Abstract
Gram-negative plant pathogenic bacteria possess a type III secretion system (T3SS) to inject bacterial proteins, called type III effectors (T3Es), into host cells through a specialized syringe structure. T3Es are virulence factors that can suppress plant immunity but they can also conversely be recognized by the plant and trigger specific resistance mechanisms. The T3SS and injected T3Es play a central role in determining the outcome of a host-pathogen interaction. Still little is known in plant pathogens on the assembly of the T3SS and the regulatory mechanisms involved in the temporal control of its biosynthesis and T3E translocation. However, recent insights point out the role of several proteins as prime candidates in the role of regulators of the type III secretion (T3S) process. In this review we report on the most recent advances on the regulation of the T3S by focusing on protein players involved in secretion/translocation regulations, including type III chaperones (T3Cs), type III secretion substrate specificity switch (T3S4) proteins and other T3S orchestrators.
Keywords: bacterial plant pathogens, type III secretion system (T3SS), type III chaperones (T3Cs), type III secretion substrate specificity switch (T3S4), secretion of type III effectors (T3Es), control of secretion/translocation
INTRODUCTION
The type III secretion system (T3SS) is a major determinant of pathogenicity characterized in numerous Gram-negative animal and plant pathogenic bacteria (Cornelis and van Gijsegem, 2000; He et al., 2004; Tampakaki et al., 2010). It allows type III effector (T3E) delivery into the host cells thanks to a complex and ordered type III secretion (T3S) process (Büttner, 2012). T3Es are virulence factors that can suppress plant immunity or they can also conversely be recognized by the plant and trigger the so called effector-triggered immunity (Jones and Dangl, 2006; Feng and Zhou, 2012). This review is devoted to proteins that affect regulation of the secretion/translocation of T3Es in the two main groups of T3SS in plant pathogens, i.e., Hrp1- (Hrp for hypersensitive response and pathogenicity) and Hrp2-T3SS phytopathogenic bacteria (see Tampakaki et al., 2010, and Büttner, 2012; for reviews on the genomic organization and architecture of T3SS). We give here an overview of the specialized type III chaperones (T3Cs) and of other T3S control proteins characterized in Hrp1 T3SS bacteria (Pseudomonas syringae and Erwinia amylovora), and in Hrp2 T3SS bacteria (Xanthomonas spp. and Ralstonia solanacearum).
TYPE III CHAPERONES
Type III chaperones can be defined as helper proteins, mainly acting through direct interactions with T3Es, required for the delivery of effectors into the host cell. T3Cs are small (15–20 kDa), acidic, cytoplasmic proteins, and harbor a predicted α-helical secondary structure in their C-terminal part (Feldman and Cornelis, 2003; Parsot et al., 2003). T3Cs can be strictly or partially required to prevent cytoplasmic proteolysis and premature aggregation of T3Es, and/or maintain T3Es in a secretion competent status (Feldman and Cornelis, 2003; Parsot et al., 2003). T3Cs do not share amino acid sequence similarity, nevertheless, a classification has been proposed dividing them into three classes according to their cognate substrates. Class I is subdivided into classes IA and IB, corresponding to chaperones binding to one or several T3Es, respectively (Cornelis and van Gijsegem, 2000; Parsot et al., 2003). Class II chaperones that are specialized translocator-chaperones and class III chaperones, flagellar-associated T3SS chaperones, are up to now only described in animal pathogenic bacteria (see Büttner, 2012, as a review). We will focus on class I T3Cs as it is the main class described in phytopathogenic bacteria (Figure 1).
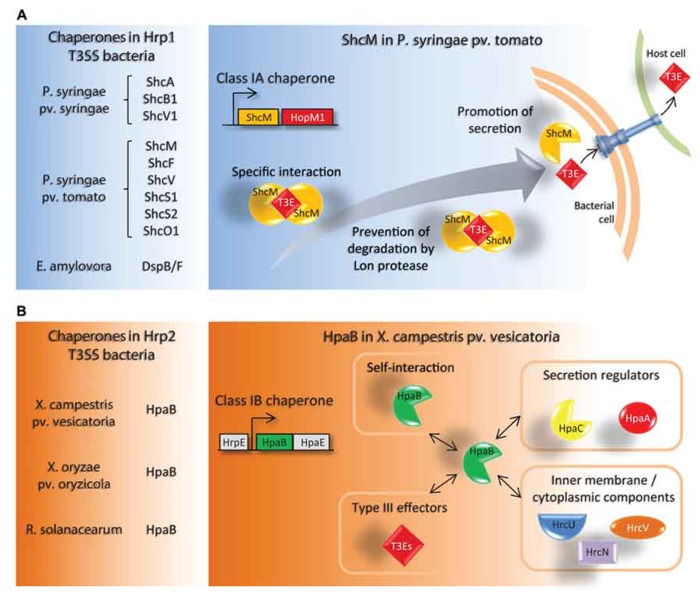
FIGURE 1.
Schematic representation of specific features of class IA and class IB T3Cs in plant pathogenic bacteria. (A) List of known T3Cs in Hrp1 T3SS bacteria (left panel). All are class IA T3Cs. A model depicting the role of the T3C ShcM is shown as a representative example (right panel). ShcM is co-transcribed with its cognate T3E HopM1. ShcM specifically interacts with HopM1, prevents its degradation by Lon proteases in the bacterial cytoplasm and promotes its secretion/translocation via the T3SS. (B) List of known T3Cs in Hrp2 T3SS bacteria (left panel). All are class IB T3Cs. A schematic representation of known binding partners of HpaB in X. campestris pv. vesicatoria is shown (right panel). HpaB is encoded within the hrp cluster and without any T3E in the vicinity. HpaB interacts with itself and with multiple T3SS-related proteins, comprising the secretion regulators HpaC and HpaA (see also Figure 2), the inner membrane components HrcU and HrcV, the cytoplasmic ATPase HrcN and T3Es. HpaB is required for efficient secretion of T3Es.
