SUMMARY
A regulatory network comprised of “core” (Oct4, Sox2, Nanog) and other transcription factors maintains embryonic stem (ES) cells in a self-renewing and pluripotent state. To develop an expanded framework with which to understand how these properties of ES cells are controlled, we have employed a modification of ChIP-Chip approaches, termed bioChIP-Chip, to identify target promoters of nine factors, including somatic cell reprogramming factors (Oct4, Sox2, Klf4, c-Myc) and others (Nanog, Dax1, Rex1, Zpf281, and Nac1), on a global scale in mouse ES (mES) cells. Targets fall into two classes, correlating with the extent of factor occupancy. Targets bound by one or few factors tend to be inactive, or repressed, in ES cells. Remarkably, numerous genes bound by multiple (>4) factors, encoding several proteins within a protein interaction network associated with pluripotency, are largely active and then repressed on differentiation. In addition, we propose a transcriptional hierarchy for reprogramming factors in which Klf4 lies upstream of feed-forward circuits involving Oct4 and Sox2, and also broadly distinguish targets of c-Myc versus other factors. Our data provide a resource for further exploration of the complex network maintaining pluripotency in ES cells.
INTRODUCTION
Pluripotency, the capacity to generate all cell types, is a defining property of embryonic stem (ES) cells, cultured cells derived from the inner cell mass of the mammalian blastocyst (Evans and Kaufman, 1981; Martin, 1981). In addition, ES cells can be maintained in a proliferative state for prolonged periods, the phenomenon of self-renewal. Pluripotency may be imposed on somatic cells following their fusion with ES cells (Cowan et al., 2005), and at least one transcription factor specifically expressed in ES cells, the homeodomain protein Nanog, facilitates fusion-induced pluripotency (Silva et al., 2006). Moreover, forced expression of other transcriptional factors (Oct4, Sox2, Klf4, and c-Myc) reprograms mouse fibroblasts to ES-like cells (called iPS cells)(Takahashi and Yamanaka, 2006), the quality of which is further enhanced upon selection of cells that express endogenous Oct4 or Nanog (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007b). Recently, it has been shown that the same factors reprogram human fibroblasts to a pluripotent state (Park et al., 2007; Takahashi et al., 2007; Yu et al., 2007) How pluripotency is established and maintained in ES cells is of great interest, as an improved understanding of the transcription factors and epigenetic modifications operating in a regulatory network will facilitate both directed programming of ES cells to specific lineages and the reprogramming of somatic cells to an ES-like state.
Until recently, attention has focused almost exclusively on a small set of transcription factors, Oct4, Sox2, and Nanog as “core” pluripotency factors for human or mouse ES cells (Orkin, 2005). Oct4 has long been recognized to be essential in vivo and in vitro for early development and maintenance of ES cell pluripotency (Nichols et al., 1998). Indeed, the dosage of Oct4 is crucial: reduced expression permit trophoectoderm development, whereas enhanced expression drives primitive endoderm differentiation (Niwa et al., 2000). Sox2 is generally considered a transcriptional partner of Oct4 (Avilion et al., 2003). Rather than directly interacting with Oct4 protein, Sox2 assembles on target regulatory elements with Oct4 to collaborate in transcriptional control. Nanog promotes ES cell self-renewal and alleviates the requirement for Leukemia Inhibitory Factor (LIF) in tissue culture (Chambers et al., 2003; Mitsui et al., 2003). While considerable evidence speaks to the importance of these factors in maintaining the properties of ES cells, evidence also points to the involvement of additional transcription factors in the control of pluripotency (Ivanova et al., 2006; Wang et al., 2006; Zhou et al., 2007).
To account for the unique properties of pluripotent ES or iPS cells at the molecular level, it will be necessary to understand the transcriptional networks responsible for maintaining pluripotency. Studies to this end have entailed the search for additional transcription factors and delineation of a protein-protein interaction network highly enriched for factors involved in the control of pluripotency (Ivanova et al., 2006; Wang et al., 2006), and preliminary global target gene assessment for the initial “core” factors (Boyer et al., 2005; Loh et al., 2006). Starting with the identification of protein partners of Nanog through protein complexes purification and microsequencing, coupled with iterative affinity purification of interacting proteins, we generated a network that includes additional factors required for maintenance of pluripotency (Wang et al., 2006). Among this latter class, we encountered Sall4 (Sakaki-Yumoto et al., 2006; Wu et al., 2006; Zhang et al., 2006), Dax1 (Niakan et al., 2006), and Rif1 (Loh et al., 2006), factors identified independently by others as involved in maintenance of ES cell pluripotency. The protein network is connected to complexes, such as NuRD remodeling complex and PRC1, implicated in transcriptional repression (Wang et al., 2006). In parallel, other groups have used new methods for global target mapping (ChIP-Chip and ChIP-PET) to predicted target genes regulated by Oct4, Sox2, and Nanog in mouse and human ES cells. These studies revealed combinatorial occupancy of target gene promoters by these “core” factors and both autoregulatory and feed-forward transcriptional circuits (Boyer et al., 2005; Loh et al., 2006). The discovery of the original 4-factor reprogramming set (Takahashi and Yamanaka, 2006) brought Klf4 and c-Myc to the forefront as additional proteins to be integrated into the network inducing and/or maintaining pluripotency. In parallel, other work has suggested that histone modification signatures, specifically histone 3 lysine 4 and histone 3 lysine 27 trimethylation (H3K4me3 and H3K27me3, respectively), are important in controlling gene regulation in ES cells (Bernstein et al., 2006; Boyer et al., 2006; Lee et al., 2006; Pan et al., 2007; Zhao et al., 2007).
Genome-wide mapping of transcription factor targets by ChIP, combined with microarrays or sequencing methods, is a powerful tool for laying a foundation for understanding transcriptional networks (Iyer et al., 2001; Kim et al., 2005; Loh et al., 2006; Ren et al., 2000; Roh et al., 2004). Expanding the number of transcription factors analyzed by ChIP-based methods should be especially informative in dissecting system level biological processes, as ~10% of annotated mammalian genes are predicted to encode DNA-binding proteins (Messina et al., 2004). A practical limitation to current ChIP approaches is the availability of suitable “ChIP quality” antibodies.
Here, we report the application of in vivo biotinylation mediated ChIP (bioChIP) to global target mapping (bioChIP-Chip) of an expanded set of factors associated with pluripotency of mES cells. This approach, which relies on streptavidin affinity capture of tagged proteins, is comparable to conventional ChIP-Chip but circumvents issues related to antibody availability. Using bioChIP-Chip, we have identified target promoters of nine transcription factors, including the somatic cell reprogramming factors, on a global scale in mES cells. We have constructed an expanded transcriptional regulatory network containing the previously known three core factors, as well as additional factors. Our data argue that differential regulation of target genes correlates with the extent of promoter occupancy by multiple factors. Moreover, we propose a transcriptional hierarchy for the somatic reprogramming factors and broadly distinguish targets of c-Myc versus the other factors. Our data provides a resource with which to probe mechanisms of pluripotency control and differentiation by the complex transcriptional regulatory network in ES or iPS cells.
RESULTS
Global mapping of target genes by biotin-mediated chromatin immunoprecipitation (bioChIP)
Prior genome-wide chromatin immunoprecipitation (ChIP) analyses have been performed in mouse and human ES cells with “core” factors (Oct4, Sox2, and Nanog) involved in maintaining pluripotency. These studies relied on antibodies suitable for ChIP and were carried out with different platforms (Boyer et al., 2005; Loh et al., 2006). Our first goal was to define the targets of a larger set of pluripotency factors with greater consistency in the experimental platform. To this end, we assessed the suitability of streptavidin affinity-capture of in vivo biotin-tagging of proteins (de Boer et al., 2003; Sanchez et al., 2007; Wang et al., 2006) as an alternative to antibody-based ChIP.
Earlier studies proposed application of the biotin-tagging method for ChIP assay (de Boer et al., 2003; van Werven and Timmers, 2006), but did not explore its utility in combination with microarrays (bioChIP-Chip) in complex mammalian systems (Figure 1A and Experimental Procedures). As a proof of concept, we performed bioChIP and conventional ChIP reactions in mES cells for both Nanog and c-Myc (Myc). bioChIP and ChIP samples were hybridized onto Affymetrix mouse promoter arrays with appropriate references to map the target loci of each factor. We compared targets predicted by the two methods for both factors. As shown in Figure 1C and 1D, the majority of targets predicted for each factor by the two methods were shared. 67% and 81% of bioChIP targets of Nanog and Myc, respectively, were identified with the conventional ChIP approach. The overall shapes of binding peaks of Nanog and Myc across the genome were also nearly identical for the two methods (Figure S1). Furthermore, the correlation of target loci from the two different methods was 0.896 for Nanog (Figure S3, see Experimental Procedure), suggesting that bioChIP is comparable to conventional antibody ChIP.
