Abstract
α-Hydroxynitrosamine metabolites of nitrosamines decompose to a reactive diazohydroxide and an aldehyde. To test the hypothesis that the aldehydes contribute to the harmful effects of nitrosamines, the toxic and mutagenic activity of three model methylating agents were compared in Chinese hamster ovary cells expressing human O6-alkylguanine DNA alkyltransferase (AGT) or not. N-Nitrosomethylurethane (NMUr), acetoxymethylmethylnitrosamine (AMMN) and 4-(methylnitrosamino)-4-acetoxy-1-(3-pyridyl)-1-butanone (NNK-4-OAc) are all activated by ester hydrolysis to methanediazohydroxide. NMUr does not form an aldehyde whereas AMMN generates formaldehyde and NNK-4-OAc produces 4-oxo-1-(3-pyridyl)-1-butanone (OPB). Since these compounds were likely to alkylate DNA to different extents, the toxic and mutagenic activity of these compounds was normalized to the levels of the most cytotoxic and mutagenic DNA adduct, O6-mG, to assess if the aldehydes contributed to the toxicological properties of these methylating agents. Levels of 7-mG indicated that the differences in cytotoxic and mutagenic effects of these compounds resulted from differences in their ability to methylate DNA. When normalized against the levels of O6-mG, there was no difference between these three compounds in cells that lacked AGT. However, AMMN and NNK-4-OAc were more toxic than NMUr in cells expressing AGT when normalized against O6-mG levels. In addition, AMMN was more mutagenic than NNK-4-OAc and MNUr in these cells. These findings demonstrate that the aldehyde decomposition products of nitrosamines can contribute to the cytotoxic and/or mutagenic activity of methylating nitrosamines.
Keywords: nitrosamines, aldehyde, mutagenicity, toxicity, DNA repair
Introduction
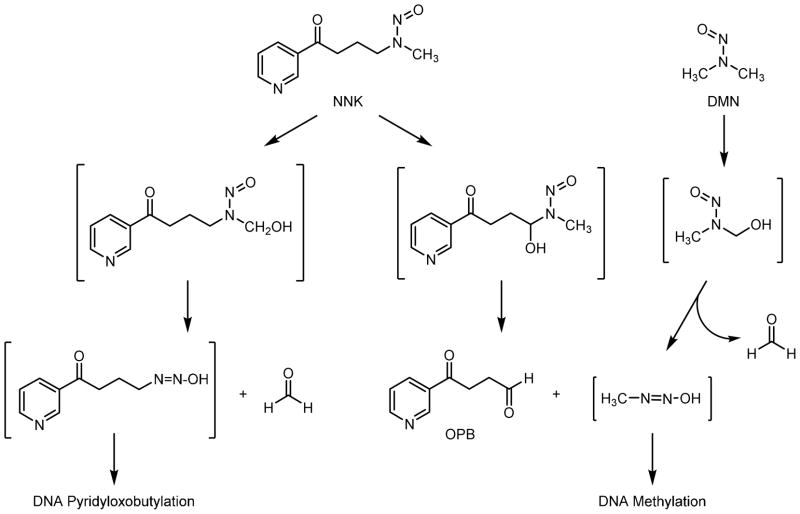
N-Nitrosodimethylamine (DMN) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) are two nitrosamines that are present in tobacco smoke.1,2 These compounds are potent carcinogens in laboratory animals. DMN induces liver, respiratory tract, kidney and blood vessel tumors by several different routes of exposure and is reasonably anticipated to be a human carcinogen (Group 2A).3–5 NNK is a powerful lung-specific carcinogen in animal models2 and has been designated as a human carcinogen (Group 1).6
Both DMN and NNK are activated by cytochrome P450-catalyzed oxidation of the carbon adjacent to the nitrosamine (Scheme 1).2,7–9 The resulting α-hydroxylnitrosamine metabolite then spontaneously decomposes to an alkanediazohydroxide and an aldehyde. The alkanediazohydroxide alkylates DNA to form DNA adducts that miscode during DNA replication, causing mutations that lead to tumor formation. The fate and biological activity of the aldehyde decomposition product is less well studied.
Scheme 1.
Pathways leading to DNA adduct and aldehyde formation for NNK and DMN.
Alpha-hydroxylation of DMN generates N-nitroso(hydroxymethyl)methylamine that decomposes to methanediazohydroxide and formaldehyde (Scheme 1). Methanediazohydroxide leads to a methyldiazonium ion that reacts with DNA to form 7-methylguanine (7-mG) and O6-methylguanine (O6-mG) along with other adducts.10 NNK undergoes two different α-hydroxylation reactions (Scheme 1). Methylene hydroxylation leads to the formation of methanediazohydroxide and 4-oxo-1-(3-pyridyl)-1-butanone (OPB), whereas methyl hydroxylation leads to the formation of 4-oxo-(3-pyridyl)-butanediazohydroxide and formaldehyde. As with DMN, the methylation pathway generates 7-mG and O6-mG.2 The pyridyloxobutylation pathway leads to a variety of adducts, four of which are 7-[4-3-(pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine, O2-[4-3-(pyridyl)-4-oxobut-1-yl]-2′-deoxycytosine, O2-[4-3-(pyridyl)-4-oxobut-1-yl]thymidine, and O6-[4-3-(pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine.11–14 Adducts from both pathways have been detected in tissue DNA from NNK-treated rodents.2,15–17
Simultaneously formed with the alkanediazohydroxide, nitrosamine-derived aldehydes may contribute to the toxicological properties of a nitrosamine by at least two pathways. First, they can be genotoxic. OPB induced sister chromatid exchanges in V79 cells and caused DNA single strand breaks in V79 cells and hepatocytes.18–20 Similarly, formaldehyde caused DNA single strand breaks and induced DNA-protein crosslinks in cells.20,21 Formaldehyde also reacted with dA, dC, and dG generating DNA adducts.22–26 Significant to their possible importance in nitrosamine derived cancer, formaldehyde DNA adducts are generated from DMN and NNK in vivo and the corresponding α-acetoxynitrosamines in vitro.27,28 This DNA damage may lead to mutations. Formaldehyde was mutagenic in human cells, as well as Chinese hamster ovary (CHO), V79 and mouse lymphoma cells.8,29–32 OPB has not been tested for its mutagenic properties.
