Abstract
More than half of the human genome is made of Transposable Elements. Their ongoing mobilization is a driving force in genetic diversity; however, little is known about how the host regulates their activity. Here, we show that the Microprocessor (Drosha-DGCR8), which is required for microRNA biogenesis, also recognizes and binds RNAs derived from human LINE-1 (Long INterspersed Element 1), Alu and SVA retrotransposons. Expression analyses demonstrate that cells lacking a functional Microprocessor accumulate LINE-1 mRNA and encoded proteins. Furthermore, we show that structured regions of the LINE-1 mRNA can be cleaved in vitro by Drosha. Additionally, we used a cell culture-based assay to show that the Microprocessor negatively regulates LINE-1 and Alu retrotransposition in vivo. Altogether, these data reveal a new role for the Microprocessor as a post-transcriptional repressor of mammalian retrotransposons acting as a defender of human genome integrity.
Transposable elements have been a major force in shaping mammalian genomes. At least four distinct classes of mobile DNA elements are dispersed through the human genome, comprising half of its genomic mass1. Most transposable elements are defective fossil copies accumulated through human evolution; although a small fraction of non-LTR (long terminal repeat) retrotransposons are currently active in the germline and somatic human genomes. Long INterspersed element class 1 (LINE-1 or L1) is a family of non-LTR retroelements in mammals that represent a fifth of the human genome1. Although more than 99% of LINE-1 copies are inactive, an average human genome contains 80-100 retrotransposition competent L1s (RC-L1s) capable of mobilization1,2. In addition, RC-L1 encoded proteins are responsible for the mobilization of non-autonomous non-LTR retrotransposons (Short INterspersed Elements -SINEs- including Alu and SVA)3–5, cellular mRNAs6,7 and selected non-coding cellular RNAs8, which together comprise more than a tenth of the human genome. Thus, the activity of RC-L1s, Alus and SVAs continue to impact the human genome and new mobilization events are a constant source of human DNA variation1,9–11 as well as contribute to human disease1.
All characterized active or RC-L1s belong to the youngest subfamily of L1 elements denoted L1Hs or L1-PA112. RC-L1s are 6-kb long elements that contain a 5′untranslated region (UTR) with an internal promoter, followed by two non-overlapping Open Reading Frames (ORFs) and end in a short 3′UTR containing a poly (A) tail1. LINE-1 encoded proteins bind in cis to the same mRNA from which they were translated generating a RiboNucleoprotein Particle (RNP) that is a proposed retrotransposition intermediate13. Both encoded proteins are required for LINE-1 retrotransposition14, although little is known about how cellular host factors affect and regulate LINE-1 retrotransposition. By contrast, Alu retrotransposons are 0.3-kb long elements derived from the 7SL RNA and are transcribed by RNA polymerase III15,16. Primate specific Alu elements contain two monomers separated by an A-rich tract and end in a poly (A) tail required for their trans-retrotransposition by LINE-1 encoded proteins6. An average human genome contains more than a million Alu elements and there are more than 6,000 active Alu core sequences per average genome (which belong to the Y and S subfamilies)16.
Due to the potential negative impact of newly inserted transposable elements, it is highly likely that restriction mechanisms that operate at different levels in the gene expression cascade may have evolved to control a high rate of transposition-mediated mutagenesis. Indeed, several restriction cellular factors have been described that affect mobilization in mammals of transposable elements1. Among the factors known to modulate retrotransposition, several RNA-mediated processes have been previously identified17,18. Notably, piRNAs have been shown to control the expression of LINE-1s specifically in the germline of mammals and insects1,18.
MicroRNAs (miRNAs) are small regulatory RNAs (21-24 nucleotides) that act as key regulators of many biological processes19. They are generated from longer RNA transcripts known as primary miRNAs (pri-miRNAs), which are characterized for containing hairpin RNA secondary structures. A complex known as the Microprocessor, comprising the RNase III type enzyme Drosha and its partner DGCR8, catalyzes the nuclear step of miRNA biogenesis20–22. DGCR8 recognizes the RNA substrate in the nucleus through two double-stranded RNA binding motifs, whereas Drosha functions as the endonuclease generating stem loop precursors, termed pre-miRNAs, which are further processed by Dicer in the cytoplasm to produce mature miRNAs23.
In this study we set out to characterize a role for the Microprocessor in controlling the activity of mammalian retroelements. Our results demonstrate that the Microprocessor recognizes and processes structured regions within RC-L1 RNAs, leading to a decrease in RC-L1 mRNA levels. In addition, we demonstrate that the Microprocessor directly controls the retrotransposition rate of engineered mammalian LINE-1 and Alu elements in vivo. Thus, we have unraveled a new post-transcriptional mechanism that limits the mobilization of endogenous retrotransposons in mammals.
RESULTS
The Microprocessor binds RNAs from Transposable Elements
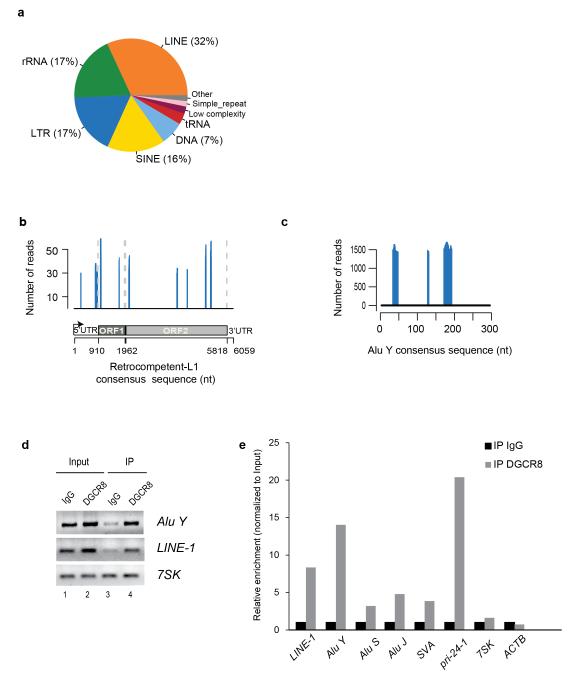
To characterize potential novel RNA targets of the Microprocessor, we recently performed high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP) of endogenous DGCR8 protein in human HEK293T cells. We found that DGCR8 binds not only to pri-miRNAs but also to many cellular RNAs. Additionally, we observed that 25% of the binding sites mapped to messenger RNAs (mRNAs) whereas 4% of binding sites corresponded to long non-coding RNAs (lncRNAs), suggesting an expanded role for DGCR8 in gene expression regulation24 (Supplementary Fig. 1a). Of particular interest, we noted that a large fraction of the RNA targets (31% of the total) corresponded to human repetitive sequences. Notably, the largest group within this class corresponded to transposable elements (73%), with most reads mapping to LINE-1 elements (32%), short interspersed elements (SINEs) (16%), LTR-containing retrotransposons (17%) and DNA-Transposons (7%) (Fig.1a, and Supplementary Table 1). To further characterize this interaction, we only included DGCR8 CLIP reads mapping to annotated transposable elements that display high similarity to consensus sequences derived from active LINE-1 and SINEs (Alu and SVA) elements (identity >= 99% and coverage>= 90%). Notably, upon annotation of reads we found several binding sites on the sense and antisense strands of an RC-L1 mRNA consensus sequence (Fig. 1b and Supplementary Fig. 1b). Similarly, we also found a substantial number of reads mapping to sense and antisense sequences from the most active family of Alu elements (Alu Y) and SVA elements (Fig. 1c, Supplementary Fig. 1c,d and Supplementary Table 2, 3). Previous studies have demonstrated that cultured cells are characterized for expressing a wide constellation of LINE-1 and Alu RNAs25,26. Thus, we examined the repertoire of LINE-1 mRNAs expressed in HEK293T cells by Reverse Transcription (RT)-PCR and sequencing. Despite not being a quantitative assay, this analysis revealed that HEK293T cells express L1Hs/L1PA-1 mRNAs as well as mRNAs derived from evolutionary older LINE-1 subfamilies (L1PA2-L1PA612, Supplementary Fig. 1e). Of interest, we also found a small but significant fraction of reads mapping to L1PA4 elements (Supplementary Fig.1f). Reducing the minimum identity to consensus sequences (>= 90%) also revealed the presence of significant clusters in L1PA2 and L1PA3 elements (Supplementary Fig. 1g,h, respectively, and Supplementary Table 3). Altogether, these data demonstrate binding of DGRC8 to a constellation of LINE-1, Alu and SVA derived RNAs. It is noticeable that we found DGCR8 binding sites to sense and antisense RNA sequences derived from transposable elements. Binding to antisense sequences might be caused by the presence of Alu and LINE-1 derived sequences within 3′UTRs of human genes27, by the presence of a conserved antisense promoter within the LINE-1 5′UTR26,28and it is also likely that permissive transcription29 might lead to the generation of sense and antisense RNA sequences derived from transposable elements.
