further confirmed using the traditional method of radiolabeling the RNA and examining the cleavage patterns using gel electrophoresis (Figure 1D). Thus, presence of a GT wobble base pair at the human RNase H1 preferred cleavage site changes the cleavage pattern for the matched (A1/RNAmu) duplex versus the mismatched (A1/RNAwt) duplex. This observation provided an explanation for the modest selectivity exhibited by ASO A1 in cell culture.
Rationale for improving allele selectivity by using chemical modifications to suppress minor RNase H cleavage sites
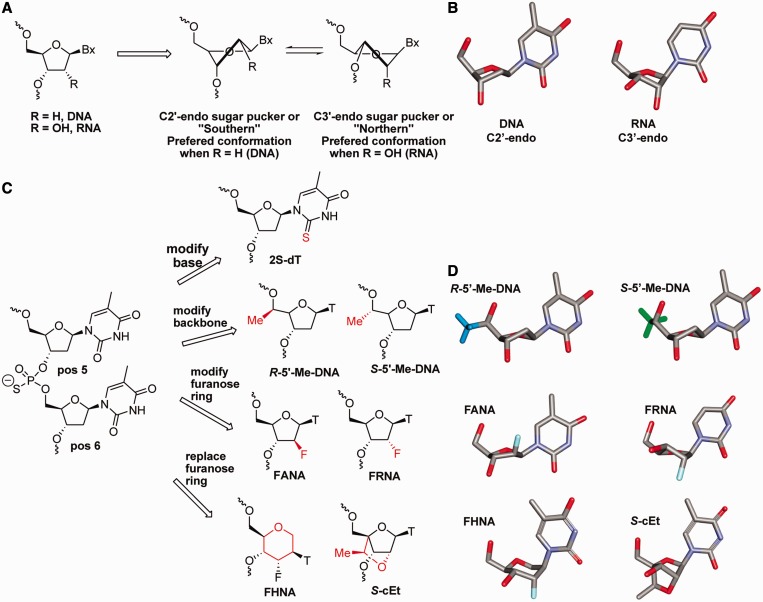
We next hypothesized that the allele selectivity could be enhanced by introducing chemical modifications in the ASO across from the minor cleavage sites (b, c and d) that either do not support or interfere with RNase H cleavage (Figure 2). As the minor cleavage sites on RNAwt were distinct and separated from the preferred cleavage site (a) on RNAmu, we anticipated that improved selectivity could be achieved without affecting activity versus the mutant allele. Chemical modifications, which bias the conformation of the furanose ring toward the C3′-endo sugar pucker (Figure 2A and B) across from the cleavage site on the RNA, do not support RNase H activity (37). The effect of ASO chemical modifications on RNase H activity is also dependent on the exact position of incorporation relative to the cleavage site (5′ or 3′) on the RNA and is transmitted up to two nucleotides on either side of the chemical modification (37).
Figure 2.
Chemical strategies to improve allele selectivity. (A) Structural equilibrium and (B) conformational models of the furanose rings in DNA and RNA (C) Strategies for introducing chemical modifications at positions 5 and 6 of ASO A1 to modulate RNase H1 cleavage at minor cleavage sites b, c and d. (D) Conformational models showing the furanose rings of R-5′-Me-DNA and S-5′-Me-DNA in the C2′-endo sugar pucker, FANA in the O4′-endo and FRNA in the C3′-endo sugar puckers and the hexitol ring of FHNA and the ‘locked’ sugar conformation of S-cEt nucleotides.
Given that there were multiple minor cleavage sites on RNAwt, the optimal location for introducing chemical modifications into the ASO could not be predicted a priori. As a result, we sequentially replaced the dT nucleotides at positions 5 and 6 in the ASO with chemical modifications (Figure 2C and D). The candidate modifications included (i) 2S-dT where the 2-oxygen on the nucleobase is replaced with sulfur (34); (ii) backbone modifications like R-5′-Me-DNA and S-5′-Me-DNA, which place minimal steric bulk adjacent to the phosphate backbone but are not known to impact the sugar pucker of the furanose ring (41); (iii) 2′-modified furanose analogs such as 2′-fluoro arabino nucleic acid (FANA), which improves RNase H activity at the site of incorporation (42), and 2′-fluoro ribonucleic acid (FRNA), which exists predominantly in the RNA-like C3′-endo sugar pucker (43); and (iv) FHNA (35) and S-cEt (44), which mimic or lock the nucleoside furanose ring in the C3′-endo sugar pucker while greatly improving affinity for complementary RNA.
For selected modifications, we characterized the effect of the new ASO designs on the thermal stability of the ASO duplexes with the matched and mismatched RNA complements RNAmu and RNAwt, respectively (Supplementary Figure S4). Expectedly, we found no significant differences in the ability of these chemical modifications to improve thermal discrimination for the GT wobble base pair.
