Abstract
Vampire amoebae (vampyrellids) are predators of algae, fungi, protozoa and small metazoans known primarily from soils and in freshwater habitats. They are among the very few heterotrophic naked, filose and reticulose protists that have received some attention from a morphological and ecological point of view over the last few decades, because of the peculiar mode of feeding of known species. Yet, the true extent of their biodiversity remains largely unknown. Here we use a complementary approach of culturing and sequence database mining to address this issue, focusing our efforts on marine environments, where vampyrellids are very poorly known. We present 10 new vampyrellid isolates, 8 from marine or brackish sediments, and 2 from soil or freshwater sediment. Two of the former correspond to the genera Thalassomyxa Grell and Penardia Cash for which sequence data were previously unavailable. Small-subunit ribosomal DNA analysis confirms they are all related to previously sequenced vampyrellids. An exhaustive screening of the NCBI GenBank database and of 454 sequence data generated by the European BioMarKs consortium revealed hundreds of distinct environmental vampyrellid sequences. We show that vampyrellids are much more diverse than previously thought, especially in marine habitats. Our new isolates, which cover almost the full phylogenetic range of vampyrellid sequences revealed in this study, offer a rare opportunity to integrate data from environmental DNA surveys with phenotypic information. However, the very large genetic diversity we highlight within vampyrellids (especially in marine sediments and soils) contrasts with the paradoxically low morphological distinctiveness we observed across our isolates.
Keywords: BioMarKs, environmental clones, Penardia, SSU rDNA, Thalassomyxa, Vampyrellida
Introduction
Heterotrophic naked filose and reticulose amoebae are common in soils and in freshwater and marine sediments, and are suspected to have an important role in most microbial ecosystems (for example, Old and Chakraborty, 1986; Bonkowski et al., 2000). Yet they remain very poorly understood; for the vast majority of described species, molecular data are still missing, and very few lineages have been studied extensively morphologically and/or ecologically. Among them, vampire amoebae (vampyrellids) stand out as an exception. Known since the second half of the nineteenth century (Cienkowski, 1865, 1876; Zopf, 1885), they have attracted significantly more attention than most other naked filose or reticulose amoebae because of their peculiar mode of feeding on algae and fungal spores and hyphae. Vampyrellids typically bore holes in the prey's cell wall and suck up the content of the cell or enter the cell wall to digest the prey from the inside (for example, Lloyd, 1929; Hülsmann, 1993). An alternation of feeding amoebae and digestive cysts is observed, and ultrastructural data confirm the taxonomic unity of the group (Röpstorf et al., 1994). Freshwater vampyrellids such as Vampyrella (Cienkowski, 1865) and Lateromyxa (Hülsmann, 1993) comprise medium to large amoebae (25 up to 100 μm), with a usually round cell body from which filose pseudopodia arise. They display the cell wall-boring behaviour typical for the group. Larger soil and freshwater forms generally assigned to the genera Leptophrys (Hertwig and Lesser, 1874), Arachnula (Cienkowski, 1876) or Theratromyxa (Zwillenberg, 1953) can adopt a branched or even reticulate morphology and grow to sizes of up to a few millimetres (Old and Darbyshire, 1980). They may also engulf a whole prey (algae, fungi, protozoa or even small metazoans) by phagocytosis (for example, Dobell, 1913; Old and Darbyshire, 1980), and have been suggested to have a significant role in suppression of plant disease-causing organisms such as fungi and nematode worms in soils (Old, 1978; Old and Chakraborty, 1986).
It is only recently that the first small-subunit ribosomal DNA gene (SSU rDNA) sequences became available for any vampyrellids (Bass et al., 2009). Three large, branched, naked soil amoebae, one tentatively identified as Arachnula impatiens, and the others attributed to a new genus Platyreta, were shown to be closely related and part of a diverse lineage known up to then only from environmental sequences (‘Novel Clade 8' in Bass and Cavalier-Smith, 2004). More recently, Hess et al. (2012) isolated and sequenced three more species: Vampyrella lateritia, V. pendula and Leptophrys vorax. The clade containing all of these sequenced vampyrellids branches as sister to the phytomyxid plant and stramenopile parasites/symbionts within the Endomyxa part of phylum Cercozoa (Cavalier-Smith, 1998; 2002). Bass et al. (2009) also reported the results of cercozoan-wide environmental SSU rDNA library studies from a range of marine and non-marine habitat types. The environmental sequences that fell into the vampyrellids were mostly non-marine, with the exception of some basal lineages. The genus Vampyrelloides (Schepotieff, 1912), comprising large branched amoebae of up to 1000 μm, has been proposed to be the only described marine vampyrellid (for example, Patterson, 1999; Patterson et al., 2000), but was not included in the ultrastructural study by Röpstorf et al. (1994). Therefore, all genera currently accepted as vampyrellids based on life cycle, feeding behaviour, ultrastructural data (Röpstorf et al., 1994), and molecular phylogenetic analyses (Bass et al., 2009; Hess et al., 2012) seem to inhabit non-marine environments only. In the absence of microscopy evidence to the contrary and concordant with the available sequences in GenBank, Hess et al. (2012) suggested that vampyrellids may thus be largely confined to non-marine habitats. By contrast, we now used a combined approach of culturing and sequence database mining and present new results suggesting a marine origin for the whole vampyrellid clade, and revealing that the diversity and abundance of vampire amoebae in marine habitats is huge.
Materials and methods
Culturing methods and DNA extraction
Localities and dates of collection of samples for isolation are given in Table 1. In the culture lab, marine samples were placed in Artificial Sea Water or F2 medium (CCAP, Argyll, Scotland, UK) and freshwater samples in Volvic mineral water (Danone, Paris, France). For each sample, some dishes were supplemented with a selected food source grown separately on its own (the diatom Phaeodactylum sp. for marine samples, and baker's yeasts or the diatom Achnanthes sp. for freshwater samples). Dishes were checked regularly for vampyrellid-looking amoebae. For differential interference contrast light micrographs, we used a Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan) with × 40 (NA 0.6) or × 60 (NA 1.0) differential interference contrast water immersion lenses. Phase contrast micrographs were taken from a Leica DM IRB microscope (Leica, Wetzlar, Germany) and a Nikon Eclipse TS100 microscope (Nikon). Images were recorded on a Sony HD HDR-XR155 camcorder or HDV 1080i Handycam (Sony, Tokyo, Japan). Frames were captured using the software PMB version 5.2 (Sony) or Final Cut Express HD 3.5.1 (Apple Inc., Cupertino, CA, USA). As soon as vampyrellid-looking amoebae reached a reasonably high density in mixed cultures and had been photographed, DNA was extracted to avoid losing them altogether while trying to isolate them from all other protozoa in the sample. In most cases, this was done by picking individual cells (1 to ca 50) with a micropipette. DNA extraction was then performed either by using the UltraClean Soil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA) following the Maximum Yield Protocol, or by placing them in guanidium thiocyanate buffer and using a protocol described in Tkach and Pawlowski (1999). In a few cases where the vampyrellid population in the dish was already declining, total DNA extractions were performed on the whole mixed culture. Most of the culture medium was decanted off, and using a sterile scraper cells were collected from the bottom of the culture dish. Total DNA was then extracted from the pellet of organic material using the UltraClean Soil DNA Isolation Kit as above.
