Abstract
miRNAs are small non-coding RNAs able to modulate target-gene expression. It has been postulated that miRNAs confer robustness to biological processes, but a clear experimental evidence is still missing. Using a synthetic biology approach, we demonstrate that microRNAs provide phenotypic robustness to transcriptional regulatory networks by buffering fluctuations in protein levels. Here we construct a network motif in mammalian cells exhibiting a “toggle - switch” phenotype in which two alternative protein expression levels define its ON and OFF states. The motif consists of an inducible transcription factor that self-regulates its own transcription and that of a miRNA against the transcription factor itself. We confirm, using mathematical modeling and experimental approaches, that the microRNA confers robustness to the toggle-switch by enabling the cell to maintain and transmit its state. When absent, a dramatic increase in protein noise level occurs, causing the cell to randomly switch between the two states.
Introduction
miRNAs are small non-coding RNAs able to modulate protein expression by pairing to cognate sites at the 3′ untraslated region (UTR) of target mRNAs, thus promoting mRNA degradation and/or translation inhibition. The role of miRNAs is still under debate because, unlike transcription-factors, the phenotype induced by their perturbation (i.e. knock-down or overexpression) is usually subtle1. For example, in C. elegans, systematic knockout of single miRNAs, or even double or triple mutants, fails to elicit an observable phenotype2,3,4.
It has been postulated that miRNAs play a pivotal role in conferring robustness to biological processes against enviromental fluctuations during development5,6,7 and, more generally, in buffering fluctuations in gene expression6,5,1,8,9,10,11,12,13,7.
Biological robustness has been defined as the ability to correctly perform a biological function against endogenous and exogenous perturbations14, such as stochastic fluctuations in gene expression. This “biological noise” has long been observed in both prokaryotes and eukaryotes, where it increases phenotypic variability among individual cells.15,16,17,18 Increased cell-to-cell variability has been found to be either benificial, such as during stress or differentiation, or detrimental such as during embryonic development of multicellular organisms, where it can decrease robustness if not properly controlled19.
For example, correct development of sensory organs in Drosophila depends on the presence of the microRNA miR-7, but only in the face of environmental perturbations5. Indeed, miR-7-mutant animals subjected to fluctuating environmental temperature fail to form complete antennae, whereas wild-type larvae develop normally5.
So far, the ability to elucidate the role of miRNAs in conferring robustness to a biological process has been hampered by the difficulty in clearly distinguishing the contribution of the miRNA from the cross-talk of all the other regulatory pathways that control the same biological process. Indeed, miRNA-mediated negative feedback loops are often found coupled to multiple transcriptional feedback loops in gene regulatory networks across species and collaborate with transcription factors to regulate their targets13,5,20.
Synthetic biology can be used to assess, in a controlled setting, the contribution of the miRNA to the overall phenotype exhibited by an engineered biological circuit12,21. This is a complementary approach to the investigation of miRNAs in an endogenous setting and it can uncover the design principles underlying biological robustness.
We construct a simple but non-trivial network motif (Positive Negative Feedback Loop - PNFL) exhibiting a “toggle-switch” phenotype created by two alternative protein expression levels (ON and OFF). The motif consists of a Transcription Factor (TF) that regulates its own transcription (Positive Feedback Loop - PFL) and that of a naturally occurring human miRNA directed against the TF itself (miRNA-mediated Negative Feedback Loop - NFL). The main phenotype of a toggle-switch is the ability to maintain an ON (high protein level) or OFF (low protein level) state indefinitely across multiple cell divisions and to switch only when a specific signal is transiently applied. Mathematical and experimental analyses demonstrate that the PNFL motif is a robust toggle-switch, whereas the motif lacking the miRNA (i.e. the PFL motif) it is not. Indeed, the PFL motif will spontaneously switch ON because of a higher noise level, thus clearly demonstrating the role of miRNAs in conferring phenotypic robustness to transcriptional networks.
Results
A synthetic Positive Negative Feedback Loop network motif
We constructed a PNFL motif in Chinese Hamster Ovary (CHO) made up of well known and characterized biological parts, orthogonal to the endogenous ones, assembled as depicted in Fig. 1.