A second way that aldehydes can contribute to the harmful effects of nitrosamines is by inhibiting the repair of DNA adducts formed from the alkanediazohydroxides. Formaldehyde inhibited the removal of O6-mG from DNA by the repair protein, O6-alkylguanine DNA alkyltransferase (AGT), and potentiated the toxicity and mutagenicity of N-methylnitrosourea in normal human pulmonary fibroblasts.8 OPB but not formaldehyde reduced the activity of AGT in hepatocytes.33 Both formaldehyde and OPB synergistically increased the number of single strand breaks induced by N-methylnitrosourea.34
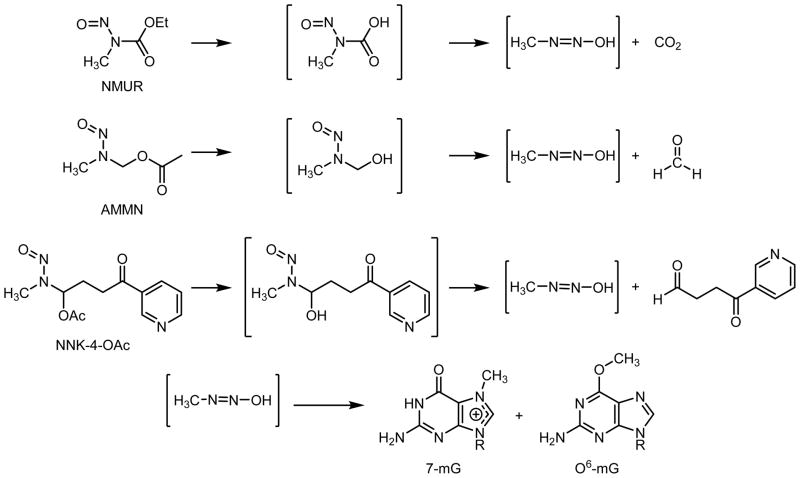
In this report, we test the hypothesis that aldehydes contribute to the toxicological properties of nitrosamines by generating mutagenic and/or toxic DNA adducts and/or by interfering with repair of alkyl DNA adducts. Our overall strategy was to compare the toxic and mutagenic activity of N-methylnitrosourethane (NMUr) to that of N-acetoxymethyl-N-methylnitrosamine (AMMN) and 4-(methylnitrosamino)-4-acetoxy-1-(3-pyridyl)-1-butanone (NNK-4-OAc) in CHO cells expressing or not expressing human AGT. These model compounds are all activated by ester hydrolysis to generate the reactive methanediazohydroxide (Scheme 2). No aldehyde is formed during the decomposition of NMUr. AMMN hydrolyzes to generate the methylating agent and formaldehyde35 whereas NNK-4-OAc is expected to generate methandiazohydroxide and OPB. Since these compounds were likely to have different rates of hydrolysis within the cell, methyl DNA adduct levels were measured to determine how efficient each model compound was at generating DNA damage. The most abundant adduct, 7-methylguanine, was employed as a dosimeter of the total amount of methanediazohydroxide that reached nuclear DNA. Levels of O6-mG were utilized to determine if the aldehyde modulated the toxicological properties of the methylating agent since this adduct is the major cytotoxic and mutagenic adduct generated by methanediazohydroxide.36,37 A comparison of the toxic and mutagenic properties of AMMN or NNK-4-OAc to NMUr at concentrations that yielded similar initial levels of O6-mG provided evidence that these aldehydes may contribute to the toxicological properties of methylating nitrosamines.
Scheme 2.
Pathway of decomposition for NMUr, AMMN and NNK-4-OAc to methandiazohydroxide and subsequent DNA adduct formation.
Materials and Methods
Chemicals
NMUr and AMMN were obtained from the National Cancer Institute’s Chemical Carcinogen Reference Standards Repository (Midwest Research Institute, Kansas City, MO). 3-[2-(3-Pyridyl)-1,3-dithian-2-yl]propan-1-al was synthesized as previously described.38 Porcine liver esterase, crystal violet and 6-thioguanine were purchased from Sigma (St. Louis, MO). Calf thymus DNA (Sigma) was cleaned up with phenol-chloroform extraction prior to use. All other chemicals were purchased from Aldrich (Milwaukee, WI).
Instrumentation
1H NMR spectra were obtained either at 300 MHz or at 500 MHz on a Varian Inova-300 or 500 NMR spectrometer (Chemistry Department of the University of Minnesota, Minneapolis) in CDCl3.
2-[3-Acetoxy-3-(methylnitrosamino)propan-1-yl]-2-(3-pyridyl)-1,3-dithiane
A solution of 3-[2-(3-pyridyl)-1,3-dithian-2-yl]propan-1-al (390 mg, 1.54 mmol) in glacial acetic acid (5 mL) was added to a solution of methylamine hydrochloride (104 mg, 1.54 mmol) in glacial acetic acid (7 mL). After the reaction mixture was cooled to 10–15 °C, an aqueous solution of sodium nitrite (215 mg, 3.0 mmol) was added. The reaction was stirred at 10–15 °C overnight. Acetic acid was removed in vacuo and a saturated solution of NaHCO3 (30 mL) was added to the residue. The solution was extracted with chloroform (2 × 30 mL). The chloroform layer was washed with water (3 × 30 mL), dried over anhydrous MgSO4, and evaporated to dryness. The residue was then purified by preparative TLC on silica gel 150 Å (size: 20 × 20 cm, 1000 μm thickness, 60F254) which was eluted with EtOAc/hexanes (50:50) to give the product as a light yellow solid (33 mg, 0.09 mmol, 6% yield). 1H NMR (300 MHz, CDCl3) δ 9.1 (s, 1H), 8.5 (d, 1H), 8.2 (d, 1H), 7.3 (m, 1H), 6.96 (t, 1H), 2.87 (s, 3H), 2.69 (m, 4H), 1.8–2.2 (m, 9H). MS m/z = 356 [M + H]+, 296, 267, 161.
4-Acetoxy-4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK-4-OAc)
N-Chlorosuccinimide (400 mg, 3.0 mmol) was added to a solution of 2-[3-acetoxy-3-(methylnitrosamino)propan-1-yl]-2-(3-pyridyl)-1,3-dithiane (110 mg, 0.30 mmol) in acetonitrile containing 20% 20 mM sodium phosphate, pH 7.0 (10 mL). The reaction was stirred for 30 minutes at room temperature. The product was extracted with ether. The extract was dried over anhydrous MgSO4, and evaporated to dryness. The residue was purified by preparative TLC on silica gel 150 Å (size: 20 × 20 cm, 1000 μm thickness, 60F254) eluting with ether to give the product as a light yellow solid (39 mg, 0.15 mmol, 50 % yield). 1H NMR (500 MHz, CDCl3) δ 9.1 (s, 1H), 8.8 (d, 1H), 8.2 (d, 1H), 7.4 (m, 1H), 7.2 (t, 1H), 3.1 (m, 2H), 3.0 (s, 3H), 2.6 (m, 2H), 2.1 (s, 3H). MS m/z= 265[M + H]+, 206, 177, 164, 146.
In Vitro DNA Alkylation Reactions
The extent of DNA methylation in vitro was determined using a previously published method.39 Briefly, 0 or 10 μM NMUr, AMMN or NNK-4-OAc was incubated with calf thymus DNA (2 mg) in the presence of 5.5 units porcine liver esterase (17 units/mg) in 15 mM sodium citrate buffer, pH 7, at room temperature (total volume = 1 mL). After 1 h, NaCl (2 mM) was added for a final concentration of 0.33 mM and the DNA was precipitated with ethanol. It washed three times with 70% ethanol, then three times with 100% ethanol and dried under nitrogen for storage at −20 °C until DNA adduct analysis.
Cell lines
These experiments were performed with Chinese hamster ovary cell line, AA8, stably transfected with the pCMV-neo-Bam vector without an inserted cDNA sequence (CHOpcDNA3) or the same plasmid expressing human AGT (CHOAGT); the parental cell line (AA8) does not express the repair protein AGT.40 Western blot analysis confirmed the expression of AGT in CHOAGT cells.40 The AGT activity in these cells was approximately 450 fmol/mg protein.40 The cell lines were maintained in Minimum Essential Medium Alpha Modification (αMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 1 mg/ml geneticin (Invitrogen) and 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 incubator. Pre-existing HPRT mutants were eliminated by growing the cells for three days in complete medium supplemented with 100 μM hypoxanthine, 0.4 μM aminopterin and 16 μM thymidine as previously described.41
Cytotoxicity and mutagenicity assay
Cytotoxicity and mutation frequency at the hprt locus were determined according to published methods.42,43 Twenty four h prior to treatment, 6-well or 60 mm plates were seeded with 2 × 105 cells or 5 × 105 cells/plate, respectively. The cells were exposed to NMUr (CHOpcDNA3: 0–8 μM; CHOAGT: 0–250 μM), AMMN (CHOpcDNA3: 0–4 μM; CHOAGT: 0–60 μM) or NNK-4-OAc (CHOpcDNA3: 0–2.5 μM; CHOAGT: 0–40 μM) for 1 h. CMU and AMMN were delivered as concentrated aqueous solutions; NNK-4-OAc was dissolved in DMSO. The control cells in the NNK-4-OAc experiments were exposed to DMSO alone (120 μL DMSO/4 mL media). Each treatment was performed in duplicate (six-well plates) or triplicate (60 mm plates). The cells were washed with PBS and further grown in fresh medium for 16–24 h. The cells were then harvested and counted, and were re-plated for measurement of colony forming ability (cytotoxicity) and mutant frequency. Each experiment was repeated three-four times.