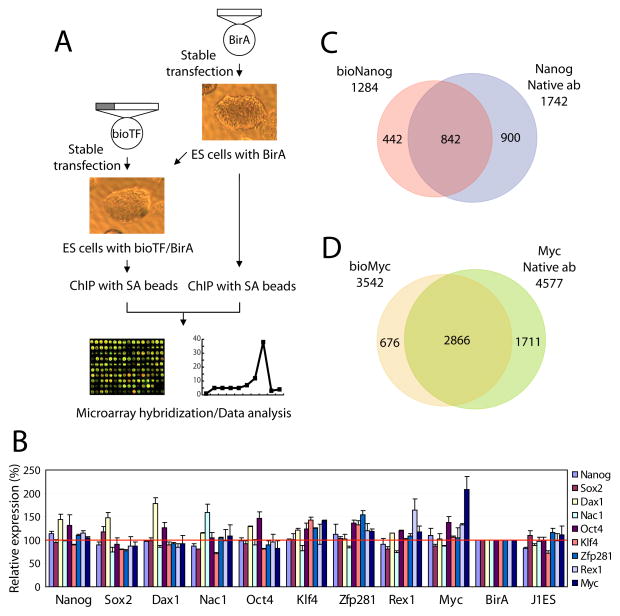
Figure 1. Strategy of in vivo biotinylation-mediated chromatin immunoprecipitation and microarray (bioChIP-chip).
(A) Schematic representation of biotin-mediated chromatin immunoprecipitation. The gray bar represents BirA target sequence (MSGLNDIFEAQKIEWHEGAPSSR).
(B) Expression analysis of nine genes using cell lines expressing biotin-tagged proteins. Biotin-tagged cell lines are indicated on the horizontal axis and transcript levels are presented as color bars.
(C)–(D) Overlap of target promoters between bioChIP-chip and conventional ChIP-chip experiments for Nanog (C) and Myc (D). Predicted overlap might be underestimated due to a fixed statistical threshold (see also Figure 2, and Figure S3).
Interestingly, on comparison with previously published mES ChIP-PET data (Loh et al., 2006), we observed only limited overlap of common promoter targets (48% of ChIP-PET targets were also defined as bioNanog targets). Given that ChIP-PET predicted only 434 promoter targets (337 comparable RefSeq promoters) for Nanog in contrast to our ChIP data (1284 bioChIP targets and 1742 conventional ChIP targets, Figure 1C), we asked whether the lower number reflects partial coverage, perhaps due to the depth of sequencing and/or tiling array repeat masking. We tested conventional antibody ChIP material on one of the seven Affymetrix whole genome mouse arrays (Mouse Tiling 2.0R F Array) that covers ~15% of the entire genome and ~14% of well annotated genes and found 223 promoter targets ( >= 50% number of ChIP-PET promoter targets) with a similar proportion of non-promoter targets as predicted by ChIP-PET (12.8%). This implies that the ChIP-PET data set may lack targets due to inadequate depth of sequencing (Euskirchen et al., 2007). In addition, we identified 137 genes in common in our bioChIP, and published human Nanog ChIP data that are not represented in ChIP-PET (Table S4)(Boyer et al., 2005). Although differing platforms and/or depth of sequencing in ChIP-PET may account for partial overlap, the degree of overlap we have observed is greater than observed in other studies (Loh et al., 2006; Zeller et al., 2006).
We proceeded to determine the global target promoters for nine transcription factors in ES cells, including previously examined “core” factors (Nanog, Oct4, and Sox2)(Boyer et al., 2005; Loh et al., 2006), somatic cell reprogramming factors (Klf4 and Myc, in addition to Oct4 and Sox2) (Maherali et al., 2007; Okita et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007b), and protein-interacting partners of Nanog and Oct4 (Dax1, Nac1, Zfp281 and Rex1) (Wang et al., 2006). In each instance, we employed mES cells expressing a sub-endogenous level of the respective biotin-tagged factor, so as to avoid perturbing the existing network (Figure S2). The levels of which exogeneous proteins were expressed fail to elicit subsequent change in the transcript levels of the nine factors (Figure 1B) (Wang et al., 2006).
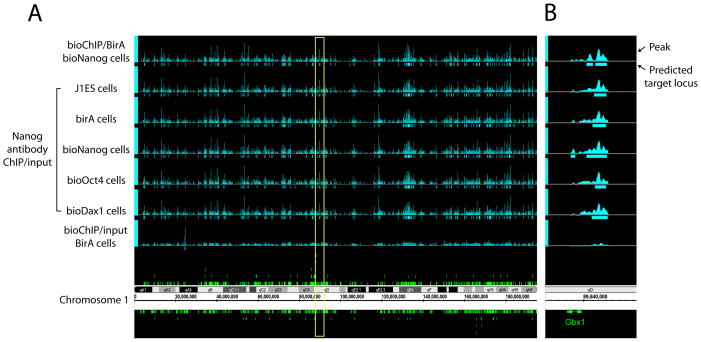
Before analyzing the targets of the various factors, we performed additional validation experiments to assess whether low expression of a biotin-tagged protein might perturb chromatin occupancy by untagged proteins. In these experiments we performed conventional Nanog antibody ChIP reactions using wild-type mES cells, mES cells expressing BirA alone, and mES cells expressing BirA plus tagged versions of Dax1, Oct4, and Nanog. Figure 2 shows that the overall patterns of Nanog binding peaks among these different cell lines are virtually indistinguishable. Target correlations across the cell lines were also very strong (most were > 0.960 and the correlation between bioChIP data and antibody ChIP data across multiple cells is > 0.880, Figure S3). These data exclude significant effects of sub-endogenous levels of expressed protein on factor occupancy. To exclude artifacts due to biotinylation of endogeneous proteins by expressed BirA, we performed ChIP-chip using BirA expressing cells with input genomic DNA as a reference (Figure 2). We observed only 18 specific peaks among all promoter regions. Thus, non-specific effects due to biotinlyation of endogenous DNA-binding proteins are insignificant compared with the number of targets for each factor (>500).
Figure 2. Chromosomal view of Nanog occupancy detected by bioChIP-chip and conventional ChIP-chip.
(A) Comparison of Nanog binding patterns using multiple cell lines is displayed using Affymetrix Integrated Genome Browser. In addition to bioNanog ChIP (top), antibody ChIP-chip data from control cell lines (J1 ES and BirA expressing cells) and cells expressing ectopic biotin-tagged protein (bioDax1 and bioOct4 cells) are tested. Non-specific biotinylation by BirA enzyme was also tested (bottom). Yellow box indicates the chromosomal loci harboring Gbx1.
(B) Representative view of Nanog occupancy to its target Gbx1 upstream promoter.
Taken together, our findings demonstrate that the bioChIP-chip method is a valid alternative to the conventional ChIP-chip method. A distinct advantage of the approach is that the inherent variability in the quality of antibodies available for different proteins is avoided. A single experimental platform can then be applied to all factors of interest.
Promoter occupancy of nine transcription factors in mES cells
In addition to the nine transcription factors noted above, we mapped two histone modifications, H3K4me3 and H3K27me3, by antibody ChIP-chip. Information from approximately 8kb upstream and 2kb downstream of 19,253 well-characterized transcription start site (TSS) of RefSeq genes from UCSC genome browser was used for analysis (see Experimental Procedure) (Kuhn et al., 2007).
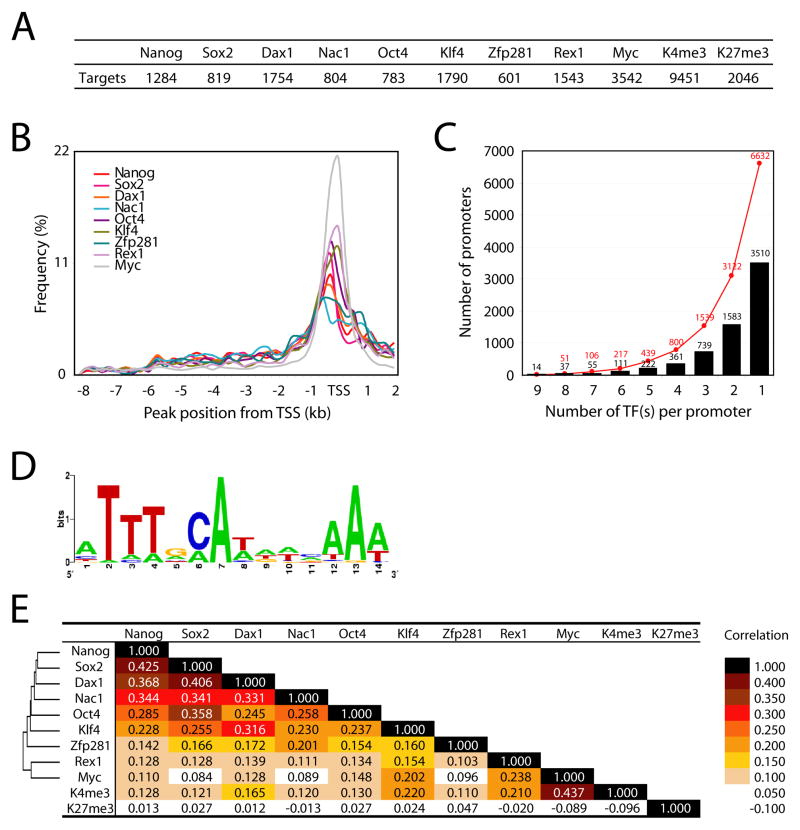
The number of target promoters occupied by each factor is shown in Figure 3A. A compilation of target genes for each factor and binding peak positions are provided in Table S1 and Table S2, respectively. As might be expected, the number of targets occupied by the different factors varies greatly (Figure 3A). Notably, Myc occupies many more target promoters (18% of all promoters) than the other factors we tested. This result is in accordance with prior observations in other cell types (Fernandez et al., 2003; Li et al., 2003; Mao et al., 2003). We also found that approximately 50% and 10% of promoters bear H3K4me3 and H3K27me3 marks, respectively (discussed below). We observed that the vast majority of binding sites for each factor were in close proximity to the TSS (Figure 3B), and more than a third of mouse promoters were occupied by at least one of the 9 transcription factors we tested (6632 promoters, Figure 3C).