CLASS IA T3C ARE MOSTLY INVOLVED IN THE SECRETION OF A SPECIFIC T3E
Class IA T3Cs were described in P. syringae pv. syringae, P. syringae pv. tomato and E. amylovora, all three pathogens harboring a Hrp1 T3SS (Figure 1A). The corresponding genes are located next to the cognate effector genes in the bacterial genome. These T3Cs were named Shc for Specific hop chaperones in Pseudomonas species as they help secretion of T3Es named Hops (hrp-dependent outer proteins), and to Dsp for disease specific protein, i.e., chaperone or effector in E. amylovora. For all class IA T3Cs studied, a direct interaction T3C-cognate T3E has been demonstrated. To date, three Shc proteins have been characterized in P. syringae pv. syringae, including ShcA, which controls the secretion and translocation of HopA1 (van Dijk et al., 2002), and ShcB1 and ShcV1 that have been demonstrated to stabilize their cognate T3Es HopB1 and HopV1, respectively (Charity et al., 2003; Losada and Hutcheson, 2005). They prevent the T3E degradation mediated by the Lon protease, which exerts a negative regulatory effect on the T3S in P. syringae (Bretz et al., 2002). Three typical class IA T3Cs have been identified in P. syringae pv. tomato. ShcM plays several roles to promote efficient HopM1 translocation: it is required for the secretion and the translocation of HopM1 (Badel et al., 2003), but also protects HopM1 from Lon degradation, interacting with the effector through a chaperone binding domain (CBD) which is different from the Lon-targeting domain (Losada and Hutcheson, 2005; Figure 1A). ShcF is required for HopF stabilization but was shown dispensable for HopF secretion (Shan et al., 2004), whereas ShcV is required for HopV1 secretion (Wehling et al., 2004). Three other chaperones, SchS1, SchO1 and SchS2, harbor the typical features of T3Cs, but have atypical properties for class IA T3Cs as they could facilitate translocation of homologous T3Es (Kabisch et al., 2005). In another study, it was shown that SchS1, SchO1 and SchS2 were additionally able to bind to each other and to other’s cognate effectors (Guo et al., 2005). These three chaperones facilitate the secretion/translocation of their cognate T3E (HopS1, HopO1-1 and HopS2, respectively), and SchS1 and SchS2 can substitute ShcO1 to help HopO1-1 type III delivery (Guo et al., 2005). Other putative chaperones have been identified, located upstream a putative T3E target, but even if they show the physical properties of class IA T3Cs, their role still needs to be demonstrated. This is the case for the T3C-T3E pairs ShcN-HopN1 (Lopez-Solanilla et al., 2004) and ORF2-AvrE1 (Badel et al., 2006).
In E. amylovora, a putative T3C-T3E pair was characterized, the couple DspB/F-DspA/E (Gaudriault et al., 1997). DspB/F interacts with, stabilizes, and is important for DspA/E secretion (Gaudriault et al., 2002). Two studies identified the DspB/F binding sites in the N-terminal part of DspA/E, however with some differences in the regions of interactions (Triplett et al., 2009; Oh et al., 2010). This may indicate the presence of multiple CBD to help T3E translocation.
Class IA chaperones from phytopathogenic bacteria share common structural and functional features with class IA chaperones in animal pathogenic bacteria (He et al., 2004). The binding of both T3E and T3C partners is described in more details in the latter systems, notably through structural analysis of the T3C-T3E complex (Büttner et al., 2005; Lillington et al., 2011; Vujanac and Stebbins, 2013). Recently, co-crystal structure of the complex ShcA-HopA1 from P. syringae has also been obtained, both proteins sharing a fold and interacting through a conserved structural motif (called the β-motif) where HopA1 shows an extended non-globular conformation critical for the stability of the complex (Janjusevic et al., 2013). This β-motif is strongly conserved from animal to plant pathogens and was also described with the DspB/F chaperone of E. amylovora (Triplett et al., 2010), suggesting that it may be widely implicated in T3C-T3E complexes.
Another particular case of class IA chaperone is the HrpG protein which was described to act as a suppressor of a negative regulator of the T3SS (Wei et al., 2005). HrpG was characterized in P. syringae pv. syringae as a chaperone-like protein, as HrpG is structurally close to SicP, a class IA chaperone of Salmonella typhimurium, and harbors the characteristics of T3Cs (Wei et al., 2005). Wei et al. (2005) identified HrpG as an interactor of HrpV, a conserved component of the P. syringae T3SS, previously demonstrated to down-regulate the expression of the hrp gene cluster (Preston et al., 1998). Hence, the assembly of the HrpG–HrpV protein complex leads to the suppression of the hrpV-dependent negative regulation of the hrp gene cluster. More recently, Ortiz-Martin et al. (2010) studied HrpG in P. syringae pv. phaseolicola and highlighted for HrpG an additional HrpV-independent role in virulence.
