Background: Mutations in the NEMO ZF cause various forms of anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID).
Results: The NEMO ZF contains a larger ubiquitin binding surface, which is required for IKK/NF-κB activation.
Conclusion: This study provides an understanding of how defective mutations of NEMO ZF in NF-κB signaling leads to the EDA-ID pathology.
Significance: This study provides novel insights in the NEMO ZF-dependent mechanisms for IKK activation in NF-κB signaling.
Keywords: Fluorescence, Immunodeficiency, NF-κB, Ubiquitin, Zinc Finger, EDA-ID, IKK Complex, NEMO
Abstract
Hypomorphic mutations in the X-linked human NEMO gene result in various forms of anhidrotic ectodermal dysplasia with immunodeficiency. NEMO function is mediated by two distal ubiquitin binding domains located in the regulatory C-terminal domain of the protein: the coiled-coil 2-leucine zipper (CC2-LZ) domain and the zinc finger (ZF) domain. Here, we investigated the effect of the D406V mutation found in the NEMO ZF of an ectodermal dysplasia with immunodeficiency patients. This point mutation does not impair the folding of NEMO ZF or mono-ubiquitin binding but is sufficient to alter NEMO function, as NEMO-deficient fibroblasts and Jurkat T lymphocytes reconstituted with full-length D406V NEMO lead to partial and strong defects in NF-κB activation, respectively. To further characterize the ubiquitin binding properties of NEMO ZF, we employed di-ubiquitin (di-Ub) chains composed of several different linkages (Lys-48, Lys-63, and linear (Met-1-linked)). We showed that the pathogenic mutation preferentially impairs the interaction with Lys-63 and Met-1-linked di-Ub, which correlates with its ubiquitin binding defect in vivo. Furthermore, sedimentation velocity and gel filtration showed that NEMO ZF, like other NEMO related-ZFs, binds mono-Ub and di-Ub with distinct stoichiometries, indicating the presence of a new Ub site within the NEMO ZF. Extensive mutagenesis was then performed on NEMO ZF and characterization of mutants allowed the proposal of a structural model of NEMO ZF in interaction with a Lys-63 di-Ub chain.
Introduction
The NF-κB pathway represents a prototypic example of a signaling pathway in which integration of multiple types of ubiquitination is required to activate NF-κB transcription factors (1, 2). In unstimulated cells, NF-κB transcription factors interact with inhibitors of the IκB family, and the resulting complexes are inactive in the cytoplasm. Stimulation of cells with a variety of stimuli, including inflammatory cytokines (TNF, IL-1) and toll-like receptor (TLR) ligands, leads to the activation of the IκB kinase (IKK)4 complex composed of two kinases (IKK-α and IKK-β) and a non-catalytic regulatory protein called NEMO (NF-κB essential modulator, also known as IKK-γ). The active IKK complex phosphorylates the IκB proteins, promoting their Lys-48-linked ubiquitination and degradation by the proteasome. This allows NF-κB to enter into the nucleus and induce the transcription of target genes involved in immunity, inflammation, cell survival, and apoptosis regulation.
Although efforts over the past few years have yielded considerable structural data related to the IKK complex, the in vivo NEMO-dependent mechanism of IKK activation remains ambiguous. This is likely due to the fact that NEMO may use multiple mechanisms to orchestrate IKK activation. NEMO-dependent IKK activation can be achieved by IKK trans-autophosphorylation and/or by an upstream kinase such as TGF-β-activated kinase 1 (TAK1). However, TAK1 does not represent the sole upstream kinase involved in IKK activation (3), and its requirement depends on the cell type (4).
In all models of NEMO-dependent IKK activation, (trans-autophosphorylation, TAK1-induced phosphorylation, and a concerted mechanism) full NF-κB and IKK activation depend on the ability of NEMO to interact with ubiquitin chains such as linear (5), Lys-63 chains (6), and Lys-63/linear hybrid chains (7) as well as its ability to form higher order, oligomeric signaling complexes (1, 8). The ubiquitin binding activity requires NEMO dimerization and depends on two ubiquitin binding domains (UBD); one is located in the C-terminal coiled-coil domain, referred to as NOA/UBAN domain (also called NUB or CC2-LZ); the other is located in the C-terminal zinc finger (ZF) domain. Both NEMO UBD are separated by an unstructured Pro rich linker of 50 residues. Although numerous reports have sought to characterize the NOA domain, the properties of the NEMO ZF UBD are still poorly understood. Recently, it has been shown that the NEMO ZF, but not the NOA/UBAN domain, plays a crucial role in IKKβ activation through the binding to mono-ubiquitylated PKCϵ in EGF signaling (9).
The critical role of NEMO ZF in the NF-κB pathway has been established by showing that NEMO ZF mutants are neither able to restore TNF-induced-NF-κB activation in NEMO-deficient mouse embryonic fibroblasts (MEF) (10) nor NF-κB activation triggered by genotoxic stress in NEMO-deficient Pre-B lymphocytes (11). Importantly, this NF-κB activation could be restored in NEMO-deficient T or MEF cells in response to several stimuli when the NEMO ZF domain was replaced with other ZFs known to interact with ubiquitin such as the ubiquitin-binding zinc finger (UBZ) domain of Werner helicase-interacting protein 1 (WRNP1), providing strong evidence that the essential, if not unique, function of NEMO-ZF is to interact with ubiquitin (12). Recently, a new role of NEMO ZF was reported in which the ZF functions as a docking site to enhance the IκBα substrate specificity for IKKβ. However, it is difficult to envision how ZF can achieve this function at a structural level given the dimeric or tetrameric crystal structure of IKKβ (13).
UBDs based on small ZF modules can structurally be divided into three classes: Npl4 zinc finger (NZF), A20-like zinc finger (A20 Znf), and ubiquitin-binding zinc finger (UBZ). The structure of the NEMO ZF domain has been solved by NMR (14), and a model has been proposed for the interaction with mono-ubiquitin based on NMR chemical shift perturbations with 15N-labeled mono-ubiquitin and mutagenesis data (15). Like many UBD domains, the NEMO ZF binds the mono-ubiquitin via the hydrophobic patch around Ile-44 of ubiquitin. This interaction occurs via the hydrophobic side of the ZF α-helix in a similar manner as ZF of the ubiquitin binding zinc finger (UBZ) class (16).
Several crystal structures of NZF and A20-like zinc finger domains in interaction with a di-Ub chain or three mono-ubiquitins have recently emerged. Among them, NZF domains of TAB2/TAB3 (17, 18) and A20 ZnF4 bind to Lys-63-linked ubiquitin chains (19), whereas the NZF domain of the HOIL-1L subunit of LUBAC complex and A20 Zn7 exhibits linkage specificity for linear ubiquitin chains (20, 21). For all of them, linkage specificity is not provided by a direct recognition of ZF binding determinants with the Lys-63 or linear linkage but rather by linkage-dependent spatial conformations taken by different di-ubiquitins. Interestingly, although all ZF UBDs interact with the canonical Ile-44-based surface of one ubiquitin, they use different binding surfaces on adjacent ubiquitins to achieve their linkage specificity.
A few years after its discovery, mutations within the gene encoding NEMO were found to be associated with the X-linked human diseases incontinentia pigmenti and immunodeficiency with or without anhydrotic ectodermal dysplasia (ID or EDA-ID) (2). To decipher the NEMO-dependent mechanism of IKK and NF-κB activation, we previously investigated in detail how EDA-ID and incontinentia pigmenti mutations affect NEMO function. None of these mutations leading to defects of NF-κB activation affects NEMO expression. We showed that A288G and Glu-315 EDA-ID mutations as well as A323P incontinentia pigmenti mutation in CC2-LZ, whether or not in the NOA/UBAN binding site, strongly reduce folding and stability of NEMO oligomers (dimers and higher order oligomers), thereby leading to a defect in its ubiquitin binding activity (22) as well as in its TRAF6-induced Lys-63 ubiquitination (23). In the case of the C417F EDA-ID mutation in ZF domain, the solution structure of the mutant by NMR revealed a disordered region with an important loss of stability, providing a structural basis to explain the EDA-ID phenotype (14). Nevertheless, all pathogenic mutations do not affect the stability of NOA or ZF domains. New patient mutations in the NOA site were recently identified that, contrary to previously studied mutations, do not affect folding but specifically impair interactions with Lys-63-linked and linear poly-Ub chains (24).
Here we investigated the effect of the D406V mutation previously reported in the NEMO ZF of an EDA-ID patient. This D406V patient exhibited typical signs of EDA (absence of sweat glands, paucity of hair follicles, and dry scaly skin) with a deficiency in B-cell switching in response to CD40 activation (25). We showed that this mutation does not alter in vitro the folding of ZF or mono-Ub binding, although the mutation strongly reduced the NF-κB activation in Jurkat T lymphocytes and to a lesser extent in MEF fibroblasts. A thorough analysis of the ubiquitin properties of WT and mutant NEMO ZF showed that the mutation in vitro alters the binding with Lys-63 and linear di-ubiquitin, which correlates with its ubiquitin binding defect in vivo. Extensive mutagenesis on the ZF was then undertaken to localize a new ubiquitin binding site to which Asp-406 belongs, and a model of ZF·Lys-63 di-Ub complex was proposed. Finally, we further verified the functional importance of the second Ub site of NEMO ZF for NF-κB activation.
EXPERIMENTAL PROCEDURES
Plasmids, ZF Peptides, Antibodies, and Reagents
pcDNA3 plasmids (Invitrogen) encoding human FLAG-WT NEMO and its mutants ΔZF (ZF deletion), D406V, F395S, and F395W and the pGEX4T2-di-Ub plasmid (GE Healthcare) encoding linear di-ubiquitin were prepared using conventional molecular cloning and PCR mutagenesis methods as described previously (22, 26). pLenti6.4/R4R2/V5-DEST-based plasmids (Invitrogen) encoding FLAG-WT and FLAG-D406V NEMO and a pET52b3C/LIC plasmid (Merck Millipore) encoding StrepTag II-tagged human ubiquitin-conjugating enzyme E2–25K-C170S were prepared using Gateway Technology and Ligation Independent Cloning methods, respectively, following the manufacturer's instructions. pGEX-Ubc13 (18894) and pET16b-Mms2 (18893) were provided by Addgene (Cambridge, MA). NF-κB luciferase reporter and pcDNA3-FLAG-NEMO-ΔZF plasmids as well as pEF1-βgal plasmid to normalize transfection have been previously described (15). Unlabeled and N-terminal fluorescein-labeled-NEMO ZF (residues 392–419), ABIN2 ZF (residues 399–429), and NRP ZF (residues 549–577) peptides (>98% purity) were purchased from Covalab (Villeurbane, France). MALDI mass spectroscopy confirmed the fluorescein/ZF peptide molar ratio of 1 to 1 for all fluorescein-labeled peptides. Antibodies against the following proteins or tags were: anti-ubiquitin (U0508, Sigma), anti-FLAG M2 (F3165, Sigma), anti-StrepTag II (Merck Millipore), anti-IκBα (#4814, Cell Signaling), and anti-GST (ABN116, Merck Millipore). Cells were incubated with Polybrene (Sigma), TNF-α (BD Pharmingen), IL-1β (BD Pharmingen), or LPS from Salmonella abortus (Sigma). Homemade antibodies against NEMO (B24) and nucleoside diphosphate kinase B (NDPK-B) were previously described in Ref. 14.
Cell Culture, Transient Transfections, Retroviral Transduction, Western Blot, and Coimmunoprecipitation
Human 293T cells (American Type Culture Collection) and NEMO-deficient MEFs (27) were grown in DMEM, and Jurkat T cells (JM4.5.2; Ref 28) were cultured in RPMI supplemented with 10% fetal bovine serum, penicillin (50 units/ml), and streptomycin (50 μg/ml) as previously described (22, 29). NEMO-deficient Jurkat T and 293T cells were transiently transfected using FuGENE HD (Roche Applied Science) and Lipofectamine 2000 (Invitrogen), respectively, according to the manufacturer's instructions. Retroviral transduction of NEMO-deficient MEFs with pLenti6.4 NEMO constructs was described elsewhere (26). Immunoprecipitation, Western blotting, and NF-κB luciferase reporter assay were performed as previously described (15).
