Abstract
Endogenous S-nitrosylation of proteins, a principal mechanism of cellular signaling in eukaryotes, has not been observed in microbes. We report that protein S-nitrosylation is an obligate concomitant of anaerobic respiration on nitrate in Escherichia coli. Endogenous S-nitrosylation during anaerobic respiration is controlled by the transcription factor OxyR, previously thought to operate only under aerobic conditions. Deletion of OxyR resulted in large increases in protein S-nitrosylation, and S-nitrosylation of OxyR induced transcription from a regulon that is distinct from the regulon induced by OxyR oxidation. Furthermore, products unique to the anaerobic regulon protected against S-nitrosothiols, and anaerobic growth of E. coli lacking OxyR was impaired on nitrate. Thus, OxyR serves as a master regulator of S-nitrosylation, and alternative posttranslational modifications of OxyR control distinct transcriptional responses.
Nitric oxide (NO) influences eukaryotic cellular processes in large part through protein S-nitrosylation, and aberrant S-nitrosylation generates a nitrosative stress that is a component of multiple diseases (1–3). Although microorganisms may be exposed to NO, which is both a component of host defense mechanisms (4) and a minor byproduct of anaerobic metabolism (5), endogenous S-nitrosylation has not been reported. We demonstrate that S-nitrosylation is a prominent feature of anaerobic respiration in Escherichia coli and describe a regulon that regulates endogenous S-nitrosylation.
Wild-type (WT) E. coli were grown anaerobically in minimal media containing either 10 mM fumarate or nitrate, and S-nitrosylation was analyzed in lysates by using photolysis-chemiluminescence (6), which measures NO displaced selectively from Cys residues, and the biotin-switch method (6), in which biotinylation identifies S-nitrosylated cysteines (S-nitrosothiols, SNOs). Whereas SNOs were not detectable in fumarate-grown cells, cells respiring anaerobically in the presence of nitrate (ARN) accumulated S-nitrosylated proteins (SNO-proteins) (Fig. 1, A and B). When E. coli were grown on fumarate and then switched to medium containing nitrate (10 mM), SNOs accumulated progressively ( fig. S1). Bacteria respiring anaerobically on nitrate can catalyze nitrosation of amino dyes when supplemented with high concentrations of exogenous nitrite (5, 7), but in our experiments, endogenous proteins underwent S-nitrosylation without exogenous nitrite.
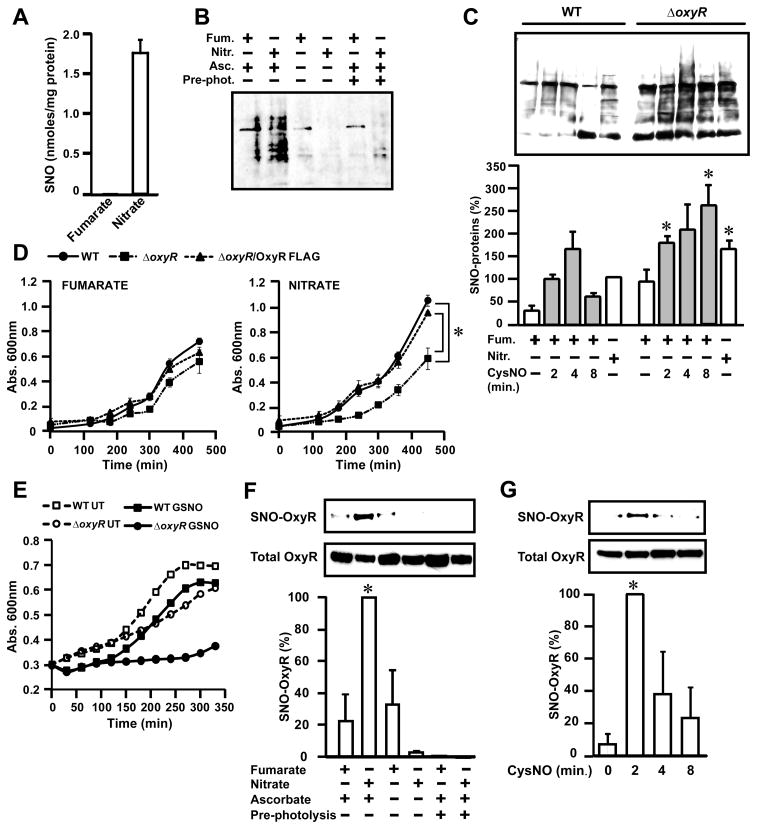
Figure 1. Accumulation of SNO-proteins and nitrosative stress in E. coli respiring anaerobically on nitrate.