CLASS IB T3Cs ARE INVOLVED IN THE SECRETION OF SEVERAL T3Es
Genes which encode for class IB chaperones, binding several T3Es, are typically located within the hrp cluster. Class IB T3Cs were identified in several Hrp2 T3SS phytopathogenic bacteria, with one conserved protein named HpaB (for hrp-associated; Figure 1B). HpaB has been particularly well characterized in X. campestris pv. vesicatoria, as a T3C with a wide specificity of substrates. It was shown that HpaB could self-interact (Büttner et al., 2006) as well as with the inner membrane proteins HrcU and HrcV (Lorenz and Büttner, 2011; Hartmann and Büttner, 2013), with the ATPase HrcN (Lorenz and Büttner, 2009), with T3Es (AvrBs1 and AvrBs3; Büttner et al., 2004), and with other proteins involved in the T3S control (HpaC and HpaA; Büttner et al., 2004; Lorenz et al., 2008a; Figure 1B). HpaB promotes the secretion of many Xanthomonas T3Es (XopC, XopJ, AvrBst, AvrBs1, AvrBs3, and XopF1; Büttner et al., 2004, 2006). Like typical chaperones, HpaB is a small acidic protein, with Yersinia, Shigella flexneri or Salmonella enterica T3Cs predicted similarities (Büttner et al., 2006), and with a rich content in leucine amino acids (Büttner et al., 2004), which may contribute to its ability to interact with various proteins. A HpaB homolog identified in X. orizae pv. oryzicola (Zou et al., 2006) was demonstrated to be involved in efficient translocation of 15 T3Es out of the 16 T3Es identified in this strain (Furutani et al., 2009). In R. solanacearum, HpaB was demonstrated to be required for efficient translocation of more than 66 T3Es (Mukaihara et al., 2004, 2010) but no data concerning putative interactions with T3Es is available. However, HpaB was shown to be dispensable for the translocation of harpin, pilin, and translocon proteins (Mukaihara et al., 2010). All these data support the view of a broader involvement of HpaB in T3S control rather than just a T3E escort function. This hypothesis is comforted by the hrp- phenotype (no disease, no hypersensitive response, HR) of hpaB mutants, despite evidence for functional pili production in X. campestris pv. vesicatoria (Weber et al., 2005) or in R. solanacearum (Mukaihara et al., 2004). It also suggests that chaperone-mediated control for T3E delivery may be different according to the microorganism. The fact that specialized class II chaperones, which are responsible for the secretion of the translocon proteins in animal pathogenic bacteria, were not identified in phytopathogenic bacteria shows a noticeable difference compared to T3S regulators from phytopathogenic bacteria. Therefore, how secretion of translocon proteins is promoted in absence of class II T3Cs in plant pathogenic bacteria remains to be addressed.
T3S4 PROTEINS
Based on the current knowledge of T3SS assembly, it is presumed that secretion of the Hrp pilus subunits must proceed before secretion of translocon proteins and effectors. This hierarchical process led to the classification of secreted proteins as “early” and “late” T3SS substrates. The involvement of proteins controlling T3S export process were first described as early as 1995 with the identification of the InvJ protein from S. typhimurium (Collazo et al., 1995). However, the link between functionally related proteins and the presence of a conserved T3S4 (type III secretion substrate specificity switch) domain was first described by Agrain et al. (2005a), with the characterization of the YscP protein from Yersina species. Members of the YscP family have little or no homology at the amino acid level, but are all proline-rich and harbor the so-called T3S4 domain in their C-terminal part. This structurally conserved domain is globular and composed of seven hydrophobic clusters of amino acids that define predicted β-strands and α-helices (Agrain et al., 2005a). Yersinia YscP protein has been particularly well characterized, highlighting a dual function: YscP is required to switch secretion from early to late substrates, including Yop (Yersinia outer proteins) T3Es (Edqvist et al., 2003; Agrain et al., 2005a) and controls the needle length, the nice molecular ruler model describing an attachment of the C-terminal part of YscP to the base of the secretion apparatus while the N-terminal part travels the inner channel of the growing needle. The needle is completed when its size corresponds to a fully stretched YscP protein (Journet et al., 2003; Agrain et al., 2005b). Since then, T3S4 proteins have been also intensively studied in other animal pathogenic bacteria, mainly in Salmonella (Kubori et al., 2000; Marlovits et al., 2006) and Shigella (Magdalena et al., 2002; Botteaux et al., 2008) species.
In plant pathogenic bacteria, the most studied T3S4 proteins are HpaC from X. campestris pv. vesicatoria (Büttner et al., 2006; Lorenz et al., 2008b) and HrpP from P. syringae (Morello and Collmer, 2009). Both proteins are important factors for bacterial pathogenicity. A hpaC mutant was shown to trigger reduced disease symptoms and reduced HR on susceptible and resistant pepper, respectively (Büttner et al., 2006). HrpP was found to be required for virulence on tomato and HR elicitation on Nicotiana tabacum (Morello and Collmer, 2009). Studies on HpaC and HrpP revealed that both proteins are involved at different levels in the regulation of T3S. In X. campestris, while the secretion of the Hrp pilin is not affected in a hpaC mutant, secretion of the translocon proteins (HrpF and XopA) and several T3Es (AvrBs3, XopC, XopJ and XopF1) is abolished (Büttner et al., 2006). On the other hand, in P. syringae, a hrpP mutant is severely impaired for pilin secretion but also for translocation of several T3Es such as AvrPto and therefore appears to behave almost like a T3SS-defective mutant (Ramos et al., 2007; Morello and Collmer, 2009). Quite surprisingly, HrpP itself is translocated into plant cells; however it was shown that the translocation of AvrPto is not dependent on the translocation of HrpP (Morello and Collmer, 2009).