Colony forming ability was determined by plating 100 cells from each treatment into 6-well plates.42 Two wells were plated per treatment. The plating efficiencies were similar for both cell lines. After 6–7 days growth in normal medium, colonies were stained with 0.05% crystal violet and were counted with an aCOLyte colony counter (Synbiosis, Frederick, MD). Cell survival was normalized to the number of colonies from untreated cells.
For the mutagenesis assay, each treated culture was plated into 60 mm dishes and grown in normal medium, with sub-culturing, to allow phenotypic expression of induced mutants. After 7 days, the cells were harvested, counted and plated for determination of cloning efficiency and mutant frequency. The cloning efficiency was determined by seeding 200 cells into 60 mm dishes in normal medium. Colonies were stained with crystal violet after 6–7 days growth and counted. The mutant frequency was determined by plating 100,000 cells into 100 mm dishes in medium containing 5 μg/ml 6-thioguanine to select for mutant colonies. Ten dishes per treatment were plated. The medium was changed every 3 to 4 days. After 10 days, the colonies were stained and counted with an aCOLyte colony counter. The mutant frequency was expressed as the number of 6-thioguanine resistant colonies per 100,000 viable cells.
DNA isolation
Cells (2 × 106) were plated onto 150 mm culture dishes. Three days later, they were exposed to NMUr (CHOpcDNA3: 0.5, 1, or 2 μM; CHOAGT: 50, 100 or 150 μM), AMMN (CHOpcDNA3: 0.25, 0.5, or 1 μM; CHOAGT: 10, 30 or 60 μM) and NNK-4-OAc (CHOpcDNA3: 0.05, 0.25, or 0.5 μM; CHOAGT: 5, 10 or 20 μM) for 1 h. These concentrations represented low, medium and high toxic concentrations of methylating agent. Immediately after the 1 h treatment, cells from 2–3 plates were pooled for DNA isolation. Triplicate samples were generated at each concentration. DNA was extracted using a Qiagen Genomic DNA Midi Kit (Valencia, CA) following the manufacturer’s protocol with minor modifications. Briefly, the cells were lysed and applied to a Genomic-tip 100/G column. The column was washed with buffer QC three times, and eluted with buffer QF. The eluates were then precipitated with isopropanol and the precipitated DNA was washed three times with 70% ethanol, and then three times with 100% ethanol. DNA was dried under nitrogen and stored at − 20 °C until analysis.
DNA Adducts Analysis
DNA (50–100 μg) was dissolved in 10 mM sodium phosphate, pH 7 (100 μL) and spiked with [13C12H3]7-methylguanine (0.6–10 pmol) and [2H3]O6-mdG (30 fmol). The solution was heated at 80°C for 60 min. A portion of the hydrolysates (75 μL) was reserved for the determination of guanine concentration. After cooling on ice for 30 min, samples were neutralized with sodium hydroxide. The remainder of the acid hydrolysate was applied to a Strata-X C18 cartridge (30 μm, 30 mg, Phenomenex, Torrance, CA). The cartridge was sequentially eluted with 1 mL water and 1 mL 100% methanol. The 100% methanol fraction contained the adducts; it was collected and concentrated to dryness under reduced pressure. The residue was resuspended in 25 mM ammonium acetate (16 μL) for LC/MS/MS analysis.
Capillary LC/ESI-MS/MS analyses were carried out on a TSQ Vantage mass spectrometer (Thermo Electron, Bellafonte, PA) interfaced with an Agilent 1100 series capillary HPLC (Agilent Technologies, Palo Alto, CA). Analyses were performed in the positive ion mode with N2 as the nebulizing and drying gas. The voltage setting was 3.2 kV and the heated capillary was 250 °C. Samples (injection vol: 8 μL) were separated on a 150 × 0.5 mm, 3 μm Hypersil GOLD PFP column (Thermo Fischer, Torrance, CA). The column was eluted with a 25 min linear gradient from 25 mM ammonium acetate containing 2% of 3:1 methanol-acetonitrile to 25 mM ammonium acetate containing 25% 3:1 methanol-acetonitrile at a flow rate of 10 μL/min. Quantitation was done with selected reaction monitoring (SRM). The mass transitions (parent to product) monitored were 166→149 for 7-mG and O6-mG, 169→152 for [2H3]O6-mG, and 170→153 for [13C12H3]7-mG. The retention times for 7-mG and O6-mG were 12.7 and 18.5, respectively. The amount of 7-mG and O6-mG in each sample was determined from the product of the area ratio (unlabeled DNA adduct/labeled standard) and the amount of internal standard added. Calibration curves were constructed from known amounts of authentic standards spiked with known amounts of internal standard. Guanine concentrations were determined as previously described.44 The amount of DNA adducts were normalized to the amount of the guanine in each sample.
Statistical methods
The statistical analysis related endpoints of interest, either cytotoxicity (fraction of cells surviving) or mutagenicity (number of mutant colonies) to (a) the three methylating agents of interest (b) levels of O6-mG produced by exposure to the methylating agents and (c) interactions between (a) and (b). In addition, the analysis sought to determine if cell line AGT expression (+/-) was related to cytotoxicity or mutagenicity. Tests for the main effects of O6-mG and the interactions between O6-mG and either compound or AGT expression were of primary interest in the analyses. We tested the null hypothesis that these effects and interactions were zero using standard regression/analysis of variance methods as built into the program R (using the lm procedure).
Results
Synthesis
These studies required the synthesis of NNK-4-OAc. This compound was achieved via the route shown in Scheme 3. Attempts to synthesize the compound by combining OPB, methylamine and sodium nitrite in acetic acid were unproductive. Protection of the ketone group of OPB with 1,3-dithiane allowed for the successful preparation of the intermediate, 2-[3-acetoxy-3-(methylnitrosamino)propan-1-yl]-2-(3-pyridyl)-1,3-dithiane, albeit in low yield. Deprotection of the ketone was achieved with N-chlorosuccinimide in buffered aqueous acetonitrile. The NMR and mass spectra of the product were consistent with the desired structure.
Scheme 3.
Synthesis of NNK-4-OAc; NCS = N-chlorosuccimide
In vitro DNA alkylation reactions
To confirm that the three methylating agents generate methyl DNA adducts following ester hydrolysis, calf thymus DNA was reacted with 10 μM of each methylating agent in vitro in the presence of esterase according to published methods.39 All three compounds generated similar levels of 7-mG and O6-mG under these reaction conditions (Table 1). In addition, the ratio of 7-mG/O6-mG was not impacted by the chemical structure of the methylating nitrosamines. Therefore, we will use a ratio of 7 as the expected value for this ratio in the cell-based studies when there is no repair of O6-mG.
Table 1.
Levels of 7-mG and O6-mG detected in calf thymus DNA following 1 h treatment with 10 μM NMUr, AMMN or NNK-4-OAc in vitro.a
| 7-mG | O6-mG | 7-mG/O6-mG | |
|---|---|---|---|
|
| |||
| (pmol adduct/μmol guanine) | |||
| NMUr | 138 ± 14 | 21 ± 4 | 7.2 ± 1.8 |
| AMMN | 121 ± 6 | 17 ± 4 | 6.8 ± 1.3 |
| NNK-4-OAc | 106 ± 4 | 17 ± 1 | 6.3 ± 0.4 |
Each methylating agent (10 μM) was incubated with calf thymus DNA (2 mg/mL) in the presence of 5.5 units of esterase in 15 mM sodium citrate, pH 7, for 1 h at RT. DNA was isolated by precipitation and adducts were measured by LC-MS/MS analysis.
