Abstract
Human histidine triad nucleotide binding protein 1 (hHint1) is a member of a ubiquitous and ancient branch of the histidine triad (HIT) protein superfamily. hHint1 is a homodimeric protein that catalyzes the hydrolysis of model substrates, phosphoramidate (TpAd) and acyl adenylate (AIPA), with a high efficiency. Recently, catalytically inactive hHint1 has been identified as the cause of inherited peripheral neuropathy (1). We have carried out the first detailed kinetic mechanistic studies of hHint1 and have found that the reaction mechanism is consistent with a double displacement mechanism, in which the active site nucleophile His112 is first adenylylated by the substrate, followed by hydrolysis of the AMP-enzyme intermediate. A transient burst phase followed by a linear phase from the stopped-flow fluorescence assay indicated that enzyme adenylylation was faster than the subsequent intermediate hydrolysis and product release. Solvent viscosity experiments suggested that both chemical transformation and diffusion-sensitive events (product release or protein conformational change) limit the overall turnover. The catalytic trapping experiments and data simulation indicated that the true koff rate of the final product AMP is unlikely to control the overall kcat. Therefore, a protein conformational change associated with product release is likely rate limiting. In addition, the rate of Hint1 adenylylation was found to be dependent on two residues with pKas of 6.5 and 8, with the former pKa agreeing well with the NMR titration results for the pKa of the active site nucleophile His112. When compared to the uncatalyzed rates, hHint1 was shown to enhance acyl-AMP and AMP phosphoramidate hydrolysis by 106 to 108- fold. Taken together, our analysis indicates that hHint1 catalyzes the hydrolysis of phosphoramidate and acyl adenylate with high efficiency, through a mechanism that relies on rapid adenylylation of the active residue, His-112, while being partially rate limited by intermediate hydrolysis and product release associated with a conformational change. Given the high sequence homology of Hint proteins across all kingdoms of life, it is likely that their kinetic and catalytic mechanisms will be similar to those elucidated for hHint1.
Introduction
Histidine triad nucleotide binding proteins (Hints) are members of the histidine triad (HIT) protein superfamily of nucleotidyl transferases and hydrolases that contain a highly conserved active site motif of His-X-His-X-His-XX, where X is a hydrophobic amino acid (2) (Fig.1a). The Hint branch is the most ancient and is ubiquitously expressed in both prokaryotes and eukaryotes; however, their biological roles have not been fully characterized. Although initially Hint1 was reported as a protein kinase C inhibitor (3), subsequent studies have not supported thisobservation(4-6). Hint1 also was suggested to be a zinc binding protein due to the presence of four histidines in the active site (3), however, a rationale for this potentiality has not been forthcoming.
Figure 1.
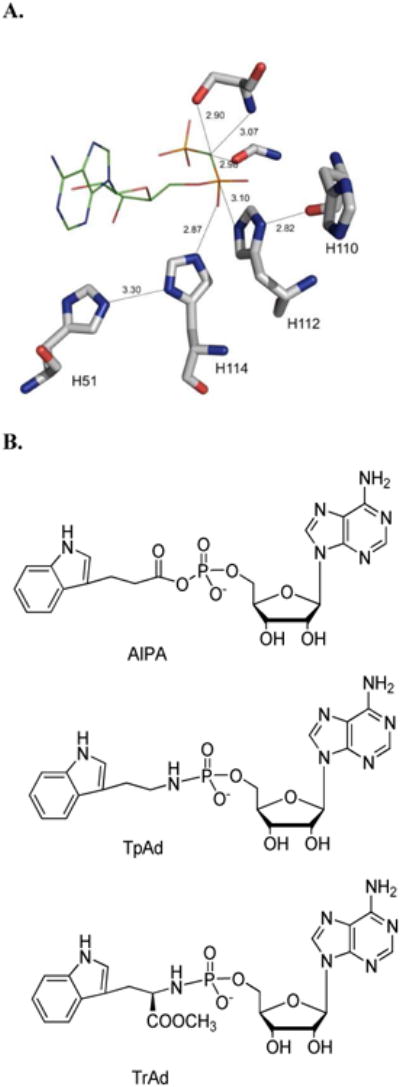
(A) The active site of hHint1 with AMP binding (PDB: 1KPF). (B) Structure of substrates.
Recently, Hint1 has emerged as a tumor suppressor through multiple molecular mechanisms involving the regulation of apoptosis (7), gene transcription (8-11), and the cell cycle (12, 13). Hint1 deleted mice have an increased susceptibility to the carcinogenic DMBA (7, 12-dimethylbenzanthracene) induced ovarian and mammary tumors, as well as the increased occurrence of spontaneous tumors (14). Hint1 expression appears to correlate with the down-regulation of p53 and the proapoptotic factor Bax (7). Consistent with this finding and a potential role in tumorgenesis, Hint1 has been found to be associated with the Bax promoter and is a component of the Tip60 histone acetyltransferase complex (15). Two hybrid screening experiments revealed that Hint1 was associated with cyclin-dependent kinase 7 (Cdk7); however, knock-out mice studies fail to reveal a phenotype consistent with the regulation of cdk7, indicating that Hint1 is not required for Cdk7 function (16). Evidence has also accumulated that eukaryotic Hint1 is associated with several gene transcription factors, such as TFIIH (8), MITF(9, 10), and USF2(11). In addition to their emerging role in cancer, a link between Hint1 and the regulation of postsynaptic dopamine transmission (17, 18) and the development of hepatic ischemia reperfusion injury has been discovered (19).
While a variety of interactions with Hint1 have been discovered, little is known about the enzymatic activity and the underlying catalytic mechanism, as well as the possible linkage between enzymatic activity and intracellular function(s). It has been suggested that Hint enzymatic activity is inhibited via heterodimerization with Asw (avian sex-specific W-linked) protein, a potentially sex-determining protein in birds (20, 21). Brenner and coworkers have reported that rabbit Hint1 and yeast HNT hydrolyze adenosine 5′-monophophoramidate (AMP-NH2) to AMP and ammonia, and adenosine lysine phophoramidate to AMP and lysine(22), while Wagner and coworkers demonstrated by 31P-NMR that human Hint1 and E. coli HinT (ycfF) are efficient purine nucleoside phosphoramidases (23-25). Substrate specificity studies have revealed a preference by Hint1s for nucleoside phosphoramidate substrates containing purine ribonucleosides coupled to unhindered alkyl amine(24). In addition, acyl nucleoside monophosphates were found to be modestly better substrates for both ecHinT and hHint1with kcat/Km values ranging from 106-108 M−1s−1. Since acyl-AMP generated by lysyl t-RNA synthetase has also been shown to be a good substrate of both bacterial and human Hint1, Chou and co-workers have proposed that acyl-AMP prepared by AARS's may be natural substrate of Hints (26). Intriguingly, Wang and co-workers recently have determined the crystal structures of Hint1 in complex with aminoacyl adenylate analogs, and revealed that Hint1 broadly recognizes only the common main chain of the aminoacyl moiety, without specific interacting with the amino acid side chain(27). Although Hints are unable to carry out phosphate hydrolysis, Krakowiak and co-workers have observed that Hint1 is capable of cleaving the P-S bond of nucleoside 5′-O-phosphorothiolates, suggesting that the production of H2S during desulfuration reaction catalyzed by Hint1 may biologically significant (28).
While efficient phosphoramidases and acyl-AMP hydrolases, until recently a direct connection between Hint1 catalysis and a biological phenotype has proved elusive. Yeast genetic and growth studies by Breiganowski and co-workers established that the growth of yeast on galactose is dependent on Hnt activity (29), while Chou and co-workers demonstrated that Hint activity is necessary for the growth of E. coli under high salt conditions (23). Bardaweel and co-workers have further demonstrated that the activity of E.coli D-alanine dehydrogenase, and therefore growth on alanine, is dependent on E. coli Hint (ecHinT) catalysis (30). Recently, hHint loss of function has been proposed as a cause of inherited peripheral neuropathy, a condition that affects 1 in 2500 individuals (1). A genome wide SNP analysis of 33 families exhibiting inherited peripheral neuropathy founda strong correlation between the loss of catalytically functional hHint1 and autosomally recessive axonal neurophathy with neuromyotonia, thus providing evidence for the first phenotype linked to hHint1 enzymatic activity (1).
Although the HIT superfamily is comprised of five and possibly six distinct subfamilies, investigations of their catalytic and kinetic mechanisms of action have only been carried out on GalT and Fhit. In both cases, catalysis proceeds by formation of a histidine-NMP intermediate with inversion of the phosphorous configuration, followed by transfer to either water (Fhit) or galactose (GalT). For GalT, uridylylation of the nucleophilic histidine by UDP-glucose and subsequent deuridylylation by galactose1-phosphate were found to be rapid with rates of 281 s−1, and 166 s−1 at 4°C, respectively (31). Since these two rates were higher than the kcat value (62 s−1 at 4°C), it has been proposed that either product release or a conformational change may be rate limiting (31). Most importantly, structural analysis of the GalT active site revealed that two of the conserved histidines and a proximal cysteine formed a coordinate complex with an atom of Fe (32). For Fhit, while conversion of Ap3A to ADP and AMP proceeds through a double displacement mechanism, less is known about the specific catalytic steps (33). When the putative nucleophilic histidine was replaced by glycine, free histidine was found to rescue enzymatic activity (34). In addition, AMP-imidazole and AMP-N-methylimidazole were found to be substrates (35). Results of steady state kinetic and mutagenesis studies have implied that one member of the catalytic triad, His-98, is likely responsible for the donation of a proton to the leaving group, as well as substrate binding (34). This is in marked contrast to GalT, in which the corresponding histidines are coordinated to Fe. Since there is no evidence of significant Fe or metal coordination to the active site histidines of either Fhit or Hints, Hint1 appears to be catalytically similar to Fhit, and not GalT. Nevertheless, a detailed kinetic mechanism for either enzyme has yet to be elucidated.
The catalytic mechanism of Hint1 has been proposed to proceed through a two-step double displacement mechanism, analogous to Fhit, in which His-112 is thought to form a covalent Hint-AMP intermediate followed by hydrolysis and the release of AMP (26, 36) (Fig. 1A). However, a detailed picture of the catalytic and kinetic mechanism of Hint1 has yet to be elucidated. Phosphoramidates, such as TpAd and TrAd, and amino acyl mimics such as AIPA (Fig.1B) can undergo P-N bond and P-O bond cleavage by Hint1, respectively. Due to the ability of nucleosides to quench the fluorescence of the covalently linked indole moiety of tryptamine (Fig.1B), we have developed a sensitive continuous fluorescence assay using these model substrates, TpAd and AIPA(24, 37). With these tools, we have been able to delineate the kinetic mechanism and features of the catalytic mechanism of Hint1 by site-directed mutagenesis, pre-steady state and steady state kinetic analyses, solvent viscometric studies and kinetic model fittings. The results of these studies shed light on the parameters influencing Hint1 catalysis in particular and the HIT superfamily in general.
