Abstract
In vitro growth experiments have demonstrated that aromatic compounds derived from lignin can be metabolized and represent a major carbon resource for many soil bacteria. However, the proteins that mediate the movement of these metabolites across the cell membrane have not been thoroughly characterized. To address this deficiency, we used a library representative of lignin degradation products and a thermal stability screen to determine ligand specificity for a set of solute-binding proteins (SBPs) from ATP-binding cassette (ABC) transporters. The ligand mapping process identified a set of proteins from Alphaproteobacteria that recognize various benzoate derivatives. Seven high-resolution crystal structures of these proteins in complex with four different aromatic compounds were obtained. The protein–ligand complexes provide details of molecular recognition that can be used to infer binding specificity. This structure–function characterization provides new insight for the biological roles of these ABC transporters and their SBPs, which had been previously annotated as branched-chain amino-acid-binding proteins. The knowledge derived from the crystal structures provides a foundation for development of sequencebased methods to predict the ligand specificity of other uncharacterized transporters. These results also demonstrate that Alphaproteobacteria possess a diverse set of transport capabilities for lignin-derived compounds. Characterization of this new class of transporters improves genomic annotation projects and provides insight into the metabolic potential of soil bacteria.
Keywords: ABC transporters, lignin degradation, Rhodopseudomonas palustris, solute-binding protein, benzoate
Introduction
Lignin is found in vascular plants and provides structure, rigidity, and protection from microbial pathogens and insects. This material is the secondmost abundant polymer on Earth after cellulose and comprises up to 30% of dry plant mass. The lignin macromolecule is produced from the random polymerization of phenylpropanoid units and the resulting three-dimensional structure is chemically heterogeneous.1 This structural complexity confers resistance to biological degradation and consequently the deconstruction of the polymer typically requires multiple classes of oxidative enzymes.2 The biodegradation process results in a highly heterogeneous pool of compounds that are challenging to metabolize and consequentially impact the composition and organization of microbial communities, soil carbon turnover, and the selection and utilization of biofuel feedstocks.
Although lignin is abundant in soil, the biological mechanisms for its degradation and utilization by microorganisms are only partially understood. Lignin breakdown has been investigated in white rot and brown rot fungi,3 which are capable of degrading polymeric structures into soluble lignin fragments, multi-aromatic ring compounds, and small monomeric derivatives. Some bacterial species have been shown to possess enzymes capable of degrading lignin in vitro4–6 and bacteria from the Actinobacteria and Proteobacteria phyla can metabolize aromatic compounds derived from lignin.7–11 For example, Rhodopseudomonas palustris, a purple non-sulfur bacterium in the Alphaproteobacter branch that inhabits various environments, is able to utilize lignin derivatives under both aerobic and anaerobic conditions.12,13
Thus far, identification of bacterial and fungal enzymatic pathways for aromatic compound degradation has been a major focus for investigations of the role of lignin in soil ecosystems. The transport of aromatic small molecules across cellular membranes has received less attention, although it is an essential step in metabolic utilization of lignin degradation products. Specific transporters for the aromatic substances produced by the biological breakdown of lignin have not been identified in many soil microorganisms known to utilize this carbon resource. Aromatic compounds derived from lignin are assumed to be imported by both ATP-dependent and ATP-independent mechanisms. Transporters in the major facilitator superfamily (MFS)14 or tripartite ATP-independent periplasmic superfamily15 are both ATP-independent transporters. Several MFS family transporters for aromatic compounds have been experimentally characterized.16–18 Most of these proteins import lignin monomers, such as vanillate (VLA) and benzoate (BEZ), but some have been shown to recognize xenobiotic aromatic compounds as well.19
ATP-binding cassette (ABC) transporters are ubiquitous integral membrane proteins with diverse biological functions. These proteins couple ATP hydrolysis to the transport of various molecules across cellular membranes. The prototypical bacterial ABC importer is composed of a transmembrane permease, a cytoplasmic ATPase subunit, and a periplasmic solute-binding protein (SBP).20 SBPs mediate nutrient import and are primarily responsible for determining ligand specificity of the transporter complex.21,22 Known ABC transporters with specificity for BEZ-like chemicals are encoded by the patDABC operon, involved in the active transport of phthalate in Rhodococcus jostii RHA123 and by the hmgDEFGH operon, proposed to transport homogentisic acid in Pseudomonas putida.24 We recently identified several SBPs from R. palustris that specifically bind small aromatic molecules typically produced during lignin degradation.25 These SBPs are not genetically linked, exhibit low (<40%) sequence identity to each other, and were originally annotated as branched-chain amino-acid-binding proteins.26 A BLAST search of the GenBank sequence database reveals hundreds of homologs of these proteins that are annotated as SBPs but with a biological role other than the transport of aromatic compounds.
To improve our ability to identify transporters that import compounds derived from lignin degradation, we have cloned and purified multiple SBPs for high-throughput ligand screening assays and crystallographic studies. We used solute-binding subunits of the ABC transporter family as surrogates to determine the specificity of the entire transporter complex.21,22 Preliminary analysis of these proteins allowed us to identify several distinct clusters of SBPs that recognize various products of lignin degradation. In this article, we focus on a group designated as “cluster I” while an companion article describing the structure-function studies of additional clusters will be published separately. Cluster I representatives are homologs of R. palustris SBPs (RPA0668, RPA0985, and RPA4029) that exhibit a preference for binding BEZ derivatives. Although microorganisms can utilize a variety of lignin degradation products, the coenzyme A ester of BEZ is a key intermediate for the anaerobic metabolism of aromatic substrates prior to ring cleavage and subsequent conversion into intermediate metabolites used for microbial growth. In the presence of oxygen, BEZ is often metabolized by oxygenases that introduce hydroxyl groups to promote ring cleavage.28 The number and diversity of pathways for utilization of BEZ indicate that this compound is a central metabolite and many bacterial species likely possess efficient mechanisms to import these compounds.
Results and Discussion
Target selection for structure-function studies
SBPs for biochemical and structural characterization were selected based on their sequence similarity to previously characterized SBPs from R. palustris CGA009 (Fig. 1) that bound BEZ derivatives relevant to lignin degradation products. Genomes utilized for comparative sequence searches included six R. palustris ecotypes and several related organisms from Alphaproteobacteria. The R. palustris ecotypes not only share a core of ∼2500 genes but also exhibit a high level of genetic disparity and variable abilities to utilize aromatic compounds for growth.26 This genetic and phenotypic diversity is the basis of a proposal by some researchers to classify these ecotypes as unique species.26,30 Similarly, although they are considered closely related by 16S rRNA sequencing, Bradyrhizobium japonicum and R. palustris strains have very little genomic overlap, with even less in common with the more distantly related Sinorhizobium meliloti (syn: Ensifer meliloti) and Nitrobacter winogradskyi bacteria. Even though all these representative species are chosen from Alphaproteobacteria, they exhibit diverse metabolic capabilities and inhabit several unique niches in the soil ecosystem.
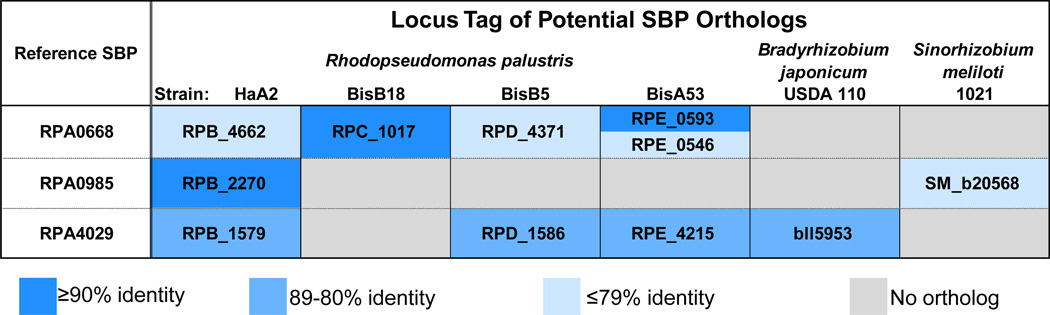
Fig. 1.
Sequence comparisons of ABC transporter SBPs. Previously identified SBPs with the capacity to bind aromatic molecules were used to identify proteins with sequence similarity in related bacterial species. Reference protein sequences were aligned with potential orthologs using the BLAST algorithm27 to search the genomic set of open reading frames from the indicated organisms: R. palustris strain HaA2 (RPB), BisB18 (RPC), BisB5 (RPD), BisA53 (RPE), DX-1, and TIE-1; B. japonicum USDA 110 (bll, blr); S. meliloti 1021 (SM_a, SM_b, SM_c); and N. winogradskyi Nb-255 (Nwi). Reference SBPs are from R. palustris CGA009 (RPA). Coding regions of prospective orthologs were obtained from the National Center for Biotechnology Information microbial genome data repository. The distribution of SBPs in R. palustris strains DX-1 and TIE-1 matched the SBP profile for the reference CGA009 strain with corresponding proteins exhibiting greater than 90% sequence identity. Prospective orthologs from these strains were omitted from this chart for simplicity. N. winogradskyi did not contain any protein with sequence identity >50% and was also omitted.
The genome of R. palustris CGA009 encodes over 100 ABC transporters with 21 genes annotated as encoding branched-chain amino-acid-binding SBPs. Experimental characterization of the binding ligands for these proteins indicated many incorrect assignments29 and suggested that functional assignments based on sequence analysis must be interpreted with caution when predicting biologically relevant and specific ligands. Cluster I targets selected for ligand screening and structural studies (Fig. 1) were generally restricted to a primary sequence identity cutoff of greater than 50% to improve prospects for selection of SBPs that bind BEZ derivatives and decipher the details of specific ligand recognition. In some cases, there were multiple potential orthologs that met these criteria. Based on previous studies that examined the relationship between sequence identity and conservation of function,31,32 we anticipated that all cluster I SBPs would exhibit binding profiles similar to the reference proteins from the R. palustris CGA009 strain.
Small-molecule screening library
A broad-spectrum chemical screening library29 was modified to increase the representation of aromatic compounds relevant to lignin biodegradation products and improve the ability to profile chemical selectivity. The structural heterogeneity of lignin polymers coupled with microbial deconstruction mechanisms that use free-radical-based random depolymerization2 results in numerous structurally diverse breakdown products. Although the identity and relative distribution of these metabolites depend on the plant source, they are represented by a mixture of phenylpropanoid monomers and multimers as well as small aromatic molecules with various ring substituents (i.e., hydroxyl, methoxyl, aldehyde, keto, and carboxyl groups). Our selection strategy to generate the screening library containing a representative set of compounds was based on literature mining as well as previous experimental data that identified ligands for SBPs likely involved in the utilization of lignin degradation products.