Ubiquitin Expression and Purification of Various Proteins
Ubc13 and linear di-Ub expressed as a glutathione S-transferase (GST) fusion protein and His-Mms2 were purified from soluble bacterial extracts with glutathione Sepharose 4B (GE Healthcare) and nickel-nitrilotriacetic acid resin (Qiagen) depending on the tag according to the manufacturer's instructions. StrepTagII-E2–25K-C170S was purified from insoluble bacterial extracts, solubilized by 6 m urea in 100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, and 1 mm dithioerythritol, and centrifuged at 17,000 × g for 15 min at room temperature. The desired protein was then refolded by dilution (0.8 m urea) and purified using a 10-ml streptactin column according to the manufacturer's instructions (Qiagen). After appropriate protease cleavage (thrombin or HRV 3C protease), the GST and StrepTagII fragments were removed by repeating the affinity purification step. After purification, all proteins were concentrated and stored at −80 °C. Lys-63- and Lys-48-di-Ub were enzymatically produced according to the protocol derived from the one described in Komander et al. (29). Briefly, 1 mm mono-Ub (Boston Biochem) was incubated for 4 h at 37 °C with 0.1 μm E1 and 4–30 μm E2 (E25K-C170S for Lys-48-linkage and Mms2-Ubc13 for Lys-63-linkage) in reaction buffer containing 50 mm Tris-HCl, pH 8, 5 mm MgCl2, 10 mm creatine phosphate, 0.6 units/ml inorganic phosphate, 0.6 units/ml creatine phosphokinase, 2.5 mm ATP, and 0.5 mm dithioerythritol. The reaction mixtures were then stopped by the addition of 5 mm dithioerythritol and 1 mm EDTA and acidified to pH 4.5 with 2 n acetic acid before loading on a Resource S column (6-ml GE Healthcare) equilibrated in buffer 50 mm ammonium acetate buffer, pH 4.5 containing 1 mm EDTA and 1 mm dithioerythritol. The desired chain length of Ub chains was recovered by elution using a 240-ml gradient of NaCl (0–0.6 m). Fractions containing the desired di-Ub were concentrated by ultrafiltration (Amicon Ultra, Millipore) and extensively dialyzed twice against 50 mm Tris-HCl, pH 7.5, containing 150 mm NaCl before freezing at −80 °C. All protein concentrations were determined either by the method of Bradford or by absorbance at 280 nm using extinction coefficients deduced from the amino acid sequence.
NEMO ZF Polyubiquitin Binding Assays
293T cells (2.5 × 106) transiently transfected with pcDNA3 plasmids containing the WT or mutant FLAG-NEMO or with an empty plasmid were lysed at 4 °C in 50 mm Tris-HCl buffer, pH 7.5, containing 200 mm NaCl, 1 mm dithioerythritol, and 1% Triton X-100 (LB buffer) supplemented with an EDTA-free protease inhibitor mixture. After centrifugation, crude extracts were incubated at 4 °C for 90 min with anti-FLAG M2 agarose beads, and the beads were washed 3 times with the LB buffer before elution with the FLAG peptide (2 mg/ml). The recovered supernatant was then incubated overnight at 4 °C with His-DARPin (1D5) immobilized on nickel-nitrilotriacetic acid magnetic agarose beads (100 μl, Qiagen). After 2 washes with LB buffer containing 0.2% Triton X-100, nickel-nitrilotriacetic acid beads were incubated for 90 min at 4 °C with linear, Lys-48 or Lys-63 tetra-ubiquitin chains (each at a concentration of 36 μm) in 50 mm Tris-HCl, pH 7.5, containing 200 mm NaCl, 0.1 mm Tris(2-carboxyethyl)phosphine, 10% glycerol, and 20 mm imidazole (buffer B). After extensive washing in buffer B, the ubiquitin complexes were eluted with 0.5 m imidazole, and the amount of ubiquitin bound was analyzed by immunoblotting with an anti-ubiquitin antibody.
Size Exclusion Chromatography
Before size exclusion chromatography analysis, all fluorescein-labeled ZF peptides were allowed to fold in binding assay buffer A, consisting of 50 mm Tris-HCl buffer, pH 7.5, 150 mm NaCl, 0.1 mm dodecyl maltoside, 1 mm Tris(2-carboxyethyl)phosphine, and 0.6 mm ZnCl2. To analyze the interaction between the WT F-ZF (20 μm) and its D406V mutant (20 μm) with either mono-Ub (20 μm) or various forms of di-Ub (20 μm Lys-48-, Lys-63, or linear), size exclusion chromatography was performed at 4 °C by loading 400-μl samples on a Superdex 75 10/300 HR column (GE Healthcare) equilibrated at a flow rate of 0.4 ml/min in the binding assay buffer A without ZnCl2. The molecular mass calibration curve of the column was determined in the same buffer using the following standard globular proteins: BSA, ovalbumin, chymotrypsinogen A, ribonuclease A, cytochrome, and aprotinin. The protein elution volume was monitored at 280 nm, whereas the fluorescein-labeled WT and mutant ZFs (free and bound) was monitored at 485 nm. The respective elution volume of F-ZF and various forms of ubiquitin (mono-Ub, Lys-63-, Lys-48-, and linear di-Ub) were determined by loading the protein sample alone. When the ZF·ubiquitin complex is well-separated from free ZF and free mono-or di-Ub, binding stoichiometry of the complex could be accurately determined by calculating the absorbance ratio of the complex at 485 and 280 nm using extension coefficients of 41,000 m−1·cm−1 at 485 nm and pH 7.5 for the fluorescein-labeled ZF and of 17,940, 1,370, and 2,740 m−1·cm−1 at 280 nm for F-ZF, mono-Ub, and di-Ub, respectively.
Sedimentation Velocity (SV) Analytical Ultracentrifugation (AUC)
Sedimentation velocity experiments were carried out at 20 °C in a Beckman Coulter XL-I analytical ultracentrifuge equipped with double UV and Rayleigh interference detection. After refolding the ZF peptide with a molar excess of zinc, all protein samples were extensively dialyzed overnight against buffer A without ZnCl2. Data were collected at 42,000 rpm for 6 h using an AN50 Ti rotor containing 12-mm path-length epoxy double-sector cell filled with 400 μl of sample and 410 μl of reference buffer. Sedimentation profiles were recorded from absorbance measurements at 280 nm for individual proteins, whereas those of samples containing a mixture of ZF and different forms of ubiquitin (mono-ubiquitin, Lys-63-, Lys-48-, or linear di-ubiquitin) were recorded from the specific absorbance of the fluorescein-labeled ZF at 440 nm in combination with interferometric measurements obtained from the on-board Rayleigh interferometer. The partial specific volume (vbar) as well as the extinction coefficient (ϵ) at 280 nm of ZF and different di-ubiquitins were calculated at 20 °C from their amino acid composition using Sednterp software Version 1.09. Similarly, the density and the viscosity of the buffer were calculated with the same software. Differential sedimentation coefficient distributions c(s) were determined using the software SEDFIT 13.0b (31). The stoichiometry of different ubiquitin·ZF complexes were determined using both interferometric and absorbance data as previously described (32). The ϵ value for the fluorescein-labeled ZF at 440 nm and at pH 7.5, calculated from its absorbance spectra, was 4,700 m−1·cm−1 and 0 for all ubiquitins, whereas the refractive increment for all proteins was 0.185 ml·g−1.
Binding Affinity Measured by Fluorescence Polarization
Fluorescence polarization (FP) measurements were recorded on a Tecan microplate reader infiniteR F500 (Tecan France S.A.S) at 25 °C using a 485-nm excitation filter and a 535-nm emission filter (fluorescein) in binding assay buffer A in the presence or absence of 0.6 mm ZnCl2. For the fluorescein-labeled WT and D406V F-ZF, titrations were carried out using three independent concentrations of peptides (0.1, 1, and 10 μm) and increasing concentrations of different types of ubiquitin (mono-Ub, Lys-63-, Lys-48-, and linear di-Ub) from 0 to 1000 μm. For all other ZF mutant peptides, binding assays were performed using a F-ZF concentration of 0.1 μm and a variable concentration of ubiquitin from 0 to 500 μm. Each experiment was performed at least in duplicate. We verified that no correction of the FP data was needed because no significant alteration of the fluorescein quantum yield was observed upon binding. To assess nonspecific binding (NS), similar experiments were performed using a fixed concentration of the fluorescein alone, or a fixed concentration of the fluorescein-labeled ZF without ZnCl2 (unfolded F-ZF). In these latter experiments the weak deviation of the FP signal, which varies linearly with ubiquitin concentration, can be fitted according to a linear function with a slope of 0.01–0.04 millipolarization units (mP)/μm depending on the type of ubiquitin. Curve-fitting of the FP data represented as the fraction of bound labeled ZF (26) includes this NS component and was done by nonlinear regression using the equation Pobs = Pmax·Ubi/(KD + Ubi) + NS·Ubi + P0 to solve the dissociation constant (KD) given Pmax (polarization after saturation with ubiquitin), Ubi (ubiquitin concentration), NS (nonspecific binding component), P0 (polarization in the absence of ubiquitin), and Pobs (polarization at a given concentration of ubiquitin). It should be noted that a binding model with two similar, non-interacting binding sites is like a single-site binding model except for the inclusion of the complex stoichiometry. The binding Gibbs free energy of each ZF·ubiquitin complex was calculated at 20 °C by the equation ΔG = −RTln(1/KD), where R is the gas constant, and T is the temperature in degrees Kelvin. The effect of the mutation was assessed from the difference in binding Gibbs free energy between the mutant and the WT as ΔΔGmu-wt = ΔGmut − ΔGwt. In competition experiments by unlabeled NEMO ZF, experimental data were fitted according to the binding equation Pobs = P0 − (P0 − Pmin)·[ZF]/(IC50 + [ZF]). Note that the IC50, which can be simplified to IC50 = KI (1 + fluorescein-labeled [ZF]0/KD), is approximately equivalent to the inhibition constant of unlabeled peptide (KI) under these conditions as the ratio fluorescein-labeled [ZF]0/KD is ≈0.
Zinc Ion Colorimetric Titration Assay
To determine the zinc content of the WT and D406V ZF, a colorimetric 4-(2-pyridylazo)resorcinol metal binding assay was used as described in Ref. 33. In brief, the WT and D406V synthetic peptides were first refolded for at least 1 h in buffer A containing 0.6 mm ZnCl2, and extensively dialyzed against the same buffer without ZnCl2 to remove unbound zinc. Protein samples (5–20 μm) were then incubated for 30 min at room temperature with 10 mm hydrogen peroxide to completely release the zinc ion before adding 0.1 mm 4-(2-pyridylazo)resorcinol. The amount of the released zinc was determined by quantitating the (4-(2-pyridylazo)resorcinol)2-Zn2+ complex at 492 nm from a calibration curve, calculated from the known concentrations of ZnCl2.
Fluorescence Spectroscopy
Tryptophan fluorescence measurements of both WT and D406V ZF(W) were recorded on a PTI multimodular QuantaMaster spectrofluorimeter (PTI, Lawrenceville, NJ) at 20 °C in buffer A with or without 0.6 mm ZnCl2. The emission spectra and the fluorescence quantum yields were obtained using an N-acetyl tryptophanamide (Sigma) standard as described previously (15).
Structural Modeling
The ZF·Lys-63 di-Ub complex shown in Fig. 7, A and B, was generated using the standard protocols implemented in HADDOCK 2.0 (34) with initial atom coordinates of NEMO ZF and Lys-63 di-Ub (Protein Data Bank (PDB) entries 2JVX and 2WWZ, respectively). The ambiguous restraints were defined on the basis of the mutagenesis data on NEMO ZF shown in this study and of the NMR data on ubiquitin described previously (15). NEMO residues were considered as “active residues” in the HADDOCK procedure when mutations induce a difference of binding free energy >1 kJ·mol−1. During docking calculations, side chains and backbone flexibility (semi-flexible segments) were included both in the proximal and distal ubiquitins (6–12, 32–39, 42–48, 66–72). In addition, the C-terminal tails and Lys-63 of the proximal ubiquitin were allowed to move during all stages of simulated annealing (fully flexible segment). The Gly-76–Lys-63 isopeptide was modeled by including a set of distance restraints (unambiguous restraint distance) based on published interatomic distances for a peptide bond.
