Abstract
To learn more about cancer-associated fibroblasts (CAFs), we have isolated fibroblasts from different stages of breast cancer progression and analysed their function and gene expression. These analyses reveal that activation of the YAP transcription factor is a signature feature of CAFs. YAP function is required for CAFs to promote matrix stiffening, cancer cell invasion and angiogenesis. Remodelling of the ECM and promotion of cancer cell invasion requires the actomyosin cytoskeleton. YAP regulates the expression of several cytoskeletal regulators, including ANLN, and DIAPH3, and controls the protein levels of MYL9/MLC2. Matrix stiffening further enhances YAP activation, thus establishing a feed-forward self-reinforcing loop that helps to maintain the CAF phenotype. Actomyosin contractility and Src function are required for YAP activation by stiff matrices. Further, transient ROCK inhibition is able to disrupt the feed-forward loop leading to a long-lasting reversion of the CAF phenotype.
Introduction
Tumours contain a complex mixture of cell types and matrix components1, 2. Non-cancerous cells and matricellular molecules within the tumour can promote dissemination3. In addition, the physical characteristics of the extracellular matrix (ECM) enhance invasion and metastasis4-8. Increased matrix stiffness is a feature of most solid tumours9. Matrix stiffness can be sensed by many cell types. Force exerted on integrin-mediated adhesions can lead to the activation of FAK, Src family kinases and RhoA7, 10, 11. More recently, it has been shown that stiff matrices promote the activity of the YAP and TAZ transcriptional regulators12. Elevated YAP and TAZ function is associated with increased cancer stem cell properties and metastasis13, 14. These observations help to explain how matrix stiffness might be linked to more aggressive tumour phenotypes. However, the interplay between the tumour stroma and matrix stiffness is not well understood.
Cancer-associated fibroblasts (CAFs) are found in many solid tumours, including breast and squamous cell carcinoma, and promote invasion and metastasis through the production of soluble factors and matrix remodelling15. In some cases, matrix remodelling leads to the generation of tracks through the ECM that enable subsequent cancer cell invasion6. Markers such as αSMA and S100A4 are associated with CAFs, however they are often not specific for CAFs and may not relate directly to the functional properties of CAFs16. This has hampered understanding of the pro-tumorigenic signals in the stroma that are required to generate and maintain CAFs. In this study, we uncover a positive feedback loop involving YAP function and matrix stiffening that is critical for the function of CAFs.
Results
Characterisation of fibroblasts from different stages of mammary tumour progression
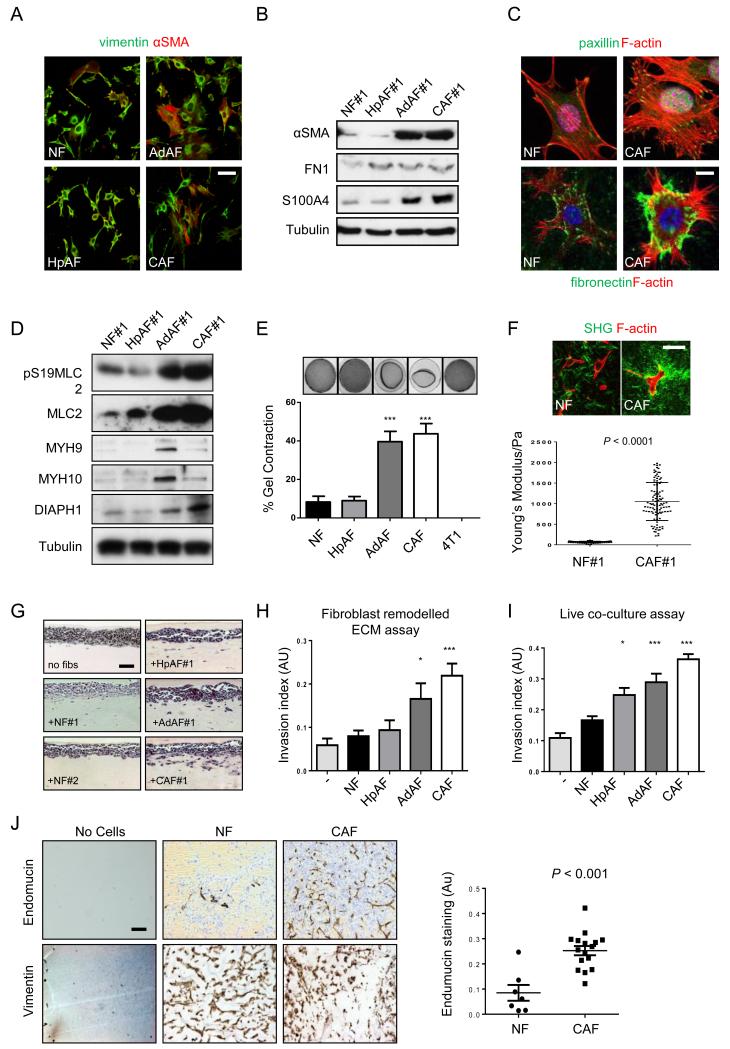
To learn more about CAFs we isolated fibroblasts from normal mouse mammary glands and hyperplastic tissue, mammary adenoma and mammary carcinoma in mice containing the MMTV-PyMT transgene17 (Supplementary Figure 1a). This model was chosen because it contains a fibroblastic stroma with increased collagen fibres (Supplementary Figure 1b), and its progression can be modulated by genetic manipulation of fibroblasts18, 19. To facilitate further experiments, the fibroblasts we isolated were immortalised using HPV-E620. In total we isolated three sets of normal mammary fibroblast (NFs), two hyperplasia-associated fibroblasts (HpAFs), two adenoma associated fibroblasts (AdAFs), and four sets of CAFs. The multi-focal nature of the PyMT model17, 21 enabled us to establish a hyperplasia-, an adenoma- and a carcinoma-associated fibroblast line from different regions of the same mouse; these were designated HpAF#1, AdAF#1 and CAF#1. We confirmed that they were positive for the expected fibroblast markers (Figure 1a). Expression of CAF markers, αSMA and S100A4, was increased in the fibroblasts isolated from adenoma and carcinoma (Figure 1b). Quantification revealed increased αSMA in all four CAF isolates, FAP levels were highest in CAF#1 and CAF#2, while S100A4 was elevated in CAF#1 and CAF#4 (Supplementary Figure 2c). At the single cell level there was significant heterogeneity in the expression of αSMA, S100A4, and FAP, both in vivo and in vitro: not all fibroblasts in tumours were αSMA positive, and various combinations of αSMA, S100A4 and FAP positivity were observed (Supplementary Figure 2d and also Supplementary Figure 1b). Similar results were obtained on the human SCC CAF lines CerCAF, HNCAF and VCAF (Supplementary Figure 2e). Thus, the diversity of CAF marker expression16 is represented in the CAFs that we isolated (Supplementary Figure 2f). CAFs exhibited cytoskeletal changes that suggested increased isometric tension. They had more pronounced stress fibres, more focal adhesions, and increased fibronectin fibrillogenesis (Figure 1c). MLC2 phosphorylation and total protein levels were strongly increased in AdAF#1 and CAF#1, and several other proteins implicated in cytoskeletal regulation (MYH9, MYH10, and DIAPH1) were more modestly up-regualted in AdAF#1 and CAF#1 (Figure 1d). More significantly, the ability of the fibroblasts to contract collagen gels, a measure of their matrix remodelling capacity, increased with disease stage (Figure 1e and Supplementary Figure 2a). Cancer cells were unable to remodel the matrices. Contraction of the gels by CAFs was associated with the appearance of thicker collagen fibres (upper panels Figure 1f), and an eight-fold increase in the stiffness of the matrix from ~120Pa to >1kPa (Figure 1f). This is highly consistent with the change in tissue stiffness observed between normal mammary tissue and tumours9. We next tested whether CAF remodelled matrices were rendered permissive for the invasion of breast cancer cells. Fibroblasts were allowed to remodel the matrix for five days before being killed. The ability of cancer cells to invade into these matrices was then evaluated. CAF#1 and, to a lesser extent AdAF#1, were able to promote cancer cell invasion (Figure 1g&h). NFs and HpAFs did not generate matrices permissive for invasion. Similarly, in organotypic co-culture models, CAFs were the most effective at promoting the invasion of breast cancer cells, both (Figure 1i and Supplementary Figure 2b). These results were confirmed using the syngeneic breast cancer cell line MMTV-PyMT TS222 (Supplementary Figure 1c). Finally, we demonstrated that CAFs were able to promote angiogenesis in the absence of any cancer cells in vivo (Figure 1j – endomucin stains endothelial cells, vimentin stains fibroblasts). These data demonstrate the successful isolation of fibroblasts from different stages of a murine breast cancer model. Further, we show that their matrix remodelling and invasion promoting abilities increase with the disease stage.