Figure 3. Summary of nine transcription factor occupancy and histone modification status.
(A) Number of target promoters bound by each factor or associated with H3K4 or H3K27 trimethylation.
(B) Relative position of chromosomal target loci of each factor to the TSS.
(C) Number of common targets of multiple factors. Y-axis represents the number of target promoters occupied by transcription factor(s). Red dots represent the accumulated number of target promoters.
(D) Predicted consensus binding motif of multiple factor target loci using MEME.
(E) Correlation between each factor targets and hierarchical cluster of nine factors based on their target similarity.
A previous study of human ES (hES) cells showed that Nanog, Oct4, and Sox2 share many targets (353 genes) (Boyer et al., 2005). Surprisingly, our bioChIP-chip data reveal that many more promoters are co-occupied by multiple factors. Specifically, we have observed that >100 promoters are occupied by at least 7 factors, and ~800 promoters are occupied by at least 4 of the 9 factors examined (Figure 3C). More interestingly, actual binding loci of multiple factors within the target promoters are virtually coincident, suggesting that factors work as protein complexes or within compact cis-regulatory elements when multiple factors occupy the same target locus (Figure S4). We also observed numerous target loci occupied by fewer factors (Figure S4). To define a consensus motif that might be utilized by the multiple factors, we tested ±100bp genomic sequence information from the center position of predicted common target loci using MEME (Bailey et al., 2006). Interestingly the consensus motif (ATTTGCAT) predicted from MEME (e-value 1.4E-50, Figure 3D) was very similar to sequences previously predicted by different algorithms as Oct4 or Sox2-Oct4 target sequences (Loh et al., 2006; Macisaac et al., 2006).
We also validated predicted target loci by quantitative ChIP-PCR using primer pairs specific to the predicted target loci that are occupied by either multiple, or fewer, factors with various MAT p-values (see Experimental Procedure and Table S3) to confirm that our target cutoff was appropriate to minimize false positives. As shown in Figure S5, most of target loci we tested for each factor show substantial enrichment over the BirA control. With these results and additional quantitative PCR confirmation, we estimate an average false positive rate of < ~5%. These estimates are comparable to those in any previous studies in which antibodies were employed (Boyer et al., 2006; Lee et al., 2006). While our global dataset may miss some authentic targets due to our cutoff criteria, few irrelevant loci are likely to be present.
Interestingly, among the 6632 targets bound in aggregate by the 9 factors, 50% are occupied by only one of the nine factors (Figure 3C). Clustering of the 9 transcription factors was performed based on their target correlations and is summarized in Figure 3E. Of the 9 tested factors, Nanog, Sox2, Dax1, Nac1, Oct4, Klf4, and Zfp281 exhibit overall similarity in their targets. In contrast, targets of Myc and Rex1 segregate to a distinct cluster (Figure 3E, cluster). Functional classification of the presumptive targets of each factor using the PANTHER classification tool also demonstrates separation of factors in two classes (Mi et al., 2007). In general, target genes of each of the tested factors are enriched in genes involved in nucleic acid metabolism and transcriptional control. Interestingly, targets of Myc or Rex1 are implicated in protein metabolism, rather than in developmental processes, whereas targets of the other factors are enriched in genes for developmental processes (Figure S6).
Histone modification signatures
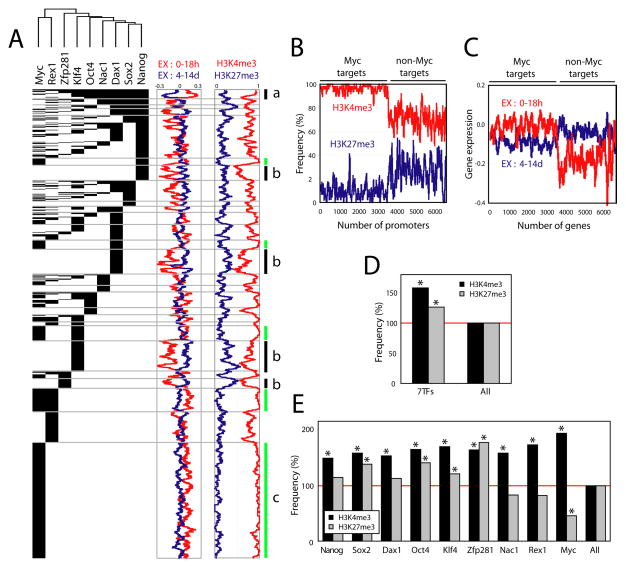
Core pluripotency factors are thought to be involved in both gene activation and repression in ES cells (Boyer et al., 2005; Loh et al., 2006). However, mechanisms that account for this differential regulation are not understood. To address this question, we performed supervised clustering (Figure 4A, see Experimental Procedure) to reveal the relationship, if any, between targets of various combinations of transcription factors and corresponding H3K4me3 and H3K27me3 marks, as well as gene expression profiles.
Figure 4. H3K4me3 and H3K27me3 status and factor occupancy of the promoters.
(A) A supervised cluster image showing 6632 target promoters occupied by different factor combinations (see Experimental Procedure). Corresponding H3K4me3 (red) and H3K27me3 (blue) histone marks (presence: 1; absence: 0) as well as gene expression profiles (log2) upon J1 ES cell differentiation (0–18h: red, 4–14d: blue, see Experimental Procedure) are shown as moving window averaged lines (bin size 50 and step size 1). Bar ‘a’ represents the promoters occupied by multiple factors including at least Nanog, Sox2, Dax1, Nac1 and Oct4 (left panel) and corresponding gene expression changes upon differentiation (middle panel) as well as their histone marks (right panel). Bars ‘b’ represent the clusters of promoters occupied by a single factor Nanog, Dax1, Klf4, and Zfp281 respectively (see also Figure 5E and 5H). Green lines (bars c) represent Myc target promoters with corresponding gene expression profiles and histone mark status.
(B) H3K4me3 (red line) and H3K27me3 (blue line) status for Myc target promoters.
(C) Expression profiles of Myc target genes at different time points upon differentiation (0–18h: red, 4–14d: blue). Total 6632 target genes of any of 9 factors are shown, and moving window average (bin size 50 and step size 1) was applied (B–C).
(D) Factor target promoters are both H3K4me3 and H3K27me3 rich over all promoters. ‘7TFs’ represent the targets of any of 7 factors (Nanog, Sox2, Dax1, Nac1, Oct4, Klf4, Zfp281), and ‘All’ represents all promoters. Asterisk indicates hypergeometiric probability < 0.0001.
(E) Histone marks on the target promoters of each factor. Asterisk indicates p-value < 0.0001.
Surprisingly, the H3K4me3 and H4K27me3 marks of Myc target promoters exhibit a unique distribution in comparison with promoters bound by the other factors (Figure 4A, green bars c). Specifically, 96% and 5% of Myc target promoters bear H3K4me3 and H3K27me3, respectively (Figure 4B). A previous study based on quantitative PCR suggested a relationship between Myc occupancy and various histone marks (Guccione et al., 2006). Our data confirm this observation on a genome-wide scale, and establish the correlation within ES cells for the first time. As anticipated by these histone marks, Myc target genes are more frequently expressed as compared with targets of other factors in ES cells (Figure 4C). Our findings provide evidence that Myc occupancy is associated with large-scale, global alteration of chromatin at Myc targets, and that such effects are qualitatively different from those associated with the core pluripotency factors.