Table 1. Origin of the 10 new vampyrellid strains isolated in this study.
| Isolate | Collection date | Geographic origin | Habitat | Sample type | Phylogenetic positiona |
|---|---|---|---|---|---|
| NVam1 (Penardia sp.) | Spring 2010 | North Carolina, USA | Brackish | Muddy sediment, Cape Fear River, near the estuary | Clade C, subclade P |
| CAraX | Spring 2009 | California, USA | Marine | Sandy sediment, San Francisco Ocean Beach | Clade B, lineage B5 |
| MVa1x (Thalassomyxa sp.) | Summer 2011 | Majorca, Spain | Marine | Diatom-rich coastal sandy sediment | Clade B, subclade T |
| KibAr | Autumn 2007 | Cumbria, UKb | Marine | Coastal sediment and rock scrapings, Walney Island | Clade B, subclade T |
| En42C | Autumn 2007 | Cumbria, UKb | Marine | Coastal sediment and rock scrapings, Walney Island | Clade B, subclade T |
| V1ld4 c | Autumn 2010 | Cumbria, UKb | Marine | Coastal sediment and rock scrapings, Walney Island | Clade B, subclade T |
| V2ld4 | Autumn 2010 | Cumbria, UKb | Marine | Coastal sediment and rock scrapings, Walney Island | Clade B, subclade T |
| V1ld9 d | Autumn 2010 | Cumbria, UKb | Marine | Coastal sediment and rock scrapings, Walney Island | Clade B, subclade T |
| WaAra | Spring 2008 | Wales, UK | Freshwater | Mixture of moss and lichens from a garden in Gregynog | Clade A, Leptophryidae |
| BAra1 | Summer 2009 | Amazon basin, Brazil | Freshwater | Mixture of aquatic plants from a small Rio Negro tributary | Clade A, Leptophryidae |
Amplification and sequencing of the SSU rDNA
The complete SSU rDNA sequence was amplified from each isolate in two or three overlapping fragments, using different possible combinations of universal and lineage-specific primers listed in Supplementary Table S1. PCR amplifications were done in a total volume of 30 μl with an amplification profile typically consisting of 35 cycles with 30 s at 95 °C, 30 s at 56 °C, and 90 s at 72 °C, followed by 5 min at 72 °C for the final extension. PCR products of the appropriate length were excised from agarose gels, and cleaned following the protocol of the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). Purified PCR products were sequenced directly, or when needed, amplicons were first cloned into StrataClone SoloPack Competent Cells using the StrataClone PCR Cloning Kit (Stratagene, Agilent Technologies, Santa Clara, CA, USA). White colonies were screened using the primers M13for (5′-CGTTGTAAAACGACGGCCAGT-3′) and M13rev (5′-CACAGGAAACAGCTATGACCA-3′). Sequencing was performed with the BigDye Terminator v1.1 Cycle Sequencing Kit, and analysed with an ABI 3730xl DNA sequencer (Applied Biosystems, Life Technologies, Carlsbad, CA, USA). The new SSU rDNA sequences reported in this paper have been deposited in the NCBI GenBank database (accession numbers KC779511–KC779520). They were edited and aligned manually using the BioEdit software (Hall, 1999), following the secondary structure model proposed by (Wuyts et al., 2000).
Database mining by BLASTn searches and construction of the sequence data sets
Exhaustive BLASTn searches (Altschul et al., 1990) were performed using both our new and existing vampyrellid SSU rDNA sequences to seed searches to identify all vampyrellid clone sequences from environmental libraries present in the GenBank database (Supplementary Table S2). Further BLASTn searches were performed using manually truncated sequences (both at the 5′ and 3′ ends) as queries. This allowed retrieval of shorter clone sequences that would escape identification when using complete sequences as queries because of lower overall similarity scores compared with more distantly related but full-length sequences. All identified vampyrellid sequences were used as additional queries in further rounds of BLASTn searches until no more new unambiguous vampyrellid sequences could be found. Clones shorter than 400 bp were not considered. The presence of sequence chimeras was assessed by visual screening of the alignment in search for contradictory sequence signatures, and potential chimeric clones were confirmed as such by distance analyses based on different subsets of unambiguously aligned regions (as described in Berney et al., 2004). In total, six environmental clones were identified as chimeric (Supplementary Table S3). The data set we constructed for phylogenetic analyses contains 1470 unambiguously aligned positions and 120 sequences, and includes all sequences from vampyrellid isolates, a selection of identified vampyrellid environmental clones, members of all known Cercozoa lineages, and 20 outgroup sequences. Only environmental clones longer than 1000 bp were included, and clones >98% identical to another, longer one were discarded to avoid over-populating the figure. Missing data in partial environmental sequences were encoded as such (Ns). The non-vampyrellid part of any chimeric sequence included in this data set was also encoded as missing data.
We also had access to the 454 sequence data generated by the European BioMarKs consortium (http://www.BioMarKs.org/), of which all authors are members. Briefly, DNA- and complementary DNA (cDNA)-derived SSU rDNA amplicon sets were generated from samples taken from surface waters, the deep chlorophyll maximum layer, and sediment at six European offshore coastal stations. The SSU rDNA fragment targeted spans the longest and most phylogenetically informative hypervariable region of the gene (V4). Detailed information about the sampling sites, DNA and RNA extraction and PCR amplification protocols, and the extensive bioinformatic curation of the sequence data are available in Supplementary File 1 (see also Logares et al., 2012 and Rodríquez-Martínez et al., 2012). Vampyrellid reads were extracted from the BioMarKs sequence data and clustered into distinct V4 SSU rDNA types (‘SSU-types') as explained in detail in Supplementary File 1. The clusters are provided in fasta format in Supplementary File 2 and the reads are available as a sequence read archive in the European Nucleotide Archive (http://www.ebi.ac.uk/ena/data/view/ERP002516). All vampyrellid SSU-types were assembled into a second data set restricted to the V4 region of the SSU rDNA (360 unambiguously aligned positions and 534 sequences), also including some of our new isolates and relevant environmental clones from GenBank spanning the V4 region.