Base modification 2S-dT has a position dependent effect on allele selectivity
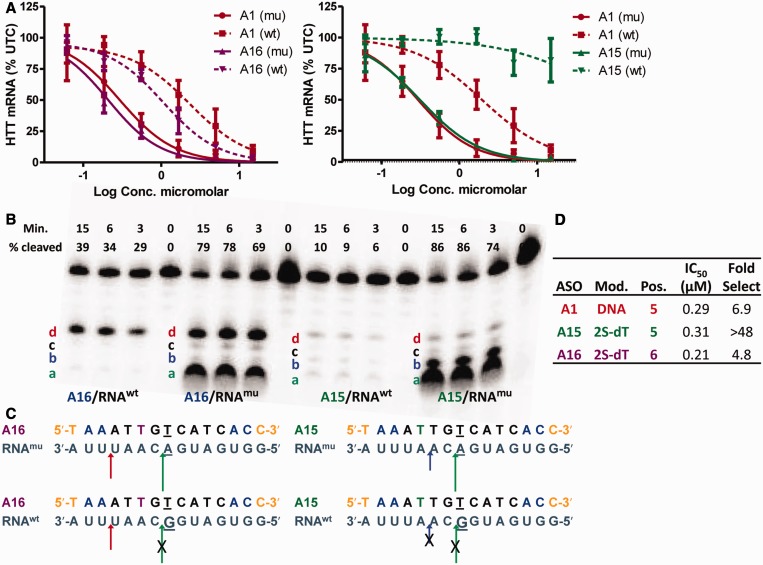
Introducing 2S-dT at position 5 or 6 in the ASO had a position-dependent effect on allele selectivity (Figure 3). Replacing dT with 2S-dT at position 5 (ASO A15) resulted in >48-fold improvement in allele selectivity, whereas the analogous substitution at position 6 (ASO A16) had no effect on allele selectivity relative to A1 (Figure 3A). To understand this behavior, we examined the RNase H cleavage patterns of the matched and mismatched duplexes of ASOs A15 and A16. ASO A16 with 2S-dT at position 6 showed two major cleavage sites (a and d) for the matched duplex (A16/RNAmu) (Figure 3B). Only one cleavage site (d) was observed for the mismatched duplex (A16/RNAwt) where the G:T wobble base pair ablates the major cleavage site (a). Thus, 2S-dT at is unable to suppress RNase H cleavage two nucleotides upstream from the site of incorporation (d), and even a single cleavage site on the mismatched duplex is sufficient to blunt increased selectivity (Figure 3C).
Figure 3.
2S-dT has a positional effect on allele selectivity. (A) Dose-dependent reduction of muHTT and wtHTT mRNA in human GM04022 fibroblasts using ASOs A15 and A16. (B) Human RNase H1 cleavage patterns for A16 with 2S-dT at position 6 and A15 with 2S-dT at position 5. (C) Pictorial representation of human RNase H cleavage patterns for A16 and A15. 2S-dT at position 5 blocks cleavage sites (d and c), whereas 2S-dT at position 6 blocks cleavage sites (b and c) on the matched and mismatched duplexes. The GT base pair ablates cleavage sites (a and b) on the mismatched duplex for ASOs A15 and A16. Underlined nucleotide indicates position across from the SNP site. (D) Summary of potency and selectivity profile of A15 and A16. Error bars are in ±SD.
In contrast, ASO A15 with 2S-dT at position 5 showed two cleavage sites (a and b) for the matched duplex but no cleavage sites for the mismatched duplex (Figure 3B). In this case, the combination of 2S-dT at position 5 and the GT wobble base pair blocks cleavage sites (a and b) resulting in greatly improved allele selectivity. In ASO A15, the S-cEt nucleotides in the 5′-wing suppress RNase H cleavage two nucleotides upstream from the site of 2S-dT incorporation (Figure 3C). As expected, introducing 2S-dT at position 5 or 6 (A15 and A16, respectively) had no impact on thermal discrimination of the GT wobble base pair relative to control ASO A1 (Supplementary Figure S4).
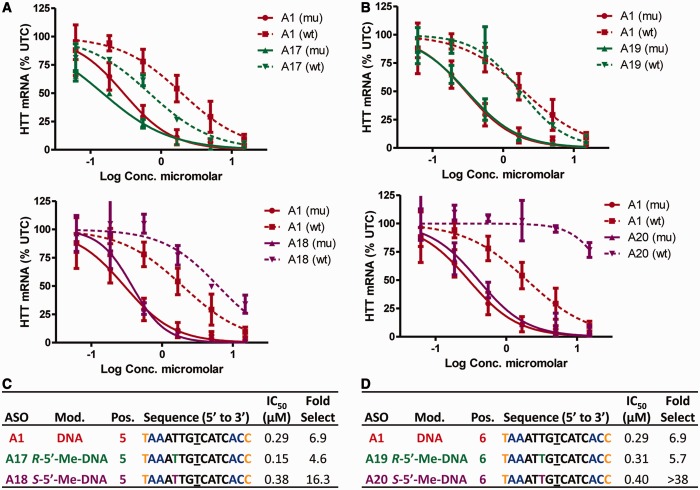
Backbone modifications R-5′-Me-DNA and S-5′-Me-DNA have a configuration and position-dependent effect on allele selectivity
The effect of the backbone modifications on allele selectivity was dependent on position and on the absolute configuration of the 5′-methyl group (Figure 4). ASO A17 with R-5′-Me-DNA at position 5 showed no improvement in allele selectivity relative to control ASO A1, whereas A18 modified with S-5′-Me-DNA at position 5 showed 16-fold selectivity (Figure 4A and C). Introducing R-5′-Me-DNA at position 6 (A19) also showed no improvement in allele selectivity, whereas introducing S-5′-Me-DNA at position 6 (A20) showed >38-fold selectivity (Figure 4B and D).
Figure 4.