To pursue these correlations further, we examined the relationship of the target promoters of 7 factors (excluding Rex1) with the H3K4me3 and H3K27me3 marks. We removed Myc and Rex1 from this analysis, because the predicted targets of these factors reveal functional segregation (discussed above). Interestingly, the predicted target promoters are enriched overall in both H3K4me3 and H3K27me3 marks as compared with all promoters (58% and 26% respectively, Figure 4D). However, closer examination of the correlation of targets of individual factors with these histone marks reveals three different classes (Figure 4E). Target promoters of Nanog, Sox2, Dax1, Oct4, and Klf4 bear enriched marks for both H3K4me3 and H3K27me3. However, predicted targets of Zfp281 show considerable enrichment for the repressive H3K27me3 mark, consistent with a role of Zfp281 in gene repression. Similar to target promoters of Myc, Rex1 and Nac1 targets show less H3K27me3 marks, indicating possible roles of these factors in gene activation. Interestingly, physical interaction between Rex1, Nac1 and Baf155 (one of the Swi/Snf complex proteins) was observed in mES cells (Wang et al., 2006).
Recent data obtained with hES cells showed that most H3K27me3 marks overlap H3K4me3 marks (Pan et al., 2007; Zhao et al., 2007). In our data we have observed somewhat less extensive overlap as 35% of H3K27me3 marks (725 among 2046) overlap with H3K4me3 marks. The apparent quantitative difference relates to the threshold level used in assigning target genes. As we reduce the stringency of target selection, we observe a 39% increase in genes with H3K27me3 marks (2046 to 2843). Bivalent signatures increased 138% (725 to 1729) and 61% of H3K27me3 marks then lie within H3K4me3 marks on the promoters. Our results are in accord with the prior observation that H3K27me3 signals are in general weaker than H3K4me3 signals (Pan et al., 2007; Zhao et al., 2007).
Interestingly, previously identified clusters of gene promoters devoid of H3K4me3 and H3K27me3 marks on their promoters in hES cells (Guenther et al., 2007; Zhao et al., 2007) are also not bound by any of nine factors we tested (Figure S7). Presumably, the mechanism of repression is unique, as H3K27 methylation is one of the principal histone modification marks associated with gene repression.
Regulation of target gene expression by transcription factor occupancy
A striking observation emerges from supervised clustering analysis in considering potential mechanisms that might account for the differential regulation of transcription factor targets in ES cells. The genes whose promoters are occupied targets by multiple factors, including Nanog, Sox2, Dax1, Nac1, Oct4, and Klf4, are generally active in ES cells, and dramatically repressed upon cellular differentiation (Figure 4A, bar a and Figure S8). On the other hand, the clusters of genes that are inactive or repressed in ES cells, but are expressed upon differentiation, are comprised largely of those gene promoters bound by a single factor (for example, Nanog, Dax1, Klf4, or Zfp281) as shown in Figure 4A, bars b. This observation suggests that the roles of the pluripotency factors are highly sensitive to their immediate context. A single factor may bind to targets that are “poised” and inactive, or may act to repress its targets, presumably in association with corepression complexes, whereas it may participate in gene activation when bound to a promoter region in concert with several other pluripotency regulators. Prior protein network analysis revealed multiple connections between several core factors and repressive chromatin remodeling complexes, including NuRD and Polycomb (PRC1), in mES cells (Wang et al., 2006).
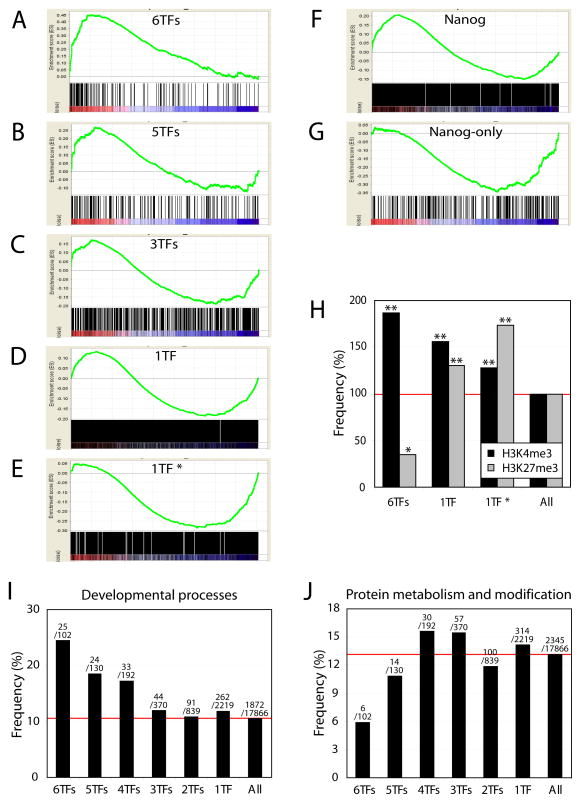
To pursue this observation further, we classified target genes based on the co-occupancy of transcription factors on their promoters. Since Myc and Rex1 have distinct sets of targets (Figure 3E and Figure 4A), we focused on 6 others factors for further analysis (Nanog, Sox2, Dax1, Nac1, Oct4 and Klf4; Zfp281 was excluded due to less target gene overlap). We observed significant differences between common targets of all 6 factors and targets of single factors in their gene expression profiles, as revealed by gene set enrichment analysis (GSEA) (Subramanian et al., 2005) (Figure 5A–5E). The majority of common targets of 6 factors are highly active (Figure 5A). In the case of targets bound by fewer factors, both active and repressed genes are nearly balanced (Figure 5C and Figure S8). Surprisingly, targets occupied by any single factor were predominantly inactive or repressed in ES cells (Figure 5D) and this is even more apparent with the targets of Nanog, Dax1, Klf4 and Zfp281 as shown in Figure 4A, bars b (Figure 5E, 1TF*). The relationship between target promoter occupancy and gene expression level is in excellent accordance with the observed histone marks. The common target promoters of 6 factors show an 80% increase of the H3K4me3 signature and a 60% decrease of H3K27me3 signature, as compared to all promoters. On the other hand, unique targets of only one factor exhibited an increased level of the H3K27me3 signature (Figure 5H, 1TF and 1TF*).
Figure 5. Target gene expression and transcription factor occupancy on their promoters.
(A)–(E) GSEA analyses showing the relationship between target gene expression and factor occupancy. Target promoters were classified based on the number of co-occupying factors and corresponding gene expression upon differentiation was tested. Common targets of 6 factors (A) are enriched in active genes in ES cells, whereas single factor only targets are more repressed (D). ‘1TF*’ represents a subset of ‘1TF’ which includes promoters solely occupied by either Nanog, Dax1, Klf4 or Zfp281 as described in Figure 4A, bars b (E).
(F)–(G) Nanog targets are both active and repressed in ES cells (F), however targets only occupied by Nanog are repressed (G).
(H) Common target promoters of 6 factors (Nanog, Sox2, Dax1, Nac1, Oct4 and Klf4) are enriched for H3K4me3 marks and reduced for H3K27me3 marks. Promoters occupied by only one factor show an increase in H3K27me3 marks (1TF and 1TF*). Double asterisk indicates p-value < 0.0001, and single asterisk indicates p-value = 0.006.
(I)–(J) Genes of multiple factor targets (at least 4TFs) are enriched in developmental processes.
The above findings argue that pluripotency factors act in a highly combinatorial fashion to activate or maintain expression of a subset of target genes, while they are inactive or function more often to repress genes when acting alone, or with only one or few other factors. Distinguishing the “on”-”off” state of targets based on the extent of promoter occupancy may provide a mechanism by which a relatively small set of factors controls two complementary aspects of transcription required for maintenance of pluripotency. While pluripotency factors hold differentiation-promoting genes in check, they must also function together to drive expression of genes encoding proteins required for self-renewal.
In accord with this interpretation, the predicted targets of multiple factors and single factors differ in gene ontology (GO) categories (Figure 5I and 5J). The 6 factor common target genes are implicated more frequently in developmental processes than targets occupied by fewer factors. The enrichment for genes involved in developmental processes is correlated positively with the number of bound factors from 3–6 (Figure 5I).
Furthermore, we tested roles of each factor in their target gene regulation to determine if any single factor is more associated with either gene activation or repression. Surprisingly, except for Myc and Rex1, all the remaining factors occupy promoters of both non-expressed and expressed genes (Figure S9). An example is shown in Figure 5F. Among all Nanog target promoters, genes are roughly equally expressed or repressed, whereas among Nanog-only targets, genes are predominantly inactivated or repressed (Figure 5G). This observation is common to all the other factors, except Myc and Rex1 (Figure S10).