Figure 1. Isolation and characterisation of fibroblasts from different disease stages.
A) Vimentin (green) and αSMA (red) staining of fibroblasts isolated from different disease stages. Scale bar, 50μm. B) Western blots of αSMA, fibronectin, and S100A4 levels in fibroblasts isolated from different disease stages. C) Upper panels show F-actin (red), paxillin (green) and DAPI (magenta) staining in NFs and CAFs. Scale bar, 10μm. Lower panels show F-actin (red), fibronectin (green) and DAPI (blue) staining in NFs and CAFs. D) Western blots showing pS19-MLC2/MYL9, MLC2/MYL9, MYH9, MYH10, and DIAPH1 levels in fibroblasts. E) Images show gel contraction by fibroblasts and 4T1 breast cancer cells. Histogram shows mean of ± s.e.m. (n = number of gels (NF, 15; HpAF 13; AdAF, 15; CAF, 35; 4T1, 4), assessed over multiple experiments (NF, 5; HpAF, 5; AdAF, 5; CAF, 10; 4T1, 2). F) Images show F-actin staining of fibroblasts and collagen second harmonic signal. Scale bar, 50μm. Histogram shows elastic modulus of matrices remodelled by NFs or CAFs. Each data point represents a single measurement, n=100 measurements in total. Line and error bars indicate mean ± s.d. G) Representative images showing the invasion of 4T1 cells cultured in the presence of different fibroblasts. Scale bar, 50μm. H) Quantification of 4T1 invasion into matrices previously remodelled by different fibroblasts. Bars represent mean ± s.e.m. n= organotypic assays (-, 4; NF, 18; HpAF, 5; AdAF, 6; CAF, 24), assessed over multiple experiments (-, 2; NF, 4; HpAF, 2; AdAF, 2; CAF, 6). I) Quantification of 4T1 invasion when co-cultured with different fibroblasts. Bars represent mean ± s.e.m. n= organotypic assays (-, 29; NF, 53; HpAF, 19; AdAF, 22; CAF, 56), assessed over multiple experiments (-, 6; NF, 12; HpAF, 4; AdAF, 5; CAF, 12). J) Images show endomucin and vimentin staining of matrix plugs with or without different fibroblasts injected sub-cut in mice. Scale bar, 50μm. Graph shows quantification of endomucin staining relative to fibroblast number (vimentin staining). Each data point represents a different plug (NF, n=7; CAF, n=16). * P < 0.05; ** P < 0.01; *** P < 0.001 - unpaired t-test.
YAP is activated CAFs
We then performed global mRNA analysis on the fibroblasts isolated from different disease stages. RNA samples were prepared from the fibroblasts cultured on the collagen-rich matrices used for the functional assays. Gene Set Enrichment Analysis23 (GSEA) was performed to identify changes in co-ordinated gene expression programmes (Supplementary Table 1). Strikingly, we noted that the genes up-regulated in our murine CAFs showed significant overlap with gene expression in the stroma of human breast cancer with poor prognosis24, 25 ENREF 17 (Supplementary Figure 3a). These observations suggest that our experimental system is likely to have significant relevance for human disease. As expected, GSEA revealed changes in inflammatory signalling26-28, although these changes were typically more pronounced in HpAFs and AdAFs (Supplementary Table 1). We also noted significantly increased expression of genes associated with two mechano-sensitive pathways, YAP/TAZ29-32 ENREF 25 (Table 1 and Supplementary Figure 2b) and SRF33-35 (Supplementary Table 1). We chose to investigate whether YAP or TAZ, closely related paralogues, were involved in CAF function.
Table 1. Enrichment analysis of YAP gene sets in different fibroblasts.
Table showing GSEA of multiple YAP and YAP/TAZ gene sets in different fibroblasts. Comparisons based on pooled gene expression profiles of 3 NFs, 1 HpAF, 1 AdAF and 4 CAFs. Table shows the P values for enrichment of different gene sets in hyperplasia, adenoma and carcinoma-associated fibroblasts from MMTV-PyMT mice when compared to normal fibroblasts. Green boxes indicate significant positive enrichment and red text indicates negative enrichment. Reference to each gene set is indicated. n.s., non-significant.
| Gene set | HpAF | AdAF | CAF |
|---|---|---|---|
| YAP/TAZ30 | n.s. | 0.026915 | <0.001 |
| YAP liver32 | 0.003521 | <0.001 | <0.001 |
| YAP common MCF10 and 3T331 | 0.001919 | <0.001 | 0.040493 |
| Induced by YAP in 3T331 | 0.004505 | <0.001 | 0.00266 |
| Induced by YAP29 | 0.004796 | 0.002747 | 0.010526 |
| Induced by YAP and TAZ29 | 0.038375 | <0.001 | 0.021866 |
| Tead dependent YAP and TAZ29 | n.s. | n.s. | <0.001 |
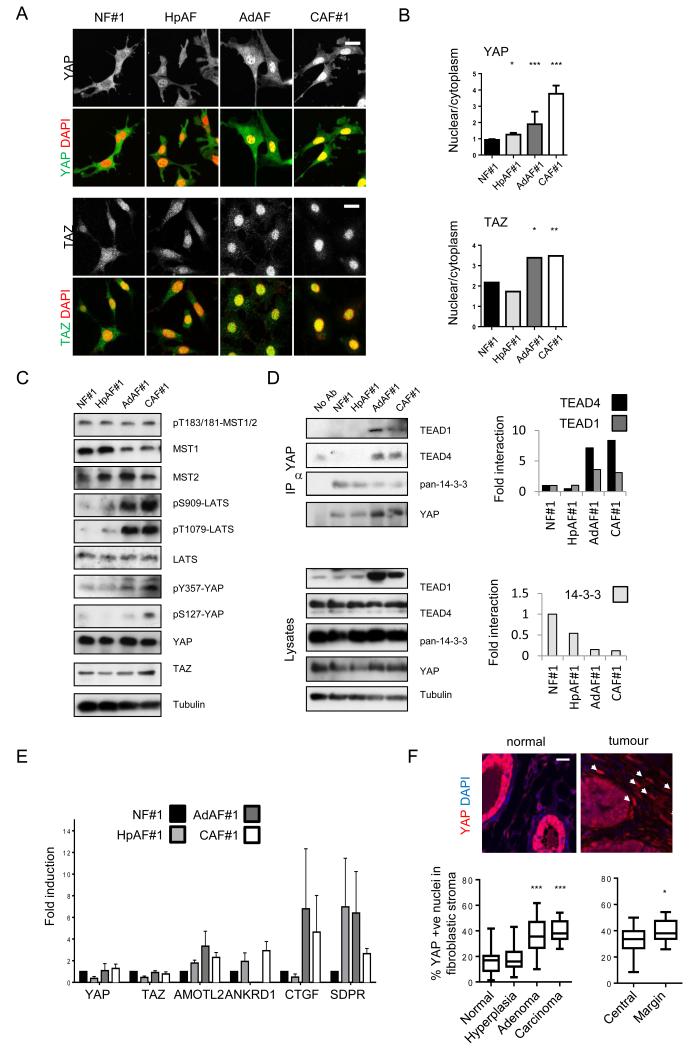
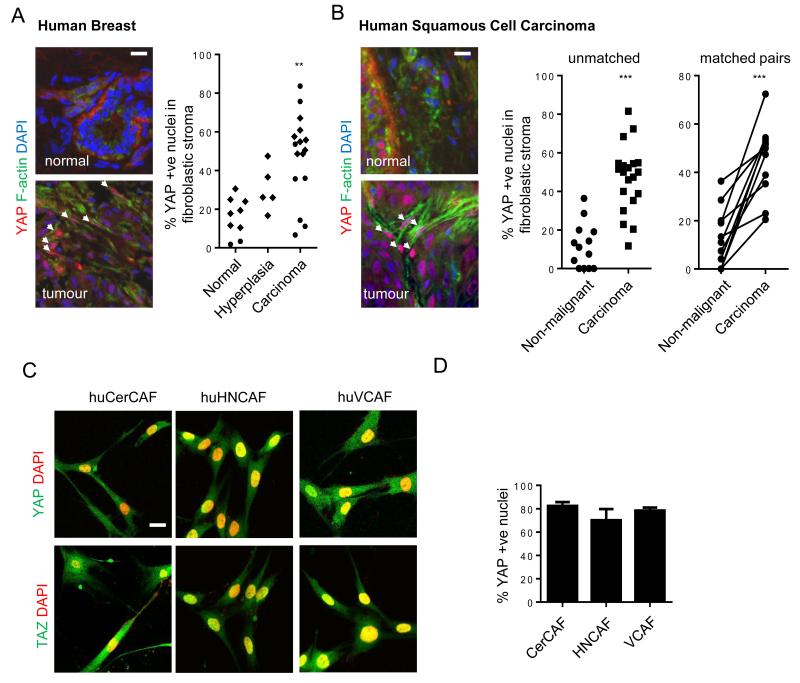
YAP/TAZ are negatively regulated by phosphorylation by LATS1&2, that are in turn regulated by MST1&236, 37. Phosphorylation of YAP and TAZ is associated with their sequestration in the cytoplasm38. Reduced MST1/2 and LATS1/2 activity leads to nuclear accumulation, binding to TEAD and other proteins, and activation of transcription. YAP activation is well known to be oncogenic within epithelial cells36, but its role in the tumour stroma has not been explored. Recently, YAP/TAZ activation has been reported in response to mechanical stress and perturbation of the actin cytoskeleton12, 39-42. Figure 2a shows that YAP is pre-dominantly cytoplasmic in NFs but accumulates in the nucleus in CAFs; HpAFs and AdAFs show intermediate levels of nuclear YAP (alternative quantification of % cells with predominantly nuclear YAP is shown in Supplementary Figure 3c). Nuclear localisation of YAP was a universal feature of CAFs, regardless of their expression levels of αSMA, FAP, or S100A4 (Supplementary Figure 