The current screening library (Supplemental Table 1) includes primarily monoaromatic carboxylic acids. The aromatic rings differ in the number, position, and identity of ring substituents. Compounds with substituents at the 3 and 4 ring positions were preferred as the monolignol precursors (p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol) contain hydroxyl and methoxyl groups at the 4 and 3 benzene ring positions. However, some compounds with 3 and 2 benzyl ring hydroxylations and/or methoxylations are included in the library since they are found in lignin breakdown products.33 Some organisms can metabolize aromatic compounds with nitrogen and halogen substituents, and the transport capability for these xenobiotics may be due in part to the family of binding proteins for aromatic compounds derived from lignin degradation. Therefore, the screening library was also supplemented with a limited number of nitrogen-containing aromatic molecules such as benzamide and benzonitrile (Supplemental Table 1, categorized as “small aromatic molecule”). Non-aromatic molecules were included in the library as controls for specificity and because they may also represent relevant ligands (Supplemental Table 1, “non-aromatic”). Quinic acid and shikimic acid are cyclic precursors of aromatic amino acid/phenylpropanoid synthesis; cis,cis-muconate is a dicarboxylic acid product of aerobic aromatic ring cleavage. Saturated shortchain fatty acids with C3 to C8 aliphatic tails were also screened to evaluate dependence on aromatic ring structures for ligand binding. An analysis of fungal lignin degradation products by mass spectrometry indicates many aromatic compounds similar in size and structure to the small molecules contained in our library.33 We restricted our library to monocyclic ligands since enzymes for biphenyl compound metabolism (such as 5,5′-dehydrovanillate) have not been identified by biochemical methods or genomic sequence analysis of the screened organisms.7
Ligand-binding profiles
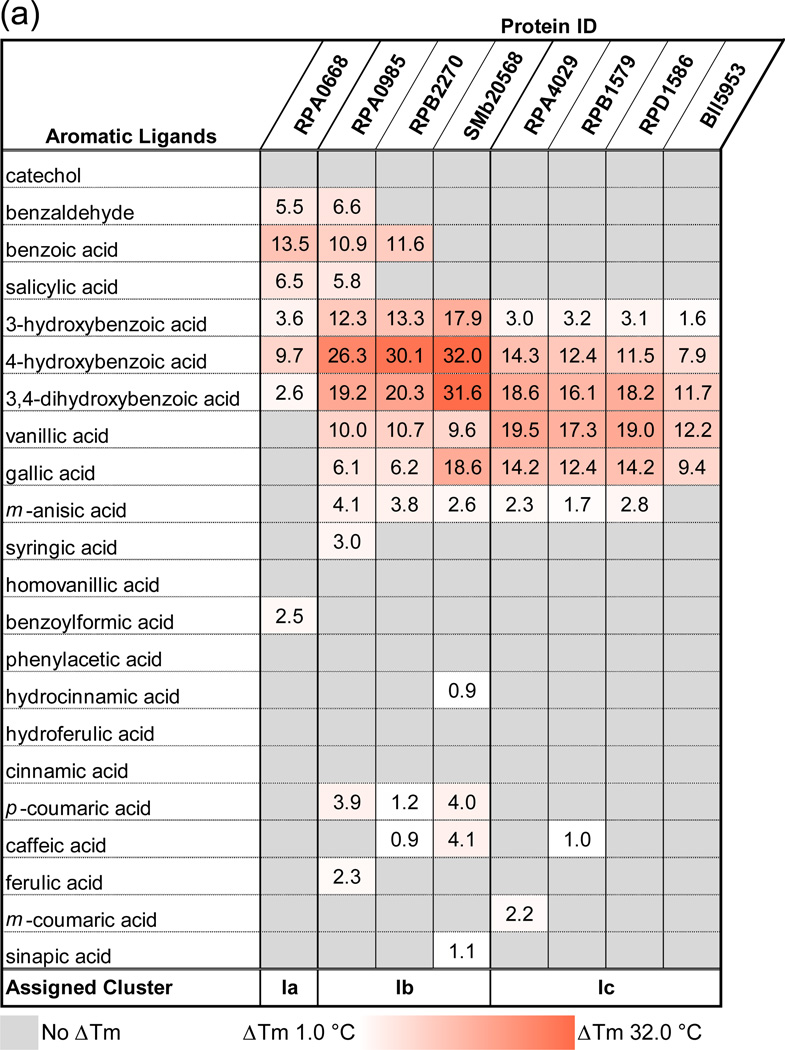
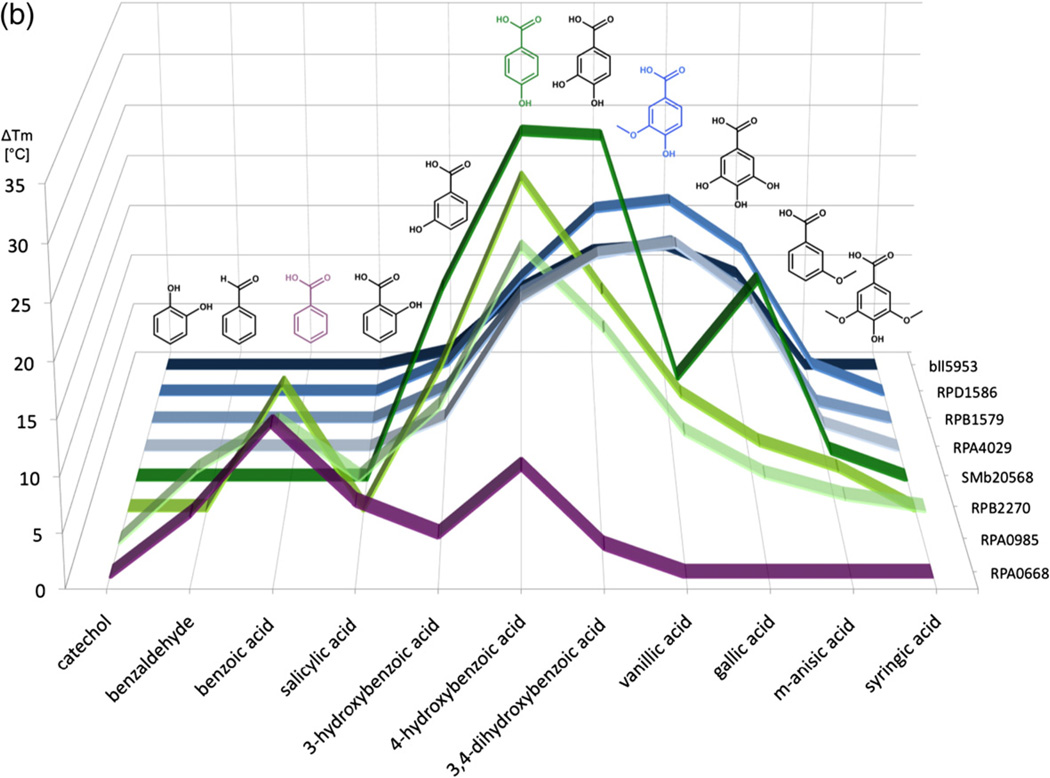
A fluorescence thermal shift (FTS) assay was used to generate ligand thermal stabilization profiles for eight SBPs that have been previously shown or predicted to bind compounds derived from lignin degradation. FTS assays are widely used for ligand screening but do not provide sufficient thermodynamic parameters to directly infer binding constants from a thermal stabilization profile.34 However, studies have shown that the FTS assay results typically reflect ligand-binding profiles obtained through alternative experimental approaches25 such as isothermal titration calorimetry, and the rank order of ligand binding generally correlates with affinity constants.25,34,35 A summary of calculated differences in melting temperatures (ΔTm) obtained via the FTS screen is presented in Fig. 2a, with the full listing of results presented in Supplemental Table 2. Organization by common ligand chemical features suggests a distinct pattern of protein–ligand interactions. Subgroups Ia, Ib, and Ic were defined by both the ability to bind compounds with a specific number and type of ring substituents and by the sequence identity to previously characterized SBPs (RPA0668, RPA0985, and RPA4029) from R. palustris CGA009 (Fig. 1).
Fig. 2.
FTS assay results for uncharacterized SBPs. The calculated ΔTm represents the difference in the Tm of protein mixed with prospective ligands and the Tm of protein-only reactions. Values are an average of at least three replicates. Average standard deviations for subgroup Ia protein–ligand experimental measurements is 0.50°C (0.15–1.33°C range), that for subgroup Ib is 0.65°C (0–2.94°C), and that for subgroup Ic proteins is 0.36°C (0–1.76°C). (a) This chart lists the ΔTm values of all proteins with a selection of lignin derivatives. Cells are colored in a whiteto-red gradient based on magnitude of the ΔTm. Ligands with increasing ring substitution are arranged in the top section (catechol to syringic acid) followed by molecules with both increasing chain lengths and ring substitutions (homovanillic acid to sinapic acid). (b) Graph illustrating overlap in ligand-binding profile between subgroups. The y-axis shows ΔTm values induced by ligands listed on the x-axis. Protein IDs are arranged along the z-axis. Subgroup Ia lines are in purple, Ib lines are in green, and Ic lines are in blue. Ligands that induced a maximal ΔTm in a subgroup are shown in the same color as the subgroup line.
For subgroup Ia (Fig. 2b, purple line), excellent thermal stabilization is observed for aromatic compounds with an aldehyde or carboxyl group (e.g., benzaldehyde and BEZ, respectively) with or without an additional hydroxyl group. Thermal stability of the complex was decreased or eliminated with a longer carbon chain at the carboxylic group (i.e., phenylacetic acid, hydrocinnamic acid). A single hydroxyl group at any ring position is acceptable, but thermal stabilization is reduced by the addition of a second hydroxyl group [3,4- dihydroxybenzoate (DHB)] and eliminated by the addition of a methoxyl group (m-anisic acid, VLA).
Proteins in subgroups Ib and Ic exhibit some overlap in thermal stabilization profile with subgroup Ia but there are novel binding capabilities for each group (Fig. 2b). All targets in cluster I subgroups a–c preferentially bind compounds containing a benzene ring substituted with a carboxylic group and thermal stabilization is negatively affected by increasing the carbon chain length of the acid. The Ib subgroup represented by RPA0985, RPB2270, and SMb20568 is highly stabilized by ligands with one or two hydroxyl groups in the 3 and 4 ring position but is penalized by a methoxyl group. The ligand-binding profile of the SMb20568 protein is overall similar to the R. palustris SBPs, but unlike RPA0985 and RPB2270, it shows an equivalent thermal stabilization with 4-hydroxybenzoate (PHB) and DHB and exhibits a greater ΔTm with gallic acid (Fig. 2b). This altered profile suggests that the ligand-binding pocket is larger and can accommodate Tri-hydroxylated rings more easily. These observations are consistent with previous studies that mapped the “solute-binding protein-dependent transportome” and the function of the associated transcriptional regulator37 of the ABC transport system containing the SMb20568 protein. The authors noted that expression of genes encoding key components of this transport system is induced by addition of aromatic compounds (DHB, PHB, and quinic acid) to the growth media. This induction supports the role of SMb20568 in binding aromatic compounds and as part of an ABC transporter complex for the import of breakdown products derived from lignin. SMb20568 has a 61% sequence identity to the reference RPA0985 transporter and has the lowest sequence identity to a reference SBP of any protein in this set.
In comparison, the Ic subgroup also interacts with ligands substituted in 3 or 4 ring positions but has some preference for 3-methoxyl substituents. For example, the SBP set containing RPA4029, RPB1579, RPD1586, and Bll5953 binds similar molecules, and the preferred ligands (DHB, VLA) are the same for each protein. The overall Tm shifts for Bll5953 are smaller but follow the same pattern observed for proteins from other R. palustris strains.
Since SBPs can be purified with pre-bound ligands, we attempted refolding of select targets from each subgroup to assess the impact of adventitious ligands in the interpretation of FTS screening results. Although not all proteins are amenable to this procedure, refolding was successful for a number of the samples with retention of ligand-binding functionality. Comparison of treated and untreated samples reveals that for this set of proteins, a pre-bound ligand does not prevent the identification of preferred compounds (Fig. S1).
Structure solution
All seven crystal structures reported here were solved by the single-wavelength anomalous diffraction (SAD) method from selenomethionine (SeMet)-labeled protein crystals that diffracted up to 1.30–2.22Å (Table 1). For each target, native protein was screened for crystallization in the presence or absence of a ligand. Two ligands for co-crystallization were selected from a rank ordering of compounds that exhibited the highest level of thermal stabilization (Fig. 2) based on the FTS assay results. Structures of four different SBPs from R. palustris (RPA0668, RPA0985, RPB2270, and RPD1586) were determined with four ligands [BEZ, PHB, DHB, and 4-hydroxy-3-methoxybenzoate (VLA)] (Table 1). Structures of all four proteins were determined in complex with PHB; RPA0668 was also solved in complex with BEZ, and RPD1586 was determined in complex with three different ligands (PHB, DHB, and VLA). These structures provide rich data for ligand coordination patterns and structure–function comparisons.
Table 1.