Cytotoxicity and mutagenesis
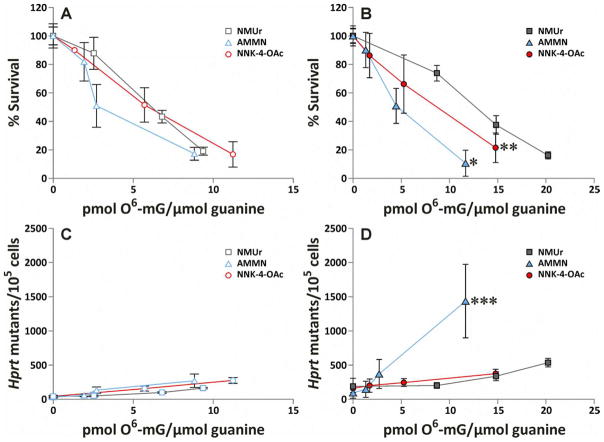
The toxic and mutagenic activity of NMUr, AMMN and NNK-4-OAc were determined in CHOpcDNA3 and CHOAGT cell lines. The cells were treated with increasing concentrations of the methylating agents for 1 h and then plated to assess cytotoxicity or mutagenicity. Cell survival was determined via a colony forming assay. All three compounds were much more toxic in the cells lacking hAGT (CHOpcDNA3) (Figure 1A–C). The EC50s for cytotoxicity indicated that NNK-4-OAc was the most toxic compound in either cell line whereas NMUr is the least toxic (Table 2). The difference between the three compounds is greater in the CHOAGT cell line (8 fold difference in EC50) than in the CHOpcDNA3 cell line (4 fold difference in EC50).
Figure 1.
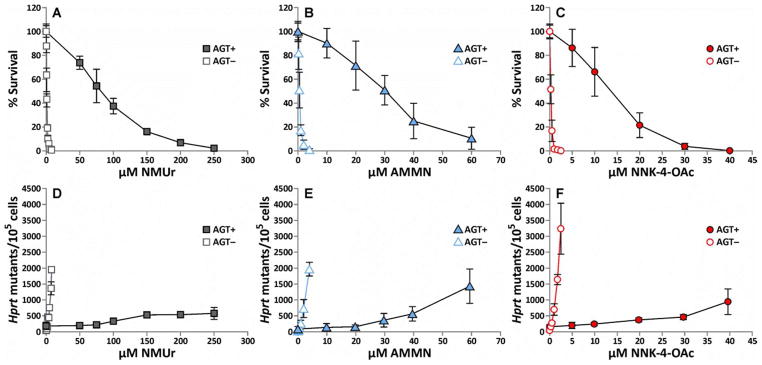
Cytotoxicity (A–C) and hprt mutagenic activity (D–F) in CHOpcDNA3 (AGT−) and CHOAGT (AGT+) cell lines following a 1 h treatment with increasing concentrations of NMUr (A, D), AMMN (B, E) or NNK-4-OAc (C, F). The values represent the average of 3–4 separate experiments performed with duplicates or triplicates. The error bars represent the standard deviation. The lines were generated by connecting the data points. Data used to generate this figure are presented in Supplemental Data.
Table 2.
EC50 values determined for each methylating agent in CHO cell lines.a
| Compound | EC50 (μM) | EC50 Ratio CHOAGT/CHOpcDNA3 |
|
|---|---|---|---|
|
| |||
| CHOpcDNA3 | CHOAGT | ||
| NMUr | 0.95 ± 0.03 | 80 ± 0.2 | 80 |
| AMMN | 0.51 ± 0.01 | 30 ± 1 | 60 |
| NNK-4-OAc | 0.26 ± 0.01 | 13 ± 0.4 | 50 |
CHO cell lines were incubated with NMUr (CHOpcDNA3: 0–8 μM; CHOAGT: 0–250 μM), AMMN (CHOpcDNA3: 0–4 μM; CHOAGT: 0–60 μM) or NNK-4-OAc (CHOpcDNA3: 0–2.5 μM; CHOAGT: 0–40 μM) for 1 h. Colony forming ability was determined 6–7 days following treatment. Data in Figure 1 was used to calculate EC50 values. The numbers are the average ± S.D.
To determine the ability of the methylating agents to induce point mutations, the mutation frequency in the hprt gene was measured by selecting mutants with 6-thioguanine. All three methylating agents caused a dose dependent increase in mutation frequency with the cells lacking AGT being considerably more sensitive to their mutagenic effects (Figure 1D–F). Similar to the cytotoxicity results, the order of mutagenic activity at equal concentrations was NNK-4-OAc > AMMN > NMUr in both cell lines.
DNA adducts analysis
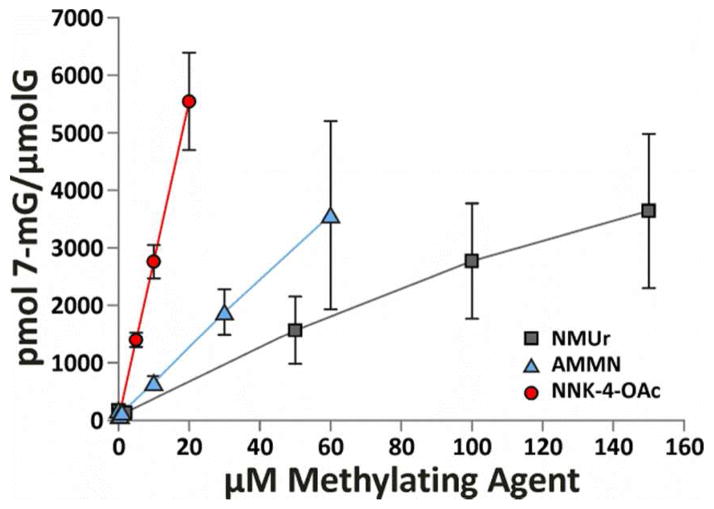
To normalize the toxic and genotoxic effects of these methylating agents to their ability to alkylate DNA, levels of 7-mG and O6-mG were measured in DNA isolated from cells immediately following a 1 h exposure to the methylating agents. For each compound, three concentrations from the toxicity studies were selected for DNA adduct measurements in each cell line. For each compound and cell line, the concentrations selected represented a range of cytotoxicity (low, medium and high). Neutral thermal and subsequent acid hydrolysates were analyzed by LC-MS/MS to determine the levels of 7-mG and O6-mG, respectively (Table 3). The analysis of 7-mG levels in the cells lacking AGT (CHOpcDNA3) was complicated by inconsistent levels of a co-eluting peak so these levels are not reported. This contamination did not significantly affect the analysis of this adduct in the DNA from CHOAGT cells because these cells were treated with much higher concentration of methylating agents. Consequently, the levels of 7-mG were much higher than the co-eluting contamination. A concentration dependent increase in 7-mG levels was observed for each compound in these cells (Figure 2). NNK-4-OAc generated the most DNA adducts followed by AMMN and, then, NMUr. As expected, these data parallel the observed toxicity differences between the three methylating agents in each cell line.
Table 3.
Levels of 7-mG and O6-mG in CHO cells following 1 h treatment with NMUr, AMMN or NNK-4-OAc.a
| μM | CHOpcDNA3
|
μM | CHOAGT
|
||
|---|---|---|---|---|---|
| O6-mGb | 7-mG | O6-mG | 7mG/O6-mG | ||
|
|
|
||||
| (pmol/μmol guanine) | (pmol/μmol guanine) | ||||
|
|
|
||||
| NMUr | |||||
| 0.5 | 2.5 ± 0.7 | 50 | 1570 ± 590 | 8.7 ± 3.0 | 188 ± 73 |
| 1 | 6.8 ± 2.1 | 100 | 2770 ± 1000 | 15 ± 5 | 207 ± 110 |
| 2 | 9.4 | 150 | 3640 ± 1340 | 20 ± 6 | 192 ± 90 |
| AMMN | |||||
| 0.25 | 1.9 ± 0.6 | 10 | 653 ± 112 | 1.3 ± 0.0 | 494 ± 82c |
| 0.5 | 2.7 ± 1.0 | 30 | 1880 ± 400 | 4.5 ± 0.5 | 430 ± 131c |
| 1 | 8.8 ± 3.7 | 60 | 3570 ± 160 | 12 ± 4 | 393 ± 359d |
| NNK-4-OAc | |||||
| 0.05 | 1.3 ± 0.5 | 5 | 1400 ± 120 | 1.7 ± 0.2 | 823 ± 130c |
| 0.25 | 5.7 ± 1.9 | 10 | 2760 ± 290 | 5.3 ± 1.2 | 537 ± 89c |
| 0.5 | 11 ± 6 | 20 | 5540 ± 850 | 15 ± 9 | 491 ± 300c |
CHO cell lines were incubated with NMUr, AMMN or NNK-4-OAc at the indicated concentrations for 1 h. The cells were collected for DNA isolation. The levels of 7-mG and O6-mG in DNA hydrolysates were quantified by LC-MS/MS analysis as described in the Materials and Methods section. The numbers are the average ± S.D.