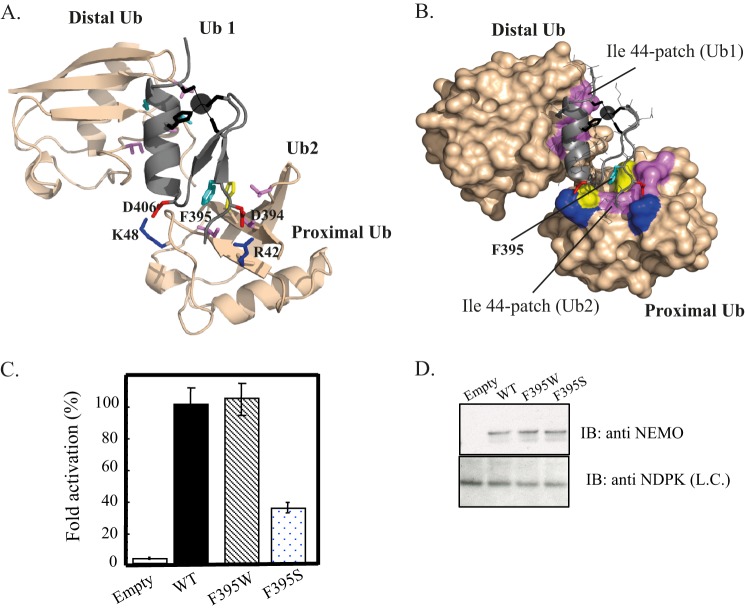
FIGURE 7.
Model of the NEMO ZF-Lys-63-di-ubiquitin complex. Ribbon (A) and surface (B) representations of the low energy structure of the NEMO ZF complex with Lys-63 di-Ub showing the second ubiquitin binding site (Ub2) and the prior ubiquitin binding site (Ub1) previously described in Ref. 15. The docking model was generated using the HADDOCK docking procedure. NEMO ZF is shown in gray, and Lys-63 di-Ub in light orange. The Phe-395, which forms the central hydrophobic core of Ub2, is colored in cyan and interacts with the Ile-44 patch of the proximal ubiquitin. Asp-406 and Asp-394 are shown in red and form salt bridges with Lys-48 and Arg-42 colored in blue. The zinc atom is represented as a black sphere and ligating residues (Cys-397, Cys-400, His-413, Cys-417) are represented as black sticks. The buried surface area of 1706 ± 56 A2 is 1.7-fold higher than the one calculated with the ZF-mono-Ub complex (1014 ± 55 Å2 (15)). C, NEMO-deficient Jurkat T cells were transiently co-transfected with the reporter plasmid Igκ-luciferase, a second reporter plasmid, EF1-β-galactosidase, to normalize transfections, and with an empty vector (Empty) or an expression vector encoding either WT, F395W, or F395S mutant. After transfection, cells were stimulated for 4 h with 20 ng/ml TNF-α. The NF-κB activation corresponding to the ratio of luciferase activity in the presence and in the absence of the stimulus is taken as 100% activity for WT. Error bars represent the S.D. values over three experiments. D, Western blot showing NEMO expression level in cell extracts of transiently transfected Jurkat T cells. Nucleotide diphosphate kinase (NDPK) was immunoblotted as a loading control (L.C.).
RESULTS
The EDA-ID-related D406V Mutation Impairs NF-κB Activation by Altering the NEMO ZF-dependent Polyubiquitin Binding Activity
Although studies on the D406V mutation found in a EDA-ID patient have already been reported (11, 25, 35), the mutation effect on NF-κB activation remains unclear as attenuated TNF-α-induced NF-κB activation was observed in one study (35), whereas another reported no impairment of NF-κB activation (25). To clarify this point, we reconstituted NEMO-deficient Jurkat T lymphocytes or NEMO-deficient MEFs with either the mutant or the WT gene, and we tested their abilities to restore the NF-κB activation. As shown in Fig. 1A, the mutant displays a severe NF-κB activation defect after TNF-α and IL-1β stimulation in Jurkat T lymphocytes transiently transfected with an NF-κB-dependent luciferase reporter construct. This mutation defect comprising 47 and 33% of activity compared with WT in response to IL-1β and TNF-α, respectively, was not due to altered NEMO expression as the mutant and WT were expressed at similar levels (Fig. 1A). No appreciable defect in NF-κB activation was observed using the same NF-κB reporter assay in stably reconstituted NEMO-deficient MEFs (not shown). Nevertheless, we could observe a slight, albeit significant reduction of IκB degradation of the mutant after TNF stimulation in reconstituted NEMO-deficient MEFs retrovirally expressing either WT or mutant NEMO at the same level (Fig. 1B, upper panel). The mutation defect was more easily detectable in an experiment in which IκB resynthesis was blocked with cycloheximide (Fig. 1B, lower panel). Hence, the D406V mutation led to a defect in NF-κB activation, but the effect appeared to be more severe in Jurkat T lymphocytes than in fibroblast cells.
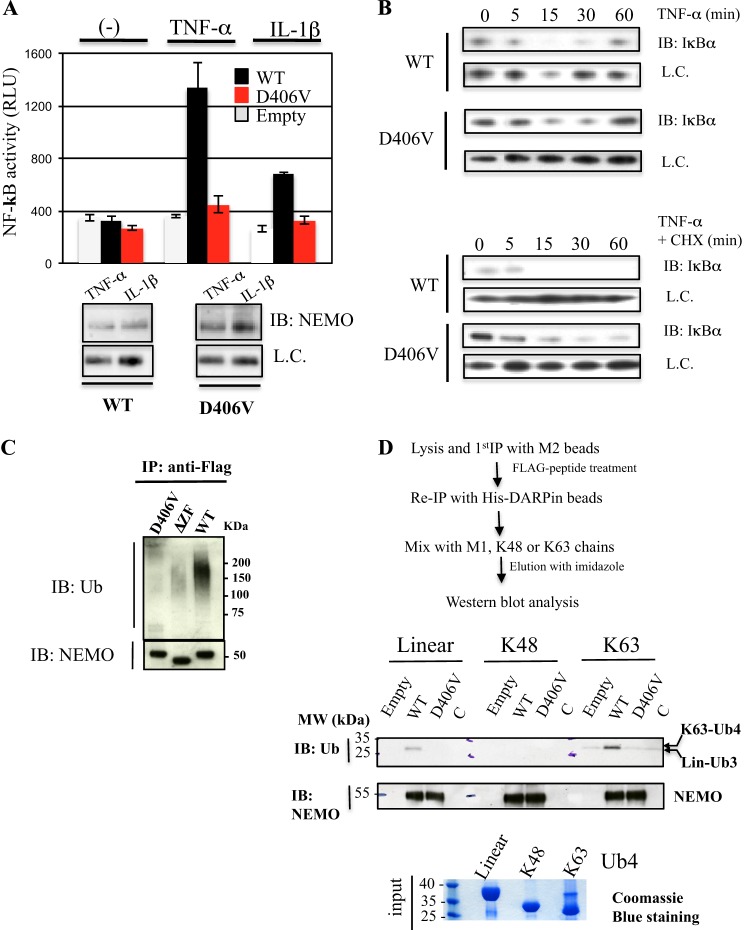
FIGURE 1.
Effect of the EDA-ID-related D406V mutation on NF-κB activation and of NEMO polyubiquitin binding activity. A, NEMO-deficient Jurkat T lymphocytes were co-transfected with an NF-κB inducible luciferase reporter plasmid and an empty plasmid or a plasmid expressing either FLAG-tagged WT or D406V mutant NEMO. Cells were then stimulated for 4 h with TNF-α (20 ng/ml) or IL-1β (10 ng/ml), and luciferase activity was measured after 4 h. Error bars represent the S.D. over two independent experiments performed in duplicate. Below are immunoblots (IB) showing NEMO expression levels in transiently transfected Jurkat T cells. B, retrovirally reconstituted NEMO-deficient MEF with either FLAG-tagged WT or D406V mutant NEMO were stimulated for the indicated times with TNF-α alone (20 ng/ml, upper blot) or in combination with cycloheximide (CHX, 25 μg/ml, lower blot), and NF-κB activity was measured by Western blot analysis of total levels of IκBα at indicated time points. L.C., loading control. C, the D406V mutation in NEMO ZF impairs the NEMO polyubiquitin binding activity. Whole cell extracts from 293T cells transfected with plasmid encoding FLAG-NEMO WT, D406V, or a ZF-deleted mutant as control were immunoprecipitated (IP) with anti-FLAG antibody. Immunoprecipitates were analyzed by immunoblot using the indicated antibodies. D, binding activity of WT and D406V mutant NEMO ZF toward linear, Lys-48, and Lys-63 ubiquitin chains. WT or mutant FLAG-NEMO transiently expressed in 293T cells were isolated following a tandem purification procedure as indicated. The DARPin molecule used for the second IP is a strong CC2-LZ binder that prevents the ubiquitin binding of the NOA domain (22). After immobilization with His-DARPin beads, the NEMO proteins were then incubated with linear, Lys-48, or Lys-63 tetra-ubiquitin chains, and the amount of ubiquitin bound was determined by immunoblotting with the indicated antibodies. Empty, empty vector; C, control with His-DARPin beads only.
Next, we addressed the question of how the D406V mutation affects NEMO function. To this end, the polyubiquitin binding activity of NEMO, which is crucial for NF-κB activation, was assessed by performing co-immunoprecipitation experiments. 293 T cells were transiently transfected with flagged-versions of the WT or D406V mutant or a ZF-deleted mutant as control, and the interaction between NEMO and the polyubiquitin chains was examined through a Western blot of the pulldown of FLAG-tagged NEMO and was revealed with an anti-ubiquitin antibody (Fig. 1C). The single-point mutant D406V as well as the ZF-deleted mutant were unable to bind free or anchored polyubiquitin chains as compared with the WT (Fig. 1C). Because NEMO ZF can function as an UBD in a NOA/UBAN-independent manner (9), we next addressed whether this ubiquitin binding defect depends on the type of ubiquitin linkage. We first isolated FLAG-WT NEMO and its D406V mutant from 293T cells after a tandem affinity purification procedure that used His-DARPin beads (Fig. 1D). The His-DARPin molecule is a strong binder of CC2-LZ, and the crystal structure of the complex showed that its interaction prevents the binding between the NOA UBD and polyubiquitin chains (22). Once immobilized on His-DARPin beads, the NEMO protein was incubated with homologous linear (also called α-amino- or Met-1-linked), Lys-63, or Lys-48 tetra-ubiquitin chains, and the amount of ubiquitin bound was analyzed by immunoblotting. As shown in Fig. 1D, the D406V impaired the binding to Lys-63 chains as no specific complex between the mutant and Lys-63 or Lys-48 chains or between the WT and Lys-48 chains was observed. In the case of the binding study with linear chains, similar binding defects were obtained as we only observed a specific interaction between the WT and linear chains. However, a detailed analysis showed that the linear chain bound corresponded to a linear Ub3, likely due to an uncontrolled deubiquitinase activity in our assay. It is worth pointing out that no deubiquitinase inhibitor based on a thiol-specific reagent such as NEM or iodoacetamide could be added in the buffer, as these compounds may alter the ubiquitin binding activity of NEMO ZF by modifying the free cysteine residue at position 396. Thus, these data indicate that the defective D406V mutation in NF-κB activation impairs ZF-dependent polyubiquitin binding activity toward Lys-63 and linear chains without any contribution of the NOA UBD.