3d, also compare Supplementary Figures 2c and 3c). The localisation of TAZ showed more subtle differences between normal mammary fibroblasts and CAFs (Figure 2a). Interestingly, the activity of the upstream negative regulators, MST1/2, was not different between NFs and CAFs. Further, the activity of LATS kinases and phosphorylation of S127 on YAP was even elevated in CAFs (Figure 2c). Together, these data argue that YAP is not activated by reduced activity of the canonical MST1/2 – LATS pathways in CAFs. Instead, YAP is regulated at the level of interaction with TEAD and 14-3-3 proteins. In NFs, YAP interacted strongly with 14-3-3 proteins, but it switched to interacting with TEAD1 and TEAD4 transcription factors in CAFs (Figure 2d). This switch in binding partners correlated with increasing Y357 phosphorylation of YAP43 (Figure 2c). We confirmed that YAP was indeed transcriptionally active in CAFs by monitoring the expression of 4 endogenous YAP/TAZ target genes in two different CAF lines. Figure 2e and Supplementary Figure 3e show a consistent pattern of activation of YAP target genes in CAFs (Supplementary Figure 3f confirms these genes as YAP targets). Histochemical analysis of the normal murine mammary tissue and PyMT-driven tumours revealed nuclear accumulation of YAP in the stroma of large adenoma and carcinoma lesions (Figure 2f). In advanced carcinoma lessions, YAP activation in the stroma was further enhanced in the tumoral margins. Moreover, human breast cancer and squamous cell carcinoma biopsies showed nuclear accumulation of YAP in stromal fibroblasts (Figure 3a&b). YAP/TAZ activation was maintained in three different isolates of human squamous cell carcinoma CAFs (Figure 3c&d).
Figure 2. YAP is activated in cancer-associated fibroblasts.
A) Images show YAP or TAZ localisation (greyscale and green) and DAPI (red) in different fibroblasts. Scale bar, 20μm. B) Quantification of nuclear relative to cytosolic fluorescent intensity of YAP or TAZ. Bars represent mean ± s.e.m for YAP data (NF, n=4 experiments with ~15 20× fields of view analysed in each experiment; HpAF, n=3; AdAF, n=3; CAF, n=4). Bars represent mean for TAZ data (one representative experiment shown, with ~15 20× fields of view analysed. Additional data in Table S2 (Statistics Source Data). C) Western blots show active MST1/2 (judged by pT183/T180), MST1 and MST2 levels, active LATS1/2 (judged by pS909 and pT1079), LATS1/2 levels, pS127-YAP, pY357-YAP, TAZ levels, YAP levels and tubulin loading control in NF#1, HpAF#1, AdAF#1, and CAF#1. D) Western blots show immune-precipitation of TEAD1, TEAD4 and 14-3-3 with YAP in NF#1, HpAF#1, AdAF#1, and CAF#1. Graphs represent quantification of the blots showing the amount of TEAD1, TEAD4 and 14-3-3 proteins bound to YAP in HpAF#1, AdAF#1, and CAF#1 relative to NF#1. E) qRT-PCR analysis of gene expression in different fibroblasts. Data from three independent experiments performed in triplicate (normalized toGapdh). Bars represent mean ± s.e.m. of the average fold induction compared to NF#1 in each experiment. F) Images show YAP localisation (red) and DAPI (blue) staining of normal and cancerous mammary tissue. White arrows indicate nuclear accumulation of YAP in the stroma. Scale bar, 20μm. Charts show quantification from different stages of MMTV-PyMT disease and from different tumoural regions on carcinomas. n= number of images analysed (normal, n=27; hyperplasia, n=41; adenoma, n=20; carcinoma, n=18; central, n=19; margin, n=18). Images were taken from multiple tissue samples (normal, 3; hyperplasia, 2; adenoma, 7; carcinoma, 7; central, 7; margin 7). In the box-and-whisker plot, the central box represents values from the lower to upper quartile. The middle line represents the mean. The horizontal line extends from the minimum to the maximum value. Values represent the % of YAP positive stromal cells per 20× field of view. * P < 0.05; ** P < 0.01; *** P < 0.001 – unpaired t-test.
Figure 3. YAP is activated in cancer-associated fibroblasts in human disease.
A) Images show YAP localisation (red), F-actin (green), and DAPI (blue) staining of human mammary tissue. Scale bar is 20μm. White arrows indicate nuclear accumulation of YAP in the stroma. Chart shows the proportion of non-leukocytic stroma with nuclear YAP localisation in normal, hyperplastic, and malignant breast tissue (round symbols are from analysis of frozen tissue, diamonds are from analysis of formalin-fixed paraffin embedded tissue). Each point represents a different tumour or normal tissue sample; normal, n=9; hyperplasia, n=5; carcinoma, n=16. B) Images show YAP localisation (red), F-actin (green), and DAPI (blue) staining of human squamous epithelium. White arrows indicate nuclear accumulation of YAP in the stroma. Chart shows the proportion of non-leukocytic stroma with nuclear YAP localisation in cancerous human head and neck squamous cell carcinoma and adjacent non-malignant mucosa (both in unmatched and matched pairs from the same patient). Each point represents a different tumour or normal tissue sample; normal, n=13; carcinoma, n=20 (including 11 matched pairs). Scale bar is 20μm. C) Images show YAP or TAZ localisation (green) and DAPI (red) in human CerCAFs,HNCAFs and VCAFs. Scale bars is 20μm. D) Histogram shows the % of CerCAF, HNCAF and VCAF with predominantly nuclear YAP. Bars represent mean ± s.e.m of 5 independet experiments. ** P < 0.01, *** P < 0.001 – unpaired t-test.