Expansion of core transcriptional regulatory network in ES cells
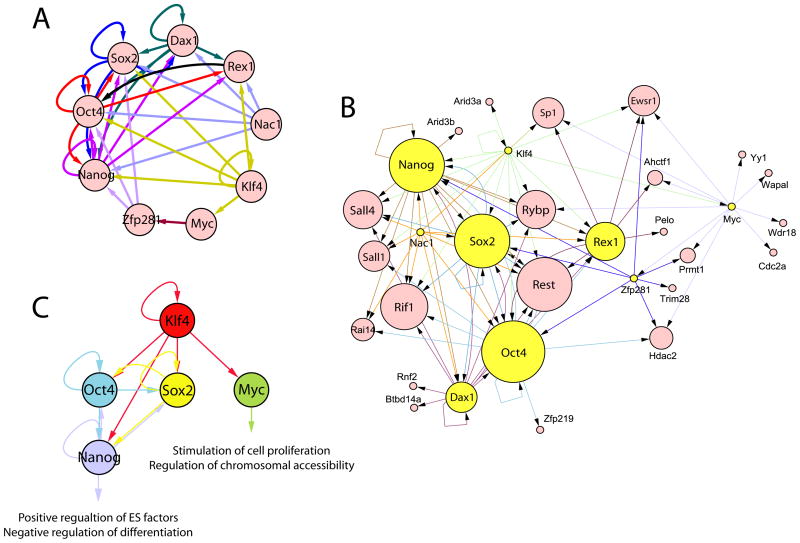
Previous studies suggested that key transcription factors in ES cells participate in several transcriptional regulatory circuits, including auto-regulation, feed-forward regulation and interconnectivity (Boyer et al., 2005). To explore this on a larger scale, we visualized transcriptional interconnectivity of the 9 factors we tested (Figure 6A). Our data describe highly intertwined, complex regulatory circuits exhibiting all three regulatory mechanisms. In addition to auto-regulatory mechanisms involving Nanog, Oct4, and Sox2, we observe that Dax1 and Klf4 also display potential auto-regulatory loops. Among these five genes, Nanog, Oct4, Sox2, and Dax1 are target hubs of at least 4 of the 9 tested factors (and 4 of 6 factors: Nanog, Sox2, Dax1, Nac1, Oct4, and Klf4).
Figure 6. Expanded transcriptional regulatory network and regulatory circuit within 4 somatic cell reprogramming factors and Nanog.
(A) Transcriptional regulatory circuit within nine factors. Five factors (Nanog, Oct4, Sox2, Dax1, and Klf4) show auto-regulatory mechanism.
(B) Expanded transcriptional regulatory network showing target hubs of multiple factors within the previously identified protein interaction network in ES cells. Yellow circles represent nine factors we examined. The size of each circle reflects the degree of factor co-occupancy. Arrowhead indicates the direction of transcriptional regulation (A–B). Sox2, Klf4 and Myc were not in the original protein interaction network (Wang et al., 2006).
(C). Transcriptional regulatory circuit within 4 somatic cell reprogramming factors and Nanog
Combining transcription regulatory networks and protein interaction networks facilitates a comprehensive understanding of differential gene expression regulation in complex genomes (Walhout, 2006). We asked if the interconnectivity of 9 factors might be useful to expand the core transcriptional network by combining target data with protein-protein interaction data. Accordingly, we merged our transcriptional regulatory network with the protein interaction network we previously reported (Figure 6B) (Wang et al., 2006). The initial protein network is comprised 35 proteins, the majority of which are essential to ES cell pluripotency and/or early development. Surprisingly, promoters of 77% of the protein network genes are occupied by at least one of the 9 factors tested (27 of 35, p-value < 2.4 × 10−7). Eleven of 35 genes are occupied by at least 4 factors (of any 9 factors, p-value < 9.1 × 10−8). More interestingly, 9 of 35 are occupied by at least 4 of 6 factors (Nanog, Sox2, Dax1, Nac1, Oct4, and Klf4, p-value < 5.3 × 10−8). In Figure 6B, target interactions are depicted with the size of each circle reflecting the degree of factor co-occupancy of the promoter of the gene encoding each factor. In this manner, we identify additional target hubs (by 4 of 9 factors), including Dax1, REST, Rif1, Rex1, Sall4, Rybp, Sall1, Ewsr1, and SP1 within the interactome, in addition to previously accepted target hubs (Nanog, Oct4, and Sox2) (Figure 6B). Independent evidence demonstrating the importance of several of these factors [Sall4 (Sakaki-Yumoto et al., 2006; Zhang et al., 2006), Rif1 (Loh et al., 2006), Rybp (Pirity et al., 2005)] in maintaining ES pluripotency is consistent with this network architecture. Taken together, our data reveal that the actual “core” factor set in ES cells is much larger and more highly interconnected than previously suspected (Loh et al., 2006; Wang et al., 2006; Zhang et al., 2006).
In addition to the target hubs within the expanded core transcription factor unit, we have identified many additional targets of multiple factors. These targets are highly likely to be important in ES cell self-renewal and lineage commitment (Jeong et al., 2001). Table 1 lists DNA-binding (or chromatin-associated) proteins whose promoters are occupied by multiple factors (at least 5 of 6 factors, Nanog, Dax1, Sox2, Nac1, Oct4, and Klf4). The predicted targets encode a set of proteins involved in regulation of development decisions, signaling pathways, and chromatin remodeling. Although functional assessment is necessary to determine the roles of many of these proteins in ES cell pluripotency and self-renewal, validation of several target genes (Nanog, Oct4, REST, Sall4, Sox2, Rex1) and identification of several others within the protein interaction network (Wang et al., 2006) make it highly likely that many others in this set will be shown subsequently to be functionally relevant. Moreover, among targets encoding non-DNA associated proteins, many are important in an ES cell context, including Tcl1, which participates in the PI3K/Akt signaling pathway and promotes ES cell proliferation (Ivanova et al., 2006); Il6st (gp130), which is involved in the LIF/STAT3 pathway (Ernst et al., 1996; Yoshida et al., 1994); and BMP4, a critical signaling molecule for early differentiation and ES cells. Indeed, Fbxo15, the locus first employed as a marker for somatic reprogramming (Takahashi and Yamanaka, 2006), is also a multifactor target gene.
Table 1.
Examples of DNA binding proteins that are common targets of multiple transcription factors (at least 5 of 6 factors, Nanog, Dax1, Sox2, Nac1, Oct4, and Klf4)
| Symbol | Accession No. | Gene Name |
|---|---|---|
| 6030445D17Rik | NM_177079 | Riken cDNA 6030445d17 gene |
| Ankrd10 | NM_133971 | Ankyrin repeat domain 10 |
| Asxl1 | NM_001039939 | Additional sex combs like 1 (drosophila) |
| Cbx1 | NM_007622 | Chromobox homolog 1 (drosophila hp1 beta) |
| Cbx7 | NM_144811 | Chromobox homolog 7 |
| Cdx1 | NM_009880 | Caudal type homeo box 1 |
| Chd9 | NM_177224 | Chromodomain helicase dna binding protein 9 |
| Dido1 | NM_175551 | Death inducer-obliterator 1 |
| E2f4 | NM_148952 | E2F transcription factor 4 |
| Evx1 | NM_007966 | Even skipped homeotic gene 1 homolog |
| Fubp3 | NM_001033389 | Far upstream element (fuse) binding protein 3 |
| Gbx2 | NM_010262 | Gastrulation brain homeobox 1 |
| Grhl3 | NM_001013756 | Grainyhead-like 3 (drosophila) |
| H2afx | NM_010436 | H2A histone family, member x |
| Hist1h2an | NM_178184 | Hypothetical protein 1190022l06 |
| Hist1h3i | NM_178207 | Histone 1, h3g |
| Hnrpdl | NM_016690 | Heterogeneous nuclear ribonucleoprotein d-like |
| Hoxb13 | NM_008267 | Homeo box b13 |
| Jarid2 | NM_021878 | Jumonji, at rich interactive domain 2 |
| Klf2 | NM_008452 | Kruppel-like factor 2 (lung) |
| Klf9 | NM_010638 | Kruppel-like factor 9 |
| Max | NM_008558 | Max protein |
| Mllt6 | NM_139311 | Myeloid/lymphoid or mixed lineage-leukemia translocation to 6 homolog (drosophila) |
| Msh6 | NM_010830 | Muts homolog 6 (e. coli) |
| Msx2 | NM_013601 | Homeo box, msh-like 2 |
| Mybl2 | NM_008652 | Myeloblastosis oncogene-like 2 |
| Myst2 | NM_177619 | Myst histone acetyltransferase 2 |
| Mzf1 | NM_145819 | Myeloid zinc finger 1 |
| Nanog | NM_028016 | Nanog homeobox |
| Nkx2-2 | NM_010919 | Nk2 transcription factor related, locus 2 (drosophila) |
| Otx2 | NM_144841 | Orthodenticle homolog 2 (drosophila) |
| Pax6 | NM_013627 | Paired box gene 6 |
| Phc1 | NM_007905 | Polyhomeotic-like 1 (drosophila) |
| Pou5f1 | NM_013633 | Pou domain, class 5, transcription factor 1 |
| Rarg | NM_001042727 | Retinoic acid receptor, gamma |
| Rax | NM_013833 | Retina and anterior neural fold homeobox |
| Rbbp5 | NM_172517 | Riken cDNA 4933411j24 gene |
| Rest | NM_011263 | Re1-silencing transcription factor |
| Rnf12 | NM_011276 | Ring finger protein 12 |
| Sall4 | NM_175303 | Testis expressed gene 20 |
| Sox13 | NM_011439 | Sry-box containing gene 13 |
| Sox2 | NM_011443 | Sry-box containing gene 2 |
| Spic | NM_011461 | Spi-c transcription factor (spi-1/pu.1 related) |
| T | NM_009309 | Brachyury |
| Tbx3 | NM_198052 | T-box 3 |
| Tcea3 | NM_011542 | Transcription elongation factor a (sii), 3 |
| Tcfap2c | NM_009335 | Transcription factor ap-2, gamma |
| Tcfcp2l1 | NM_023755 | Riken cDNA 4932442m07 gene |
| Tgif | NM_009372 | TG interacting factor |
| Trib3 | NM_144554 | Induced in fatty liver dystrophy 2 |
| Trib3 | NM_175093 | Induced in fatty liver dystrophy 2 |
| Trp53bp1 | NM_013735 | Transformation related protein 53 binding protein 1 |