Effect of R-5′-Me-DNA and S-5′-Me-DNA on allele selectivity is dependent on position of incorporation and on the absolute configuration of the 5′-methyl group. Dose-dependent reduction of muHTT and wtHTT mRNA in human GM04022 fibroblasts for (A) ASOs A17 (R-5′-Me-DNA at position 5) and A18 (S-5′-Me-DNA at position 5) and (B) ASOs A19 (R-5′-Me-DNA at position 6) and A20 (S-5′-Me-DNA at position 6). Summary of sequence, activity and selectivity for (C) ASOs A1, A17 and A18 (D) ASOs A1, A19 and A20. Underlined nucleotide indicates position across from the SNP site. Error bars are in ±SD.
Examination of the RNase H1 cleavage patterns for the matched and mismatched duplexes showed that R-5′-Me-DNA does not disrupt the minor cleavage sites (d and c for A17 and c for A19, Supplementary Figure S5A–C) on the mismatched duplex. In contrast, S-5′-Me-DNA reduced (position 5) or eliminated (position 6) RNase H cleavage (Supplementary Figure S5D and E), resulting in improved allele selectivity.
Interestingly, we also observed that the configuration of the 5′-methyl group changes the RNase H1 cleavage preference on the RNA (Supplementary Figure S5). For example, although S-5′-Me-DNA allows RNase H1 cleavage across and on the 5′-side of the RNA from its site of incorporation, it does not allow RNase H1 cleavage on the 3′-side of the RNA. In contrast, R-5′-Me-DNA enhances the intensity of RNase H1 cleavage one nucleotide over on the 3′-side of the RNA from the site of incorporation.
The precise orientations of the R- and S-5′-Me groups in the heteroduplex are not properly understood, given that high-resolution nuclear magnetic resonance or crystal structures of 5′-Me-DNA modified duplexes are not available. Assuming canonical orientations around the backbone torsion angles α, β and γ, the S-5′-Me group is expected to lie in the minor groove side of the modified duplex, which is an important recognition site for RNase H1 (38). In contrast, the R-5′-Me group is expected to lie toward the edge of the minor groove and may not be as disruptive to the recognition and processing of the heteroduplexes by RNase H1.
Configuration of the 2′-fluorine atom can modulate allele selectivity
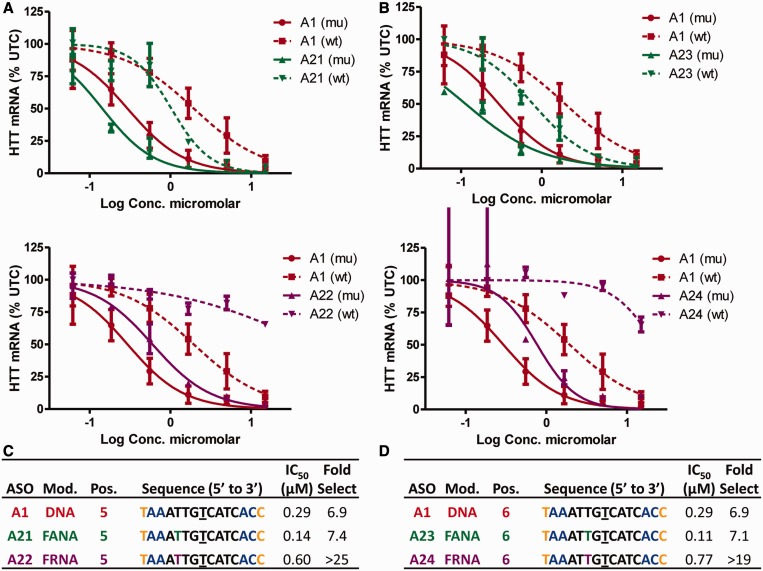
Introducing highly electronegative fluorine atoms at the 2′-position of the nucleoside building blocks can have remarkable effects on the conformation of the furanose rings and on the biological properties of the modified oligonucleotides (35). ASO A21 with FANA at position 5 showed no improvement in allele selectivity relative to control ASO A1, whereas A22 modified with FRNA at position 5 showed >25-fold selectivity (Figure 5A and C). Introducing FANA at position 6 (A23) also showed no improvement in allele selectivity relative to A1, whereas introducing FRNA at position 6 (A24) showed >19-fold selectivity (Figure 5B and D). Analysis of the RNase H cleavage patterns further confirmed that introducing FANA at positions 5 or 6 did not ablate the minor cleavage sites on the mismatched duplex (Supplementary Figure S6A–C). In contrast, FRNA at positions 5 or 6 ablated the minor cleavage sites on the mismatched duplex (Supplementary Figure S6D and E).
Figure 5.
Effect of FANA and FRNA on allele selectivity is dependent on position of incorporation and on the absolute configuration of the 2′-fluorine atom. Dose-dependent reduction of muHTT and wtHTT mRNA in human GM04022 fibroblasts for (A) ASOs A21 (FANA at position 5) and A22 (FRNA at position 5) and (B) ASOs A23 (FANA at position 6) and A24 (FRNA at position 6). Summary of sequence, activity and selectivity for (C) ASOs A1, A21 and A22 (D) ASOs A1, A23 and A24. Underlined nucleotide indicates position across from the SNP site. Error bars are in ±SD.
The differential effect of FANA and FRNA on RNase H cleavage can be rationalized by the differences in furanose sugar pucker of these modifications. The furanose ring of FANA exists in the O4′-endo sugar pucker (Figure 2D), which is known to facilitate RNase H1 activity (42). In contrast, the furanose ring of FRNA exists in the C3′-endo sugar pucker (Figure 2D), which is detrimental to RNase H activity. It is also noteworthy, that R-5′-Me-DNA showed almost identical effect on RNase H cleavage (pattern and intensity) as FANA. This suggests that the R-5′-Me group could potentially modulate the furanose sugar pucker toward the O4′-endo conformation.
