Abstract
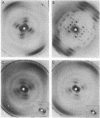
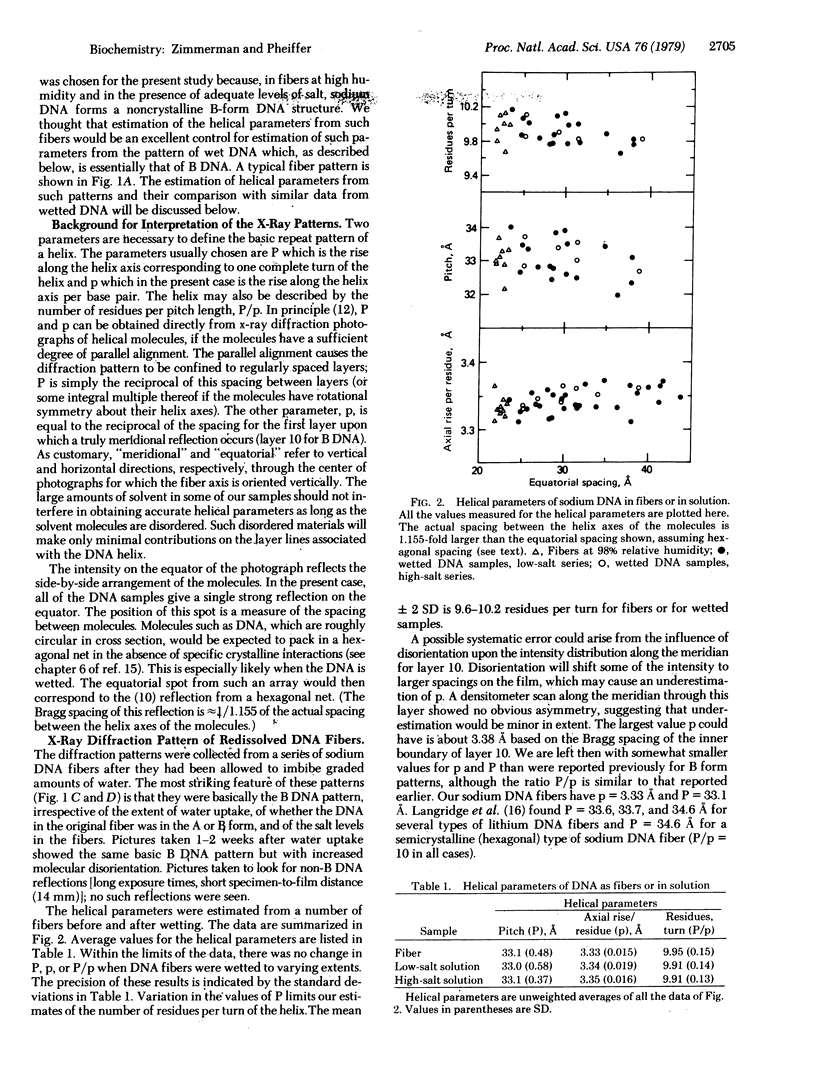
We have measured the helical parameters of DNA in concentrated solutions by x-ray fiber diffraction methods. Fibers of the sodium salt of DNA were swollen with water within capillaries; the capillary served to limit water uptake, slowing dissolution. Samples containing up to 80% water gave essentially a B-form diffraction pattern and had virtually the same helical parameters [9.91 base pairs per turn (SD = 0.14); 3.34 A axial rise per residue (SD = 0.019)] as did the initial fibers [9.95 base pairs per turn (SD = 0.15); 3.33 A axial rise per residue (SD = 0.015)]. Hence, under highly solvated conditions in which the interactions between molecules should be greatly decreased, DNA maintains its classical B-form structure.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291x(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Bram S. The secondary structure of DNA in solution and in nucleohistone. J Mol Biol. 1971 May 28;58(1):277–288. doi: 10.1016/0022-2836(71)90246-4. [DOI] [PubMed] [Google Scholar]
- Cooper P. J., Hamilton L. D. The A-B conformational change in the sodium salt of DNA. J Mol Biol. 1966 Apr;16(2):562–563. doi: 10.1016/s0022-2836(66)80193-6. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover S. D. Symmetry and packing in B-DNA. J Mol Biol. 1977 Mar 15;110(4):699–700. doi: 10.1016/s0022-2836(77)80085-5. [DOI] [PubMed] [Google Scholar]
- Eisenberg H., Cohen G. An interpretation of the low-angle X-ray scattering of DNA solutions. J Mol Biol. 1968 Oct 28;37(2):355–362. doi: 10.1016/0022-2836(68)90274-x. [DOI] [PubMed] [Google Scholar]
- FRANKLIN R. E., GOSLING R. G. Molecular configuration in sodium thymonucleate. Nature. 1953 Apr 25;171(4356):740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- FULLER W., WILKINS M. H., WILSON H. R., HAMILTON L. D. THE MOLECULAR CONFIGURATION OF DEOXYRIBONUCLEIC ACID. IV. X-RAY DIFFRACTION STUDY OF THE A FORM. J Mol Biol. 1965 May;12:60–76. doi: 10.1016/s0022-2836(65)80282-0. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. DNA structure: evidence from electron microscopy. Science. 1978 Aug 11;201(4355):525–527. doi: 10.1126/science.663672. [DOI] [PubMed] [Google Scholar]
- Hogan M., Dattagupta N., Crothers D. M. Transient electric dichroism of rod-like DNA molecules. Proc Natl Acad Sci U S A. 1978 Jan;75(1):195–199. doi: 10.1073/pnas.75.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARVIN D. A., SPENCER M., WILKINS M. H., HAMILTON L. D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J Mol Biol. 1961 Oct;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- Neville D. M., Jr, Davies D. R. The interaction of acridine dyes with DNA: an x-ray diffraction and optical investigation. J Mol Biol. 1966 May;17(1):57–74. doi: 10.1016/s0022-2836(66)80094-3. [DOI] [PubMed] [Google Scholar]
- Vollenweider H. J., James A., Szybalski W. Discrete length classes of DNA depend on mode of dehydration. Proc Natl Acad Sci U S A. 1978 Feb;75(2):710–714. doi: 10.1073/pnas.75.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- WILKINS M. H. F., STOKES A. R., WILSON H. R. Molecular structure of deoxypentose nucleic acids. Nature. 1953 Apr 25;171(4356):738–740. doi: 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B. Polynucleotide models. II. Prediction of the radial position and tilt of the bases from the helical parameters. Biopolymers. 1977 Apr;16(4):749–763. doi: 10.1002/bip.1977.360160405. [DOI] [PubMed] [Google Scholar]