Maximum likelihood and Bayesian phylogenetic analyses
All phylogenetic analyses were performed using the GTR model of substitution (Lanave et al., 1984; Rodriguez et al., 1990), taking into account a gamma-shaped distribution of the rates of substitution among variable sites, with eight rate categories. All necessary parameters were estimated from the data sets. For both data sets, a maximum likelihood (ML) tree (Felsenstein, 1981) was determined with the program RaxML, version 7.0.3 (Stamatakis, 2006), using 200 inferences from distinct maximum parsimony starting trees. For the first data set, the reliability of internal branches was assessed with the bootstrap method (Felsenstein, 1985) using 200 non-parametric bootstrap replicates with 10 inferences for each from distinct maximum parsimony starting trees (option –b –# 200 –u 10). In addition, Bayesian analyses were performed with MrBayes version 3.1 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). Two runs of four simultaneous chains were run for 2 500 000 generations (heat parameters set to default), and trees were sampled every 100 generations. For each run 25 000 trees were sampled, 5000 of which were discarded as the burn-in. Posterior probabilities of the branching pattern were estimated from the 40 000 remaining trees and mapped onto the ML tree. The posterior probability 50% majority-rule consensus tree was fully compatible with the corresponding ML tree.
Results
New isolates and a revised vampyrellid phylogeny
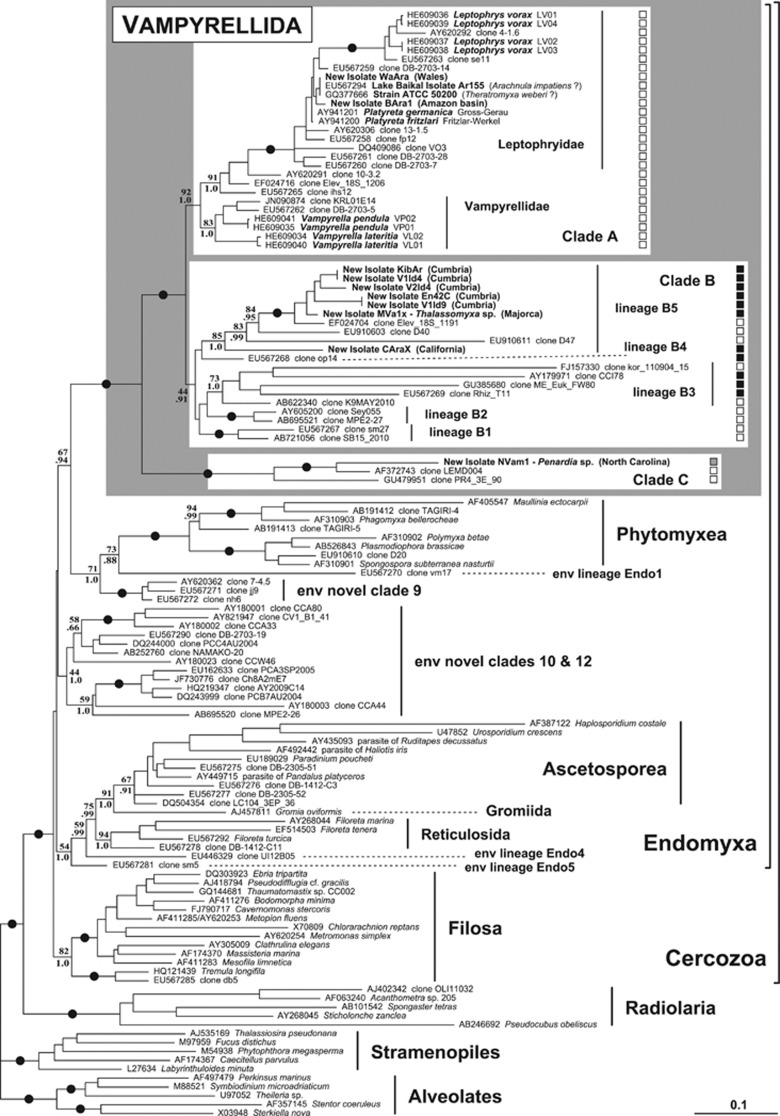
We isolated and sequenced ten new strains of vampyrellid-like naked, filose or reticulose amoebae (Table 1). SSU rDNA analyses showed that these represented eight distinct sequence types, six from marine or brackish sediments and two from soil or freshwater sediment. The light microscopic morphology of our isolates is illustrated in Figures 1, 2, 3; it is consistent with them belonging to Vampyrellida (Hess et al., 2012). As Figures 1, 2, 3 show the cells are very plastic and variable in shape and size, particularly in response to different levels and types of available food. The phylogenetic position of our new isolates is shown in Figure 4. They cluster together with all previously sequenced vampyrellid isolates and 28 putative vampyrellid environmental clones in a very robust clade with maximal statistical support. Our expanded data set confirms the position of vampyrellids as a sister group to Phytomyxea within the Endomyxa part of phylum Cercozoa (Bass et al,. 2009; Hess et al., 2012). Three main clades of vampyrellids are identified in Figure 4(A, B and C).
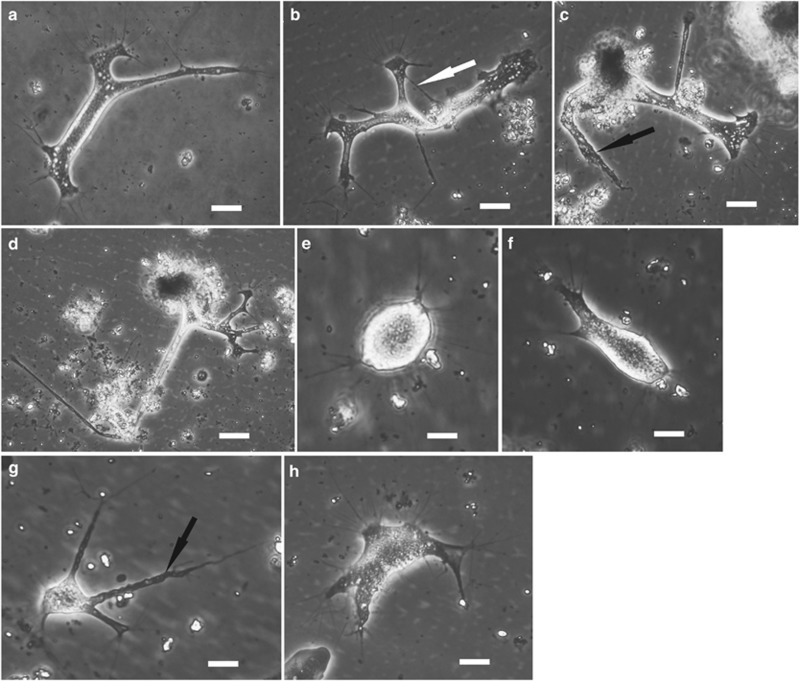
Figure 1.
Phase contrast light micrographs of our terrestrial isolates. (a) Isolate from Lake Baikal published previously (Bass et al., 2009). (b–d) Isolate WaAra (Wales, soil/ moss sample). Two different individuals (c and d are the same); this isolate was undistinguishable morphologically from the Baikal isolate, and with identical SSU rDNA sequence. (e–h) Isolate BAra1 (Amazon basin, associated with aquatic plants). Three different individuals (e and f are the same); cell body generally more compact than in the other soil isolates we observed. In all of these isolates, filopodia tend to be concentrated at the ends of cytoplasmic arms. The white arrow in b highlights a temporary anastomosis between two arms, and the black arrows in c and g show the typical pattern of rapid resorption of these arms when the cell changes direction. Scale bars: 20 μm in a–d; 10 μm in e–h.