(A) Endogenous S-nitrosylation in E. coli growing anaerobically on nitrate. E. coli were grown in minimal medium containing either fumarate or nitrate (10 mM) and harvested during mid-log phase (absorbance at 600 nm of 0.4). The SNO content of lysates was determined by mercury-coupled photolysis-chemiluminescence (n=3, ± SEM).
(B) SNO-proteins generated during anaerobic growth. E. coli were grown as in (A), and lysates were subjected to biotin-switch and Western blotting with NeutrAvidin-horseradish peroxidase. Ascorbate (±) and pre-ultraviolet photolysis (pre-phot.) controls were included.
(C) Regulation by OxyR of protein S-nitrosylation and denitrosylation. WT and ΔoxyR E. coli were grown anaerobically as in (A). As indicated, cells growing on fumarate were treated with CysNO (200 μM) and SNO-proteins in lysates were visualized by biotin-switch as in (B). Results represent three to five separate experiments (± SEM).* P< 0.05 WT versus ΔoxyR.
(D) Impaired growth of OxyR-null bacteria on nitrate. WT, ΔoxyR and ΔoxyR/OxyR-FLAG (ΔoxyR overexpressing FLAG-tagged OxyR) were grown as in (A) (n=5 at each time point; *P < 0.05).
(E) Protection by OxyR against nitrosative stress. Growth of WT and of ΔoxyRE. coli following single addition of GSNO (125 μM)at time 0( n=3). UT, untreated.
(F) S-nitrosylation of OxyR during anaerobic growth on nitrate. ΔoxyR /OxyR-FLAG E. coli were grown as in (A). SNO-OxyR in lysates was analyzed by SNO-RAC (n=4, ± SEM). *Differs from fumarate control by analysis of variance (ANOVA) with Dunnett’s test, P < 0.05. Control samples were generated by omission of ascorbate or by pre-photolysis.
(G) Dynamic S-nitrosylation and denitrosylation of OxyR. ΔoxyR/OxyR-FLAG E. coli grown anaerobically on 10 mM fumarate as in (A) were treated with a single addition of CysNO (200 μM). Lysates were analyzed by SNO-RAC (n=4, ± SEM). *Differs from 0 min control by ANOVA with Dunnett’s test, P < 0.05.
Although S-nitrosylation chemistry is classically O2–dependent (8), anaerobic S-nitrosylation after addition of NO to eukaryotic cells has been shown, which involves the participation of transition metals (9, 10). Anaerobic exposure of bacteria to the NO donor diethylamine NONOate (DEANO) (100 μM, 30 min) resulted in the accumulation of SNOs (8.9 ± 2.0 nmol SNO per mg protein), and prior treatment of cells with a chelator of divalent metal cations, 1, 10-phenanthroline (Phen) (1 mM, 30 min) decreased the formation of SNOs and of other photolabile nitrosylated species (XNOs, known to include transition metal-NO species) (fig. S2A). Pretreatment of bacteria with Phen also decreased formation of SNOs during ARN (fig. S2A). NrfA (nitrite reductase) and NarG (nitrate reductase) are potential sources of NO (5, 7, 11) implicated in N-nitrosation of exogenous probes (5, 12). Amounts of SNO were slightly lower in ΔnrfA versus WT E. coli but were reduced by about 80% in the ΔnarG strain (fig. S2B); nitrite levels were somewhat reduced but remained at about 4 mM in ΔnarG cells (fig. S2C). Thus, the NarG nitrate reductase appears to be a major source of S-nitrosylating activity in E. coli under anaerobic conditions, and S-nitrosylation is largely independent of nitrite concentration per se and of nitrite reduction by NrfA.