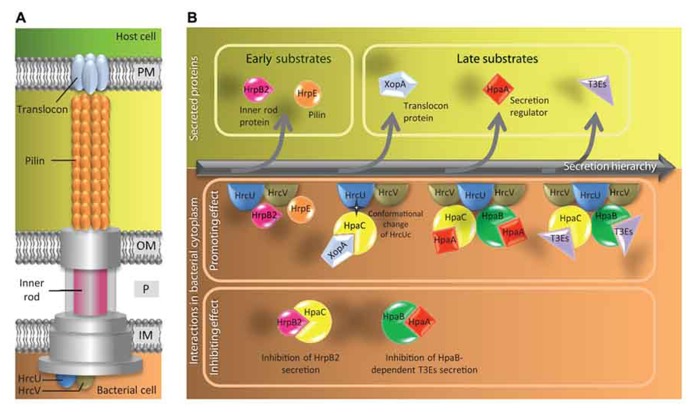
To date, our understanding of T3S4 proteins is mostly based on the analysis of HpaC, a putative secretion switch factor regulating early (HrpB2 and HrpE pilin) and late (XopA translocator and T3Es) substrate secretion (Figure 2). HpaC appears to be a crossroad control protein with several interactors associated to different steps in the T3S process. Indeed, HpaC is able to self-interact and direct interactions with HpaC were identified with multiple partners including the inner membrane proteins HrcU and HrcV (Lorenz et al., 2008b; Hartmann and Büttner, 2013), the class IB chaperone HpaB (Büttner et al., 2006), the regulator HpaA (Lorenz et al., 2008a), the translocator XopA (Büttner et al., 2006), the ATPase HrcN (Lorenz and Büttner, 2009), and with T3Es (AvrBs3 and XopF1; Büttner et al., 2006; Lorenz et al., 2008b). HpaC was also shown to interact with HrpB2, an early T3S substrate required both for pilin subunit and translocators HrpF and XopA secretion (Rossier et al., 2000; Lorenz et al., 2008b; Hartmann et al., 2012). HrpB2 is part of the inner rod of the T3SS (Hartmann et al., 2012) and an interaction with the T3SS core component HrcU is necessary for its secretion (Lorenz and Büttner, 2011). HrpB2 is over-secreted in a hpaC mutant and in the same time, secretion of translocon proteins and T3Es is reduced (Büttner et al., 2006; Lorenz et al., 2008b). The HpaC-dependent substrate specificity switch requires the interaction between HpaC and the C-terminal domain of HrcU (HrcUc; Schulz and Büttner, 2011), that presumably induces a conformational change altering substrate specificity of the T3SS (Lorenz and Büttner, 2011). This change of conformation might be due to an autocatalytic cleavage of HrcU at a NPTH motif conserved between HrcU homologs in pathogenic bacteria. Mutations in the NPTH amino-acid motif of HrcU alter its interactions with both HpaC and HrpB2, suggesting that they may share the same binding site on HrcU (Lorenz and Büttner, 2011). Thus HpaC and HrpB2 could compete for their interaction with HrcU. It has been therefore proposed that HpaC could prevent efficient interaction between HrpB2 and HrcU and allow the access of the docking site of HrcU for secretion of translocon proteins and T3Es. But no interaction has been uncovered to date between HrcU and the T3SS substrates tested (XopA and XopF1; Lorenz et al., 2008b). However, it is noteworthy that the class IB HpaB was found to interact with HrcU (Lorenz and Büttner, 2011) and HrcV (Büttner et al., 2006; Hartmann and Büttner, 2013). It is conceivable that the docking of T3Es to HrcU and HrcV could be mediated by HpaB (Figure 2).
FIGURE 2.
Model of the control of type III secretion hierarchy in X. campestris pv. vesicatoria. (A) Schematic representation of the T3SS highlighting components mentioned in (B). The basal body of the T3SS spans the inner membrane (IM), the periplasm (P) and the outer membrane (OM) of the bacterium. The extracellular part of the T3SS comprises pilin subunits that form the pilus and translocon proteins that form a pore in the host plasma membrane (PM). The outer membrane/periplasmic inner rod is presumably the intracellular prolongation of the pilus. HrcU and HrcV are inner membrane components with a cytoplasmic C-terminal part. (B) Schematic view of the secreted proteins (upper part) and the protein interactions involved (lower part) during the hierarchical secretion process. HrcU and HrcV probably act as a docking platform for secretion. The first substrates that travel through the T3SS are the pilin HrpE and the inner rod protein HrpB2. Secretion of HrpB2 is required for secretion of HrpE. The switch of substrate specificity between early and late substrates (translocon proteins and T3Es) depends on a HpaC-mediated conformational change of the HrcU C-terminal domain (HrcUc). HpaC is a T3S4 protein that interacts with HrpB2 and the late substates. The switch between secretion of translocon proteins and T3Es requires the regulator HpaA. Secretion of HpaA releases the class IB chaperone HpaB. HpaB can bind to T3Es to promote their secretion.
In R. solanacearum, the putative T3S4 domain protein HpaP is known to play a role in pathogenicity, since the corresponding mutant was reduced both for HR elicitation on tobacco and disease symptom production on tomato (van Gijsegem et al., 2002). In addition, HpaP was also required for an efficient secretion of the T3E PopA. Recent work characterized HpaP as a putative non-secreted T3S4 protein involved in the modulation of the secretion of early and late type III substrates, HrpY pilin and AvrA and PopP1 T3Es, respectively (Lohou et al., unpublished manuscript). As for HpaP homologs (Morello and Collmer, 2009; Schulz and Büttner, 2011), the T3S4 domain of R. solanacearum was demonstrated to be essential for HpaP’s role in virulence on tomato and Arabidopsis thaliana (Lohou et al., unpublished). Two other candidate T3S4 proteins studied in X. oryzae pv. oryzae and X. oryzae pv. oryzicola, both named HpaP, were shown to be important for pathogenicity on rice and for HR (Cho et al., 2008; Guo et al., 2012). In X. oryzae pv. oryzae, 16 T3Es have been found out to be T3SS substrates and all of them but one are dependent on HpaP for efficient translocation into tomato leaves (Furutani et al., 2009).
ADDITIONAL T3S-ASSOCIATED REGULATORS: HELPERS, EFFECTORS OR BOTH?