YAP is required for CAF function
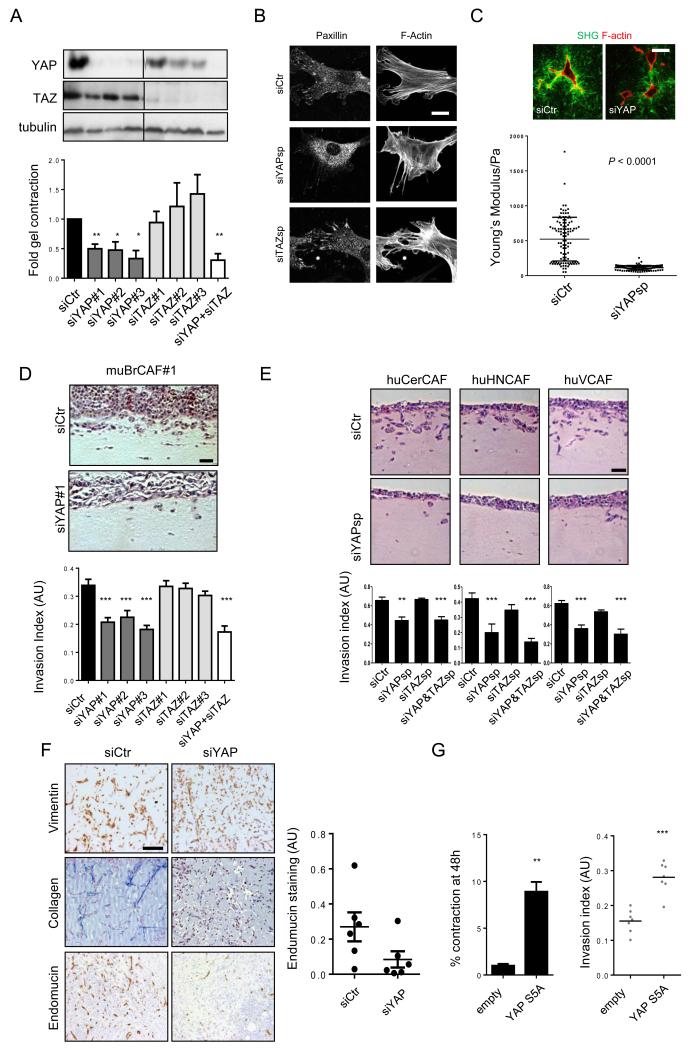
We next tested whether YAP or TAZ function was important for the functional properties of CAFs. Depletion of YAP with three different siRNA reduced the ability of the CAFs to physically contract collagen-rich matrices (Figure 4a). YAP-depleted cells also had fewer focal adhesions, essential structures for force transmission between the cytoskeleton and matrix (Figure 4b). These defects were associated with fewer thick collagen fibres within matrices containing YAP-depleted cells together with a lower elastic modulus (Figure 4c). In contrast, depletion of TAZ had little effect (Figure 4a&b). The ability of CAFs to promote cancer cell invasion was also significantly dependent on YAP function, but TAZ was not required (Figure 4d and Supplementary Figure 4a). We confirmed these observations in three different human CAF lines derived from squamous cell carcinoma (Figure 4e). Depletion of YAP also reduced the ability of CAFs to form fibrous collagen networks and promote angiogenesis in vivo (Figure 4f – Masson’s Trichrome and endomucin staining, respectively). Conversely, activation of YAP signalling by either over-expression of an active mutant or depletion of MST1&2 increased invasion promoting ability of NFs (Supplementary Figure 4b and Figure 4g). Together, these data demonstrate that YAP is critical for many aspects of the tumour-promoting function of CAFs.
Figure 4. YAP is required for cancer associated fibroblast function.
A) Western blots showing depletion of YAP or TAZ with three different siRNA. Chart shows matrix remodelling by CAF#1 following transfection with different YAP or TAZ siRNA. Bars represent mean ± s.e.m (n=20 organotypic assays assessed over 4 independent experiments). B) Panels show paxillin and F-actin staining in control, siYAP, and siTAZ transfected CAF#1. Scale bar, 10μm. C) Images show F-actin staining of control and siYAP-transfected fibroblasts and collagen second harmonic signal. Scale bar is 50μm. Histogram shows elastic modulus of matrices remodelled by CAF#1 transfected with control or YAP siRNA. Each data point represents a single measurement, n=96 measurements in total. Line and error bars indicate mean ± s.d. D) Representative images and quantification of 4T1 invasion when co-cultured with murine breast CAF#1 (muBrCAF#1) transfected with control, YAP or TAZ siRNA. Bars represent mean ± s.e.m. (n=15 organotypic assays assessed over 3experiments). Scale bar, 50μm. E) Images show squamous cell carcinoma invasion (SCC12 cells) in organotypic co-culture models including human CerCAFs, HNCAFs) and VCAFs. Histogram shows quantification of carcinoma invasion when co-cultured with indicated human CAFs transfected with control, YAP or TAZ siRNA smart-pools. Scale bar, 50μm. Bars represent mean ± s.e.m. of n=10 organotypic assays assessed over 3 independent experiments. F) Images show endomucin, Masson’s Trichrome, and vimentin staining of matrix plugs with murine breast CAF#1 transfected with control or YAP siRNA and injected sub-cut in mice. Scale bar, 100μm. Chart shows quantification of endomucin staining in 6 control and 6 YAP siRNA plugs, each plug is from a different mouse. Line and error bars indicate mean ± s.e.m. G) Charts show the effect of YAP S5A expression in NF#1 on gel contraction and ability to promote cancer cell invasion. Bars represent mean ± s.e.m. of three independent experiments (gel contraction). For organotypic invasion, every organotypic measurement is shown individually (pooled from 2 independent experiments) ** P < 0.01, *** P < 0.001 – unpaired t-test. Additional data for Fig. 4e, 4g in Table S2 (Statistics Source Data).
YAP regulates the contractile actomyosin cytoskeleton
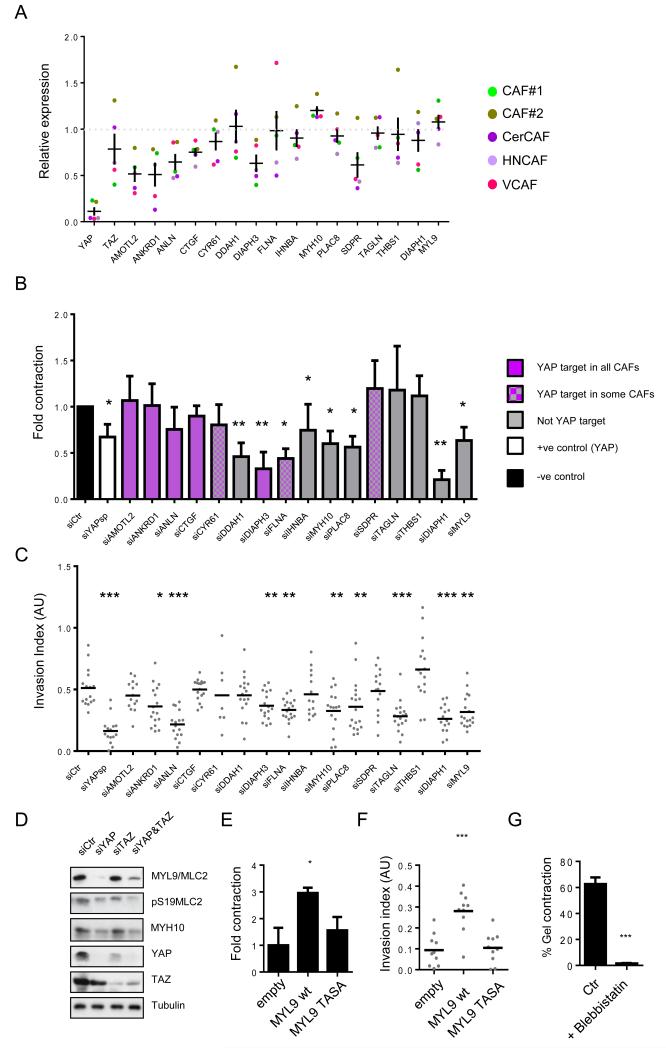
We then sought to identify YAP regulated genes that were required for matrix remodelling by CAFs. We focused on 14 genes that our microarray analysis had indicated were up-regulated in CAFs (see Figure 2e and Supplementary Figure 3e for validation of specific genes) that had been implicated as YAP targets in genomic analyses29, 31. We additionally tested DIAPH1 and MYL9/MLC2 because we had noted their elevated protein levels in CAFs (Figure 1d). Figure 5a shows the effect of YAP siRNA on the mRNA expression levels of the indicated genes (based on YAP siRNA in CAF#1, CAF#2, CerCAF, HNCAF and VCAF). This revealed that 6 out of the 14 genes with elevated mRNA levels in CAFs were found to be consistently transcriptionally regulated by YAP (AMOTL2, ANKRD1, ANLN, CTGF, DIAPH3, SDPR). Others were YAP targets in the minority of the five CAF isolates that we tested (DIAPH1, FLNA, THBS1). MYL9/MLC2 and MYH10 were not transcriptionally regulated by YAP. Amongst these genes, we identified several that are required for CAF-driven matrix remodelling and invasion (Figure 5b&c – see Supplementary Figure 4c for knock-down efficiency). Significantly, ANLN, DIAPH3 and FLNA were required for CAFs to both remodel the ECM and promote invasion (Figure5b&c, invasion data shown with multiple siRNA in Supplementary Figure 4d). This establishes a role for at least two YAP transcriptional targets in enabling the CAF phenotype. This is in agreement with previous reports showing YAP and TEAD binding to the ANLN promoter31. We also found that MYL9 and MYH10 are required for CAF functionality (Figures 5b&c, see Suppementary Figures 4e for multiple siRNA). These genes are not transcriptionally regulated by YAP, nonetheless we explored whether their protein levels might depend on YAP. Figure 5d shows that depletion of YAP, but not TAZ, reduces both the total levels of MYL9 and MYH10 and the level of active S19 phosphorylated MYL9/MLC2. Similar data were obtained in CAF#2 and human vulval SCC CAFs (Supplementary Figure 4f). The critical importance of MYL9/MLC2 for CAF function was confirmed by over-expressing either wild-type MYL9/MLC2 or an inactive mutant in NFs. Only wild-type MYL9/MLC2 over-expression resulted in a gain of matrix remodelling and invasion promoting functions (Figure 5e&f). Finally, blockade of MYL9/MLC function (using blebbistatin) reduced matrix remodelling by CAFs (Figure 5g). Together, these data establish regulation of MYL9/MLC2 by YAP as a critical factor for generating CAFs with matrix remodelling and invasion promoting abilities.