Figure 1.
DGCR8 binds a constellation of transcripts from repetitive elements. (a) Pie chart showing distribution of reads mapping to repetitive elements in sense and antisense orientation. 73% of the significant clusters map to transposable elements including DNA-Transposons, LINE-1s, LTR-containing retrotransposons and SINEs. (b) Distribution of DGCR8 binding sites in a human RC-L1Hs consensus sequence (FDR <0.01). Only sense peaks with a minimum of 29 reads are represented. Schematic representation of the RC-L1 element (bottom). UTR, untranslated region; ORF, open reading frame. (c) Distribution of significant DGCR8 binding sites in a human Alu Y consensus sequence in sense orientation (FDR <0.01). (d) Amplification of Alu Y, LINE-1 and 7SK mRNAs by RT-PCR upon immunoprecipitation of endogenous DGCR8 protein. (e) Real-time Reverse Transcriptase (RT)-PCR analysis of several transposable elements (LINE-1, Alu Y, Alu S Alu J and SVA) upon immunoprecipitation of endogenous DGCR8 of one representative experiment. Grey bars represent the relative enrichment over control IgG immunoprecipitation (black bars) and data is expressed as a percentage of input for normalization purposes. In this assay, a well-known target of the Microprocessor (pri-miR-24-1) was used as an internal positive control, whereas 7SK and ACTB were used as negative controls. IP,immunoprecipitation.
We next analyzed whether DGCR8 directly binds sense RNAs derived from transposable elements by immunoprecipitation of endogenous DGCR8 protein followed by analysis of the associated RNAs by semi-quantitative and quantitative RT-PCR. Notably, we confirmed binding of endogenous DGCR8 to sense RNA sequences derived from L1Hs/L1PA1, Alu Y and pri-miR-24-1 transcripts (Fig. 1d,e). We also observed minor binding to Alu S, Alu J and SVA derived RNAs (Fig. 1e). Additional controls revealed lack of DGCR8 binding to 7SK and β-actin derived RNAs. Altogether, these data revealed that DGCR8, the RNA-binding component of the Microprocessor, binds LINE-1, Alu and SVA derived RNAs. This data suggests that the Microprocessor may have a role in controlling the abundance of transposable elements mRNAs.
The Microprocessor regulates the abundance of L1 mRNAs
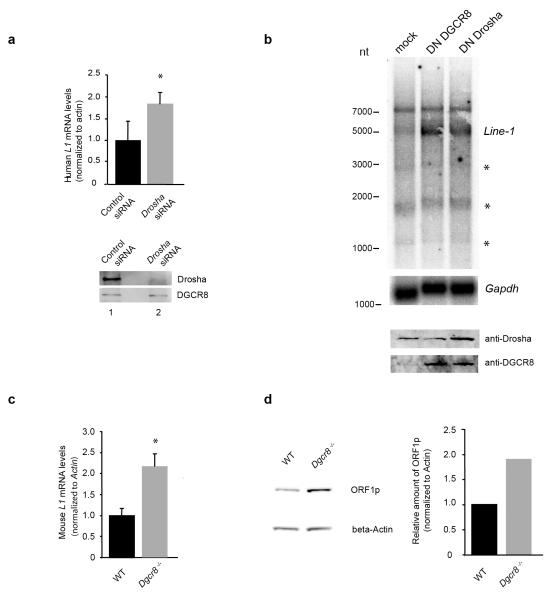
In order to study the physiological effect of this interaction, we determined LINE-1 mRNA steady-state levels upon a reduction of Microprocessor components in human teratocarcinoma PA-1 cells, which are known to naturally overexpress L1 mRNAs and L1-encoded ORF1p30. Notably, we observed a two-fold increase in the levels of human RC-L1 mRNA upon siRNA-mediated depletion of Drosha (Fig. 2a). This was further confirmed by overexpressing dominant negative (DN) forms of DGCR8 or Drosha proteins in PA-1 cells22,31, which resulted in a strong accumulation of sense full-length L1 mRNA (Fig. 2b and Supplementary Fig. 2a), while inducing accumulation of pri-miRNAs, as expected (Supplementary Fig. 2b). The detected smaller L1 RNA transcripts (Fig. 2b) likely arise from cryptic splicing or by the use of premature polyadenylation signals, as previously described32–34(marked with asterisks on Fig. 2b). In agreement with previous studies, we also observed that expression of DN-Drosha induces an increase in DGCR8 protein levels (Fig. 2b), as the DGCR8 mRNA itself is a substrate of the Microprocessor35. Previous studies have demonstrated that DNA-methylation of the mammalian L1 promoter (which contains a canonical CpG island) negatively correlates with L1 expression levels34,36,37. Thus, we analyzed L1 promoter methylation upon transient Drosha depletion in PA-1 cells using bisulfite DNA conversion, but found no significant changes at a genome wide level (Supplementary Fig. 3a,b) or at specific RC-L1s loci (Supplementary Fig. 3c)2. Thus, the observed changes in L1 mRNA levels upon Microprocessor inactivation are likely not to be caused by alterations in the methylation level of the LINE-1 promoter. We next examined LINE-1 expression levels in mouse DGCR8-deficient ES cells (Dgcr8−/−) and parental ES cells38. Notably, Dgcr8−/− ES cells displayed a two-fold increase in the amount of L1 mRNA for all three known active LINE-1 subfamilies ((TF, AF and GF elements)39–41 (Fig. 2c and S.R.H. and S.M., unpublished data). Consistently, we also observed elevated levels of L1-ORF1p from TF elements in Dgcr8/− (Fig. 2d), which correlated with elevated levels of TF LINE-1 mRNA levels (Fig. 2c). These observations have led us to hypothesize that DGCR8 binds to the L1 mRNA and that the activity of Microprocessor negatively regulates L1 RNA abundance at a post-transcriptional level.
Figure 2.
The Microprocessor regulates the abundance of L1 mRNA and L1-encoded ORF1p protein. (a) Real time RT-PCR analysis of endogenous L1 mRNA in PA-1 cells upon depletion of Drosha. Western blot analysis confirms Drosha depletion (bottom panel). (b) Northern blot analysis of endogenous polyadenylated L1 mRNA in PA-1 cells upon overexpression of Dominant negative (DN) forms of both DGCR8 (lane 2) and Drosha (lane 3 (asterisks depict smaller fragments arising from cryptic splice sites or truncated versions of L1 mRNAs). Gapdh hybridization was used as a loading control. Western blot analysis shows the level of expression of the DN forms (bottom panel). (c) Real time RT-PCR analysis of mouse TF L1 mRNA in Dgcr8−/− mouse ES cells and parental ES cells (WT). (d) Western blot analysis of L1-ORF1p in Dgcr8−/− mouse ES cells and parental ES cells (WT). Western-blot was quantified using ImageQuant TL software (GE) plotted data are the average of 3 technical replicates. (a) and (c) values are averages of three independent biological replicates; *P< 0.05 (t test). Error bars, s.d. Uncropped versions of the blots are shown in Supplementary Fig. 8.