Finally, there are reports in the literature of T3SS-associated proteins with an apparent dual role, being involved both in the control of the export process, being crucial for T3E secretion, but also being themselves translocated into the plant cell and described as virulence factors. The first example is X. campestris pv. vesicatoria HpaA, originally described as a hrp-associated protein, since the corresponding mutant was not able to trigger disease symptoms on susceptible pepper and tomato, but retained the ability to elicit a reduced HR on several resistant pepper and tomato lines (Huguet et al., 1998). Lorenz et al. (2008a) characterized HpaA as important for the secretion of different T3SS substrates (the pilin HrpE, the translocon proteins HrpF and XopA, and the T3Es AvrBs3, XopJ and XopC), as well as for the translocation of these T3Es. Two nuclear localization signals (NLS) were identified in HpaA, which was demonstrated to be secreted and translocated to the plant nucleus in a T3SS-dependent manner (Huguet et al., 1998; Lorenz et al., 2008a). Translocation of HpaA is HpaB-dependent, in agreement with the identified interaction between the two proteins (Alegria et al., 2004; Lorenz et al., 2008a).
The second example is HrpJ, a Hrp-associated protein found in P. syringae pathovars and E. amylovora. HrpJ was described as a T3S regulator required for E. amylovora pathogenicity as a hrpJ mutant displayed a reduced HR phenotype on N. tabacum and produced no disease symptoms on pear fruit (Nissinen et al., 2007). Bocsanczy et al. (2008) identified a direct interaction between HrpJ and the two harpins HrpN and HrpW. HrpJ was proposed to act as a chaperone to allow HrpN secretion, and both proteins are also required for DspA/E translocation, suggesting that HrpJ acts indirectly on the translocation of DspA/E by facilitating the secretion of HrpN (Nissinen et al., 2007; Bocsanczy et al., 2008). In P. syringae pv. tomato, the hrpJ mutant triggers a reduced HR in non-host tobacco plants and is also unable to provoke disease symptoms on A. thaliana (Fu et al., 2006). HrpJ was found to be required for the translocation of several T3Es (AvrB1, AvrRpt2, AvrPto1, AvrPtoB and HopB1; Fu et al., 2006). The secretion of the three harpins HrpZ1, HrpW1 and HopAK1, and of the translocon protein HrpK1 is HrpJ dependent (Fu et al., 2006; Crabill et al., 2012), whereas the pilus subunit HrpA1 is over-secreted in a hrpJ mutant (Crabill et al., 2012). These observations suggest that HrpJ controls the switch from secretion of the pilin to secretion of the harpins and of translocon proteins that are subsequently both involved in the interaction with the plant cell membrane. A P. syringae T3SS-deficient strain is able to grow better when inoculated on transgenic A. thaliana expressing HrpJ than on wild-type plants, and triggers two-fold less callose deposition (Crabill et al., 2012). This result suggested a role in planta for HrpJ as a T3E plant innate immunity suppressor (Crabill et al., 2012; Wei and Collmer, 2012) distinct from its known regulatory role during the biogenesis of the T3SS. The fact that HpaA is addressed to plant cell nucleus during infection could also be an indication that such proteins may have dual roles, being both involved in the control of the translocation process in the bacterium and in a more specific function as a T3E once inside the plant cell.
CONCLUSIONS AND PERSPECTIVES
The current knowledge on T3Cs in plant pathogenic bacteria makes apparent a major difference between T3SS export control in Hrp groups 1 and 2: control of T3E export mainly proceeds in group 1 through class IA chaperones whereas group 2 seems to require class IB chaperones (Figure 1). It remains to be determined if this dichotomy reflects fundamental differences in T3SS export control mechanisms or just results from evolutionary divergence. T3S4 domain proteins appear as key players in the control of the secretion process. Future progress on the mechanistic T3S control will rely on more systematic identification of hetero-oligomeric protein complexes between the various T3SS control proteins identified, and on the evaluation of the different affinities between the interacting partners.
It is more than likely that T3S in bacterial plant pathogens is an ordered and sequential process, as demonstrated for T3SS of animal and human pathogens (Kubori and Nagai, 2011; Hicks and Galán, 2013). To date, observations supporting this view only relate on the assembly of the T3SS based on the discrimination between early and late T3SS substrates. However, there is still no direct evidence of a secretion hierarchy among T3Es in plant pathogens. This question of whether T3Es act in a coordinated spatial and temporal manner during host infection is particularly relevant for bacterial plant pathogens considering that they harbor large repertoires of translocated T3Es (Cunnac et al., 2009; Poueymiro and Genin, 2009). Future studies addressing this point should have important implications on T3E action inside host cells, as already demonstrated in other pathogens (Kubori and Galán, 2003). The recent development of microscopy-based assays opens exciting perspectives to provide a comprehensive description of dynamics of effector translocation and translocation temporal order (Mills et al., 2013).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Nemo Peeters for critical reading of the manuscript. David Lohou was funded by a grant from the French Ministry of National Education and Research. Fabienne Vailleau work was supported by a French Agence Nationale de la Recherche grant (ANR-2010-JCJC-1710-01). Our work is performed at the LIPM that is part of the Laboratoire d’Excellence (LABEX) entitled TULIP (ANR-10-LABX-41).