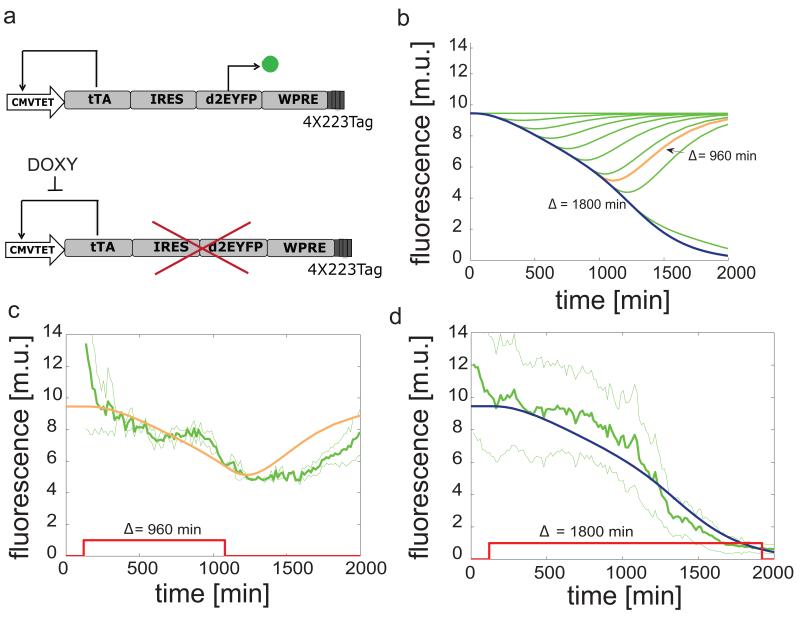
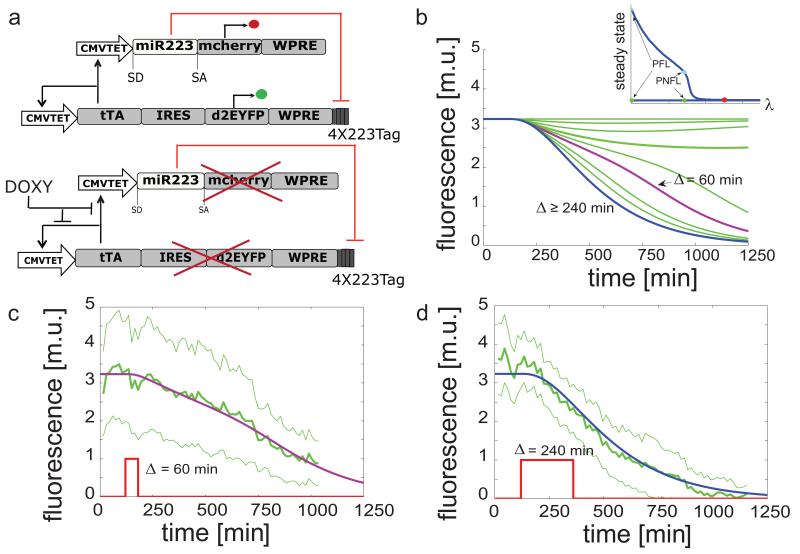
Figure 1. Schematics of the PNFL motif and “switch OFF” experiments following pulses of Doxycycline of different duration.
(a): the tetracycline-controlled transactivator (tTA) is self-regulated, in the absence of doxycycline, by binding the tTA-responsive CMV-TET promoter, thus generating a positive feedback loop (PFL - black lines), whose dynamics is tracked by a destabilized EYFP (d2EYFP). The same CMV-TET promoter drives the transcription of the human microRNA miR-223 embedded in the first intron of the low affinity nerve growth factor receptor (ΔLNGFR)23, followed by a reporter gene encoding for the mCherry fluorescent protein (Negative Feedback Loop - NFL - red lines). miR223 in turn down-regulates the tTA mRNA levels through 4-repeated target sequences perfectly complementary to the miR-223 seed sequence, placed at the 3′UTR of the PFL gene expression cassette. Doxycycline interrupts the tTA-mediated transcriptional activation. WPRE, woodchuck hepatitis virus post-transcription regulatory element. (b): simulated d2EYFP fluorescence of PNFL cells following simulated treatment with Doxycycline of different duration Δ. In the inset, the bifurcation diagram when varying the miRNA strength (λ) is shown. (c),(d): experimental d2EYFP fluorescence using the microfluidics device (solid green line) following treatment with Doxycycline (red line) at time 120 min and removal after Δ = 60 min (c) or 240 min (d); standard deviation is among at least three replicates (thin green lines); simulations (blue and purple lines) are rescaled to the experimental data and also represented in (b) (same colors).
The PNFL motif consists of a synthetic transcription factor (tTA) binding its own promoter (CMV-TET) thus self-activating its own expression. The binding of the transcription factor to the promoter can be inihibited by Doxycycline. The transcription factor also binds another promoter (CMV-TET) driving expression of a natural occuring mammalian microRNA (miR-223). miR-223 is specifically expressed in the myeloid lineage, where it targets multiple genes inducing a low magnitude of repression, mainly acting as a rheostat to adjust protein output22. The microRNA, once expressed, targets the mRNA encoding for the transcription factor, thanks to four sequence tags at the 3′ end of the transcript perfectly complementraty to the miR223 seed sequence23 (see Supplementary Note 1).
We embedded the positive feedback motif and negative feedback motif in two separate lentiviral vectors; we then performed sequential integration in CHO cells. The ability of lentiviruses to randomly integrate in the genome of the host cells, with different expression levels of the transgene according to the site of integration, allowed us to generate nine monoclonal populations of CHO cells (PFL cells) carrying the PFL motif shown in Fig. 2a. PFL clones were then used to generate fourteen matched PNFL clonal populations of CHO cells (PNFL cells), carrying both the PFL and the NFL cassettes shown in Fig.1a (see also Supplementary Fig. S1, Supplementary Fig. S2 and Supplementary Note 2).
Figure 2. Schematics of the PFL motif and “switch OFF” experiments following pulses of Doxycycline of different duration.
(a): the tetracycline-controlled transactivator (tTA) is self-regulated, in the absence of doxycycline, by binding the tTA-responsive CMV-TET promoter, thus generating a positive feedback loop (PFL - black lines), whose dynamics is tracked by a destabilized EYFP (d2EYFP). (b): simulated d2EYFP fluorescence of PFL cells following simulated treatment with Doxycycline of different duration Δ. (c),(d): experimental d2EYFP fluorescence using the microfluidics device (solid green line) following treatment with Doxycycline (red line) at time 120 min and removed after Δ = 960 min (c) or 1800 min (d); standard deviation (thin green lines) is among at least three replicates; simulations (blue and orange lines) are rescaled to experimental data and also represented in (b) (same colors).