Experimental and Methods
Materials
AMP, AMP-agarose, ampicillin, chlorophenicol, HEPES, MgCl2, sucrose, were purchased from Sigma-Aldrich (St. Louis, MO). TpAd and AIPA were synthesized following the procedures published in (24) and (37), respectively. Steady state kinetic data were collected on a Varian Cary Eclipse fluorimeter with the kinetic program equipped with a thermostated cuvette holder (Palo Alto, CA). Transient kinetic data were obtained on a single-wavelength stopped-flow apparatus (model SX.18MV, Applied Photophysics, Leatherhead, UK). Kinetic data were analyzed with the JMP IN 7 software (SAS Institute, Inc., Cary, NC, USA)
Site-directed mutagenesis, protein expression, and purification
The active site mutations, H51A, H110A, H112G, and H114A, were generated using human Hint1-pSGA02 vector with QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotide primer used were 5′-CAA GCA CCA ACA GCT TTT CTG GTG ATA CC-3′, 5′-CAG TCT GTC TAT GCC GTT CAT CTC CAT G-3′, 5′-GTC TAT CAC GTT GCT CTC CAT GTT CTT GG-3′, and 5′-CAC GTT CAT CTC GCT GTT CTT GGA GG-3′, respectively. The wild-type and mutated human Hint1-pSGA02 plasmids were transformed into E. coli hinT disrupt ted strain BB2 as previously described (23). The expression and purification of the mutants was similar to those for wild-type, which followed a previously published procedure (23). Briefly, AMP-agarose column was applied after the cell lysis by lysozyme and sonication. Hint protein was eluted by 25mM AMP solution. A PD-10 column was employed to desalt the protein samples. Protein concentrations were determined with the Bradford protein assay (38).
Stopped flow fluorescence analysis of adenylylation of human Hint1
As mentioned above, we have developed a continuous and sensitive fluorescence assay using two fluorogenic substrates, TpAd and AIPA. Due to the magnitude of fluorescence quenching (over 20 fold), hydrolysis of TpAd or AIPA by Hint1, respectively, leads to an increase in indole fluorescence from tryptamine, the product for the first half reaction. This fluorescence increase can be monitored with time in stopped flow kinetic experiments, and can be used to measure the reaction rates of the adenylylation of human Hint1 as well, if the hydrolysis of adenylylated Hint1 is much slower than formation of adenylylated Hint1 intermediate. Therefore, we monitored the reaction rates of the adenylylation of Hint1 at 25°C using a fluorescence stopped flow apparatus (Applied Photophysics, model SX.18MV). AIPA (5-40μM) or TpAd (5-60μM) in HEPES buffer (20mM; 1mM MgCl2, pH 7.2) was transferred to one stopped flow syringe, and human Hint1 (WT, 69μg/ml, 5μM) in HEPES buffer (20mM; 1mM MgCl2, pH 7.2) was transferred to the other syringe. Each time equal volumes (50μL) of the enzyme solution and the substrate were injected and mixed rapidly. The excitation wavelength was set to 280nm and the fluorescence was monitored through a cutoff filter of 320nm. The time-course curves were fit by a single exponential equation with a steady-state term (eq1) using JMP IN 7 software, where A is the amplitude of the burst, kl is the linear rate of increase of fluorescence, and kb is the burst rate, or the observed pseudo-first-order rate constant. The results represent the average of six experiments. The kinetic parameters k2, Kadenylyl, k2/Kadenylyl for AIPA or TpAd reaction were obtained by non-linear fitting to the data by eq1 and eq 2, and [S] is the substrate concentration.
Active site titration
In the active site titration experiment, human Hint1 (1, 2, 3, 4, 5μM) in HEPES buffer was mixed with AIPA (10μM) and the fluorescence intensity was monitored. The time course data were fit into eq 1. The amplitude, A, represents a measure of the moles of adenylylated enzyme in the first catalytic cycle, can be plotted linearly against the Hint1 monomer concentration, and the slope indicates the active site percentage per monomer.
Steady state fluorescence assay of human Hint1
The steady state fluorescence assay was performed at 25°C using Varian/Cary Eclipse Fluorimeter as described previously (24). For the standard assay, the hydrolysis of the fluorogenic substrate by human Hint1 was carried out in degassed HEPES buffer (20mM HEPES, 1mM MgCl2, pH 7.2) in quartz cuvettes. The excitation wavelength was set to 280nm and the emission fluorescence was measured at 360nm. The excitation and emission slits were set at 5nm. Initial velocity of hydrolysis were measured with hH1 or mutant (6nM-50nM) and varied concentrations of either phosphoramidate (0-10μM) or acyl adenylate (0-10μM). The curves were fit to eq 3 to yield the first order rate constant kcat and apparent Michaelis-Menten constant Km using JMP7 software, where v is the initial velocity, [E]t is the total enzyme concentration, and [S] is the substrate concentration.
Solvent viscosity effect experiments
The viscosity of the solution was controlled by the addition of appropriate amounts of microviscosogen, sucrose, or macroviscosogen, PEG8000. The relative viscosities (ŋrel) of the solution were taken from the reference (39). The steady state first order rate constants (kcat) and apparent Michaelis-Menten constants (Km) were determined as described above in HEPES buffer (20mM HEPES, 1mM MgCl2, pH 7.2) containing the appropriate amounts of sucrose (0, 14, 24, 32%) or PEG8000 (5%). A minimal 4-step mechanism including a kinetically significant product release is shown in scheme 1, in which the observed kcat is a combination of two chemistry steps (adenylylation and hydrolysis) and one diffusive step (eq 6). Data were fit using eq 9-11. The 0kcat or 0(kcat/Km), and kcat or (kcat/Km), represent the kinetic parameters in the absence and presence of microviscosogen, respectively.
Scheme 1.
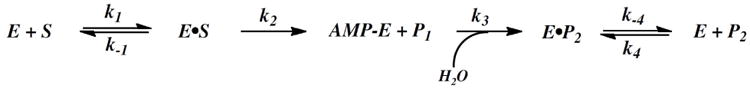
Proposed minimal 4-step mechanism of hHint1.
pH dependence of pre-steady state parameters
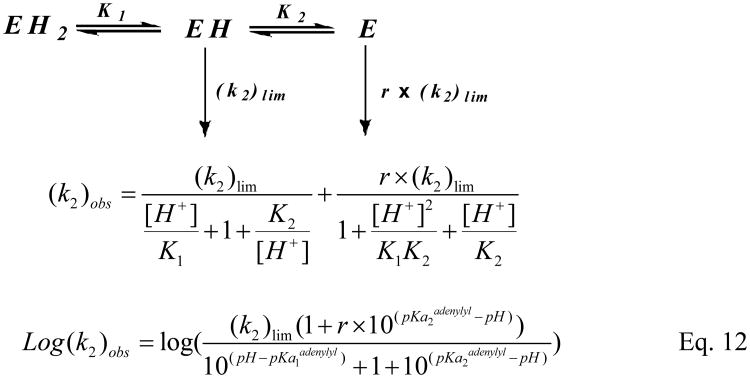
At pH values ranging from 5.8-9.0, the adenylylation kinetic parameters for the first half reaction, k2 and k2/Kadenylyl, were determined using stopped-flow apparatus with human Hint1 (final concentration WT 2.5μM) and TpAd (5-60μM) or AIPA (2.5-20 μM) in phosphate buffer (100mM). Assays were performed as described for the pre-steady state kinetic experiments (vide supra). The time course curves were first fit with eq 1 to obtain the values for kb, and then k2 and k2/Kadenylyl were obtained for each pH from the fitting with eq 2. To fit the k2vs. pH data for WT, eq 12 was used (Scheme 2).
Scheme 2.
pH dependence of steady state kinetic parameters
The pH-rate profiles of steady state hydrolysis of phosphoramidate by hint1 and mutants were determined with saturated concentration of substrate (50μM) and appropriate enzyme concentration (0.0625μM-0.25μM) in phosphate buffer (100mM, pH 5.8-9). The kcat vs. pH data were fit with eq 13.
Dissociation constant (Kd) determination by tryptaphan fluorescence titration
Titration with AMP was performed using hHint1 (6μM) in 1.5ml HEPES buffer (pH 7.2, 20mM HEPES, MgCl2). The AMP stock was added and the mixture was stirred for 30s, then the emission fluorescence intensity ranging from 300-450nm was measured for 30s at a scan speed of 300nm/min (excitation 280nm). The peak fluorescence intensity at about 351nm was obtained from the average intensities 349-353nm. Appropriate corrections were made for dilution effects (never exceeding 10%). Data were fit into eq 14, where F is the fluorescence intensity, Fe is the fluorescence intensity of the protein without ligand binding, Fel is fluorescence intensity of the protein with ligand binding, Et is total concentration of the protein, Lt is total concentration of the ligand, and Kd is the dissociation constant.
Catalytic trapping experiments
The WT human Hint1 (5μM) was pre-incubated with AMP (0-5000μM) in one stopped flow syringe for 10min, then mixed with TpAd (40μM) in the other syringe. The time course of the reaction was monitored as previously described. To minimize the inner filter effect, the concentration of AMP was controlled not to exceed 5% of the absorbance.
NMR spectroscopy to analyze histidine pKa change in human Hint1
Wild type hHint1 and H112A were purified by an AMP-agarose column from echinT deletion stain (BB2) as described previously (23). Purified protein was exchanged to phosphate buffer in H2O at different pHs (0.1M sodium phosphate, pH 6.00-9.00) and concentrated to 8mg/ml. 300μL of protein sample was transferred to a new tube, frozen, and lyophilized. After lyophilization, the samples were reconstituted in 300μL of 99.8% D2O. Further proton exchange was carried out by storing the samples at 4 °C for 40hr. Before NMR analysis, the samples were centrifuged at 13,000 g for 10 min, and the supernatant was transferred to a Shigemi 5 mm NMR microtube assembly (matched with D2O, bottom length 15 mm, type BMS-3 from Shigemi Inc., Allison Park, USA). The 1D proton NMR and TOCSY spectra were acquired on 600MHz Varian Inova 6001 spectrometers equipped with a Bioselect probe at 25 °C. Histidine C-2 proton resonances were well resolved and assigned by comparison of spectra under the continuous closely spaced values of pHs. The pKa values were obtained by non-linear fitting of the titration data to the modified Henderson-Hasselbalch equation (eq 15) (40), where δobs, δ H+, and δH are the chemical shifts for the observed, protonated, and neutral imidazoles, respectively.