A co-eluting contaminant at levels up to 200 pmol/μmol guanine interfered with the quantification of 7-mG in DNA from cells treated with the lower concentrations of methylating agents. As a result, the 7-mG levels in the CHOpcDNA3 cells are not shown. They ranged from 66–200 pmol/μmol guanine.
Statistically different from NMUr, p < 0.001
Statistically different from NMUr, p < 0.05
Figure 2.
Levels of 7-mG detected immediately following a 1 h treatment with NMUr, AMMN or NNK-4-OAc in CHOAGT cells. Data points are the average of three numbers. Error bars represent standard deviation. The lines were generated by connecting the data points.
Despite the higher levels of 7-mG, the levels of O6-mG detected in the CHOAGT cells were similar to those observed in the CHOpcDNA3 cells as a consequence of the extensive AGT-mediated repair in the CHOAGT cells. The ratio of 7-mG to O6-mG is a measure of the extent of O6-mG repair since 7-mG is only slowly removed from DNA. The high ratio of 7-mG to O6-mG in the DNA isolated from CHOAGT cells is evidence for the extensive repair of O6-mG (Table 3). These ratios are more than 20 times those observed in the in vitro treated DNA (Table 1). The amount of O6-mG repair was greater in CHOAGT cells treated with NNK-4-OAc or AMMN than in cells treated with NMUr as indicated by the higher ratios of 7-mG/O6-mG in the NNK-4-OAc- or AMMN-treated cells (Table 3).
To determine the influence of the aldehyde on the toxicity of the methylating nitrosamine, the levels of O6-mG were plotted relative to the cytotoxic effects of each methylating agent. Since O6-mG is the most cytotoxic adduct formed by these methylating agents, the expectation is that the slope of the line will be steeper (i.e., the compound will be more toxic relative to the levels of O6-mG) for the compounds that also generate aldehydes if the aldehyde contributes to the cytotoxic effects of the methylating agent. In cells lacking AGT, there was no difference in the cytotoxicity versus O6-mG plots for any of the methylating agents (Figure 3A). However, there was a significant difference between the plots of cytotoxicity versus O6-mG for each compound in the cells expressing AGT. In this case, AMMN was more toxic than NNK-4-OAc which was more toxic than NMUr at concentrations that generated comparable levels of O6-mG (Figure 3B). The differences between the plots were significant: AMMN versus NNK-4-OAc, p = 4 × 10−4; AMMN versus NMUr, p = 4 × 10−9; and NNK-4-OAc versus NMUr, p = 0.035.
Figure 3.
Cytotoxicity (A, B) or mutagenicity (C, D) compared to levels of O6-mG detected in CHOpcDNA3 (A, C) and CHOAGT cells (B, D) immediately after a 1 h treatment with NMUr, AMMN or NNK-4-OAc. Levels of O6-mG are averages of three replicates. Cytotoxicity and mutagenicity data are averages from 3–4 separate experiments performed with duplicates or triplicates. Error bars represent standard deviation. The lines were generated by connecting the data points. *Significantly different from NNK-4-OAc, p = 4 × 10−4, and NMUr, p = 4 × 10−9. **Significantly different from NNK-4-OAc, p = 0.035. ***Significantly different from NMUr and NNK-4-OAc.
To explore the ability of AGT to influence the cytotoxic effects of each methylating agents, the plots of cytotoxicity versus O6-mG for each compound were compared between the two cell lines (Figure S1A–C). If there was extensive repair of O6-mG such that other minor less toxic adducts start to contribute to the overall cytotoxic effects of the methylating agents, the slope of the line is expected to decrease. In fact, this is what was observed with NMUr. The expression of AGT resulted in a significant decrease in the slope of the line for the toxicity relative to O6-mG levels for NMUr (Figure S1 A, p = 2 × 10−10). This shift was less pronounced for NNK-4-OAc where there was a small but significant influence of AGT expression on the slope of the cytotoxicity versus O6-mG plot (Figure S1B, p = 0.03). AGT expression did not affect the plot of cytotoxicity versus O6-mG levels caused by AMMN (Figure S1C).
To determine the influence of the aldehyde on the mutagenicity of the methylating agent, the levels of O6-mG were plotted relative to the mutagenic effects of each methylating agent. Since O6-mG is the most mutagenic adduct formed by these methylating agents, the expectation is that the slope of the line will be increased (i.e., the compound will be more mutagenic relative to the levels of O6-mG) for the compounds that also generate aldehydes if the aldehyde contributes to the mutagenic effects of the methylating agent. In the absence of AGT, there were only minor differences between the plot of mutation frequency versus O6-mG plots with AMMN and NNK-4-OAc were slightly more mutagenic relative to O6-mG than NMUr (Figure 3C, p = 0.02 and 0.0002, respectively). In the presence of AGT, the plot of mutation frequency versus O6-mG for AMMN was substantially different than those obtained with NMUr or NNK-4-OAc (Figure 3D, p = 6.5 × 10−11 and 1.7 × 10−12, respectively). There was no difference between NMUr and NNK-4-OAc in this cell line.
To explore the ability of AGT to influence the mutagenic effects of each methylating agents, the plots of hprt mutation frequency versus O6-mG for each compound were compared between the two cell lines (Figure S1 D–F). If there was extensive repair of O6-mG such that other adducts start to contribute to the overall mutagenic effects of the methylating agents, the slope of the line for mutation frequency versus O6-mG versus is expected to increase in the cells expressing AGT (more mutations per O6-mG). There was no change in the slope of the line for mutation frequency versus O6-mG for NMUr in the cells expressing AGT (Figure S1D), indicating that the residual levels of O6-mG in these cells were likely responsible for the mutations observed in NMUr-treated CHOAGT cells. Similarly, the plot of mutation frequency and O6-mG levels for NNK-4-OAc were similar for the two cell lines (Figure S1F). On the other hand, AMMN was more mutagenic relative to O6-mG in CHOAGT cells than in CHOpcDNA3 cells (Figure S1E, p = 3 × 10−7), consistent with the presence of other adducts contributing to the mutagenic properties of AMMN in the CHOAGT cells.
Discussion
In this study we demonstrated that the toxic and mutagenic effects of compounds that generate methandiazohydroxide can be influenced by the presence of aldehyde decomposition products but only under conditions where O6-mG is repaired. In the cell line not expressing AGT (CHOpcDNA3), the differences in the cytotoxic and mutagenic effects between the three compounds could be explained by their relative ability to alkylate DNA. This conclusion is supported by the similarity of the plots of cytotoxicity or mutation frequency versus O6-mG levels in these cells (Figure 3A and C). In the cells expressing AGT (CHOAGT), there were compound specific differences in the plots of cytotoxicity or mutation frequency versus O6-mG levels (Figure 3B and D). The largest effects were observed with AMMN, suggesting that formaldehyde contributes both to the cytotoxic and mutagenic effects of this methylating agent. Smaller effects were observed with NNK-4-OAc where the plot of cytotoxicity versus O6-mG levels was modestly different from that obtained with NMUr, suggesting that the aldehyde OPB enhances the cytotoxic effect of the methylating agent.