PNFL network motif is a toggle-switch with miRNA-dependent dynamics
In order to examine the dynamic behaviour of the PNFL motif, we developed a mathematical model of eight nonlinear differential equations (for details refer to Supplementary Note 3). This model is an extension of a model we previously published for the PFL motif to which we added four extra equations describing the miR-223 and the mCherry protein24. The extra parameters, specific to the PNFL model, were chosen to minimize the error between model predictions and experimental observations (Supplementary Fig. S3 and Supplementary Table S1).
The PNFL model consists of the following ordinary differential equations:
| (1) |
| (2) |
| (3) |
| (4) |
where Tm =tTA mRNA, Tp = tTA protein, Mi =inactive form of the microRNA, Ma = active form of the microRNA. The first equation represents the transcription rate of the tTA mRNA (Tm) as the sum of a production term, proportional to a function f(Tp, D) of the tTA protein level (Tp) and of doxycycline (D), and a degradation term, proportional to the mRNA degradation rate (d1) plus the effect of the microRNA (λ · g(Ma)). The parameter λ represents the strength of the microRNA. By setting λ = 0, the effect of the microRNA is abolished thus obtaining the model of the PFL motif, as described by Eqs. (1) and (2). The second equation describes the protein translation rate of the tTA protein as proportional to the amount of mRNA (Tm) and to the protein degradation rate (d2). The third and fourth equations describe the transcription rate of the microRNA from the tTA inducible promoter and its maturation. Specifically, in the third equation, the transcription rate of the microRNA (Mi) is proportional to a production term, G2f(Tp, D), which is a function of the tTA protein Tp binding to the microRNA promoter, and to a degradation (d3) plus maturation (KD) term. The fourth equation describes the maturation rate of the mature microRNA (Ma) as a function of the immature microRNA and of the degradation rate of the mature microRNA. For simplicity, we omitted four additional linear differential equations that are needed to model the translantion and maturation of the two fluorescent reporters (Supplementary Note 3 for the full model).
We explored the behaviour of the PNFL motif and its robustness to model parameters by numerical bifurcation analysis. As shown in the inset of Figure 1b and in Supplementary Figure S4, the PNFL can behave as a bistable system (toggle-switch), which, in the absence of Doxycycline, can either be ON (high expression of the d2EYFP) or OFF (low expression of the d2EYFP), as long as the repression of the miRNA is not too strong. Specifically, when the level of the TF is above the promoter activation threshold, the PFL motif will switch ON and stay ON due to its autocatalytic activity; if, however, the TF activity is transiently blocked (i.e. by a small molecule, or signaling event) then its protein product will decrease below the promoter activation threshold and the PFL will switch OFF and stay OFF even after the transient block has been removed11,25,26,27,24,28.
In order to explore the dynamic properties of the PNFL motif, we simulated in silico a series of “switch OFF” experiments. The switch OFF simulations were performed starting from the ON steady-state and then simulating treatment with Doxycycline for a limited time interval (Δ).
Numerical simulations of the d2EYFP fluorescence level in Figure 1b for the PNFL model, and in Figure 2b for the PFL model (used as a control) show that, in both cases, when the duration of Doxycycline treatment is below a threshold value and , d2EYFP expression initially decreases (switches off) and then increases back ON once Doxycycline is removed. However, when the Doxycycline treatment lasts longer than the threshold duration Δth, d2EYFP expression switches OFF and it does not increase again, even after Doxycycline has been removed. This is the telltale feature of a bistable “toggle switch”.
To experimentally probe the toggle-switch behaviour of the PNFL circuit, and to confirm numerical simulations, we used an innovative microfluidics platform coupled to an inverted epi-fluorescence microscope to administer Doxycycline and to follow in real time d2EYFP fluorescence in clonal populations of PFL and PNFL cells29 (Supplementary Note 4 and Supplementary Note 5 for details on the microfluidic device). We selected a clonal population of PFL cells (PFL7 in Supplementary Fig. S2) and a matched clonal population of PNFL cells (PNFL7-2 in Supplementary Fig. S2) derived from the same PFL clone. We then performed in vitro “switch OFF” experiments by treating PNFL and PFL cells with Doxycycline (at a concentration of 1μg/ml) for a limited time interval (Δ). The duration Δ of Doxycycline treatments was chosen according to the in silico analysis performed with the PNFL and PFL models.
d2EYFP fluorescence measurements (Fig. 1c,d for the PNFL circuit, and Fig. 2c,d for the PFL) confirmed the results of the in silico analysis, showing that the PNFL is bistable: indeed treatment with a pulse of Doxycycline lasting 60 min (Fig. 1c), or 240 min (Fig. 1d) permanently switched the circuit OFF, whereas a pulse of 20 min did not cause any visible effect (Supplementary Figure S5). The PFL had the same overall behaviour but its dynamics were much slower (Fig. 2c,d). Moreover, as detailed in the next section, we further probed the bistable behaviour of additional PFL and PNFL clones by Fluorescent Activated Cell Sorter (FACS), as reported in Supplementary Figure S6 and Figure 3.
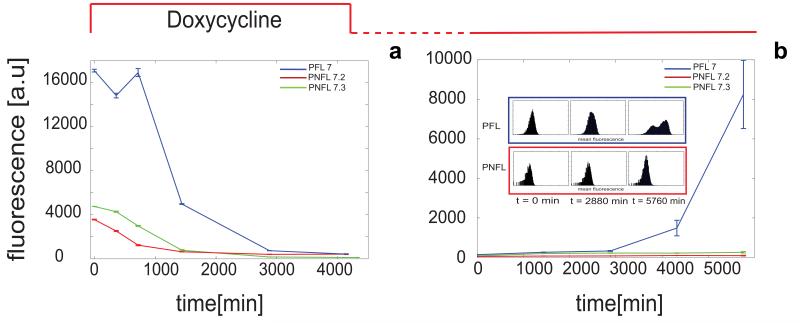
Figure 3. Fluorescence-Activated Cell Sorting (FACS) switch off experiment of PFL and PNFL cells.