The D406V Mutation Does Not Interfere with Either Zinc-induced Folding of NEMO ZF or with Mono-ubiquitin-NEMO ZF Binding
Because alteration of ubiquitin binding activity may result from impairment of the ZF structure, we next investigated whether the D406V mutation affects NEMO ZF folding. We previously showed that the F395W substitution in the ZF (ZF(W)) mutant peptide acts as a sensitive probe to monitor zinc-induced folding of ZF (15). The WT and D406V ZF(W) peptides were then generated, and their fluorescence emission spectra were recorded in the presence or absence of zinc (Fig. 2A). Both WT and D406V ZF(W) peptides exhibit an increase in the fluorescence signal upon the addition of zinc with similar fluorescence quantum yield ratios of 1.4 and 1.3, respectively. To further characterize the zinc-induced folding, the metal binding stoichiometries were also evaluated using a 4-(2-pyridylazo)resorcinol (PAR) colorimetric assay. At a concentration of 5 μm, both WT and D406V ZF peptides bound a single zinc ion with a ZF:Zn2+ stoichiometry of 1.2 ± 0.2 and 0.9 ± 0.1, respectively. Taken together, this indicates that the Asp-406 mutation does not dramatically affect the overall structure of ZF.
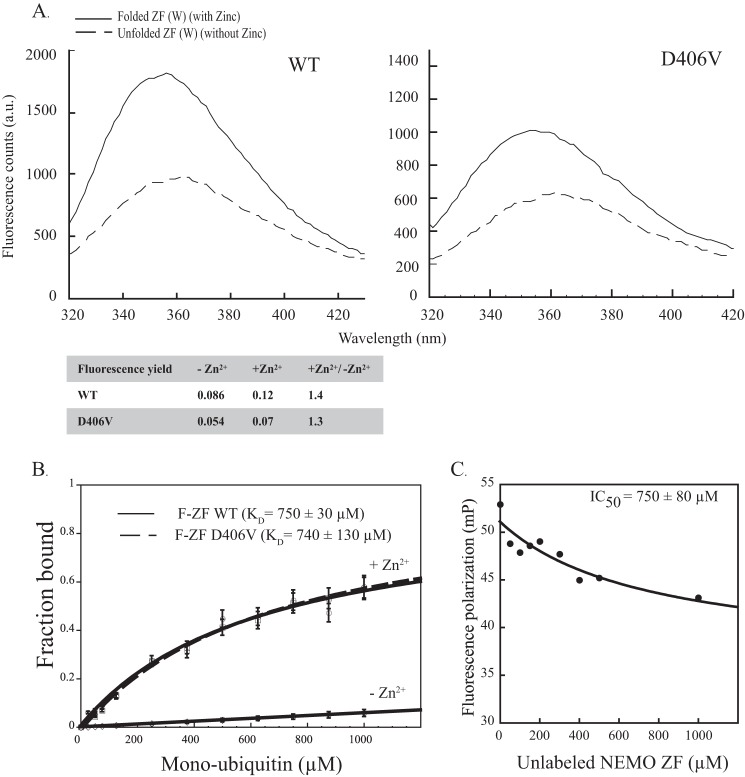
FIGURE 2.
Neither the zinc-induced folding of NEMO ZF nor its mono-ubiquitin binding property are affected by the D406V mutation. A, the emission spectra (305–450 nm) of ZF (W) mutant peptides. WT and D406V mutant, subtracted from buffer signal were recorded upon tryptophan excitation at 295 nm. ZF (W) peptides at 25 μm were tested in the absence or presence of zinc. The addition of zinc led to a similar fluorescence yield for the two peptides WT and D406V, as indicated in the table below. a.u., absorbance units. B, affinities of fluorescein-labeled NEMO ZF (F-ZF) and its D406V mutant for mono-ubiquitin measured by fluorescence polarization. The dissociation constants (KD) were determined using ZF concentrations of 0.1 μm and increasing concentrations of mono-ubiquitin as described under “Experimental Procedures.” No significant binding was observed between unfolded ZF (−Zn2+) and mono-ubiquitin (negative control). Error bars represent S.D. over two independent experiments. C, competition by unlabeled NEMO ZF of the binding of fluorescein-labeled NEMO ZF to mono-Ub. Fluorescein-labeled NEMO ZF (0.1 μm, 50 μl) was incubated for 30 min at 25 °C with mono-Ub (400 μm) in the presence of variable concentrations of unlabeled NEMO ZF (0–1000 μm). mP, millipolarization units.
Next we generated the N-terminal fluorescein-coupled WT and D406V ZF peptides, denoted hereafter as F-ZF peptides, to determine the mono-ubiquitin binding affinity of the WT and D406V mutant by FP. Titration of these two F-ZF peptides with increasing concentrations of mono-ubiquitin resulted in a gradual increase in fluorescence polarization that is only observed in the presence of Zn2+, indicating that a proper zinc-induced folding of ZF is required for specific binding to mono-ubiquitin (Fig. 2B). Of note, no significant increase of FP signal was observed upon titration of F-ZF peptides with unlabeled ZF peptides or upon titration of the fluorescein alone with mono-ubiquitin. This indicates that the FP signal change of F-ZF upon ubiquitin titration is not due to any ubiquitin-dependent ZF:ZF interaction or nonspecific interaction between fluorescein and mono-ubiquitin. A dissociation constant (KD) of 750 ± 30 μm was obtained for WT F-ZF at 25 °C and pH 7.0, which is close to the value observed for D406V F-ZF at the same pH and temperature. Moreover, competition experiments of WT F-ZF with unlabeled ZF for the mono-ubiquitin binding gave an IC50 value of 750 ± 80 μm similar to the KD of WT F-ZF (Fig. 2C), again ruling out the possibility that the N-terminal fluorescein probe interferes with mono-ubiquitin binding. Hence, these data indicate that D406V mutation has no detrimental effect on mono-ubiquitin binding to NEMO ZF. This is consistent with the structural model of the ZF complex with mono-ubiquitin (15).
The ZF UBD Contains Two Ubiquitin Binding Sites That Result in Preferential Binding to a Di-ubiquitin Chain
The lack of correlation between in vivo (Fig. 1, C and D) and in vitro (Fig. 2B) effects of the D406V mutation on ubiquitin binding led us to pursue further studies of the interaction between the ZF and polyubiquitin chains of different lengths and linkages. Fig. 3A shows the binding curves of WT F-ZF for the mono-ubiquitin and Lys-63 di-ubiquitin with a binding affinity for Lys-63 di-ubiquitin (KD = 38 ± 3 μm), which is 20-fold increased as compared with mono-ubiquitin (KD = 750 ± 30 μm). Affinity increases of only 2.5- and 3.5-fold were found for Lys-63 tri- and Poly3–7-Ub, respectively (not shown), suggesting that di-ubiquitin contains all determinants required to achieve an optimal interaction with ZF.
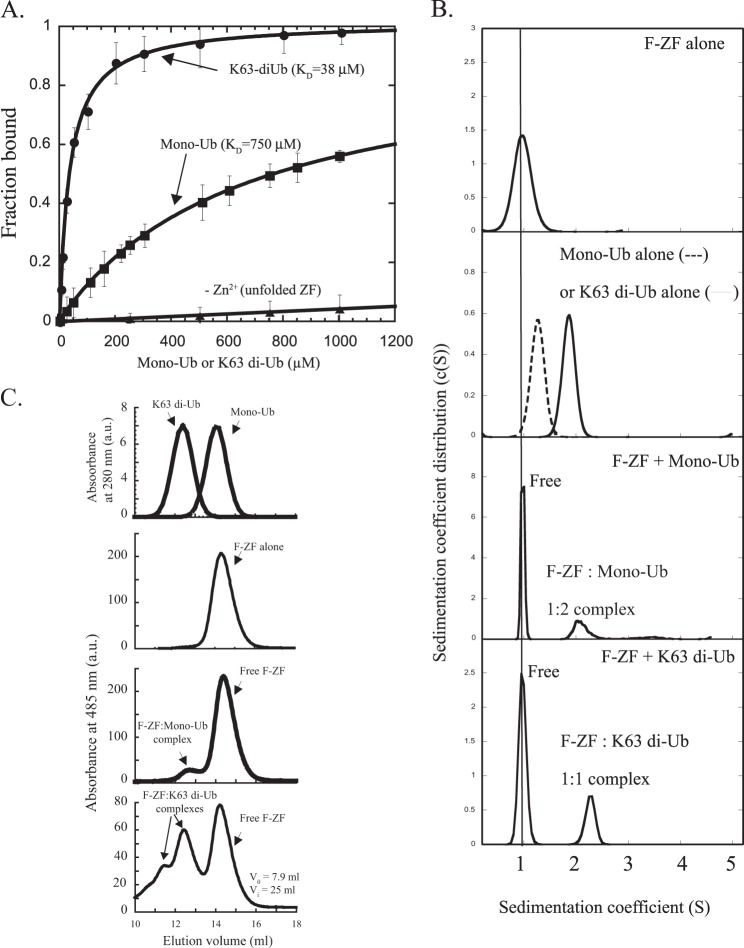
FIGURE 3.
The ZF UBD contains two ubiquitin binding sites that result in higher binding affinity to di-ubiquitin as compared with mono-ubiquitin. A, fluorescence polarization data for fluorescein-labeled NEMO ZF (F-ZF, 0.1 μm) interacting with mono-ubiquitin or Lys-63-di-ubiquitin. The dissociation constants (KD) were determined as described under “Experimental Procedures.” Error bars represent S.D. over two independent experiments. B, SV-AUC measured size distributions of fluorescein-labeled NEMO ZF in the presence or absence of mono-ubiquitin or Lys-63 di-ubiquitin. Sedimentation profiles of the free and bound states of NEMO ZF were monitored at 20 °C by absorbance at 485 nm, whereas those of mono- and di-ubiquitin alone were monitoring by absorbance at 280 nm or Rayleigh interference as described under “Experimental Procedures.” a.u., absorbance units. C, size exclusion chromatography analysis of NEMO F-ZF interactions with mono-ubiquitin or Lys-63-linked di-Ub. Fluorescein-labeled ZF (20 μm) was incubated in the absence or presence of mono-ubiquitin (Mono-Ub, 20 μm) or Lys-63-linked di-ubiquitin (K63 di-Ub, 20 μm), and the mixture was subjected to gel filtration analysis on a Superdex 75 HR 10/30 column at 4 °C. The amount of free or bound ZF formed upon incubation with mono-Ub or Lys-63 di-Ub was monitored at 485 nm by recording specific absorbance of the fluorescein-labeled ZF, whereas the elution profiles of mono-Ub and Lys-63 di-Ub alone were recorded at 280 nm as indicated. The void (V0) and internal (Vi) volumes of the column are indicated.
The affinity gain for Lys-63 di-ubiquitin as compared with mono-ubiquitin could result from distinct or equivalent binding stoichiometries. This could not be accurately measured by fluorescence polarization, especially due to the low affinity of ZF for mono-ubiquitin. We, therefore, investigated the composition of the mono-ubiquitin·F-ZF and Lys-63 di-ubiquitin·F-ZF complexes by sedimentation velocity AUC (Fig. 3B and Table 1) and gel filtration (Fig. 3C) using higher molar concentrations. A complex formation with either the mono-ubiquitin or Lys-63 di-ubiquitin was easily observed by recording the specific absorbance of the labeled ZF (F-ZF) at 485 nm in the presence or absence of different forms of ubiquitin (Fig. 3, B and C). Strikingly, the mono-ubiquitin·ZF complex behaves as the Lys-63 di-ubiquitin·ZF complex with a sedimentation coefficient (S*) of 1.9 ± 0.1 S (Table 1), close to the value of 2.3 ± 0.1 obtained with the Lys-63 di-ubiquitin·ZF complex and significantly different from the mono-ubiquitin alone (1.3 ± 0.1 S) or F-ZF alone (1 ± 0.1 S). The calculated molecular mass of the mono-ubiquitin·ZF complex was 20.8 ± 2.3 kDa, and its binding stoichiometry calculated from absorbance measurements at 280 and 485 nm and interferometry (see “Experimental Procedures”) corresponds to two mono-ubiquitins bound per ZF peptide. On the other hand, the binding stoichiometry of ZF·Lys-63 di-ubiquitin complex was quite different with one di-ubiquitin chain bound to one ZF molecule (experimental molecular mass = 19.0 ± 2.3 kDa). Similar binding stoichiometries were observed at 4 °C in gel filtration as the elution volume of mono-ubiquitin·ZF complex, which is markedly different from elution volume of the ZF or mono-ubiquitin alone, was almost the same as that of Lys-63 di-ubiquitin·ZF complex (Fig. 3C). Other minor and transient complexes corresponding to heavier Lys-63-di-ubiquitin·ZF complexes were observed only in gel filtration, possibly due to temperature and hydrostatic pressure differences between gel filtration and SV-AUC (see “Discussion”). Binding studies of F-ZF were also performed in SV-AUC with Lys-48-linked or linear di-ubiquitin (Table 1). A binding stoichiometry of 1:1 was observed with either Lys-48- or linear di-ubiquitin, indicating that the same complex stoichiometry was found for different forms of di-ubiquitin (Lys-48, Lys-63, and linear). Taken together, theses results indicate that NEMO ZF contains two ubiquitin binding sites, which give to the NEMO ZF a higher binding affinity to di-ubiquitin than to mono-ubiquitin.