Strategic incorporation of S-cEt or FHNA in the gap improves allele selectivity
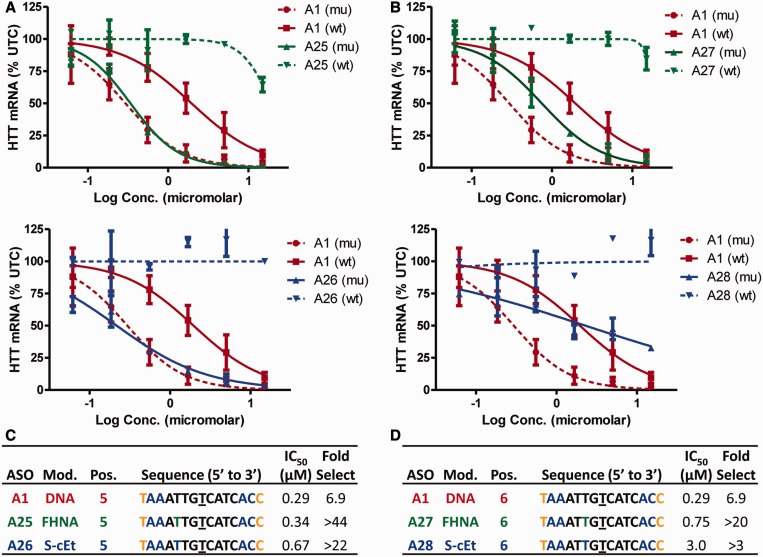
FHNA and S-cEt at positions 5 or 6 resulted in an improvement in allele selectivity, but the effect on activity versus the mutant allele was mixed. ASOs A25 and A26 with FHNA or S-cEt at position 5, respectively, exhibited activity comparable with A1 but significantly improved allele selectivity (Figure 6A and C). The improved allele selectivity was confirmed in the RNase H cleavage assay where both ASOs showed negligible cleavage of the mismatched duplex (Supplementary Figure S7A–C).
Figure 6.
FHNA and S-cEt improve allele selectivity when incorporated at positions 5 and 6 of ASO A1. Dose-dependent reduction of muHTT and wtHTT mRNA in human GM04022 fibroblasts for (A) ASO A25 (FHNA at position 6) and A26 (S-cEt at position 5) (B) ASO A27 (FHNA at position 6) and A28 (S-cEt at position 6). Summary of sequence, activity and selectivity for ASOs (C) A1, A25, A26 and (D) A27 and A28. Underlined nucleotide indicates position across from the SNP site. Error bars are in ±SD.
The effect of incorporating FHNA (A27) or S-cEt (A28) at position 6 was less successful. Both ASOs exhibited good allele selectivity, but ASO A28 exhibited significant loss in potency versus muHTT relative to ASO A1 (Figure 6B and D). These observations were confirmed in the biochemical assay where both ASOs showed reduced intensity in cleavage at site (a) along with reduced number of total cleavage sites (Supplementary Figure S7D–E).
In contrast to FRNA and FHNA where the furanose ring exists predominantly in the C3′-endo sugar pucker, the sugar ring of cEt (44) is slightly bulkier and locked in the C3′-endo sugar pucker. Consequently, introducing FHNA at position 6 (within two nucleotides of the major cleavage site a) appears to have a smaller detrimental effect on RNAse H activity in contrast to S-cEt, which reduced activity considerably.
Shortening the PS DNA gap in ASO A1 improves allele selectivity
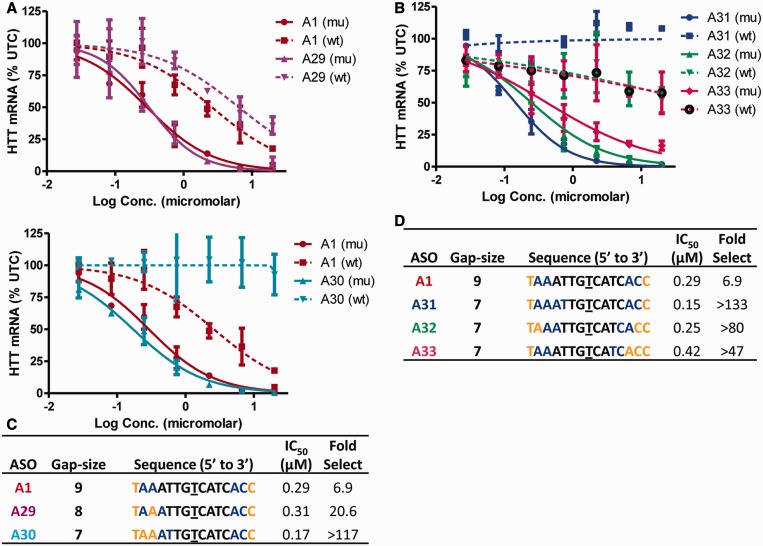
ASOs A25 and A26 have essentially a 7-nt PS DNA gap; therefore, we hypothesized that shortening the PS DNA gap in ASO A1 could improve allele selectivity while maintaining activity. To this end, we evaluated ASOs A29 and A30, which have PS DNA gaps of 8 and 7 nt, respectively. We found that although both ASOs showed excellent potency for reducing muHTT, ASO A29 exhibited a 3-fold improvement in allele selectivity relative to A1, whereas ASO A30 showed profoundly improved selectivity versus wtHTT (>100-fold selectivity) (Figure 7A and C).
Figure 7.