(a): d2EYFP fluorescence levels in PFL cells (blue line) and PNFL 7-2 and 7-3 cells (red and green line respectively) were measured at 0 hrs, 6 hrs (360 min), 12 hrs (720 min), 24 hrs (1440 min), 48 hrs (2880 min) and 72 hrs (4320 min) following treatment with Doxycycline (1 μg/ml) at time 0 hrs. (b): d2EYFP fluorescence levels in PFL cells (blue line) and PNFL cells (red line) cells were measured at 0 hrs, 24 hrs (1440 min), 48 hrs (2880 min), 96 hrs (5760 min) following removal of Doxycycline at time 0 hrs. Prior to time 0 hrs, both PFL and PNFL cells were grown in the presence of Doxycycline for 72 hrs. (Subpanel): histogram displaying FACS data for PFL 7 (blue frame) and PNFL 7-2 (red frame) at times t = 0, 2880 min and 5760 min; CVs computed at 0 hrs, 24 hrs, 48 hrs, 72 hrs are respectively 74.5, 105.1, 108, 202.6 for PFL and 81.9, 88.4, 85.75, 96 for the PNFL, thus revealing the higher variability for PFL clones compared to PFL. Error bars represent the standard deviation among three replicates.
miRNA confers phenotypic robustness by buffering noise
Ideally, once the toggle-switch is OFF, it should stay OFF indefinitely. However, due to the auto-catalytic nature of both the PFL and PNFL motifs (i.e. a transcription factor self-activating its own promoter), it should be much easier for both motifs to stay ON than to stay OFF. Indeed, when the switch is OFF, small fluctuations in the transcription factor protein levels may be amplified by the positive feedback loop, causing the system to switch back ON.16
In order to analyse the effect of the microRNA on noise propagation in the PNFL network motif, we considered a simplified stochastic gene expression model with transcription, translation and degradation as probabilistic events occurring at exponentially distributed time intervals30,31 (Supplemental Fig. S7).
Specifically, in the model, mRNAs are produced in transcriptional bursts from the promoter, in agreement with the recent modeling and experimental evidences concerning noise in eukaryotic gene expression32,16,15.
We then derived, under a set of simplifying assumptions (Supplementary Note 6), a system of differential equations describing the dynamics of the statistical moments in four different scenarios (Supplementary Figure S7 and Supplementary Table S2): (1) unregulated transcription, i.e. the mRNA is transcribed from a constitutive promoter30 (2) regulated transcription where the protein activates its own transcription (i.e. the PFL motif); (3) a negative feedback loop mediated by the microRNA inducing degradation of the target mRNA (i.e. a NFL motif); (4) the PNFL motif when both positive feedback loop and the microRNA mediated negative feedback loop are present (Methods and Supplementary Note 6).
Finally, we computed an analytical expression for the Coefficient of Variation (CV), i.e. the relative deviation of protein expression in each cell compared to the population average, which can be used as a measure of noise. Indeed, a small CV corresponds to a tight distribution centered around the mean, hence a small cell-to-cell variability; a large CV corresponds to a loose distribution, indicating large cell-to-cell variability.
From the analytical expression of the CV in the four scenarios (Supplementary Note 6), it can be deduced that, independently of the model parameter values, noise increseas when a transcriptional positive feedback loop is present, compared to unregulated transcription, in agreement with previous modelling and experimental results16,15,27. On the contrary, noise decreases, compared to unregulated transcripiton, when a microRNA-mediated negative feedback loop is present, again in agreement with previous modelling and experimental results on negative feedbacks16,15,33. In the case of the PNFL motif, when both negative and positive feedbacks are present, noise is reduced compared to the PFL motif, but is greater than the unregulated transcription.
In order to investigate the validity of the stochastic model predicitions, we measured the distribution of fluorescence levels across the cells in each of the monoclonal cell populations (9 for the PFL and 14 for the PNFL) as shown in Fig. 4a. We then quantified the protein noise level by computing the (CV) as the relative deviation of d2EYFP fluorescence in each cell compared to the clonal population average. Figure 4b reports the theoretical CV s derived for both the PFL and the PNFL stochastic models, superimposed to the experimentally measured CV s in the 23 clones (9 PFL and 14 PNFL) (see also Supplementary Note 7 and Supplementary Fig. S8 and Supplementary Table S3).