Amounts of endogenous SNO-proteins observed in WT cells during ARN were comparable to those in WT cells after exposure to 200 μM S-nitrosocysteine (CysNO) (Fig. 1C), which results in protein S-nitrosylation sufficient to generate a growth-impairing nitrosative stress (13). Amounts of SNO-proteins were also higher in ΔoxyR cells than in WT cells during ARN (Fig. 1C). The ΔoxyR cells grew more slowly than WT cells on nitrate, and this growth defect was rescued by introduction of a plasmid overexpressing OxyR (whereas OxyR rescue had no effect on growth on fumarate) (Fig. 1D). Thus, the well-known growth-promoting effect of nitrate vis-à-vis fumarate (14) is dependent on OxyR. The ΔoxyR strain was also significantly more sensitive to growth inhibition by exogenous S-nitrosoglutathione (GSNO)(Fig. 1E and fig. S3). Amounts of SNO-protein were higher in ΔoxyR cells than in WT cells after exposure to exogenous CysNO, which reflected reduced denitrosylation (SNO clearance)(Fig. 1C). Thus, protein S-nitrosylation during ARN generates a nitrosative stress that is ameliorated by OxyR, and OxyR (and possibly other genes in its regulon) appears to protect against nitrosative stress at least in part by augmenting protein denitrosylation.
Under aerobic conditions, OxyR can be activated in vitro by oxidation or nitrosylation of a critical regulatory Cys thiol (15), but S-oxidation (which is mediated by hydrogen peroxide) would be inconsistent with anaerobiosis. To test for endogenous S-nitrosylation of OxyR, we expressed FLAG-tagged OxyR in ΔoxyR cells and assayed SNO-OxyR-FLAG with the SNO-RAC (resin-assisted capture) method (6), which captures SNO-proteins on resin. SNO-OxyR-FLAG was detected in cells growing anaerobically on nitrate but not on fumarate (Fig. 1F). The extent of OxyR S-nitrosylation during ARN was comparable to that resulting from exposure to 200 μM CysNO (Fig. 1G and fig. S4, A and B), and this S-nitrosylation by CysNO was also rapidly reversed by endogenous denitrosylation (Fig. 1G). Treatment with chloramphenicol to block translation before addition of CysNO did not affect total amounts of OxyR (fig. S4C), which ruled out preferential degradation of SNO-OxyR. We found no apparent differences in rates of oxyR transcription between WT cells grown anaerobically on nitrate or fumarate (fig. S5). Thus, nitrosative stress that is a concomitant of ARN is sensed by OxyR S-nitrosylation.
To identify OxyR-dependent genes that showed increased transcription under conditions that induce S-nitrosylation, we performed a global transcriptome analysis of WT versus ΔoxyR E. coli grown anaerobically on either nitrate or fumarate (10 mM). Stringency cutoffs were set to identify genes in WT E. coli that were induced at least twofold on nitrate versus fumarate (tables S1 and S2). The 145 genes identified were clustered by using dChip microarray analysis software (16) (Fig. 2A). A majority (129 genes) were OxyR-dependent– that is, they did not show increased transcription during ARN in ΔoxyR cells (Fig. 2, A and B; fig. S6A; and tables S1 and S2) or, in a small subset, were more highly expressed on fumarate in ΔoxyR versus WT (fig. S6B). Thus, the phenotype that distinguishes anaerobic growth on nitrate from anaerobic growth on fumarate is in large part OxyR-dependent. The remaining genes showed increased expression in both WT and ΔoxyR (Fig. 2, A and B, and tables S1 and S2) or only in ΔoxyR (table S3). The 20 genes with the greatest increase in transcription during ARN in WT E. coli were further validated by quantitative real-time polymerase chain reaction (QPCR) (Fig. 2B); they include many activated in response to nitrosative stress, including hmp, hcp-hcr, yeaR-yoaG, and ytfE (17–21). The genes with greatest transcriptional change in the ΔoxyR strain (table S3) are directly responsive to NO concentration (22, 23), including those encoding the NO-metabolizing enzymes hmp (24) and norV/norW (25); further, enhanced transcription of norV/norW was observed only in ΔoxyR. Thus, WT E. coli exhibit a genetic signature of nitrosative stress during ARN, and an additional set of protective genes are highly expressed in the absence of OxyR.
Figure 2. Microarray analysis of gene expression in E. coli growing anaerobically.