Precise sequence of the PS DNA gap region is important for optimal allele selectivity. Dose-dependent reduction of muHTT and wtHTT mRNA in human GM04022 fibroblasts for (A) ASOs A29 (8-base PS DNA gap) and A30 (seven-base PS DNA gap) (b) ASOs A31, A32 and A33 (seven-base PS DNA gaps frame-shifted by one nucleotide). Summary of sequence, activity and selectivity for (C) ASOs A1, A29, A30 and (D) A31, A32 and A33. Underlined nucleotide indicates position across from the SNP site. Error bars are in ±SD.
To understand the importance of the precise sequence of the 7-base gap on activity and selectivity, we evaluated ASOs A31, A32 and A33 where the sequence of the gap was frame shifted by 1 nt. Although all the ASOs showed >50-fold selectivity, ASO A31, which has the same gap-sequence as ASO A30, showed the best activity and selectivity profile (Figure 7B and D).
Modified ASOs are well tolerated after direct injection into the CNS of wt mice and rats
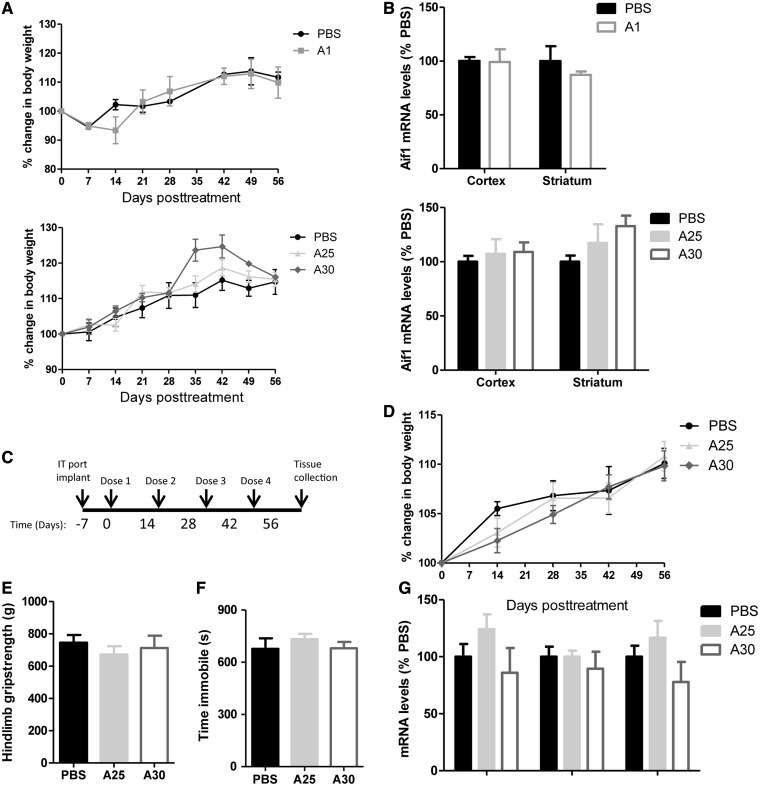
To determine the safety and tolerability of the modified ASOs, non-transgenic mice were treated with either the benchmark compound A1, the optimized cEt (A30), the FHNA ASO (A25) or vehicle (PBS). Mice received a single 300 µg ICV bolus injection into the right lateral ventricle, and mice were monitored for 8 weeks post-treatment. No adverse events were observed in any of the animals in any of the treatment groups, and all animals maintained a normal body weight relative to PBS-treated controls (Figure 8A). To determine whether these compounds caused microglial activation, striatal and cortical tissue, adjacent to the injection site was collected and Aif1 mRNA, a microglial activation marker was quantified. Aif1 mRNA levels were not significantly increased in any of the treatment groups in either tissue type (Figure 8B).
Figure 8.
Tolerability profile of selected ASOs in the CNS of wt mice and rats. (A and B) Non-transgenic mice (n = 4/group) were administered 300 µg of A1, A25, A30 or PBS by ICV injection and were monitored for tolerability for 8 weeks post-treatment. ASO treatment in non-transgenic mice did not alter (A) body weight or (B) striatal or cortical Aif1 mRNA levels. (C–G) Repeated IT dosing in rats every 2 weeks for 8 weeks did not alter (D) body mass, (E) hindlimb grip-strength, (F) time immobile in the open-field or (G) spinal cord mRNA levels of Aif1, Gfap or Mpeg1. Error bars are in±SEM.
To further assess the tolerability of these new modifications, we repeatedly injected the optimized S-cEt (A30) or the FHNA (A25) ASOs IT into Sprague–Dawley rats. Rats were dosed IT every 2 weeks for four doses, and tissues were collected 2 weeks after the last injection (Figure 8C). No adverse events were observed in any of the rats, for any of the treatment groups, and all rats maintained normal body weight (Figure 8D). Two weeks after the fourth and final injection, hindlimb grip-strength (Figure 8E) and ambient motor activity (Figure 8F), as scored by time immobile in the open-field, were similar in ASO-treated animals and PBS controls. Finally, mRNA for markers of microglial activation and inflammation (Aif1, Gfap and Mpeg1) in spinal cord tissue, directly adjacent to the injection site, were not significantly altered in ASO-treated animals (Figure 8G).