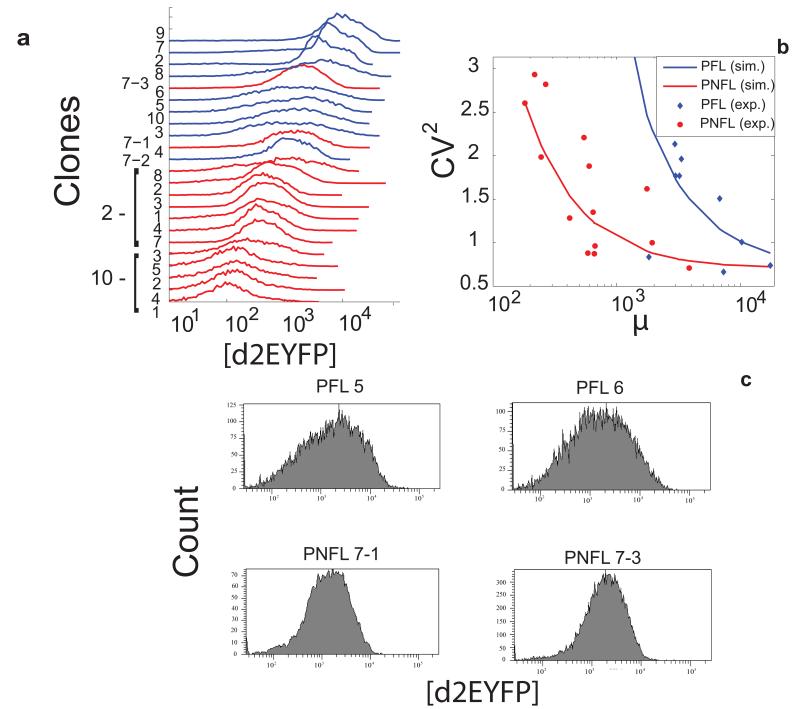
Figure 4. The miRNA-mediated negative feedback loop reduces fluctuations in protein expression.
(a): FACS-derived histograms of the distribution of d2EYFP fluorescence for the 9 clonal populations of PFL cells (in blue) and the 14 clonal populations of PNFL cells (in red). (b): Experimental (dots) and simulated (solid lines) values of CV2 as a function of mean fluorescence for PFL clones (in blue) and PNFL clones (in red); the simulated CV2 values were computed as described in Supplementary Notes (Eqs 10 - 19). (c) FACS-derived histograms of the distribution of d2EYFP fluorescence in two representative clonal populations of PFL and PNFL cells with similar average fluorescent intensities (PFL 5,6 and PNFL 7-1, 7-3).
Figure 4c shows representative examples of the d2EYFP fluorescence distribution of two PFL clones and two PNFL clones, with similar average fluorescence intensities; Figure 4b shows the CV for all the PFL and PNFL clones.
The experimental results clearly demonstrate a reduced protein noise level (CV) in the PNFL cells as compared to the PFL cells, indicating a specific effect of the miRNA-mediated negative feedback in buffering protein expression fluctuations.
The PFL motif is predicted to have a higher noise level than the PNFL motif because fluctuations in the number of transcribed mRNAs are translated into fluctuations in the number of proteins, which in turn control the number of transcribed mRNAs. This results in a net amplification of noise (Supplementary Note 7). On the other hand, when the miRNA-mediated negative feedback is present, noise is reduced because fluctuations in the number of mRNA molecules amplified by the PFL are counterbalanced by the presence of the miRNA.
We decided to evaluate the phenotypic robustness of both the PNFL and PFL motifs in terms of how long they were able to stay OFF in the absence of Doxycycline.
We performed a long-term experiment by treating monoclonal population of PNFL cells and their matched control population of PFL cells with Doxycycline for 72 hrs (4320 min); we then washed Doxycycline out, and monitored d2EYFP fluorescence in cells for 96 hrs (5760 min) by Fluorescence Activated Cell Sorting (FACS). If both motifs exhibit a robust phenotype, then they should not reexpress d2EYFP and stay permanently OFF.
As shown in Figure 3a for the PFL 7 clone and the matched PNFL 7-2 and 7-3 clones, following Doxycycline treatment, both the PNFL and PFL cells switched d2EYFP expression OFF, with dynamics similar to those estimated using the microfluidics setup.
Strikingly, following Doxycycline washout, PFL cells started switching back ON at 72 hrs (4320 min) and fully expressed d2EYFP at 96 hrs (5670 min) (Fig. 3b). On the contrary, PNFL cells stayed OFF (Fig. 3b).
As further confirmation of the miRNA role, we performed the same experiment on PNFL cells but this time inhibiting the action of the miRNA by transfection with a Locked Nucleic Acid complementary to the miR223 sequence (LNA 223)34. PNFL cells treated with the LNA increased d2YFP expression at 72 and 96 hrs, whereas PNFL cells treated with a control LNA maintained their OFF state, confirming our previous results (details and data shown in Supplementary Note 8 and Fig. S9).
We repeated the long-term experiment on additional PFL and matched PNFL clones, in order to exclude that the difference in phenotypic robustness we observed was clone-specific. As reported in Supplementary Figures S6a, S6b for the PFL 2 and PNFL 2-2, 2-3 and 2-7 clones, and in Supplementary Figures S6c, S6d for the PFL 10 and PNFL 10-2 clones, the presence of the miRNA-mediated feedback loop consistently confers robustness to the PFL motif, allowing the PNFL cells to maintain their OFF state for longer as compared to the matched PFL cells. The main difference among the three PFL clones (PFL-7, PFL-2 and PFL-10) was observed in the time taken to switch back ON following Doxycyline removal (Fig. 3b and Supplementary Figures S6b, S6d). This difference is likely due to the different insertion site of the PFL cassette in the genome of the three clones.
Discussion
The combination of computational analysis coupled with experimental biology, at the core of synthetic biology, has been used as a powerful tool to elucidate complex cellular processes, such as cell polarization35 and transcriptional regulation12. Here we used the synthetic biology paradigm to explore the role of miRNAs in conferring robustness to a fundamental network motif, the Positive Feedback Loop.
A Positive Feedback Loop enables a gene regulatory network to encode a “cellular memory” due to its switch-like behaviour created by two different stable states36,37. Transitions across the states (i.e. switching) can be triggered by a specific endogenous or exogenous event11,8, thus enabling the control of key decision processes such as cell differentiation and cell fate in response to specific clues38.
