Abstract
The XCEDE (XML-based Clinical and Experimental Data Exchange) XML schema, developed by members of the BIRN (Biomedical Informatics Research Network), provides an extensive metadata hierarchy for storing, describing and documenting the data generated by scientific studies. Currently at version 2.0, the XCEDE schema serves as a specification for the exchange of scientific data between databases, analysis tools, and web services. It provides a structured metadata hierarchy, storing information relevant to various aspects of an experiment (project, subject, protocol, etc.). Each hierarchy level also provides for the storage of data provenance information allowing for a traceable record of processing and/or changes to the underlying data. The schema is extensible to support the needs of various data modalities and to express types of data not originally envisioned by the developers. The latest version of the XCEDE schema and manual are available from http://www.xcede.org/
Keywords: XML, Schema, Database, Biomedical Technology
I. INTRODUCTION
Managing and sharing the increasingly large and diverse datasets that are generated in high-throughput and multi-site human clinical and imaging studies requires substantial infrastructure. Large-scale collaborative studies offer significant technical challenges, requiring the collection, storage, and dissemination of large amounts of data originating from a variety of data collection equipment and using data formats that may not be standardized even within a particular site. Overcoming these challenges is paramount for the success of research projects that rely on data shared between otherwise independent research groups.
XCEDE is an XML (Extensible Markup Language) (Bray, Paoli et al. 2006) data schema designed to provide a standard storage and communication mechanism for the many types of data generated by a scientific study. Using the structure specified by XCEDE facilitates the exchange of experimental data between tools, such as data analysis software, databases and web services. The schema is specified using the W3C standard XML Schema 1.0 language (Thompson, Beech et al. 2004).
The power of XML in typical usage derives mainly from three simple syntactic components: user-named elements that can contain other elements, user-named attributes that are attached to elements, and namespaces (Bray, Hollander et al. 2006) that provide a global context in which to interpret elements or attributes. These constructs, as well as widely-used interfaces such as DOM (Hors, Hégaret et al. 2004) and SAX,1 and add-on technologies like XPath (Clark and DeRose 1999) and XSLT (Clark 1999), enable the development of data representation languages and software tools for an increasingly wide range of applications.
XCEDE was originally designed in the context of neuroimaging studies and is one component of the BIRN (Biomedical Informatics Research Network) (Keator, Grethe et al. 2008) infrastructure. XCEDE complements the BIRN Human Imaging Database (HID) (Keator, Grethe et al. 2008) and the eXtensible Neuroimaging Archive Toolkit (XNAT) (Marcus, Olsen et al. 2007), extensible systems for the management, discovery, retrieval, and analysis of clinical and biomedical imaging data. This close coupling allows for an interchangeable source-sink relationship between data management systems and XML files. The XML files serve as a standardized mechanism for transport and interchange of experimental data, facilitating import/export between heterogeneous databases, development of specialized web services, local storage of experimental information within data collections, and creation of human and machine readable descriptions of the actual data. Within the BIRN project for example, a multi-center project to collect functional MRI (fMRI) and associated clinical data on schizophrenic and control subjects from over a dozen institutions has served as an important use case for XCEDE.
Despite its origins in the neuroscience community, XCEDE is designed to be extensible and applicable to other biomedical fields. XCEDE components are fairly generic, and represent concepts that occur widely in various scientific disciplines. For those concepts that are not adequately expressed using the generic XCEDE core framework, any XCEDE element may be extended with relevant data items using type derivation within XML Schema.
II. BACKGROUND
XML, a text-based format, has seen much use in the scientific community to store structured data. Version 1 of XCEDE (Keator, Gadde et al. 2006), introduced in 2006, was in itself a merging of concepts and structures introduced in earlier independent XML-based projects such as the BIAC XML Header (BXH) and an XML schema originally developed for the fMRI Data Center. XCEDE in its current form (version 2) represents a complete restructuring, employing reusable abstract data types, consistent schema design rules, additional components such as analysis and terminologies, and a formally-defined experiment hierarchy. In particular, rather than exposing the relationships between levels of the experiment hierarchy through nested XML elements, XCEDE 2 now exposes their relationships (as with the other major XCEDE components) through the use of a flat list of elements linked by ID fields, making it easier to integrate with existing relational databases and also allowing applications to stream arbitrary subsets of the hierarchy structure as needed. As such, the current version of XCEDE reflects many of the lessons learned from the initial deployment of XCEDE within the BIRN.
The CCLRC Scientific Metadata Model (CSMDM) (Sufi and Mathews 2004) provides a general model for storing scientific metadata, and was designed to support a common data portal indexing the data of several independent research centers, each representing data in their own structures. CSMDM, which has been implemented in XML Schema form, is then a lingua franca to represent the underlying experimental structure and metadata of each center’s studies, using a hierarchical experimental model similar to the one used by XCEDE, with the notable exception of a subject (i.e. experimentee) level. This limitation for some scientific disciplines is noted by (Lohrey, Killeen et al. 2009) in their review of several existing scientific metadata models (including an earlier version of XCEDE), in designing their own XML-based object models and data management services as a Framework for “subject-centric” research.