Crystallization conditions, data collection, and refinement statistics
| RPA0668/BEZ | RPA0668/PHB | RPA0985/PHB | RPB2270/PHB | RPD1586/PHB | RPD1586/DHB | RPD1586/VLA | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P212121 | P21212 | P212121 | P212121 | C2221 | C2221 | P21 |
| Cell dimensions | |||||||
| a (Å) | 42.3 | 52.4 | 41.9 | 42.1 | 46.8 | 46.6 | 46.9 |
| b (Å) | 57.8 | 183.9 | 61.9 | 61.2 | 132.6 | 131.9 | 130.5 |
| c (Å) | 133.9 | 76.1 | 126.2 | 130.2 | 131.0 | 130.0 | 70.4 |
| β (°) | 109.2 | ||||||
| Protein molecules/ASU | 1 | 2 | 1 | 1 | 1 | 1 | 2 |
| Se sites/monomer | 6 | 6 | 12 | 13 | 12 | 12 | 12 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Radiation source | APS, 19-ID | APS, 19-ID | APS, ID-19 | APS, 19-ID | APS, 19-ID | APS, 19-ID | APS, 19-ID |
| Wavelength (Å) | 0.9793 | 0.9793 | 0.9794 | 0.9793 | 0.9792 | 0.9792 | 0.9792 |
| Resolution (Å)a | 40.0–1.84 | 50.0–1.40 | 50–1.45 | 50–1.30 | 50.0–2.20 | 50.0–1.90 | 50.0–1.86 |
| Unique reflections | 28,325 (1383) | 145,161 (7195) | 59,333 (2906) | 81,874 (3298) | 19,925 (488) | 31,855 (1595) | 66,219 (2789) |
| Rmergeb | 0.098 (0.774) | 0.090 (0.737) | 0.107 (0.834) | 0.100 (0.424) | 0.149 (0.541) | 0.095 (0.533) | 0.100 (0.501) |
| 〈I〉/〈σI〉 | 17.3 (2.1) | 22.9 (2.8) | 24.5 (2.1) | 30.3 (2.2) | 16.3 (1.8) | 38.2 (3.3) | 25.1 (2.2) |
| Completeness (%) | 97.6 (99.7) | 99.9 (100) | 99.7 (100) | 97.5 (79.3) | 98.5(96.3) | 99.9 (100) | 99.9 (83.9) |
| Redundancy | 5.3 (5.2) | 5.8 (5.6) | 6.1 (5.9) | 6.3 (2.9) | 4.6 (4.1) | 6.9 (6.3) | 3.6 (2.8) |
| Refinement | |||||||
| Resolution (Å) | 22.6–1.84 | 35.15–1.40 | 39.78–1.45 | 30.58–1.30 | 36.61–2.22 | 36.22–1.90 | 36.41–1.86 |
| Reflections: work/test set | 26,840/1428 | 143,574/1460 | 57,865/1202 | 77,705/4085 | 18,832/1025 | 30,205/1611 | 62,791/3349 |
| Rwork/Rfreec | 0.150/0.185 | 0.139/0.175 | 0.134/0.164 | 0.122/0.153 | 0.182/0.234 | 0.133/0.182 | 0.135/0.191 |
| No. of atoms: protein/ ligands/water |
2639/9/174 | 5546/73/641 | 2837/45/263 | 2994/64/380 | 2748/10/112 | 2821/15/252 | 5557/40/524 |
| Average B factor (Å2): protein/ligands/water |
32.6/18.2/34.4 | 14.0/23.1/26.6 | 20.8/39.2/34.4 | 11.3/26.6/27.5 | 50.6/35.8/49.5 | 36.3/27.8/45.4 | 29.8/26.4/38.4 |
| rmsd from ideal | |||||||
| Bond lengths (Å) | 0.015 | 0.015 | 0.016 | 0.015 | 0.005 | 0.009 | 0.009 |
| Bond angles (°) | 1.23 | 1.615 | 1.585 | 1.716 | 0.983 | 1.269 | 1.218 |
| Ramachandran statistics of φ/ψ angles (%) | |||||||
| Most favored | 97.77 | 97.80 | 98.09 | 98.09 | 98.1 | 98.12 | 98.5 |
| Outliers | 0 | 0.28 | 0.27 | 0.27 | 0 | 0 | 0 |
| PDB code | 4EVR | 4EVQ | 4EVS | 4F06 | 4EY3 | 4EYK | 4EYG |
| Crystallization conditions | |||||||
| 0.1M sodium acetate, 25% PEG 4000, 8% 2-propanol, 5mM sodium BEZ, 24°C |
0.2M Li2SO4, 0.1M sodium acetate/acetic acid, pH4.5, 30% PEG 8000, 5mM sodium PHB, 16°C |
2M (NH4)2SO4 0.1M citric acid/ NaOH, pH3.5, 16°C |
2M (NH4)2SO4 0.1M citric acid/ NaOH, pH3.5, 16°C |
0.1 M sodium citrate/ HCl, pH5.6, 20% 2-propanol, 20% PEG 4000, 24°C |
0.1M sodium citrate/ HCl, pH5.6, 20% 2-propanol, 20% PEG 4000, 5mM sodium VLA, 24°C |
0.1M sodium citrate/ HCl, pH5.6, 20% 2-propanol, 20% PEG 4000, 5mM sodium VLA, 24°C |
|
APS, Advanced Photon Source; ASU, asymmetric unit.
Values in parentheses correspond to the highest-resolution shell.
Rmerge=∑h∑j|Ihj−〈Ih〉|/∑h∑jIhj, where Ihj is the intensity of observation j of reflection h.
R=∑h|F0|−|Fc|/∑h|F0| for all reflections, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is calculated analogously for the test reflections, randomly selected, and excluded from the refinement.
RPA0668 in complex with BEZ (RPA0668/BEZ) crystallizes in the orthorhombic P212121 space group, with one protein molecule in the asymmetric unit (residues Gly32–Leu391). Additional components of the crystal structure include 1 BEZ molecule and 174 water molecules.
The crystals of RPA0668 in complex with PHB (RPA0668/PHB) are orthorhombic with the P21212 space group. The asymmetric unit contains two protein chains, A and B. In molecule A, residues Gly32–Leu391 have been modeled into the corresponding electron density. Molecule B is slightly better ordered and composed of residues Gly21– Leu391. In addition to the polypeptide chains, the model consists of 2 PHB molecules, 641 water molecules, 5 sulfate ions, 2 glycerol molecules, 2 ethylene glycol molecules, and 2 acetate ions.
The crystals of RPA0985 in complex with PHB (RPA0985/PHB) belong to the orthorhombic system, P212121 space group. The asymmetric unit is occupied by one fully ordered protein molecule containing residues Asp23–Gln390. In addition to the polypeptide chain, the final atomic model includes 1 PHB molecule, 263 water molecules, and 7 sulfate ions.
The crystals of RPB2270 in complex with PHB (RPB2270/PHB) belong to the orthorhombic system, P212121 space group. The asymmetric unit is occupied by one polypeptide chain containing residues Asp24–Gln391. In addition to the protein molecule, the final atomic model includes 2 PHB molecules [1 in the ligand-binding site and 1 on the protein surface (data not shown)], 380 water molecules, 5 sulfate ions, 3 glycerol molecules, and 1 sodium ion.
The crystals of RPD1586 in complex with PHB (RPD1586/PHB) and with DHB (RPD1586/DHB) belong to the same C-centered orthorhombic space group C2221 and demonstrate similar unit cell parameters (Table 1). The crystal structure of RPD1586/PHB has 1 polypeptide chain with residues Ala27–Thr390, 1 PHB ligand, and 112 water molecules. The asymmetric unit of the RPD1586/ DHB structure also contains 1 protein molecule (residues Glu28–Thr390), 1 DHB ligand, 1 ethylene glycol molecule, and 252 water molecules. The crystals of RPD1586 in complex with VLA (4-hydroxy-3-methoxybenzoate, RPD1586/VLA) are monoclinic with space group P21. The atomic model of the RPD1586/VLA structure contains 2 polypeptide chains, A and B, of residues Glu28– Thr390 and Glu28–Lys391, respectively, 2 VLA ligands, 2 ethylene glycol molecules, 2 isopropyl alcohol molecules, and 524 water molecules.
The quality of the crystallographic models was assessed by the MolProbity server,38 revealing generally appropriate stereochemistry. The RPA0668/PHB, RPA0985/PHB, and RPB2270/ PHB structures contain a few residues outside the expected Ramachandran plot regions. These outliers, however, are well defined in the electron density maps and clearly represent unusual structural features rather than a misinterpretation of the crystallographic data.
Overall structure
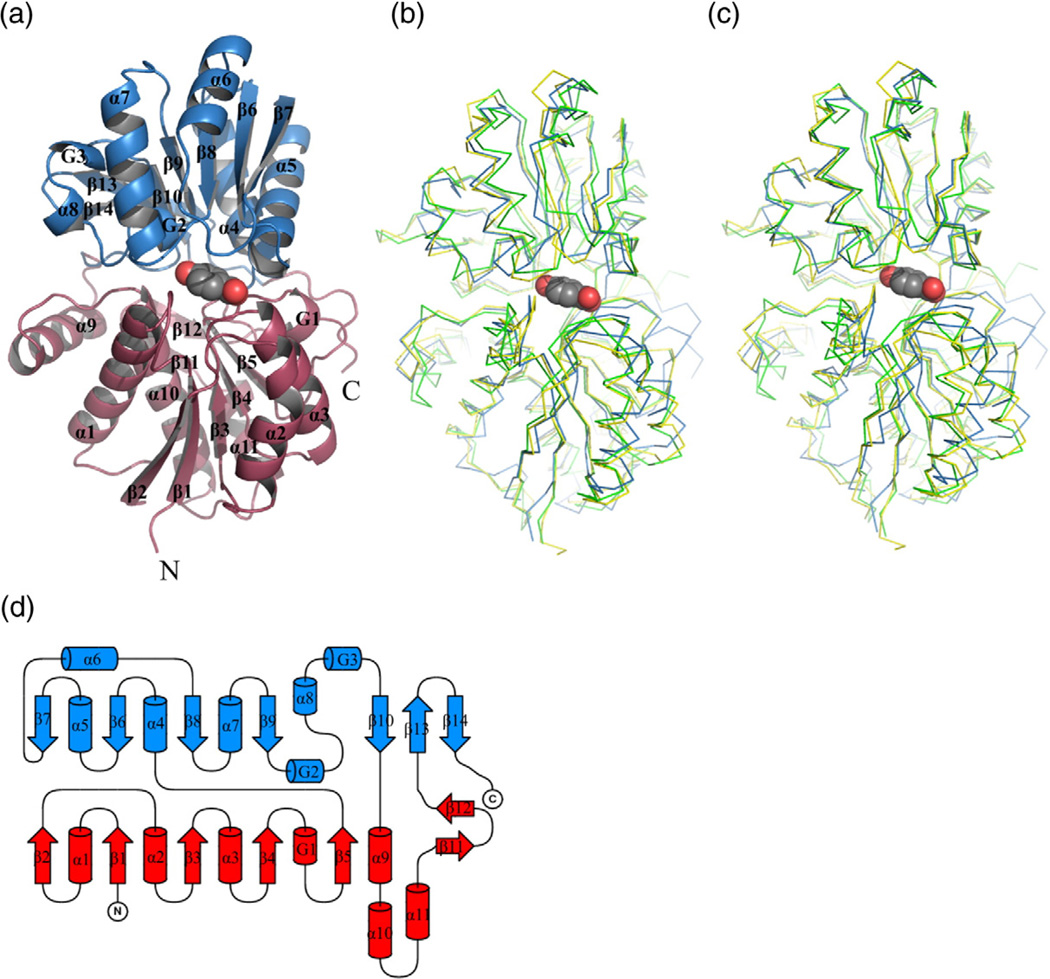
The SBPs of ABC transporters with known structures exhibit many common features39 that are also present in the structures of these R. palustris SBPs. Namely, each protein is composed of two domains (lobes), I and II (or N- and C-terminal), separated by a deep cleft that hosts a ligand-binding site. Each of the domains contains a central β-sheet with similar topology, flanked on both sides by helices (Fig. 3). The core motif of domain I consists of five β-strands arranged in a parallel fashion with topology β2β1β3β4β5. This β-sheet is surrounded by six α-helices (α1, α2, α3, α9, α10, and α11) and one 310-helix (G1). In addition, domain I contains a satellite β-hairpin, β11–β12. In domain II, the central parallel β-sheet (β7β6β8β9β10) is extended by an additional β-hairpin, β13–β14. As a consequence of the β- hairpin insertion, the complete core motif from domain II is a mixed β-sheet. The flanking elements include five α-helices (α4, α5, α6, α7, and α8) and two 310 helices (G2 and G3).
Fig. 3.
Structure of subgroup I SBPs from R. palustris. (a) Overall structure of RPA0668. Domain I is colored red; domain II is in blue. The PHB molecule is shown in van der Waals representation. (b) Superposition of RPA0668/PHB (blue), RPA0985/PHB (green), and RPD1586/PHB (yellow). (c) Superposition of RPA0668/ PHB (blue), RPA0985/PHB (green), and RPD1586/PHB (yellow) with domain II from RPA0668/PHB as a reference. (d) Topology diagram. Color code as in (a).
The two lobes are linked by three segments that correspond to loops β5–α4, β10–α9, and β12–β13. This type of connection between the domains as well as the topology of the central β-sheets classify these proteins as Class I SBPs, according to the classification proposed by Fukami-Kobayashi et al.39,40 Class I proteins fall into cluster B in the classification proposed by Berntsson et al.39 Other proteins belonging to the same class/cluster include those that bind carbohydrates and the branched-chain amino acids leucine, isoleucine, and valine.