The switch-like behaviour of the PFL motif, however, comes at a cost: first, the PFL may considerably slow down the transcriptional response of the circuit to the triggering event24; this can be acceptable, or necessary, in some cases, but detrimental in others; the second and biggest issue is that the PFL motif increases biological noise because small fluctuations in the expression levels of the transcription factor are amplified by the self-regulatory loop. The increased cell-to-cell variability in the transcription factor concentration may cause the transcription factor level to be higher than the promoter activation treshold, inducing transition of the switch from the OFF state to the ON state, even in the absence of the triggering event36,8.
Here, we experimentally demonstrated that the presence of a miRNA suffices to solve both issues at once, by conferring phenotypic robustness to the toggle-switch, thanks to the reduction of biological noise, and by enabling the circuit to respond faster.
Therefore, the difference in robustness between the PNFL and PFL motifs can be explained by taking into account the “stochastic switching” phenomenon. Stochastic fluctuations in gene expression, due to biological noise, will induce random transitions between the two states of the toggle-switch36,39,40.
Interestingly, if the miRNA repression on the target mRNA is too strong, then the PNFL will no longer be able to function as a toggle-switch because of a loss of bistability (Supplementary Figure S4). This intuitively means that the miRNA will force the circuit to always be OFF by inhibiting the transcription factor expression. In addition, the stronger the miRNA repression, the smaller the difference between the OFF state and the ON state in terms of protein expression levels. These constraints may have contributed to the evolution of the weak transcriptional effect exerted by the majority of the known miRNAs.
A recent experimental and computational analysis investigated the properties of the incoherent feedforward loop motif (FFL) consisting a transcription factor inducing expression of both a reporter gene and a miRNA directed against the same reporter gene12. The authors demonstrated that the FFL made the reporter gene expression level insensitive to the motif-carrying plasmid copy number. Moreover, they observed that the presence of the miRNA led to a reduced variability in reporter protein expression among cells transfected with the motif-carrying plasmid. Deans et al used a small short-hairpin RNA (shRNA) to enhance the robustness and performance of an inducible mammalian genetic switch.41
Our results add further support to the role of miRNAs as noise suppressor. In addition, we demonstrated that reducing noise is essential to maintain a robust phenotype by preventing cells from unwanted stochastic switching.
We believe that our findings on the role of the miRNA explain the abundance of miRNA-mediated negative feedbacks in gene regulatory networks across species13,20. Indeed, it was previously suggested13 that miRNA-mediated NFLs may be needed to reduce intracellular noise, thus conferring phenotypic robustness to the pathways they are part of. Here, we demonstrated that indeed this is the case.
One of the best studied examples of PNFL in vivo is the E2F1/miR-17-92 network42,13,43. Here the E2F1 transcription factor, a main regulator of cell cycle, regulates its own transcription and that of the miR-17-92 cluster targeting E2F1 itself. According to our results, and as previously suggested, the role of the miRNA-mediated NFL should be of preventing stochastic activation of E2F1 leading to unwanted proliferation or apoptosis13,43.
In addition, several developmental transcription factors are regulated by positive feedback loops38. These PFLs could be frequently coupled to miRNA-mediated NFLs to prevent untimely activation of the transcription factor, which would likely be lethal for the developing embryo13. For example the muscle regulatory factor MYOD activates its own transcription44,45 as well as transcription of miRNAs predicted to target MyoD itself43,46.
Another example is provided by intestinal cell specification in the nematode C. elegans during embryonic development. This process is regulated by a positive feedback loop involving elt-2, the master regulatory gene of intestinal differentiation19. Raj et al19 demonstrated that stochastic activation of the elt-2 PFL underlies incomplete penetrance of mutant embryos, leading to pronounced phenotpyic variability.
A limitation of our synthetic network approach is that the miRNA seed sequence is perfectly complementary to the four recognition sites we engineered at the 3′ ttA_IRES_d2EYFP of the mRNA (Fig. 1a). A perfectly complementary binding site causes the miRNA to induce a catalytic-like RNA interference process, rather than the more subtle gene expression “thresholding” effect observed in endogenous miRNA binding sites47. This effect causes the miRNA-induced target mRNA repression to decrease, and eventually be completely abolished, if the target mRNA increases above a certain threshold saturating the miRNA pool47. Our results are relevant to endogenous miRNA regulation as long as the mRNA expression remains below the threshold since, in this situation, endogeneous miRNA binding sites and perfectly complementary binding sites have very similar effects.47
Our findings are a direct proof of the essential role of microRNAs in providing phenotypic robustness to a biological process. Our results are also relevant to the growing field of synthetic biology, in that they demonstrate how to synthetise robust circuits by employing miRNA-mediated feedback loops.
Methods
Construction of PNFL network in Chinese Hamster Ovary cells
The synthetic circuit was implemented in a lentiviral vector using the ViraPower Promoterless Lentiviral Gateway Expression System (Invitrogen). The pMAtTA-IRES-EGFP-WPRE vector was synthesised by GENEART. The d2EYFP encoding gene from pd2EYFP-1 (Clontech) was substituted in place of the EGFP to generate a new vector termed pMAtTA-IRES-d2EYFP. The pMAtTA-IRES-d2EYFP was recombined with the pDONR221 (Invitrogen) to yield the pENTRtTA-IRES-d2EYFP vector. The pMA-miR223-destRFP-WPRE vector48 was synthesised by GENEART. An mCherry fluorescent protein-encoding gene was substituted for the RFP, in order to generate the pMA-miR223-mCherry-WPRE. The CMV-TET promoter was amplified from pTRE2 (Clontech) by PCR and inserted into a pENTR5′-TOPO vector to obtain the pENTR5′-TOPO-CMV-TET with specific recombination sites. We performed recombination reactions between the pENTRtTA-IRES-d2EYFP/pENTR-miR223-mCherry, pENTR5′-TOPO-CMV-TET and the pLenti/R4R2/V5-DEST according to manufacturer instructions.