Determination of uncatalyzed rate of Tpad and AIPA hydrolysis
The uncatalyzed rates of hydrolysis of TpAd and AIPA were determined by monitoring the reaction in a quartz sealed tube as previously described with minor modifications.(41) Reaction mixtures containing the anhydride AIPA (0.05 M) in aqueous potassium phosphate buffer (0.1 M, pH 6.8) were incubated at temperatures from 10 to 47 °C for various time intervals. Reactions were then stopped by freezing, and the product mixtures analyzed by proton NMR (500 MHz) after 10-fold dilution with D2O solutions containing added pyrazine (0.01 M, δ 8.60 ppm) as an integration standard. Hydrolysis of the anhydride linkage of AIPA was accompanied by the disappearance of a doublet at 5.88 ppm and the appearance of a doublet associated with the product AMP at 6.08 ppm (confirmed by addition of the authentic molecule), and also by the disappearance of a doublet at 7.35 ppm and the appearance of a doublet at 7.45 ppm associated with the product indolepropionic acid (confirmed by addition of the authentic molecule). Reaction mixtures containing TpAd (0.05 M) in aqueous potassium phosphate buffer (0.1 M, pH 6.8) were incubated at temperatures from 59 to 121 °C for various time intervals. Reactions were then stopped by cooling, and the product mixtures analyzed by proton NMR (500 MHz) after 10-fold dilution with D2O solutions containing added pyrazine (0.01 M, δ 8.60 ppm) as an integration standard. Hydrolysis of the phosphoramide linkage of TpAd was accompanied by the disappearance of a doublet at 5.90 ppm and the appearance of a doublet associated with the product AMP at 6.08 ppm (confirmed by addition of the authentic molecule).
Results
Transient State Kinetics
We have observed that the hydrolysis of TpAd or AIPA results in a significant increase in fluorescence intensity, due to release of tryptamine, allowing us to monitor spectrophotometrically the steady-state and transient state kinetics of the reaction (42, 43). Stopped flow fluorescence of human Hint1 using the phosphoramidate substrate, TpAd, or acyl adenylate substrate, AIPA, both displayed an initial “burst” phase followed by a linear steady state phase (Fig. S1A). The “burst” phase suggested rapid formation of an intermediate within the first catalytic cycle, while the linear steady state phase (kl) suggested a slow rate-limiting step following intermediate formation. Data fitting analysis revealed kl values of 2.0 ± 0.4 s−1 and 2.7 ± 0.8 s−1 for TpAd and AIPA, respectively (Table 1). These values agreed well with the observed steady state kcatvalues 2.1 ± 0.1 s−1 and 1.98 ± 0.02 s−1 for TpAd and AIPA (Table 2), respectively, thus suggesting that intermediate hydrolysis and/or product release are likely rate limiting.
Table 1. Pre-steady state Kinetics of hHint1 using AIPA or TpAd as substrate in HEPES buffer (20mM HEPES, 1mM MgCl2, pH 7.2) at 25°C.
| AIPA | TpAd | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| k2 (s−1) | K adenylyl (μM) | k2/Kadenylyl (M−1s−1) | k2 (s−1) | Kadenylyl (μM) | k2/Kmadenylyl (M−1s−1) | |
| WT | 443 ± 38 | 5 ± 1 | (8.8 ± 2.8) × 107 | 203 ± 13 | 13 ± 2 | (1.6 ± 0.4) × 107 |
| H114A | 851 ± 45 | 35 ± 5 | (2.4 ± 0.5) × 107 | 34 ± 1 | 57 ± 7 | (0.6 ± 0.1) × 106 |
| H110A | 965 ± 83 | 15 ± 4 | (6.4 ± 2.0) × 107 | 29 ± 1 | 12 ± 2 | (2.5 ± 0.4) × 106 |
| H51A | 313 ± 12 | 17 ± 2 | (1.8 ± 0.3) × 107 | 23 ± 1 | 27 ± 2 | (0.9 ± 0.1) × 106 |
Table 2. Steady state Kinetics of hHint1 in HEPES buffer (pH 7.2, 20mM HEPES, MgCl2) at 25°C.
| AIPA | TpAd | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| kcat (s−1) | *kl (s−1) | Km (μM) | kcat/Km (M−1s−1) | kcat (s−1) | *kl (s−1) | Km (μM) | kcat/Km (M−1s−1) | |
| WT | 1.98±0.02 | 2.7±0.8 | 0.04±0.002 | (5.3±0.5) × 107 | 2.1±0.1 | 2.0±0.4 | 0.13±0.02 | (1.5±0.3) × 107 |
| H114A | 0.69±0.05 | 0.23±0.05 | 0.12±0.01 | (0.6±0.1) × 107 | 0.29±0.02 | 0.14±0.01 | 0.27±0.05 | (1.1±0.3) × 107 |
| H110A | 0.60±0.02 | 0.16±0.02 | 0.04±0.004 | (1.6±0.2) × 107 | 0.24±0.01 | 0.18±0.02 | 0.08±0.01 | (2.9±0.4) × 106 |
| H51A | 0.98±0.06 | 0.33±0.04 | 0.05±0.03 | (1.9±0.2) × 107 | 0.53±0.02 | 0.27±0.03 | 1.8±0.2 | (0.3±0.04) × 106 |
kl represents linear rate calculated from pre-steady state measurement
Due to a single tryptophan (Tryp-123) in the C-terminus, hHint1 exhibits fluorescence (ex 280, em 340). To rule out the possibility that the observed fluorescence “burst” might originates from a change inhHint1 fluorescence, we prepared the Tryp-123 to Ala mutant, demonstrated that the protein fluorescence had been abolished, then examined the hydrolysis of AIPA by W123A with stopped flow kinetics. When compared to wild-type hHint1, the W123A mutant exhibited a similar burst rate and slightly increased Kadenylyl (k2=301s−1, Kadenylyl=8μM), compared to wild-type hHint1 (k2=443s−1, Kadenylyl =5μM). The reduced rate for the burst also agrees with the previous notion that W123 affects Hint enzymatic activity. Furthermore, fluorescence quenching, presumably from the enzyme, was observed when the catalytically inactive mutant, H112A, was mixed with substrate, which is consistent with the observed fluorescence increase during the reaction arising from substrate hydrolysis.
Next, we characterized the “burst” kinetics for TpAd and AIPA. As can be seen in Figure S1B and S1C, the burst rate was found to be hyperbolic and substrate concentration dependent for both TpAd and AIPA. The upper limits of the burst rate (kb) suggest that the adenylylation rates (k2) of Hint1 for AIPA and TpAd are 443 s−1 and 203 s−1, respectively (Table 1). Notably, the second order rate constants, k2/Kadenylyl, for AIPA and TpAd are 8.8 ± 2.8 × 107 M−1s−1 and 1.6 ± 0.4) × 107 M−1s−1, respectively, which approach the diffusion limit (108-1010M−1s−1). Thus, the efficiency of the enzyme during the first step (adenylylation) is quite high and the substrate is likely to be “sticky” (k−1<<k2), limiting dissociation of the substrate from enzyme prior to intermediate formation. With sticky substrates the slopes of the linear portion at low substrate concentrations approached the association rate constants (k1)(44) for AIPA and TpAd of 6 × 107 M−1s−1 and 1 × 107 M−1s−1, respectively. The correspondence of these values for k1 with the steady-state kcat/Km values (Eq.8) supports this assertion.
To assess the degree of cooperativity between the two active sites of hHint1, we examined the burst amplitude, since it is indicative of the number of product molecules turned over in the first catalytic cycle and thus the active enzyme concentration. As can be seen in Figure 2B, the burst amplitude was found to be dependent on enzyme concentration.
Figure 2.
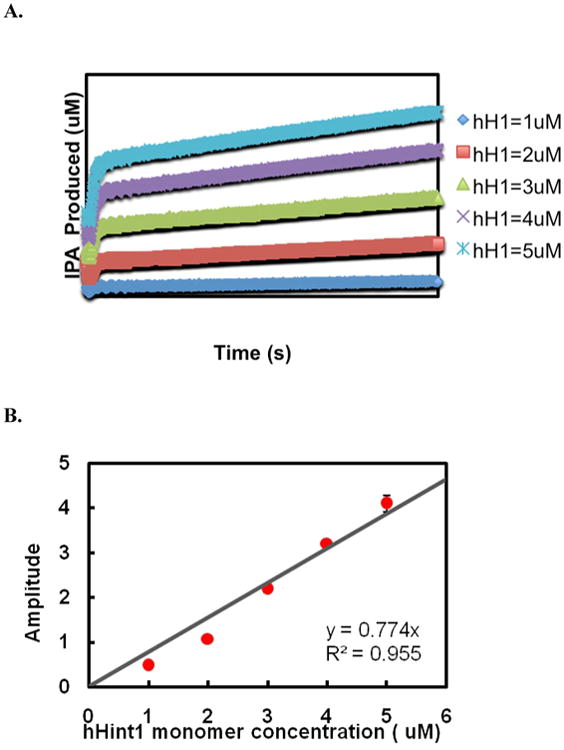
Active site titration. (A) The WT human Hint1 (1, 2, 3, 4, 5μM) was mixed with AIPA (10 μM) in stopped flow cell and the time course of IPA production was monitored at 360nm. (B) The amplitude of “burst” vs. human Hint1 monomer concentration was plotted. A slope of 0.77 with R2 =0.955 indicated that 77% of the active site catalyzes the reaction in the first catalytic cycle.
Plotting the amplitude verses hHint1 monomer concentration yielded a linear correlation with a slope of 0.77 (Fig. 2B, R2=0.955), indicating employment of 77% of the active sites in the first catalytic cycle. Thus, the amplitude of the “burst” is stoichiometric with the calculated Hint monomer concentration, indicating little cooperativity between the two active sites.
Inspection of the active site of hHint1 has revealed that in addition to the active site nucleophile, His-112, His-51, His-110 and His-114 are also strictly conserved (Fig.1A). Although not close enough to hydrogen bond to His-112, His-114 has been shown to be in close proximity to the phosphate oxygens of bound AMP. His-51 appears to form a close hydrogen bond to His-114, while the backbone carbonyl of His-110 forms a hydrogen bond to His-112. We addressed the role of each of these residues on the burst phase of the reaction with AIPA and TpAd by site-directed mutagenesis. Regardless of the mutant or the substrate, a rapid burst phase was observed. For AIPA, the value of the burst phase increased approximately 2- fold, from 443 s−1 for wild-type, to 851 s−1 and 965 s−1 for the mutants His-114-Ala and His-110-Ala, respectively. In contrast, each of the mutants reduced the burst phase rate when TpAd was the substrate from203 s−1 to 23-34 s−1. This discrepancy suggests that somewhat different catalytic mechanisms may be employed for TpAd and AIPA hydrolysis by these mutants. As a potential proton donor, H114 could possibly transfer a proton to the phosphoryl oxygen, which is 2.87Å away, which could be transferred to the amine of the amino group facilitating TpAd P-N bond cleavage. In contrast, dissociation of the acyl product generated by the AIPA reaction may be accelerated in the absence of H114 and proton transfer, assuming charge repulsion is a sufficient driving force. Since stabilization of H114 by H51 may promote the proton transfer, it is possible that in the absence of H51, hydrolysis of TpAd is more affected than AIPA hydrolysis. Since crystal structure analysis demonstrates that the backbone carbonyl of His110 hydrogen bonds to the imidazole of H112, but not H114, His110 is likely to stabilize deprotonated H112, which would be required for both TpAd and AIPA hydrolysis. The similar impact of the H110A mutant on the rate of adenylation by both substrates supports this assertion.