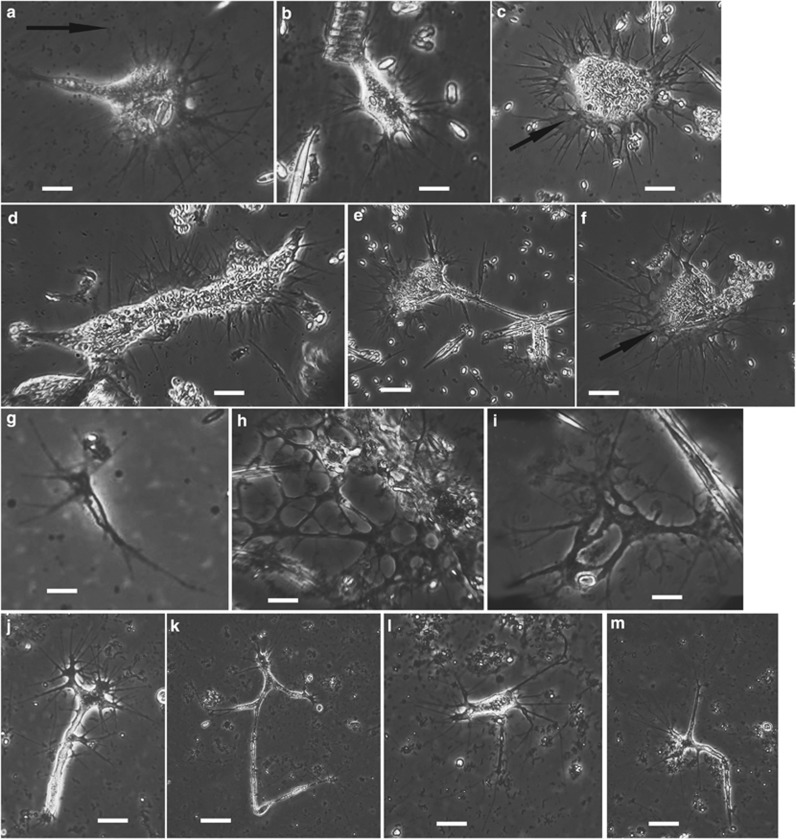
Figure 2.
Phase contrast light micrographs of isolates KibAr, V1ld4, En42C and V1ld9 (Cumbria, UK, marine sediment) and CAraX (California, USA, marine sediment). (a, b) Isolate KibAr (two different individuals). (c–f) Isolate V1ld4 (four different individuals). These two isolates have identical SSU rDNA sequences and were undistinguishable morphologically, with filopodia radiating from all around a generally compact cell body, only rarely assuming an elongated form (as in d). The closely related isolate V2ld4 looked very similar but died before good pictures could be taken. The cell shown in a illustrates the typical pattern of active movement for this morphotype, with regular resorption of the trailing cytoplasm (the arrow indicates the direction of movement). In b, the cell is shown attacking a cluster of diatoms. Occasionally two parts of a cell move in different directions, and instead of one being resorbed they can end up dividing into two viable smaller individuals by cell fission (illustrated in e). Arrows in c and f highlight the very thin cytoplasmic veil from which filopodia emerge. Temporary anastomoses at the base of filopodia are visible in f. (g) Isolate En42C (very small individual). This isolate died soon after initial observation. (h, i) Isolate V1ld9 (two different individuals). At maximum size the cells were highly reticulate with many lacunae; SSU rDNA sequence almost identical to that of isolate En42C. (j–m) Isolate CAraX (four different individuals). Highly branching morphotype similar to that observed in terrestrial isolates, with filopodia concentrated at the end of cytoplasmic arms. Scale bars: 20 μm in c, e, f, k; 15 μm in d, h, j, m; 10 μm in a, b, i, l; 5 μm in g.
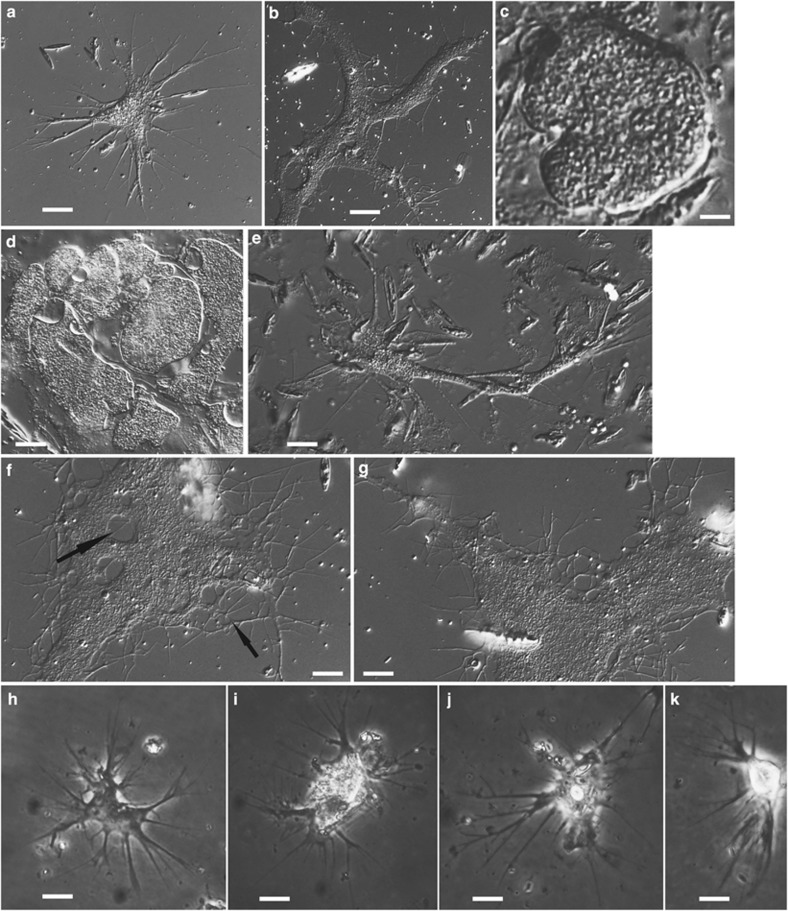
Figure 3.