The Microprocessor cleaves the 5′UTR of LINE-1 mRNAs
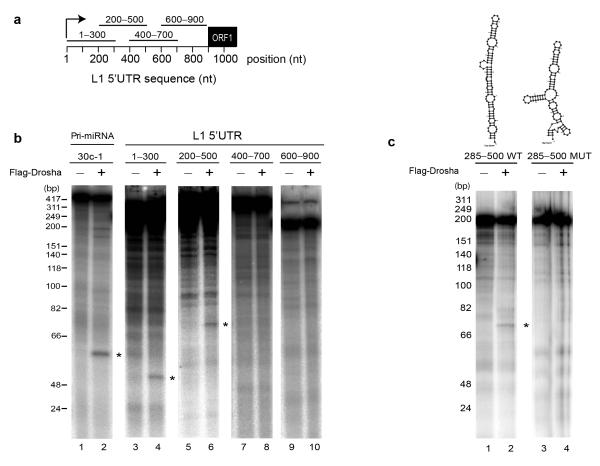
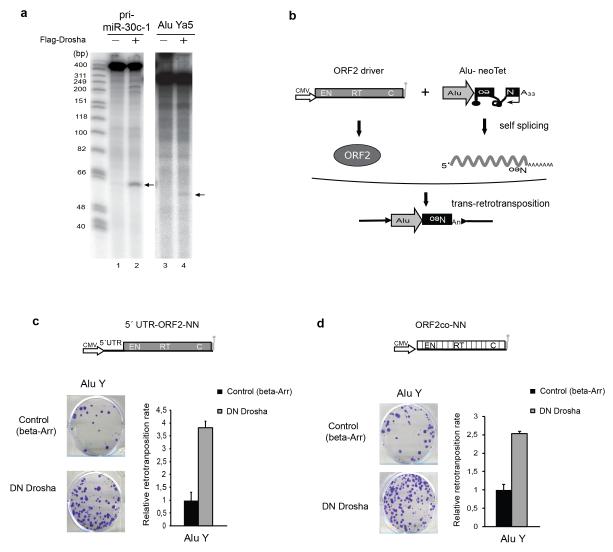
We next tested whether LINE-1 derived RNAs could be directly processed by the endonucleolytic activity of Drosha in vitro. We focused on the 5′UTR region of human RC-L1s, based on the number of CLIP reads mapping to this region (Fig. 1b), its predicted secondary structure (Supplementary Fig. 4a,b) and relevance for DGCR8 binding (Supplementary Fig 4c). We generated four overlapping radiolabeled RNA transcripts corresponding to the L1PA1/L1Hs 5′UTR (Fig. 3a, 300 nucleotides each and Supplementary Fig 4d-g). Remarkably, in vitro incubation of these RNAs with immunopurified Microprocessor from cultured cells (using Flag-Drosha) revealed specific cleavage of the regions ranging from 1 to 300 and from 200 to 500 nucleotides within the L1 5′UTR (Fig. 3b, lanes 4, 6), as well as of pri-miR-30c-1 radiolabeled RNA that was used as positive control for the activity of the Microprocessor (Fig. 3b, lane 2). By contrast, we failed to detect processing of the other 5′UTR derived RNAs despite the presence of a predicted stable secondary structure (400-700 and 600-900, Fig. 3b, lanes 8, 10 and Supplementary Fig. 4f,g). We speculate that processing of these regions by the Microprocessor requires additional sequences that might affect RNA folding and/or recognition. Alternatively, it is possible that DGCR8 binds these regions in a Drosha-independent manner avoiding processing. Importantly, in vitro cleavage of the 1-300 and 200-500 L1-derived RNAs was abolished when using a mutant immunopurified Microprocessor unable to bind RNA (Flag-DN (Dominant Negative) Drosha, Supplementary Fig. 5a, lanes 3, 6 and 9). Controls revealed that Flag-Drosha and Flag-DN Drosha tagged proteins still bind endogenous DGCR8 to a similar extent as revealed by co-immunoprecipitation analyses (Supplementary Fig. 2c). Further analyses revealed that the 285-500 region of the RC-L1 5′UTR could adopt a predicted structure that could be efficiently processed by Drosha in vitro (Fig. 3c). Notably, disruption of this structure by mutagenesis abolished processing (Fig. 3c, lane 4 and Supplementary Fig. 4h). In sum, these data revealed that the Microprocessor binds and processes RC-L1-derived RNAs. We next used primer extension analysis to characterize the sites of Drosha-mediated in vitro processing within the 1-300 L1-derived RNA. Notably, we detected processing at the positions +200 to +270 in L1.342 (Supplementary Fig. 5b). Additionally, we mapped the in vitro processing site of the 285-500 L1-derived RNA at positions +354 to +438 in L1.3 (Supplementary Fig. 5c). To confirm these processing sites in vivo, we used a 5′Phosphate-dependent 5′RACE assay using total RNA derived from cultured cells (Supplementary Fig. 5d, see Methods). Notably, we detected in vivo processing sites near position +438 (+/− 1/2 bp) in the RNA fraction derived from PA-1 and HEK293T cells (66% and 77% of sequenced clones respectively), confirming our in vitro results (Supplementary Fig. 5e). Altogether, these data indicate that the Microprocessor recognizes and processes at least two regions of the 5′UTR of the RC-L1 mRNA and suggests that this activity may interfere with the ability of the RC-LI mRNA to mobilize.
Figure 3.
The 5′UTR of L1 mRNA is cleaved by immunopurified Drosha in vitro. (a) Schematic representation of four 300-nucleotide in vitro transcribed fragments spanning the sense L1 5′UTR region used in (b) for in vitro processing. Transcripts were incubated (+) or not (−) with immunopurified Microprocessor (using FLAG-Drosha). Cleavage products are indicated with asterisks (lanes 2, 4, 6). An RNA ladder marker indicates sizes in base-pairs on the left. (c) In vitro processing of 285-500 L1 5′UTR region (lane 2). Top panel shows a predicted pri-miRNA-like structure of this region. Cleavage is abolished upon introducing mutations that disturb this structure (lane 4 and top panel).
The Microprocessor regulates L1 retrotransposition in vivo
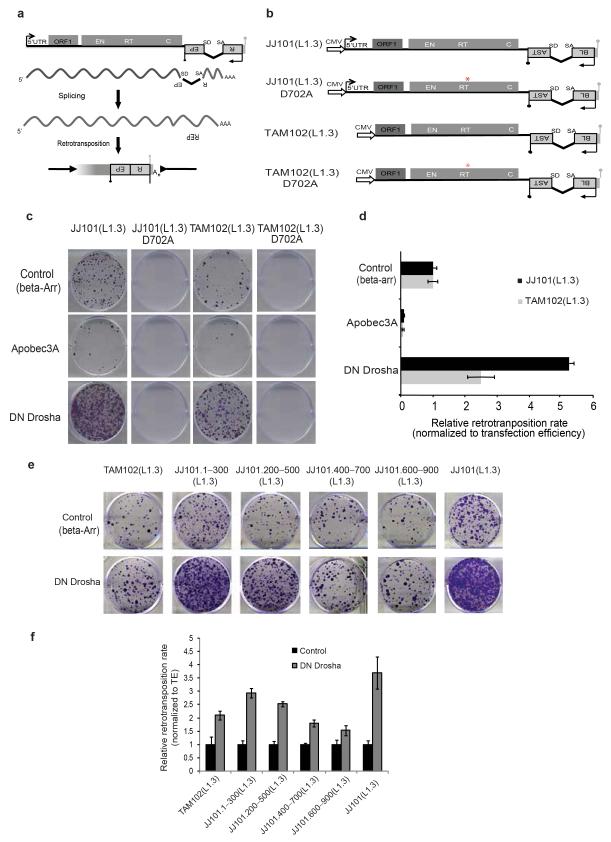
We next used a retrotransposition assay to analyze the role of the Microprocessor in LINE-1 mobility in cultured human cells. This assay is based on the activation of a reporter gene upon a round of L1 retrotransposition14. We used a retrotransposition indicator cassette that consists of a Blasticidin resistance gene (BSD) in the antisense orientation relative to a full-length LINE-1 element that is disrupted by an intron in the sense orientation. Upon transfection into cells, the Blasticidin resistance gene can only be activated after a round of LINE-1 retrotransposition allowing the selection and quantification of cells harboring new retrotransposition events in culture (Fig. 4a)14,43. Since the LINE-1 5′UTR has internal promoter activity1, we included an exogenous strong promoter (CMV) to correct for differences in the expression of the tagged L1 construct, as this could affect the overall rate of mobilization (Fig. 4b). Notably, the retrotransposition rate of a full-length L1 construct (JJ101(L1.3)) increased by 5-fold upon overexpression of DN-Drosha (Fig. 4c,d). Interestingly, removal of the 5′UTR from the engineered construct (TAM102(L1.3)) partially abolished the increase in retrotransposition upon Microprocessor depletion (Fig. 4c,d). Furthermore, we observed severely reduced retrotransposition levels with constructs containing an RT-mutated (reverse transcriptase, RT) engineered LINE-1 allelic construct (Fig.4b,c). In agreement with a proposed role for APOBEC proteins in inhibiting both LINE-1 and Alu retrotransposition44, we observed an inhibitory effect upon APOBEC3A overexpression in this assay (Fig. 4c). Additionally, co-transfection of a DN-DGCR8 expression vector also increased retrotransposition, confirming the results observed with DN-Drosha (Supplementary Fig. 6a,b). Importantly, we determined that expression of either DN-Drosha or DN-DGCR8 was not toxic to cultured cells (Supplementary Fig. 6c). Finally, we confirmed that engineered L1 retrotransposition increased 5-fold upon overexpression of a DN-DGCR8 expression vector using a different retrotransposition assay based on the activation of a Neomycin resistance gene (mneoI14, Supplementary Fig.6d,e). In sum, these data correlate with the processing data and further suggest that the Microprocessor controls LINE-1 retrotransposition.