REFERENCES
- Agrain C., Callebaut I., Journet L., Sorg I., Paroz C., Mota L. J., et al. (2005a). Characterization of a Type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 56 54–67 10.1111/j.1365-2958.2005.04534.x [DOI] [PubMed] [Google Scholar]
- Agrain C., Sorg I., Paroz C., Cornelis G. R. (2005b). Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol. Microbiol. 57 1415–1427 10.1111/j.1365-2958.2005.04758.x [DOI] [PubMed] [Google Scholar]
- Alegria M. C., Docena C., Khater L., Ramos C. H., da Silva A. C., Farah C. S. (2004). New protein-protein interactions identified for the regulatory and structural components and substrates of the type III Secretion system of the phytopathogen Xanthomonas axonopodis Pathovar citri. J. Bacteriol. 186 6186–6197 10.1128/JB.186.18.6186-6197.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel J. L., Nomura K., Bandyopadhyay S., Shimizu R., Collmer A., He S. Y. (2003). Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol. Microbiol. 49 1239–1251 10.1046/j.1365-2958.2003.03647.x [DOI] [PubMed] [Google Scholar]
- Badel J. L., Shimizu R., Oh H. S., Collmer A. (2006). A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant Microbe Interact. 19 99–111 10.1094/MPMI-19-0099 [DOI] [PubMed] [Google Scholar]
- Bocsanczy A. M., Nissinen R. M., Oh C. S., Beer S. V. (2008). HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol. Plant Pathol. 9 425–434 10.1111/j.1364-3703.2008.00471.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botteaux A., Sani M., Kayath C. A., Boekema E. J., Allaoui A. (2008). Spa32 interaction with the inner-membrane Spa40 component of the type III secretion system of Shigella flexneri is required for the control of the needle length by a molecular tape measure mechanism. Mol. Microbiol. 70 1515–1528 10.1111/j.1365-2958.2008.06499.x [DOI] [PubMed] [Google Scholar]
- Bretz J., Losada L., Lisboa K., Hutcheson S. W. (2002). Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 45 397–409 10.1046/j.1365-2958.2002.03008.x [DOI] [PubMed] [Google Scholar]
- Büttner C. R., Cornelis G. R., Heinz D. W., Niemann H. H. (2005). Crystal structure of Yersinia enterocolitica type III secretion chaperone SycT. Protein Sci. 14 1993–2002 10.1110/ps.051474605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D. (2012). Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76 262–310 10.1128/MMBR.05017-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D., Gurlebeck D., Noel L. D., Bonas U. (2004). HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol. Microbiol. 54 755–768 10.1111/j.1365-2958.2004.04302.x [DOI] [PubMed] [Google Scholar]
- Büttner D., Lorenz C., Weber E., Bonas U. (2006). Targeting of two effector protein classes to the type III secretion system by a HpaC- and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 59 513–527 10.1111/j.1365-2958.2005.04924.x [DOI] [PubMed] [Google Scholar]
- Charity J. C., Pak K., Delwiche C. F., Hutcheson S. W. (2003). Novel exchangeable effector loci associated with the Pseudomonas syringae hrp pathogenicity island: evidence for integron-like assembly from transposed gene cassettes. Mol. Plant Microbe Interact. 16 495–507 10.1094/MPMI.2003.16.6.495 [DOI] [PubMed] [Google Scholar]
- Cho H. J., Park Y. J., Noh T. H., Kim Y. T., Kim J. G., Song E. S., et al. (2008). Molecular analysis of the hrp gene cluster in Xanthomonas oryzae pathovar oryzae KACC10859. Microb. Pathog. 44 473–483 10.1016/j.micpath.2007.12.002 [DOI] [PubMed] [Google Scholar]
- Collazo C. M., Zierler M. K., Galan J. E. (1995). Functional analysis of the Salmonella typhimurium invasion genes invl and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol. Microbiol. 15 25–38 10.1111/j.1365-2958.1995.tb02218.x [DOI] [PubMed] [Google Scholar]
- Cornelis G. R, van Gijsegem F. (2000). Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54 735–774 10.1146/annurev.micro.54.1.735 [DOI] [PubMed] [Google Scholar]
- Crabill E., Karpisek A., Alfano J. R. (2012). The Pseudomonas syringae HrpJ protein controls the secretion of type III translocator proteins and has a virulence role inside plant cells. Mol. Microbiol. 85 225–238 10.1111/j.1365-2958.2012.08097.x [DOI] [PubMed] [Google Scholar]
- Cunnac S., Lindeberg M., Collmer A. (2009). Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12 53–60 10.1016/j.mib.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Edqvist P. J., Olsson J., Lavander M., Sundberg L., Forsberg A., Wolf-Watz H., et al. (2003). YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J. Bacteriol. 185 2259–2266 10.1128/JB.185.7.2259-2266.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M. F., Cornelis G. R. (2003). The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219 151–158 10.1016/S0378-1097(03)00042-9 [DOI] [PubMed] [Google Scholar]
- Feng F., Zhou J. M. (2012). Plant-bacterail pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15 469–476 10.1016/j.pbi.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Guo M., Alfano J. R. (2006). Pseudomonas syringae HrpJ is a type III secreted protein that is required for plant pathogenesis, injection of effectors, and secretion of the HrpZ1 Harpin. J. Bacteriol. 188 6060–6069 10.1128/JB.00718-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani A., Takaoka M., Sanada H., Noguchi Y., Oku T., Tsuno K., et al. (2009). Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 22 96–106 10.1094/MPMI-22-1-0096 [DOI] [PubMed] [Google Scholar]
- Gaudriault S., Malandrin L., Paulin J. P., Barny M. A. (1997). DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol. Microbiol. 26 1057–1069 10.1046/j.1365-2958.1997.6442015.x [DOI] [PubMed] [Google Scholar]
- Gaudriault S., Paulin J. P., Barny M. A. (2002). The DspB/F protein of Erwinia amylovora is a type III secretion chaperone ensuring efficient intrabacterial production of the Hrp-secreted DspA/E pathogenicity factor. Mol. Plant Pathol. 3 313–320 10.1046/j.1364-3703.2002.00124.x [DOI] [PubMed] [Google Scholar]