Improved allele selectivity in cell culture translates to brains of transgenic mice
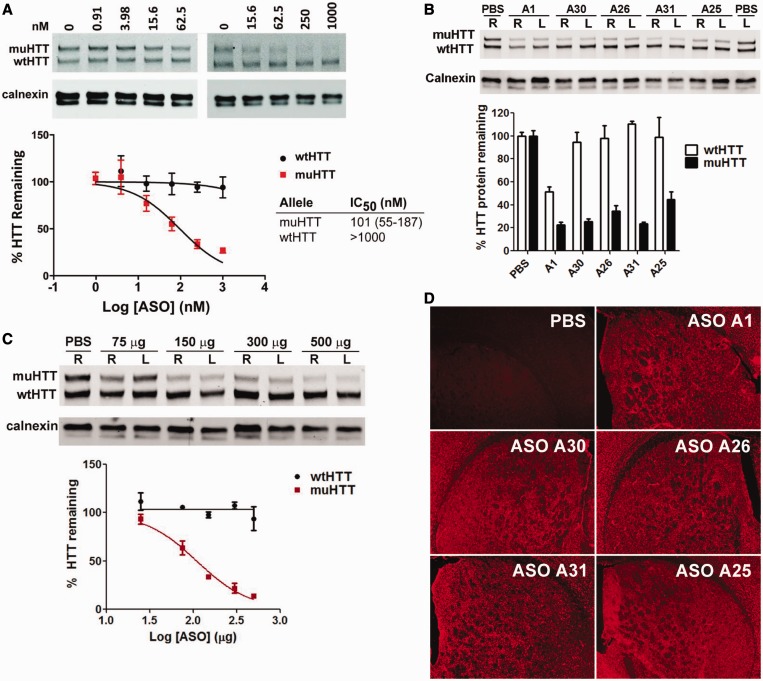
We evaluated selected ASOs in Hu97/18 mice (45), a completely humanized mouse model of HD (Figure 9). This mouse model contains both the mutant human HTT allele, with the associated SNPs, and the wt human HTT allele. We first evaluated the ability of ASO A30 to reduce muHTT protein in primary neuronal cells derived from Hu97/18 mouse embryos in the absence of any transfection agent to facilitate delivery of the ASO across the cell membrane (Figure 9A). The ASO reduced muHTT expression in a dose-dependent manner (IC50 = 101 nM). No reduction of wtHTT was observed even at the highest dose of ASO A30 (1000 nM) tested. Next, mice (n = 4/group) were injected in the right lateral ventricle (ICV) with a single bolus injection (300 µg) of ASO (A1, A25, A26, A30 or A31) or vehicle control (PBS), and tissue was collected 28 days later. Brains were harvested, and HTT protein was quantified in a 2 mm coronal slab from each hemisphere by allelic separation immunoblotting for mu and wtHTT protein; the results were normalized to calnexin (Figure 9B). The posterior portions of treated brains were then post-fixed for immunohistochemical assessment of ASO distribution.
Figure 9.
Activity and selectivity of ASOs in the rodent CNS. (A) Allele selective knockdown of muHTT protein with ASO A30 in neuronal cells derived from cortical and striatal tissues of Hu97/18 mouse embryos under free-uptake conditions. (B–D) Hu97/18 mice (n = 4/group) were injected ICV with a single dose of 300 µg of ASO (B and D) or the indicated dose (C) in PBS. Mice were sacrificed after 28 days, the brain was harvested, and a 2 mm coronal slab from each hemisphere (R,L) was analyzed by allelic separation immunoblotting for muHTT and wtHTT protein, and the results were normalized to calnexin protein. (B) Optimized ASOs A25, A26, A30 and A31 show similar activity but improved allele selectivity relative to control ASO A1. (C) Dose response for allele-selective knockdown of muHTT protein following ICV injection of ASO A30 in Hu97/18 mice (D) Immunohistochemical staining for ASO (red) illustrates distribution to all parts of the brain following a single ICV bolus injection.
As observed in the cell culture experiment, all the ASOs showed 60–80% reduction of muHTT protein. However, although ASO A1 showed a 52% reduction of the wtHTT protein, we observed essentially no reduction of the wtHTT protein for the mice treated with the allele-selective ASOs A25, A26, A30 and A31. We also measured the ability of ASO A30 to reduce muHTT protein in a dose-response experiment (Figure 9C). Mice (n = 4/group) were injected (ICV bolus) with increasing doses (75, 150, 300 and 500 µg) of ASO A30 or vehicle control and brain tissue was harvested after 28 days using the procedures described earlier in the text. The ASOs produced a dose-dependent reduction in muHTT (ED50 = 110 µg) with no reduction in wtHTT even at the highest dose tested (500 µg). All ASOs also showed similar broad and even bilateral localization after ICV bolus injection, indicating that the modifications had no effect on distribution (Figure 9D). Thus, the animal model recapitulated the potency and selectivity observations from the cell culture experiments. Collectively, these studies illustrate the potency, selectivity and tolerability of these compounds, and support their development as potential therapeutics for HD.
DISCUSSION
Oligonucleotide therapeutics can be useful for the treatment of autosomal dominant diseases where selective suppression of the transcript derived from the disease allele is desirable. We describe here an approach that allows for the highly selective targeting of a single base difference between wt and mutant HTT alleles using chemically modified ASOs. Furthermore, we show that the ASOs can silence the targeted allele throughout the CNS in a safe manner and at therapeutically relevant doses after direct injection into the CNS of rodents.
wtHTT is critical in early stages of life (6,9) but the precise roles of the protein in adulthood remain unclear. Because of this, two strategies for ASO mediated therapeutic repression of HTT have been proposed. The first is to partially reduce both alleleic variants. The second is to selectively reduce the mutant allele while preserving the wt variant. This second, more discriminating allele-selective approach, would ameliorate any theoretical concerns of long-term reduction of wtHTT in HD patients (46). Allele selectivity can be achieved by either targeting SNPs associated with the CAG as described here or by targeting the CAG repeat directly.