It is not unexpected that the three methylating agents alkylated DNA with different efficiencies since the mechanism of ester hydrolysis will be dependent on chemical structure. AMMN and NMUr are likely hydrolyzed by a different subset of cellular esterases since the carboxylesterase inhibitor, paraoxon, reduced the cytotoxicity of AMMN but not NMUr in P388 cells.45 Cellular thiols have also be implicated in the decomposition of NMUr.46 AMMN and NNK-4-OAc may also decompose via intermediary formation of a nitrosiminium ion.47 Independent of the mechanism of hydrolysis, the net result is the formation of methanediazohydroxide for all three compounds and an aldehyde in the cases of AMMN and NNK-4-OAc. The chemical stability of the intermediates following ester hydrolysis is another factor that will affect the levels of DNA alkylation by these three compounds. The calculated half-life of N-methyl-N-nitrosocarbamic acid is about 1.5 min45 whereas AMMN-derived N-α-hydroxymethyl-N-methylnitrosamine had a half-life of ~ 9 seconds at pH 7.4.48,49 The stability of corresponding intermediate from NNK-4-OAc has not been investigated.
As predicted, O6-mG was the main toxic and mutagenic adduct formed by all three compounds, as evidenced by the ability of AGT to protect against their cytotoxic and mutagenic effects (Figure 1). This observation confirms numerous literature reports that O6-mG is the dominant miscoding and toxic adduct formed by SN1-type alkylating agents.36,37 This adduct miscodes during DNA replication, causing GC to AT transition mutations50 and AGT is known to protect against the majority of hprt mutations induced by methylating agents in CHO cells.51,52 It is also the major toxic methyl DNA adduct formed as a result of the detection of O6-mG-T mismatches by mismatch repair.37,51–54
The mechanism by which the aldehydes contribute to the cytotoxic effects of AMMN and NNK-4-OAc will require further study. The toxicity of formaldehyde has been reported and at least some of its toxicity can be ascribed to the formation of DNA adducts.55,56 On the other hand, OPB was not toxic in either cell line up to 500 μM (data not shown). It is possible that OPB is delivered to intracellular locations more efficiently by the nitrosamine and that, under those circumstances, OPB may be directly toxic to the cell.
One possible mechanism by which the aldehydes increase the toxicity of the methylating agents is through an indirect mechanism such as interfering with the repair of a toxic methyl DNA adduct. For example, aldehyde-mediated inhibition of AGT would enhance the cytotoxicity of a methylating agent. This possibility is suggested by the influence of AGT on the plots of O6-mG versus cytotoxicity for the three compounds (Figure S1). In the case of NMUr, there is a large difference between the slopes obtained for O6-mG versus cytotoxicity in the two cell lines. The more gradual slope of the line from the cells expressing AGT suggests that other DNA adducts are contributing to the cytotoxic effects of this compound, such as 3-methyladenine and other minor adducts.57 In contrast, the plots for O6-mG versus cytotoxicity for AMMN and NNK-4-OAc were less influenced by AGT expression. One possible explanation for these observations is that the aldehydes are mildly interfering with AGT-mediated repair of O6-mG. However, the repair of O6-mG seemed to be more efficient in the cells treated with AMMN or NNK-4-OAc as evidence by the more than 2 fold increase in the 7-mG/O6-mG ratios in the cells treated with NNK-4-OAc and AMMN relative to those exposed to NMUr (Table 3). Since this measurement of repair occurs at 1 h, it is possible that the aldehydes interfere with the long term repair of O6-mG. The active site cysteine is very reactive and could react directly with aldehydes, resulting in inactivation.8,58,59 Alternatively, aldehydes can reduce the overall sulfhydryl reducing potential of a cell by reacting with GSH, leading to a loss of AGT activity.59
The reason for the more rapid initial repair of O6-mG in the NNK-4-OAc- and AMMN-treated cells is unknown. It may be that kinetics of DNA alkylation by NNK-4-OAc and AMMN is faster relative to NMUr. If the NNK-4-OAc and AMMN-mediated alkylation reactions are completed much earlier in the 1 h incubation period, it will appear as if more O6-mG repair occurred with these two nitrosamines. Another possibility is that the aldehydes increased the rate of degradation of the alkylated AGT protein. It is known that the alkylated protein can block O6-mG repair by active AGT60 so increased degradation of the protein is expected to increase the rate of AGT mediated repair. However, there was no apparent difference in the rate of AGT degradation as measured by immunoblot analysis between the three compounds (data not shown) suggesting that this is not an explanation for the enhanced ratio of 7-mG/O6-mG in the AMMN and NNK-4-OAc-treated cells. Additional experiments will be required to resolve this question.
Formaldehyde contributes to the mutagenic activity of AMMN, as indicated by the dramatically increased mutagenic activity relative to O6-mG as compared to that observed with NMUr or NNK-4-OAc in the cells expressing AGT (CHOAGT) (Figure 3D). It is likely that AMMN-derived formaldehyde DNA adducts are formed in these cells. Formaldehyde DNA adducts have been observed in DNA reacted with AMMN as well as in liver and lung DNA from DMN-treated rats.27,28 This influence of formaldehyde on the mutagenic activity of AMMN was muted in the cells lacking AGT since O6-mG is likely the major mutagenic adduct formed from this compound. Interestingly, the mutation spectrum of AMMN is more complicated than that observed with other methylating nitrosamines.61 In addition to GC to AT transitions as expected for the formation of O6-mG, mutations at AT base pairs (AT to TA and AT to CG mutations) as well as frameshift mutations were observed. The contribution of these transversion mutations decreased as the concentration of AMMN increased. Formaldehyde is a likely source of these transversion mutations since it induced AT to TA, AT to CG and GC to TA mutations in CHO cells.30 These results combined with our data suggest that formaldehyde adducts may make major contributions to the mutagenic and carcinogenic properties of DMN, particularly when O6-mG is actively repaired. Future studies will explore the importance of formaldehyde DNA adduct formation in the genotoxic properties of AMMN and DMN.
In conclusion, our results demonstrate that OPB and formaldehyde can contribute to the cytotoxic effects of NNK-4-OAc and AMMN and formaldehyde contributes to the mutagenic activity of AMMN under conditions where there has been extensive repair of O6-mG. Future experiments will be required to determine how these aldehydes are exerting their effects. However, these findings indicate that the aldehyde should be considered when determining the toxicological properties of nitrosamines.
Supplementary Material
Acknowledgments
Funding Sources
This research was supported by the National Cancer Institute [CA-059887 (LAP), CA-115309 (LAP), CA-138338 (LAP) and CA-018137 (AEP)]. The Masonic Cancer Center Analytical Biochemistry and Biostatistics Shared Resources are funded in part by the National Cancer Institute [P30 CA-77598].
The authors thank Peter Villalta in Masonic Cancer Center Analytical Biochemistry Shared Resource at the University of Minnesota for assistance with mass spectrometry as well as Alyssa Fish for technical assistance. The authors also thank Bob Carlson for assistance with graphic design.