The HL7 Clinical Document Architecture (CDA) (Dolin, Alschuler et al. 2006) is an XML-based specification that facilitates the exchange of medical records using the HL7 reference terminologies. It shares with XCEDE the use of existing terminologies to annotate data. XCEDE’s focus is quite different, being particularly suitable for data and metadata associated with large-scale automated biomedical image data analysis, whereas the CDA is tailored more towards representing an entire health service workflow (both clinical and administrative). However, the use of common or related terminologies enables integration of data coming from these diverse sources.
The Clinical Data Interchange Standards Consortium (CDISC2) has developed a number of standards to support patient care and clinical research. The group’s Operational Data Model (ODM3) overlaps substantially with XCEDE and a direct transformation between some or all of the two standards may be possible. Key differences, however, are apparent in their relative emphasis on different aspects of biomedical data. The ODM provides substantial support for questionnaire-based clinical trials, whereas XCEDE’s origin in the biomedical imaging community has led to a greater emphasis on binary file representation and post-processing.
XCEDE can be layered atop DICOM4, the radiology standard for image communication. Where DICOM describes image acquisition parameters and attributes of particular imaging devices, XCEDE adds experimental context about projects, investigators, and non-imaging assessments together with protocol prescriptions that relate and constrain them. Although DICOM supports proprietary extensions, it is not an XML standard and thus lacks the general purpose extensibility and wealth of off the shelf tools available to users of XCEDE.
A major component of XCEDE (and its earliest incarnations) is an interface to binary data streams stored in external files. Several other projects target this aspect of data analysis, including the XML-based Extensible Scientific Interchange Language (XSIL) (Blackburn, Lazzarini et al. 1999), the Binary Format Description (BFD) Language (Myers, Chappell et al. 2003) (an extension of XSIL), the Medical Reality Markup Language (Gering, Nabavi et al. 1999) (MRML), and the non-XML-based Nearly Raw Raster Data (NRRD) format5. XCEDE and NRRD, furthermore, formalize certain types of metadata associated with the streams, e.g. to place image data in a particular coordinate space. The binary data component of XCEDE encompasses all these goals, and goes a step further, handling idiosyncracies of data ordering produced by some data collection equipment (e.g. Siemens DICOM “mosaic” images, which encode 3-D images into a 2-D plane), and, importantly, placing these data streams in context of a scientific experiment hierarchy whose components can be individually addressed and linked with the binary data.
The Annotation and Image Markup (AIM) Project (Rubin, Mongkolwat et al. 2008) of the National Cancer Institute’s Cancer Biomedical Informatics Grid (caBIG) (Fenstermacher, Street et al. 2005) is building an ontology and schema for image annotation. The effort provides a comprehensive substantial data model for addressing the radiologist’s need to annotate findings when performing qualitative reads of clinical images. In addition to the AIM XML format, the project includes applications to transform AIM documents to DICOM structured reports (Clunie 2000) and HL7 CDA XML documents. AIM and XCEDE are largely complementary in their aims, with XCEDE focusing more directly on data acquisition and analysis. We therefore expect that as each project continues to develop, key integration points will be identified.
III. XCEDE STRUCTURE AND COMPONENTS
An XCEDE dataset is a repository of data (extracted from data collection equipment, paper-based forms, databases, etc.) represented as a collection of one or more XML documents, each of which validates against the XCEDE schema. An XCEDE document consists of a root <XCEDE> element, which stores an optional list of annotations and a revision history (encoding revision time and creator), and which encapsulates any number of top-level XCEDE components which collectively represent a full XCEDE dataset or a subset thereof. The XCEDE specification does not prescribe any particular mechanism by which these documents are located, stored, grouped, or linked, though XCEDE allows certain XML elements to link to other target elements and to optionally specify URIs as hints about the location of the documents containing these targets.
For example, a given XCEDE dataset may be stored as a single XML document, or a collection of files in a single directory on a file system, or may be distributed within a hierarchical directory structure (which may or may not reflect the semantic structure of the data within), or may be stored within a database accessible by query through a web interface. However, the semantics of the dataset should be fully reflected in the XML representation, and should not be dependent on how the dataset is stored.
All major XCEDE elements are associated with a type in the schema, with the same name as the element, but with the addition of the suffix ‘_t’. For example the structure of the <project> element is defined by the project_t type. Several types are labeled with the abstract_prefix, indicating that they are not intended to be used directly as the content model for an XML element; however, types derived from these abstract types may be used to define such elements.
XML elements are optionally associated with a namespace, which is a URI that provides a context in which to interpret the elements. All XCEDE version 2 elements are defined within the http://www.xcede.org/xcede-2 XML namespace, allowing schema users to extend the schema with elements in a different namespace. Because the meanings of XCEDE elements are defined only within the context of the appropriate XCEDE namespace, they are protected from schema extensions whose elements in other namespaces have the same names as existing XCEDE elements.
XCEDE consists of several major components:
Several experimental hierarchy levels, such as “project”, “subject”, “visit”, etc., which can be annotated with level-specific metadata.
External documents and references to binary data in external files are represented by resources.