The crossover elements of SBPs serve as a hinge that enables the lobes to move. It is proposed that, in the absence of a ligand, the protein can more readily adopt an open conformation with the ligandbinding site exposed to solvent.41 Upon ligand binding, the individual domains are shifted with respect to each other, trapping a ligand molecule at the domain interface. This phenomenon has been compared to the mechanism by which the Venus flytrap operates.42 Ligand-induced conformational change leads to the so-called closed state. It has also been noted that SBPs may exist as open-liganded and closed-unliganded forms.39,43–45 As all structures of SBPs from R. palustris have been determined as complexes with their respective ligands and the binding pocket is inaccessible to solvent, we postulate that they represent the closed state.
Despite low identity at their amino acid sequence level (Table 2), the tertiary structures of RPA0668, RPA0985, RPB2270, and RPD1586 are very similar. Superposition of representative subgroup I structures (Fig. 3b) indicates that the secondary-structure elements described above are conserved in all proteins examined in this study. Pairwise superpositions give rmsds within a 0.35–2.11Å range, with the best agreement between the RPA0985/PHB and RPB2270/PHB molecules (Table 2). The worst match is observed for the RPA0668/PHB– RPA0985/PHB pair (Table 2). A more detailed analysis shows, however, that this relatively large discrepancy is mostly due to differences in domain I of the respective proteins as illustrated by superposition of the structures using domain II as the reference. Domain I superposition of the RPA0668/PHB–RPA0985/PHB pair gives an rmsd of 2.18Å, while domain II superposition gives an rmsd of 1.32Å. The three RPD1586 structures are nearly identical with an rmsd as low as ∼0.17Å. The two RPA0668 structures superpose slightly worse, with 0.43Å rmsd.
Table 2.
Similarity between R. palustris SBPs and their structural relatives
| RPA0668 PHB |
RPA0985 PHB |
RPB2270 PHB |
RPD1586 PHB |
RPD1586 DHB |
RPD1586 VLA |
PDB 3i45 |
PDB 3td9 |
PDB 3sg0 |
OX2-CX- C1-C6 torsion angle |
Cavity volume |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| RPA0668/BEZ | 0.43 (356) | 1.89 (341) | 1.88 (340) | 1.67 (346) | 1.67 (346) | 1.66 (344) | 2.20 (335) | 1.97 (336) | 2.18 (321) | −27.7 | 138.8 |
| RPA0668/PHB | — | 2.11 (340) | 1.89 (344) | 1.66 (342) | 1.65 (342) | 1.67 (342) | 2.27 (334) | 2.00 (329) | 2.14 (323) | −25.3 | 141.8 |
| RPA0985/PHB | 27.8/42.6 | — | 0.35 (368) | 1.47 (353) | 1.58 (355) | 1.56 (355) | 2.28 (343) | 2.07 (333) | 1.93 (314) | −20.9 | 154.1 |
| RPB2270/PHB | 27.7/43.1 | 90.0/94.1 | — | 1.47 (355) | 1.54 (354) | 1.52 (355) | 2.19 (342) | 2.15 (334) | 1.89 (322) | −24.2 | 153.6 |
| RPD1586/PHB | 32.2/49.6 | 43.3/61.7 | 43.8/62.7 | — | 0.17 (363) | 0.15 (363) | 2.32 (344) | 2.33 (336) | 2.19 (323) | −33.7 | 179.7 |
| RPD1586/DHB | 32.2/49.6 | 43.3/61.7 | 43.8/62.7 | 100 | — | 0.17 (363) | 2.34 (344) | 2.35 (337) | 2.09 (315) | −25.7 | 165.4 |
| RPD1586/VLA | 32.2/49.6 | 43.3/61.7 | 43.8/62.7 | 100 | 100 | — | 2.33 (346) | 2.33 (335) | 2.20 (323) | −27.4 | 175.1 |
| PDB 3i45 | 22.7/38.0 | 22.2/37.8 | 22.6/37.3 | 24.5/42.4 | 24.5/42.4 | 24.5/42.4 | 128.0 | ||||
| PDB 3td9 | 20.5/33.6 | 24.3/35.5 | 25.8/40.5 | 24.9/45.3 | 24.9/45.3 | 24.9/45.3 | 235.0 | ||||
| PDB 3sg0 | 23.1/36.3 | 23.5/40.4 | 23.4/39.6 | 24.8/42.0 | 24.8/42.0 | 24.8/42.0 | 228.9 |
The molecules were superposed in Coot.46 If multiple protein chains exist in the asymmetric unit, only chain A was compared. Sequence identity was determined using the EMBOSS Needle program.47 Cavity volumes were calculated in SUFRNET with 1.2Å radius for gap spheres.48 For calculations, double conformations were removed and conformer A was used. Hydrogen atoms were not included. Selenium atoms were treated as sulfur atoms. The volumes refer to molecules listed in the first column. The upper right side of the table shows rmsd (in angstroms) with number of superimposed Cα atoms (in parentheses); the lower left side of the table shows sequence identity and similarity (in percentage). The torsion angles defining conformation of BEZ derivatives are provided (in degrees, chain A ligand). The far right column shows cavity volume (in cubic angstroms).
Ligand binding
The ligand-binding site is located in a cleft between domains I and II. In each of the structures, a ligand molecule is well defined in the electron density maps, leaving no doubt about its identity and orientation (Fig. 4). All the structures are assumed to represent ligand-bound closed protein conformations. The smallest cavity volume is observed for RPA0668 while RPD1586 contains the largest cavity volume (Table 2). The aromatic molecules are coordinated by a combination of hydrogen bonds (H-bonds) and van der Waals interactions. It is of note that in all structures, the aromatic ring position and orientation are virtually the same and the carboxyl group of a ligand moiety is twisted out of the ring plane (Fig. 4). The average OX2-CX-C1-C6 torsion angle for BEZ derivatives is −26.4° (Table 2; for atom labeling scheme, see Fig. 4g). Theoretical calculations predict these conformers as high-energy forms of 3-hydroxybenzoate and PHB.49 In the more favorable conformer, the carboxyl group is coplanar with the aromaticmoiety, which is the conformation observed in crystal structures of BEZ and its derivatives.50–52
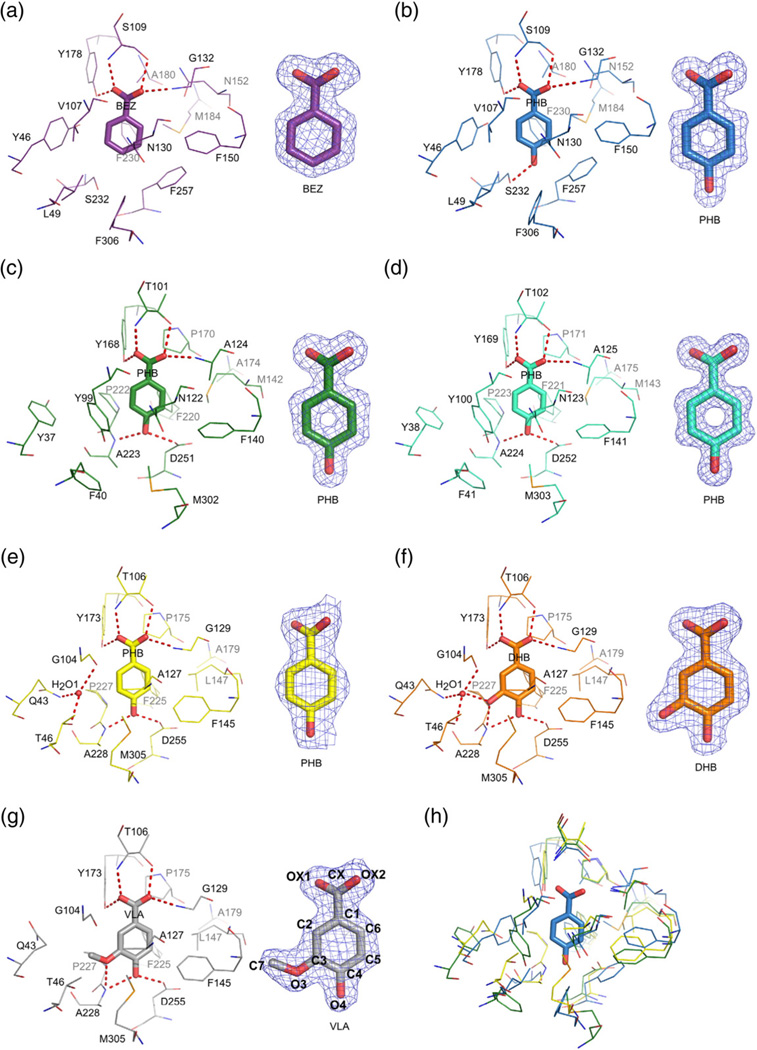
Fig. 4.
Ligand binding in R. palustris cluster I SBPs. The ligand molecules are shown in a stick representation. Potential hydrogen bonds are depicted as broken lines. On the right, 2mFo–DFc electron density map contoured at the 1σ level for each ligand is shown. (a) RPA0668/BEZ; (b) RPA0668/PHB; (c) RPA0985/PHB; (d) RPB2270/PHB; (e) RPD1586/PHB; (f) RPD1586/DHB; (g) RPD1586/VLA; (h) superposition of RPA0668/PHB (blue), RPA0985/PHB (green), and RPD1586/ PHB (yellow). For clarity, only the PHB ligand from the RPA0668 complex is shown (in a stick representation). The numbering of ligand atoms used throughout the text corresponds to the labeling scheme shown for VLA.
Subgroup Ia
The structure of RPA0668 has been determined in complex with the two most preferred ligands BEZ and PHB. The protein anchors the BEZ moiety through four H-bonds; two bonds are provided by the protein main chain and two are contributed by side chains (Table 3 and Fig. 4a). Ser109 participates in two of these interactions, through binding to the OX1 atom via its main-chain amide group and to OX2 via its side chain. The remaining hydrogen bonds established by the OX1 and OX2 atoms engage the side chain of Tyr178 and the main-chain amide group of Gly132, respectively. Although the binding pocket is lined with several aromatic side chains, they participate neither in face-to-face nor in T-shaped edge-to-face interactions with the phenyl ring. There are, however, edge-to-face interactions with less perfect geometries, for example, with Phe230. Moreover, an NH–π interaction involves the Asn130 side chain (NH–π/benzene centroid distance ∼3.3Å). The binding mode of PHB by the RPA0668 protein is essentially identical with that observed in the RPA0668/BEZ structure (Table 3 and Fig. 4b). The only difference is the presence of an additional hydrogen bond formed between the 4-OH group and the side chain of Ser232.
Table 3.
Hydrogen bonds between R. palustris SBPs and their ligands
| Residue/Atom/Distance (Å) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligand atom |
Domain | RPA0668/ BEZ |
RPA0668/ PHB |
RPA0985/ PHB |
RPB2270/ PHB |
RPD1586/ PHB |
RPD1586/ DHB | RPD1586/ VLA | |||||||
| OX1 | I | S109 | 2.89 | S109 | 2.82 | T101 | 2.81 | T102 | 2.79 | T106 | 2.85 | T106 | 2.79 | T106 | 2.84 |
| N | N | (2.77) | N | N | N | N | N | (2.75) | |||||||
| OX1 | II | Y178 | 2.57 | Y178 | 2.62 | Y168 | 2.71 | Y169 | 2.73 | Y173 | 2.87 | Y173 | 2.75 | Y173 | 2.74 |
| OH | OH | (2.62) | OH | OH | OH | OH | OH | (2.76) | |||||||
| OX2 | I | S109 | 2.55 | S109 | 2.62 | T101 | 2.59 | T102 | 2.65 | T106 | 2.60 | T106 | 2.56 | T106 | 2.50 |
| OG | OG | (2.64) | OG1 | OG1 | OG1 | OG1 | OG1 | (2.58) | |||||||
| OX2 | I | G132 | 3.03 | G132 | 3.01 | A124 | 2.94 | A125 | 2.96 | G129 | 2.77 | G129 | 2.81 | G129 | 2.80 |
| N | N | (3.02) | N | N | N | N | N | (2.84) | |||||||
| O3 | II | — | — | — | — | — | A228 Na |
2.96 | A228 Na |
3.05 (3.03) | |||||
| O3 | — | — | — | — | — | Wat1 | 2.66 | — | |||||||
| O4 | II | — | S232 | 2.74 | A223 | 3.12 | A224 | 3.11 | A228 | 2.86 | A228 | 3.03 | A228 | 2.90 | |
| OG | (2.78) | N | N | N | N | N | (2.97) | ||||||||
| O4 | II | — | — | D251 | 2.64 | D252 | 2.67 | D255 | 2.55 | D255 | 2.54 | D255 | 2.51 | ||
| OD2 | OD1 | OD1 | OD1 | OD2 | (2.49) | ||||||||||
For structures with two molecules in the asymmetric unit, the numbers in parentheses refer to monomer B.