| Zfp13 | NM_011747 | Zinc finger protein 13 |
| Zfp206 | NM_001033425 | Zinc finger protein 206 |
| Zfp36l1 | NM_007564 | Zinc finger protein 36, c3h type-like 1 |
| Zfp42 | NM_009556 | Zinc finger protein 42 |
| Zfp704 | NM_133218 | Zinc finger protein 704 |
| Zic2 | NM_009574 | Zic finger protein of the cerebellum 2 |
| Zic5 | NM_022987 | Zinc finger protein of the cerebellum 5 |
Regulatory network within 4 somatic cell reprogramming factors
Fibroblasts of either mouse or human origin can be reprogrammed to a pluripotent ES-like state (iPS cells) upon forced expression of 3 or 4 (or more) factors, including Klf4, Oct4, Sox2, Nanog, and c-Myc (Park et al., 2007; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007b; Yu et al., 2007). iPS cells appear highly similar to conventional ES cells. The regulatory relationships among the reprogramming factors are, therefore, of particular interest in an attempt to account for the potency of this cocktail of factors. The transcriptional hierarchy within the original 4 reprogramming factors is depicted in Figure 6C. In addition to previously identified feed-forward regulation within Oct4, Sox2 and Nanog, our results argue that Klf4 is an upstream regulator of larger feed-forward loops containing Oct4, Sox2, and other common downstream targets, such as Nanog, and also occupies the c-Myc promoter. In addition to the histone marks of Myc targets (discussed above), our findings from target categorization and the predicted regulatory network also support distinct functions of Myc in ES cells. These functions are likely to include positive regulation of proliferation, negative regulation of differentiation, and regulation of chromosomal accessibility of other factors, as previously suggested (Niwa, 2007). The findings described above regarding histone marks associated with Myc occupancy provide experimental evidence in support of these inferences.
DISCUSSION
Here we demonstrate the utility of in vivo biotinylation of tagged proteins and streptavidin affinity capture to identify global targets of multiple factors involved in the transcriptional control of pluripotency in ES cells. Our approach provides a degree of consistency in the experimental platform generally not attainable in ChIP-Chip experiments that rely on diverse antibodies of unknown specificity and sensitivity. We suggest that the bioChIP-Chip method may serve as a useful tool for assessing the quality of native antibodies, given that there is no simple a priori method for determining the suitability of a given antibody for ChIP procedures. As cell lines expressing tagged proteins may also be employed to study protein-protein interactions, the generation of two independent data-rich resources can be achieved with a single cell line “reagent” and similar procedures. While we have focused on promoter arrays (−8 kb to +2 kb), we have found that the vast majority of factor binding associated with each gene lies in the immediate vicinity of the transcriptional start sites (Figure 3B). Expansion of our bioChIP study by use of whole genome arrays or sequencing based methods would provide an even broader genomic view of chromatin occupancy. However, the inherent difficulty in assigning distant binding sites with respective target genes will hamper interpretation of these additional data.
Our studies not only suggest a more comprehensive and complex view of the pluripotency network in ES cells than prior work (Boyer et al., 2005; Loh et al., 2006), but also provide additional insights into specific regulatory features and an extensive database for further exploration. First, by mapping promoter occupancy of 9 factors, including the original 4 somatic cell reprogramming factors, we have uncovered remarkable combinatorial binding at many targets: 800 gene promoters are bound by 4 or more transcription factors of those tested (Figure 3C). Second, whereas numerous targets are shared by an extended set of pluripotency factors (Oct4, Sox2, Nanog, Klf4, Dax1, Zfp281, and Nac1), the targets of c-Myc (and also Rex1) largely fall into a different cluster (Figure 3E and Figure 4A). Third, we have discovered a striking correlation between the number of bound factors and the likelihood that a target gene is expressed in wild-type ES cells and then repressed on differentiation (Figure 4A and Figure 5). Thus, when acting alone or with few other factors, the pluripotency regulators largely occupy promoters of genes that are inactive or repressed. Distinguishing between these two possible mechanisms at individual target genes requires additional studies. When bound with multiple other factors, target genes tend to be expressed. Although the mechanistic bases for these context-dependent differences are yet to be elucidated, these observations provide a means for direct involvement of these factors in promoting self-renewal by activating expression of those genes (including the pluripotency factors themselves) and simultaneously inhibiting expression of differentiation-promoting genes. One possibility is that the pluripotency factors individually serve as weak activators, and that multi-factor binding augments activator function. A priori, the converse situation might have applied; that is, multifactor binding would predominantly be associated with repression. Despite the presence of pluripotency factors in protein complexes with corepressor components (Wang et al., 2006), our findings are inconsistent with this possibility. Another notable observation regarding histone marks is that the Polycomb targets are largely different from common targets of multiple core transcription factors. We demonstrate that common targets of multiple factors are active in ES cells and their histone marks show distinct patterns (Figure 5H). Fourth, by combining target promoter occupancy data with our prior protein interaction network, we identified additional regulatory hubs, defined as those gene promoters bound by multiple factors (Figure 6B). These new hubs include Sall4, Rif1, Rest, and Dax1, all of which have been shown to be important for ES cell properties in independent studies. Fifth, our studies suggest a hierarchy within the 4 somatic reprogramming factors, such that Klf4 serves as an upstream regulator of feed-forward circuits involving Oct4 and Sox2, as well as more downstream effectors (e.g. Nanog), and is predicted to regulate c-Myc based on promoter occupancy (Figure 6C).
In addition to the inferences gained regarding the pluripotency factors themselves, our data provide insight into how c-Myc differs from these core factors in regulating its targets. In addition to sharing few targets, c-Myc occupancy is associated with striking differences in associated histone marks, such that H3K4me3 is highly enriched and H3K27me3 is exceedingly low (Figure 4A, 4B and 4E), and with enrichment for expressed genes (Figure 4C). These findings are consistent with the view that c-Myc occupancy is associated with broad changes in chromatin accessibility. This unique target regulation by Myc may account for its capacity to enhance reprogramming, while also being dispensable as an exogenous factor (Nakagawa et al., 2007; Wernig et al., 2007a).
The discovery of a class of predicted targets bound by multiple (>4) pluripotency transcription factors (Figure 4A, Figure 5A, 5B and Table 1) is of particular interest as these genes are largely expressed in ES cells and repressed on differentiation. As this class includes several genes within the protein interaction network (e.g. Nanog, Oct4, REST, Sall4, Sox2), it is highly likely that additional genes within this set, that have not as yet been evaluated for potential roles in ES cells, will prove to be critical to the maintenance of pluripotency. The recognition that human skin cells can be reprogrammed to iPS cells with the same factors that are active in mouse cells (Park et al., 2007; Takahashi et al., 2007; Yu et al., 2007) provides strong evidence in favor of common networks in pluripotent mouse and human cells, despite differences in the growth factor requirements and behavior of cultured mouse and human ES cells. Our data constitute a framework for further exploration of the complex transcriptional network dedicated to establishment and preservation of pluripotency.