TABLE 1.
Sedimentation velocity analysis of NEMO ZF interaction with mono-Ub or various forms or di-Ub (linear and Lys-48- and Lys-63-linked)
| Protein | Sedimentation coefficient | f/f0 | Molecular mass | Complex stoichiometry ZF-Ubi |
|---|---|---|---|---|
| S* | kDa | |||
| F-ZF (150 μm) | 1.0 ± 0.1 | 1.2 | 4.7 ± 1 | |
| Mono-Ub (100 μm) | 1.3 ± 0.1 | 1.06 | 8.4 ± 0.4 | |
| Lys-48-di-Ub (100 μm) | 1.8 ± 0.1 | 1.07 | 15.4 ± 2 | |
| Lys-63-di-Ub (100 μm) | 1.9 ± 0.1 | 1.06 | 15.0 ± 2 | |
| Met-1-di-Ub (100 μm)a | 1.9 ± 0.1 | 1.12 | 16.1 ± 1 | |
| F-ZF + mono-Ub (300:100 μm)b,c | 1.9 ± 0.1 (1.05) | 1.5 | 20.8 ± 2.3 | 1:2 |
| F-ZF + Lys-48-di-Ub (150:100 μm)b | 2.3 ± 0.1 (1.0) | 1.22 | 20.3 ± 2.2 | 1:1 |
| F-ZF + Lys-63-di-Ub (150:100 μm)b | 2.3 ± 0.1 (0.95) | 1.16 | 19.0 ± 2.3 | 1:1 |
| F-ZF + Met-1-di-Ub (150:100 μm)b | 2.2 ± 0.1 (1.01) | 1.26 | 20.8 ± 2.0 | 1:1 |
a A heavier species with a sedimentation coefficient of 4.4 ± 0.5 S and a frictional coefficient (f/f0) of 1.12 was observed when Met-1-diUb was analyzed alone. This species of 59 ± 8 kDa was compatible with a trimer of Met-1-diUb.
b The sedimentation coefficients measured for the free state of fluorescein N-terminal-labeled NEMO are indicated in parentheses.
c A minor complex of ZF·monoUb with a stoichiometry of 2:4 (44.3 kDa) was also detected under these experimental conditions.
Other ZF Domains Exhibit Ubiquitin Binding Properties Similar to That of NEMO ZF
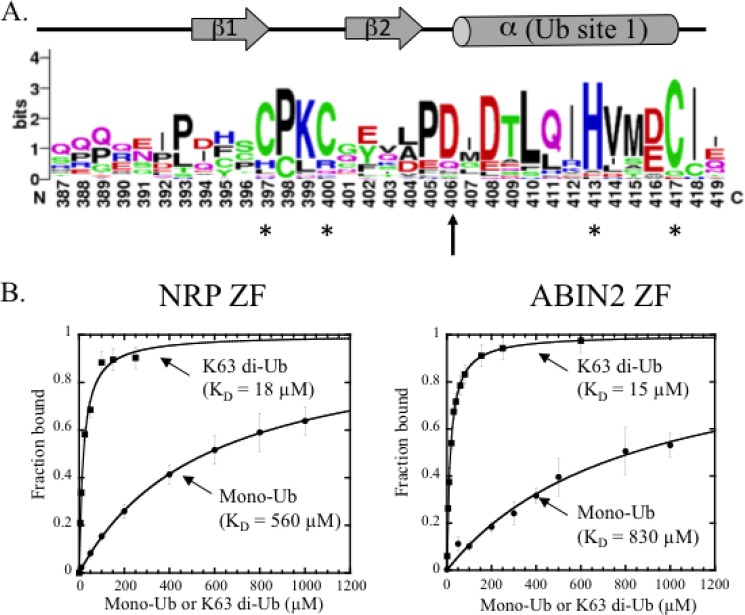
Previous studies showed that the NEMO ZF can be functionally replaced with the ZF of Optineurin (also called NRP) or ABIN2, suggesting that the three ZFs display similar ubiquitin binding properties. A pleiotropic role of Optineurin has been reported in protein secretion (36), PRR (pattern recognition receptor)-dependent innate antiviral response (37), autophagy (38), and cell cycle (38), whereas the role of ABIN2 is poorly understood despite its clear co-involvement with the TPL-2 MEK kinase in the Erk MAPK pathway (39). Sequence alignment of NEMO ZF with NRP and ABIN2 ZF counterparts from several species predicts that both NRP and ABIN2 ZFs adopt a classical ββα-fold characteristic of the CCHC-type zinc finger family (Fig. 4A). Interestingly, the aspartic acid residue at position 406 in NEMO ZF is strictly conserved in NRP and ABIN ZFs, highlighting the functional importance of this residue within ZF domains. To examine the ubiquitin binding properties of NRP and ABIN2 ZFs, we generated two N-terminal fluorescein-labeled ZF peptides and tested their abilities to bind mono-ubiquitin or Lys-63-di-ubiquitin by fluorescence polarization. Like NEMO ZF, zinc-dependent ubiquitin binding was observed with both NRP and ABIN2 peptides (not shown). Moreover, the ZF of NRP and ABIN2 also exhibit a strong preference for Lys-63 di-ubiquitin binding with affinities that are 31- and 55-fold increased, respectively, as compared with affinities with mono-ubiquitin (Fig. 4B). Hence, the preferential di-ubiquitin binding of NEMO ZF is not unique. This property is shared with ABIN2 and NRP ZFs, which belong to the same NEMO-related ZF family.
FIGURE 4.
ABIN2 and NRP ZFs share similar ubiquitin binding properties with NEMO ZF. A, conservation of the aspartic acid residue at position 406 in NEMO, ABIN2, and NRP ZFs that form classical CCHH and CCHC fingers. Residue conservation at each position is calculated using WebLogo and results from the structure-based sequence alignment of the NEMO ZF domains with NRP and ABIN2 ZFs (available upon request). The conserved aspartic acid residue, which is mutated to valine in the EDA-ID patient, is noted with an arrow, whereas the four conserved zinc-coordinating residues are noted with asterisks. The secondary structure of NEMO ZF identified from its NMR structure (PDB 2JVX) is indicated above the consensus sequence logo. The prior ubiquitin binding site (Ub site 1) previously described in Ref. 14) is also indicated. B, mono-Ub (circles) and Lys-63 di-Ub (squares) titrations of fluorescein-labeled NRP ZF (0.1 μm) and ABIN2-ZF (0.1 μm) monitored by fluorescence polarization. The experimental conditions are as in Fig. 3A. Error bars represent S.D. over two independent experiments performed in duplicate.
The Functional Defect Induced by the D406V Mutation Is Due to a Specific Binding Defect with Lys-63 and Linear Di-ubiquitin
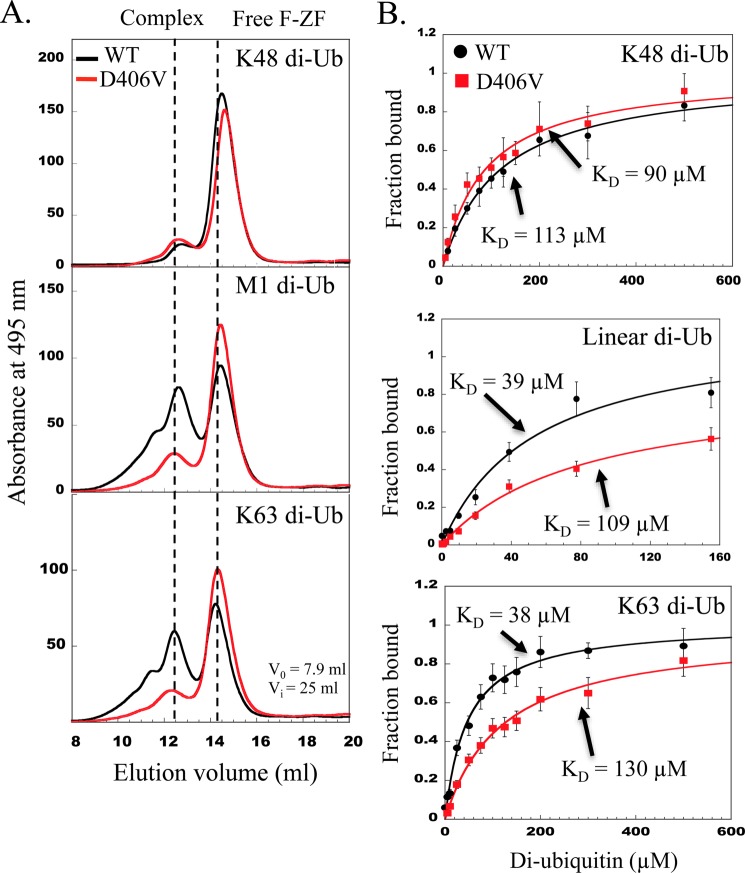
To quantitatively determine whether the D406V mutation affects binding to a di-ubiquitin chain of different linkage, the complex formation between the fluorescein-labeled WT and D406V F-ZF peptides and various forms of di-ubiquitin (Lys-48, Lys-63, and linear) were analyzed by gel filtration (Fig. 5A), and the dissociation constants (KD) were determined by fluorescence polarization (Fig. 5B), albeit performed at different temperatures. In gel filtration, higher amounts of linear and Lys-63 di-ubiquitins interact with the WT F-ZF compared with Lys-48 di-ubiquitin. This little preference of F-ZF for Lys-63 and linear di-ubiquitin compared with Lys-48 di-Ub was confirmed by fluorescence polarization data, as similar KD (38 and 39 μm, respectively) were obtained for Lys-63 and linear di-ubiquitin, whereas the KD for Lys-48 di-ubiquitin was 113 ± 11 μm. More importantly, when we compared the WT F-ZF with its D406V mutant upon gel filtration, an impairment of the binding to Lys-63 and linear di-ubiquitin was observed with the mutant. By contrast, no detectable effect on the binding to the Lys-48 di-ubiquitin was observed as similar amounts of complex with the WT and mutant were detected. All these gel filtration data were further confirmed by a more quantitative binding assay using fluorescence polarization. Indeed, the mutation preferentially alters the Lys-63 and linear binding as their KD values are 3.4- and 2.8-fold increased, respectively, as compared with the WT, and this effect appears to be specific as similar KD values of 113 ± 11 and 90 ± 8 μm for Lys-48-di-ubiquitin were observed with the WT and the mutant. Our in vitro data with the isolated ZF hence correlate with those with the NEMO full length. This provides an explanation of why the D406V mutation induces such an alteration of the NF-κB pathway, causing the EDA-ID pathology. The functional defect is not due to an incapacity to fold but rather is due to a specific binding defect with Lys-63 and linear di-ubiquitins but not with mono-or Lys-48 di-ubiquitins.