Although the distance between proton donor and acceptor suggests the presence of a hydrogen bond, the putative position of a hydrogen atom favors interaction with the O4 atom rather than with O3.
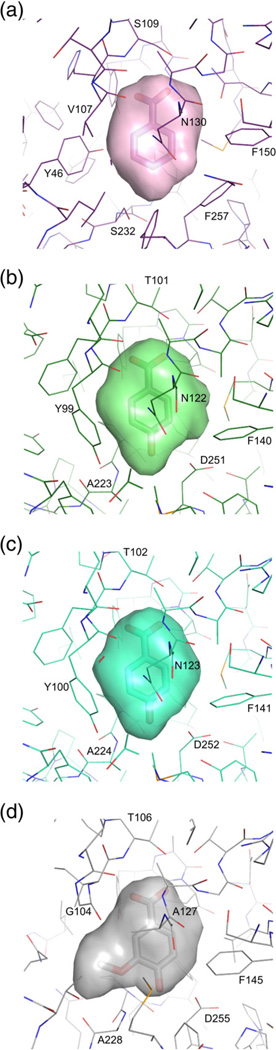
The cavity volume and its environment are consistent with the ligand preference indicated by thermal stabilization screens. Optimal thermal stabilization of the RPA0668 protein is observed for compounds with minimal ring substitutions. They are limited to an aldehyde or a carboxyl group and a single additional hydroxyl group. These constraints on the number and size of ring substituents likely reflect the small cavity volume of subgroup Ia (Table 2 and Fig. 5a). The highest level of thermal stabilization was observed with BEZ with a slight decrease observed by introduction of a hydroxyl group in position 4. Substitutions in other locations on the benzyl ring were observed to decrease the thermal stability of the complex (Fig. 2), which is consistent with the structural data. The chemical environment of the 3- and 4-OH binding region is influenced by the proximity of the side chains of either Phe257 and Phe306 or Leu49 and Tyr46, depending on the orientation of the aromatic ring. For the 4-OH substitution, the ligand is stabilized via an H-bond with Ser232. Analysis of the structural data indicates that the 3-OH substitution can be stabilized via an H-bond with Ser232 if this residue adopts a different conformation. The observed decrease in thermal stability induced by the 3-OH substitution relative to the 4-OH substitution may result from a nonoptimal interaction or disruption of hydrophobic packing. This prediction is also consistent with the disruptive effects of additional hydrox-yl groups (e.g., DHB), which further decrease the level of thermal stabilization (Fig. 2).
Fig. 5.
The binding pockets of R. palustris cluster I SBPs. The cavities shown in a surface representation were calculated in SURFNET.53 (a) RPA0668/BEZ; (b) RPA0985/PHB; (c) RPB2270/PHB; (d) RPA1586/VLA.
Subgroup Ib
The structures of two subgroup Ib representatives, RPA0985 and RPB2270, have been determined in complex with PHB. These SBPs are strongly stabilized in the presence of this ligand (ΔTm ∼30°C) and much less in the presence of BEZ (∼10°C). This observation is consistent with a binding analysis using isothermal calorimetry,54 which showed that RPA0668 affinity for BEZ is eightfold higher than the affinity of RPA0985 for this ligand. Despite the differences in amino acid sequence, the overall binding site architecture and ligand coordination features of two subgroup Ib representatives are similar to those observed for RPA0668, subgroup Ia. The RPA0985 and RPB2270 proteins have high sequence identity (Table 2) and their structures superpose with a low rmsd (0.35Å). Not surprisingly, their ligand-binding modes are practically identical (Fig. 4c and d). The hydrogen bond coordination of the carboxyl group of the PHB is essentially equivalent to RPA0668/PHB (Fig. 4a). The only difference is that Ser109 of RPA0668 is substituted with threonine (Thr101/Thr102 in RPA0985/RPB2270, respectively). Both proteins also contain an asparagine residue (Asn122/ Asn123) that forms an NH–π interaction with the aromatic ring of the ligand molecule. A notable difference between the structures of Ia and Ib representatives is that in Ib, the 4-hydroxyl group of PHB forms two hydrogen bonds. One of them engages the side chain of an aspartate residue (Asp251/Asp252) and the other one involves the main-chain amide group of an alanine residue (Ala223/Ala224). The latter residue corresponds to Ser232 in RPA0668, while Asp251 (or Asp252) corresponds to the Phe257 residue in subgroup Ia and contributes to a more polar binding environment.
The structural features are consistent with the observed differences in the thermal stability profile. RPA0985 and RPB2270 display a similar ligand stabilization pattern to RPA0668 but exhibit enhanced thermal stability for the BEZ derivatives with 3- or 4-hydroxyl ring substitutions over BEZ (Fig. 2). In addition to a slightly larger cavity volume (Table 2 and Fig. 5b and c), the presence of Asp251/Asp252 (RPA0985/RPB2270) versus Phe257 (in RPA0668) enhances the H-bonding of the 4-hydroxyl group of PHB. Incorporation of an additional charged residue into the pocket also explains the observed marked decrease in the level of thermal stability in the presence of BEZ relative to the RPA0668 protein. Tyr99 (replacing V107 in RPA0668) might be responsible for increased affinity for the 3-hydroxy-substituted compound.
Subgroup Ic
Subgroup Ic shows a ligand-binding pattern very similar to Ib, with the largest stabilizing effect provided by VLA (4-hydroxy-3-methoxybenzoate). The RPD1586 protein from the Ic subgroup was crystallized with PHB, DHB, and VLA. In all complexes, the basic network of hydrogen bonds is remarkably similar to each other and to those observed for subgroups Ia and Ib (Fig. 4e – g). In particular, the binding mode of PHB by RPD1586 closely resembles the organization found in the RPA0985/PHB and RPB2270/PHB complexes. The notable exception is a lack of the NH–π interaction due to an Asn→Ala substitution (Fig. 4e). Moreover, in contrast to the solvent-free cavities of subgoups Ia and Ib proteins, a single water molecule is present in the RPD1586/PHB and RPD1586/DHB structures (H2O1, Fig. 4e and f). The water molecule is coordinated by residues Gln43, Thr46, and Gly104 (equivalent to Tyr46/Tyr37/ Tyr38, Leu49/Phe40/Phe41, and Val107/Tyr99/ Tyr100 in RPA0668/RPA0985/RPB2270, respectively) but is not involved in hydrogen bonding with the PHB ligand. It does, however, make an H-bond with the 3-hydroxyl group of the DHB ligand. When the protein is complexed with VLA, this water molecule is expelled from the binding cavity to provide room for the methoxyl group. In addition, Gln43 adopts a different conformation to achieve a more hydrophobic environment for the aliphatic substituent. In the DHB and VLA complexes, the distance between the O3 atom and the Ala228 mainchain amide group allows for an H-bond. However, the putative position of the hydrogen atom suggests that the interaction with the O4 atom is more likely. We cannot completely exclude, however, the possibility that the hydrogen bond is bifurcated.
The cavities of all ligand-complexed forms of the RPD1586 protein are larger and less symmetrical than those of the RPA0668, RPA0985, and RPB2270 protein–ligand structures (Table 2 and Fig. 5d). This larger pocket size is attributed to the need to accommodate more complex ring substituents and in some cases a water molecule. The RPD1586 protein and characterized homologs are stabilized by BEZ derivatives with ring substitutions at the 3 and 4 ring positions and shows a preference for the 3-methoxyl substituent (present, for example, in VLA) (Fig. 2). In contrast to the proteins from subgroups Ia and Ib, subgroup Ic proteins exhibit increased thermal stability with DHB relative to PHB. The stabilizing effect associated with the incorporation of the 3-hydroxyl group can be attributed to the fact that it is hydrogen bonded to the protein via a water molecule. Interestingly, this anticipated stabilization resulting from coordination of the 3-hydroxyl substituent is not reflected in the FTS data for the 3-hydroxybenzoate/subgroup Ic complexes. The observed stabilization for 3-hydroxybenzoate/subgroup Ic complexes is much lower than those observed for subgroup Ib complexes with the same compound. This could be explained by the assumption that thermal input during the FTS assay accelerates the release of water from the subgroup Ic pocket. This decreased occupancy would disrupt coordination of the ligand 3-hydroxyl group and likely decrease protein thermal stability. In contrast, if RPA0985 forms a hydrogen bond with the 3-hydroxyl group, it would be through a permanent section of the binding pocket and not a labile water molecule that can be expelled from the binding site. According to the FTS assay observations, VLA provides maximal stabilization for subgroup Ic proteins. This is in agreement with the structural data indicating that the VLA molecule fills all the available volume, offering numerous anchoring interactions, and matches the chemical environment of the pocket.
Similarity to other SBPs
The structures of many SBPs with and without ligands have been deposited in the Protein Data Bank (PDB). A BLAST search of the PDB for proteins with sequence similarity to the targets described in this article returns proteins with relatively low sequence identity. The best hits for each SBP examined had between 24% and 28% sequence identity to a previously solved protein. A search for structural homologs of RPA0668 with the PDBe Fold server55 identified SBPs that bind hydrophobic amino acids such as phenylalanine (PDB code 3td9, unpublished; 1usi56), leucine (1z16,57 1usk,56 3ipc,58 3lop, unpublished), isoleucine (1z1757), alanine (3ip558), proline (3ip658), and valine (1z18,57 3lkb, unpublished). All of these proteins identified using the PDBe Fold server exhibit less than 23% sequence identity (range, 19–22.6%; similarity range, 33.6–38.9%) with the RPA0668 protein. In most cases, these structures correspond to one protein complexed with various ligands. The rmsd range between these hits and RPA0668/PHB is between 2.05 and 2.41Å. Additional structural relatives include AmiC protein,59 complexed with acetamide (1pea) or butyramide (1qnl, 1qo0), and various proteins that bind aromatic compounds (other than phenylalanine). Among those is a protein from Rhodospirillum rubrum that has nicotinic acid bound in the pocket (3i45, unpublished). The latter group also contains other R. palustris SBPs that interact with derivatives of phenylacetic and phenylpyruvic acids (3uk0, 3ukj, 3tx6, 3sg0, to be described elsewhere).
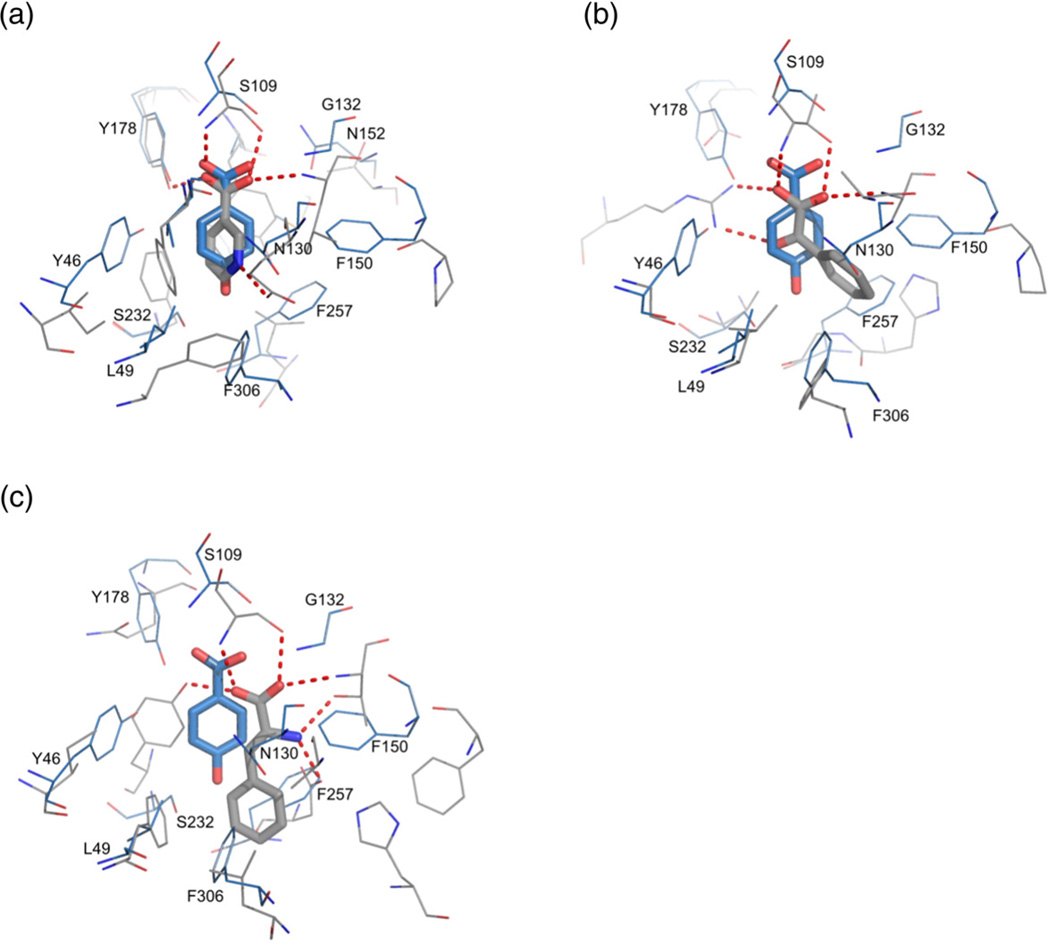
We have compared the structures and ligandbinding modes of R. palustris SBPs (with RPA0668/ PHB as the main reference) with three distant homologs, namely, 3i45, 3sg0, and 3dt9 (Fig. 6 and Table 2), with structures determined in complex with aromatic molecules: nicotinic acid, benzoylformic acid, and phenylalanine, respectively. Structurally, nicotinic acid is the closest analog of the BEZ derivatives identified as cluster I ligands. In the two latter compounds, the aliphatic chains are longer by one or two carbon atoms, respectively. In this context, it is not surprising that the best superposition of the binding sites is obtained with the 3i45 structure (Fig. 6a). However, even though the overall similarity between RPA0668 and 3sg0 or 3dt9 is poor, some of the protein–ligand interactions are shared.