Protocols are templates for a time-ordered sequence of steps, and can be referred to by data that follows the protocol (such as a series of MR scans, or a clinical assessment).
Storage of structured raw data within the XML itself. Time-based event data and clinical assessment values are two types of data supported by built-in XCEDE types.
A generic data type to store the results of a data analysis, also supporting links to other XCEDE elements representing the inputs and other output data (if any).
Hierarchical catalogs store simple name-value pair data, which can be used for messaging or for other application-specific uses.
Data processing history is stored in provenance elements.
Terminology annotations, allowing users to further describe data by linking to terms in standard ontologies.
Each of these components are described in further detail below.
A. Experiment Hierarchy
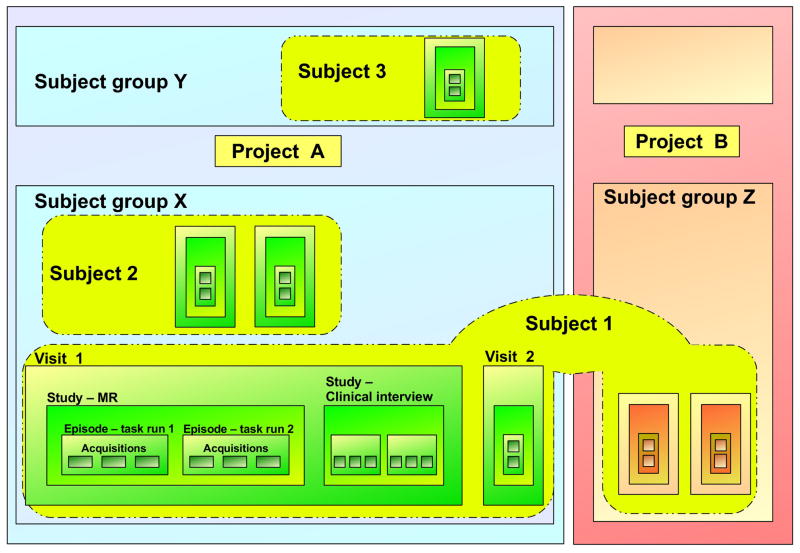
The examples in the rest of this document are drawn from a simple fictitious case study constructed to demonstrate several XCEDE features. The organization of this case study is illustrated in Figure 1. As shown, the XCEDE experiment hierarchy consists of several levels representing divisions of experiment data at various granularities. One of XCEDE’s goals is to apply to diverse research frameworks. As such, the linking mechanism between levels is flexible enough to support the omission of levels if the schema user finds them unnecessary. Figure 2 shows how the hierarchy illustrated in Figure 1 might be represented in XCEDE.
Figure 1. XCEDE Hierarchy Levels.
A conceptual representation of the levels of the XCEDE hierarchy, in the context of a research study with clinical and imaging data.
Figure 2. XCEDE Hierarchy example.
Hierarchy “level” elements are bold-faced for clarity. Note that level elements include attributes linking the elements to related elements in the hierarchy (e.g. ‘subjectID’ in ‘visit’ links to ‘subject’). The linked item is not required to be defined within the same document.
Elements at each level may contain level-specific “info” elements (like <subjectInfo>), whose schema types may be extended to store experiment-specific or data modality-specific (MR, clinical, etc.) metadata. As a model for similar extensions, some magnetic resonance imaging (MRI) -specific acquisition parameters are explicitly codified within a separate schema that provides extensions to the generic “acquisition info” type in the core XCEDE schema. The benefit of using an extension (as opposed to using a generic core XCEDE element) in this particular case is that the mrAcquisitionInfo_t type embeds terminology/definition information for each element directly into the schema using fixed values, so dataset creators do not have to explicitly add these “metadata about the metadata” themselves to their datasets (see “Terminologies”).
In the typical intended usage, a project is the top-level division of experiment data, and represents a research project which collects and analyzes data from one or more subjects which are divided (within the project) into subject groups. A subject may be a member of multiple research projects, and it is the subject group that maintains and distinguishes the mappings between subjects and research projects. In this example, there are two projects A and B, both of which share a subject 1, who participates in project A as a member of its subject group X.
A visit may represent a subject’s appearance at an experiment site (for collaborative projects, this could be the institution or lab at which the data is being collected or analyzed). A visit may be further subdivided into one or more studies, each consisting of one or more data collection episodes. The example shows subject 1 involved in a clinical interview followed by an fMRI scanning session.
Visit and study are intended to represent common divisions found in many experimental designs, in which a research subject undergoes relatively distinct experimental tests (MRI scan and clinical evaluation in this example) on a single site visit. An episode represents the time interval during which data is collected by one or more instruments. Each set of data collected over this time interval are represented by an acquisition. Multiple acquisitions within an episode represent data acquired simultaneously over the time interval represented by the episode. So, as in the above example, an episode in an fMRI study may encapsulate the acquisition of a time-series of volume images from an MR scanner, as well as other acquisitions of behavioral or physiological data; all these (simultaneously collected) data would be stored as individual acquisitions and stored as part of the same episode.