Cell culture and clones generation
Chinese Hamster Ovary (CHO) cells were maintained at 37 °C in a 5% CO2-humidified incubator, and cultured in α-MEM (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen), 1% L-glutammine and 1% antibiotic/antimycotic solution (GIBCO BRL).
To transduce cells with the virus produced 500,000 CHO cells were plated and incubated overnight. On the day of transduction the medium was removed and 1 mL of the virus was added to the cells together with polybrene (Invitrogen) to a final concentration of 6 μg/mL. After an overnight incubation the medium containing the virus was removed and replaced with complete culture medium containing Blasticidin (Sigma) to a final concentration of to select for stably transduced cells.
Cells were sorted for fluorescence intensity using a BD FACSAria Cell Sorting System (Becton Dickinson). d2EYFP was excited at 488 nm, and emission was detected using a 525 nm bandpass filter. Serial dilutions of stably transduced cells (up to 0.05 cells/mL) were plated in 96-well microtitre plates, and dilutions containing only one cell per well were selected. Monoclonal colonies were cultured and amplified as described, to obtain monoclonal populations24. Monoclonal CHO-PFL cell lines were transduced with the virus carrying the NFL with the same procedure. Monoclonal population of infected cells were isolated as described above to generate the PNFL cell lines.
Fluorescence Activated Cell Sorting
Cells from 35mm culture plates were trypsinised, filtered and subjected to Fluorescence-Activated Cell Sorting (FACS) analysis in a Becton Dickinson FACSAria, using excitation and emission filters described above. Each phenotypical analysis was performed in biological duplicates.
Switch-OFF and switch-ON experiments
PFL7 and PNFL 7.2 cells were plated in a 12-well culture plate in order to obtain 400,000 cells per well, and then treated (or mock-treated) with 1 μg/ml Doxycycline. Fluorescence was measured by Fluorescence-Activated Cell Sorting (FACS) analysis at 0, 6, 12, 24, 48 and 72 hrs after treatment. The Doxycycline-containing medium was replaced 48 hrs after treatment. For the switch-ON experiments, PFL7 and PNFL 7.2 cells were pre-treated with 1 μg/ml Doxycycline for 72 hrs and then plated as described above, either in medium containing Doxycycline or not (control). Fluorescence was measured by FACS at 0, 24, 48, 72 and 96 hrs after treatment. For both switch-OFF and switch-ON experiments, each FACS measurement was made on a sample of 20.000 events in duplicate.
Microfluidics and Microscopy
We used the microfluidic device for mammalian cell culture described in49. Before cell loading, the channels in the device were coated with 100 μg/ml fibronectin for 1 hour at room temperature. After coating, the fibronectin solution was removed using a vacuum manifold, and a high density cell suspension (107 cells/ml) was loaded in the dedicated inlet. PFL and PNFL cells were loaded into the cell traps at a seeding density of 10 to 20 cells per trap, and the device was placed in a cell culture incubator to allow attachment of cells. The device was then placed under a Nikon Eclipse TI-E inverted epifluorescence microscope, equipped with an incubation chamber (H201-OP R2, Okolab). The two syringes connected to the inlets were filled with untreated medium, or medium containing 1 μg/ml doxycycline. Sulforhodamine 101 (Sigma-Aldrich, Germany), a fluorescent dye, at a concentration of 1 μM was added to the medium containing doxycycline. The flow was adjusted to provide doxycycline for a given time interval (40 min, 60 min, 180 min for PNFL cells and 960 min or 1800 min for PFL cells). Bright field and fluorescence images of 3 to 20 traps per experiment were acquired with a 40× objective at intervals of 15 min. Fluorescence images were taken using FITC (for d2EYFP, excitation 460/40 nm, emission 510/50 nm) and TRITC (for Sulforhodamine 101: excitation 530/30 nm, emission 590/60 nm) filters. Images were acquired using a Peltier-cooled Andor Clara camera controlled by Nikon Instrument Software AR v.3.22.14. Image acquisition and processing was performed as described in24 (Supplementary Note 9).
Dynamical model of the PNFL
In order to model the behaviour of the PNFL motif, we develop a mathematical model of eight non linear ODEs as detailed in Supplementary Note 3. Numerical simulations were run using Matlab 2010b (Mathworks Inc.). For the parameter identification, we used the PottersWheel toolbox50 implemented in MATLAB, which is also described in24.
Stochastic models of the PFL and PNFL circuits
Following a stochastic model of gene expression30, we derived the Squared Coefficient of Variation CV2 = (σ/μ)2 for the PFL and PNFL circuits, as well as of a control circuit without any feedback (NOPFL), as described in Supplementary Note 6 and 7.
Supplementary Material
Acknowledgements
We are grateful to Graciana Diez-Rouex, Andrea Ballabio and Antonella De Matteis for their critical reading of the manuscript. We would also like to thank Nicoletta Moretti for helping with cell culture and Laura Pisapia for performing the FACS analysis. This work was supported by the Italian Institute of Technology Project SEED, by the HFSP Program Grant RGP0020/2011-DIBERNARDO, and by the Fondazione Telethon Grant TGM11SB1.