The advantages of a pan-allele approach is that a single ASO drug could be used to treat the entire HD population and that suppression of mutant and wtHTT in rodent models of HD provides a clear therapeutic benefit (10). However, although no adverse effects from lowering wtHTT in the rodent brain have been reported, more work is needed to assess the long-term safety of lowering wtHTT in the CNS of adult patients. Thus, an approach that selectively lowers the mutant allele is attractive, as it limits potential liabilities from partial suppression of the wt allele.
Targeting the CAG expansion directly with chemically modified oligonucleotides is an allele selective approach that can target the entire HD population (16), as well as other diseases caused by expanded CAG repeats (47). Targeting the CAG expansion directly is also appealing because it prevents the translation of all variants containing the toxic expanded polyglutamine tract (48). However, this approach suffers from the potential for downregulating other CAG repeat containing genes (15). Moreover, the majority of HD patients contain <45 CAG repeats. This could reduce the efficiency of allele-selective inhibition, which relies on co-operative binding of multiple ASOs to the repeat expansion to achieve target suppression. Furthermore, relatively high doses of the oligonucleotide were needed to produce ∼50% reduction of muHTT in the CNS of rodents (17), suggesting that further optimization is required to improve the in vivo pharmacodynamics of these compounds for use as a human therapy. In comparison, a single injection of ASO A30 (ED50 = 110 µg) was able to reduce muHTT in the entire mouse brain.
The SNP-targeting approach is a method that can selectively target the mutant allele, without the potential for downregulating other CAG containing transcripts. A disadvantage of the allele selective SNP-targeting approach is that multiple ASOs will be needed to treat the entire HD population owing to heterozygosity of SNPs in the patient population (22,49). For example, the ASOs described in this report could provide an allele-specific treatment option for ∼49% of the Caucasian HD population. This poses clinical and regulatory burdens to develop multiple safe and efficacious ASOs to treat the entire HD population in an allele-selective manner. However, the ASOs described in this report are well tolerated after direct injection into the CNS of rodents, have a similar distribution and activity patterns as those reported previously (10) and can selectively lower muHTT without the added complications of lowering wtHTT. Ongoing studies with the current SNP-targeting ASOs in animal disease models will help ascertain the importance of selectively lowering muHTT and support their further development as potential allele selective inhibitors of muHTT for treatment of HD in humans.
In a broader context, the improved allele selectivity observed with ASO designs containing short PS DNA gap regions suggests that it should be possible to achieve efficient allele-selective inhibition for other gene targets by using two strategies (i) screening SNP associated mRNA sequence space with shorter (seven- and eight-base) gap ASOs and (ii) by moving the position of the SNP across the gap to ascertain the precise gap position for optimal activity and allele selectivity. It should also be highlighted that the ability to produce profound changes in ASO behavior with subtle structural changes like replacing one oxygen atom with sulfur (2S-dT), adding a methyl group at the 5′-position of DNA or inverting the configuration of one fluorine atom (FANA to FRNA) in an oligonucleotide with a molecular weight of ∼5500 is remarkable and unprecedented.
With advances in personalized medicine, understanding the genetic differences between patient sub-groups will be important for the diagnosis and treatment of human diseases. The advent of next-generation sequencing technologies will ensure that this information becomes available for large segments of the human population. Against this background, the ability to selectively downregulate expression of individual alleles by targeting SNPs in a therapeutically relevant manner could have enormous implications. In this context, our work lays the foundation for the rational design of SNP targeting ASOs for treating autosomal dominant diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Isis Pharmaceuticals.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank John Wu, Josh Nichols and Walt Lima at for assistance with the human RNase H1 experiments and Hans Gaus for assistance with the LCMS experiment.