Figure 4.
The Microprocessor negatively regulates L1 retrotranspositionin vivo. (a) Cartoon depicting the LINE-1 based retrotransposition assay in cultured cells. The transcription start site at the L1 5′ UTR (black arrow), the L1 open reading frames (ORF1 and ORF2; gray rectangles), and the L1 poly (A) site (grey lollipop) are indicated. The relative locations of the endonuclease (EN), reverse transcriptase (RT), and cysteine-rich (C) domains of ORF2 are also indicated. (b) Schematic representation of the JJ101(L1.3) vector containing a full-length human RC-L1 sequence tagged with a mblastI cassette (Blasticidin-resistance gene). The 5′UTR of L1.3 was removed to generate construct TAM102(L1.3). Alleles containing a missense mutation in the Reverse Transcriptase (RT) domain of L1-ORF2 (red asterisk, JJ101 (L1.3) D702A and TAM102 (L1.3) D702A) were used as an internal control. CMV denotes the presence of a Cytomegalovirus promoter. Black and grey lollipops represent polyadenylation signals. (c,e) Cell culture-based LINE-1 retrotranposition assay. (c) HeLa cells were co-transfected with JJ101(L1.3), TAM102(L1.3) and corresponding control vectors or the indicated engineered LINE-1 construct containing an individual fragment derived from the 5′UTR region plus the indicated expression plasmids (β-arrestin (β-arr), Apobec3A, or DN Drosha), as indicated on the left. Each image shows representative data from L1 retrotransposition assays conducted in triplicate. (d,f) Quantification of LINE-1 retrotranposition (from panel c and e respectively). Blasticidin-resistant foci were manually counted and quantified. Data is presented as the proportion of the activity seen in cultures co-transfected with the plasmid expressing the negative control (β-arrestin) and normalized using transfection efficiency and toxicity. Shown are averages of 3 independent biological replicates ± s.d.
To further explore this regulation, we next tested LINE-1 constructs that lack a full-length 5′UTR but contain the four overlapping regions assayed for in vitro processing (see Fig. 3a). Notably, we observed increased L1 retrotransposition with constructs containing the 1-300 and 200-500 regions upon Microprocessor depletion, consistent with the in vitro processing data (Fig. 4e,f). Accordingly, when we removed the 5′UTR and included the predicted stem-loop structure contained in region 285-500 in the engineered construct (Supplementary Fig. 6f and Fig. 3c) we observed a 2-fold increase in the retrotransposition rate upon DN-Drosha co-transfection (Supplementary Fig. 6g,h). Importantly, a mutated version of this predicted stem-loop structure (see Fig. 3c and Supplementary Fig. 4h) or a scrambled version did not significantly change retrotransposition efficiency (Supplementary Fig. 6g,h). Thus, these data agree with the in vitro and in vivo processing data and further suggests that the 5′UTR of RC-L1s is a target of the Microprocessor during the retrotransposition cycle.
The Microprocessor regulates Alu trans-retrotransposition
We next tested whether Alu-derived RNAs could be processed in vitro by the Microprocessor. To do that, we first generated a 290 nucleotide long radiolabeled RNA transcript corresponding to the core sequence derived from an active Alu Ya5 element16. Remarkably, upon in vitro incubation of this RNA with immunopurified Microprocessor (using Flag-Drosha) we observed processing of the Alu Ya5-derived RNA, not as effectively as observed with the pri-miR-30c-1 radiolabeled RNA used as a positive control (Fig. 5a, lanes 4 and 2 respectively). Thus, these data further support that the Microprocessor can bind and process RNAs derived from active Alu transposable elements.
Figure 5.
Alu is in vitro processed and its retrotransposition is regulated by the Microprocessor. (a) In vitro processing of Alu Ya5. An Alu Ya5 core sequence was in vitro transcribed and incubated with Flag-Drosha immunoprecipitates. Upon addition of Drosha (+) Alu Ya5 transcripts were cleaved (lane 4), processing of pri-miR-30c-1 is shown as a positive control (lane 2). (b) Rationale of the Alu trans-mobilization assay. In the scheme, a cartoon depicts the ‘ORF2 driver’ containing an exogenous promoter (white arrow, CMV) and the coding sequence for LINE-1 ORF2 (grey rectangle). In addition, a tagged Alu (grey arrow) with a neoTet retrotransposition indicator cassette (black boxed backwards Neo) containing an encoded poly (A) tail (A33) is depicted. Note that the neoTet cassette contains a self-spliceable group I intron (black curvy lane). Lollipops represent polyadenylation signals. With this configuration, expression of a Neomycin resistant gene can only occur upon a round of trans-retrotransposition (bottom part of the panel). (c) Trans-retrotransposition experiments using 5′UTR-ORF2-NN and (d) ORF2co-NN as a driver (scheme shown). Each image shows representative data from Alutrans-retrotransposition assays conducted in duplicate. β-arrestin is used as a control as it does not affect Alu retrotransposition. The relative retrotransposition activity is quantitated on the right. Data was normalized using transfection efficiency and toxicity. Shown is average of three independent biological replicates ± s.d.
Next, in order to determine whether the Microprocessor can control the mobilization of Alu in cultured cells, we used a previously established Alu trans-retrotransposition assay3,7 (Fig. 5b). To do that, we tagged the Alu Ya5 core with a neoTet cassette7. As Alu are pol-III derived transcripts, the neoTet cassette is based on a self-spliceable intron that activates a Neomycin resistant reporter upon insertion into genomic DNA. In this assay, cells are co-transfected with a LINE-1 derived construct lacking ORF1p (‘ORF2 driver’) that is able to produce ORF2p mediating the mobilization in trans of the tagged spliced Alu RNA (Fig. 5b). The number of G418-resistant foci is used to determine the level of trans-retrotransposition of the Alu core. Notably, the retrotransposition rate of the Alu Ya5 construct increased by 3.5-fold upon overexpression of DN-Drosha in cells co-transfected with a driver that contains the 5′UTR and ORF2 sequences of a RC-L1 (5′UTR-ORF2-NN, Fig. 5c). Controls revealed severely reduced trans-retrotransposition in cells transfected with only the tagged Alu, as expected (S.R.H. and S.M., unpublished data). As the Alu mobilization assay requires the presence of a driver L1 and a tagged Alu, it is likely that the observed increase in trans-retrotransposition upon Microprocessor depletion reflects both Alu and L1-driver in vivo RNA processing, making these assays difficult to quantify. To overcome these limitations, we removed the L1-5′UTR region and used a RC-L1 ORF2 codon optimized sequence in the driver construct45 (plasmid ORF2co-NN, Fig 5d). As expected, the codon optimized LINE-1 driver produced more mRNA upon transfection into cultured cells as determined by RT-qPCR45,46 (Supplementary Fig.6i). Remarkably, we observed a 2.5-fold increase in the mobilization rate of an Alu Ya5 construct upon co-transfection with the ORF2co-NN driver and DN-Drosha (Fig. 5d). Notably, assays conducted in the presence of DN-DGCR8 produced similar results (Supplementary Fig. 6j). Thus, our data suggest that the Microprocessor negatively regulates the mobilization of Alu retrotransposons in cultured cells. These results are consistent with the existence of pol-III derived pri-miRNAs and with previously characterized miRNAs that reside within Alu sequences47. Additionally, these data further support that DGCR8 can bind a constellation of transposable elements-derived RNAs despite having different sequences or changes in sequences within distinct subfamilies (Fig. 1 and Supplementary Table 2, 3).