XCEDE elements describing the experimental hierarchy, as with all XCEDE data, may be created on-the-fly by a tool or service backed by data in another form, or can be stored in XML files. For data stored in XML files, the BXH/XCEDE Tools6 provides a tool xcede2edit to easily annotate, from the command-line, any relevant XCEDE elements with the appropriate experiment level linking IDs. Thus lower-level applications that are typically unaware of the global experimental context can create XCEDE data and leave it to higher-level tools to insert the resulting data into the hierarchy.
B. Resources
XCEDE elements can associate data with several types of external entities, collectively known as resources. These entities, derived from the base type resource_t, are categorized as either information resources or data resources.
1. Information Resource
Information resources are references to documents or publications that may provide additional insight on the associated data items, but are not expected to provide computer-readable data themselves. Lists of information resources might include pointers to equipment manuals, peer-reviewed publications, websites or any resources that can be described by the 15 fields of the Dublin Core Metadata Element Set (Kunze and Baker 2007) via the derived type dcResource_t. XCEDE data browsers could, for example, represent these information resources as clickable links for those users who desire more information.
2. Data Resource
Data resources are pointers to actual files or remotely-addressed datasets (using URIs) from which actual data and metadata may be extracted by software tools. The binary data resource component is one example of a data resource, and provides a generic interface to a binary data stream stored in one or more external files. XCEDE provides multiple layers of derived types to store more specialized information about the binary data. These derived types can be used, for example, to represent multi-dimensional data and to further map the multi-dimensional data within an arbitrary coordinate system. Wrapping raw binary data streams with an XCEDE binary data resource makes the data more accessible, as software tools that implement this component are insulated from having to understand the peculiarities of various binary data formats.
The binary data resource shown in Figure 3 is an example of a mapped, dimensioned, binary data resource as created using the BXH/XCEDE Tools, pointing to image data collected during subject 1’s fMRI scanning session. Not only does the resource specify the data type, byte order, and locations of binary data within files, it provides one temporal dimension and three spatial dimensions for the data which are mapped to a coordinate space (in this case, a RAS patient coordinate space as typically used in MR imaging).
Figure 3. Binary Data Resource example.
The children of the <resource> element are shaded to indicate the (ancestor) type providing those elements. Note that the element name is “resource” but the xsi:type attribute indicates that the content is of the derived type mappedBinaryDataResource_t, which inherits from and adds to the content model of its parent types.
C. Protocols
The protocol_t type provided with XCEDE describes an ordered, perhaps hierarchical, sequence of experimental steps. A protocol defined using this structure can be used to validate, after the fact, that a particular series of experimental procedures were followed, including order relationships and time constraints. The protocol definition consists of ordered steps, each step consisting of items. Items are specified using either an <itemRange> tag allowing for the precise definition of valid ranges (e.g. the field-of-view for an MRI data collection protocol shown in Figure 4) or <itemChoice> tags providing facilities for specifying options in the protocol (e.g. specifying the pulse sequence option for an MRI data collection in Figure 4). For example, a schema user may wish to indicate that a given clinical study must collect a particular list of assessments no later than 3 weeks after a particular MRI scan or specify a time window for data collection. The order of questions in those clinical assessments can also be represented in the protocol_t type (see “Assessment definitions”).
Figure 4. Protocol example.
The two components in this (reduced) example are a clinical assessment (Edinburgh Handedness Inventory) and a T1-weighted MR scan, with constraints on timing between the components and on acquisition parameters.
Figure 4 shows a simple instance of type protocol_t representing the desired sequence of steps for the first visit in project A. This protocol requires that a clinical assessment based on the Edinburgh Handedness Inventory (Oldfield 1971) should be collected at the very beginning of the protocol, and an MRI scan (T1) with given acquisition parameters that should be acquired ideally the same day, but no later than 7 days after the clinical assessment.
1. Assessment definitions
In XCEDE, the formal definition of an assessment and the cataloging of the actual data values collected for that assessment are specified in different parts of the schema. The formal description of the assessment questions and possible answer choices are specified in the protocol section of the schema using the type protocol_t whereas the actual assessment data for an acquisition are stored using the assessment_t type (see “Assessment data”).
Assessments that are composed of multiple items (like the Edinburgh Handedness clinical assessment described above) can be formally described using the protocol component of XCEDE, where each item is annotated with the text that defines the item (in a clinical assessment, this might include the text of the question that would be asked of the patient).
Figure 5 shows an individual item that might have been added to the example in Figure 4 to describe one of the assessment questions. These descriptions might be used to give hints to a user interface for assessment data entry.
Figure 5. Assessment Item Definition example.
The upper section shows how the XCEDE assessment item (shown in the lower section) might be presented in a form. The highlighted portion shows one of five valid options for the edhandinv_writing item (a stored instance of this particular choice is shown in Figure 6).
D. Data
Whereas XCEDE data resources point to data stored outside the XCEDE framework, data can also be represented in XML and stored within an XCEDE dataset itself, using the top-level <data> element of type abstract_data_t. XCEDE provides two built-in derivations of this type to support assessment and event data (described below).