Figure 5. Identification of YAP regulated mRNA and proteins required for CAF function.
A) Histogram shows the expression of the 14 genes of interest 72hours after YAP siRNA transfection of CAF#1, CAF#2, CerCAF, HNCAF and VCAF. Expression levels are relative to Lamc2. Each data point represents a single measurement performed in triplicate. Line and error bars indicate mean ± s.e.m of average of the triplicate values from the 5 cell lines analysed. B) Histogram shows the effect of siRNA mediated depletion of the indicated genes on the ability of CAF#1 to contract collagen-rich gels. Bars represent mean ± s.e.m. (n=7 independent experiments for siYAP, n=5 independent experiments for siMYL9 and siMYH10, n=4 independent experiments for all other siRNA). C) Plot shows the effect of siRNA mediated depletion of the indicated genes on the ability of CAF#1 to promote cancer cell invasion. Every organotypic measurement is shown individually, bar shows the mean pooled from 2 independent experiments. D) Western blots show levels of MYL9/MLC2, pS19MLC2/MYL9, MYH10, YAP, and TAZ in CAF#1 transfected with either control, YAP, TAZ or YAP&TAZ siRNA. E) Chart shows the effect of MYL9/MLC2 and MYL9/MLC2TASA (T18A, S19A – unphosphorylable mutant) over-expression in NF#1 on gel contraction. Bars represent mean ± s.e.m. of 3 experiments. F) Chart shows the effect of MYL9/MLC2 and MYL9/MLC2TASA over-expression in NF#1 on ability to promote cancer cell invasion. Every organotypic measurement is shown individually (pooled from 2 independent experiments). G) Graph shows the effect of 25μM blebbistatin on matrix remodelling by CAF#1. Bars represent mean ± s.e.m. of 3 independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 – unpaired t-test. Additional data for Fig. 5b, 5c, 5e, 5f, 5g in Table S2 (Statistics Source Data).
A positive feedback loop involving actomyosin contractility sustains YAP activation
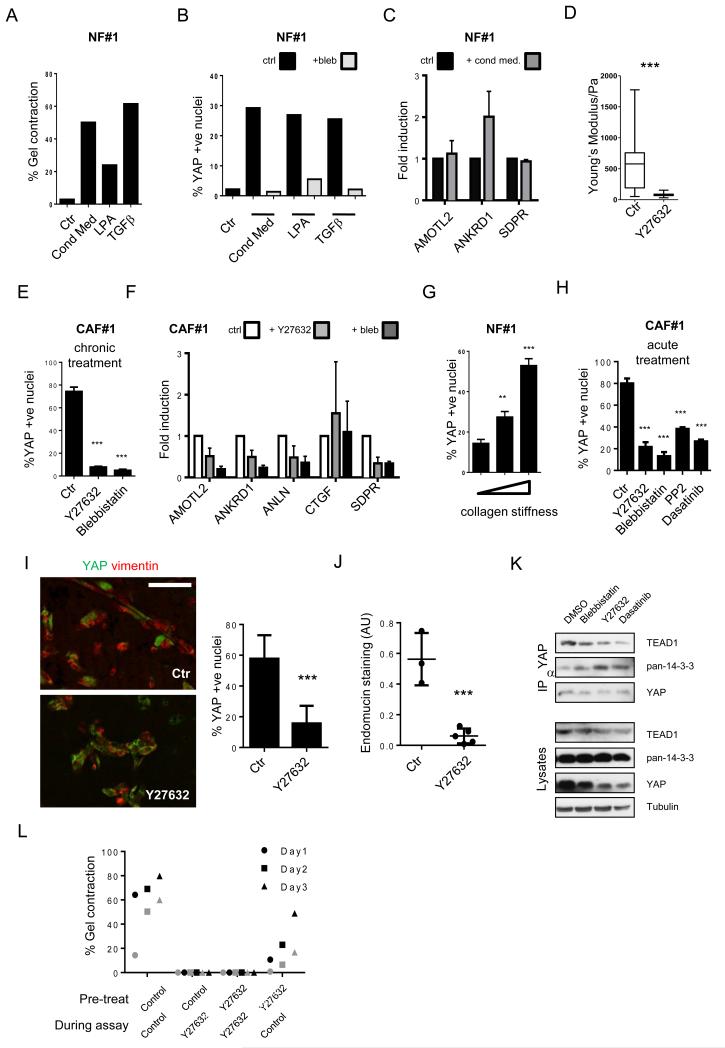
To understand what drives YAP activation in CAFs, we investigated how YAP activity was induced in normal fibroblasts. Conditioned media from cancer cells promotes matrix remodelling by normal fibroblasts (Figure 6a) and increases their ability to support cancer cell invasion (Supplementary Figure 5a-c). This change in behaviour was also associated with the nuclear translocation of YAP and the activation of some YAP/TAZ target genes (Figure 6b&c). The degree of YAP nuclear localisation triggered by conditioned media, and the range of target genes activated was less extensive than in CAFs (compare Figures 2 and 6b&c). We also noted that other pro-contractile stimuli, such as LPA and TGFβ, promoted nuclear accumulation of YAP (Figure 6a&b). YAP can respond to matrix stiffness12, therefore the ability of soluble factors to activate YAP could be the indirect result of their ability to promote matrix remodelling and concomitant stiffening. Such a mechanism of YAP activation would be reduced by blockade of matrix remodelling. We prevented matrix remodelling by chronically blocking actomyosin function (using blebbistatin). Actomyosin inhibition blocked the matrix remodelling and nuclear accumulation of YAP induced by conditioned media, TGFβ, and LPA (Figure 6a&b). These data show that YAP can be activated by various soluble factors; however the mechanism may be indirect because a functional contractile cytoskeleton is required in all cases. We next focused on the high steady state level of YAP signalling in CAFs. This elevated level of YAP signalling could be prevented if the stiffening of the ECM was blocked. Figure 6d shows that ROCK inhibition reduces the elastic modulus of matrices containing CAFs from >500Pa to ~100Pa. This is associated with reduced nuclear localisation of YAP and reduced expression of several, but not all, YAP target genes (Figure 6e&f). Similar results were obtained if actomyosin function was blocked using blebbistatin. Finally, we confirmed previous findings12 that increasing collagen matrix stiffness is sufficient to drive the nuclear accumulation of YAP in NFs (Figure 6g).
Figure 6. Actomyosin and Src-dependence of YAP activation in CAFs.
A) Effects of 4T1 conditioned media, LPA and TGFβ on matrix remodelling by NFs. One of two independent experiments is shown. B) Effects of 4T1 conditioned media, TGFβ, and LPA on YAP localisation in NFs. Grey bars indicate presence of blebbistatin. One of two independent experiments is shown. C) Effect of 4T1 conditioned media on gene expression in NFs normalised to Gapdh. Mean and ± S.E.M. n=3 experiments. D) Elastic modulus of matrices remodelled by CAF#1 transfected with control siRNA with or without Y27362 for 3 days. Central box represents the lower and upper quartiles, middle line represents the mean, and horizontal lines show the minimum to the maximum value (n=100 independent elastic modulus measurements). E) YAP localisation in CAF#1 with or without Y27362 and blebbistatin for 3 days. Mean ± S.E.M. n=3 experiments. F) Effects of Y27632 and blebbistatin on gene expression in CAF#1 normalized to Gapdh expression. Mean of n=4 experiments, ± s.e.m. G) YAP localisation in NFs cultured on 1.5mg/ml collagen I gel (~100Pa), 8mg/ml collagen (~300Pa) and collagen-coated glass (>10kPa). Mean ± S.D. n=8 experiments. H) YAP localisation in CAF#1 with or without Y27362, blebbistatin, PP2, and Dasatinib for 2 hours. Mean ± S.E.M. n=4 experiments. I) YAP and vimentin staining in CAF#1 matrix plugs grown sub-cutaneously in mice, where indicated mice received 50mg/kg Y27632. Scale bar, 100μm. Average number of YAP +ve nuclei shown for each mouse as an individual data point nuclei (4 control and 5 Y27632-treated mice in one cohort). Bars represent mean ± s.e.m. J) Quantification of endomucin. Each point is an independent plug (3 control and 5 Y27632-treated mice in one cohort). Bars represent mean ± s.e.m. K) Immune-precipitation of TEAD1, TEAD4 and 14-3-3 with YAP in control, blebbistatin-, Y27632-, and Dasatinib-treated CAF#1. L) Matrix remodelling by CAF#2 previously cultured with Y27632, or cultured with Y27632 during the assay. Data from two experiments, distinguished in black and grey. * P < 0.05, ** P < 0.01, *** P < 0.001 – unpaired t-test. Additional data for Fig. 6a,e,&l in Table S2 (Statistics Source Data).