FIGURE 5.
The EDA-ID-associated D406V mutation impairs the binding for Lys-63- and linear di-ubiquitin. A, size exclusion chromatography analysis of WT and D406V mutant ZF interactions with various forms of di-Ub chains (linear (Met-1) and Lys-48- and Lys-63-linked di-Ub). The experimental conditions were as in Fig. 3C, including the void (V0) and internal (Vi) volumes of the column. B, binding curves of WT F-ZF (black circles) and its D406V mutant (red squares) with linear or Lys-48- or Lys-63-linked di-ubiquitin measured by fluorescence polarization at 25 °C. Binding constants (KD) extracted from non-linear least-squares fitting (solid line) are indicated for each di-ubiquitin taking into account a complex stoichiometry of 1:1. Error bars represent S.D. from three independent experiments performed with 0.1, 1, or 10 μm WT and mutant F-ZF.
Structure-guided Mapping and Mutagenesis to Elucidate the Second Ubiquitin Binding Site in NEMO ZF
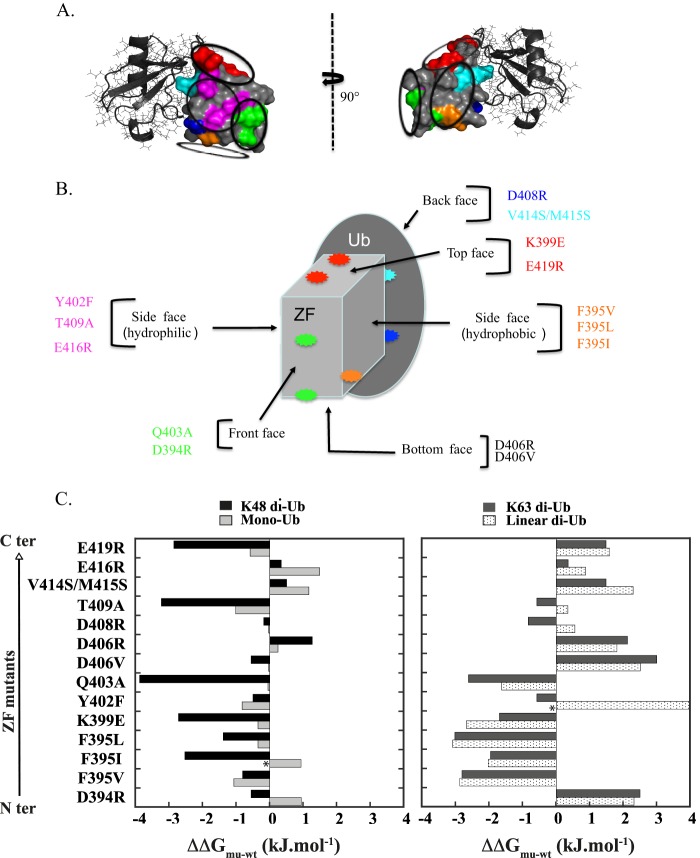
Complete characterization of the EDA-ID-related D406V mutant prompted us to pursue the exact localization of the second ubiquitin binding site within the ZF domain. Despite the use of extensive crystallization conditions and of numerous ZF mutants with a reduced surface entropy (40), no diffraction quality co-crystal could be obtained with the ZF and mono-ubiquitin or any di-ubiquitin (linear, Lys-48, or Lys-63 chains). Extensive mutagenesis was then conducted on the NEMO ZF to map the second ubiquitin interface. The rationale for the design of mutants depicted in Fig. 6, A and B, was structurally guided by (i) our prior mutagenesis and NMR studies showing one ubiquitin binding site along the ZF α-helix and (ii) the fact that no residue forming the hydrophobic core or zinc-tetra-coordination could be mutated without dramatic destabilization of the ZF structure. We reasoned that if the ZF structure can be compared with a rectangular box with one face representing the previously characterized mono-ubiquitin binding site, we could implement a non-biased strategy to map the second binding site by considering all mutations on the other five faces. Out of 28 residues forming the ZF, we generated 13 novel single-point and double ZF mutants such that each mutant represented a potential binding face. The double V414S/M415S mutant was used as a control for the prior ubiquitin binding site located along the α-helix. All mutants were N-terminal fluorescein-labeled and tested by fluorescence polarization for their ability to bind mono-ubiquitin, Lys-48, Lys-63, or linear di-ubiquitin. Fig. 6C summarizes the binding free energy difference of all these mutants relative to the WT (ΔΔGmut-wt), calculated from the dissociation constants shown in Table 2. All mutations, except V414S/M415S (control), D394R, F395I, and E416R mutants, have negligible effects on mono-ubiquitin binding. For instance, the D406R mutant, which bears an opposite charge compared with WT, displays a ΔΔGmu-wt of only 0.25 ± 0.1 kJ·mol−1, calculated from KD values of 830 ± 70 and 750 ± 30 μm for the D406R mutant and WT, respectively. Two mutants, F395I and Y402F, behaved differently as compared with other mutants, as the binding curves of F395I for mono-Ub and of Y402F for linear di-Ub were not hyperbolic, reflecting cooperative binding. Interestingly, the mutants that induced a positive (reduced affinity) or negative (increased affinity) ΔΔGmu-wt for binding to Lys-63 di-ubiquitin gave similar effects upon binding to linear di-ubiquitin, suggesting a similar binding mode. On the other hand, the binding mode of Lys-48 di-ubiquitin appears to be different from Lys-63 or linear chains as some mutants exhibit opposite effects upon Lys-48 or Lys-63 di-ubiquitin binding. This is the case for instance of the D394R and E419R mutations, which give, respectively, negative ΔΔGmu-wt values of −0.5 and −2.8 kJ·mol−1 for Lys-48 binding and positive values of +2.5 kJ·mol−1 and +1.5 kJ·mol−1 for Lys-63 binding. The greatest effects on Lys-63 or linear binding were for the three mutants, F395V, F395L, and F395I, which bear mutations spatially close to Asp-406 and Asp-394 in the ZF structure. Besides, these mutation effects remain minor on mono-ubiquitin binding, indicating a specific effect on di-ubiquitin binding. Taken together, our mutagenesis data provide evidence that the second ubiquitin binding site is centered around the triad of acidic and hydrophobic residues Asp-394, Phe-395, and Asp-406.
FIGURE 6.
Rationale for the design of mutants in NEMO ZF. A, different views of the model of the NEMO-ZF-mono-ubiquitin complex previously described in Ref. 15. The ββα structure of NEMO ZF is shown in a surface representation, and the mono-ubiquitin is shown in ribbon representation. Residues of NEMO ZF that were mutated for structure-function analysis are indicated in different colors. B, schematic view of the complex, with the ZF depicted as a rectangular box, and residues color-coded as in A. The back face represents the previously characterized binding site of ZF that contacts ubiquitin along the α-helix. All mutations designed for identification of the second binding site lie on each potential ubiquitin binding face. C, binding free energy differences of mutants relative to the WT for mono-Ub and different forms of di-ubiquitin (linear, Lys-48-linked, and Lys-63-linked). Binding free energy differences (ΔΔGmu-wt) extrapolated at 20 °C were calculated from binding affinity values provided in Table 2. Binding affinities of WT ZF and its mutants were determined at 25 °C by fluorescence polarization. Positive ΔΔGmu-wt values indicate that the mutation reduces the complex affinity, whereas negative values indicate that the mutation induces an affinity increase. Asterisks denote the two mutants, F395I and Y402F, that exhibit sigmoidal binding curves for mono-ubiquitin and linear di-Ub, respectively.
TABLE 2.
Binding constants of the WT and its mutants for mono-Ub and various forms of di-Ub (linear and Lys-48-linked and Lys-63-linked)
| ZF | Mono-Ub |
Lys-48-di-Ub |
||
|---|---|---|---|---|
| KD | ΔΔG mu-wt | KD | ΔΔG mu-wt | |
| μm | kJ·mol−1 | μm | kJ·mol−1 | |
| WT | 750 ± 30 | 0 | 113 ± 40 | 0 |
| D394R | 1105 ± 50 | +0.95 | 90 ± 20 | −0.5 |
| F395V | 484 ± 40 | −1.05 | 81 ± 10 | −0.8 |
| F395I | 1100 ± 80a | +0.9 | 40 ± 8 | −2.5 |
| F395L | 650 ± 50 | +0.35 | 64 ± 8 | −1.4 |
| K399E | 650 ± 50 | +0.35 | 37 ± 16 | −2.7 |
| Y402F | 535 ± 35 | −0.8 | 92 ± 14 | −0.5 |
| Q403A | 734 ± 50 | −0.05 | 23 ± 7 | −3.9 |
| D406V | 740 ± 130 | −0.03 | 90 ± 15 | −0.5 |
| D406R | 830 ± 70 | +0.25 | 190 ± 17 | + 1.3 |
| D408R | 740 ± 60 | −0.03 | 105 ± 20 | −0.1 |
| T409A | 494 ± 40 | −1.0 | 30 ± 4 | −3.2 |
| V414S/M415S | 1210 ± 70 | +1.1 | 139 ± 28 | + 0.5 |
| E416R | 1380 ± 150 | +1.5 | 130 ± 20 | + 0.3 |
| E419R | 590 ± 60 | −0.6 | 35 ± 5 | −2.8 |
| ZF | Lys-63-di-Ub |
Linear di-Ub |
||
|---|---|---|---|---|
| KD | ΔΔGmu-wt | KD | ΔΔGmu-wt | |
| μm | kJ.mol−1 | μm | kJ.mol−1 | |
| WT | 38 ± 3 | 0 | 39 ± 6 | 0 |
| D394R | 106 ± 20 | +2.5 | 101 ± 25 | +2.3 |
| F395V | 12 ± 3 | −2.8 | 12 ± 3 | −2.9 |
| F395I | 17 ± 4 | −2 | 17 ± 4 | −2.0 |
| F395L | 11 ± 4 | −3 | 11 ± 3 | −3.1 |
| K399E | 19 ± 4 | −1.7 | 13 ± 3 | −2.6 |
| Y402F | 30 ± 7 | −0.6 | 340 ± 66a | +5.2 |
| Q403A | 13 ± 3 | −2.6 | 20 ± 5 | −1.6 |
| D406V | 130 ± 6 | +3 | 109 ± 6 | +2.5 |
| D406R | 91 ± 20 | +2 | 82 ± 20 | +1.8 |
| D408R | 27 ± 8 | −0.8 | 49 ± 11 | +0.5 |
| T409A | 30 ± 4 | −0.6 | 45 ± 9 | +0.3 |
| V414S/M415S | 70 ± 7 | +1.5 | 100 ± 27 | +2.3 |
| E416R | 44 ± 11 | +0.35 | 56 ± 8 | +0.9 |
| E419R | 70 ± 18 | +1.5 | 75 ± 16 | +1.6 |
a Binding curves were fitted to the Hill equation for these mutants. The Hill coefficients for F395I and Y402F were 1.7 and 4.2, respectively.