Differential interference contrast light micrographs of isolate MVa1x (Thalassomyxa sp., Majorca, Spain, marine sediment) and phase contrast light micrographs of isolate NVam1 (Penardia sp., North Carolina, USA, brackish sediment). (a–g) Thalassomyxa sp. from Majorca; six different individuals (f and g are the same). Younger, more rapidly moving cells have a relatively compact (a) but sometimes elongated (e) morphology, similar to that of isolates KibAr and V1ld4 (see Figure 2). They can fuse into very large, slowly moving plasmodia (b, f, g) with lacunae in the cytoplasm and anastomoses between the peripheral filopodia (highlighted with arrows in f). Both smaller cells and giant plasmodia go into digestive cysts at regular intervals (c, d); a single plasmode can break up into many cysts of variable shapes and sizes (d). (h–k) Isolate NVam1 (Penardia sp.; four different individuals). Cell body generally intermediate between the more compact morphotype of some marine isolates and the branched morphotype of the terrestrial ones, but on average smaller than in all other isolates. Filopodia emerge from all around the cell; they are usually broader at the base and branching into thinner extensions. Temporary anastomoses frequently observed at the base of filopodia. Scale bars: 30 μm in a, e–g; 25 μm in b; 20 μm in d; 10 μm in h, i; 5 μm in c, j, k.
Figure 4.
SSU rDNA phylogeny of vampyrellids in a cercozoan context, showing the position of our new isolates with respect to previously published isolates and identified environmental clones from GenBank. Alveolates and stramenopiles were used to root the tree. The ML topology is shown; bootstrap support values after 200 replicates and Bayesian posterior probabilities (see Materials and methods section) are indicated at nodes when above 50% and/or 0.70, respectively. Black blobs represent support values at or above 95%/0.95. Vampyrellids cluster in three main clades A, B and C. New and previously published isolates are highlighted in bold. Black squares identify marine sequences and white squares identify terrestrial (soil or freshwater) sequences; isolate NVam1 was found in brackish sediment. More details about the ecological provenance of environmental sequences from GenBank are given in Supplementary Table S2.
Clade A is strongly supported by both ML and Bayesian methods and so far comprises only sequences derived from non-marine environmental libraries, and all existing isolates from non-marine (soil, freshwater and moss-associated) habitats. Morphologically, this clade includes amoebae ranging from highly branched and sometimes reticulate forms like Leptophrys and Platyreta to the globular Vampyrella spp. We did not notice any morphological differences between our new terrestrial isolate WaAra from Wales (Figures 1b–d) and our previously published isolate from Lake Baikal (Figure 1a). Freshwater isolate BAra1 (Amazon basin) was very similar but less often assumed an elongated form with several cytoplasmic arms extending simultaneously in many directions (Figures 1e–h). Sequences from both isolates are indeed very similar to existing sequences of the Lake Baikal isolate (EU567294), Platyreta spp. (AY941200 and AY941201), and an ATCC strain (now lost) tentatively identified as Theratromyxa weberi (Wegener Parfrey et al., 2010) with <2% overall sequence divergence. At the exception of genus Leptophrys, resolution within the terminal part of clade A (corresponding to family Leptophryidae) is poor, with very little variation between SSU rDNA sequences outside of the most variable regions. Detailed observations on new similar isolates would be necessary to confirm the identification of any of these organisms as either Arachnula or Theratromyxa. For now, we prefer to consider both isolates WaAra and Bara1 as unidentified vampyrellids.
Clade B is a weakly supported assemblage of many lineages from both marine and terrestrial environmental libraries, some from basically freshwater sites with high levels of dissolved minerals. It also contains all our new marine isolates, all of which are distinct from any sequence existing in GenBank. These isolates robustly branch together in a lineage (B5) including a few freshwater environmental clones. Isolate CAraX (California) forms the most basal branch of that lineage; it has a highly branching morphology, reminiscent of soil members of clade A (Figures 2j–m). Isolate MVa1x matches the original description of Thalassomyxa (Grell, 1985, 1992), with cells able to fuse into a giant plasmodium and forming digestive cysts at regular time intervals (Figures 3a–g); we believe it belongs to that genus. Isolates En42C and V1ld9 and isolates KibAr and V1ld4 turned out to be pairs of isolates with, respectively, identical or almost identical SSU rDNA sequences (<0.3% overall sequence divergence); they were found in samples collected in close locations in 2007 and 2010. Isolates En42C and V1ld9 displayed an ability to form a complex reticulum by anastomosis with many lacunae (Figures 2g–i). The remaining isolates from this clade tend to have a more compact cell shape, with filopodia emerging more evenly from all around the cell than is the case for most clade A representatives. They can sometimes also adopt a more elongate shape (Figures 2a–f).
Finally, clade C is strongly supported and a robustly basal sister lineage to A+B. It contains our new isolate NVam1, found in brackish sediment from North Carolina (Figures 3h–k). This isolate is slightly smaller than all others and displays an intermediate morphotype between the more compact cell body of the marine isolates KibAr and V1ld4 and the branched morphology of the terrestrial ones. Filopodia emerge from all around the cell; they are usually broader at the base and branching into thinner extensions. Temporary anastomoses are frequently observed at the base of filopodia. Isolate NVam1 matches the morphological and behavioural characteristics of Penardia (Cash, 1904), but the type species P. mutabilis was isolated from a Sphagnum bog, so our isolate must represent a distinct species in that genus. Clade C also includes two freshwater sequences, derived from a peat bog environmental survey (Lara et al., 2011) and a survey of eukaryote diversity in sediments from an anoxic lake (Dawson and Pace, 2002).
Ecological diversity of vampyrellids with emphasis on marine habitats
A comprehensive search of GenBank revealed 62 environmental SSU rDNA clones unambiguously attributable to vampyrellids (as of 31 October 2012), ranging from about 400 bp to full-length SSU rDNA sequences, mostly from libraries constructed using a variety of general eukaryotic primers (Supplementary Table S2). These clones were obtained in surveys of a wide variety of both marine and freshwater environments, from the water column to sediments ranging from oxic to anoxic (see Supplementary Table S2 for all references). Only clones longer than 1000 bp are included in Figure 4, whereas relevant shorter SSU rDNA fragments spanning the V4 region were included in the second data set (see below). However, Supplementary Table S2 lists the lineage affiliation of all other identified vampyrellid environmental clones following the labelling used in Figure 4.