The data above establish a link between the contractile cytoskeleton, matrix stiffness, and YAP activity. However these phenomena could be linked in a number of different, but not mutually exclusive, ways. Actomyosin function could simply be required to generate a stiff ECM, but not to respond to it. Similarly, the contractile actin network might be required for sensing of matrix rigidity. To explicitly test the latter hypothesis, we allowed CAFs three days to remodel the collagen-rich matrix and then acutely treated with inhibitors (Figure 7a). These experiments reveal that ROCK, myosin and Src function (blocked by PP2 and Dasatinib) are required to maintain the nuclear localisation of YAP in CAFs, even after the ECM has been remodelled. Consistent with this, treatment of mice with Y27632 reduced the nuclear localisation of YAP in CAFs, and prevented the CAFs from promoting angiogenesis (Figure 7b&c). Examination of cell morphology revealed that ROCK inhibition disrupted contractile stress fibres, whereas Src inhibition did not (Supplementary Figure 5d). Further, ROCK inhibition reduced Src activity (Supplementary Figure 5e). Thus, we propose that Src acts downstream44 from cytoskeletal tension to regulate YAP. Inhibition of ROCK and Src did not affect phosphorylation of MST1/2 and S127-YAP (Supplementary Figure 5e). However, blebbistatin, Y27632, and Dasatinib all reduced the association of YAP with TEAD1 and TEAD4, but increased its association with 14-3-3 proteins (Figure 7d). Together, these data suggest that matrix stiffness promotes actomyosin-dependent regulation of Src-family kinases leading to a switching of YAP away from binding 14-3-3 proteins in favour of association with TEAD1 and TEAD4. This change does not appear to be linked to changes in MST activity or S127-YAP phosphorylation (Figure 2c and Supplementary Figure 5e).
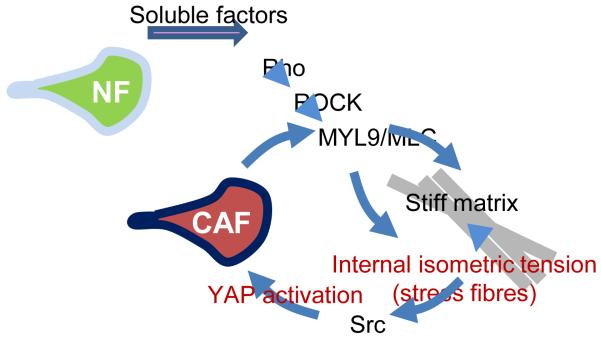
Figure 7.
Model outlining the role of YAP in the generation and maintenance of CAFs. A range of soluble factors can promote matrix remodelling by normal fibroblasts. This requires some level of ROCK and actomyosin function. As the matrix becomes stiffer, isometric tension within the cell increases leading to Src activation and the association of YAP with TEAD transcription factors. Active YAP then promotes the expression of ANLN and DIAPH3 and stabilises actomyosin proteins. This leads to further matrix stiffening, thereby generating a positive feedback loop.
YAP is required for elevated levels of MYL9/MLC2 and MYH10 and matrix stiffening by CAFs (Figure 5). Conversely, stiff matrices and the contractile actin cytoskeleton work together to activate YAP (Figure 6). Together these elements generate a positive feedback loop. Depending on the level of damping, this loop of matrix stiffening and YAP activation might be able to be ‘broken’ by inhibition of the loop for a period of time. We explored this hypothesis by culturing CAFs in the presence of Y27632 for a prolonged period and then assayed their phenotype after removal of the inhibitor. Figure 7e shows that Y27632 effectively reduced matrix remodelling when included during the assay. More notably, cells that had previously been cultured in Y27632, but were assayed in normal media, showed significantly defective matrix remodelling abilities 3-5 days after removal of the drug (Figure 7e). This demonstrates that if the feed-forward loop between the matrix and YAP can be blocked, then a stable reversion in cell behaviour arises.
Discussion
Here we demonstrate a new role for the YAP oncoprotein in the tumour stroma. YAP function is critical for the establishment and maintenance of CAFs. We propose that soluble factors secreted by cancer cells, initially promote an intermediate level of YAP and TAZ activity45, 46. YAP activation occurs as a consequence of two inter-related events: first, the matrix remodelling and stiffening induced by factors such as LPA and TGFβ, and, second, changes in the contractile actin cytoskeleton within fibroblasts. Stiff matrices require actomyosin function for their generation; however they also enable actomyosin to generate isometric tension within cells (Figure 7). This leads to stress fibre formation, and the activation of Src-family kinases at focal adhesions47. Src-family kinase function is required for YAP activity. YAP function is required for the expression of MYL9, matrix stiffening, and many pro-tumorigenic properties of fibroblasts. The matrix stiffening resulting from YAP activation generates a positive feedback loop, leading to the more robust and sustained activation of the YAP pathway observed in CAFs. Once this feedback is established it can become self-sustaining, and this could explain the stability of the CAF phenotype in the absence of co-culture with cancer cells. In vivo, it is likely that this mechanism co-operates with epigenetic changes induced by soluble factors in the tumour micro-environment. It is also possible that YAP activation could be initiated by soluble factors in the absence of cancer cells, these may act by triggering matrix stiffening or through more direct G-protein coupled receptor signalling to YAP45. It will be interesting to determine if YAP plays a role in matrix stiffening in pre-cancerous lesions or, more generally, in fibrosis. YAP-dependent matrix stiffening driven by CAFs may also lead to pro-tumorigenic YAP activation in cancer cells in close proximity to the stiff matrix. Consistent with this, we and others have noted high levels of nuclear YAP in cancer cells adjacent to the stroma13. Similarly, matrix stiffening may lead to the propagation of YAP activity to nearby normal fibroblasts. To conclude, we identify a key role for YAP in the tumour stroma. It is required for many of the pro-tumorigenic functions of CAFs, including matrix stiffening, invasion, and angiogenesis. High levels of YAP function in the stroma are sustained through positive feedback between CAF-driven matrix stiffening and mechano-transduction leading to YAP activation.
EXPERIMENTAL PROCEDURES
Mouse strains
Transgenic FVB/n mice expressing the Polyoma Middle T antigen oncogene under the Mouse Mammary Tumour Virus promoter (MMTV-PyMT)17 were kept in accordance with UK regulations under project licence PPL/80/2368. Wild type FVB/n mice were used as recipients for the fibroblast/matrix plug injections. Established syngeneic FVB/n MMTV-PyMT breast cancer cell line TS222 was a kind gift from J. Joyce (Memorial Sloan-Kettering Cancer Center, New York, USA).
Isolation and immortalization of fibroblasts from different tumoural stages
Normal mammary glands, hyperplastic mammary tissue (generally from 6 week old females), mammary adenoma/early carcinoma tissue (from 8-10 week old females) and mammary carcinoma/late carcinoma/invasive carcinoma tissue (older than 12 weeks) were dissected out and part taken for histological analysis and part for fibroblast isolation. Two approaches were followed for fibroblast isolation. In order to isolate fibroblasts from normal tissue, which is very soft and fatty, we placed the tissue into dishes where it was compressed under a 20mm coverslip to prevent fatty tissue floating. We then covered it with DMEM (Invitrogen), 10% FCS (PAA Labs), 1% ITS (insulin–transferrin–selenium; #41400-045; Invitrogen) supplement and changed the media daily. After 7-10 days, fibroblasts started to grow out of the tissue into the coverslip and the dish. Then, the tissue was removed and the fibroblast population expanded. For hyperplastic, adenomal and cancerous tissue, the sample was cut into small pieces and collagenase/dispase digested. After filtering the undigested tissue, the solution was serial centrifuged and the final pellet re-suspended in DMEM (Invitrogen) 10% FBS, 1% ITS and seeded on a culture dish. After 30 min the fibroblasts had already adhered to the dish while other cellular types remained in suspension. In both cases, fibroblasts were subsequently grown and immortalised with HPV-E6 retrovirus20. They were then selected using 2.5 μg mL-1 puromycin.