Abbreviations
- O6-mG
O6-methylguanine
- 7-mG
7-methylguanine
- AGT
O6-alkylguanine DNA alkyltransferase
- AMMN
N-acetoxymethyl-N-methylnitrosamine
- CHO
Chinese hamster ovary
- CHOpcDNA3
Chinese hamster ovary cells stably transfected with the pCMV-neo-Bam vector without an inserted cDNA sequence
- CHOAGT
Chinese hamster ovary cells stably transfected with the pCMV-neo-Bam vector expressing human AGT
- DMN
dimethylnitrosamine
- hprt
hypoxanthine-guanine phosphoribosyl transferase
- NMUr
N-nitrosomethylurethane
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNK-4-OAc
4-methylnitrosamino-4-acetoxy-1-(3-pyridyl)-1-butanone
- OPB
4-oxo-1-(3-pyridyl)-1-butanone
Footnotes
Supporting Information Available. Table S1 displays the cytotoxicity and mutagenicity data obtained for NMUr, AMMN and NNK-4-OAc in CHOpcDNA3 and CHOAGT cells and Figure S1 displays the influence of AGT expression on the cytotoxic and mutagenic activity of the methylating agents relative to O6-mG levels. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rhoades JW, Johnson DE. N-Dimethylnitrosamine in tobacco smoke condensate. Nature. 1972;236:307–308. doi: 10.1038/236307b0. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N- nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 3.IARC. Some inorganic substances, chlorinated hydrocarbons, aromatic amines, N-nitroso compounds and natural products. International Agency for Research on Cancer; Lyon: 1972. N-nitrosodimethylamine; pp. 95–106. [Google Scholar]
- 4.IARC. Some N-Nitroso Compounds. International Agency for Research on Cancer; Lyon: 1978. N-Nitrosodimethylamine; pp. 125–175. [Google Scholar]
- 5.National Toxicology Program. 12th Report on Carcinogens. US Department of Health and Human Services; Washington DC: 2011. [Google Scholar]
- 6.International Agency for Research on Cancer. Smokeless tobacco and tobacco- specific nitrosamines. Vol. 89. IARC; Lyon, France: 2007. [Google Scholar]
- 7.Keefer LK, Lijinsky W, Garcia H. Deuterium isotope effect on the carcinogenicity of dimethylnitrosamine in rat liver. J Natl Cancer Inst. 1973;51:299–302. doi: 10.1093/jnci/51.1.299. [DOI] [PubMed] [Google Scholar]
- 8.Grafström RC, Curren RD, Yang LL, Harris CC. Genotoxicity of formaldehyde in cultured human bronchial fibroblasts. Science. 1985;228:89–91. doi: 10.1126/science.3975633. [DOI] [PubMed] [Google Scholar]
- 9.Hong J, Yang CS. The nature of microsomal N-nitrosodimethylamine demethylase and its role in carcinogen activation. Carcinogenesis. 1985;6:1805–1809. doi: 10.1093/carcin/6.12.1805. [DOI] [PubMed] [Google Scholar]
- 10.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Spratt TE, Liu XK, Hecht SS, Pegg AE, Peterson LA. Pyridyloxobutyl adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine is present in 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone-treated DNA and is a substrate for O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 1997;10:562–567. doi: 10.1021/tx9602067. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Cheng G, Sturla SJ, McIntee EJ, Villalta PW, Upadhyaya P, Hecht SS. Identification of adducts formed by pyridyloxobutylation of deoxyguanosine and DNA by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a chemically activated form of tobacco specific carcinogens. Chem Res Toxicol. 2003;16:616–626. doi: 10.1021/tx034003b. [DOI] [PubMed] [Google Scholar]
- 13.Sturla SJ, Scott J, Lao Y, Hecht SS, Villalta PW. Mass spectrometric analysis of relative levels of pyridyloxobutylation adducts formed in the reaction of DNA with a chemically activated form of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem Res Toxicol. 2005;18:1048–1055. doi: 10.1021/tx050028u. [DOI] [PubMed] [Google Scholar]
- 14.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem Res Toxicol. 2006;19:674–682. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomson NM, Kenney PM, Peterson LA. The pyridyloxobutyl DNA adduct, O6-[4-oxo-4-(3-pyridyl)butyl]guanine, is detected in tissues from 4- (methylnitrosamino)-1-(3-pyridyl)-1-butanone-treated A/J mice. Chem Res Toxicol. 2003;16:1–6. doi: 10.1021/tx025585k. [DOI] [PubMed] [Google Scholar]
- 16.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, Wang M, Villalta PW, Lindgren BR, Upadhyaya P, Lao Y, Hecht SS. Analysis of pyridyloxobutyl and pyridylhydroxybutyl DNA adducts in extrahepatic tissues of F344 rats treated chronically with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2009;22:926–936. doi: 10.1021/tx900015d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alaoui-Jamali MA, Gagnon R, El Alami N, Castonguay A. Cytotoxicity, sister-chromatid exchanges and DNA single-strand breaks induced by 4-oxo-4-(3-pyridyl)butanal, a metabolite of a tobacco-specific N-nitrosamine. Mutat Res. 1990;240:25–33. doi: 10.1016/0165-1218(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Alaoui-Jamali MA, Alami NE, Castonguay A. Metabolism and DNA single strand breaks induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and its analogues in primary culture of rat hepatocytes. Cancer Res. 1990;50:1810–1816. [PubMed] [Google Scholar]
- 20.Demkowicz-Dobrazanski K, Castonguay A. Comparison of DNA alkali-labile sites induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and 4-oxo-4-(3-pyridyl)butanal in rat hepatocytes. Carcinogenesis. 1991;12:2135–2140. doi: 10.1093/carcin/12.11.2135. [DOI] [PubMed] [Google Scholar]
- 21.Grafström RC, Fornace J, Autrup H, Lechner JF, Harris CC. Formaldehyde damage to DNA and inhibition of DNA repair in human bronchial cells. Science. 1983;220:216–218. doi: 10.1126/science.6828890. [DOI] [PubMed] [Google Scholar]
- 22.Chaw YFM, Crane LE, Lange P, Shapiro R. Isolation and identification of cross-links from formaldehyde-treated nucleic acids. Biochemistry. 1980;19:5525–5531. doi: 10.1021/bi00565a010. [DOI] [PubMed] [Google Scholar]
- 23.Beland FA, Fullerton NF, Heflich RH. Rapid isolation, hydrolysis and chromatography of formaldehyde-modified DNA. J Chromatogr. 1984;308:121–131. doi: 10.1016/s0021-9673(01)87539-7. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, Solomon MS, Hopkins PB. Formaldehyde preferentially interstrand cross-links duplex DNA through deoxyadenosine residues at the sequence 5′-d(AT) JACS. 1992;114:9240–9241. [Google Scholar]
- 25.Huang H, Hopkins PB. DNA interstrand cross-linking by formaldehyde: Nucleotide sequence preference and covalent structure of the predominant cross-link formed in synthetic oligonucleotides. JACS. 1993;115:9402–9408. [Google Scholar]
- 26.Cheng G, Shi Y, Sturla SJ, Jalas JR, McIntee EJ, Villalta PW, Wang M, Hecht SS. Reactions of formaldehyde plus acetaldehyde with deoxyguanosine and DNA: formation of cyclic deoxyguanosine adducts and formaldehyde cross-links. Chem Res Toxicol. 2003;16:145–152. doi: 10.1021/tx025614r. [DOI] [PubMed] [Google Scholar]
- 27.Wang M, Cheng G, Villalta PW, Hecht SS. Development of liquid chromatography electrospray ionization tandem mass spectrometry methods for analysis of DNA adducts of formaldehyde and their application to rats treated with N-nitrosodimethylamine or 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem Res Toxicol. 2007;20:1141–1148. doi: 10.1021/tx700189c. [DOI] [PubMed] [Google Scholar]
- 28.Cheng G, Wang M, Upadhyaya P, Villalta PW, Hecht SS. Formation of formaldehyde adducts in the reactions of DNA and deoxyribonucleosides with alpha-acetates of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), and N-nitrosodimethylamine (NDMA) Chem Res Toxicol. 2008;21:746–751. doi: 10.1021/tx7003823. [DOI] [PubMed] [Google Scholar]