Footnotes
Competing Financial Interests The authors declare no competing financial interests.
References
- [1].Inui M, Martello G, Piccolo S. Microrna control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- [2].Alvarez-Saavedra E, Horvitz HR. Many families of c. elegans micrornas are not essential for development or viability. Current Biology. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Miska EA, et al. Most Caenorhabditis elegans microRNAs Are Individually Not Essential for Development or Viability. PLoS Genet. 2007;3:e215+. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Abbott AL, et al. The let-7 microrna family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in caenorhabditis elegans. Developmental Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li X, Cassidy JJ, Reinke CA, Fischboeck S, Ca RW. A microrna imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ebert MS, Sharp PA. Roles for micrornas in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl 1) doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- [8].Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes & Development. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chang X, Liu Z, Chen L, Wang R. Bistability and oscillations in gene regulation mediated by small noncoding RNAs. PLoS ONE. 2011;6:10. doi: 10.1371/journal.pone.0017029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li Y, Li Y, Zhang H, Chen Y. MicroRNA-Mediated positive feedback loop and optimized bistable switch in a cancer network involving miR-17-92. PLoS ONE. 2011;6:e26302. doi: 10.1371/journal.pone.0026302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yosef N, Regev A. Impulse control: Temporal dynamics in gene transcription. Cell. 2011;144:886–896. doi: 10.1016/j.cell.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bleris L, et al. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Molecular Systems Biology. 2011;7 doi: 10.1038/msb.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tsang J, Zhu J, Oudenaarden AV. Microrna-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–67. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kitano H. Biological robustness. Nature Reviews Genetics. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- [15].Arjun R, van Oudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nature Reviews Genetics. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- [17].Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. Journal of Molecular Biology. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- [18].Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nature genetics. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- [19].Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Martinez NJ, et al. A c. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes and Development. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lim WA, Lee CM, Tang C. Design principles of regulatory networks: searching for the molecular algorithms of the cell. Molecular cell. 2013;49:202–212. doi: 10.1016/j.molcel.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotech. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- [24].Siciliano V, et al. Construction and modelling of an inducible positive feedback loop stably integrated in a mammalian cell-line. PLoS Comput Biol. 2011;7:e1002074. doi: 10.1371/journal.pcbi.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in escherichia coli. Nature. 2000;403:339–42. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- [26].Maeda YT, Sano M. Regulatory dynamics of synthetic gene networks with positive feedback. Journal of Molecular Biology. 2006;359:1107–1124. doi: 10.1016/j.jmb.2006.03.064. [DOI] [PubMed] [Google Scholar]
- [27].Becskei A, Séraphin B, Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–35. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kramer BP, et al. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- [29].Bennett MR, Hasty J. Microfluidic devices for measuring gene network dynamics in single cells. Nature reviews. Genetics. 2009;10:628–38. doi: 10.1038/nrg2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Singh A. Negative feedback through mrna provides the best control of gene-expression noise. NanoBioscience, IEEE Transactions on. 2011;10:194–200. doi: 10.1109/TNB.2011.2168826. [DOI] [PubMed] [Google Scholar]
- [31].Osella M, Bosia C, Cora’ D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput Biol. 2011;7:e1001101. doi: 10.1371/journal.pcbi.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- [33].Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Accurate prediction of gene feedback circuit behavior from component properties. Molecular Systems Biology. 2007;3:143. doi: 10.1038/msb4100185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Petersen M, Wengel J. Lna: a versatile tool for therapeutics and genomics. Trends in Biotechnology. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- [35].Chau AH, Walter JM, Gerardin J, Tang C, Lim WA. Designing synthetic regulatory networks capable of Self-Organizing cell polarization. Cell. 2012;151:320–332. doi: 10.1016/j.cell.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Acar M, Becskei A, Oudenaarden AV. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–32. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- [37].Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Prediction and measurement of an autoregulatory genetic module. Proceedings of the National Academy of Sciences. 2003;100:7714–7719. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mehta P, Mukhopadhyay R, Wingreen NS. Exponential sensitivity of noise-driven switching in genetic networks. Phys. Biol. 2008;5:026005+. doi: 10.1088/1478-3975/5/2/026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Walczak AM, Onuchic JN, Wolynes PG. Absolute rate theories of epigenetic stability. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18926–18931. doi: 10.1073/pnas.0509547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- [42].O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- [43].Sylvestre Y, et al. An e2f/mir-20a autoregulatory feedback loop. The Journal of Biological Chemistry. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- [44].Fong AP, et al. Genetic and epigenetic determinants of neurogenesis and myogenesis. Developmental cell. 2012;22:721–735. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Thayer MJ, et al. Positive autoregulation of the myogenic determination gene myod1. Cell. 1989;58:241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- [46].Gagan J, Dey BK, Dutta A. MicroRNAs regulate and provide robustness to the myogenic transcriptional network. Current Opinion in Pharmacology. 2012 doi: 10.1016/j.coph.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mukherji S, et al. Micrornas can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Amendola M, et al. Regulated and multiple miRNA and siRNA delivery into primary cells by a lentiviral platform. Molecular therapy : the journal of the American Society of Gene Therapy. 2009;17:1039–1052. doi: 10.1038/mt.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kolnik M, Tsimring LS, Hasty J. Vacuum-assisted cell loading enables shear-free mammalian microfluidic culture. Lab on a chip. 2012 doi: 10.1039/c2lc40569e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Maiwald T, Timmer J. Dynamical modeling and multi-experiment fitting with potterswheel. Bioinformatics. 2008;24:2037–2043. doi: 10.1093/bioinformatics/btn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.