DISCUSSION
Most EDA-ID patients bearing hypomorphic mutations in the NEMO ZF showed various levels of impairment of NF-κB activation in response to multiple stimuli, including LPS, TNF-α, IL-1β, and CD40L. In the case of a patient bearing the C417F EDA-ID mutation, the relevant functional defect was shown to be due to the alteration of the ZF structure as the mutation impedes the proper tetrahedral coordination of the zinc atom (14). Similar structural perturbations may be found in C417R/Y EDA-ID patients exhibiting impairments of NF-κB activation (25, 41) due to the inability of the Arg/Tyr residues to properly chelate a zinc atom. Here, we show that the D406V EDA-ID mutation does not alter the ZF structure as judged by fluorescence spectroscopy and metal binding colorimetric assay. Nevertheless, this single-point mutation is sufficient to reduce NF-κB activation. Furthermore, we show that the level of impairment of NF-κB activation is cell- and stimulus-specific, with a greater defect in Jurkat T lymphocytes than in fibroblasts and a more reduced-NF-κB activation by TNF-α compared with IL-1β (Fig. 1A) or LPS (not shown). This is consistent with previous data attributing a stimulus-specific role of the NEMO ZF in NF-κB activation (10, 11, 14, 42). The defective D406V mutation is due to a specific impairment in binding to free or anchored polyubiquitin chains (Fig. 1C), and both gel filtration and fluorescence polarization methods using the isolated ZF reveal that the D406V mutation preferentially impairs linear and Lys-63 di-Ub binding over Lys-48 one or mono-ubiquitin (Fig. 4, A and B). A similar ubiquitin binding defect of D406V mutant was also observed with the full length NEMO in a NOA-independent manner (Fig. 1D). This mutational effect observed in vitro with the individual ZF UBD could appear modest (3-fold in KD) in comparison to the strong effect observed in vivo in the context of the full length NEMO (Fig. 1, C and D). One possible explanation could be in the ability of the ZF-containing NEMO to dimerize via its coiled-coil domains. Indeed, NEMO dimerization could spatially bring two ZF UBDs together in a more closed space, which may result in enhanced poly-Ub binding by an avidity-based mechanism. In line with this, it is interesting to note that heavier complexes with a binding stoichiometry of two ZF bound to two di-Ub chains were observed in gel filtration even with the individual ZF. These ubiquitin complexes, which could represent a cellular scenario with the ZF-containing NEMO, were only observed with Lys-63 and linear linkages and were totally disrupted by the D406V mutation.
Another important point to consider is the different temperatures used in all ubiquitin binding experiments. Indeed, a standard GST pulldown assay showed that a GST-tagged version of ZF (residues 363–419) forms a stable complex with a linear tetra-ubiquitin chain at 4 °C, but not at room temperature, indicating that the in vitro stability of the poly-Ub·ZF complex dramatically decreases at room temperature (not shown). Thus, this temperature-dependent binding of NEMO ZF likely provides a simple explanation for the differences in affinity that have been observed in gel filtration or tetra-ubiquitin pulldown assays at 4 °C and at 25 °C in fluorescence polarization.
Comparative binding studies of mono-Ub or di-Ub to ZF by SV-AUC and gel filtration clearly demonstrate the existence of two ubiquitin sites within one ZF molecule. The binding stoichiometry of the ZF·mono-Ub complex formerly considered 1:1 on the basis of fluorescence titration and 15N NMR relaxation (15) could not be accurately determined for the following reasons: (i) the binding affinity of ZF for mono-Ub measured by fluorescence spectroscopy is too low to ideally perform ZF titrations under stoichiometric conditions (i.e. [ZF] ≈ KD), and (ii) the isotropic rotational correlation time of the mono-Ub·ZF complex (τc) calculated from 15N NMR relaxation can only yield a rough estimate of the molecular mass as it is also related to overall molecular shape. To identify the second Ub binding site, extensive crystallization assays of NEMO ZF with different di-Ub linkage or mono-Ub were performed. Unfortunately, no useful crystal could be obtained. Even the use of ZF variants with reduced surface entropy such as K399A/E419A failed to produce any crystals. A HADDOCK model was then generated on the basis of our previous NMR titration with 15N-enriched mono-Ub (15) and extensive mutagenesis data on NEMO ZF (Fig. 6 and Table 2). Fig. 7 shows a low energy structure of the NEMO ZF complex with a Lys-63 di-Ub, taking into account that only mutations that induce a difference of binding free energy (ΔΔGmu-wt) greater than 1 kJ·mol−1 at 293 K point out the interfacing residues of ZF involved in the interaction (active residues in HADDOCK docking procedure).
The hydrophobic patch of the distal ubiquitin, which is formed by Leu-8, Ile-44, and Val-70, interacts with the hydrophobic side of the ZF α-helix in a manner close to the binding with mono-Ub (15). This hydrophobic patch is recognized by the first Ub binding site of ZF, which is mainly composed of the hydrophobic side chains of Val-414 and Met-415. The second Ub binding site, which is formed by the Phe-395-centered patch of the ZF, interacts with the hydrophobic cleft of the proximal ubiquitin, consisting of Ile-44, Leu-6, Val-70, His-68, and Gly-47. The insertion of Phe-395 into this cleft makes its aromatic side chain completely hidden from the solvent. In addition to the Phe-395-centered hydrophobic core, the second ubiquitin binding site is composed of negatively charged residues such as Asp-406 and Asp-394 at its periphery. The Asp-406 side chain forms a salt bridge with Lys-48 of the proximal ubiquitin, whereas Asp-394 forms a salt bridge with Arg-42 of the same ubiquitin moiety.
To further determine the functional importance of the ZF second ubiquitin binding site, we mutated the Phe-395 residue forming the hydrophobic core of the ZF second ubiquitin binding site by either the polar residue, Ser, or the aromatic residue, Trp. NEMO-deficient Jurkat T cells were then transiently reconstituted with either the WT or the F395W or F395S NEMO mutant, and we tested their abilities to restore the TNF-α-induced NF-κB activation using a NF-κB gene reporter assay (Fig. 7, C and D). Both F395W and F395S mutants were expressed with expression levels comparable to that of WT. Consistent with our structural model, the F395W mutant exhibits a NF-κB activation level similar to the WT. On the other hand, the F395S mutant did not completely restore the NF-κB activation, displaying only 38 ± 5% of activity compared with the WT in response to TNF-α, indicating that the Phe-395-centered hydrophobic patch on NEMO ZF should be preserved for a full NF-κB activation. Moreover, these data provide strong evidences that the Phe to Trp substitution at position 395 can act as a sensitive probe for monitoring the folding or ubiquitin binding by fluorescence (Fig. 2A) as the F395W mutant is able to restore the NF-κB activation as efficiently as WT NEMO.
Finally, our thorough analysis of the NEMO ZF confirms that it individually forms a functional UBD domain. Some linkage preference with Lys-63 and linear di-Ub over Lys-48 di-Ub was observed by the use of three quantitative biochemical methods such as SV-AUC, gel filtration, and fluorescent polarization. These latter methods appear to be more appropriate to detect linkage preference of any UBD for poly-Ub because they do not generate linkage-preference artifacts such as the one produced in GST pulldown assays or SPR assays when a bivalent antibody is used (43). The slight linkage preference for Lys-63 or linear di-Ubs is likely due to the different conformations between Lys-48 di-Ub and linear or Lys-63 di-Ub (44). Indeed, crystal or solution structures demonstrate more favorable access of hydrophobic patches of the distal and proximal ubiquitins in Lys-63 or linear di-Ub compared with the hydrophobic patch-to-patch contact observed between Ub monomers in Lys-48 di-Ub. Moreover, the salt bridge between the ZF Asp-406 and Lys-48 of the proximal ubiquitin of a Lys-63 di-Ub chain (Fig. 7, A and B) may contribute to this linkage preference with Lys-63 or linear di-Ub over Lys-48 di-Ub, as the Lys-48 residue is engaged in the isopeptide bond in the Lys-48 di-Ub and can no longer form a salt bridge with Asp-406. Nevertheless, it is important to note that even if a small linkage preference for linear and Lys-63 di-Ub over Lys-48 di-Ub is reported here (only 3-fold in KD), it is not sufficient to consider the individual ZF UBD as a specific UBD domain for linear or Lys-63 chains, particularly when compared with other linkage-specific ZF modules such as TAB2/TAB3 NZF (Lys-63) or HOIL-1L NZF (linear).
In addition to its distal ZF UBD, NEMO contains the well characterized NOA/UBAN UBD, which individually exhibits an intrinsic linkage specificity for linear chains compared with Lys-48 and Lys-63 di-Ub (5, 45). We previously showed that these tandem UBDs contained in the NEMO C terminus, denoted NOAZ (residues 241–419 in human), enhance affinity and specificity for Lys-63 poly-Ub over linear poly-Ub (12), consistent with recent work on multiple UBD-containing ubiquitin receptors (46). However, two recent reports were unable to observe this linkage-preference avidity using conventional GST/maltose-binding protein (MBP) pulldown assay (47) or microscale thermophoresis (48). Beside artificial linkage-preference generated by GST or maltose-binding protein dimerization in pulldown assays (43, 49), one explanation could be in the binding stoichiometry differences of the NOA/UBAN UBD for linear and Lys-63 chains. Indeed, the dimeric NOA UBD binds two linear di-Ub chains as judged from the crystal structure or AUC-SV (50), whereas it only interacts with one Lys-63 di-Ub or longer Lys-63 chain in ITC (45) or AUC.5 This stoichiometry difference could have an important impact on the results, as higher amounts of linear chain versus Lys-63 can be pulled down by the same amount of NOA UBD, possibly yielding an apparent linkage selectivity for linear chains in pulldown assays. In microscale thermophoresis, binding measurements were made using a variable concentration of unlabeled NEMO and a modified version of di-Ub, which was labeled in a non-site-specific manner with a fluorescent dye containing an amine-reactive group. These latter conditions could preclude avid binding because modification of lysine residues or α-amino group on ubiquitin by the reactive fluorescent dye could disrupt the ZF UBD binding. Moreover, it is important to consider that the reverse titration using a fixed concentration of NEMO and variable concentrations of ubiquitin could be a more sensitive approach to monitor the contribution of each UBD upon avid binding. Hence, conclusions about the role of tandem NEMO UBDs will require careful structural and biochemical studies to determine the precise mechanism of recognition to different poly-Ub topologies within polyubiquitin chains.
Another complication making it challenging to observe the linkage-specific avidity resides in the fact that a Pro-rich linker of about 40 amino acids separates NOA and ZF UBDs. This linker does not adopt a rigid structure as judged from our circular dichroism measurements. Recent work on tandem UIM UBDs found in Vps27 and Rap80 proteins has proven the importance of the α-helical linker structure to bring binding units together in space and to provide the maximum opportunity for high affinity and highly linkage-selective interactions (51). In the flexible NEMO linker, optimum structure for tightly coupling ZF and NOA binding units in space is presumably governed by the trans or cis conformation of proline residues. Incorrect cis or trans population of some proline residues produced by overexpression in Escherichia coli (52) or the lack of a specific eukaryotic binding partner could make them inefficient for avid binding. Our recent work supports this notion as different conformations of NEMO linkers based on CD and multiangle light scattering (MALS) experiments could be observed.5 Structural determination of both ZF and NOA UBDs in complex with linear or Lys-63 polyubiquitin chains, albeit challenging, will undoubtedly provide a molecular basis for explaining how NEMO achieves chain-length preference, linkage preference, and affinity to ubiquitins.
Acknowledgments
We are grateful to M.Kaminska for the GST-(Ub)2 construct and to V. Dubosclard for the pcDNA3-F395W NEMO mutant. We thank Prof. Alain Israël and Samuel Levy for critical reading of the manuscript and Patrick England for access to the Proteopole of Institut Pasteur.
This work was supported, in whole or in part, by Foundation BNP Paribas and the Institut de Recherches SERVIER (Croissy sur Seine, France).
V. Dubosclard, E. Fontan, and F. Agou, unpublished information.
V. Dubosclard et al., unpublished information.
- IKK
- IκB kinase
- NF-κB
- nuclear factor κB
- NEMO
- NF-κB essential modulator
- LZ
- leucine zipper
- CC2
- coiled-coil 2
- NOA
- NEMO Optineurin ABIN
- UBD
- ubiquitin binding domain
- ZF
- zinc finger
- Ub
- ubiquitin
- UBAN
- ubiquitin binding in ABIN and NEMO domain
- NRP
- NEMO-related protein
- SV-AUC
- sedimentation velocity analytical ultracentrifugation
- MEF
- mouse embryonic fibroblast
- NZF
- Npl4 zinc finger
- ID
- immunodeficiency
- EDA
- ectodermal dysplasia
- FP
- fluorescence polarization
- NS
- nonspecific binding
- F-ZF peptide
- D406V ZF peptide.