(A) Expression of OxyR -regulated genes on nitrate versus fumarate. Genes in WT cells induced >2.0 -fold on nitrate (left panel); a large majority are OxyR-dependent. Genspring GX software (Agilent) was used to analyze microarray results, and genes were clustered by using dChip.
(B) Validation by QPCR of the top 20 genes regulated anaerobically on nitrate in WT E. coli and the role of OxyR. Cells were grown anaerobically on nitrate or fumarate before QPCR. Fold expression of each gene on nitrate was calculated relative to expression on fumarate using the comparative Ct method. Expression in each sample was normalized to that of 16s rRNA(n=2).
We used E. coli Entry Point (http://coli.berkeley.edu/cgi-bin/ecoli/coli_entry.pl) to group the ~ 150 OxyR-dependent genes identified by transcriptome analysis into known or predicted operons. The transcription of ~60 genes or operons was dependent on OxyR under anaerobic conditions (tables S1 and S2). OxyR-dependent genes identified in this analysis may include genes that respond directly to OxyR, as well as genes or operons that may be activated by other OxyR-dependent genes (and does not preclude codependence on other transcriptional regulators). Most of the genes or operons had not been previously identified as being OxyR-dependent. Thus, OxyR controls the expression of a large set of genes that form a nitrosative stress regulon that is distinct from the OxyR regulon activated by oxidative stress.
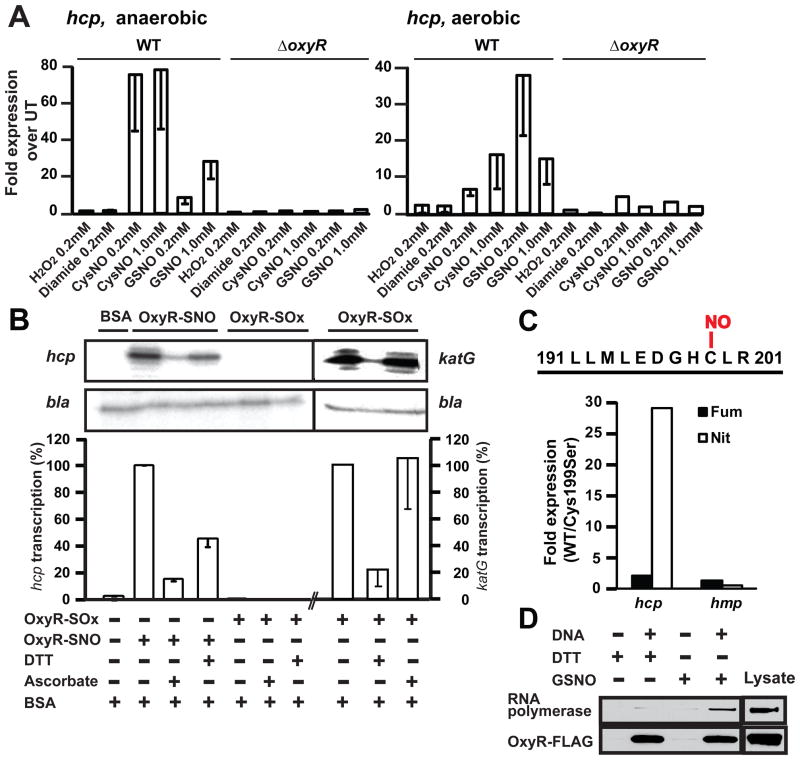
To elucidate the mechanism of OxyR activation during ARN, we focused on hcp (encoding a hybrid cluster protein), which is highly induced in an OxyR-dependent manner (40-fold versus 5-fold in WT versus ΔoxyR) (table S1). In WT cells, transcription of hcp was increased under both aerobic and anaerobic conditions by treatment with CysNO, GSNO, or DEANO but not by hydrogen peroxide(H 2O2), diamide, or oxidized glutathione (Fig. 3A and fig. S7A). Transcription was not increased in ΔoxyR cells (Fig. 3A). By contrast, genes in the known oxidative stress regulon of OxyR (katG, sufA and grxA) (26) showed increased transcription in response to oxidants (fig. S7B). These results indicate that hcp responded to OxyR activated by S-nitrosylation rather than S-oxidation (S-OH, S-S, S-SG). Modification-specific transcriptional activation by OxyR of the hcp promoter was confirmed by in vitro transcription assay with purified redox forms of OxyR (Fig. 3B). Reduced and S-oxidized OxyR had, respectively, one and no free thiol per molecule by dithionitrobenzoic acid assay. SNO-OxyR had one SNO per molecule as shown by photolysis-chemiluminescence that was localized by mass spectrometry to Cys199, and all other thiols were in the reduced form( figs. S8 to S11, supplementary text).