The observed linear steady state phase (kl) observed for each of the mutants when AIPA is the substrate was reduced from 6.4 to 13- fold, while for TpAd the rate was reduced from 7.3 to 14-fold (Table 2). Similar to wild-type, the values of the linear rate are in agreement with the observed kcat values for the mutants and each of the substrates (Table 2).
Examination of the k2/Kadenylyl values for AIPA revealed that while the k2 value either increased for both the His-110-Ala and His-114-Ala mutants or slightly decreased for the His-51-Ala mutant, the overall efficiencies of the mutants declined relative to wild-type because of the increase in Kadenylyl. With the exception of the His-110-Ala mutant, the Kadenylyl values for the mutant hHints increased with the substrate TpAd. Consequently, comparable lower values for k2 resulted in significantly lower k2/Kadenylyl values for each of the active site mutants with TpAd.
Steady state kinetic parameters
The Michaelis-Menten parameters for hHint1 with the substrates AIPA and TpAd were found to be nearly identical resulting in similar kcat/Km values (Table 2), suggesting that the enzyme proceeds through similar rate limiting steps for each substrate. Since the concentration of the second substrate (water) for a double displacement mechanism (Ping Pong mechanism) can be regarded as fixed and saturated, the steady state kcat/Km approximates the pre-steady state k2/Kadenylyl. Indeed, the measured values of kcat/Km and k2/Kadenylyl are within a similar range. Each of the active site histidine mutations were found to lower the kcat by 2 to 2.9- fold for AIPA and similarly 2.8 to 8.8- fold for TpAd. With the exception of the H51A mutant Km for TpAd, which was 13.8- fold greater than the wild-type Km, the Kms for the mutants were either identical or within 3- fold of the value for wild-type hHint1.
Viscosity effect studies
Previous kinetic studies of Hints with phosphoramidate substrates have proposed that hydrolysis of the active site histidine-adenylate is the likely rate-determining step (24). Nevertheless, it is quite possible that product release may also be at least partially rate limiting. Since the molecular diffusion rates vary inversely with the viscosity of the solvent, a diffusion-controlled rate limiting step can be probed by a viscosity variation assay. By increasing the viscosity of the solution with a microviscosogen such as sucrose, the rate of diffusion is lowered whereas the unimolecular reaction rate remains unperturbed. Thus, the diffusion-controlled steps in enzymatic pathways can be isolated and the extent to which the diffusion-controlled steps limit the reaction rate can be estimated.
The steady state hydrolysis rate of TpAd by wild-type Hint1 was measured in the presence and absence of microviscosogen, sucrose, in HEPES buffer at pH 7.2. From the kinetic mechanism proposed in Scheme 1, increasing solvent microviscosity would be predicted to affect k1, k−1, k4 and k−4, but not k2 and k3, with the overall kcat being dependent on a combination of k2, k3, and k−4 (eq. 6).
According to eq. 9, the linear plot of the ratio of the second-order rate constant without viscosogen over the second-order rate constant in the presence of viscosogen, (kcat/Km)0/(kcat/Km), vs. relative viscosity, ŋrel, yields the partition ratio for the ES complex, k−1/k2, as the reciprocal of the slope k2/(k−1+k2) subtracting unity. For a slope close to 1 (slope=0.9) (Fig. 3A), then k−1<k2 (k−1/k2 = 0.1), indicating that the first irreversible step along the reaction pathway is considerably faster than substrate dissociation. Thus, in agreement with the transient state kinetics, the k2/Kadenylyl1 value for TpAd with hHint1 is near the diffusion-controlled limit (vide supra). In contrast, the relative first order rate constant, 0kcat/kcat, was only 60% dependent on the solvent viscosity (Fig. 3B, slope =0.61). Thus employing the expression for 0kcat/kcat (eq. 10) the overall reaction rate was found to be nearly equally limited by an internal presumably chemistry (k3) step and a diffusion-dependent (k−4) step.
Figure 3.
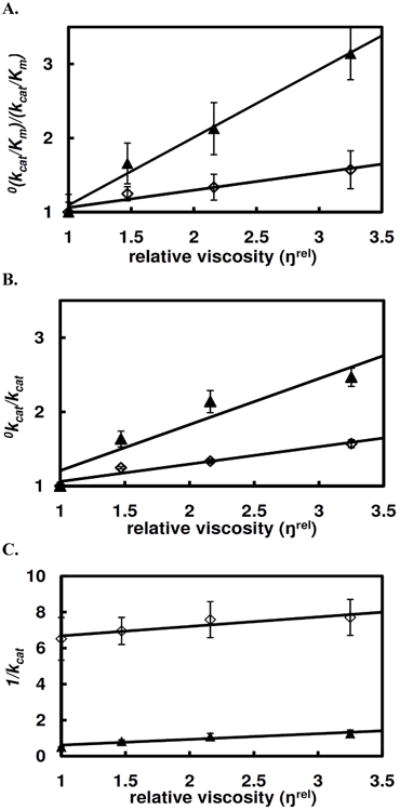
Solvent viscosity effects on the steady state hydrolysis of TpAd (▲) and TrAd (◊) in HEPES buffer at pH 7.2. (A) plot of the relative second-order rate constant as a function of relative viscosity (ŋrel); (B) plot of the relative kcat vs. relative viscosity (ŋrel); (C) plot of 1/kcat vs. relative viscosity (ŋrel).
When a plot of (1/kcat) vs. relative viscosity, ŋrel, was examined, a slope of 0.31 and an intercept of 0.30 were observed (Fig. 3C). Employing eq 11 and a k2 value of 203s−1 determined from pre-steady state studies (vide supra), a value of 3.33 s−1 could be calculated for k−4 and a value of 3.48 s−1 for k3, thus indicating that the steady state rate of hHint1 is partially rate limited by an internal presumably chemical step and an external product release step.
To ensure that the observed decrease in kcat/Km and kcat values are due to the increased solvent microviscosity and not interactions with the protein, control experiments were conducted in the presence of macroviscosogen, PEG 8000, which increases the macroviscosity but not the microviscosity of the solvent (45). The addition of a macroviscosogen will not perturb rates of small molecule diffusion. The kcat and kcat/Km values for TpAd in the presence of 5% PEG-8000 (ŋrel =3) were found to be unaffected (kcat=1.97 ± 0.10 s−1, kcat/Km = (1.6 ± 0.4) × 107 M−1s−1) relative to the values for wild-type hHint1 determined without the macroviscogen.
In addition, to rule out possible artifactual effects of viscosogen on the enzyme reaction due to interactions between the viscosogen and enzyme/substrate, the effect of viscosity on the hydrolysis of a poor substrate, TrAd (Fig 1, kcat= 0.27 s−1, Km = 4.1 μM, kcat/Km = 7.0 × 104 M−1s−1), was determined (24). Given that a burst phase was not detected by pre-steady state kinetic analysis for TrAd, a chemical transformation rate, rather than the diffusional rate, can be reasonably assumed to govern the overall reaction rate. As can be seen in Figure 3B, the slope of the plot of 0kcat/kcat vs. relative viscosity was found to be 0.08 or close to zero, indicating little or no effect of the viscosogen on the chemical steps of the reaction.
Catalytic Trapping Experiment And Simulation
The pre-steady-state kinetic data suggest that the acylation step for hHint1 is fast and does not limit maximum turnover (Fig.2). To establish whether the deacylation step or possibly a product release step (AMP dissociation) could limit turnover, we performed a catalytic trapping experiment. In this experiment, the enzyme is preequilbrated with the product, AMP, in one syringe of the stopped-flow machine before the reaction is started with the substrate TpAd in the second syringe (Fig. 5A). In the absence of AMP, a burst phase followed by a slower linear phase is observed (Fig.5B). These data are best simulated using the two-step kinetic scheme in Fig. 5a where k1′ and k2′ are set at 7.2 × 106 M−1s−1 and 0.5 s−1. The former corresponds to the rate constant for intermediate formation and is close in value to steady state kcat/Km for TpAd (Table 2). The latter corresponds to the net rate constant for deacylation and includes not only the breakdown of the intermediate but also the release of the product AMP and any potential conformational steps. When the enzyme is preequilibrated with 2500 μM AMP, the kinetic transient begins to deviate from the simple biphasic transient observed in the absence of AMP (Fig.5C). A rapid “burst” in fluorescence is followed by a slow exponential phase and a linear phase that corresponds to steady-state product formation. The initial rapid “burst” has a similar rate to the control, albeit with a lower amplitude, suggestive of a small amount of free enzyme in the cuvette prior to reaction start with TpAd. We measured a Kd of 200 μM for AMP to Hint1 in fluorescence titration experiments (Fig.S2) indicating that some of the enzyme is likely to exist in an uncomplexed form in the syringe prior to mixing. The slow exponential phase is consistent with rapid exchange of AMP at the active sites, although the dissociation of AMP could be fast. To analyze this kinetic transient, we, therefore, simulated the trapping data with a small amount of free enzyme (0.7 μM) along with a larger amount of the E•A complex (1.8 μM). Using fixed values of k1 and k2 from the control simulation lacking product equilibration, a best fit to the kinetic trapping data were obtained using a koff of 2.7 s−1 (Fig.5C). Expected kinetic transients assuming that koff is either equal to or 100-fold larger than k2′ are shown for comparison in Fig.5C demonstrating the capability of sensitively monitoring changes in AMP release rate. Overall, the catalytic trapping data illustrate that AMP release is not a significant factor controlling the deacylation step and is thus likely to only play a partial role in defining kcat.
Figure 5.