DGCR8-dependent regulation of LINE-1s is miRNA-independent
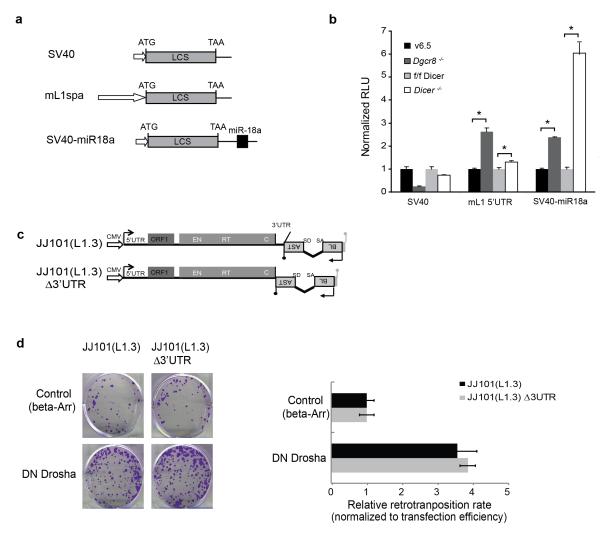
To further analyze the regulation of transposable elements by the Microprocessor, we used luciferase-based reporter constructs containing the 5′UTR of an RC-L1 element to monitor the in vivo processing of this region in cultured cells. We focused on the 5′UTR region of human RC-L1s as we showed that it could be processed by the Microprocessor in vitro and in vivo (Fig. 3, 4). Interestingly, reporters containing the human RC-L1 5′UTR element displayed a small but significant increase in luciferase activity upon Drosha depletion, suggesting in vivo processing by the Microprocessor (Supplementary Fig. 7a,b). Parallel assays using a luciferase reporter containing a canonical miR-18a binding site in its 3′UTR were used as a control of miRNA levels and thus of Drosha function. As expected, the miR-18a-containing reporter showed increased levels of luciferase activity upon Drosha depletion48 (Supplementary Fig. 7a,b). Transfection of reporters containing the 5′UTR of a human RC-L1 element with plasmids expressing DN-DGCR8 or DN-Drosha produced similar results (S.R.H. and S.M., unpublished data). Additionally, evolutionarily older LINE-1 5′UTRs generated similar results upon Microprocessor depletion (S.R.H. and S.M., unpublished data) consistent with the presence of CLIP reads mapping to evolutionary older LINE-1s (L1PA2-PA4, Supplementary Fig. 1). Similarly, when we used a luciferase construct containing the 5′UTR of an active mouse LINE-1 (TF class, L1spa39), which also adopts a potential secondary ordered structure (Fig. 6a and Supplementary Fig. 4i), we observed a three-fold increase in reporter activity in Dgcr8−/−ES cells when compared to parental cells (Fig. 6b). These data also suggest that the Microprocessor can negatively regulate RC-L1 expression in mammalian cells at the post-transcriptional level. Controls using a luciferase reporter containing a miR-18a binding site in Dgcr8−/− ES cells revealed a significant increase in luciferase activity when compared to wild-type cells. On the other hand, previous studies have shown that Dicer depletion results in increased levels of LINE-1 mRNAs17. As both the Microprocessor and Dicer are involved in miRNA biogenesis, we next tested whether changes in gene expression of luciferase constructs containing the 5′UTR of an active mouse LINE-1 are a consequence of lacking canonical miRNAs. To do that, we transfected mouse Dicer−/− ES cells49 with the same reporter containing the 5′UTR of an active mouse LINE-1. Importantly, the same reporter displayed a small increase in luciferase activity in the absence of Dicer when compared to parental cells (Fig. 6a,b), confirming a modest role for Dicer-dependent small RNAs in LINE-1 mRNA abundance17. Notably, the effect in luciferase expression was greater in the absence of DGCR8 than in the absence of Dicer, further suggesting a direct regulation of transposable elements-derived RNAs by the Microprocessor rather than regulation due to defects in small RNA biogenesis. As expected, parallel experiments revealed a significant increase in luciferase activity produced by the reporter containing the miR-18a in Dicer−/− ES cells. Additional Northern-blot controls confirmed lack of canonical miRNAs in Dicer−/− and Dgcr8−/− ES cells (S.R.H. and S.M., unpublished data.). These data suggest a miRNA-independent role for DGCR8 in regulating LINE-1 expression/retrotransposition, rather than an indirect effect due to the absence of miRNAs.
Figure 6.
LINE-1 Regulation by the Microprocessor is Dicer and miRNA-independent. (a) Schematic representation of firefly luciferase reporters containing the 5′UTR from a mouse RC-L1-L1spa- (mL1spa) and controls, simian virus 40 promoter (SV40) and SV40-miR18a, containing a target site for miR-18a in the 3′UTR. LCS; firefly luciferase open reading frame. (b) Dgcr8−/−, Dicer−/− ES cells, as well as parental wild-type cells (v6.5 and f/f Dicer, respectively) were transfected with vectors described in (a). Error bars indicate standard deviation (n=3 biological replicas) and *P< 0.05 (t test). (c) Cartoon depicting the LINE-1 engineered constructs assayed in cultured cells (following nomenclature used in Fig. 4). Construct JJ101(L1.3)Δ3′UTR is a derivative of plasmid JJ101(L1.3) that lacks LINE-1 3′UTR. (d) HeLa cells were co-transfected with JJ101(L1.3) or JJ101(L1.3)Δ3′UTR vectors plus the indicated expression plasmids (β-arrestin (β-arr) or DN Drosha), as indicated on the left side. Each image shows representative data from L1 retrotransposition assays conducted in triplicate. The graph shows a quantification of the assay, and data is presented as the proportion of the activity seen in cultures co-transfected with the plasmid expressing the negative control (β-arrestin) and normalized using transfection efficiency and toxicity. Shown are averages of 3 independent biological replicates ± s.d.
To further support these findings, we generated a human RC-L1 retrotransposition construct lacking the 3′UTR region (Fig.6c, construct JJ101(L1.3)Δ3′UTR) and determined its retrotransposition rate upon Microprocessor depletion. We reasoned that as most miRNAs interact with 3′UTRs19,50, this construct would allow testing whether the effects of Microprocessor depletion on engineered retrotransposition were mediated by cellular miRNAs. Importantly, previous studies have demonstrated that the 3′UTR of RC-L1s is not required for engineered LINE-1 retrotransposition14 and we confirmed these findings (Fig.6d, samples co-transfected with β-Arr). Notably, we observed a 4-fold increase in the rate of engineered LINE-1 retrotransposition using the construct that lacks the 3′UTR upon DN-Drosha co-transfection (Fig. 6d, JJ101(L1.3)Δ3′UTR). Parallel controls revealed a similar increase in retrotransposition for the wild-type construct that contains the 3′UTR region (Fig.6d, JJ101(L1.3)). In sum, these data are consistent with L1 retrotransposition being regulated by direct processing of RC-L1 mRNAs rather than by absence of inhibitory miRNAs upon Microprocessor depletion. Altogether, our data suggest that the Microprocessor can negatively regulate RC-L1 expression in mammalian cells at the post-transcriptional level in a likely miRNA independent manner.