- Guo M., Chancey S. T., Tian F., Ge Z., Jamir Y., Alfano J. R. (2005). Pseudomonas syringae type III chaperones ShcO1, ShcS1, and ShcS2 facilitate translocation of their cognate effectors and can substitute for each other in the secretion of HopO1-1. J. Bacteriol. 187 4257–4269 10.1128/JB.187.12.4257-4269.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Cai L. L., Zou H. S., Ma W. X., Liu X. L., Zou L. F., et al. (2012). Ketoglutarate transport protein KgtP is secreted through the type III secretion system and contributes to virulence in Xanthomonas oryzae pv. oryzae. Appl. Environ. Microbiol. 78 5672–5681 10.1128/AEM.07997-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann N, Büttner D. (2013). The inner membrane protein HrcV from Xanthomonas is involved in substrate docking during type III secretion. Mol. Plant Microbe Interact. 26 1176–1189 10.1094/MPMI-01-13-0019-R [DOI] [PubMed] [Google Scholar]
- Hartmann N., Schulz S., Lorenz C., Fraas S., Hause G, Büttner D. (2012). Characterization of HrpB2 from Xanthomonas campestris pv. vesicatoria identifies protein regions that are essential for type III secretion pilus formation. Microbiology 158 1334–1349 10.1099/mic.0.057604-0 [DOI] [PubMed] [Google Scholar]
- He S. Y., Nomura K, Whittam T. S. (2004). Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694 181–206 10.1016/j.bbamcr.2004.03.011 [DOI] [PubMed] [Google Scholar]
- Hicks S. W, Galán J. E. (2013). Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat. Rev. Microbiol. 11 316–326 10.1038/nrmicro3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet E., Hahn K., Wengelnik K., Bonas U. (1998). hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol. 29 1379–1390 10.1046/j.1365-2958.1998.01019.x [DOI] [PubMed] [Google Scholar]
- Janjusevic R., Quezada C. M., Small J., Stebbins C. E. (2013). Structure of the HopA1(21-102)-ShcA chaperone-effector complex of Pseudomonas syringae reveals conservation of a virulence factor binding motif from animal to plant pathogens. J. Bacteriol. 195 658–664 10.1128/JB.01621-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444 323–329 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Journet L., Agrain C., Broz P., Cornelis G. R. (2003). The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302 1757–1760 10.1126/science.1091422 [DOI] [PubMed] [Google Scholar]
- Kabisch U., Landgraf A., Krause J., Bonas U., Boch J. (2005). Type III secretion chaperones ShcS1 and ShcO1 from Pseudomonas syringae pv. tomato DC3000 bind more than one effector. Microbiology 151 269–280 10.1099/mic.0.27491-0 [DOI] [PubMed] [Google Scholar]
- Kubori T, Galá J. E. (2003). Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115 333–342 10.1016/S0092-8674(03)00849-3 [DOI] [PubMed] [Google Scholar]
- Kubori T., Nagai H. (2011). Bacterial effector-involved temporal and spatial regulation by hijack of the host ubiquitin pathway. Front. Microbiol. 2:145 10.3389/fmicb.2011.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T., Sukhan A., Aizawa S. I., Galan J. E. (2000). Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U.S.A. 97 10225–10230 10.1073/pnas.170128997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillington J. E., Lovett J. E., Johnson S., Roversi P., Timmel C. R., Lea S. M. (2011). Shigella flexneri Spa15 crystal structure verified in solution by double electron electron resonance. J. Mol. Biol. 405 427–435 10.1016/j.jmb.2010.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Solanilla E., Bronstein P. A., Schneider A. R., Collmer A. (2004). HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 54 353–365 10.1111/j.1365-2958.2004.04285.x [DOI] [PubMed] [Google Scholar]
- Lorenz C, Büttner D. (2009). Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 191 1414–1428 10.1128/JB.01446-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Büttner D. (2011). Secretion of early and late substrates of the type III secretion system from Xanthomonas is controlled by HpaC and the C-terminal domain of HrcU. Mol. Microbiol. 79 447–467 10.1111/j.1365-2958.2010.07461.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C., Kirchner O., Egler M., Stuttmann J., Bonas U, Büttner D. (2008a). HpaA from Xanthomonas is a regulator of type III secretion. Mol. Microbiol. 69 344–360 10.1111/j.1365-2958.2008.06280.x [DOI] [PubMed] [Google Scholar]
- Lorenz C., Schulz S., Wolsch T., Rossier O., Bonas U, Büttner D. (2008b). HpaC controls substrate specificity of the Xanthomonas type III secretion system. PLoS Pathog. 4:e1000094 10.1371/journal.ppat.1000094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada L. C., Hutcheson S. W. (2005). Type III secretion chaperones of Pseudomonas syringae protect effectors from Lon-associated degradation. Mol. Microbiol. 55 941–953 10.1111/j.1365-2958.2004.04438.x [DOI] [PubMed] [Google Scholar]
- Magdalena J., Hachani A., Chamekh M., Jouihri N., Gounon P., Blocker A., et al. (2002). Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J. Bacteriol. 184 3433–3441 10.1128/JB.184.13.3433-3441.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits T. C., Kubori T., Lara-Tejero M., Thomas D., Unger V. M., Galan J. E. (2006). Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441 637–640 10.1038/nature04822 [DOI] [PubMed] [Google Scholar]
- Mills E., Baruch K., Aviv G., Nitzan M., Rosenshine I. (2013). Dynamics of the type III secretion system activity of enteropathogenic Escherichia coli. MBio 4 4 10.1128/mBio.00303-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello J. E., Collmer A. (2009). Pseudomonas syringae HrpP Is a type III secretion substrate specificity switch domain protein that is translocated into plant cells but functions atypically for a substrate-switching protein. J. Bacteriol. 191 3120–3131 10.1128/JB.01623-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaihara T., Tamura N., Iwabuchi M. (2010). Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 23 251–262 10.1094/MPMI-23-3-0251 [DOI] [PubMed] [Google Scholar]