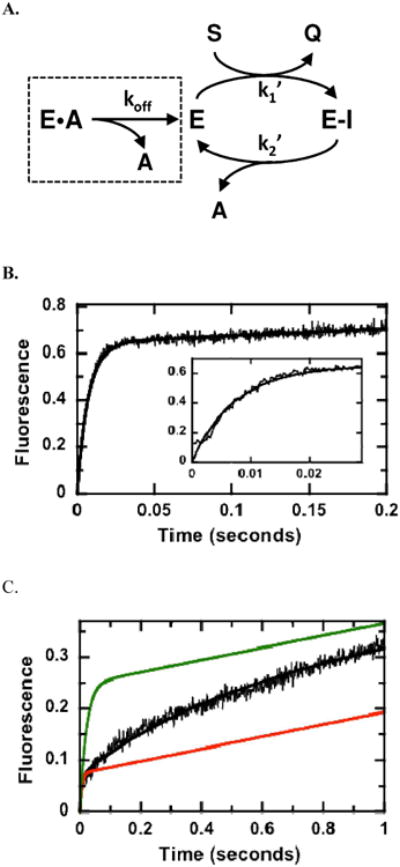
Catalytic Trapping of AMP.(A) Kinetic mechanism. Data in the absence and presence of AMP are simulated using the program DynaFit and a two-step acylation/deacylation mechanism. (B) Kinetic transient in the absence of AMP. Hint1 (5 μM) is mixed with TpAd (20μM) in the stopped flow instrument and the fluorescence changes are simulated using the kinetic scheme in panel A where k1′ and k2′ are 7.2 μM−1s−1 and 0.5 s−1 and the output for Q is 0.26 volts/mM. (C) Kinetic transient in the presence of 2500 mM AMP. Conditions are the same as in panel B except 2500 mM AMP is preequilbrated with the enzyme prior to mixing. The fluorescence changes are simulated using fixed values for k1 ′ and k2′ from panel B, 0.7 and 1.8 mM E and E•A, an output for Q of 0.1 volts/mM and a koff of 2.7 s−1 (black line). The data were also simulated using koff values of 50 (green line) and 0.5 s−1 (red line).
pH dependence of hHint1 adenylylation
To address the ionization state of residues which are critical for hHint1 adenylylation, the pH effect on the pre-steady state kinetics was determined over the pH range of 5.8-9.0 using either AIPA (2.5-20μM) or TpAd (5-30μM) as substrates. For both substrates, the data were best fit with equation 12, according to a mechanism involving three protonic forms, only two (EH and E) of which are active (Scheme 2). The adenylylation rate constants for EH and E were represented as (k2)lim and r ×(k2)lim, where r represents the ratio relative to rate for EH(0 <r<1). The pH vs.k2 profile indicated a dependence of two ionizable groups with pKas around 6.6 and 8.0 (for AIPA, 6.68±0.09 and 8.03±0.16, and for TpAd, 6.56 ±0.13and 7.90±0.24). Similar values of (k2)lim were fit for AIPA (673 ± 50s−1) and TpAd (634 ± 86s−1) (Fig. 6A). A ratio (r) of k2 values at two ionization states (E vs. EH) of 0.72 ± 0.11was fit for AIPA, indicating the deprotonated form(E) is 72% active compared to the EH form. In contrast, an r value of 0.16 ± 0.07 (E vs. EH) was fit for TpAd, suggesting the E form is only 16% active compared to the EH form. This accounts for the rapid decrease in k2 at the basic limb of the pH profile for TpAd.
Figure 6.
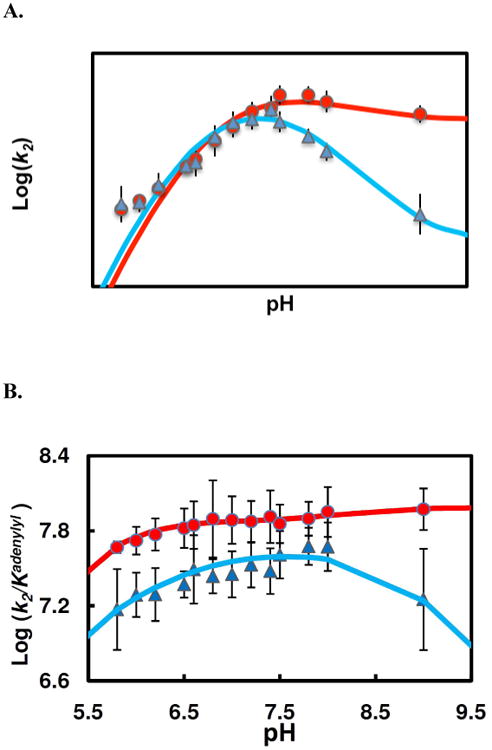
pH dependence of pre-steady state kinetic parameters of the enzyme adenylylation by TpAd (▲) and AIPA(●). (A) Plots of adenylylation rate (k2) vs. pH. The data fitting (Scheme 2, eq 12) for AIPA reaction (red line) yields following values of the characterizing parameters: pK1= 6.68 ± 0.09, pK2= 8.03 ± 0.16, r= 0.72 ± 0.11, (k2)lim= 673±50 s−1; for TpAd reaction (blue line), pK1= 6.56 ± 0.13, pK2= 7.90 ± 0.24, r= 0.16 ± 0.08, (k2)lim= 634 ± 86 s−1. (B) Plots of k2/Kadenylyl vs. pH. The pH profiles for AIPA reaction (red line) and for TpAd (blue line) were relatively flat, indicating substrates are sticky.
On the other hand, the acid limbs of pH vs.k2/Kadenylyl for AIPA and TpAd showed relatively flat curves, indicating the substrates are sticky and the apparent pKas are displaced to lower pH (Fig. 6B). Since both curves exhibited a change of only a half log unit in the value of k2/Kadenylyl over a single pH unit, the apparent pKas are unlikely to represent the titration of a single ionizable group in the adenylylation reaction.
pH dependence of steady state kinetic parameters
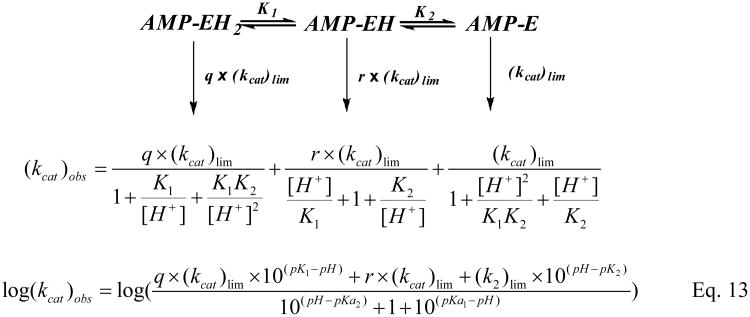
The dependence of the steady-state substrate hydrolysis on pH was determined with saturating concentrations of AIPA or TpAd (Fig.7). Since the observed steady state rates for AIPA and TpAd are similar at each pH, the pH vs. kcat profile for the hHint1 catalyzed reaction was found to be best fit by a minimal two-pKa model with three active forms (Scheme 3), yielding a pKa1 of 7.52 ± 0.18 and a pKa2 of 8.15 ± 0.10, and kcat values for the three states, EH2, EH, and E of 1.44±0.08 s−1, 6.28±0.56 s−1 and 0.85±0.05 s−1, respectively. Since the hydrolysis of adenylylated Hint1 and product release are partially rate limiting in steady state, the pH profile presumably reflect the overall pH effects on both steps(46).
Figure 7.
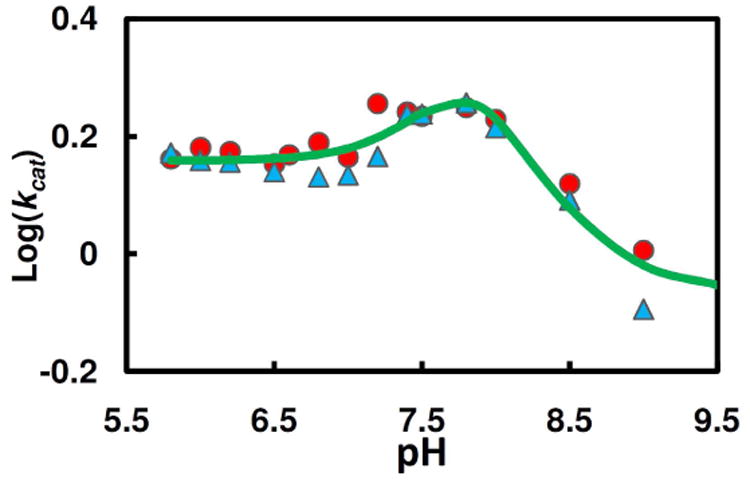
Comparison of pH (▲, TpAd; ●, AIPA) dependence of steady state kinetic parameters (kcat). The pH profile data were fit by eq 13 (Scheme 3), yielding pK1= 7.52 ± 0.18, pK2= 8.15 ± 0.10, r= 7.3 ± 2.0, q= 1.68 ± 0.16, and (kcat)lim for the three active forms (AMP-EH2, AMP-EH, and AMP-E) are 1.44±0.08 s−1, 6.28±0.56 s−1, and 0.85±0.05 s−1, respectively.
Scheme 3.
Determination of the uncatalyzed rate of TpAd and AIPA hydrolysis
The hydrolysis of AIPA, and TpAd, as monitored by the disappearance of the peak at 5.88 ppm, proceeded to completion with first order kinetics. The results yielded a linear Arrhenius plots with k25 = 1.1 × 10−6 s−1 at 25 °C and ΔH‡ = 20.1 kcal/mol at pH 6.8 for AIPA and k25 = 2.0 × 10−8 s−1 at 25 °C and ΔH‡ = 16.9 kcal/mol at pH 6.8 for TpAd (Fig.8). For AIPA, the rate of hydrolysis increased monotonically with increasing pH, by a factor of ∼3 in the region between pH 4.8 (0.1 M potassium acetate) and 8.4 (0.1 M potassium borate), while for TpAd the rate decreased by a factor of 3 over the same pH range.
Figure 8.
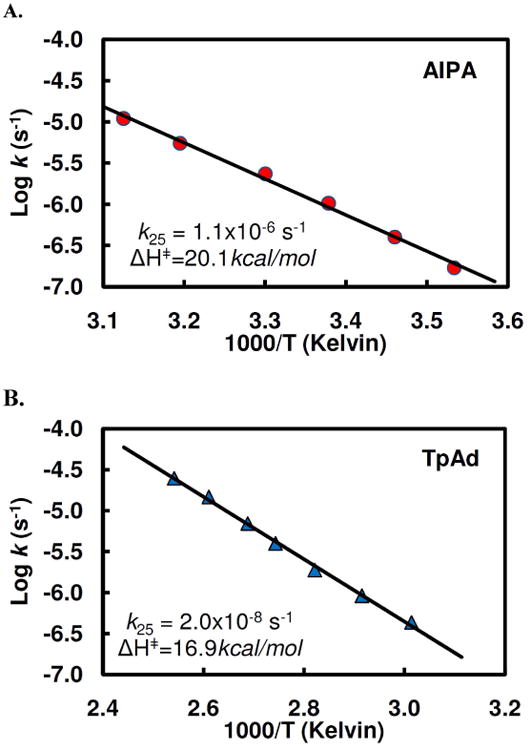
Arrhenius plots for hydrolysis of AIPA (A) and TpAd (B) at 25°C at pH 6.8.
pH dependence of the chemical shifts of active site hHint1 histidines
To assist in understanding the pH dependence of hHint1 pre-steady-state and steady-state kinetics, the effect on the chemical shifts of the active site histidines was determined. Representative 1D proton TOCSY spectra for hHint1 in D2O are shown in Fig. S4. Of the six well resolved histidine C-2 proton resonances in region of 7.5-9 ppm, one titratable signal was identified. By comparison with the spectra of the mutant His-112-Gly and His-112-Ala at pH 7.5, this side-chain resonance could be assigned to His-112. Fitting with the modified Henderson-Hasselbalch equation (eq 15) of the pH vs. C-2 chemical shift (δ) plot yielded a pKa value for His-112 of 6.50 ± 0.06.