EXPERIMENTAL PROCEDURES
ES cell lines and culture
Mouse J1 ES cell lines were maintained in ES medium (DMEM; Dulbecco’s modified Eagle’s medium) supplemented with 15% fetal calf serum, 0.1 mM ß-mercaptoethanol, 2 mM L-glutamine, 0.1 mM non-essential amino acid, 1% of nucleoside mix (100X stock, Sigma), 1000U/ml recombinant leukemia inhibitory factor (LIF; Chemicon) and 50 U/ml Penicillin/Streptomycin. ES cells expressing biotin-tagged Nanog, Dax1, Nac1, Oct4, Zfp281, and Rex1 were described previously (Wang et al., 2006). For biotin-tagged Klf4, Sox2 and c-Myc, cDNAs for each gene were amplified via PCR from an embryonic cDNA library (Clontech) and incorporated into the pEF1α-FLBIO vector. Each vector was stably transfected into the ES cells stably expressing Escherichia coli biotin ligase BirA enzyme. Positive clones were selected by growth in Puromycin and the level of ectopic expression was detected by quantitative RT-PCR or Western blotting assay with anti-streptavidin-HRP and factor specific antibodies. (Wang et al., 2006).
Chromatin immunoprecipitation and antibodies
For biotin-mediated ChIP, approximately, 5 × 107 mES cells expressing both BirA and biotinylated proteins were used. Briefly, cells were cross-linked by addition of final 1% formaldehyde for 7 min at room temperature. Cross-linking was terminated by adding final 125 mM glycine and cells were washed with cold phosphate-buffered saline (PBS) containing PMSF, scraped off the plates, collected by centrifugation and washed again. Collected cell pellet was resuspended in SDS ChIP buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl pH 8.1, 150 mM NaCl, and protease inhibitors). Cells were sonicated and fragmented DNA was visualized on an agarose gel (average size 0.5–1 kb). The sample was centrifuged at 12000 rpm at 4°C for 10 min and supernatant was collected. Sample was pre-cleared with protein A beads at 4°C for 1 hr and incubated with streptavidin beads (Dynabeads® MyOne™ Streptavidin T1) at 4°C overnight. For reference sample, J1 ES cells expressing BirA enzyme without biotinylated protein were used. Immunoprecipitated complexes were successively washed with buffer I (2% SDS), buffer II (0.1% Deoxycholate, 1% Triton X-100, 1 mM EDTA, 50 mM HEPES pH 7.5, 500 mM NaCl), buffer III (250 mM LiCl, 0.5% NP-40, 0.5% Deoxycholate, 1 mM EDTA, 10 mM Tris-Cl pH 8.1) and TE buffer (10 mM Tris-Cl pH 7.5, 1 mM EDTA). All washes were for 8 min at room temperature. SDS elution buffer was added and incubated at 65°C overnight to reverse crosslink protein-DNA complexes. The sample was treated with RNase A and Proteinase K, extracted with phenol:chloroform and precipitated. The pellet was resuspended in 25μl of water.
Conventional ChIP reaction was performed as described previously (Kim et al., 2005) with 1:100 dilution of following antibodies; anti-Nanog (ab21603, Abcam), anti-c-Myc (sc-764x, Santa Cruz), anti-H3K4me3 (ab8580, Abcam) and anti-H3K27me3 (07–449, Upstate). Input genomic DNA was used for the reference sample. For western blotting assays shown in Figure S2, anti-Nanog, anti-Sox2, anti-Dax1, anti-Oct4, anti-Klf4, and anti-c-Myc antibodies were purchased from Santa Cruz Biotechnology. Anti-Nac1 was a generous gift from Scott Mackler (University of Pennsylvania School of Medicine).
Microarray and data processing
At least three biological replicates of hybridization were performed on Affymetrix GeneChip Mouse promoter 1.0R arrays. ChIP samples were amplified by ligation-mediated PCR (LM-PCR), as described previously (Ren et al., 2000). Subsequent DNA fragmentation and biotin labeling steps were performed according to the manufacturer’s instructions (http://www.affymetrix.com/support/downloads/manuals/chromatin_immun_ChIP.pdf). Microarray hybridization, washing, and scanning were performed at Microarray Core Facility -Dana-Farber Cancer Institute. MAT (Model-based Analysis of Tiling-array) was applied to predict the target loci (Johnson et al., 2006), and targets were predicted at the MAT with p-value=1.00E-6. Genomic regions between 8 kb upstream and 2 kb downstream of transcription start site (TSS) of well annotated genes from mouse genome annotation released in March 2006 (mm8) was used, and for the genes that have multiple transcripts with same TSS presented in RefSeq, we used information from only one transcript the analysis (total 19,253 genes).
For gene expression analysis, we obtained expression data from a previous study (Perez-Iratxeta et al., 2005) where a triplicate time course experiment of in vitro differentiation of mES cells was performed (11 time points; day 0, 6 hours, 12 hours, 18 hours, 24 hours, 36 hours, 48 hours, 4 days, 7 days, 9 days, and 14 days). The CEL files were imported into dChIP software(Schadt et al., 2001) for data normalization, extraction of expression values, and generating GTC file for GSEA analysis (Subramanian et al., 2005).
For the supervised clustering image shown in Figure 4A, we first performed un-supervised hierarchical clustering across the transcription factors based on their target correlation using Cluster software (Figure 3E and Figure 4A, cluster) (Eisen et al., 1998). To get a simple view of common targets of multiple factors and unique targets of single factor, we first randomized the order of 6632 genes, then sequentially sorted targets of each factor from Myc (the factor that has the most different set of targets as shown in Figure 2D hierarchical cluster) to Nanog. For visualization of target gene expression profile in Figure 4A, expression values of each gene during ES cell differentiation time course of 0 hours, 6 hours, 12 hours, and 18 hours (EX: 0–18h; red line), and day 4, day 7, day 9, and day 14 (EX: 4–14d; blue line) were averaged based on the similarity of expression profiles before moving window average was applied. Examples of the individual time point data (0 hours, 12 hours, 36 hours, 48 hours, 9 days, and 14 days) are shown in Figure S8.
Quantitative real-time PCR
Quantitative PCR was performed with a Bio-Rad iCycler in a 25μl SYBR Green reaction with approximately 2% of ChIP sample. PCR parameters were: 95°C for 3 min and 40 cycles of 95°C 20 sec, 60°C 30 sec, and 68°C 30 sec. The amount of each amplification product was determined relative to a standard curve, and fold enrichment was calculated by comparison of amplified product from bioChIP sample and ChIP samples from BirA containing ES cells. Primer pairs for quantitative ChIP-PCR were designed using ±150bp genomic sequence information specific to the predicted target loci to generate 100bp to 125bp amplified products. All primer sequences used in Figure 1B and Figure S5 are listed in Table S3.
Visualization of Regulatory Network
Cytoscape software version 2.3 (Shannon et al., 2003) was used for the visualization of transcription regulatory networks shown in Figure 6.
Supplementary Material
Figure S1. Comparison of bioChIP-chip and antibody ChIP-chip
Global patterns of Nanog and Myc binding to the target promoters of genes on chromosome 8 are displayed using Affymetrix Integrated Genome Browser and screen captured. ‘bioNanog’, and ‘bioMyc’ represent the data from bioChIP-chip. ‘Nanog antibody’, and ‘Myc antibody’ represent the data from antibody ChIP-chip
Figure S2. Sub-endogeneous expression of biotin tagged proteins in biotin-tagged cell lines.
Nuclear extracts from ES cells expressing each biotin-tagged protein were subjected to Western blot analyses using the indicated antibodies. Both the biotin-tagged and endogenous levels of the protein are shown from bioTF cells but BirA expressing cells express only endogeneous proteins. n.s indicates non-specific.
Figure S3. Correlation of Nanog occupancy among different cell lines based on target peak similarity shown in Figure 2.
Figure S4. Representative examples of target promoter occupancy by each factor.
Multiple factors bind to their common target promoters in close proximity (Sox2 and Evx1). Some targets are co-occupied by fewer factors (Fgfr2 and Cdx4). Examples of Rex1 and Myc common targets are also shown (Crat/Ppp2r4 and Anp32b).
Figure S5. Target validation by quantitative PCR
Predicted target loci of each factor were validated by site-specific PCR analysis. Predicted targets and non-targets are separated in different colors (black and grey, respectively). Y-axis represents a relative fold enrichment of predicted target loci tested from three independent bioChIP reactions over reference samples from BirA expressing cell and normalized to Gfi1b. Primers used in this study are listed in Table S3.
Figure S6. Functional classification of targets of each factor
Percent of gene hit against total number of function hits for targets of each transcription factor was calculated from PANTHER (www.pantherdb.org). The obtained percent value was divided by the value calculated for all mouse genes, and multiplied by 100. Values above 100 indicate enrichment and values below 100 indicate depletion for each GO term. Targets of Myc or Rex1 are implicated in protein metabolism, whereas targets of the other factors are in developmental processes.
Figure S7. Cluster of genes not occupied by any of nine transcription factors
(A) Schematic representation of whole genome distribution of H3K4me3, H3K27me3, and nine factors. X-axis represents all RefSeq genes based on their chromosomal positions. Predicted histone marks H3K4me3 (red), H3K27me3 (blue) and transcription factor binding (green) on the promoters of each gene were initially assigned 1 (presence) or 0 (absence). Moving window average (bin size 100 and step size 1) was applied across the genes. Red dots represent some clusters of genes devoid of any of the nine transcription factor occupancy, H3K4me3 and H3K27me3 marks on their promoters.