DISCUSSION
The activity of LINE-1 retrotransposons may pose a risk for the genome of an individual. Previous studies have demonstrated that heritable LINE-1 and SINE insertions accumulate mostly during embryogenesis34,51,52.Additionally, recent evidence has also demonstrated that human LINE-1s and Alus are active in selected somatic cells and in several types of tumors9,37,53–56. Thus, different mechanisms may have evolved to reduce retrotransposition in somatic cells. Indeed, DNA-methylation of mammalian LINE-1 promoters is a well-known mechanism to down-regulate expression and subsequent retrotransposition of LINE-1 elements36. Failure to transcriptionally repress LINE-1 expression may negatively impact the human genome in a myriad of ways. Besides DNA-methylation36, few host factors have been previously described to negatively regulate L1/Alu retrotransposition, including APOBEC proteins44, proteins associated with the L1 ORF1 protein and its ribonucleoprotein57, the exonucleaseTrex 158, RNAi-like mechanisms driven by LINE-117or piRNAs18 and MOV1059,60.
Here, we describe a new role for the Microprocessor in controlling the activity of mammalian LINE-1s and SINEs. We have shown that the core components of the Microprocessor directly bind a constellation of mammalian transposable element-derived RNAs, including known active LINE-1s, Alu and SVAs. Additionally, we have demonstrated that the Microprocessor can process structures within the 5′UTR of RC-L1s in vitro and likely in vivo, and potentially, other factors that copurify with the Microprocessor might be able to modulate this activity in vivo21. Similarly, we have demonstrated that the core from an active Alu Ya5 is a substrate for the Microprocessor. Furthermore, the use of luciferase-based reporters and engineered retrotransposition constructs lacking 3′UTR sequences strongly suggests that Microprocessor regulation of LINE-1 mRNAs is both miRNA and Dicer-independent (Fig.6). Additionally, we have demonstrated that the mobilization of full-length engineered L1s in cultured cells can be controlled by the Microprocessor in vivo, most likely by binding and processing their 5′UTRs. However, we also speculate that other sites of Microprocessor processing may exist within coding LINE-1 sequences based on identified binding sites. It is worth mentioning that additional binding sites for DGRC8 were found in the antisense strand of RC-L1, Alu and SVA sequences, although their role remains to be determined. As the whole genome is transcribed and LINE-1s are dispersed through the genome56, we propose that these binding sites might be present within antisense LINE-1/SINE sequences located in gene transcripts (likely within 3′UTR sequences) and/or long-non-coding RNAs29. However, additional experiments are required to determine whether these derived antisense RNAs are processed by the Microprocessor and if they have a role in transposable element biology or genome biology. Although LINE-1 retrotransposons are transcriptionally silenced by DNA methylation in germ cells and most somatic human tissues1,36, we propose that the Microprocessor restricts L1 retrotransposition at a post-transcriptional level, acting against endogenous retrotransposons that escape transcriptional silencing in mammalian cells. Furthermore, the ability of the Microprocessor to restrict L1 retrotransposition in somatic cells may also have an impact on other transposable elements that require L1-encoded proteins for their mobilization; such is the case for Alu and SVA elements.
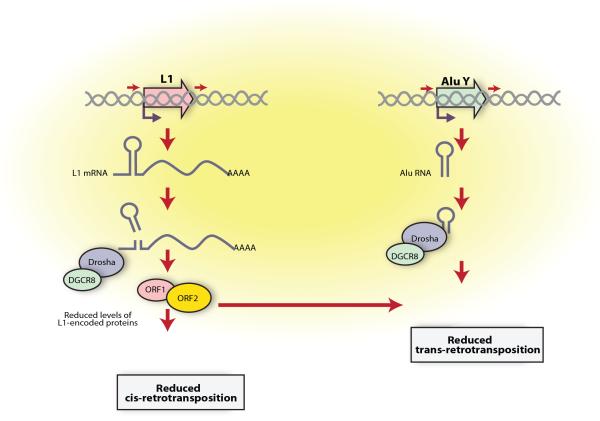
In sum, we propose a model whereby the Microprocessor complex binds the RC-L1 mRNA within the nucleus, most likely co-transcriptionally, and cleaves hairpin structures contained in the L1 mRNA (Fig. 7). We speculate that this cleavage results in LINE-1 mRNA destabilization, with a concomitant decrease in LINE-1 encoded proteins and ultimately reducing retrotransposition rates, which in turns also might affect the capacity of Alu elements to retrotranspose (Fig. 5). Thus, we suggest that the regulation of Alu mobilization may occur at two different levels (see Fig. 7). Altogether, we have demonstrated that the Microprocessor restricts mammalian retrotransposition at a post-transcriptional level, acting against endogenous retrotransposons that escape transcriptional silencing in mammalian cells.
Figure 7.
A model for the control of LINE-1 and Alu retrotransposition by the Microprocessor. Details are provided within the text.
ONLINE METHODS
Crosslinking immunoprecipitation
High-throughput sequencing and crosslinking immunoprecipitation (HITS-CLIP) was performed, as previously described24. HITS-CLIP data available at GEO database under the accession number GSE39086 was used to profile the distribution of DGCR8 CLIP reads on retrotransposons24. Reads were mapped to the genome using bowtie61.
Small RNA reads mapping
To profile the distribution of reads on retrotransposons, only those overlapping annotated retrotransposons were used. This was done to maximize the amount of reads considered as there may be indels and/or nucleotide substitutions in the genome compared to the consensus sequence that may reduce the amount of mapped reads. In the case of L1Hs and Alu elements, the annotation of the repetitive element was taken from the repeatmasker table of hg18 from UCSC (http://genome.ucsc.edu/)62 and the consensus sequences from Repbase63. In the case of older LINE-1 elements (L1PA2-PA5), the consensus sequences from12 were used to annotate L1PA2-5 elements in the human genome. In the case of SVA elements, we used two known active SVA sequences as queries (SVA-MAST2 and SVA.2)4. For each retrotransposon sequence, the overlap between the mapped reads and the annotated retrotransposons was calculated using fjoin64. Next, each of the annotated retrotransposons in the genome that had overlapping reads was aligned to its corresponding consensus sequence using exonerate (parameters -m a:l –score 1 –n 1)65. For L1Hs and Alu elements only those reads mapping to annotated retrotransposons that had an alignment coverage >= 90% and a sequence identity >=99% with its corresponding consensus sequence where further used. Due to the lower conservation among older LINE-1 subfamily members, those reads that had an alignment coverage >= 90% and a sequence identity >=90% were also considered in the analysis. Finally, the position of HITS-CLIP reads was transferred on the consensus sequence using the alignment as a guide. If a read mapped to more than one position in the consensus sequence and/or in the genome, one was selected at random to build the profile. To identify significant positions in the consensus sequences covered by reads the False Discovery Rate (FDR) was calculated as described in66 by computing the background frequency after randomly placing the same reads on the consensus sequences for 1000 iterations. Only those positions with an FDR < 0.01 were considered significant and displayed in the final plots. This procedure was performed independently for sense and antisense reads. All mapping information is provided in the Supplementary File.
Analysis of expressed L1 mRNAs
Total RNA was extracted from HEK293T cells using Trizol (Invitrogen) and 1μg was treated with DNase I (Invitrogen), Reverse Transcribed using M-MLV RT (Promega) and used in RT–PCRs using LINE-1 ORF1 primers as described26,34. Amplified products were cloned in pGEMT-Easy (Promega) and at least 25 independent clones sequenced. Sequences were analyzed with RepeatMasker (http://repeatmasker.org.).
RNA extraction and quantitative RT-PCR
Total RNA was isolated from cells using Trizol (Invitrogen) and treated with DNAse (RQ1 DNAse, Promega, M601A) and checked for DNA contamination by PCR. 500 ng of total RNA was used for quantitative RT-PCR analyses with SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen, 11736-051). Data was analyzed with Bio-Rad CFX Manager software. All experiments show the average and standard deviations of at least three independent biological replicas. Primers for qRT-PCR analysis are listed in Supplementary Table 4.