Discussions
There has been an emerging interest in Hint proteins since it has been discovered that they are involved in a variety of biological processes across all kingdoms of life (47). Hints have been shown to be highly efficient nucleoside phosphoramidases and acy-AMP hydrolases (37). Nevertheless, the biological relevance of the catalytic activity of Hints has remained obscure. In three cases, the Hint catalysis has been clearly shown to be essential. Brenner and co-workers have demonstrated that yeast deficient in catalyically active Hint1 homolog, HNT1, are unable to grow on galactose at elevated temperatures (48). In addition, they found that HNT1 was a regulator of the TFIIH kinase subcomplex of the transcription complex, TFIIH, and the Cdk7 ortholog, Kin28 (48). Our laboratory has demonstrated that E. coli also encode only one Hint protein, ecHinT. We have also shown that catalytically active ecHinT is necessary for the growth of E. coli under high salt conditions (23). In addition, the ability of E. coli to grow on alanine was shown to depend on the catalytic regulation of D-amino acid dehydrogenase activity by ecHinT(30). In addition, recently, the first example association of hHint1 with a human phenotype has been discovered by the demonstration that inherited peripheral neuropathy is caused by the loss of hHint1 function due to at least 8 different mutations(1). Mutations at two of these residues are known to either affect catalytic activity (His-51) or result in the loss of catalytic activity (His-112)(1, 49).
Detailed catalytic and kinetic mechanistic studies have been carried out with only the DcpS subfamily of the HIT superfamily, while catalytic studies have been pursued with GalT, Fhit and Hint1. Preliminary studies of the catalytic mechanism of Hints has revealed that 1) of the four conserved active site histidines, one is essential for catalytic activity (26), 2) the reaction proceeds through an active site His-AMP intermediate (26), and 3) the pre-steady state kinetics are characterized by a burst phase followed by a linear rate (50). To begin to elucidate the kinetic mechanism of Hint1s, we chose to characterize the kinetic mechanism of hHint1.
The transient burst followed by a linear phase observed during earlier stopped flow experiments for ecHinT was also observed for hHint1(50). This observation is consistent with our hypothesis that hHint1 proceeds by rapid formation of the AMP-Hint intermediate, followed by at least partial rate limiting hydrolysis of the intermediate. The rate of the burst phase was found to be substrate dependent (k2= 443s−1 and 203s−1 for AIPA and TpAd, respectively), whereas nearly identical values for the overall turnover numbers were observed for both substrates. Moreover, the different burst rates for TpAd and AIPA and the inability to observe at least one additional exponential phase suggests that if a conformational change does occur upon substrate binding, it is quite rapid and therefore not kinetically significant. In addition, the transient burst was found to correspond to 77% of the hHint1 monomer concentration. Although the substrate specificity of hHint1 has been shown to be dependent on interactions contributed by the C-terminus of each monomer to the active site of the adjacent monomer (51), the observation of a burst corresponding to nearly 80% of the hHint1 monomer concentration indicates that the two active sites of hHint1 independently catalyze substrate hydrolysis. The hyperbolic, rather than a sigmoidal relationship, of the steady state rate vs. substrate concentration plot further confirmed that kinetically significant cooperative interactions do not exist between the two active sites of hHint. In contrast, the active sites of homodimeric DcpS exhibited negative cooperativity (52).
When compared to TpAd, burst kinetics was not observed for the less efficient substrate, TrAd, and the linear rate corresponded to the steady-state kcat value. In addition, the kcat for TrAd (kcat = 0.27 s−1) was found to be 7.7- fold lower than the kcat for TpAd(24). Thus, the kcat for the poor substrate, TrAd, is likely due to rate limiting active site adenylylation, since the rate of enzyme-AMP intermediate hydrolysis and subsequent AMP dissociation would be independent of the alkyl amine leaving group. The rate of hHint1 adenylylation by TpAd is thus shown to be 17,000- foldgreater than the rate observed for TrAd. Consequently, rate of the first step of the kinetic mechanism is profoundly affected by the alkyl amine of the nucleoside phosphoramidate substrate.
Because enzyme adenylylation by TpAd is rapid and not rate-limiting, then either intermediate hydrolysis, product release, or a combination of these two steps should govern the overall substrate turnover for hHint1. Since the rates of kinetically significant steps can be dependent on the nature of the solvent, the effect of solvent viscosity on enzyme catalysis has been extensively evaluated. For example, Townsend and co-workers have elegantly applied a series of solvent viscosity studies to elucidate the rate-limiting steps of carbapenam synthetase and β-lactam synthetase (53). Consequently, we chose to characterize the rate-limiting steps of hHint1 with a series of solvent effect studies. Steady-state kinetic studies conducted with increasing concentrations of the microviscogen, sucrose, revealed that the second-order rate constant (kcat/Km) was significantly affected, while the overall reaction rate (kcat) was only modestly affected. By plotting the relative kcat/Km values vs. relative viscosity, the dependence of all the reaction processes on diffusional control preceding and including the first irreversible step can be determined (39, 54). The significant sensitivity of the kcat/Km for TpAd to viscosity (slope of 0.9) suggests that the steps preceding the first irreversible step, hHint1 adenylylation, are under diffusional control. Likewise, the viscosity dependence of kcat, provides an estimate of the extent to which the turnover number is limited by product diffusional release. The observed kcat for TpAd was found to be moderately sensitive to solvent viscosity (a slope of about 0.6), implying that product release accounts for approximately half of the overall rate (kcat). Based on these results, we propose that hHint1 catalysis proceeds minimally through a four step kinetic mechanism (Scheme 1).
To further characterize the release of AMP by hHint1, catalytic trapping experiments and fluorescence titration experiments were carried out. The burst amplitude was indeed shown to be dependent on the AMP concentration, which is consistent with AMP release being at least partially rate-limiting. Simulation of catalytic trapping data showed the true dissociation rate of AMP (koff) may not be significant in determining the overall kcat. Examination of viscosity effects on kcat using Kramer's model (eq. 16) indicated that the association of a conformational change and product release contributed to the viscosity effect. Nevertheless, hHint1 has very low affinity for AMP. Since both the Ki determined by the catalytic trapping studies (Fig. S3) and the Kd determined by fluorescence titration experiments for AMP (Fig. S2) were found to be approximately 200 μM, the association rate constant for AMP is quite low.
Given the results of the steady-state and pre-steady-state kinetics and the viscosity experiments, the rate constants for the proposed minimal kinetic mechanism for hHint1 can be assembled based on Scheme 1. First, based on the slopes of the plot of (kcat/Km)0/(kcat/Km) verses relative viscosity for AIPA and TpAd (Fig. 3a) and the burst rates for each substrate (k2), equation 9 can be employed to calculate values of 111 s−1 and 22.6 s−1 for the rate of substrate dissociation of (k−1) for AIPA and TpAd, respectively (Scheme 4). Knowing the values of k2 and k−1, the values for k1 can then be calculated from the equation 4 for KAdenylyl, yielding second order rate constants of 111μM−1s−1 and 17.4 μM−1s−1 for AIPA and TpAd, respectively. Next, employing equations 11 and the slope and intercept of the plot of 1/kcat vs. the relative viscosity for TpAd (Fig. 3c), the values 3.23 s−1 and 3.38 s−1 could be determined for k3 and k−4, respectively. Lastly, given a Kd for AMP of 200 μM, and the value of k−4, a value of 0.0162 μM-1s−1 could be calculated for the association rate constant for AMP.
Scheme 4.

Proposed hHint1 kinetic mechanism with the substrates AIPA and TpAd.
With the derived rate constants in hand, kcat values of 1.63 ± 0.01s−1 and 1.65 ± 0.03s−1 for TpAd and AIPA, respectively, and Km values of 0.131 ± 0.004 μM and 0.039 ± 0.005 μM for TpAd and AIPA, respectively, were derived. The simulated kcat and Km values correspond closely to the experimental values (Table 2) for kcat of 1.98 ± 0.02 s−1 and 2.0 ± 0.1 s−1 for TpAd and AIPA, respectively, and Km values of 0.13 ± 0.02 μM and 0.040 ± 0.002 μM for TpAd and AIPA, respectively, thus providing support for the proposed kinetic mechanism. In addition, although the dissociation of AMP from the enzyme was found by kinetic viscosity studies to be coupled to hHint1 conformational changes, the rates of these steps are not observable and therefore appear not to be kinetically significant.
We hypothesize that the AMP binding to hHint1, as well as AMP release, are potentially coupled to conformational changes induced by the flexible C-terminus. Deletion of the C-terminus of hHint1 results in a 200-400- fold decrease in the rate of hHint1 adenylylation and a 100-200- fold decrease in the overall rate. Future studies, delineating the role ofthe dynamics of the C-terminus on hHint1on product release should help address this hypothesis.
The kinetic mechanism for hHint1 outlined has been established for pH 7.0. The active site of hHint1, however, is composed of four histidines, including a nucleophilic histidine (His-112), which is adenylylated by both AIPA and TpAd. As expected, pH vs. rate studies for enzyme adenylylation exhibited strong pH dependence with observable pKa's of 6.68 and 8.03 for AIPA and 6.51 and 7.90 for TpAd. In both cases, the first pKa was found to correspond to the pKa of 6.5 determined by NMR titration of His-112. We noticed that the pH vs. k2 profile for TpAd differs from that for AIPA at the basic limb, indicating that the deprotonated form (E) is less able to catalyze TpAd reaction (r =0.16) than AIPA hydrolysis (r =0.72) (scheme 2 and Fig 6A). This discrepancy can be explained by the dependence of the rate of adenylylation on an ionizable group with a pKa of approximately 8. Deprotonation of this group at pH values greater than 8 would result in a decrease in the rate of adenylylation for TpAd, with little effect on the adenylation rate for AIPA. We propose that H114 could be this ionizable group and act as a proton donor in Hint1 adenylylation (Fig. 1A) by facilitating protonation of either the tryptamine or indole proprionic acid leaving groups during hydrolysis of TpAd and AIPA, respectively.
Two pKa's of 7.52 and 8.15 were observed for the pH vs. kcat profile. Since both hydrolysis of the adenylylated enzyme and product release are partially rate limiting, neither pKa should correspond to free His-112. The pKa of adenylylated imidazole has been shown to be 5.4, significantly reducing the efficiency of P-N bond cleavage at physiological Ph (55). It is more likely that the second pKa corresponds to the protonated imidazole for adenylylated His-112, while the first pKa corresponds to an active site acid, possibly His-114.