- 29.Liber HL, Benforado K, Crosby RM, Simpson D, Skopek TR. Formaldehyde-induced and spontaneous alterations in human hprt DNA sequence and mRNA expression. Mutat Res. 1989;226:31–37. doi: 10.1016/0165-7992(89)90089-4. [DOI] [PubMed] [Google Scholar]
- 30.Graves RJ, Trueman P, Jones S, Green T. DNA sequence analysis of methylene chloride-induced HPRT mutations in Chinese hamster ovary cells: comparison with the mutation spectrum obtained for 1,2-dibromoethane and formaldehyde. Mutagenesis. 1996;11:229–233. doi: 10.1093/mutage/11.3.229. [DOI] [PubMed] [Google Scholar]
- 31.Merk O, Speit G. Significance of formaldehyde-induced DNA-protein crosslinks for mutagenesis. Environ Mol Mutagen. 1998;32:260–268. doi: 10.1002/(sici)1098-2280(1998)32:3<260::aid-em9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 32.Speit G, Merk O. Evaluation of mutagenic effects of formaldehyde in vitro: detection of crosslinks and mutations in mouse lymphoma cells. Mutagenesis. 2002;17:183–187. doi: 10.1093/mutage/17.3.183. [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Castonguay A, Gerson SL. Lack of correlation between DNA methylation and hepatocarcinogenesis in rats and hamsters treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butatone. Carcinogenesis. 1992;13(#11):2137–2140. doi: 10.1093/carcin/13.11.2137. [DOI] [PubMed] [Google Scholar]
- 34.Demkowicz-Dobrazanski K, Castonguay A. Modulation by glutathione of DNA strand breaks induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and its aldehyde metabolites in rat hepatocytes. Carcinogenesis. 1992;13:1447–1454. doi: 10.1093/carcin/13.8.1447. [DOI] [PubMed] [Google Scholar]
- 35.Roller PP, Shimp DR, Keefer LK. Synthesis and solvolysis of methyl(acetoxymethyl)nitrosamine. Solution chemistry of the presumed carcinogenic metabolite of dimethylnitrosamine. Tetrahedron Letters. 1975;#25:2065–2068. [Google Scholar]
- 36.Bignami M, O’Driscoll M, Aquilina G, Karran P. Unmasking a killer: DNA O6-methylguanine and the cytotoxicity of methylating agents. Mutat Res. 2000;462:71–82. doi: 10.1016/s1383-5742(00)00016-8. [DOI] [PubMed] [Google Scholar]
- 37.Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst) 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Lin JM, Amin S, Murphy SE, Solomon JJ, Hecht SS. Synthesis of [3,3-D2]4-hydroxy-1-(3-pyridyl)-1-butanone, an internal standard for analysis of tobacco- specific nitrosamine hemoglobin and DNA adducts. J Label Compounds Radiopharm. 1993;33:285–292. [Google Scholar]
- 39.Peterson LA, Liu XK, Hecht SS. Pyridyloxobutyl DNA adducts inhibit the repair of O6-methylguanine. Cancer Res. 1993;53:2780–2785. [PubMed] [Google Scholar]
- 40.Loktionova NA, Pegg AE. Point mutations in human O6-alkylguanine DNA alkyltransferase prevent the sensitization of O6-benzylguanine to killing by N,N′-bis(2-chloroethyl)-N-nitrosourea. Cancer Res. 1996;56:1578–1583. [PubMed] [Google Scholar]
- 41.Coryell VH, Stearns DM. Molecular analysis of hprt mutations induced by chromium picolinate in CHO AA8 cells. Mutat Res. 2006;610:114–123. doi: 10.1016/j.mrgentox.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 42.O’Neill JP, Brimer PA, Machanoff R, Hirsch GP, Hsie AW. A quantitative assay of mutation induction at the hypoxanthine-guanine phosphoribosyl transferase locus in Chinese hamster ovary cells (CHO/HGPRT system): development and definition of the system. Mutat Res. 1977;45:91–101. doi: 10.1016/0027-5107(77)90047-1. [DOI] [PubMed] [Google Scholar]
- 43.O’Neill JP, Hsie AW. Phenotypic expression time of mutagen-induced 6-thioguanine resistance in Chinese hamster ovary cells (CHO/HGPRT system) Mutat Res. 1979;59:109–118. doi: 10.1016/0027-5107(79)90196-9. [DOI] [PubMed] [Google Scholar]
- 44.Hecht SS, Trushin N, Castonguay A, Rivenson A. Comparative tumorigenicity and DNA methylation in F344 rats by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and N-nitrosodimethylamine. Cancer Res. 1986;46:498–502. [PubMed] [Google Scholar]
- 45.Weinkam RJ, Plakunov I. A kinetic model of the cell culture cytotoxicity of metabolically activated agents: N-methyl-N-(acetoxymethyl)nitrosamine, methylnitrosourethane, and (methylazoxy)methanol acetate. Chem Res Toxicol. 1989;2:288–294. doi: 10.1021/tx00011a004. [DOI] [PubMed] [Google Scholar]
- 46.Schoental R, Rive DJ. Interaction of N-alkyl-N-nitrosourethanes with thiols. Biochem J. 1965;97:466–474. doi: 10.1042/bj0970466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai H, Fishbein JC. α-(Acyloxy)dialkylnitrosamines: effect of structure on the formation of N-nitrosiminium ions and a predicted change in mechanism. JACS. 1999;121:1826–1833. [Google Scholar]
- 48.Mochizuki M, Anjo T, Okada M. Isolation and characterization of N-alkyl-N-(hydroxymethyl)nitrosamines from N-alkyl-N-(hydroperoxymethyl) nitrosamines by deoxygenation. Tetrahedron Letters. 1980;21:3693–3696. [Google Scholar]
- 49.Mesic M, Revis C, Fishbein JC. Effects of structure on the reactivity of α-hydroxydialkynitrosamines in aqueous solutions. JACS. 1996;118:7412–7413. [Google Scholar]
- 50.Toorchen D, Topal MD. Mechanisms of chemical mutagenesis and carcinogenesis: effects on DNA replication of methylation at the O6-guanine position of dGTP. Carcinogenesis. 1983;4:1591–1597. doi: 10.1093/carcin/4.12.1591. [DOI] [PubMed] [Google Scholar]
- 51.Kaina B, Fritz G, Mitra S, Coquerelle T. Transfection and expression of human O6-methylguanine-DNA methyltransferase (MGMT) cDNA in Chinese hamster cells: the role of MGMT in protection against the genotoxic effects of alkylating agents. Carcinogenesis. 1991;12:1857–1867. doi: 10.1093/carcin/12.10.1857. [DOI] [PubMed] [Google Scholar]
- 52.Cai Y, Wu MH, Xu-Welliver M, Pegg AE, Ludeman SM, Dolan ME. Effect of O6-benzylguanine on alkylating agent-induced toxicity and mutagenicity. In Chinese hamster ovary cells expressing wild-type and mutant O6-alkylguanine-DNA alkyltransferases. Cancer Res. 2000;60:5464–5469. [PubMed] [Google Scholar]
- 53.Dolan ME, Norbeck L, Clyde C, Hora NK, Erickson LC, Pegg AE. Expression of mammalian O6-alkylguanine-DNA alkyltransferase in a cell line sensitive to alkylating agents. Carcinogenesis. 1989;10:1613–1619. doi: 10.1093/carcin/10.9.1613. [DOI] [PubMed] [Google Scholar]
- 54.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. PNAS. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, Van Houten B, Iijima K, Tauchi H, Ide H. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumari A, Lim YX, Newell AH, Olson SB, McCullough AK. Formaldehyde-induced genome instability is suppressed by an XPF-dependent pathway. DNA Repair (Amst) 2012;11:236–246. doi: 10.1016/j.dnarep.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pegg AE. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem Res Toxicol. 2011;24:618–639. doi: 10.1021/tx200031q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krokan H, Grafström RC, Sundqvist K, Esterbauer H, Harris CC. Cytotoxicity, thiol depletion and inhibition of O6-methylguanine-DNA methyltransferase by various aldehydes in cultured human bronchial fibroblasts. Carcinogenesis. 1985;6:1755–1759. doi: 10.1093/carcin/6.12.1755. [DOI] [PubMed] [Google Scholar]
- 60.Edara S, Kanugula S, Pegg AE. Expression of the inactive C145A mutant human O6-alkylguanine-DNA alkyltransferase in E.coli increases cell killing and mutations by N-methyl-N′-nitro-N-nitrosoguanidine. Carcinogenesis. 1999;20:103–108. doi: 10.1093/carcin/20.1.103. [DOI] [PubMed] [Google Scholar]
- 61.Horsfall MJ, Glickman BW. Mutational specificities of environmental carcinogens in the lacI gene of Escherichia coli. I. The direct-acting analogue N-nitroso-N-methyl-N-à-acetoxymethylamine. Carcinogenesis. 1989;10:817–822. doi: 10.1093/carcin/10.5.817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.