REFERENCES
- 1. Hayden M. S., Ghosh S. (2012) NF-κB, the first quarter-century. Remarkable progress and outstanding questions. Genes Dev. 26, 203–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Courtois G., Israël A. (2011) IKK regulation and human genetics. Curr. Top. Microbiol. Immunol. 349, 73–95 [DOI] [PubMed] [Google Scholar]
- 3. Yamazaki K., Gohda J., Kanayama A., Miyamoto Y., Sakurai H., Yamamoto M., Akira S., Hayashi H., Su B., Inoue J. (2009) Two mechanistically and temporally distinct NF-κB activation pathways in IL-1 signaling. Sci. Signal. 2, ra66. [DOI] [PubMed] [Google Scholar]
- 4. Ajibade A. A., Wang Q., Cui J., Zou J., Xia X., Wang M., Tong Y., Hui W., Liu D., Su B., Wang H. Y., Wang R. F. (2012) TAK1 negatively regulates NF-κB and p38 MAP kinase activation in Gr-1+CD11b+ neutrophils. Immunity 36, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 6. Xia Z. P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., Chen Z. J. (2009) Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emmerich C. H., Ordureau A., Strickson S., Arthur J. S., Pedrioli P. G., Komander D., Cohen P. (2013) Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. U.S.A. 110, 15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu H. (2013) Higher-order assemblies in a new paradigm of signal transduction. Cell 153, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang W., Xia Y., Cao Y., Zheng Y., Bu W., Zhang L., You M. J., Koh M. Y., Cote G., Aldape K., Li Y., Verma I. M., Chiao P. J., Lu Z. (2012) EGFR-induced and PKCϵ monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell 48, 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Makris C., Roberts J. L., Karin M. (2002) The carboxyl-terminal region of IκB kinase γ (IKKγ) is required for full IKK activation. Mol. Cell. Biol. 22, 6573–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang T. T., Feinberg S. L., Suryanarayanan S., Miyamoto S. (2002) The zinc finger domain of NEMO is selectively required for NF-κB activation by UV radiation and topoisomerase inhibitors. Mol. Cell. Biol. 22, 5813–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laplantine E., Fontan E., Chiaravalli J., Lopez T., Lakisic G., Véron M., Agou F., Israël A. (2009) NEMO specifically recognizes Lys-63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 28, 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu G., Lo Y. C., Li Q., Napolitano G., Wu X., Jiang X., Dreano M., Karin M., Wu H. (2011) Crystal structure of inhibitor of κB kinase β. Nature 472, 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cordier F., Vinolo E., Véron M., Delepierre M., Agou F. (2008) Solution structure of NEMO zinc-finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation. J. Mol. Biol. 377, 1419–1432 [DOI] [PubMed] [Google Scholar]
- 15. Cordier F., Grubisha O., Traincard F., Véron M., Delepierre M., Agou F. (2009) The zinc finger of NEMO is a functional ubiquitin-binding domain. J. Biol. Chem. 284, 2902–2907 [DOI] [PubMed] [Google Scholar]
- 16. Bomar M. G., Pai M. T., Tzeng S. R., Li S. S., Zhou P. (2007) Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase η. EMBO Rep. 8, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kulathu Y., Akutsu M., Bremm A., Hofmann K., Komander D. (2009) Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 16, 1328–1330 [DOI] [PubMed] [Google Scholar]
- 18. Sato Y., Yoshikawa A., Yamashita M., Yamagata A., Fukai S. (2009) Structural basis for specific recognition of Lys-63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 28, 3903–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bosanac I., Wertz I. E., Pan B., Yu C., Kusam S., Lam C., Phu L., Phung Q., Maurer B., Arnott D., Kirkpatrick D. S., Dixit V. M., Hymowitz S. G. (2010) Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol. Cell 40, 548–557 [DOI] [PubMed] [Google Scholar]
- 20. Tokunaga F., Nishimasu H., Ishitani R., Goto E., Noguchi T., Mio K., Kamei K., Ma A., Iwai K., Nureki O. (2012) Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 31, 3856–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Verhelst K., Carpentier I., Kreike M., Meloni L., Verstrepen L., Kensche T., Dikic I., Beyaert R. (2012) A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 31, 3845–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grubisha O., Kaminska M., Duquerroy S., Fontan E., Cordier F., Haouz A., Raynal B., Chiaravalli J., Delepierre M., Israël A., Véron M., Agou F. (2010) DARPin-assisted crystallography of the CC2-LZ domain of NEMO reveals a coupling between dimerization and ubiquitin binding. J. Mol. Biol. 395, 89–104 [DOI] [PubMed] [Google Scholar]
- 23. Gautheron J., Pescatore A., Fusco F., Esposito E., Yamaoka S., Agou F., Ursini M. V., Courtois G. (2010) Identification of a new NEMO/TRAF6 interface affected in incontinentia pigmenti pathology. Hum Mol. Genet. 19, 3138–3149 [DOI] [PubMed] [Google Scholar]
- 24. Hubeau M., Ngadjeua F., Puel A., Israel L., Feinberg J., Chrabieh M., Belani K., Bodemer C., Fabre I., Plebani A., Boisson-Dupuis S., Picard C., Fischer A., Israel A., Abel L., Veron M., Casanova J. L., Agou F., Bustamante J. (2011) New mechanism of x-linked anhidrotic ectodermal dysplasia with immunodeficiency. Impairment of ubiquitin binding despite normal folding of NEMO protein. Blood 118, 926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiaravalli J., Fontan E., Fsihi H., Coic Y. M., Baleux F., Véron M., Agou F. (2011) Direct inhibition of NF-κB activation by peptide targeting the NOA ubiquitin binding domain of NEMO. Biochem. Pharmacol. 82, 1163–1174 [DOI] [PubMed] [Google Scholar]
- 26. Schmidt-Supprian M., Bloch W., Courtois G., Addicks K., Israël A., Rajewsky K., Pasparakis M. (2000) NEMO/IKK γ-deficient mice model incontinentia pigmenti. Mol. Cell 5, 981–992 [DOI] [PubMed] [Google Scholar]
- 27. Harhaj E. W., Good L., Xiao G., Uhlik M., Cvijic M. E., Rivera-Walsh I., Sun S. C. (2000) Somatic mutagenesis studies of NF-κB signaling in human T cells. Evidence for an essential role of IKK γ in NF-κB activation by T-cell costimulatory signals and HTLV-I Tax protein. Oncogene 19, 1448–1456 [DOI] [PubMed] [Google Scholar]
- 28. Vinolo E., Sebban H., Chaffotte A., Israël A., Courtois G., Véron M., Agou F. (2006) A point mutation in NEMO associated with anhidrotic ectodermal dysplasia with immunodeficiency pathology results in destabilization of the oligomer and reduces lipopolysaccharide- and tumor necrosis factor-mediated NF-κB activation. J. Biol. Chem. 281, 6334–6348 [DOI] [PubMed] [Google Scholar]
- 29. Komander D., Lord C. J., Scheel H., Swift S., Hofmann K., Ashworth A., Barford D. (2008) The structure of the CYLD USP domain explains its specificity for Lys-63-linked polyubiquitin and reveals a B box module. Mol. Cell 29, 451–464 [DOI] [PubMed] [Google Scholar]
- 30. Brown P. H., Schuck P. (2006) Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 90, 4651–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salvay A. G., Santamaria M., le Maire M., Ebel C. (2007) Analytical ultracentrifugation sedimentation velocity for the characterization of detergent-solubilized membrane proteins Ca2++-ATPase and ExbB. J. Biol. Phys. 33, 399–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hartwig A., Schwerdtle T., Bal W. (2010) Biophysical analysis of the interaction of toxic metal ions and oxidants with the zinc finger domain of XPA. Methods Mol. Biol. 649, 399–410 [DOI] [PubMed] [Google Scholar]
- 33. de Vries S. J., van Dijk M., Bonvin A. M. (2010) The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 [DOI] [PubMed] [Google Scholar]
- 34. Jain A., Ma C. A., Liu S., Brown M., Cohen J., Strober W. (2001) Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat. Immunol. 2, 223–228 [DOI] [PubMed] [Google Scholar]
- 35. Shifera A. S., Horwitz M. S. (2008) Mutations in the zinc finger domain of IKKγ block the activation of NF-κB and the induction of IL-2 in stimulated T lymphocytes. Mol. Immunol. 45, 1633–1645 [DOI] [PubMed] [Google Scholar]
- 36. Sahlender D. A., Roberts R. C., Arden S. D., Spudich G., Taylor M. J., Luzio J. P., Kendrick-Jones J., Buss F. (2005) Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 169, 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mankouri J., Fragkoudis R., Richards K. H., Wetherill L. F., Harris M., Kohl A., Elliott R. M., Macdonald A. (2010) Optineurin negatively regulates the induction of IFNβ in response to RNA virus infection. PLoS Pathog. 6:e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wild P., Farhan H., McEwan D. G., Wagner S., Rogov V. V., Brady N. R., Richter B., Korac J., Waidmann O., Choudhary C., Dötsch V., Bumann D., Dikic I. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Papoutsopoulou S., Symons A., Tharmalingham T., Belich M. P., Kaiser F., Kioussis D., O'Garra A., Tybulewicz V., Ley S. C. (2006) ABIN-2 is required for optimal activation of Erk MAP kinase in innate immune responses. Nat. Immunol. 7, 606–615 [DOI] [PubMed] [Google Scholar]
- 40. Derewenda Z. S., Vekilov P. G. (2006) Entropy and surface engineering in protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 62, 116–124 [DOI] [PubMed] [Google Scholar]
- 41. Yang F., Yamashita J., Tang E., Wang H. L., Guan K., Wang C. Y. (2004) The zinc finger mutation C417R of I-κ B kinase γ impairs lipopolysaccharide- and TNF-mediated NF-κ B activation through inhibiting phosphorylation of the I-κ B kinase β activation loop. J. Immunol. 172, 2446–2452 [DOI] [PubMed] [Google Scholar]
- 42. Schröfelbauer B., Polley S., Behar M., Ghosh G., Hoffmann A. (2012) NEMO ensures signaling specificity of the pleiotropic IKKβ by directing its kinase activity toward IκBα. Mol. Cell 47, 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sims J. J., Haririnia A., Dickinson B. C., Fushman D., Cohen R. E. (2009) Avid interactions underlie the Lys-63-linked polyubiquitin binding specificities observed for UBA domains. Nat. Struct. Mol. Biol. 16, 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fushman D., Walker O. (2010) Exploring the linkage dependence of polyubiquitin conformations using molecular modeling. J. Mol. Biol. 395, 803–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lo Y. C., Lin S. C., Rospigliosi C. C., Conze D. B., Wu C. J., Ashwell J. D., Eliezer D., Wu H. (2009) Structural basis for recognition of diubiquitins by NEMO. Mol. Cell 33, 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sims J. J., Scavone F., Cooper E. M., Kane L. A., Youle R. J., Boeke J. D., Cohen R. E. (2012) Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat. Methods 9, 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kensche T., Tokunaga F., Ikeda F., Goto E., Iwai K., Dikic I. (2012) Analysis of nuclear factor-κB (NF-κB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-κB. J. Biol. Chem. 287, 23626–23634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hadian K., Griesbach R. A., Dornauer S., Wanger T. M., Nagel D., Metlitzky M., Beisker W., Schmidt-Supprian M., Krappmann D. (2011) NF-κB essential modulator (NEMO) interaction with linear and Lys-63 ubiquitin chains contributes to NF-κB activation. J. Biol. Chem. 286, 26107–26117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Richarme G. (1982) Associative properties of the Escherichia coli galactose binding protein and maltose binding protein. Biochem. Biophys. Res. Commun. 105, 476–481 [DOI] [PubMed] [Google Scholar]
- 50. Ivins F. J., Montgomery M. G., Smith S. J., Morris-Davies A. C., Taylor I. A., Rittinger K. (2009) NEMO oligomerization and its ubiquitin-binding properties. Biochem. J. 421, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sims J. J., Cohen R. E. (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell 33, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hoffmann A., Bukau B., Kramer G. (2010) Structure and function of the molecular chaperone Trigger Factor. Biochim. Biophys. Acta 1803, 650–661 [DOI] [PubMed] [Google Scholar]