Clinical samples and human CAFs
The tumour specimens and tissue samples used in this study were obtained with informed consent from all subjects. Human SCC CAFs were isolated following similar approaches to murine CAFs and were grown and immortalised with htert lentivirus. They were selected using 400 μg mL-1 hygromycin.
Histochemical analysis
Frozen tumours were embedded in OCT and sections 6-8μm were cut. These sections were then fixed in 4% paraformaldehyde and stained using the same protocols for immunofluorescence of cultured cells (see below). Tissue fixed in 10% formal saline was paraffin embedded and sectioned using standard methods. In addition to haematoxylin and eosin (H&E) staining and Masson’s Trichrome staining, the antibodies used are listed in Supplementary Table 3. Breast cancer tissue arrays were from Biomax (References: BR802 and BR243f).
Immunofluorescence
Unless stated otherwise, all immunofluorescence experiments were performed on cells seeded on a top of a thin layer of a mixture of collagen I (#354249; BD Biosciences, Oxford, UK) and Matrigel (#354234; BD Biosciences) yielding a final collagen concentration of approximately 4.6 mg mL−1 and a final Matrigel concentration of approximately 2.2 mg mL−1 (Collagen I:Matrigel matrix hereafter). After 2 days on the gel and the pertinent treatments, cells were fixed in 4% paraformaldehyde for 1 h. Cells were permeabilised by incubation in PBS 0.5% NP-40 (Sigma) at 4°C for 20 min (twice), in PBS 0.3% Triton 100 (Sigma) at room temperature (RT) for 20 min and in PBS 0.1% Triton 100 at RT for 15 min (twice). Samples were blocked for 60 min at RT (twice) in blocking solution: 4% BSA PBS 0.05% Tween20 (Sigma). Then, cells were incubated with primary antibody in blocking solution in a wet chamber overnight at 4 °C. After 3 washes of 15 min in PBS, secondary antibody in blocking solution was added. After 3 washes of 15 min in PBS, samples were mounted and analysed using an inverted Zeiss LSM780. Antibody description and working dilutions can be found on Supplementary Table 3. To quantify YAP or TAZ nuclear localization, a measure of their activation, two approaches were employed. In the first, we calculated the percentage of cells with a clear predominant nuclear staining among the total cell number (cells with nuclear staining, cells with cytosolic staining and cells with homogeneous nuclear/cytosolic staining), counting at least 300 cells on triplicate. For the second, photographs were processed using Volocity. Briefly, we calculated for each cell the nuclear YAP/TAZ intensity (determined by the intensity within the region delimited by DAPI staining) and the cytosolic YAP/TAZ intensity (determined by the intensity without the region delimited by DAPI staining and within the region delimited by F-actin staining) for at least 25 cells in triplicate. Plots represent the ratio of nuclear versus cytosolic YAP or TAZ staining. Quantitative analysis of images of CAF markers was performed using MATLAB software. Briefly, fibroblasts were identified using automated threshold based on F-actin staining. The mean intensities of αSMA, S100A4, and FAP were then measured.
Western Blotting
Unless stated otherwise, all protein lysates were obtained from cells seeded on top of a thin layer of a mixture of collagen I:Matrigel matrices. Generally, 100 μL of the gel were spread evenly on a 6 well-plate dish. Apart from that, protein lysates were processed following standard procedures. Antibody description and working dilutions used can be found on Supplementary Table 3.
Immunoprecipitation
Endogenous YAP immunoprecipitations were performed as described48. Briefly, cells were lysed in 50 mM Tris (pH 8), 100 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM MgCl2, 2 mM PMSF, and protease and phosphatase inhibitors. The supernatants were pre-cleared by incubating 30 minutes at 4°C with protein-G beads. Supernatants were incubated with anti-YAP antibodies and protein-G beads overnight. Immunoprecipitates were washed three times with NET-buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.25% gelatine, and 0.1% Nonidet P-40) and analysed by immunoblotting. Exposures within the dynamic range were quantified by densitometry using the program NIH Image 1.60.
ECM-remodelling assay
To assess force-mediated matrix remodelling, 75 × 103 fibroblasts were embedded in 100 μL of Collagen I:Matrigel and seeded on a 35 mm glass bottom MatTek dish (P35-1.5-14-C, MatTek Co., Ashland, MA, USA). Once the gel was set, cells were maintained in DMEM + 10% FCS (unless otherwise stated) + 1% ITS. Gel contraction was monitored daily by taking photographs of the gels. Unless stated otherwise, the gel contraction value refers to the contraction observed after 3 days. To obtain the gel contraction value, the relative diameter of the well and the gel were measured using ImageJ software, and the percentage of contraction was calculated using the formula 100 × (well diameter − gel diameter) / well diameter.
Atomic force microscopy
To assess the elastic modulus of gels remodelled by fibroblasts, 75 × 103 fibroblasts were embedded in 100 μL of Collagen I:Matrigel and seeded on ultra-low attachment 96 well plate well (#3474, Costar). Once the gel was set, cells were maintained DMEM + 10% FCS + 1% ITS. After 3 days, the elastic modulus of the gels was measured. On the day of the experiment, gels were gently lifted from their well and fixed in the centre of 50 mm glass bottom Petri dishes using cyanoacrylate superglue. Once glued, Leibovitz L-15 medium (Invitrogen) supplemented with 10% FCS was added to the dish.
AFM measurements were performed with a JPK Nanowizard-I (JPK instruments, Berlin, Germany) interfaced to an inverted optical microscope (IX-81, Olympus). AFM cantilevers with pyramidal tips (MLCT, Bruker, Karlsruhe, Germany) and nominal spring constants of 0.07 Nm−1 were modified by gluing 35μm radius glass beads to the cantilever underside with UV curing glue (UV curing, Loctite, UK). Cantilever spring constants were determined prior to modification using the thermal noise method implemented in the AFM software (JPK SPM).
Prior to any indentation tests, the sensitivity of the cantilever was set by measuring the slope of force-distance curves acquired on glass regions of the Petri dish. Using the optical microscope, the tip of the cantilever was aligned over regions in the middle of the gel and, for each gel, measurements were acquired in 30-40 locations ~100μm apart. Force-distance curves were acquired with an approach speed of 5μm.s−1 until reaching the maximum set force of 3nN. After the experiment, the elastic moduli were extracted from the force-distance curves by fitting the contact portion of curves to a Hertz contact model49. For each force-distance curve, goodness of fit was evaluated by calculating r^2 values and only fits with r^2>0.80 were retained for further analyses (representing on average 80% of the acquired force-curves).
Invasion assays
Human CAFs organotypic culture system was set up as previously described6. Mouse mammary CAFs organotypic culture system was set up with some adaptations. Briefly, 106 fibroblasts, unless stated otherwise, were embedded in a mixture of collagen I:Matrigel matrix. After the gel was set at 37°C for 1 h, DMEM + 10% FCS + 1% ITS was added on the top. Sixteen hours later, 5 × 105 4T1/410.4/TS2 cells were seeded on top in DMEM + 10% FCS + 1% ITS. After 8 h, a thin layer of gel was added covering the breast cancer cells. The gel was then mounted on a metal bridge and fed from underneath with DMEM + 10% FCS + 1% ITS (changed daily). After 6 days, the cultures were fixed using 4% paraformaldehyde plus 0.25% glutaraldehyde in PBS and processed by standard methods for haematoxylin and eosin (H&E) staining. For assays involving the removal of fibroblasts (fibroblast conditioned ECMs), the fibroblasts were left to remodel the gel for 5 days, after which the gels were incubated in DMEM + 10% FCS + 1% ITS plus hygromycin (400μg mL−1) for 48 h to kill the fibroblasts and then washed three times with DMEM + 10% FCS + 1% ITS (>30 min per wash). 5 × 105 410.4/4T1 cells were then plated on top and the assays proceeded as usual. The invasion index was calculated by measuring the total area over which breast cancer cells had dispersed (including invading and non-invading cells) and the area of non-invading cells. The value shown is the average of the measurement (1 − [non-invading area/total area]) of at least ten measurements from two or more independent experiments and the error bars represent the s.e.m. MATLAB script was used to automatically identify the total cell area and the non-invading area. The M files are available on request, email robert.jenkins@cancer.org.uk
Matrigel Plugs
2 × 106 fibroblasts were resuspended in 100 μL of PBS and mixed in 300 μL of Matrigel. Then, 300 μL of the mixture were injected sub-cutaneously on the mammary fat pad of a 12 week-old FVB/n female. After 7 days, the plugs were surgically removed and taken for histological and immunofluorescence analyses. Blood vessel density was calculated by normalising the area positive for endomucin staining to the area positive for vimentin staining of all the slices processed. Each value in the plot corresponds to the blood vessel density of an individual plug.