While the natural substrate(s) for hHint1 are presently unknown, the enzyme efficiently hydrolyzes both acyl-AMP and nucleoside phosphoramidates with kcat/Km values at or near diffusion control for AIPA and TpAd. The work of Wolfenden and co-workers has demonstrated that enzymes can accelerate the rate of a reaction by factors ranging from 105-1017 –fold (56, 57). To assess the rate enhancements produced by hHint1 for substrate hydrolysis, we determined the rate constants for the uncatalyzed hydrolysis of the anhydride AIPA (k25= 1.1 × 10−6 s−1) and the phosphoric amide TpAd (k25= 2.0 × 10−8 s−1). These values are considerably smaller than the rate constants that have been reported for the uncatalyzed hydrolysis of unsubstituted acetyl phosphate (k39 = 1.9 × 10−3 s−1) and phosphoric acid amide (k37 = 1.2 × 10−4 s−1)(58, 59) presumably reflecting the accessibility of ionic species of these unsubstituted molecules that are more reactive than AIPA or TpAd. Considering that AIPA and TpAd are unnatural substrates, it is of interest that hHint1 accelerates AIPA hydrolysis by a factor of 106 and TpAd hydrolysis by a factor of 108, with enzyme efficiencies (kcat/Km) rivaling those of well-known enzymes, such as cytidine deaminase and carboxypeptidase B(60). Thus, although AIPA and TpAd are unnatural substrates, hHint1 seems highly evolved to cleave both substrates. While the recent discovery that Hints and in particular Hint1's are able to hydrolyze AARS generated amino acyl-AMPs may ultimately explain their preference for AIPA, the rationale for their phosphoramidase activity remains to be discovered.
Supplementary Material
Figure 4.
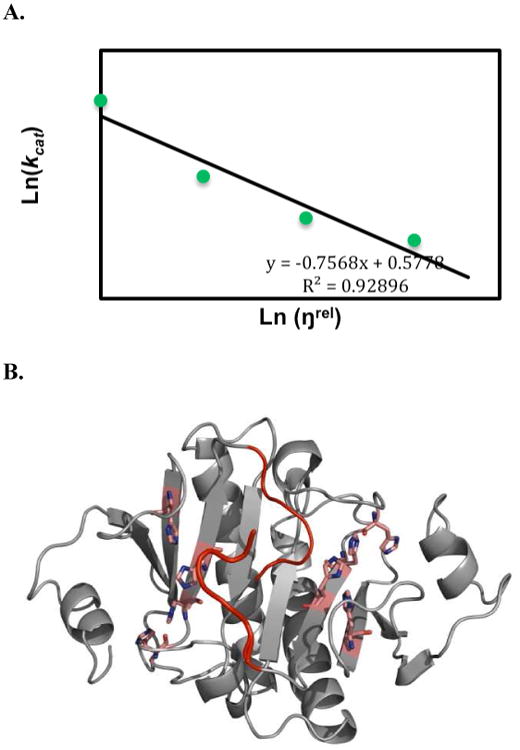
(A) Data from viscosity experiment were fit to Kramer's model (eq 16), ln(kcat) is linearly dependent on ln (ηrel) with a slope of -0.76 (R2 =0.93), indicating a strong coupling of product release with the active site conformational change. (B) The C-termini of hH1 homodimer that is proposed to undergo conformational change coupled with AMP release. The active sites are shown in pink and the C-termini are shown in red.
Acknowledgments
Partial funding for this study from the University of Minnesota Endowment and NIH grant GM-18325 (RW) is gratefully acknowledged.
Funding Statement: Support for this study from the University of Minnesota Endowment is gratefully acknowledged.
Abbreviations
- Hint
histidine triad nucleotide-binding protein
- hHint1
human histidine nucleotide-binding protein 1
- AIPA
adenosine 5′-indole-3-propionic adenylate
- GalT
Galactose-1-phosphate uridyl transferase
- HIT
histidine triad
- TpAd
tryptamine adenosine phophoramidate monoester
- TrAd
(2(R)-[Adenosyl- 5′-(phophorylamino)-3-(3-indolyl)-propionic acid methyl ester])
- Fhit
fragile histidine triad
- wt
wild type
Equation 1-18
For “Burst” kinetics,
| Eq. 1 |
| Eq. 2 |
| Eq. 3 |
For Hint1 adenylylation,
| Eq. 4 |
| Eq. 5 |
For the hydrolysis of AMP-Hint1 intermediate,
| Eq. 6 |
| Eq. 7 |
| Eq. 8 |
For viscosity experiments,
| Eq. 9 |
| Eq. 10 |
| Eq. 11 |
Fluorescence titration of ligand binding:
| Eq. 14 |
For NMR titration,
| Eq. 15 |
For Kramer's model,
| Eq. 16 |
Footnotes
Supporting Information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zimon M, Baets J, Almeida-Souza L, De Vriendt E, Nikodinovic J, Parman Y, Battaloglu E, Matur Z, Guergueltcheva V, Tournev I, Auer-Grumbach M, De Rijk P, Petersen BS, Muller T, Fransen E, Van Damme P, Loscher WN, Barisic N, Mitrovic Z, Previtali SC, Topaloglu H, Bernert G, Beleza-Meireles A, Todorovic S, Savic-Pavicevic D, Ishpekova B, Lechner S, Peeters K, Ooms T, Hahn AF, Zuchner S, Timmerman V, Van Dijck P, Rasic VM, Janecke AR, De Jonghe P, Jordanova A. Loss-of-function mutations in HINT1 cause axonal neuropathy with neuromyotonia. Nat Genet. 2012;44:1080–1083. doi: 10.1038/ng.2406. [DOI] [PubMed] [Google Scholar]
- 2.Brenner C, Garrison P, Gilmour J, Peisach D, Ringe D, Petsko GA, Lowenstein JM. Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins. Nat Struct Bio. 1997;l4:231–238. doi: 10.1038/nsb0397-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mozier NM, Walsh MP, Pearson JD. Characterization of a novel zinc binding site of protein kinase C inhibitor-1. FEBS Lett. 1991;279:14–18. doi: 10.1016/0014-5793(91)80238-x. [DOI] [PubMed] [Google Scholar]
- 4.Lima CD, Klein MG, Weinstein IB, Hendrickson WA. Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins. Proc Natl Acad Sci U S A. 1996;93:5357–5362. doi: 10.1073/pnas.93.11.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Munoz M, de la Torre-Madrid E, Sanchez-Blazquez P, Wang JB, Garzon J. NMDAR-nNOS generated zinc recruits PKCgamma to the HINT1-RGS17 complex bound to the C terminus of Mu-opioid receptors. Cell Signal. 2008;20:1855–1864. doi: 10.1016/j.cellsig.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Ajit SK, Ramineni S, Edris W, Hunt RA, Hum WT, Hepler JR, Young KH. RGSZ1 interacts with protein kinase C interacting protein PKCI-1 and modulates mu opioid receptor signaling. Cell Signal. 2007;19:723–730. doi: 10.1016/j.cellsig.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Weiske J, Huber O. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. The Journal of biological chemistry. 2006;281:27356–27366. doi: 10.1074/jbc.M513452200. [DOI] [PubMed] [Google Scholar]
- 8.Korsisaari N, Makela TP. Interactions of Cdk7 and Kin28 with Hint/PKCI-1 and Hnt1 histidine triad proteins. The Journal of biological chemistry. 2000;275:34837–34840. doi: 10.1074/jbc.C000505200. [DOI] [PubMed] [Google Scholar]
- 9.Razin E, Zhang ZC, Nechushtan H, Frenkel S, Lee YN, Arudchandran R, Rivera J. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. The Journal of biological chemistry. 1999;274:34272–34276. doi: 10.1074/jbc.274.48.34272. [DOI] [PubMed] [Google Scholar]
- 10.Lee YN, Nechushtan H, Figov N, Razin E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity. 2004;20:145–151. doi: 10.1016/s1074-7613(04)00020-2. [DOI] [PubMed] [Google Scholar]
- 11.Lee YN, Razin E. Nonconventional involvement of LysRS in the molecular mechanism of USF2 transcriptional activity in FcepsilonRI-activated mast cells. Molecular and cellular biology. 2005;25:8904–8912. doi: 10.1128/MCB.25.20.8904-8912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cen B, Li H, Weinstein IB. Histidine triad nucleotide-binding protein 1 up-regulates cellular levels of p27KIP1 by targeting ScfSKP2 ubiquitin ligase and Src. The Journal of biological chemistry. 2009;284:5265–5276. doi: 10.1074/jbc.M804531200. [DOI] [PubMed] [Google Scholar]
- 13.Cen B, Deguchi A, Weinstein IB. Activation of protein kinase G Increases the expression of p21CIP1, p27KIP1, and histidine triad protein 1 through Sp1. Cancer research. 2008;68:5355–5362. doi: 10.1158/0008-5472.CAN-07-6869. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Zhang Y, Su T, Santella RM, Weinstein IB. Hint1 is a haplo-insufficient tumor suppressor in mice. Oncogene. 2006;25:713–721. doi: 10.1038/sj.onc.1209111. [DOI] [PubMed] [Google Scholar]
- 15.Weiske J, Huber O. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. J Biol Chem. 2006;281:27356–27366. doi: 10.1074/jbc.M513452200. [DOI] [PubMed] [Google Scholar]
- 16.Korsisaari N, Rossi DJ, Luukko K, Huebner K, Henkemeyer M, Makela TP. The histidine triad protein Hint is not required for murine development or Cdk7 function. Mol Cell Biol. 2003;23:3929–3935. doi: 10.1128/MCB.23.11.3929-3935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbier E, Zapata A, Oh E, Liu Q, Zhu F, Undie A, Shippenberg T, Wang JB. Supersensitivity to amphetamine in protein kinase-C interacting protein/HINT1 knockout mice. Neuropsychopharmacology. 2007;32:1774–1782. doi: 10.1038/sj.npp.1301301. [DOI] [PubMed] [Google Scholar]
- 18.Chen Q, Wang X, O'Neill FA, Walsh D, Kendler KS, Chen XN. Is the histidine triad nucleotide-binding protein 1 (HINT1) gene a candidate for schizophrenia? Schizophrenia Research. 2008;106:200–207. doi: 10.1016/j.schres.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin J, Romanque P, Maurhofer O, Schmitter K, Hora C, Ferrand G, Dufour JF. Ablation of the Tumor Suppressor Histidine Triad Nucleotide Binding Protein 1 is Protective Against Hepatic Ischemia/Reperfusion Injury. Hepatology. 2011;53:243–252. doi: 10.1002/hep.23978. [DOI] [PubMed] [Google Scholar]