Northern blot analyses
Hybridization was performed at 42°C with an antisense riboprobe to the first 150 nucleotides of a human RC-L1 (L1.3). For normalization purposes, the membrane was stripped and re-hybridized with a Gapdh probe. Radioactive signals were analyzed using FLA-5100 Phosphorimager Fuji.
Immunoprecipitations and RT-PCR (IP/RT-PCR)
For immunoprecipitation of endogenous DGCR8, anti-DGCR8 antibody (ab90579) and Dynabeads (10001D, Invitrogen) were used. For Flag-Drosha and Flag-DN-Drosha, anti-Flag M2 affinity (Sigma, A220) was used. For T7-DGCR8, T7•Tag® Antibody Agarose (Millipore, 69026) was used. Primers used for these analyses are listed in Supplementary Table 4.
Cell lines and antibodies
HEK293T, PA-1 and HeLa cells were purchased from ATCC. Cytogenetic authentication of HeLa and PA-1 cell lines was performed by spectral karyotyping (SKY)-FISH. Cell lines used in this study were tested for mycoplasma contamination at least monthly. HEK293T and HeLa cells were grown under standard conditions in Dulbecco’s Modified Eagle Medium (DMEM). Human teratocarcinomaPA-1 cells were cultured in Minimum Essential Media (MEM) supplemented with 10% heat-inactivated fetal bovine serum. Mouse Embryonic Stem cells (mES) were grown on gelatin-coated plates (Sigma) without feeders in DMEM-high glucose supplemented with 15% (v/v) fetal bovine serum (GIBCO-Invitrogen). Dgcr8−/−mES cells were purchased from Novus Biologicals (NBA1-19349) and the parental strain (v6.5) from Thermo Scientific (MES1402). Dicer−/− and f/f Dicer were kindly provided by R. Blelloch (UCSF)49. The rabbit anti-DGCR8 was from Abcam (ab90579), the rabbit anti-Drosha antibody was from Upstate (07-717) and mouse/goat anti-β-actin antibody was from Sigma (T4026), whereas anti-mouse ORF1p antibody was kindly provided by Sandy Martin (U of Colorado)13. The working dilutions were 1:250, 1:500, 1:5000 and 1:2000 respectively. Uncropped forms of western blots are shown in Supplementary Fig.8.
siRNA depletions
Knock-down of Drosha in PA-1 cells was achieved with two rounds of siRNA transfection using Dharmafect 4 solution (Dharmacon) following manufacturer’s instructions. Briefly, cells were seeded in 6-well plates to 40% confluence and after 24 h were transfected using 25 nM of each siRNA (Drosha or control, from Dharmacon) and 10 μl of the transfection reagent. The transfection medium was replaced after 24 hr and cells were grown for another 24 hr. Cells were then re-transfected following the same protocol. siRNAs against Drosha and non-targeting siRNAs (control) were purchased from Dharmacon (L-016996-00 and D-001810-02, respectively). Cells were collected 24hr after the second transfection for analyses.
Radioactive RNA labeling and in vitro processing reactions
Templates for RNA synthesis and radiolabeling were obtained by PCR from a plasmid containing the 5′UTR from L1.3 (L1.3S-FF, primers listed in Supplementary Table 4). The forward or 5′end primer included the sequence of a T7 promoter. Additionally, the pri-miR-30c-1 was cloned in pGEMT-easy (Promega) and NdeI digested prior to transcription. Transcription reactions were performed with T7 polymerase (Ambion, AM2082) in the presence of 40 μmols of 32P-αUTP. Probes were gel-purified, phenol-extracted and ethanol precipitated following standard procedures. Approximately 50,000 cpms of each probe were incubated with 15 μl of immunoprecipitated Flag-Drosha, Flag-DN-Drosha beads or control immunoprecipitates, in the presence of buffer A (0.5mM ATP, 20mM creatine phosphate and 3.2 mM MgCl2). Reactions were incubated for 30 min at 30°C, followed by standard phenol/chloroform extraction and ethanol precipitation. RNAs were resolved in an 8-10% 1xTBE polyacrylamide urea gel. Gels were analyzed using Phosphorimager (FLA-5100 Phosphorimager Fuji).
Cell transfection and dual luciferase assays
HEK 293T cells were co-transfected with a 5′ UTR L1 luciferase construct (L1.3-S) and dominant negative forms of DGCR8 or Drosha. A complete list of plasmids used in this study can be found in Supplementary Table 5. Mouse ES cells (Dgcr8−/−, Dicer−/− and their wild-type counterparts, v6.5 and f/f dicer) were transfected with mL1spa construct and SV40 and SV40-miR18a plasmids as controls, using Lipofectamine 2000 (Invitrogen). Cells were collected 48 hr post-transfection. For siRNA studies in HeLa cells, DNA plasmids were transfected during the second round of siRNA transfection using Dharmafect DUO (Dharmacon) and cells were collected 24 hr after transfection. Efficient Drosha knockdown was monitored by co-transfecting a miR-18a reporter system (SV40-miR18a). In all assays, a plasmid expressing the Renilla luciferase gene was used as an internal control. Cells were lysed using passive lysis buffer (Promega), and the levels of firefly and Renilla luciferase activity were measured using the Promega Dual Luciferase Reaction system. The data are expressed as a ratio of firefly luciferase activity to Renilla luciferase activity and normalized to mock value (or wild-type value). Luminescence was measured with a Monolight 3010 luminometer (Pharmingen).
Mapping cleavage sites by primer extension
170 nM of unlabeled RNA was subjected to in vitro processing with or without immunopurified Drosha. RNA was phenol-purified, precipitated and denatured for 3 min at 95°C. Primer extension was carried out with 5′end-labeled oligonucleotides pairing to the 3′end of each of the RNA templates used. Primer annealing and extension was carried out in the presence of reverse transcriptase buffer (Super-Script III RT from Invitrogen, 18080), 1mM of each dNTP, RNase inhibitor and 100 U of Super-Script III RT for 30 min at 52°C. Upon completion, the RNA was hydrolyzed, and RT products were fractionated in denaturing 6% acrylamide 7M urea gels. The sequence ladder was prepared by using the same 5′end labeled oligos, the PCR templates used for in vitro transcription and the Thermosequenase cycle sequencing kit (USB, 78500).
Mapping cleavage sites in vivo using 5′ Phosphate-dependent 5′RACE
Individual cleavage products were detected using a method previously described67. An RNA linker was synthesized in vitro using Mmessage Mmachine T7 Ultra Kit (Ambion) and EcoRI- digested pBluescript KS plasmid (Stratagene) as template. 1 μg of total RNA was ligated directly to 15 pmol of the 5′ linker, requiring the presence of 5′phosphates on the ligated RNA, in a 20 μl volume (2 μl 10X T4 RNAligase buffer, 10μl PEG 8000 40% and2 μl 10X T4 RNA ligase (Ambion)). The reaction was phenol-cloroform extracted and precipitated. Reverse transcription was carried out using ThermoScript RT system (Invitrogen) and 5′UTRL1as as specific primer. PCR reactions on 4 μl of resulting cDNAs were carried out using Taq polymerase (Invitrogen) and specific primers (upSK primer and 484as primer). A nested PCR was developed using 5 μl of the first PCR product as template and two internal specific primers, SK primer and UTR447as. Products were run on a 2% agarose gel, and the products were excised, purified and cloned in pGEMT-easy vector (Promega). After blue/white screening at least 10 colonies were sequenced. The sequences corresponding to older subfamilies than L1Hs, containing G in the position 425, were not considered in the analysis.
LINE-1 retrotransposition assays
A slightly modified version of a previously established transient LINE-1 retrotransposition assay was used43. The retrotransposition indicator cassette consists of a BSD gene -Blasticidin resistance gene- in the antisense orientation (relative to LINE-1) that is disrupted by an intron (γ-globin 2) in the sense orientation, as previously described12,39,68. With this configuration, BSD expression requires LINE-1 transcription, removal of the intron by splicing, reverse transcription, and integration followed by expression of the BSD gene (Figure 4a). The number of blasticidin-resistant foci can be used to quantify retrotransposition in cultured cells. For mneoI based assays, we used a previously described protocol43. For full details of all plasmids used in this study refer to Supplementary Table 5.