RNA isolation and arrays
To obtain RNA from the different fibroblast populations, cells were seeded on top of the Collagen I:Matrigel matrix and fed with complete media. After 5 days, RNA was isolated using RNeasy Kit (#74104, Qiagen). RNA was then processed in collaboration with the Bart’s and London Centre for Genome Research (Illumina bead array platform – details available on request).
Gene-set enrichment analyses (GSEA)
Array data was processed and analysed using the Gene-set enrichment analysis software, developed by the Broad Institute of MIT and Harvard (USA) and available at www.broadinstitute.org, following the program guidelines. The specific settings applied in all analyses are: Number of Permutations (1000), Permutation Type (Gene set), Enrichment statistic (Weighted), Metric for ranking genes (t Test). The rest of the fields were left as defaulted. Values in the tables represent the nominal P value of each gene set. The list of the specific gene sets analysed and their sources are available in Supplementary Table 4. Additional references to the gene sets can be found in the Methods References section50-57. Interleukin and RANKL gene sets were obtained from www.netpath.org
qRT-PCR
Sequences of the oligonucleotides used are described in the Supplementary Table 5.
Statistical analyses
Statistical analyses were performed using Prism software (GraphPad Software Inc., La Joya, USA). Mean values and standard errors (s.e.m.) are shown in most graphs that were generated from several repeats of biological experiments. P values were obtained from t tests with paired or unpaired samples, with significance set at P < 0.05. Graphs show either the actual p value or symbols describing it (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Source data for the indicated figures can be found in Supplementary Table 2.
Transfections, inhibitors, siRNA and cDNAs
Fibroblasts were cultured in standard conditions and transfected using DharmaFECT 1 (#T-2001-03; Dharmacon, Perbio Science, Northumberland, UK). Briefly, cells were plated at 60% confluence and subjected to transfection the following day using 100 nM final concentration of siRNA. siRNA was purchased from Dharmacon and sequences are listed in the Supplementary Table 6. The following growth factors and drugs were used: TGFβ – 2ng/mL (PeproTech, Rocky Hill, NJ, USA), LPA - 2μg/mL (L7260, Sigma), Y27632 – 10μM (#1254; Tocris Bioscience, Bristol, UK), Blebbistatin – 25μm (#203391; Calbiochem/Merck), Dasatinib – 300nM (LCLabs), PP2 – 10μM (Calbiochem/Merck). Conditioned media from 4T1 cells was obtained by conditioning plain DMEM in a 75% confluent culture for 24-48h. pEGFP-MLC2 wild type and pEGFP-MLC2 T18A, S19A (TASA) mutant were a gift from Michael Olson (Beatson Institute, Glasgow, UK). pCDNA-FLAG-YAP S5A plasmid was a kind gift from Barry Thompson (London Research Institute, London, UK).
Supplementary Material
Acknowledgements
F.C., N.E., S.H., R.P.J., S.I.C., K.H., and E.S. are funded by Cancer Research UK. A.G-G. was funded by a Royal Society Newton Fellowship, E.M. is in receipt of a Dorothy Hodgkins Postgraduate Award (DHPA) from the Engineering and Physical Sciences Research Council. G.C. is in receipt of a Royal Society University Research Fellowship. We thank Nic Tapon, Barry Thompson, and lab members for help and advice throughout this work.
Abbreviations
- AdAF
Adenoma-Associated Fibroblast
- AFM
Atomic force microscopy
- αSMA
α Smooth Muscle Actin
- CAF
Carcinoma-Associated Fibroblast
- CerCAF
Cervical SCC CAF
- ECM
Extracellular matrix
- FAP
Fibroblast Activation Protein
- GSEA
Gene set enrichment analysis
- HNCAF
Head and Neck SCC CAF
- HpAF
Hyperplasia-Associated Fibroblast
- HPV
Human Papilloma Virus
- LPA
L-a-Lysophosphatidic acid
- MLC2
Myosin Light Chain
- NF
Normal Fibroblast
- LPA
Lysophosphatidic Acid
- SCC
Squamous Cell Carcinoma
- TGFβ
Transforming Growth Factor β
- VCAF
Vulval SCC CAF
Footnotes
Accession numbers. Primary accessions: Data files are available at the NCBI Gene Expression Omnibus (GEO) under GSE45256.
References
- 1.Bhowmick NA, Moses HL. Tumor-stroma interactions. Current opinion in genetics & development. 2005;15:97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:39–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvo F, Sahai E. Cell communication networks in cancer invasion. Current opinion in cell iology. 2011;23:621–629. doi: 10.1016/j.ceb.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Finak G, et al. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast Cancer Res. 2006;8:R58. doi: 10.1186/bcr1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nature cell biology. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 7.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erler JT, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Guilluy C, et al. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nature cell biology. 2011;13:722–727. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawada Y, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 13.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 14.Lamar JM, et al. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2441–2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 16.Sugimoto H, Mundel TM, Kieran MW, Kalluri R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer biology & therapy. 2006;5:1640–1646. doi: 10.4161/cbt.5.12.3354. [DOI] [PubMed] [Google Scholar]
- 17.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Molecular and cellular biology. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trimboli AJ, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JW, et al. Loss of fibroblast HIF-1alpha accelerates tumorigenesis. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vousden KH. HPV E6: ensuring all’s well at the end. Trends Microbiol. 1996;4:337–338. doi: 10.1016/0966-842x(96)30024-3. [DOI] [PubMed] [Google Scholar]
- 21.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. The American journal of pathology. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shree T, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes & development. 2011;25:2465–2479. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 24.Roepman P, et al. Dissection of a metastatic gene expression signature into distinct components. Genome biology. 2006;7:R117. doi: 10.1186/gb-2006-7-12-r117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 26.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhry SI, et al. Autocrine IL-1beta-TRAF6 signalling promotes squamous cell carcinoma invasion through paracrine TNFalpha signalling to carcinoma-associated fibroblasts. Oncogene. 2012 doi: 10.1038/onc.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojima Y, et al. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 31.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes & development. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Descot A, et al. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Molecular cell. 2009;35:291–304. doi: 10.1016/j.molcel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol Biol. 2004;5:13. doi: 10.1186/1471-2199-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGee KM, Vartiainen MK, Khaw PT, Treisman R, Bailly M. Nuclear transport of the serum response factor coactivator MRTF-A is downregulated at tensional homeostasis. EMBO Rep. 2011;12:963–970. doi: 10.1038/embor.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nature cell biology. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Molecular cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez BG, et al. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 2011;138:2337–2346. doi: 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- 40.Densham RM, et al. MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Molecular and cellular biology. 2009;29:6380–6390. doi: 10.1128/MCB.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sansores-Garcia L, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 43.Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Molecular cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 44.Azab AK, et al. RhoA and Rac1 GTPases play major and differential roles in stromal cell-derived factor-1-induced cell adhesion and chemotaxis in multiple myeloma. Blood. 2009;114:619–629. doi: 10.1182/blood-2009-01-199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamm C, Bower N, Anneren C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. Journal of cell science. 2011;124:1136–1144. doi: 10.1242/jcs.075796. [DOI] [PubMed] [Google Scholar]
- 47.Carragher NO, Frame MC. Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends in cell biology. 2004;14:241–249. doi: 10.1016/j.tcb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 48.Matallanas D, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Molecular cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris AR, Charras GT. Experimental validation of atomic force microscopy-based cell elasticity measurements. Nanotechnology. 2011;22:345102. doi: 10.1088/0957-4484/22/34/345102. [DOI] [PubMed] [Google Scholar]
- 50.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 51.Buess M, et al. Characterization of heterotypic interaction effects in vitro to deconvolute global gene expression profiles in cancer. Genome biology. 2007;8:R191. doi: 10.1186/gb-2007-8-9-r191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajski M, et al. IGF-I induced genes in stromal fibroblasts predict the clinical outcome of breast and lung cancer patients. BMC medicine. 2010;8:1. doi: 10.1186/1741-7015-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazzone M, et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5012–5017. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klapholz-Brown Z, Walmsley GG, Nusse YM, Nusse R, Brown PO. Transcriptional program induced by Wnt protein in human fibroblasts suggests mechanisms for cell cooperativity in defining tissue microenvironments. PloS one. 2007;2:e945. doi: 10.1371/journal.pone.0000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaposi-Novak P, et al. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. The Journal of clinical investigation. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park BK, et al. NF-kappaB in breast cancer cells promotes osteolytic bone metastasis by inducing osteoclastogenesis via GM-CSF. Nat Med. 2007;13:62–69. doi: 10.1038/nm1519. [DOI] [PubMed] [Google Scholar]
- 57.Farmer P, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.