Fig. 6.
Superposition of RPA0668 in complex with PHB acid (blue) with related structures (gray). Hydrogen bonds are shown as broken lines. (a) R. rubrum SBP (PDB code 3i45) in complex with nicotinic acid; (b) R. palustris SBP from cluster II (PDB code 3sg0) in complex with benzoylformic acid; (c) Thermotoga maritima SBP (PDB code 3td9) in complex with phenylalanine.
The most conservative interactions exist between the carboxyl group and a Ser/Thr residue (Ser109/ Thr101/Thr102/Thr106 in RPA0668/RPA0985/ RPB2270/RPD1586). Moreover, in all structures, the OX2 atom is hydrogen bonded to the mainchain amide group of the Gly132 equivalent (in the RPA0668 sequence). 3i45 also preserves an interaction between OX1 and a tyrosine residue (Tyr178 in RPA0668). In 3dt9, such a contact is established by a Tyr residue that corresponds to Ser232 in RPA0668 (Fig. 6c). A quite different binding pattern is observed in the 3sg0 structure, where the OX1 and C═O groups form a bidentate interaction with the side chain of an arginine residue, equivalents of which are not part of the binding pocket in cluster I R. palustris SBPs (Fig. 6b). A phenylalanine ligand from the 3td9 structure forms unique hydrogen bonds through its amino group, which binds to the side chains of threonine (the same residue interacts with OX2) and aspartate residues. The latter amino acid corresponds to Phe257 (in RPA0668) or Asp251/Asp252/Asp255 (in RPA0985/RPB2270/ RPD1586). The aromatic ring of the nicotinate moiety forms a hydrogen bond between the nitrogen atom and a glutamate residue corresponding to Asn130 in RPA0668.
Besides changes in the hydrogen-bonding pattern, there are also modifications of van der Waals interactions associated with the residue substitutions or movement of hydrophobic residues, which also influences pocket geometry. For example, instead of the phenylalanine residue that is conserved in all cluster I R. palustris SBPs (Phe150 in RPA0668), 3sg0 contains proline. In addition, another bulky residue (Phe257 in RPA0668 or its Asp equivalents in subgroups Ib and Ic) is substituted by glycine. The space acquired through the reduction of side-chain sizes better accommodates the aromatic ring of the benzoylformate ligand.
Evolutionary relationships and functional inference
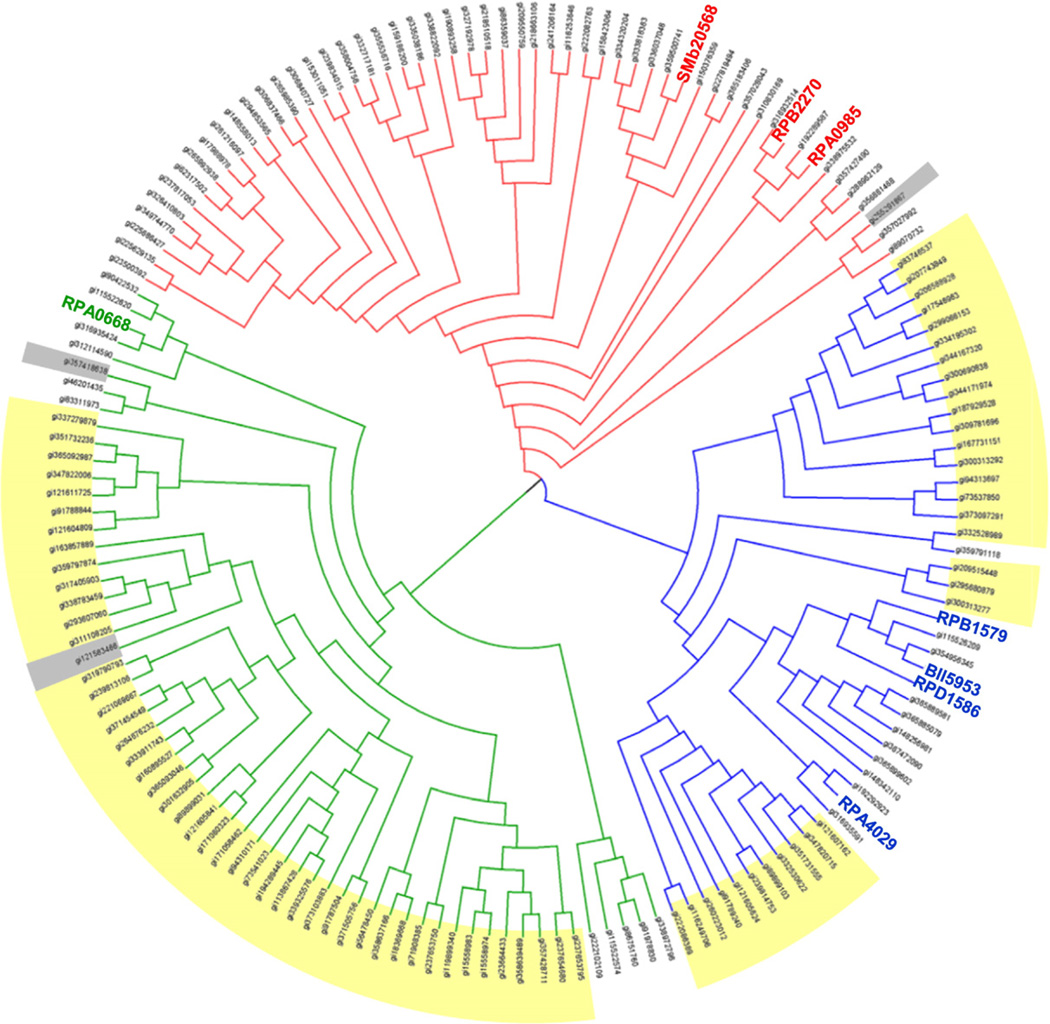
The crystal structures of the BEZ derivativebinding proteins illustrate the molecular features for this family of SBPs and provide considerable insight into the basis for ligand specificity. This knowledge can be applied to examine the relationship between the sequence and ligand-binding profiles of these proteins and provides a means for functional inference of homologous genes in other microbial species. Although a comprehensive phylogenetic analysis is beyond the scope of this article, we used BLAST27 to identify similar sequences in diverse organisms related to the original characterized SBPs from the R. palustris CGA009 strain (RPA0668, RPA0985, and RPA4029). An evaluation of the sequence relationships for high identity sequence matches (∼e−100 or lower/>50% identity with alignment length of >80%) to the original RPA proteins indicates a broad distribution organized according to their evolutionary relationships (Fig. 7). Sequences selected at this cutoff are organized in three divisions with no overlapping hits. An immediate consequence of the current results is the ability to correctly infer ligand preference for these homologs. Although these homologs are typically recognized as SBPs, none of the assigned annotations indicate a functional role associated with the import of lignin degradation products, which limits the ability to link function to biological role. In this context, it is interesting to note that the proteins in cluster I subgroup (Fig. 7) are not uniformly distributed among bacterial classes. Proteins in subgroups Ia and Ic are derived largely from Betaproteobacteria (primary classes: Burkholderiales, Rhodocyclales, and Comamonadaceae) with ∼20% representative of proteins from Alphaproteobacteria (class: Bradyrhizobiaceae and Rhizobiales). Proteins in subgroup Ib are derived exclusively from Alphaproteobacteria (primary class: Rhizobiales). Although these observations are derived from a limited sequence comparison, the nonuniform distribution of these sequences (Figs. 1 and 7) may represent functional capabilities and adaptations to different niches.
Fig. 7.
Cladogram of homologs for the RPA0668, RPA0985, and RPA4029 proteins. Sequences were selected from highidentity (>50%) matches to the R. palustris proteins, and the tree was constructed as described in Materials and Methods. Genes from Alphaproteobacteria are uncolored, Betaproteobateria are highlighted yellow, and miscellaneous sequences are in gray.
An inspection of the ClustalW alignments60 for each of the three branches indicates a widespread conservation of the ligand contact residues identified from the crystal structures with variance increasing at lower sequence identity. This general conservation of binding site residues in each subgroup provides a reliable indication of ligand characteristics for a large group of previously uncharacterized proteins. For the most part, substitutions of the ligand-binding site residues (Fig. 4) introduce only minimal changes to the binding pocket. For example, the sequence position corresponding to Ser109 (in RPA0668 sequence) is occupied predominately by threonine, as illustrated by RPA0985, RPB2270, and RPD1586. Similarly, Gly132 is sometimes replaced by an alanine without consequences for ligand binding as the ligand–residue interaction occurs via the protein main chain. Analogously, in many representatives, Phe150 is replaced by other bulky residues such as tyrosine or tryptophan. For some proteins, sequence changes in binding site residues are coupled, minimizing the overall impact on the binding site architecture. For example, Phe230 in the RPA0668 ortholog group can be replaced with a smaller proline residue, suggesting that the size of the pocket increases. However, this mutation is coupled to an Ala180→Tyr substitution, which will preserve the pocket volume. A similar situation is observed in the 3sg0 protein (see above), in which RPA0668 Phe230 is substituted by an alanine and Ala180 is replaced by a tyrosine.
In a few cases, it is difficult to determine if unconserved amino acids influence ligand specificity of the SBP because the hydrogen-bonding capabilities in the pocket are altered. In some RPA0668 homologs, Gly132 is substituted with threonine, which introduces additional hydrogenbonding potential in the ligand-binding site. Another example from RPA0668 is that Phe306 is often substituted with a tyrosine. As these residues are located at the bottom of the ligand binding pocket (Fig. 4a and b), it is tempting to speculate that for tyrosine-substituted homologs, the preferred ligand will shift from BEZ to PHB. It is unlikely that either of these single-site mutations would significantly change the cavity volume or alter the overall hydrophobic character of the RPA0668 pocket, but an altered network of hydrogen bonds could affect the complete ligand-binding profile. An example of these altered hydrogen-bonding networks might be illustrated by SMb20568 from subgroup Ib. In aligned protein sequences, this protein bears glutamine in a position equivalent to RPA0985 Phe40. Incorporation of the more polar residue near the ligand 3-hydroxyl substituent is consistent with increased protein stability of SMb20568 with 3-hydroxybenzoate or gallic acid relative to RPA0985 and RPB2270 proteins.
The purpose of overlapping transporter specificity is not currently understood, but the occurrence is not limited to R. palustris. Similar overlap is observed with aromatic transporters from the MFS family in multiple species. For example, in Acinectobacter strain ADP1, both PcaK and VanK can transport 3,4-dihydroxybenzoic acid.61 Some species contain multiple characterized transporters for benzoic acid.16,62 In this case, both genes are functional even though at least one ligand is shared between transporters. Since lignin products are diverse, the concentration of any single substrate in soil may be low enough that transporters with broad specificity confer a growth advantage. A related hypothesis is that low-level transport via a nonoptimized transporter may be necessary to induce expression of the high-affinity transporter.