REFERENCES
- 1.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 2.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 3.O'Connor TP, Crystal RG. Genetic medicines: treatment strategies for hereditary disorders. Nat. Rev. Genet. 2006;7:261–276. doi: 10.1038/nrg1829. [DOI] [PubMed] [Google Scholar]
- 4.MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 5.Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 6.Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Leavitt BR, van Raamsdonk JM, Dragatsis I, Goldowitz D, MacDonald ME, Hayden MR, Friedlander RM. Huntingtin inhibits caspase-3 activation. EMBO J. 2006;25:5896–5906. doi: 10.1038/sj.emboj.7601445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's Disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 9.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 10.Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, Shihabuddin LS, et al. Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl Acad. Sci. USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, Ojeda SR, Davidson BL. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for huntington's disease. Mol. Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boudreau RL, Spengler RM, Davidson BL. Rational design of therapeutic siRNAs: minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington's disease. Mol. Ther. 2011;19:2169–2177. doi: 10.1038/mt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, Roesch EB, Chattopadhyaya J, Damha MJ, Bennett CF, et al. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Liu J, Corey DR. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE, Lima WF, Crooke ST, Prakash TP, Corey DR. Single-stranded RNAs Use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Bilsen PH, Jaspers L, Lombardi MS, Odekerken JC, Burright EN, Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum. Gene. Ther. 2008;19:710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombardi MS, Jaspers L, Spronkmans C, Gellera C, Taroni F, Di Maria E, Donato SD, Kaemmerer WF. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Monia BP, Johnston JF, Ecker DJ, Zounes MA, Lima WF, Freier SM. Selective inhibition of mutant Ha-ras mRNA expression by antisense oligonucleotides. J. Biol. Chem. 1992;267:19954–19962. [PubMed] [Google Scholar]
- 22.Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N, Hung G, Bennett CF, Freier SM, Hayden MR. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Monte SM, Vonsattel JP, Richardson EP., Jr Morphometric demonstration of atrophic changes in the cerebral cortex, white matter, and neostriatum in Huntington's disease. J. Neuropathol. Exp. Neurol. 1988;47:516–525. doi: 10.1097/00005072-198809000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel J-P, Zamore PD, Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 26.Swayze EE, Bhat B. In: Antisense Drug Technology: Principles, Strategies, and Applications. 2nd edn. Crooke ST, editor. Boca Raton: CRC Press; 2007. pp. 143–182. [Google Scholar]
- 27.Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, Andres PL, Mahoney K, Allred P, Alexander K, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dolgin E. Getting a fix on SMA. Nat. Med. 2012;18:1605–1605. [Google Scholar]
- 29.Teplova M, Minasov G, Tereshko V, Inamati GB, Cook PD, Manoharan M, Egli M. Crystal structure and improved antisense properties of 2′-O-(2-methoxyethyl)-RNA. Nat. Struct. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 30.Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP, Wancewicz EV, Witchell D, Swayze EE. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 31.Koller E, Vincent TM, Chappell A, De S, Manoharan M, Bennett CF. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varani G, McClain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnes TW, III, Turner DH. Long-range cooperativity due to C5-propynylation of oligopyrimidines enhances specific recognition by uridine of ribo-adenosine over ribo-guanosine. J. Am. Chem. Soc. 2001;123:9186–9187. doi: 10.1021/ja0157752. [DOI] [PubMed] [Google Scholar]
- 34.Testa SM, Disney MD, Turner DH, Kierzek R. Thermodynamics of RNA-RNA duplexes with 2- or 4-thiouridines: implications for antisense design and targeting a group I intron. Biochemistry. 1999;38:16655–16662. doi: 10.1021/bi991187d. [DOI] [PubMed] [Google Scholar]
- 35.Egli M, Pallan PS, Allerson CR, Prakash TP, Berdeja A, Yu J, Lee S, Watt A, Gaus H, Bhat B, et al. Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3′-fluoro hexitol nucleic acid (FHNA and Ara-FHNA) modified oligonucleotides. J. Am. Chem. Soc. 2011;133:16642–16649. doi: 10.1021/ja207086x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H, Lima WF, Zhang H, Fan A, Sun H, Crooke ST. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 37.Lima WF, Nichols JG, Wu H, Prakash TP, Migawa MT, Wyrzykiewicz TK, Bhat B, Crooke ST. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J. Biol. Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 38.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 39.Wu H, Lima WF, Crooke ST. Properties of cloned and expressed human RNase H1. J. Biol. Chem. 1999;274:28270–28278. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
- 40.Lima WF, Rose JB, Nichols JG, Wu H, Migawa MT, Wyrzykiewicz TK, Siwkowski AM, Crooke ST. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 2007;71:83–91. doi: 10.1124/mol.106.025015. [DOI] [PubMed] [Google Scholar]
- 41.Saha AK, Caulfield TJ, Hobbs C, Upson DA, Waychunas C, Yawman AM. 5'-Me-DNA - a new oligonucleotide analog: synthesis and biochemical properties. J. Org. Chem. 1995;60:788–789. [Google Scholar]
- 42.Li F, Sarkhel S, Wilds CJ, Wawrzak Z, Prakash TP, Manoharan M, Egli M. 2′-fluoroarabino- and arabinonucleic acid show different conformations, resulting in deviating RNA affinities and processing of their heteroduplexes with RNA by RNase H. Biochemistry. 2006;45:4141–4152. doi: 10.1021/bi052322r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams DM, Benseler F, Eckstein F. Properties of 2'-fluorothymidine-containing oligonucleotides: interaction with restriction endonuclease EcoRV. Biochemistry. 1991;30:4001–4009. doi: 10.1021/bi00230a027. [DOI] [PubMed] [Google Scholar]
- 44.Pallan PS, Allerson CR, Berdeja A, Seth PP, Swayze EE, Prakash TP, Egli M. Structure and nuclease resistance of 2′,4′-constrained 2′-O-methoxyethyl (cMOE) and 2′-O-ethyl (cEt) modified DNAs. Chem. Commun. 2012;48:8195–8197. doi: 10.1039/c2cc32286b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Southwell AL, Warby SC, Carroll JB, Doty CN, Skotte NH, Zhang W, Villanueva EB, Kovalik V, Xie Y, Pouladi MA, et al. A fully humanized transgenic mouse model of Huntington disease. Hum. Mol. Genet. 2013;22:18–34. doi: 10.1093/hmg/dds397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol. Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 47.Evers MM, Pepers BA, van Deutekom JCT, Mulders SAM, den Dunnen JT, Aartsma-Rus A, van Ommen G-JB, van Roon-Mom WMC. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS One. 2011;6:e24308. doi: 10.1371/journal.pone.0024308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, Smith DL, Faull RLM, Roos RAC, Howland D, et al. Aberrant splicing of HTT generates the pathogenic Exon 1 protein in Huntington disease. Proc. Natl Acad. Sci. USA. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lombardi MS, Jaspers L, Spronkmans C, Gellera C, Taroni F, Di Maria E, Donato SD, Kaemmerer WF. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.