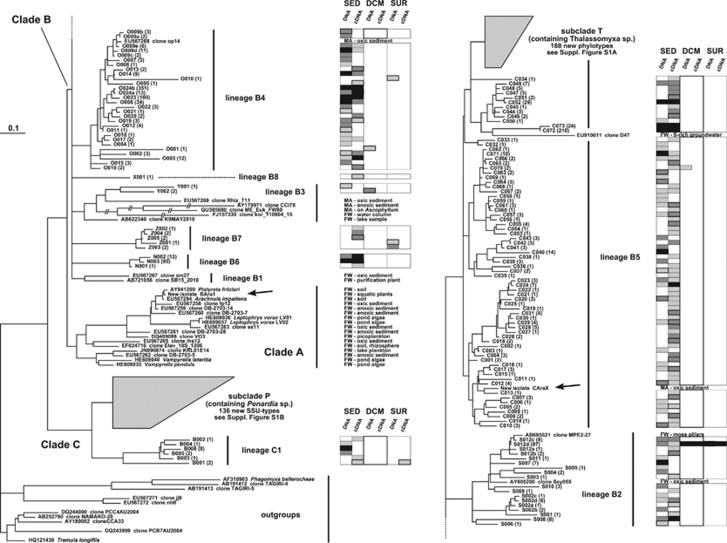
Figure 5 and Supplementary Figure S1 present the results of our extensive mining of the exclusively marine European BioMarKs consortium 454 sequence data. As explained in detail in Supplementary File 1, we identified 2766 vampyrellid reads in these data and used a stringent approach to cluster them into SSU types to ensure that we were not overestimating the uncovered genetic diversity within vampyrellids because of PCR and sequencing errors. This revealed 461 distinct vampyrellid SSU-types, the vast majority of which is novel and does not match known sequences that were presented in Figure 4. Figure 5 shows how the entirety of the identified BioMarKs vampyrellid sequences group in relation to characterised isolates and GenBank environmental clones from Figure 4. The BioMarKs sequences cluster into nine major lineages. The shortness of the V4 region made it uninformative to perform bootstrap analysis on this data set, so that the tree shown in Figure 5 should be regarded only as a graphical illustration of the uncovered vampyrellid diversity. However, the general ML tree topology obtained is remarkably similar to that obtained with the first data set (Figure 4), and specific V4 sequence signatures were observed that did support the unity and distinctiveness of all labelled lineages in Figure 5. Two of these, subclades P (within clade C, and containing our Penardia isolate) and T (within clade B, and containing our Thalassomyxa isolate) were so diverse in the BioMarKs data that they are shown separately in Supplementary Figure S1.
Figure 5.
Graphical representation of the diversity of vampyrellid SSU-types found in the BioMarKs data in relation to sequenced isolates, highlighting their habitat preferences. A ML tree is used, based on the V4 region of the SSU rDNA; it was rooted with selected members of other endomyxan lineages. Clades and lineages are labelled consistently with Figure 4. Some branch lengths within fast-evolving lineage B3 have been reduced by half. Sequences from our new isolates are highlighted with arrows. The ecological provenance of environmental sequences from GenBank is listed (see also Supplementary Table S2). That of BioMarKs V4 SSU-types is indicated in the boxes (SED, sediment; DCM, deep chlorophyll maximum; SUR, surface waters). In the labels of the BioMarKs SSU-types, the number in brackets indicates how many individual sequences belong to that SSU-type. For each SSU-type, varying shades of grey in the boxes give an indication of the number of individual sequences found in each particular sample type: light grey for unique sequences, medium grey for 2 to 5 sequences, dark grey for 6 to 20 sequences, and black for >20 sequences. The two most diverse lineages (subclades P and T) were collapsed for clarity, but the V4 diversity and ecological provenance of all SSU-types within these lineages is provided in Supplementary Figure S1.
Clade C was massively expanded by the BioMarKs sequences (136 distinct SSU-types), with many, often relatively long-branched lineages being added to the three sequences present in Figure 4 (subclade P), and a sister group to this (lineage C1) representing a highly distinct novel lineage. But the majority of the sequence types mined from BioMarKs (319 distinct SSU-types) group within clade B. Some significantly expand the diversity within lineages B2 and B4. Others represent novel but possibly rarer lineages (B6 to B8) with only few SSU-types identified. Finally, 261 SSU-types (57% of all identified BioMarKs SSU-types) group within lineage B5, together with all our new marine isolates. The majority of these are closely related to our Thalassomyxa and Cumbria isolates (subclade T), potentially making it the most diverse lineage of vampyrellids in marine habitats. In stark contrast, none of the BioMarKs sequences grouped within clade A, supporting an exclusively terrestrial origin for this clade.
The BioMarKs sequences are categorised according to their sampling provenances—surface waters, deep chlorophyll maximum layer, sediment—and whether the sequences came from DNA- or cDNA (originally RNA)-derived samples. Of the 461 SSU-types identified, 237 were found in DNA libraries and 348 in cDNA libraries, with only 124 (27% of the total) being present in both. This suggests that on average vampyrellids are more easily detected in RNA- rather than DNA-derived samples. This effect was particularly marked in the Thalassomyxa lineage, where 157 and 82 SSU-types were recovered from cDNA and DNA libraries, respectively (with 51 being present in both). However, because of potential distinct biases during extraction and PCR amplification between the two types of templates, we cannot judge whether these differences are significant or biologically informative.
Figure 5 and Supplementary Figure S1 show that vampyrellids were detected highly predominantly in sediments (87% of the vampyrellid reads). The 2766 vampyrellid reads identified represent 0.19% of the 1 476 249 eukaryotic reads in the BioMarKs data. However, when taking into account the proportion of eukaryotic reads in the data from sediments versus water column samples, vampyrellids actually account for 1.11% of all reads from sediment samples, as compared with only 0.03% of the total reads from water column samples. A few SSU-types in most lineages were found in planktonic samples as well as, or instead of in sediments (SSU-types T133 and T134b in subclade T were found multiple times in water column samples and never in the sediment, suggesting a different, unusual feeding or dispersal strategy). Also remarkable are the closely related pairs of SSU-types S012d+AB695521 (Figure 5) and T103+EF539120 (Supplementary Figure S1), which are strongly represented in all samples types (sediment, deep chlorophyll maximum, surface water; DNA and cDNA) and are also present in Genbank. It may reflect a general abundance and ubiquity of these lineages. This extends to non-marine habitats in the case of sequence AB695521, from moss pillars in an eastern Antarctic freshwater lake. Sequence EF539120 is from a subtropical Pacific picoplankton sample, which is concordant with very similar sequences showing a strong planktonic bias in the BioMarKs data.
Discussion
Phylogenetic implications of our results
Our new isolates span the entire phylogenetic range of the ‘Aconchulinida' clade on Figure 3 of Bass et al. (2009), and fully characterise Novel Clade 8 as defined in Bass and Cavalier-Smith (2004). Our findings support the designation of the whole clade as Vampyrellida (Hess et al., 2012), yet significantly expand the known diversity and phylogenetic structure of vampyrellids shown in all previous studies. In particular, we show that Grell's (1985, 1992) Thalassomyxa (isolate MVa1x, Figures 3a–g) and Cash's (1904) Penardia (isolate NVam1, Figures 3h–k), two amoebae considered up to now incertae sedis within eukaryotes (Patterson, 1999; Patterson et al., 2000) actually belong to vampyrellids. The general morphology of all vampyrellid isolates is conserved in terms of mode and tempo of cell and pseudopod movement. In sharp contrast, morphological variation within any isolate can be very important and overlap with that of distantly related isolates, so that morphology alone should not be expected to necessarily allow sufficient distinction between species, genera, or even the higher-level clades and lineages defined in Figure 4.