Conclusions
We have identified a set of SBPs from several species of soil bacteria that could potentially be responsible for the uptake of various products of lignin degradation. The FTS-based assay facilitated the classification of these proteins as BEZ derivativespecific binding proteins. The biochemically inferred specificity has been validated by seven crystal structures of cluster I proteins in complex with four lignin-derived aromatic compounds. The structures revealed a ligand-binding pocket composed of predominantly hydrophobic residues with hydrophilic groups contributing H-bonding potential placed in specific locations. Despite low primary sequence identity in all seven structures, ligand position and orientation are virtually identical. We observe the key ligand specificity determinants and they include ligand interactions with both protein main-chain and side-chain atoms. The basic ligand recognition structure for this cluster is BEZ. The aromatic ring is placed in a hydrophobic environment and, in some cases, is involved in an NH–π interaction with an Asn side chain. The carboxyl group is coordinated by two main-chain amide groups as well as hydroxyl groups of Tyr and Ser/ Thr residues. SBPs that can accommodate larger and more elaborated ligands have increased cavity volumes and strategically positioned functional groups with H-bonding potential on the surface of the pocket. This arrangement offers preference to position 4 and 3 ring substitutions and discriminates against substitutions in ring position 2. SBPs with larger cavities can bind smaller ligands by attracting a water molecule to satisfy pocket H-bonding capabilities. One can envision further expansion of this cavity to accommodate even larger compounds.
This structural–functional characterization represents a major improvement in our ability to correctly infer the ligand-binding preferences of homologs of the characterized SBPs. These molecular insights validate the relationship between sequence and function for these proteins and provide a basis for the functional inference of gene function for a significant number of sequenced organisms. Many Alpha- and Betaproteobacteria have ABC transporters with uncharacterized SBPs that have predicted lignin degradation product importation capabilities. The improved functional assignments and ability to define specific sensory and regulatory pathways will increase the predictive capability of current models and support the development of future predictive systems-level models.
Materials and Methods
Homolog identification
Potential homologs of previously characterized SBPs from R. palustris CGA009 were identified in other organisms by sequence alignments generated with the BLAST algorithm.27 Sequenced genomes from R. palustris strains HaA2 (RPB), BisB18 (RPC), BisB5 (RPD), BisA53 (RPE), TIE-1 (Rpal), and DX-1; B. japonicum USDA 110 (bll, blr); S. meliloti 1021 (SM_a, SM_b, SM_c); and N. winogradskyi Nb-255 (Nwi) were included in this analysis. Gene coding sequences were obtained from the National Center for Biotechnology Information microbial genome data repository.
Ligand library
Reagent-grade chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Alfa Aesar (Ward Hill, MA). Ligands were dissolved in one of three buffers: standard Hepes buffer at pH7.5 (100mM Hepes and 150mM NaCl), nonstandard Hepes buffer at pH12.6, or 100% dimethyl sulfoxide (DMSO). Compatible solvents and ligand stock concentrations were determined by consultation of chemical reference sources and experimental evaluation of assay performance. Stocks were stored at 4°C and examined prior to each assay for evidence of precipitation or color change. None of the aromatic small molecules in this set produced background fluorescence in the screening assays due to ligand–dye interactions. Reactions containing gallic acid and 2,3,4-trihydroxybenzoic acid discolored during the thermal melt program but did not prevent determination of melting temperatures.
FTS assay
The FTS screening method25,29 was used in a microwell plate format to identify binding ligands for the SBPs of ABC transporters. Although previous screening protocols used pooled ligands followed by deconvolution of the binding profile with individual ligands, we anticipated binding events with multiple aromatic substances in this target set. Based on previous experience and well-supported sequence-based predictions, all protein-ligand combinations were screened individually in a directed screen. For all assays, the protein concentration was 10 µM in 20-µL reactions. The final ligand concentration in initial screens was 1000 µM, resulting in a 100-fold molar excess of small molecule to protein. The protein Tm was determined by manual inspection of the first-derivative curve generated by LightCycler480 analysis software (Roche, Indianapolis, IN, USA). All reported data represent an average of at least three experimental replicates.
In our analysis, a chemical was considered a “binding ligand” if the calculated ΔTm (relative to the Tm without ligand) was greater than 1°C. For ligands in DMSO or nonstandard Hepes, control reactions containing solvent only were performed for a side-by-side comparison to identify buffer effects, but variations induced by DMSO or basic buffer are generally 1 °C or less. The ΔTm significance threshold is based on previous studies25,29 for this family of proteins as well as a qualitative assessment of protein Tm variation between assay replicates.
Cloning
B. japonicum USDA 110 cells were obtained from Gary Stacey, University of Missouri at Columbia. Genomic DNA from B. japonicum was purified from cultures grown in HM-YA media using a ZymoBead Genomic DNA kit (#D3004, Zymo Research, Irvine, CA, USA). S. meliloti 1021 genomic DNA was a gift from Michael Kahn, Washington State University. Genomic DNA purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA) was used for R. palustris strains CGA009 (ATCC# BAA-98), HaA2 (ATCC# BAA-1122D-5), and BisB5 (ATCC# BAA-1123D-5). Genes included in the cloning set included proteins encoded at the following loci: RPB_4662, RPB_1579, RPB_2270, RPD_1586, RPD4371, bll5953, and SM_b20568. All SBP coding regions were analyzed for signal peptide sequences using SignalP63 and TatP64 prediction algorithms; signal peptides were excluded from the cloned region, resulting in expression of only the predicted mature protein. Primers were designed using a high-throughput primer design tool65 and ordered from IDT (Coralville, IA, USA). Sequences were PCR amplified from genomic DNA, fused to an N-terminal hexahistidine tag in an Escherichia coli cytoplasmic expression vector, and used to generate an E. coli production strain as described previously.29,66 All targets were sequenced to verify correct insertion of the coding region. RPB4662 and RPD4371 proteins were insoluble under our expression conditions and could not be analyzed by FTS assay. Reference proteins RPA0668, RPA0985, and RPA4029 were produced from vectors constructed in previous experiments.29 Small-scale heterologous protein expression in E. coli and IMAC purification using fused hexahistidine tags was carried out as described previously to generate protein utilized in FTS assays.29
Protein expression, purification, and crystallization
The SeMet derivative for all fusion protein was prepared as described previously.67 The BL21(DE3)/ pMAGIC cells were grown at 37°C in M9 medium supplemented with 0.4% glucose, 8.5mM NaCl, 0.1mM CaCl2, 2mM MgSO4, and 1% thiamine. After the OD600 (optical density at 600nm) reached 1.0–1.5, 0.01% (w/v) each of leucine, isoleucine, lysine, phenylalanine, threonine, and valine were added to inhibit the methionine metabolic pathway and increase SeMet incorporation. SeMet was added to the culture (60mg SeMet per liter of culture), and protein expression was induced with 0.5mM IPTG at 18°C overnight. Cells were harvested, resuspended in lysis buffer, and stored at −80°C.
Fusion protein was purified according to a standard protocol.68 Lysozyme (final concentration of 1mg/mL) and one protease inhibitor cocktail tablet (cOmplete ULTRA, Roche) were added to the thawed cell suspension. The solution was incubated on ice for 20min and lysed by sonication. The lysate was clarified by centrifugation at 36,000g for 1h and filtered through a 0.44-µm membrane. Clarified lysate was applied to a 5-mL HiTrap Ni-NTA column on the ÄKTAxpress system (GE Healthcare Life Sciences, Piscataway, NJ, USA). The column was washed with lysis buffer containing 20mM imidazole and the protein was eluted with the same buffer containing 250mM imidazole. Sample was concentrated and subjected to an additional purification step on a Superdex 200 10/30 size-exclusion chromatography column (GE Healthcare Life Sciences) equilibrated with crystallization buffer containing 20mM Hepes, pH8.0, 250mM NaCl, and 2mM DTT. Fractions containing monomeric protein were identified by SDS-PAGE and then concentrated for crystallization experiments to 35–40mg/mL (i.e. ∼1mM).
Crystallization screening was set up with the help of a Mosquito liquid dispenser (TTP LabTech, Cambridge, MA, USA) using the sitting-drop, vapor-diffusion method in 96-well CrystalQuick plates (Greiner Bio-One, Monroe, NC, USA). For co-crystallization trials, ligands were used at a 5-to 10-fold molar excess over protein concentration. For each condition, 0.4µL of protein solution and 0.4µL of crystallization formulation were mixed and the mixture was equilibrated against a 135-µL reservoir. The suite of four MCSG crystallization screens (Microlytic, Woburn, MA, USA) was used and conditions yielding diffraction-quality crystals typically appeared within 3–7days. The details of the crystallization conditions are given in Table 1.
Protein refolding
A subset of proteins (RPA0668, RPA0985, RPA4029, RPD1586, and Bll5953) was subjected to small-scale denaturation and refolding experiments to determine if proteins purified from E. coli cell culture contained prebound ligands. Purified protein in 1× Hepes buffer was thawed and diluted to less than 1mg/mL with denaturation buffer (100mM Hepes, pH7.5, 150mM NaCl, and 6M urea). Denatured protein was dialyzed against the denaturation buffer with three buffer exchanges to remove pre-bound ligands. SBPs were refolded by transferring the sample to 1× Hepes buffer and removing urea with three buffer exchanges. All dialysis steps were carried out at 4°C. After buffer exchange, samples were centrifuged at 15,000rpm for 10min to pellet precipitates; supernatant was then concentrated for use in FTS assays. Refolded proteins were used immediately in FTS assays and stored for no longer than a week at 4°C. Screens were performed as described above with an untreated protein sample run side by side for comparison. RPA0668 showed extensive precipitation and loss of binding functionality but other proteins were amenable to the refolding procedure.
Data collection
Prior to flash-cooling in liquid nitrogen, all the crystals were cryoprotected in an appropriate cryoprotectant solution. Specifically, the RPA0668/BEZ and RPA0668/ PHB crystals were briefly soaked in the reservoir solution supplemented with 10% glycerol. For the RPA0985/PHB crystals, the mother liquor was saturated with sucrose. The RPB2270/PHB crystal was cryoprotected in the reservoir solution containing 25% glycerol. The RPD1586/PHB, RPD1586/DHB, and RPD1586/VLA crystals were cryoprotected in the reservoir solution containing 10% ethylene glycol. The crystals were mounted on Litholoops (Molecular Dimensions, Apopka, FL, USA). All the X-ray diffraction experiments were performed at the Structural Biology Center ID-19 beamline at the Advanced Photon Source, Argonne National Laboratory. The SAD data sets were collected at 100K near the selenium K-absorption edge. The HKL3000 suite69 was used to process the diffraction images. Intensities were converted to structure factor amplitudes in the Truncate program from the CCP4 package.70 The processing statistics are given in Table 1.
Structure solution and refinement
All the structures were solved by SAD method using selenium peak data and the HKL3000 software pipeline.69 Selenium sites were localized by SHELXD and the handedness was determined by SHELXE.71 Phasing was performed in MLPHARE72 and was followed by density modification procedure in DM.73 The initial protein models were built in ARP/wARP.74 Manual model adjustment was carried out in Coot75 and crystallographic refinement was performed in BUSTER-TNT76 (RPA0668/BEZ), PHENIX48 (RPA0668/PHB), or REFMAC546 (RPA0985/PHB, RPB2270/PHB, RPD1586/ PHB, RPD1586/DHB, and RPD1586/VLA). The RPA0668/PHB, RPA0985/PHB, and RPB2270/PHB structures were refined with individual anisotropic B-factors. The refinement protocol for the RPA0668/BEZ and RPD1586 structures included TLS (translation/ libration/screw) refinement with eight or five TLS groups per protein monomer.47 The refinement statistics are shown in Table 1.
Phylogenetic analysis
Phylogenetic trees were built using the a la carte function on the Phylogeny.fr server†. 77 Protein sequences were aligned using ClustalW and the phylogenetic distance was calculated using PhyML. The resulting tree was edited using the Drendroscope program.78
PDB accession codes
The atomic coordinates and structure factors have been deposited in the PDB with accession codes 4EVR (RPA0668/BEZ), 4EVQ (RPA0668/PHB), 4EVS (RPA0985/PHB), 4F06 (RPB2270/PHB), 4EY3 (RPD1586/PHB), 4EYK (RPD1586/DHB), and 4EYG (RPD1586/VLA).