Alu trans-retrotransposition assays
We used HeLa-HA cells and a previously published protocol4,8. Briefly, HeLa-HA cells were co-transfected with a driver ORF2 and a tagged Alu in the presence of the expression plasmid indicated. 72h post-transfection, cells were selected with 600μg/ml G418 (GIBCO-Invitrogen) for 12 days. The number of G418-resistant foci was used to quantify retrotransposition in cultured cells. For full details of all plasmids used in this study refer to Supplementary Table 5.
Toxicity assays
Toxicity of DN Drosha, DN DGCR8, β-Arrestin and A3A was assayed in HeLa cells co-transfected with a linearized plasmid (pcDNA-6-myc-His) containing a blasticidin resistant cassette and selecting BSD-resistant foci44. Briefly, 1×104HeLa cells were plated in a 100 mm tissue culture plate. After 16-18 hours, the cells were co-transfected with 2μg of the corresponding expression plasmid (DN Drosha, DN DGCR8, β-Arrestin or A3A) and 2μg of linearized pcDNA-6-myc-His vector in the presence of 24 μl of Fugene 6 transfection reagent (Roche Biochemical or Promega) as described69.Cells were cultured for 5 days and then treated with 10μg/ml Blasticidin-ClH (Invitrogen) for eight days. After fixation and staining, the number of foci was counted manually.
Bisulphite DNA sequencing
Genomic DNA from mock siRNA and Drosha siRNA-transfected PA-1 cells was extracted and purified using a Wizard DNA genomic purification kit (Promega A1120). Bisulfite conversion of genomic DNA was performed with EZ DNA methylation-Gold Kit (Zymo Research D5005) with a conversion efficiency of ~95%. The DNA-methylation status of LINE-1 was analyzed globally, as described37. The methylation status of the L1-5′UTR was analyzed from 6 previously characterized RC-L1s2,34,70 (primers used are listed in Supplementary Table 4).
mfold analyses
The potential secondary structure formed by LINE-1 RNA fragments was analyzed using the mfold software, as described71.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Hastie and J.V. Moran for comments and critical reading of the manuscript. We also are grateful to M. Madej, J. Reddington and R. Meehan for advice on DNA methylation assays and to I. Adams for discussions. We thank R. Blelloch (University of California San Francisco, San Francisco, California, USA), V.N. Kim (Seoul National University, Seoul, Korea), S.L. Martin (University of Colorado School of Medicine, Aurora, Colorado, USA), A. Roy-Engel (Tulane Cancer Center, New Orleans, LA USA), T. Heidmann (Institut Gustave Roussy, Villejuif, France and Université Paris-Sud, Orsay, France) and J. V. Moran (Howard Hughes Medical Institute, University of Michigan Medical School, Ann Arbor, Michigan, USA) for their generous gift of reagents. S.M. was the recipient of a long-term EMBO postdoctoral fellowship. S.R.H. was supported by a Marie-Curie Intra-European Fellowship and a Marie Curie CIG-Grant (PCIG10-GA-2011-303812). M.P. and E.E. were supported by the Spanish Ministry of Science (BIO2011-23920) and by the Sandra Ibarra Foundation (CSD2009-00080). M.P. is supported by the Novo Nordisk Foundation. J.L.G.P is supported by FP7-PEOPLE-2007-4-3-IRG, CICE-FEDER-P09-CTS-4980, PeS-FEDER-PI-002, FIS-FEDER-PI11/01489 and the Howard Hughes Medical Institute (IECS-55007420). J.F. C. was supported by Core funding from the Medical Research Council and by the Wellcome Trust (Grant 095518/Z/11/Z).
Footnotes
Accession codes. Gene Expression Omnibus database: GSE39086 (sequencing raw data for endogenous and overexpressed DGCR8 HITS-CLIP).
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
REFERENCES
- 1.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annual review of genomics and human genetics. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5280–5. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature genetics. 2003;35:41–8. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 4.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH. Retrotransposition of marked SVA elements by human L1s in cultured cells. Human molecular genetics. 2011;20:3386–400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raiz J, et al. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic acids research. 2012;40:1666–83. doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nature genetics. 2000;24:363–7. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 7.Wei W, et al. Human L1 retrotransposition: cis preference versus trans complementation. Molecular and cellular biology. 2001;21:1429–39. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome research. 2007;17:602–11. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–47. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–7. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. The insertional history of an active family of L1 retrotransposons in humans. Genome research. 2004;14:1221–31. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Molecular and cellular biology. 1991;11:4804–7. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–27. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 15.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nature reviews. Genetics. 2002;3:370–9. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 16.Bennett EA, et al. Active Alu retrotransposons in the human genome. Genome research. 2008;18:1875–83. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang N, Kazazian HH. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nature structural & molecular biology. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 18.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental cell. 2007;12:503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Bushati N, Cohen SM. microRNA functions. Annual review of cell and developmental biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 20.Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 21.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 22.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes & development. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews. Molecular cell biology. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 24.Macias S, et al. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nature structural & molecular biology. 2012;19:760–6. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nature genetics. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 26.Macia A, et al. Epigenetic control of retrotransposon expression in human embryonic stem cells. Molecular and cellular biology. 2011;31:300–16. doi: 10.1128/MCB.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deininger P. Alu elements: know the SINEs. Genome biology. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Molecular and cellular biology. 2001;21:1973–85. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science (New York, N.Y.) 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Perez JL, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–73. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeom K-H, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic acids research. 2006;34:4622–9. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nature genetics. 2003;35:363–6. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 33.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic acids research. 2006;34:1512–21. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wissing S, et al. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Human molecular genetics. 2012;21:208–18. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han J, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 37.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nature genetics. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naas TP, et al. An actively retrotransposing, novel subfamily of mouse L1 elements. The EMBO journal. 1998;17:590–7. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH. Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nature genetics. 1998;20:288–90. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- 41.Goodier JL, Ostertag EM, Du K, Kazazian HH. A novel active L1 retrotransposon subfamily in the mouse. Genome research. 2001;11:1677–85. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dombroski BA, Scott AF, Kazazian HH. Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6513–7. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrish TA, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature genetics. 2002;31:159–65. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 44.Bogerd HP, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8780–5. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagstaff BJ, Barnerssoi M, Roy-Engel AM. Evolutionary conservation of the functional modularity of primate and murine LINE-1 elements. PloS one. 2011;6:e19672. doi: 10.1371/journal.pone.0019672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–8. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 47.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature structural & molecular biology. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 48.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nature structural & molecular biology. 2007;14:591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 49.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & development. 2008;22:2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Perez JL, et al. LINE-1 retrotransposition in human embryonic stem cells. Human molecular genetics. 2007;16:1569–77. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 52.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes & development. 2009;23:1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 54.Lee E, et al. Landscape of somatic retrotransposition in human cancers. Science (New York, N.Y.) 2012;337:967–71. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–96. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solyom S, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome research. 2012;22:2328–38. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodier JL, Cheung LE, Kazazian HH. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Research. 2013 doi: 10.1093/nar/gkt512. doi:10.1093/nar/gkt512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–98. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology. 2012;9:53. doi: 10.1186/1742-4690-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodier JL, Cheung LE, Kazazian HH. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS genetics. 8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic acids research. 2011;39:D876–82. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and genome research. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 64.Richardson JE. fjoin: simple and efficient computation of feature overlaps. Journal of computational biology: a journal of computational molecular cell biology. 2006;13:1457–64. doi: 10.1089/cmb.2006.13.1457. [DOI] [PubMed] [Google Scholar]
- 65.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nature structural & molecular biology. 2010;17:173–9. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karginov FV, et al. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Molecular cell. 2010;38:781–8. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goodier JL, Zhang L, Vetter MR, Kazazian HH. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Molecular and cellular biology. 2007;27:6469–83. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei W, Morrish TA, Alisch RS, Moran JV. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Analytical biochemistry. 2000;284:435–8. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 70.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–70. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. Journal of molecular biology. 1999;288:911–40. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.