- 20.Pace HC, Brenner C. Feminizing chicks: a model for avian sex determination based on titration of Hint enzyme activity and the predicted structure of an Asw-Hint heterodimer. Genome Bio. 2003;l4:R18. doi: 10.1186/gb-2003-4-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parks KP, Seidle H, Wright N, Sperry JB, Bieganowski P, Howitz K, Wright DL, Brenner C. Altered specificity of Hint-W123Q supports a role for Hint inhibition by ASW in avian sex determination. Physiol Genomics. 2004;20:12–14. doi: 10.1152/physiolgenomics.00204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krakowiak A, Pace HC, Blackburn GM, Adams M, Mekhalfia A, Kaczmarek R, Baraniak J, Stec WJ, Brenner C. Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors. J Biol Chem. 2004;279:18711–18716. doi: 10.1074/jbc.M314271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou TF, Bieganowski P, Shilinski K, Cheng J, Brenner C, Wagner CR. 31P NMR and genetic analysis establish hinT as the only Escherchia coli purine nucleoside phosphoramidase and as essential for growth under high salt conditions. J Biol Chem. 2005;280:15356–15361. doi: 10.1074/jbc.M500434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou TF, Baraniak J, Kaczmarek R, Zhou X, Cheng J, Ghosh B, Wagner CR. Phosphoramidate pronucleotides: a comparison of the phosphoramidase substrate specificity of human and Escherichia coli histidine triad nucleotide binding proteins. Mol Pharm. 2007;4:208–217. doi: 10.1021/mp060070y. [DOI] [PubMed] [Google Scholar]
- 25.Wagner CR, Iyer VV, McIntee EJ. Pronucleotides: toward the in vivo delivery of antiviral and anticancer nucleotides. Med Res Rev. 2000;20:417–451. doi: 10.1002/1098-1128(200011)20:6<417::aid-med1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 26.Chou TF, Wagner CR. Lysyl-tRNA synthetase-generated lysyl-adenylate is a substrate for histidine triad nucleotide binding proteins. J Biol Chem. 2007;282:4719–4727. doi: 10.1074/jbc.M610530200. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Fang P, Schimmel P, Guo M. Side chain independent recognition of aminoacyl adenylates by the Hint1 transcription suppressor. J Phys Chem B. 2012;116:6798–6805. doi: 10.1021/jp212457w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozga M, Dolot R, Janicka M, Kaczmarek R, Krakowiak A. Histidine triad nucleotide-binding protein 1 (Hint-1) phosphoramidase transforms nucleoside 5′-O-phosphorothioates to nucleoside 5′- phosphates. J Biol Chem. doi: 10.1074/jbc.M110.162065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bieganowski P, Garrison PN, Hodawadekar SC, Faye G, Barnes LD, Brenner C. Adenosine monophosphoramidase activity of Hint and Hnt1 supports function of Kin28, Ccl1, and Tfb3. J Biol Chem. 2002;277:10852–10860. doi: 10.1074/jbc.M111480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bardaweel S, Ghosh B, Chou TF, Sadowsky MJ, Wagner CRE. coli histidine triad nucleotide binding protein 1 (ecHinT) is a catalytic regulator of D-alanine dehydrogenase (DadA) activity in vivo. PloS one. 6:e20897. doi: 10.1371/journal.pone.0020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geeganage S, Frey PA. Transient kinetics of formation and reaction of the uridylyl-enzyme form of galactose-1-P uridylyltransferase and its Q168R-Variant: Insight into the molecular basis of galactosemia. Biochemistry. 1998;37:14500–14507. doi: 10.1021/bi9815546. [DOI] [PubMed] [Google Scholar]
- 32.Wedekind JE, Frey PA, Rayment I. The structure of nucleotidylated histidine-166 of galactose-1-phosphate uridylyltransferase provides insight into phosphoryl group transfer. Biochemistry. 1996;35:11560–11569. doi: 10.1021/bi9612677. [DOI] [PubMed] [Google Scholar]
- 33.Abend A, Garrison PN, Barnes LD, Frey PA. Stereochemical retention of the configuration in the action of Fhit on phosphorus-chiral substrates. Biochemistry. 1999;38:3668–3676. doi: 10.1021/bi981895j. [DOI] [PubMed] [Google Scholar]
- 34.Huang K, Arabshahi A, Frey PA. pH-Dependence in the hydrolytic action of the human fragile histidine triad. European Journal of Organica Chemistry. 2005:5198–5206. [Google Scholar]
- 35.Huang K, Arabshahi A, Wei Y, Frey PA. The mechanism of action of the fragile histidine triad, Fhit: isolation of a covalent adenylyl enzyme and chemical rescue of H96G-Fhit. Biochemistry. 2004;43:7637–7642. doi: 10.1021/bi049762n. [DOI] [PubMed] [Google Scholar]
- 36.Brenner C. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou TF, Tikh IB, Horta BA, Ghosh B, De Alencastro RB, Wagner CR. Engineered monomeric human histidine triad nucleotide-binding protein 1 hydrolyzes fluorogenic acyl-adenylate and lysyl-tRNA synthetase-generated lysyl-adenylate. J Biol Chem. 2007;282:15137–15147. doi: 10.1074/jbc.M606972200. [DOI] [PubMed] [Google Scholar]
- 38.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 39.Brouwer AC, Kirsch JF. Investigation of diffusion-limited rates of chymotrypsin reactions by viscosity variation. Biochemistry. 1982;21:1302–1307. doi: 10.1021/bi00535a030. [DOI] [PubMed] [Google Scholar]
- 40.Markley JL, Finkenstadt WR, Dugas H, Leduc P, Drapeau GR. Proton magnetic resonance titration curves of the three histidine residues of staphylococcal protease. Biochemistry. 1975;14:998–1005. doi: 10.1021/bi00676a019. [DOI] [PubMed] [Google Scholar]
- 41.Wolfenden R, Ridgway C, Young G. Spontaneous hydrolysis of ionized phosphate monoesters and diesters and the proficiencies of phosphatases and phosphodiesterases as catalysts. Journal of the American Chemical Society. 1998;120:833–834. [Google Scholar]
- 42.Chou TF, Baraniak J, Kaczmarek R, Zhou X, Cheng J, Ghosh B, Wagner CR. Phosphoramidate Pronucleotides: A Comparison of the Phosphoramidase Substrate Specificity of Human and E. coli Histidine Triad Nucleotide Binding Proteins (Hint1) Mol Pharm. 2007 doi: 10.1021/mp060070y. In press. [DOI] [PubMed] [Google Scholar]
- 43.Bardaweel S, Pace J, Chou TF, Cody V, Wagner CR. Probing the Impact of the EchinT C-terminal Domain on Structure and Catalysis. J Mol Biol. 2010;404:672–638. doi: 10.1016/j.jmb.2010.09.066. [DOI] [PubMed] [Google Scholar]
- 44.Johnson KA. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- 45.Blacklow SC, Raines RT, Lim WA, Zamore PD, Knowles JR. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: Analysis of triose phosphate equilibria in aqueous solution by 31P NMR. Biochemistry. 1988;27:1158–1167. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- 46.Schimerlik MI, Cleland WW. pH variation of the kinetic parameters and the catalytic mechanism of malic enzyme. Biochemistry. 1977;16:576–583. doi: 10.1021/bi00623a003. [DOI] [PubMed] [Google Scholar]
- 47.Brenner C, Bieganowski P, Pace HC, Huebner K. The histidine triad superfamily of nucleotide-binding proteins. Journal of cellular physiology. 1999;181:179–187. doi: 10.1002/(SICI)1097-4652(199911)181:2<179::AID-JCP1>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bieganowski P, Garrison PN, Hodawadekar SC, Faye G, Barnes LD, Brenner C. Adenosine monophosphoramidase activity of Hint and Hnt1 supports function of Kin28, Ccl1, and Tfb3. The Journal of biological chemistry. 2002;277:10852–10860. doi: 10.1074/jbc.M111480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chou TF, Wagner CR. Lysyl-tRNA synthetase-generated lysyl-adenylate is a substrate for histidine triad nucleotide binding proteins. J Biol Chem. 2007;282:4719–4727. doi: 10.1074/jbc.M610530200. [DOI] [PubMed] [Google Scholar]
- 50.Bardaweel S, Pace J, Chou TF, Cody V, Wagner CR. Probing the impact of the echinT C-terminal domain on structure and catalysis. Journal of molecular biology. 404:627–638. doi: 10.1016/j.jmb.2010.09.066. [DOI] [PubMed] [Google Scholar]
- 51.Chou TF, Sham YY, Wagner CR. Impact of the C-terminal loop of histidine triad nucleotide binding protein1 (Hint1) on substrate specificity. Biochemistry. 2007;46:13074–13079. doi: 10.1021/bi701244h. [DOI] [PubMed] [Google Scholar]
- 52.Liu SW, Rajagopal V, Patel SS, Kiledjian M. Mechanistic and kinetic analysis of the DcpS scavenger decapping enzyme. J Biol Chem. 2008;283:16427–16436. doi: 10.1074/jbc.M800341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raber ML, Arnett SO, Townsend CA. A conserved tyrosyl-glutamyl catalytic dyad in evolutionarily linked enzymes: carbapenam synthetase and beta-lactam synthetase. Biochemistry. 2009;48:4959–4971. doi: 10.1021/bi900432n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardy LW, Kirsch JF. Diffusion-limited component of reactions catalyzed by Bacillus cereus beta-lactamase I. Biochemistry. 1984;23:1275–1282. doi: 10.1021/bi00301a040. [DOI] [PubMed] [Google Scholar]
- 55.Weber AL, Orgel LE. Amino acid activation with adenosine ′-phosphorimidazolide. J Mol Evol. 1978;11:9–16. doi: 10.1007/BF01768020. [DOI] [PubMed] [Google Scholar]
- 56.Wolfenden R. Benchmark reaction rates, the stability of biological molecules in water, and the evolution of catalytic power in enzymes. Annual review of biochemistry. 80:645–667. doi: 10.1146/annurev-biochem-060409-093051. [DOI] [PubMed] [Google Scholar]
- 57.Edwards DR, Lohman DC, Wolfenden R. Catalytic Proficiency: The Extreme Case of S-O Cleaving Sulfatases. Journal of the American Chemical Society. 2012;134:525–531. doi: 10.1021/ja208827q. [DOI] [PubMed] [Google Scholar]
- 58.Koshland DE. Effect of catalysts on the hydrolysis of acetyl phosphate. Nucleophilic displacement mechanisms in enzymatic reactions. J Am Chem Soc. 1952;74:2286–2292. [Google Scholar]
- 59.Chanley JD, Feageson E. A study of the hydrolysis of phosphoramides. II. Solvolysis of phosphoramidic acid. J Am Chem Soc. 1963;85:1181–1190. [Google Scholar]
- 60.Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Accounts of Chemical Research. 2001;34:938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.