1. Assessment data
The type assessment_t provides fields to store the values collected for each item in an assessment, and also provides ID attributes to link the data values to the individual items in the formal assessment description contained in a protocol description block as discussed in “Assessment definitions”. An example of this is shown in Figure 6. Such data is generated, for example, by handheld tablet software and sent via a web service call for import into the Human Clinical and Imaging Database (HID) (Ozyurt, Keator et al. 2010).
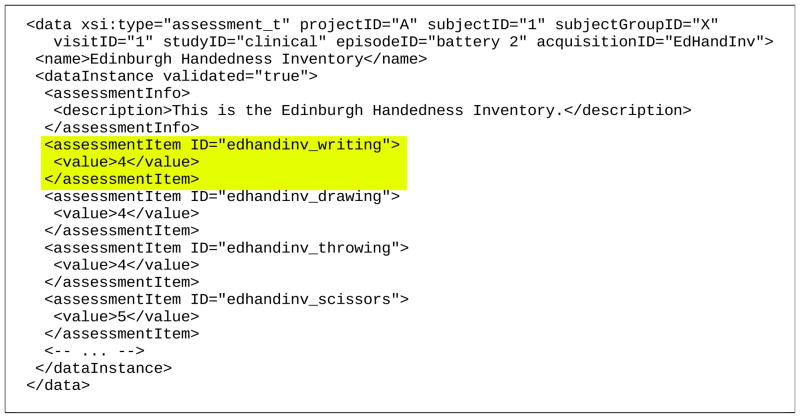
Figure 6. Assessment Data example.
This fragment encodes values from a portion of an assessment based on the Edinburgh Handedness Inventory. The highlighted assessment item labeled edhandinv_writing has a value of 4, which corresponds to the item with itemCode 4 in the edhandinv_writing assessment item definition (Figure 5), which represents “Mostly Right”.
2. Events
Events in XCEDE are merely time intervals annotated with arbitrary metadata. This component can be used to represent any metadata whose proper interpretation requires that it be associated with a particular interval in time. This component has been used to store behavioral data (e.g. stimulus/response output from stimulus presentation tools) and time-varying statistics from fMRI image quality assurance tools. XCEDE events support arbitrary numbers of descriptive fields (<value> elements), and are therefore much more expressive than standard multi-column (onset, duration, weight) timing files such as those used by popular fMRI analysis packages. The descriptive fields enable users to fully annotate events by their innate characteristics at the time of creation. During analysis, simple or sophisticated queries (perhaps using XPath or some other XML query language) can extract the timing of events of interest, including those for analysis models not anticipated at creation time.
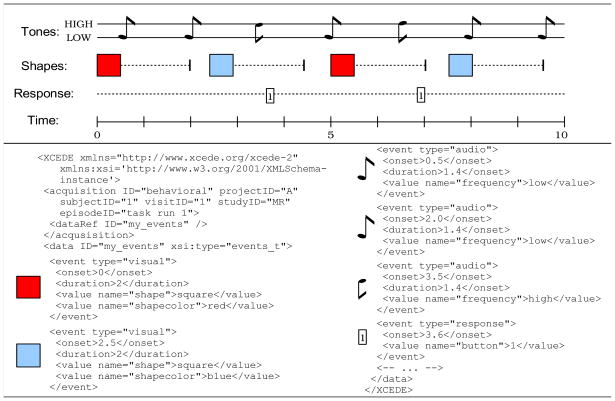
Figure 7 illustrates how stimulus/response data for the task performed by subject 1 is translated into an events_t structure as might be generated from the output files of stimulus presentation software using tools like showplay2xml, eprime2xml, presentation2xml, etc. from the BXH/XCEDE Tools suite. Each audio tone stimulus and each subject response is represented as a separate event, and is annotated with <value> elements describing all relevant characteristics about the stimuli and responses.
Figure 7. Events example.
This XCEDE fragment records audio and visual stimuli, and subject response data (several events are not represented in the XML due to space constraints).
E. Analysis
The Analysis component is a container used to document the collected output and/or results from an analysis or processing of data. An analysis is composed of the “inputs” (i.e. the files and parameters used in an analysis or processing of data), a list of the application(s) or method(s) used in the analysis (provenance), and the resultant data (i.e. values and output files).
An analysis may contain one or more measurement groups, which store information and data related to the outcome of an analysis. This allows for the annotation and documentation of measurements; for example, indicating that a measurement is the volume of a particular region of the brain (e.g. hippocampus). These annotations may point to standard terminology lists, such as brain atlases, or those used by specific applications (see Figure 8 for an example of how one might represent the output of the FreeSurfer brain segmentation application (Fischl, Salat et al. 2002)).
Figure 8. Analysis example.
FreeSurfer segmentation statistics represented in XCEDE. Each measurement group contains observations in the context of an anatomical entity (highlighted) representing the objects or concepts referenced by the given term IDs in the given nomenclature (in this case both entities are described by links to FreeSurfer and NeuroLex terminologies).
F. Catalogs
Catalogs in XCEDE provide a generic structure with simple name-value lists and optional references to local or remote data or information resources (see “Resources”). Catalogs are recursive in that they can contain a list of other catalogs.