Figure 3. Activation of hcp transcription by SNO-OxyR.
(A) Increased hcp gene expression induced by SNOs. E. coli (WT and ΔoxyR) were grown aerobically or anaerobically to mid-log phase and treated as indicated for 5 min. Expression levels of hcp RNA were determined by QPCR (WT n=3 ± SEM; ΔoxyRn=2). UT, untreated
(B) In vitro transcription at hcp promoter by S-nitrosylated but not S-oxidized OxyR. Primer extension products of in vitro transcription reactions with S-nitrosylated and S-oxidized OxyR are shown. A plasmid containing the hcp gene [and the β-lactamase (bla) gene] was used as the template. DTT and ascorbate were used at 100 mM. Results are expressed as percent induction by SNO-OxyR, normalized with respect to bla control (n=3 separate SNO-OxyR preparations, ± SEM). Activation of katG transcription by the same preparation of OxyR-SOx is also shown (grouping of images from within the same gel is indicated by vertical dividing lines). BSA, bovine serum albumin.
(C) Dependence of hcp expression during ARN on OxyR Cys199. Cys199 was identified as the sole site of S-nitrosylation in OxyR by mass spectrometry (see figs. S9 and S10, supplementary text). ΔoxyR/OxyR-FLAG and ΔoxyR/C199S-OxyR-FLAG E. coli were grown anaerobically on nitrate or fumarate. Fold expression of hcp (OxyR-dependent) and of hmp (OxyR-independent) on fumarate or nitrate in OxyR-FLAG (WT) versus C199S-OxyR-FLAG cells was calculated using the comparative Ct method. Expression levels were normalized to 16s rRNA(n = 2 experiments).
(D) Interaction of SNO-OxyR with hcp promoter and recruitment of RNA polymerase. ΔoxyR/OxyR-FLAG lysate was incubated with a biotin-labeled hcp promoter fragment in the presence of DTT or GSNO. Proteins bound to DNA were pulled down by streptavidin beads and visualized by Western blotting with antibodies to FLAG and to RNA polymerase β’. The results are representative of three experiments.
Binding of OxyR to the hcp promoter was verified by el ectrophoretic mobility shift assay, and the interaction was inhibited by DNA containing the katG OxyR binding site (fig. S12). SNO-OxyR strongly induced transcription at the hcp promoter (Fig. 3B). Transcriptional activity was decreased both by dithiothreitol (DTT) (100 mM) and by ascorbate, which denitrosylate proteins (Fig. 3B). S-oxidized OxyR did not stimulate transcription at the hcp promoter (Fig. 3B), whereas both S-oxidized and S-nitrosylated OxyR activated the katG promoter (Fig. 3B and fig. S13). An essential role for S-nitrosylation of OxyR Cys199 in hcp induction during ARN was demonstrated by the finding that transcription of hcp was not increased during ARN in ΔoxyR E. coli overexpressing a mutant form of OxyR in which Ser replaces Cys at position 199 (Cys199Ser) (Fig. 3C). Collectively, these data establish that hcp transcription is selectively activated by S-nitrosylation of OxyR in vitro and during ARN.
Oxidized OxyR activates transcription by recruiting RNA polymerase to promoters( 27). We used a biotin-labeled hcp promoter DNA fragment and evaluated its interaction with SNO-OxyR and RNA polymerase (Fig. 3D). OxyR treated with DTT or GSNO (to generate reduced and S-nitrosylated OxyR, respectively) bound to promoter DNA (Fig. 3D), but only S-nitrosylated OxyR recruited RNA polymerase, consistent with transcriptional activation by the S-nitrosylated but not reduced OxyR( 15).