- Mukaihara T., Tamura N., Murata Y., Iwabuchi M. (2004). Genetic screening of Hrp type III-related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum. Mol. Microbiol. 54 863–875 10.1111/j.1365-2958.2004.04328.x [DOI] [PubMed] [Google Scholar]
- Nissinen R. M., Ytterberg A. J., Bogdanove A. J., van Wijk K. J., Beer S. V. (2007). Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol. Plant Pathol. 8 55–67 10.1111/j.1364-3703.2006.00370.x [DOI] [PubMed] [Google Scholar]
- Oh C. S., Carpenter S. C., Hayes M. L., Beer S. V. (2010). Secretion and translocation signals and DspB/F-binding domains in the type III effector DspA/E of Erwinia amylovora. Microbiology 156 1211–1220 10.1099/mic.0.027144-0 [DOI] [PubMed] [Google Scholar]
- Ortiz-Martin I., Thwaites R., Mansfield J. W., Beuzon C. R. (2010). Negative regulation of the Hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 23 682–701 10.1094/MPMI-23-5-0682 [DOI] [PubMed] [Google Scholar]
- Parsot C., Hamiaux C., Page A. L. (2003). The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6 7–14 10.1016/S1369-5274(02)00002-4 [DOI] [PubMed] [Google Scholar]
- Poueymiro M., Genin S. (2009). Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Curr. Opin. Microbiol. 12 44–52 10.1016/j.mib.2008.11.008 [DOI] [PubMed] [Google Scholar]
- Preston G., Deng W. L., Huang H. C., Collmer A. (1998). Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J. Bacteriol. 180 4532–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. R., Morello J. E., Ravindran S., Deng W. L., Huang H. C., Collmer A. (2007). Identification of Pseudomonas syringae pv. syringae 61 type III secretion system Hrp proteins that can travel the type III pathway and contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 189 5773–5778 10.1128/JB.00435-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier O., Van den Ackerveken G., Bonas U. (2000). HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38 828–838 10.1046/j.1365-2958.2000.02173.x [DOI] [PubMed] [Google Scholar]
- Schulz S, Büttner D. (2011). Functional characterization of the type III secretion substrate specificity switch protein HpaC from Xanthomonas campestris pv. vesicatoria. Infect. Immun. 79 2998–3011 10.1128/IAI.00180-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., Oh H. S., Chen J., Guo M., Zhou J., Alfano J. R., et al. (2004). The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant Microbe Interact. 17 447–455 10.1094/MPMI.2004.17.5.447 [DOI] [PubMed] [Google Scholar]
- Tampakaki A. P., Skandalis N., Gazi A. D., Bastaki M. N., Sarris P. F., Charova S. N., et al. (2010). Playing the “Harp”: evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 48 347–370 10.1146/annurev-phyto-073009-114407 [DOI] [PubMed] [Google Scholar]
- Triplett L. R., Melotto M., Sundin G. W. (2009). Functional analysis of the N terminus of the Erwinia amylovora secreted effector DspA/E reveals features required for secretion, translocation, and binding to the chaperone DspB/F. Mol. Plant Microbe Interact. 22 1282–1292 10.1094/MPMI-22-10-1282 [DOI] [PubMed] [Google Scholar]
- Triplett L. R., Wedemeyer W. J., Sundin G. W. (2010). Homology-based modeling of the Erwinia amylovora type III secretion chaperone DspF used to identify amino acids required for virulence and interaction with the effector DspE. Res. Microbiol. 161 613–618 10.1016/j.resmic.2010.05.015 [DOI] [PubMed] [Google Scholar]
- van Dijk K., Tam V. C., Records A. R., Petnicki-Ocwieja T., Alfano J. R. (2002). The ShcA protein is a molecular chaperone that assists in the secretion of the HopPsyA effector from the type III (Hrp) protein secretion system of Pseudomonas syringae. Mol. Microbiol. 44 1469–1481 10.1046/j.1365-2958.2002.02979.x [DOI] [PubMed] [Google Scholar]
- van Gijsegem F., Vasse J., De Rycke R., Castello P., Boucher C. (2002). Genetic dissection of Ralstonia solanacearum hrp gene cluster reveals that the HrpV and HrpX proteins are required for Hrp pilus assembly. Mol. Microbiol. 44 935–946 10.1046/j.1365-2958.2002.02936.x [DOI] [PubMed] [Google Scholar]
- Vujanac M., Stebbins C. E. (2013). Context-dependent protein folding of a virulence peptide in the bacterial and host environments: structure of an SycH-YopH chaperone-effector complex. Acta Crystallogr. D Biol. Crystallogr. 69 546–554 10.1107/S0907444912051086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E., Ojanen-Reuhs T., Huguet E., Hause G., Romantschuk M., Korhonen T. K., et al. (2005). The type III-dependent Hrp pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J. Bacteriol. 187 2458–2468 10.1128/JB.187.7.2458-2468.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehling M. D., Guo M., Fu Z. Q., Alfano J. R. (2004). The Pseudomonas syringae HopPtoV protein is secreted in culture and translocated into plant cells via the type III protein secretion system in a manner dependent on the ShcV type III chaperone. J. Bacteriol. 186 3621–3630 10.1128/JB.186.11.3621-3630.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. F., Deng W. L., Huang H. C. (2005). A chaperone-like HrpG protein acts as a suppressor of HrpV in regulation of the Pseudomonas syringae pv. syringae type III secretion system. Mol. Microbiol. 57 520–536 10.1111/j.1365-2958.2005.04704.x [DOI] [PubMed] [Google Scholar]
- Wei H. L., Collmer A. (2012). Multiple lessons from the multiple functions of a regulator of type III secretion system assembly in the plant pathogen Pseudomonas syringae. Mol. Microbiol. 85 195–200 10.1111/j.1365-2958.2012.08119.x [DOI] [PubMed] [Google Scholar]
- Zou L. F., Wang X. P., Xiang Y., Zhang B., Li Y. R., Xiao Y. L., et al. (2006). Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72 6212–6224 10.1128/AEM.00511-06 [DOI] [PMC free article] [PubMed] [Google Scholar]