Our database searches highlight the extraordinary genetic diversity of vampyrellids. In Figure 4, vampyrellids comprise several dozen SSU rDNA genotypes distinct enough to be considered different genera. In fact, the order Vampyrellida appears as genetically diverse as the whole of the kingdom Fungi, as illustrated in Supplementary Figure S2. There is obviously a huge discrepancy between the abundance of vampyrellids in marine habitats, and the extreme paucity of taxonomic records of such organisms. We believe this can be mainly explained by the general lack of taxonomic knowledge on all large, naked, filose or reticulose amoebae, which means they are often simply ignored in ecological surveys because they cannot be properly identified.
All previously sequenced vampyrellids belong to clade A (Figure 4), so far detected only in soil and freshwater. Consistent with their morphology, two of our new isolates from terrestrial habitats belong to that clade and are part of a very diverse terminal lineage (family Leptophryidae in Hess et al., 2012). In an earlier study (Bass et al., 2009), available data suggested a marine origin for vampyrellids, with a single transition to freshwater habitats in the lineage leading to Leptophryidae. Our new isolates and the environmental clones identified in this study, particularly in marine environments, suggest a much more complicated evolutionary history for vampyrellids. Based on the phylogenies shown in Figures 4 and 5, a marine origin of vampyrellids is more parsimonious, but a dozen transitions from marine to freshwater life styles must be invoked to account for the observed tree topologies. Possibly this number is still an underestimation, as we are lacking data from terrestrial molecular diversity surveys of similar scope to the BioMarKs marine data analysed here. It has been suggested that marine-freshwater transitions are rare in protists (for example, Logares et al., 2009); vampyrellids appear to offer an exception to this generalisation.
Molecular and ecological diversity of vampyrellids
The high diversity of vampyrellid SSU-types in the BioMarKs V4 data (generated using general eukaryote primers) contrasts with usual observations on the relative abundance of SSU rDNA sequences of amoeboid organisms in environmental surveys compared with other protists. Many amoeboid lineages commonly observed in light microscopy surveys of various environments (in particular most lobose amoebae) are only rarely recovered in environmental DNA surveys of the same habitats. In some cases, sample preparation may lead to a physical exclusion of these organisms (or the particles with which they might be associated) before the DNA or RNA extraction, especially for water column samples. In addition, different amplification biases during environmental PCR reactions could account for this discrepancy, in relation to the sometimes unusual length and primary sequence divergence of the gene in these organisms, or the presence of possibly strong secondary structure patterns (as discussed in Berney et al., 2004). Amoeboid protist lineages such as Foraminifera (see, for example, Lecroq et al., 2009) or the recently sequenced genus Reticulamoeba (Bass et al., 2012) provide striking examples of this discrepancy. Based on their relative abundance in environmental surveys, vampyrellids seem to be less prone to such biases than other amoeboid organisms. This may be because they are multinucleated, and most have SSU rDNA sequences of relatively low divergence, matching the eukaryote-wide primers commonly used in environmental surveys. This suggests that the number of vampyrellid sequences recovered in such surveys is a significantly better indicator of their real abundance in the habitats tested than it is for most other amoeboid organisms. Our microscopy observations corroborate this suggested abundance. At least a few vampyrellid cells are invariably observed in environmental cultures derived from <0.5 g marine material (sediment, sand, algae, and so on), especially when enriched with appropriate food sources, suggesting that vampyrellids are densely distributed in nature and quick to proliferate and utilise available resources.
Clades B and C are major findings of this paper, accounting for the vast majority of vampyrellid lineages detected. In Supplementary File 1, we discuss reasons why we believe the SSU rDNA diversity we report reflects a true organismal diversity. Importantly, the level of diversity uncovered within these clades would, however, remain highly significant even if our SSU-types were overestimating the true organismal diversity they represent. Given the general physical similarity of the eight isolated strains representing the breadth of these lineages, an immediate question is: (how) do they differ? Figure 5 and Supplementary Figure S1 show that they were mostly found in marine sediments. There, they accounted for 1.11% of the total reads, suggesting they represent a significant proportion of the eukaryotic community in such habitats. A few are also planktonic, as described in the Results section, but broad-brush ecological differences cannot explain the high diversity within superficially similar sediment habitats. It is striking that such omnivorous predators seem significantly rarer in planktonic fractions, where there is much suitable food. Apparently the dispersed nature of these potential preys and the lack of extensive substrate for gliding and food capture largely protect planktonic cells from vampyrellid attack. This may, however, be more prevalent on planktonic flocs of material that would have been excluded from the BioMarKs water column samples, which were pre-filtered through a 2000 μm filter. The low coincidence of DNA- and RNA-derived signal suggests that there is differentiation between active and non-active lineages in the same samples, but these apparent differences may also be due to incomplete sampling (the sequencing depth for the BioMarKs V4 libraries did not achieve saturation) and differential ease of template amplification depending on the initial nucleic acid type. Similarly, about half of the non-marine vampyrellid environmental SSU rDNA sequences that we detected in GenBank belongs to family Leptophryidae in Clade A (see Supplementary Table S2). The huge genetic diversity within that family is consistent with ‘species'-level differences in many other protist groups (see, for example, Boenigk et al., 2012), which would suggest that terrestrial environments provide a wealth of distinct ecological niches, sufficient for scores of vampyrellid lineages with very similar morphologies but significantly distinct SSU-types to have evolved.
Food preference may be an important basis for lineage differentiation in vampyrellids. Hess et al. (2012) found some evidence for food specificity but noted that exceptions to many generalisations can be found, and too little is known about each taxon to use food preference as a taxonomic character. This agrees with observations in Bass et al. (2009) and older literature, and observations we made on our new isolates. On the other hand, some strains were difficult to maintain in culture even when a range of apparently suitable food sources was available, whereas others appeared more omnivorous. It is probable that laboratory culture conditions do not well reflect the nutritional surroundings and choices of vampyrellids in nature. The distribution of food types is likely to be patchier and generally sparser in natural habitats, and such conditions may better reveal differences in preference (rather than absolute tolerance) among vampyrellid lineages. Of particular interest is elucidating and quantifying their roles in important processes such as soil disease suppressivity and aquatic algal control, and more generally as multi-level engineers of food web structure. In conclusion, we emphasise the unique predatory mode of feeding in vampyrellids, which makes available to them a wide range of preys, in terms of phylogeny, size and lifestyle. This, considered in tandem with their diversity and abundance, suggests that they have a significant and unusual role in microbial ecosystems, transcending orthodox hierarchies in the microbial loop.