Supplementary Material
Acknowledgements
We would like to thank the members of the Structural Biology Center for their support and help in data collection. This contribution originates in part from the “Environment Sensing and Response” Scientific Focus Area program at Argonne National Laboratory. This research was supported by the U.S. Department of Energy, Office of Biological and Environmental Research (BER), as part of BER's Genomic Science Program. This research has been funded in part by a grant from the National Institutes of Health (GM094585) and by the U.S. Department of Energy, BER, under Contract DE-AC02-06CH11357.
Abbreviations used
- ABC
ATP-binding cassette
- ATCC
American Type Culture Collection
- BEZ
benzoate
- DHB
3,4-dihydroxybenzoate
- DMSO
dimethyl sulfoxide
- FTS
fluorescence thermal shift
- MFS
major facilitator superfamily
- PDB
Protein Data Bank
- PHB
4-hydroxybenzoate
- SAD
single-wavelength anomalous diffraction
- SBP
solute-binding protein
- SeMet
selenomethionine
- VLA
vanillate
Footnotes
Supplementary materials related to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2012.08.017.
References
- 1.Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, et al. Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 2004;3:29–60. [Google Scholar]
- 2.Wong DW. Structure and action mechanism of ligninolytic enzymes. Appl. Biochem. Biotechnol. 2009;157:174–209. doi: 10.1007/s12010-008-8279-z. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez C. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol. Adv. 2009;27:185–194. doi: 10.1016/j.biotechadv.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Ko JJ, Shimizu Y, Ikeda K, Kim SK, Park CH, Matsui S. Biodegradation of high molecular weight lignin under sulfate reducing conditions: lignin degradability and degradation by-products. Bioresour. Technol. 2009;100:1622–1627. doi: 10.1016/j.biortech.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 5.Bugg TD, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 2011;22:394–400. doi: 10.1016/j.copbio.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Brown ME, Walker MC, Nakashige TG, Iavarone AT, Chang MC. Discovery and characterization of heme enzymes from unse-quenced bacteria: application to microbial lignin degradation. J. Am. Chem. Soc. 2011 doi: 10.1021/ja203972q. [DOI] [PubMed] [Google Scholar]
- 7.Masai E, Katayama Y, Fukuda M. 2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71:1–15. doi: 10.1271/bbb.60437. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs G. Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 2008;1125:82–99. doi: 10.1196/annals.1419.010. [DOI] [PubMed] [Google Scholar]
- 9.Carmona M, Zamarro MT, Blazquez B, Durante-Rodriguez G, Juarez JF, Valderrama JA, et al. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol. Mol. Biol. Rev. 2009;73:71–133. doi: 10.1128/MMBR.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harwood CS, Parales RE. The betaketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 11.Phelps CD, Young LY. Microbial metabolism of the plant phenolic compounds ferulic and syringic acids under three anaerobic conditions. Microb. Ecol. 1997;33:206–215. doi: 10.1007/s002489900023. [DOI] [PubMed] [Google Scholar]
- 12.Diaz E. Bacterial degradation of aromatic pollutants: a paradigm of metabolic versatility. Int. Microbiol. 2004;7:173–180. [PubMed] [Google Scholar]
- 13.Egland PG, Pelletier DA, Dispensa M, Gibson J, Harwood CS. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl Acad. Sci. USA. 1997;94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saier MH, Jr., Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, et al. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 15.Chae JC, Zylstra GJ. 4-Chlorobenzoate uptake in Comamonas sp. strain DJ-12 is mediated by a tripartite ATP-independent periplasmic transporter. J. Bacteriol. 2006;188:8407–8412. doi: 10.1128/JB.00880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhry MT, Huang Y, Shen XH, Poetsch A, Jiang CY, Liu SJ. Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology. 2007;153:857–865. doi: 10.1099/mic.0.2006/002501-0. [DOI] [PubMed] [Google Scholar]
- 17.Paulsen IT, Nguyen L, Sliwinski MK, Rabus R, Saier MH., Jr. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 18.Paulsen IT, Sliwinski MK, Saier MH., Jr. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 19.Peng RH, Xiong AS, Xue Y, Fu XY, Gao F, Zhao W, et al. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 2008;32:927–955. doi: 10.1111/j.1574-6976.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- 20.Biemans-Oldehinkel E, Doeven MK, Poolman B. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 2006;580:1023–1035. doi: 10.1016/j.febslet.2005.11.079. [DOI] [PubMed] [Google Scholar]
- 21.Hosie AH, Allaway D, Jones MA, Walshaw DL, Johnston AW, Poole PS. Solutebinding protein-dependent ABC transporters are responsible for solute efflux in addition to solute uptake. Mol. Microbiol. 2001;40:1449–1459. doi: 10.1046/j.1365-2958.2001.02497.x. [DOI] [PubMed] [Google Scholar]
- 22.Tam R, Saier MH., Jr. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara H, Stewart GR, Mohn WW. Involvement of a novel ABC transporter and monoalkyl phthalate ester hydrolase in phthalate ester catabolism by Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2010;76:1516–1523. doi: 10.1128/AEM.02621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arias-Barrau E, Sandoval A, Naharro G, Olivera ER, Luengo JM. A two-component hydroxylase involved in the assimilation of 3-hydroxyphenyl acetate in Pseudomonas putida . J. Biol. Chem. 2005;280:26435–26447. doi: 10.1074/jbc.M501988200. [DOI] [PubMed] [Google Scholar]
- 25.Giuliani SE, Frank AM, Collart FR. Functional assignment of solute-binding proteins of ABC transporters using a fluorescence-based thermal shift assay. Biochemistry. 2008;47:13974–13984. doi: 10.1021/bi801648r. [DOI] [PubMed] [Google Scholar]
- 26.Oda Y, Larimer FW, Chain PS, Malfatti S, Shin MV, Vergez LM, et al. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc. Natl Acad. Sci. USA. 2008;105:18543–18548. doi: 10.1073/pnas.0809160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs G, Boll M, Heider J. Microbial degradation of aromatic compounds—from one strategy to four. Nat. Rev. Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 29.Giuliani SE, Frank AM, Corgliano DM, Seifert C, Hauser L, Collart FR. Environment sensing and response mediated by ABC transporters. BMC Genomics. 2011;12:S8. doi: 10.1186/1471-2164-12-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamura K, Takata K, Hiraishi A. Intrageneric relationships of members of the genus Rhodopseudomonas . J. Gen. Appl. Microbiol. 2009;55:469–478. doi: 10.2323/jgam.55.469. [DOI] [PubMed] [Google Scholar]
- 31.Addou S, Rentzsch R, Lee D, Orengo CA. Domain-based and family-specific sequence identity thresholds increase the levels of reliable protein function transfer. J. Mol. Biol. 2009;387:416–430. doi: 10.1016/j.jmb.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 32.Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 33.Martinez AT, Speranza M, Ruiz-Duenas FJ, Ferreira P, Camarero S, Guillen F, et al. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005;8:195–204. [PubMed] [Google Scholar]
- 34.Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 35.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 36.Mauchline TH, Fowler JE, East AK, Sartor AL, Zaheer R, Hosie AH, et al. Mapping the Sinorhizobium meliloti 1021 solute-binding proteindependent transportome. Proc. Natl Acad. Sci. USA. 2006;2006;103:17933–17938. doi: 10.1073/pnas.0606673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maclean AM, Haerty W, Golding GB, Finan TM. The LysR-type PcaQ protein regulates expression of a protocatechuate-inducible ABC-type transport system in Sinorhizobium meliloti . Microbiology. 2011;157:2522–2533. doi: 10.1099/mic.0.050542-0. [DOI] [PubMed] [Google Scholar]
- 38.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 40.Fukami-Kobayashi K, Tateno Y, Nishikawa K. Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 1999;286:279–290. doi: 10.1006/jmbi.1998.2454. [DOI] [PubMed] [Google Scholar]
- 41.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 42.Mao B, Pear MR, McCammon JA, Quiocho FA. Hinge-bending in L-arabinose-binding protein. The “Venus's-flytrap” model. J. Biol. Chem. 1982;257:1131–1133. [PubMed] [Google Scholar]
- 43.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 44.Flocco MM, Mowbray SL. The 1.9 Å x-ray structure of a closed unliganded form of the periplasmic glucose/galactose receptor from Salmonella typhimurium . J. Biol. Chem. 1994;269:8931–8936. [PubMed] [Google Scholar]
- 45.Oswald C, Smits SH, Hoing M, Sohn-Bosser L, Dupont L, Le Rudulier D, et al. Crystal structures of the choline/acetylcholine substratebinding protein ChoX from Sinorhizobium meliloti in the liganded and unligandedclosed states. J. Biol. Chem. 2008;283:32848–32859. doi: 10.1074/jbc.M806021200. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 48.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aarset K, Page EM, Rice DA. Molecular structures of 3-hydroxybenzoic acid and 4-hydroxy-benzoic acid, obtained by gas-phase electron diffraction and theoretical calculations. J. Phys. Chem. A. 2008;112:10040–10045. doi: 10.1021/jp804539u. [DOI] [PubMed] [Google Scholar]
- 50.Bruno G, Randaccio L. A refinement of the benzoic-acid structure at room-temperature. Acta Crystallogr., Sect. B: Struct. Sci. 1980;36:1711–1712. [Google Scholar]
- 51.Fun HK, Balasubramani K. 2,3-Diaminopyridinium 4-hydroxybenzoate. Acta Crystallogr., Sect. E: Struct. Rep. Online. 2009;65:01496–U1820. doi: 10.1107/S1600536809021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muhammad K, Rauf MK, Ebihara M, Hameed S. 4-(4-Octyloxybenzoyloxy)benzoic acid. Acta Crystallogr., Sect. E: Struct. Rep. Online. 2009;65:O424–U3042. doi: 10.1107/S1600536809003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph. 1995;13:307–308. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 54.Pietri R, Zerbs S, Corgliano DM, Allaire M, Collart FR, Miller LM. Biophysical and structural characterization of a sequence-diverse set of solute-binding proteins for aromatic compounds. J. Biol. Chem. 2012;287:23748–23756. doi: 10.1074/jbc.M112.352385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 56.Magnusson U, Salopek-Sondi B, Luck LA, Mowbray SL. X-ray structures of the leucinebinding protein illustrate conformational changes and the basis of ligand specificity. J. Biol. Chem. 2004;279:8747–8752. doi: 10.1074/jbc.M311890200. [DOI] [PubMed] [Google Scholar]
- 57.Trakhanov S, Vyas NK, Luecke H, Kristensen DM, Ma J, Quiocho FA. Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity. Biochemistry. 2005;44:6597–6608. doi: 10.1021/bi047302o. [DOI] [PubMed] [Google Scholar]
- 58.Planamente S, Vigouroux A, Mondy S, Nicaise M, Faure D, Morera S. A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens . J. Biol. Chem. 2010;285:30294–30303. doi: 10.1074/jbc.M110.140715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pearl L, Ohara B, Drew R, Wilson S. Crystal structure of amic—the controller of transcription antitermination in the amidase operon of Pseudomonas aeruginosa . EMBO J. 1994;13:5810–5817. doi: 10.1002/j.1460-2075.1994.tb06924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D'Argenio DA, Segura A, Coco WM, Bunz PV, Ornston LN. The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J. Bacteriol. 1999;181:3505–3515. doi: 10.1128/jb.181.11.3505-3515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishikawa Y, Yasumi Y, Noguchi S, Sakamoto H, Nikawa J. Functional analyses of Pseudomo-nas putida benzoate transporters expressed in the yeast Saccharomyces cerevisiae . Biosci. Biotechnol. Biochem. 2008;72:2034–2038. doi: 10.1271/bbb.80156. [DOI] [PubMed] [Google Scholar]
- 63.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 64.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoon JR, Laible PD, Gu M, Scott HN, Collart FR. Express primer tool for high-throughput gene cloning and expression. Biotechniques. 2002;33:1328–1333. doi: 10.2144/02336bc03. [DOI] [PubMed] [Google Scholar]
- 66.Zerbs S, Frank AM, Collart FR. Bacterial systems for production of heterologous proteins. Methods Enzymol. 2009;463:149–168. doi: 10.1016/S0076-6879(09)63012-3. [DOI] [PubMed] [Google Scholar]
- 67.Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: ultrafast protein structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55:1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- 68.Kim Y, Dementieva I, Zhou M, Wu R, Lezondra L, Quartey P, et al. Automation of protein purification for structural genomics. J. Struct. Funct. Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 70.The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 71.Sheldrick GM. A short history of SHELX. Acta Crystallogr., Sect. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 72.Otwinowski Z. Daresbury Study Weekend proceedings. 1991 [Google Scholar]
- 73.Cowtan K. DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994:34–38. [Google Scholar]
- 74.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 77.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.