(B) Enlarged view of chromosome 2 containing a cluster of olfactory receptor genes.
Figure S8. Transcription factor occupancy to the target promoter and corresponding gene expression during differentiation time course.
Extension of analysis shown in Figure 4A. Instead of averaging multiple time points, 6 different time points are presented in three different columns showing overall expression profiles between earlier time points (0h and 12h) or later time points (9d and 14d) are similar.
Figure S9. Target gene expression and transcription factor occupancy
Extension of analysis shown in Figure 4. Target promoters were classified based on the number of co-occupying factors onto the promoters and corresponding gene expression upon differentiation was tested using GSEA software.
Figure S10. Single factor only targets are inactivated or repressed in ES cells
Extension of analysis shown in Figure 4F and 4G. Targets of eight factors were tested in two different ways using GSEA software. Figures shown on the left column represent all the targets of each factor and their gene expression upon ES cell differentiation. For figures shown on the right column (depicted as factor-’only’) the subset of targets predicted to be occupied by only one factor were used.
Table S1. Summary of target genes of nine transcription factors
Table S2. Relative position of target loci of nine transcription factors
Table S3. Primer sequences for RT-PCR and ChIP-PCR
Table S4. Summary of predicted targets from mouse bioNanog ChIP-chip, Nanog ChIP-PET, and human Nanog ChIP-chip.
Acknowledgments
We thank the Gene Expression Microarray Core at the DFCI for sample processing. J. K. is an HHMI Research Associate. S.H.O. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental Data including seven figures, and two excel spreadsheets are available with this article at http://www.
References
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Oates A, Dunn AR. Gp130-mediated signal transduction in embryonic stem cells involves activation of Jak and Ras/mitogen-activated protein kinase pathways. J Biol Chem. 1996;271:30136–30143. doi: 10.1074/jbc.271.47.30136. [DOI] [PubMed] [Google Scholar]
- Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein MB, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array-and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bhinge AA, Morgan XC, Iyer VR. Mapping DNA-protein interactions in large genomes by sequence tag analysis of genomic enrichment. Nat Methods. 2005;2:47–53. doi: 10.1038/nmeth726. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Macisaac KD, Gordon DB, Nekludova L, Odom DT, Schreiber J, Gifford DK, Young RA, Fraenkel E. A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics. 2006;22:423–429. doi: 10.1093/bioinformatics/bti815. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mao DY, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW, Farnham PJ, Huang TH, Penn LZ. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina DN, Glasscock J, Gish W, Lovett M. An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res. 2004;14:2041–2047. doi: 10.1101/gr.2584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2007 doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Niakan KK, Davis EC, Clipsham RC, Jiang M, Dehart DB, Sulik KK, McCabe ER. Novel role for the orphan nuclear receptor Dax1 in embryogenesis, different from steroidogenesis. Mol Genet Metab. 2006;88:261–271. doi: 10.1016/j.ymgme.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Orkin SH. Chipping away at the embryonic stem cell network. Cell. 2005;122:828–830. doi: 10.1016/j.cell.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-Genome Analysis of Histone H3 Lysine 4 and Lysine 27 Methylation in Human Embryonic Stem Cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2007 doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Perez-Iratxeta C, Palidwor G, Porter CJ, Sanche NA, Huska MR, Suomela BP, Muro EM, Krzyzanowski PM, Hughes E, Campbell PA, et al. Study of stem cell function using microarray experiments. FEBS Lett. 2005;579:1795–1801. doi: 10.1016/j.febslet.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Pirity MK, Locker J, Schreiber-Agus N. Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol Cell Biol. 2005;25:7193–7202. doi: 10.1128/MCB.25.16.7193-7202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, Kodama T, Aburatani H, Asashima M, Yoshida N, et al. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development. 2006;133:3005–3013. doi: 10.1242/dev.02457. [DOI] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Li C, Ellis B, Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl. 2001;37:120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- van Werven FJ, Timmers HT. The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 2006;34:e33. doi: 10.1093/nar/gkl003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. C-Myc Is Dispensable for Direct Reprogramming of Mouse Fibroblasts. Cell Stem Cell. 2007a doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007b;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Sze SK, Lim B, Ng HH. Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J Biol Chem. 2006;281:24090–24094. doi: 10.1074/jbc.C600122200. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Chambers I, Nichols J, Smith A, Saito M, Yasukawa K, Shoyab M, Taga T, Kishimoto T. Maintenance of the pluripotential phenotype of embryonic stem cells through direct activation of gp130 signalling pathways. Mech Dev. 1994;45:163–171. doi: 10.1016/0925-4773(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, LOY, Sung WK, Shahab A, Kuzetsov VA, et al. Whole-Genome Mapping of Histone H3 Lys4 and 27 Trimethylations Reveals Distinct Genomic Compartments in Human Embryonic Stem Cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of bioChIP-chip and antibody ChIP-chip
Global patterns of Nanog and Myc binding to the target promoters of genes on chromosome 8 are displayed using Affymetrix Integrated Genome Browser and screen captured. ‘bioNanog’, and ‘bioMyc’ represent the data from bioChIP-chip. ‘Nanog antibody’, and ‘Myc antibody’ represent the data from antibody ChIP-chip
Figure S2. Sub-endogeneous expression of biotin tagged proteins in biotin-tagged cell lines.
Nuclear extracts from ES cells expressing each biotin-tagged protein were subjected to Western blot analyses using the indicated antibodies. Both the biotin-tagged and endogenous levels of the protein are shown from bioTF cells but BirA expressing cells express only endogeneous proteins. n.s indicates non-specific.
Figure S3. Correlation of Nanog occupancy among different cell lines based on target peak similarity shown in Figure 2.
Figure S4. Representative examples of target promoter occupancy by each factor.
Multiple factors bind to their common target promoters in close proximity (Sox2 and Evx1). Some targets are co-occupied by fewer factors (Fgfr2 and Cdx4). Examples of Rex1 and Myc common targets are also shown (Crat/Ppp2r4 and Anp32b).
Figure S5. Target validation by quantitative PCR
Predicted target loci of each factor were validated by site-specific PCR analysis. Predicted targets and non-targets are separated in different colors (black and grey, respectively). Y-axis represents a relative fold enrichment of predicted target loci tested from three independent bioChIP reactions over reference samples from BirA expressing cell and normalized to Gfi1b. Primers used in this study are listed in Table S3.
Figure S6. Functional classification of targets of each factor
Percent of gene hit against total number of function hits for targets of each transcription factor was calculated from PANTHER (www.pantherdb.org). The obtained percent value was divided by the value calculated for all mouse genes, and multiplied by 100. Values above 100 indicate enrichment and values below 100 indicate depletion for each GO term. Targets of Myc or Rex1 are implicated in protein metabolism, whereas targets of the other factors are in developmental processes.
Figure S7. Cluster of genes not occupied by any of nine transcription factors
(A) Schematic representation of whole genome distribution of H3K4me3, H3K27me3, and nine factors. X-axis represents all RefSeq genes based on their chromosomal positions. Predicted histone marks H3K4me3 (red), H3K27me3 (blue) and transcription factor binding (green) on the promoters of each gene were initially assigned 1 (presence) or 0 (absence). Moving window average (bin size 100 and step size 1) was applied across the genes. Red dots represent some clusters of genes devoid of any of the nine transcription factor occupancy, H3K4me3 and H3K27me3 marks on their promoters.
(B) Enlarged view of chromosome 2 containing a cluster of olfactory receptor genes.
Figure S8. Transcription factor occupancy to the target promoter and corresponding gene expression during differentiation time course.
Extension of analysis shown in Figure 4A. Instead of averaging multiple time points, 6 different time points are presented in three different columns showing overall expression profiles between earlier time points (0h and 12h) or later time points (9d and 14d) are similar.
Figure S9. Target gene expression and transcription factor occupancy
Extension of analysis shown in Figure 4. Target promoters were classified based on the number of co-occupying factors onto the promoters and corresponding gene expression upon differentiation was tested using GSEA software.
Figure S10. Single factor only targets are inactivated or repressed in ES cells
Extension of analysis shown in Figure 4F and 4G. Targets of eight factors were tested in two different ways using GSEA software. Figures shown on the left column represent all the targets of each factor and their gene expression upon ES cell differentiation. For figures shown on the right column (depicted as factor-’only’) the subset of targets predicted to be occupied by only one factor were used.
Table S1. Summary of target genes of nine transcription factors
Table S2. Relative position of target loci of nine transcription factors
Table S3. Primer sequences for RT-PCR and ChIP-PCR
Table S4. Summary of predicted targets from mouse bioNanog ChIP-chip, Nanog ChIP-PET, and human Nanog ChIP-chip.