Acknowledgments
We thank NERC for a Standard Research Grant (NE/H009426/1) (DB and CB) and a New Investigator Grant (NE/H000887/1) (DB). We are grateful to Tom Cavalier-Smith, Ema Chao, Libby Snell, Joe Carlson and Josephine Scoble for help with sampling. SS is grateful to Hiroyuki Ogata for support and advice about phylogenetic mapping, and Professor Jean-Michel Claverie for providing access to the computer facility of the PACA-Bioinfo IBISA platform. The IGS laboratory is partially supported by the French ANR grant ANT-08-BDVA-003. The BioMarKs 454 sequence data were generated as part of a study supported by the EU-FP7 ERA-net program BiodivERsA, under the project BioMarKs (2008-6530). We thank all additional members of the BioMarKs consortium who participated to field sampling and data generation (in particular Stéphane Audic and the coordinator of the project Colomban de Vargas), as well as three anonymous reviewers for very useful comments on the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bass D, Cavalier-Smith T. Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa) Int J Syst Evol Microbiol. 2004;54:2393–2404. doi: 10.1099/ijs.0.63229-0. [DOI] [PubMed] [Google Scholar]
- Bass D, Chao EEY, Nikolaev S, Yabuki A, Ishida K, Berney C, et al. Phylogeny of novel naked filose and reticulose Cercozoa: Granofilosea cl. n. and Proteomyxidea revised. Protist. 2009;160:75–109. doi: 10.1016/j.protis.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Bass D, Yabuki A, Santini S, Romac S, Berney C. Reticulamoeba is a long-branched Granofilosean (Cercozoa) that is missing from sequence databases. PLoS One. 2012;7:e49090. doi: 10.1371/journal.pone.0049090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney C, Fahrni J, Pawlowski J. How many novel eukaryotic ‘kingdoms'? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2004;2:13. doi: 10.1186/1741-7007-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boenigk J, Ereshefsky M, Hoef-Emden K, Mallet J, Bass D. Concepts in protistology: species definitions and boundaries. Eur J Protistol. 2012;48:96–102. doi: 10.1016/j.ejop.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Bonkowski M, Cheng W, Griffiths BS, Alphei J, Scheu S. Microbial-faunal interactions in the rhizosphere and effects on plant growth. Eur J Soil Biol. 2000;36:135–147. [Google Scholar]
- Cash J. On some new and little-known British freshwater Rhizopoda. J Linn Soc (Zool) 1904;24:218–225. [Google Scholar]
- Cavalier-Smith T. A revised six-kingdom system of life. Biol Rev Camb Philos Soc. 1998;73:203–266. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cienkowski L. Beiträge zur Kenntnis der Monaden. Arch Mikrosk Anat. 1865;1:203–232. [Google Scholar]
- Cienkowski L. Über einige Rhizopoden und verwandte Organismen. Arch Mikrosk Anat. 1876;12:15–50. [Google Scholar]
- Dawson SC, Pace NR. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad Sci USA. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobell C. Observations on the life-history of Cienkowski's “Arachnula”. Arch Protistenkd. 1913;31:317–353. [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Grell KG. Der Formwechsel des plasmodialen Rhizopoden Thalassomyxa australis n.g., n.sp. Protistologica. 1985;21:215–233. [Google Scholar]
- Grell KG. A species of Thalassomyxa from the North coast of Jamaica. Arch Protistenkd. 1992;142:15–33. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hertwig R, Lesser E.1874Über Rhizopoden und denselben nahestchenden Organismen Arch Mikrosk Anat 10(Suppl)61–65. [Google Scholar]
- Hess S, Sausen N, Melkonian M. Shedding light on vampires: the phylogeny of vampyrellid amoebae revisited. PLoS One. 2012;7:e31165. doi: 10.1371/journal.pone.0031165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hülsmann N. Lateromyxa gallica n. g., n. sp. (Vampyrellidae): a filopodial amoeboid protist with a novel life cycle and conspicuous ultrastructural characters. J Euk Microbiol. 1993;40:141–149. [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Lara E, Mitchell EAD, Moreira D, López García P. Highly diverse and seasonally dynamic protest community in a pristine peat bog. Protist. 2011;162:14–32. doi: 10.1016/j.protis.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Lecroq B, Gooday AJ, Cedhagen T, Sabbatini A, Pawlowski J. Molecular analyses reveal high levels of eukaryotic richness associated with enigmatic deep-sea protists (Komokiacea) Mar Biodiv. 2009;39:45–55. [Google Scholar]
- Lloyd FE. The behavior of Vampyrella lateritia, with special reference to the work of Professor Chr. Gobi. Arch Protistenkd. 1929;67:219–236. [Google Scholar]
- Logares R, Audic S, Santini S, Pernice MC, de Vargas C, Massana R. Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing. ISME J. 2012;6:1823–1833. doi: 10.1038/ismej.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R, Bråte J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 2009;17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Old KM. Perforation and lysis of fungal spores by soil amoebae. Ann Appl Biol. 1978;89:128–131. [Google Scholar]
- Old KM, Chakraborty S. Mycophagous soil amoebae: their biology and significance in the ecology of soil-borne plant pathogens. Prog Protistol. 1986;1:163–194. [Google Scholar]
- Old KM, Darbyshire JF. Arachnula impatiens Cienk., a mycophagous giant amoeba from soil. Protistologica. 1980;16:277–287. [Google Scholar]
- Patterson DJ. The diversity of Eukaryotes. Am Nat. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- Patterson DJ, Simpson AGB, Rogerson A.2000Amoebae of uncertain affinitiesIn: Lee JJ, Leedale GF, Bradbury P, (eds)The Illustrated Guide to the Protozoa2nd ednSociety of Protozoologists: Lawrence, KS, USA; 804–827. [Google Scholar]
- Rodriguez F, Oliver JL, Marin A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Rodríquez-Martínez R, Rocap G, Logares R, Romac S, Massana R. Low evolutionary diversification in a widespread and abundant uncultured protist (MAST-4) Mol Biol Evol. 2012;29:1393–1406. doi: 10.1093/molbev/msr303. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Röpstorf P, Hülsmann N, Hausmann K. Comparative fine structural investigations of interphase and mitotic nuclei of vampyrellid filose amoebae. J Euk Microbiol. 1994;41:18–30. [Google Scholar]
- Schepotieff A. Untersuchungen über niedere Organismen. III. Monerenstudien. Zool Jahrb (Abt f Anat) 1912;32:367–400. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tkach V, Pawlowski J. A new method of DNA extraction from the ethanol-fixed parasitic worms. Acta Parasitol. 1999;44:147–148. [Google Scholar]
- Wegener Parfrey L, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML, et al. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59:518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, De Rijk P, Van de Peer Y, Pison G, Rousseeuw P, De Wachter R. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res. 2000;28:4698–4708. doi: 10.1093/nar/28.23.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf W.1885Die Pilzthiere oder SchleimpilzeIn: Schenk A, (ed)Handbuch des Botanik, Enzyklopädie der Naturwissenschaften Vol. 3Eduard Trewendt: Breslau; 1–174. [Google Scholar]
- Zwillenberg LO. Theratromyxa weberi, a new proteomyxean organism from soil. Ant Leeuw (J Microbiol Serol) 1953;19:101–116. doi: 10.1007/BF02594837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.