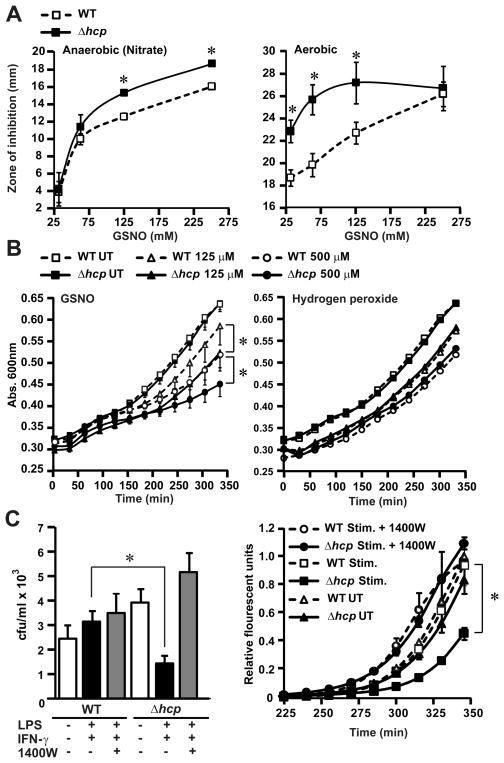
At least some OxyR-dependent genes induced during ARN serve to protect against endogenous nitrosative stress (Fig. 1D). However, the anaerobic OxyR regulon (Fig. 2) does not include genes known to ameliorate nitrosative stress (2). To evaluate the potential protective function of hcp as an exemplar, we assessed the survival of Δhcp and WT cells on agar plates in the presence of GSNO placed in cups in the agar. Under both aerobic and anaerobic conditions, the Δhcp strain had significantly (P < 0.05) larger zones of inhibition compared with those of the WT cells (Fig. 4A). Δhcp cells were also significantly more sensitive to GSNO in growth assays (Fig. 4B). By contrast, there was no statistically significant difference in the growth rates of WT and Δhcp cells treated with H2O2 (Fig. 4B). Growth inhibition by exposure to GSNO was comparable in Δhcp E. coli and E. coli lacking Hmp, a major antinitrosative enzyme (fig. S14). In addition, growth of Δhcp cells was impaired in the presence of activated macrophages, and growth differences between WT and Δhcp cells were eliminated by blocking macrophage NO production (Fig. 4C). Thus, Hcp appears to protect selectively against nitrosative stress.
Figure 4. Protection against nitrosative stress by Hcp.
(A) Hcp protects against growth inhibition by nitrosative stress. Sensitivity of WT and Δhcp cells to different concentrations of GSNO was assessed in zone-of-inhibition assays. Data represent diameter of zone of inhibition by GSNO under aerobic or anaerobic conditions (n=3 ± SEM; *P < 0.05, Student’s t test).
(B) Sensitivity of Δhcp mutant to nitrosative stress but not oxidative stress in vitro. Growth of WT and Δhcp E. coli was measured after treatment with increasing concentrations of GSNO or H2O2 (GSNO n=3, H2O2 n=5; * P< 0.01 ; WT versus Δhcp by two -way ANOVA). UT, untreated.
(C) Sensitivity of Δhcp mutant to nitrosative stress in vivo. WT or Δhcp E. coli were exposed for 30 min to cytokine-activated macrophages ± inducible NO synthase inhibitor 1400W. (Left) Internalized E. coli were plated on LB agar and colonies were counted after overnight incubation (n=4 ± SEM; *P < 0.01; WT versus Δhcp, Student’s t test). (Right) Growth of internalized E. coli was also followed by an Alamar Blue fluorescence assay (n=3 ± SEM; *P < 0.01; WT versus Δhcp, two-way ANOVA). Increased sensitivity to macrophage-derived nitrosative stress in Δhcp cells was not evident after prolonged incubations (>1 hour; not shown). IFN-γ, interferon-γ.