Catalogs are useful in a number of contexts. A catalog could be created, for example, to point to all of the acquisition resources (e.g. image data) associated with an episode. These catalogs could be contained within a parent catalog that represents each of the episodes in an MR study. Catalogs could also be used to represent the various resources generated as part of an analysis or uploaded and tagged by users. Catalogs are suitable for representing application-specific data that lies outside the XCEDE specification, such as to store working state between sessions (e.g. a list of studies that were open for browsing). Figure 9 shows how a subset of MR image segmentation and analysis might be referenced from within a catalog.
Figure 9. Catalog example.
A file listing annotated with metadata, such as data format identifiers.
Catalogs can be sent independent of the parent content to client applications that choose not to support the full XCEDE specification. In addition, because catalog documents can be quite large when they contain thousands of entries (for example, when pointing to individual DICOM files from a fMRI acquisition), separating the catalogs from their parents provides more efficient access. Alternatively, catalogs can serve as proxies for the actual data; for example, the simple structure of the catalog lends itself well to messaging, where a web-service may provide query results in a catalog, including only summaries of the requested data, but with links that can be returned to the same web service to provide more detailed data.
G. Provenance
Various elements in XCEDE refer to data or data resources that were generated from other input data by processing tools. Data provenance refers to tracking the origins of derived data and the processing steps that resulted in the output data. Besides being informative, provenance information is intended to allow a user to recreate the processing stream and replicate the output data using the same tools and inputs. Provenance information could either be written by the software tools themselves, or could be extracted by wrapper scripts that write the provenance information on the tool’s behalf (the likely scenario for external or legacy applications); see Figure 10 for an example of processing history converted from FreeSurfer segmentation output files. This simple example represents a single-step analysis, but an arbitrary number of <processStep> elements can be used to represent more complex pipelines. Provenance information can include the name of the software tool, the list of arguments passed to the software tool, the person who was initiating the processing, which machine it was run on (and the architecture of that machine), version control information for the software tool, and a global package name.
Figure 10. Provenance example.

One component of a potential multi-step pipeline.
H. Terminologies
The terminology component allows XCEDE elements to add meaning to their content by linking to terms in a terminology. There are several sources for explicit term lists, such as files that document the conventions used by a particular software tool, published atlases such as the Talairach brain atlas (Talairach and Tournoux 1988) or ontologies such as NeuroLex (previously BIRNLex, https://xwiki.nbirn.org:8443/xwiki/bin/view/%20BIRN-OTF-Public/Home), a lexically enhanced ontology designed for use in large-scale annotation of experimental results for the purposes of enabling ontology-driven data federation and knowledge discovery within the BIRN. The objects or concepts in these terminologies usually map a term ID and a label for human consumption to a definition (which, particularly in ontologies, may itself reference other terms). Content producers can associate XCEDE elements with the definition of a term in a terminology by providing a minimum of two attributes: nomenclature (which names the terminology) and termID. Additional attributes provide additional meta-information for this term, such as additional labels from the terminology (preferredLabel), labeling items for visualization (abbreviation) or to provide a textual representation of a path to the term within the ontology (termPath), if available.
The analysis results in Figure 8 are each annotated with references to the terms listed in a FreeSurfer segmentation colormap lookup table and NeuroLex. By explicitly listing the terms from both these sources in an <entity> element, the producer of this data asserts that these observations are made on the same type of object that FreeSurfer and NeuroLex assign to their respective term IDs.
Figure 11 shows how the terminology component can be used to provide automatic annotation of XML data by specifying the terminology attributes as fixed attributes in the schema itself. Schema-aware XML readers will read the XML data as if the fixed attributes had been present in the original input. Not only do these automatic annotations provide convenience for producers (who need only use the specified elements, like <tr>) and consumers (who see a richly annotated input dataset), they also serve as formal documentation for the elements themselves, as they refer to a pre-existing terminology (in this case the DICOM data dictionary and information object definitions) mapping objects to detailed definitions.
Figure 11. DICOM terminology example.
In this example, the DICOM specification is used as a terminology source via fixed attributes in the schema. Top example is the raw XML instance. Bold strings in the lower example show the fixed attributes (from the schema) that might be automatically added by schema-aware XML readers.
IV. DISCUSSION
A. Extending the schema
XCEDE is specified using the W3C standard XML Schema 1.0 definition language. One of the advantages of using XML Schema is its built-in support for explicit scoping of element names through namespaces, and fine-grained additions to the schema through type extension.
XML elements all have an URI called its namespace (which may be empty). A namespace allows common, short names to be used for XML elements, but ensures that they are not confused with other similarly-named elements in other namespaces. By specifying a non-empty namespace N for an element <X>, a producer of XML content asserts that this element <X> is not the same element <X> associated with any other namespace M. All elements defined in the XCEDE schema use the namespace http://www.xcede.org/xcede-2 and the meanings ascribed, for example, to the <events> element (described previously) apply only to those <events> elements with the XCEDE namespace. In addition, XCEDE uses language constructs within XML Schema to allow XML producers to add arbitrary XML elements in non-XCEDE namespaces to select XCEDE elements. Because the new elements are not in the same namespace, namespace-aware XML applications cannot confuse them with existing XCEDE elements, and applications that only read XCEDE data can safely ignore the externally-defined elements.
XCEDE provides several generic containers for many types of data. In cases where the provided generic types do not meet their needs, users may add elements of their own choosing using XML Schema type extension. For example, XCEDE provides an MR-specific acquisition info element (shown previously) that stores common MR acquisition parameters. This uses the XML Schema extension method to extend the XCEDE type acquisitionInfo_t with elements based on fields listed in the DICOM MR imaging module, creating a new type mrAcquisitionInfo_t.
B. Versioning
XCEDE is meant to evolve over time in order to encompass new types of data and to provide necessary adjustments or refinements to existing components. The developers of XCEDE have implemented several strategies to lessen the impact of schema updates on existing XCEDE-aware applications.
The current release of the schema is version “2.0”, with 2 being the major version number and 0 being the minor version number. Every XCEDE file supports a version attribute, indicating the version of the schema the file is expected to support. Most schema releases will involve improvements and extensions that do not significantly change the core structure of the XCEDE dataset, and each of these releases will increase the minor version number. Minor version updates to the schema are expected to be backward-compatible with (i.e. be able to validate) XCEDE datasets that were generated using earlier schemas with the same major version number. So, an XML file that validates against version “2.1” of the schema is expected to also be a valid “2.X” file, where X is greater than 1. If future schema development results in substantial changes in structure and functionality that are incompatible with earlier XCEDE datasets, it will be released with an increment in the major version number.
The namespace of all XCEDE version 2 elements is http://www.xcede.org/xcede-2, and will not change for minor version changes. The implication and intent of this strategy is that elements within the XCEDE version 2 namespace will retain their meanings through minor revisions.
Due to XML Schema 1.0’s strict restrictions on “wildcard” content, the XCEDE schema is not forward-compatible, i.e. an earlier schema is not likely to validate (using XML Schema) an XCEDE dataset generated using a future version of the schema. However, XCEDE readers can easily be made “future proof” by verifying the major revision number attribute in the XCEDE dataset, reading only elements that they understand, and ignoring unexpected content (such as that coming from later versions of the schema). Likewise, XCEDE writers should write content that strictly follows the version of the schema specified in that version attribute. If multiple schemas validate the dataset, XCEDE writers can be especially helpful by specifying the earliest version of the schema with which the file validates. This pseudo-contract between producers and consumers allows for the schema to evolve to meet future needs without rendering existing applications obsolete.
C. Validating instance documents
Because the XCEDE Schema is specified using XML Schema, many tools are available to validate that XCEDE instance documents follow the prescribed structure. Validation can be used as a post-processing step by XCEDE producers, and/or as a pre-processing step by XCEDE consumers, to ensure that generated documents are readable by XCEDE-aware applications. Though XML Schema is sufficient to check the “syntax” of an XML document, it lacks intuitive sophisticated methods for validating the content of XML elements, nor is it capable of expressing validation rules that are conditional on the presence or content of related elements in the XML instance document. For these types of validation, we recommend content-aware XML validation methods, such as Schematron (ISO/IEC 2006). This gives users of XCEDE the power to design a specific data policy for a project. For example, one might use a content-aware validator to check that every subject in the “schizophrenic” subject group had a given list of assessments. In fact, protocol validation (i.e. matching a <protocol> element to a subtree of an XCEDE experiment hierarchy) could conceivably be done entirely within Schematron.
D. Web services
Though XCEDE serves as the backbone for databases and software applications to import and export experiment data, there is scope for a technology that allows a client to query a server for subsets or summaries of the XCEDE dataset. To address this need, we are developing a companion specification, XCEDE Web Services. A server application that follows this specification will support a standard set of functions that allows client applications to query the dataset and receive XCEDE data in return. This web service is assumed to front a dataset (perhaps stored in a relational database, perhaps merely in raw XML files) that can be represented in XCEDE, and so the functions and parameters can leverage XCEDE structures and concepts in providing a generic and extensible query interface.
SUMMARY.
XCEDE is an extensible data exchange format for experimental data. The various components of XCEDE can be used individually or in concert to fully describe the structure of a scientific experiment and store raw and derived data generated by the experiment. These components cover aspects of data collection or analysis common to many scientific disciplines, including time-locked (event) metadata, experiment hierarchies, binary data interfaces, terminologies, and more. Several components of XCEDE are in active use by the fBIRN consortium, both as a file format and to facilitate network exchange of structured experimental data with research databases.
Acknowledgments
The design of XCEDE is informed by valuable input from external collaborators and BIRN-affiliated users of earlier versions of XCEDE. Many thanks to NITRC for hosting development sandboxes for XCEDE and supporting tools.
This research was supported by the National Institutes of Health (NIH) through the following NCRR grants to the Function BIRN [U24-RR021992], Morphometry BIRN [U24-RR021382], BIRN Coordinating Center [U24-RR019701], Neuroimaging Analysis Center (NAC) [P41-RR013218] and Biomedical Informatics Research Network (1 U24 RR025736-01), and the following NIH Roadmap for Medical Research grant to the National Alliance for Medical Imaging Computing (NAMIC) [U54-EB005149].