We found that endogenous protein S-nitrosylation is a concomitant of ARN, a major mode of existence in diverse microorganisms. The functioning of S-nitrosylation independent of NO synthases or O2 indicates that it may be a conserved mechanism of primordial origin for control of protein function. S-nitrosylation appears to convey an endogenous nitrosative stress during anaerobic metabolism, analogous to the oxidative stress entailed by oxidative (aerobic) metabolism. The transcription factor OxyR, activated by oxidation under aerobic conditions, is also activated by S-nitrosylation under anaerobic conditions and thereby induces expression in a distinct regulon, which reveals that a transcriptional activator can independently control two distinct regulons through alternative posttranslational modifications. Components of the regulon induced by S-nitrosylated OxyR (including hcp) protect against endogenous nitrosative stress, and protection is conferred at least in part by limiting S-nitrosylation. Thus, OxyR regulates S-nitrosylation in E. coli. The ability of proteins to respond differentially to alternative modifications of Cys thiol may represent the basis of a molecular code for redox regulation.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (RO1-HL096673, RO1-HL095463, RO1-HL059130) and Defense Advanced Research Projects Agency (N66001-10-C-2015). We thank D. Hess for helpful discussions and suggestions; P. McLaughlin for excellent technical assistance; M. Chance, G. Gokulrangan, and L. Wang for valuable assistance and discussion. The microarray data reported in this paper have been submitted in MIAME-compliant format to the Gene Expression Omnibus database (accession number GSE24524).
Footnotes
Materials and methods
References and Notes
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nat Rev Mol Cell Biol. 2005;6:150. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Benhar M, Forrester MT, Stamler JS. Nat Rev Mol Cell Biol. 2009;10:721. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 3.Foster MW, Hess DT, Stamler JS. Trends Mol Med. 2009;15:391. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacMicking J, Xie QW, Nathan C. Annu Rev Immunol. 1997;15:323. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 5.Ji XB, Hollocher TC. Appl Environ Microbiol. 1988;54:1791. doi: 10.1128/aem.54.7.1791-1794.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Materials and methods are available as supplementary material on Science Online.
- 7.Ralt D, Wishnok JS, Fitts R, Tannenbaum SR. J Bacteriol. 1988;170:359. doi: 10.1128/jb.170.1.359-364.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nedospasov A, Rafikov R, Beda N, Nudler E. Proc Natl Acad Sci U S. 2000;A97:13543. doi: 10.1073/pnas.250398197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster MW, Liu L, Zeng M, Hess DT, Stamler JS. Biochemistry. 2009;48:792. doi: 10.1021/bi801813n. [DOI] [PubMed] [Google Scholar]
- 10.Bosworth CA, Toledo JC, Jr, Zmijewski JW, Li Q, Lancaster JR., Jr Proc Natl Acad Sci U S. 2009;A106:4671. doi: 10.1073/pnas.0710416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corker H, Poole RK. J Biol Chem. 2003;278:31584. doi: 10.1074/jbc.M303282200. [DOI] [PubMed] [Google Scholar]
- 12.Calmels S, Ohshima H, Bartsch H. J Gen Microbiol. 1988;134:221. doi: 10.1099/00221287-134-1-221. [DOI] [PubMed] [Google Scholar]
- 13.Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. Cell. 1996;86:719. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 14.Stewart V. Microbiol Rev. 1988;52:190. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SO, et al. Cell. 2002;109:383. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 16.Li C, Wong WH. Proc Natl Acad Sci U S A. 2001;98:31. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pullan ST, et al. J Bacteriol. 2007;189:1845. doi: 10.1128/JB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC. Proc Natl Acad Sci U S A. 2007;104:8484. doi: 10.1073/pnas.0610888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filenko N, et al. J Bacteriol. 2007;189:4410. doi: 10.1128/JB.00080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overton TW, et al. Biochem Soc Trans. 2006;34:104. doi: 10.1042/BST0340104. [DOI] [PubMed] [Google Scholar]
- 21.Kim CC, Monack D, Falkow S. Infect Immun. 2003;71:3196. doi: 10.1128/IAI.71.6.3196-3205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruz-Ramos H, et al. EMBO J. 2002;21:3235. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner AM, Gessner CR, Gardner PR. J Biol Chem. 2003;278:10081. doi: 10.1074/jbc.M212462200. [DOI] [PubMed] [Google Scholar]
- 24.Hausladen A, Gow AJ, Stamler JS. Proc Natl Acad Sci U S A. 1998;95:14100. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes CM, et al. J Biol Chem. 2002;277:25273. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- 26.Zheng M, et al. J Bacteriol. 2001;183:4562. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kullik I, Toledano MB, Tartaglia LA, Storz G. J Bacteriol. 1995;177:1275. doi: 10.1128/jb.177.5.1275-1284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.