Footnotes
INFORMATION SHARING STATEMENT
The latest version of the XCEDE schema, manual, examples, tools, and links to an email discussion list are available from http://www.xcede.org/. Collaborative schema development is supported through a Subversion (SVN) repository hosted at NITRC (http://www.nitrc.org/projects/xcede/). Software tools to write and edit XCEDE datasets are also available from NITRC at http://www.nitrc.org/projects/bxh_xcede_tools/.
Contributor Information
Syam Gadde, Brain Imaging and Analysis Center, Duke University, Durham, NC.
Nicole Aucoin, Surgical Planning Laboratory, Brigham and Women’s Hospital, Boston, MA.
Jeffrey S. Grethe, Center for Research in Biological Systems, University of California, San Diego, La Jolla, CA.
David B. Keator, University of California, Irvine, CA.
Daniel S. Marcus, Washington University School of Medicine, St. Louis, MO.
Steve Pieper, Isomics, Inc., Cambridge, MA.
FBIRN, MBIRN, BIRN-CC, Biomedical Informatics Research Network, NIH-NCRR, Bethesda, MD, www.nbirn.net.
References
- Blackburn K, Lazzarini A, et al. XSIL: Extensible Scientific Interchange Language. 7th International Conference of High-Performance Computing and Networking; Springer; 1999. [Google Scholar]
- Bray T, Hollander D, et al. Namespaces in XML 1.0. 2. W3C; 2006. [Google Scholar]
- Bray T, Paoli J, et al. Extensible Markup Language (XML) 1.0. 4. W3C; 2006. [Google Scholar]
- Clark J. XSL Transformations (XSLT), Version 1.0. W3C; 1999. [Google Scholar]
- Clark J, DeRose S. XML Path Language (XPath), Version 1.0. W3C; 1999. [Google Scholar]
- Clunie D. DICOM Structured Reporting. Bangor, PA: PixelMed; 2000. [Google Scholar]
- Dolin RH, Alschuler L, et al. HL7 Clinical Document Architecture, Release 2. J Am Med Inform Assoc. 2006;13(1):30–39. doi: 10.1197/jamia.M1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenstermacher D, Street C, et al. The Cancer Biomedical Informatics Grid (caBIG™) Conf Proc IEEE Eng Med Biol Soc. 2005 doi: 10.1109/IEMBS.2005.1616521. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, et al. Whole brain segmentation. Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Gering DT, Nabavi A, et al. An Integrated Visualization System for Surgical Planning and Guidance using Image Fusion and Interventional Imaging. Conf Med Image Comput Comput Assist Interv (MICCAI); Cambridge, England. 1999. [Google Scholar]
- Hors AL, Hégaret PL, et al. Document Object Model (DOM) Level 3 Core Specification, Version 1.0. W3C; 2004. [Google Scholar]
- ISO/IEC. Document Schema Definition Languages (DSDL) - Part 3: Rule-based validation - Schematron. 2006. ISO/IEC19757-3:2006. [Google Scholar]
- Keator DB, Gadde S, et al. A general XML schema and SPM toolbox for storage of neuro-imaging results and anatomical labels. Neuroinformatics. 2006;4(2):199–211. doi: 10.1385/ni:4:2:199. [DOI] [PubMed] [Google Scholar]
- Keator DB, Grethe JS, et al. A National Human Neuroimaging Collaboratory Enabled by the Biomedical Informatics Research Network. IEEE Transactions on Information Technology in Biomedicine. 2008;12(2):162–172. doi: 10.1109/TITB.2008.917893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze J, Baker T. The Dublin Core Metadata Element Set. 2007 Aug; RFC 5013, from http://www.ietf.org/rfc/rfc5013.txt.
- Lohrey JM, Killeen NEB, et al. An integrated object model and method framework for subject-centric e-Research applications. Frontiers in Neuroinformatics. 2009;3:19. doi: 10.3389/neuro.11.019.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, et al. The extensible neuroimaging archive toolkit (XNAT): An informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Myers JD, Chappell AR, et al. Re-Integrating the Research Record. Computing in Science and Engineering 2003 [Google Scholar]
- Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9(1):97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Ozyurt IB, Keator DB, et al. Federated Web-accessible Clinical Data Management within an Extensible NeuroImaging Database. Neuroinformatics. 2010;8(4):231–249. doi: 10.1007/s12021-010-9078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DL, Mongkolwat P, et al. Medical Imaging on the Semantic Web: Annotation and Image Markup. 2008 AAAI Spring Symposium Series, Semantic Scientific Knowledge Integration; Stanford University; 2008. [Google Scholar]
- Sufi S, Mathews B. CCLRC Scientific Metadata Model: Version 2. CCLRC Technical Report DL-TR-2004-001. 2004 Retrieved Feb 25, 2011, from http://epubs.cclrc.ac.uk/work-details?w=30324.
- Talairach J, Tournoux P. Co-Planar Stereotaxic Atlas of the Human Brain. New York: Thieme Medical Publishers; 1988. [Google Scholar]
- Thompson HS, Beech D, et al. XML Schema 1.0. W3C; 2004. [Google Scholar]